Abstract
Nanodiamonds (NDs) are emerging carbon platforms with promise as gene/drug delivery vectors for cancer therapy. Specifically, NDs functionalized with the polymer polyethylenimine (PEI) can transfect small interfering RNAs (siRNA) in vitro with high efficiency and low cytotoxicity. Here we present a modeling framework to accurately guide the design of ND-PEI gene platforms and elucidate binding mechanisms between ND, PEI, and siRNA. This is among the first ND simulations to comprehensively account for ND size, charge distribution, surface functionalization, and graphitization. The simulation results are compared with our experimental results both for PEI loading onto NDs and for siRNA (C-myc) loading onto ND-PEI for various mixing ratios. Remarkably, the model is able to predict loading trends and saturation limits for PEI and siRNA, while confirming the essential role of ND surface functionalization in mediating ND-PEI interactions. These results demonstrate that this robust framework can be a powerful tool in ND platform development, with the capacity to realistically treat other nanoparticle systems.
Keywords: Gene delivery, Nanodiamond, Multiscale simulation, Molecular dynamics, Density function theory
Nanodiamonds (NDs) are carbon-based nanoparticles that combine properties suitable for drug delivery and imaging into a single platform.1-3 Furthermore, several studies have shown that there is no apparent toxicity in vitro and in vivo following the administration of therapeutically-relevant ND dosages.1, 4 Their versatility has allowed researchers to modify NDs with a variety of biological components, utilize them as stable fluorescent markers, and mediate among the highest reported per-gadolinium relaxivity values.5-12 NDs are capable of enhancing the efficacy of therapeutic uptake and activity by mediating the binding and release of an array of chemotherapeutics.13-14 Recently, ND-enabled gene delivery was demonstrated by polymer-functionalized NDs, effectively transfecting nucleic acids while exhibiting low toxicity.15-17
Gene delivery of plasmid DNA (pDNA) and small interfering RNA (siRNA), to enhance or knock down gene expression, has become an emerging cancer treatment strategy.18-20 However, effective gene delivery requires efficient transport of nucleic acids into target cells. siRNA, for example, is easily degraded in serum, providing a major limitation for in vivo transfection.21 Viral vectors have been challenged with immunological complications,22 while non-viral formulations, such as liposomes, polymers, and nanoparticles, have been challenged with cytotoxicity and low transfection efficiency.23-24 ND-based vectors may be a promising method for efficient/biocompatible gene delivery, having demonstrated remarkable compatibility in biological environments.25-27 NDs functionalized with the cationic polymer, polyethylenimine (PEI) (Scheme 1), have exhibited higher transfection efficiency than some polymer vectors in physiological conditions for both pDNA and siRNA.15-16 In this study we used an experimentally-validated multiscale modeling approach to understand and guide the development of ND-PEI vectors to deliver c-Myc siRNA. The knockdown in expression of the c-Myc gene has been shown to significantly reduce growth of MCF-7 breast cancer cell.18
Scheme 1.
The schematic above visualizes the synthesis of the ND-PEI-siRNA complex. The functionalization of the ND surface using the polymer, PEI, allows the conjugate to mediate strong interactions with siRNA strands. The loading of siRNA onto ND-PEI may be an effective method for gene delivery, which has many therapeutic applications including cancer treatment.
Molecular dynamics (MD) is a simulation algorithm with capabilities on the order of millions of atoms, matching the scope of gene/drug delivery systems. PEI is a common gene carrier previously investigated using MD simulations.28-30 Our model is based on standard atomistic force fields, but a key component involves the use of an empirically-derived charge distribution for NDs that is based on the measured acidity of ND solutions. This model is similar to previous work describing the binding of Doxorubicin (DOX) to NDs.31 However, our model has improved accuracy and is more comprehensive. For example, the pKa estimated in the previous work was a rough approximation since it was based on the pH at the isoelectric point rather than on titration measurements. Our model also incorporates density functional-based tight binding method (DFTB) studies to determine ND structure and charge distribution.32-34 We validated our simulation results by comparing loading trends and saturation limits for PEI binding to NDs and siRNA binding to ND-PEI to experimental results for multiple mixing ratios. This comparison demonstrated both the predictive power of this simulation technique as well as the ability to elucidate basic binding mechanisms, such as the essential role of surface functionalization in mediating strong interactions between ND and PEI.
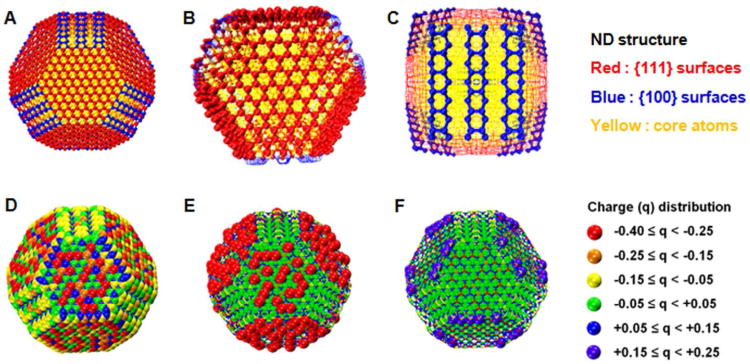
To gain a comprehensive understanding of ND structures and energetics, we developed the model from first principles, beginning with quantum mechanical calculations for the ND. DFTB is an established method for studying NDs,35-38 although past simulations have modeled smaller diameter NDs than those used in experiments (4-6 nm). To account for the size dependence of ND properties, we performed calculations using 4.1 nm NDs. Subsequently, the shape of the structure was optimized, and the state of surface graphitization and surface reconstruction was determined (See SI Materials and Methods for details). Based on these results, the ND surface charge, an essential attribute for modeling interactions with small molecules, was calculated. Figure 1A shows the truncated octahedral shape of a ND and figure 1B shows the graphitization of [111] surfaces while Figure 1C shows the 2×1 surface reconstruction revealing the lack of graphitization on [100] surfaces . The ND surface charge distribution (Figure 1D) is detailed in Figure 1E,F to illustrate the location of highly negative and positive carbon atoms revealing that carbon atoms on the [111] surface were the most negatively-charged while most positively-charged carbon atoms lay along edges where two [111] surfaces intersected. [111] surfaces (~160 atoms, average charge −2.27 ec) were typically larger than [100] surfaces (~78 atoms, average charge 0.13 ec). The strongly negative [111] and positive [100] surface electrostatic potentials were consistent with previous studies performed on smaller NDs. The mostly neutral core carbon atoms are represented in green in Figure 1E,F. This resulting ND configuration was used in all our molecular models.
Fig. 1.
The surface charge distribution of a DFTB optimized ND (C5795) structure having a diameter of 4.1 nm. The legend on the right breaks down the intervals of charge that each colored atom represents. (A) the bare optimized ND structure. (B) a close-up view of a graphitized (111) surface. (C) a close-up of the 2×1 surface reconstruction in (100) surface. (D) the complete surface charge (q) distribution. Parts (E) and (F) illustrate the regions containing only the most negatively and most positively charged atoms.
Accurate description of ND surface chemistry required simulating the ionization of surface functional groups. While several types of ND surface functionalization have been discovered, including alcohol, amine, amide, and carbonyl groups, the dominant functional group is typically believed to be carboxylic acid.39 However, neither the type nor amount of functionalization was directly available from experiments or computational procedures, so these properties were inferred indirectly from titration experiments, and modeled using effective charges located on surface carbon atoms (as described in Supporting Information). We used titrations to calculate the effective pKa which is used to determine the ionization of surface acidic groups as a function of pH. The fraction of ionizable surface sites was adjusted based on estimates provided in an earlier study of ND-DOX binding.31
Since NDs dispersed in water exhibited a pH value under 5, a base (0.01 M NaOH) was used as a titrant. Examining the shape of the titration curve in Figure S1, there was a gentle rise in pH followed by a sharp acceleration. This point was defined as the equivalence point and used to calculate the pKa (see SI Materials and Methods). Furthermore, the equivalence point occurred above pH 7, indicating that the solution behaved as a weak acid. The average pKa value for three trials is indicated in Figure S1 as 5.79 ± 0.27. This value matches closely with the pKa of carboxylic acid groups which can range from 4.5-5.5.40 While the ND surface may have complex speciation (multiple interacting ionizable sites), we used the measured pKa value to effectively model the pH dependence of the fraction of surface charges on the functionalized NDs (see SI Materials and Methods for more details).
Our ND model was used to study ND-PEI interactions, and compared with experimental results for PEI loading onto NDs as a function of mixing ratio. In past experimental studies optimizing ND vectors for transfection, various ND-PEI mixing ratios have been tested.15-16 By demonstrating compatibility between experimental and simulation results for the ND-PEI loading, we demonstrated the capacity for this simulation framework to assist in optimizing ND-PEI vectors for conditions such as mixing ratio.
Figure S2 depicts the MD model for branched PEI (800 Da) and S3 and S4 illustrate the chemical structure and charge distribution of PEI respectively, based on the polymer-consistent force field (PCFF).41 MD simulations with explicit water and counterions were performed to evaluate the significance of ND surface functionalization on PEI binding (see MD Simulations section for simulation details). The protonation ratio of PEI reported in the literatures ranges from 10%-55% at physiological pH including both linear and branched PEIs.42-43 In this manuscript, the simulated PEIs are adjusted to 20% protonation status based on previous literature on branched PEIs.44-45 The findings from this study serve as a foundation for continued investigations into the effects of protonation ratio induced by pH on the binding mechanisms between ND, PEI, and siRNA.
1 ND and 24 PEIs were placed in a simulation domain of 118 × 118 × 72 nm. The interaction of PEI with functionalized NDs was compared with non-functionalized NDs over a time scale of 8 ns to characterize ND binding. The surface charge of the non-functionalized ND was based on the values derived from DFTB simulations. The Henderson-Hasselbalch (HH) equation (see SI) was applied to calculate the expected fraction of ionized sites of functionalized ND, and 10% of the surface was assumed functionalized by ionizable acids. This degree of functionalization was at the low end of a range previously estimated (between 10-30%) based on observed drug loading.31 The 169 ionized sites on the ND surface at pH 7, as derived from the HH analysis, were randomly distributed among the ND surface atoms. The simulations were run according to the procedures and parameters outlined in the MD Simulations section.
Simulated non-functionalized NDs did not demonstrate any binding with PEI (Figure 2A and Supporting Information video S1). (All figures in this manuscript depicting molecular interactions have had water molecules and counterions removed to provide a clearer view.) This lack of binding reflects weak attractive forces between ND and PEI and reveals the significance of the charge screening effect of water. In addition, water molecules competed with PEIs to occupy positions on the ND surface since the highly polar water molecules interacted electrostatically with charged ND facets.46 In contrast, the functionalized ND was able to bind with nearly all PEI molecules (Figure 2B and Supporting Information video S2). Figure S5 shows the initial positions of NDs and PEIs in Fig 2A,B. PEIs were considered bound to a ND if they were less than 4Å from the surface at the end of the simulation. Most binding interactions occurred between protonated amines on PEI and ionized sites on ND surfaces, indicating that surface functionalization strengthens the attractive electrostatic interactions between ND and PEI. This is consistent with earlier studies of ND/DOX systems31 where it was determined that surface functionalization was crucial in promoting effective small molecule loading onto NDs.
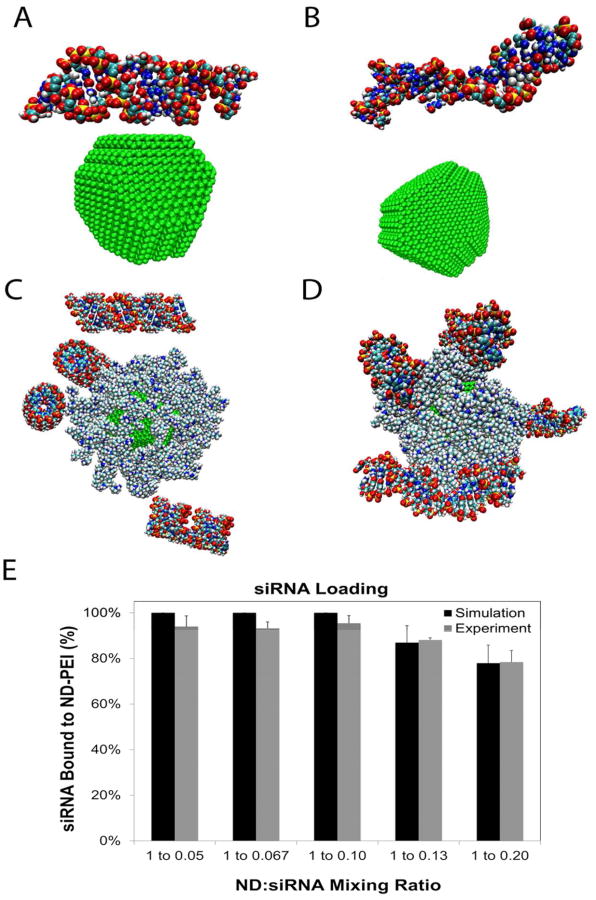
Fig. 2.
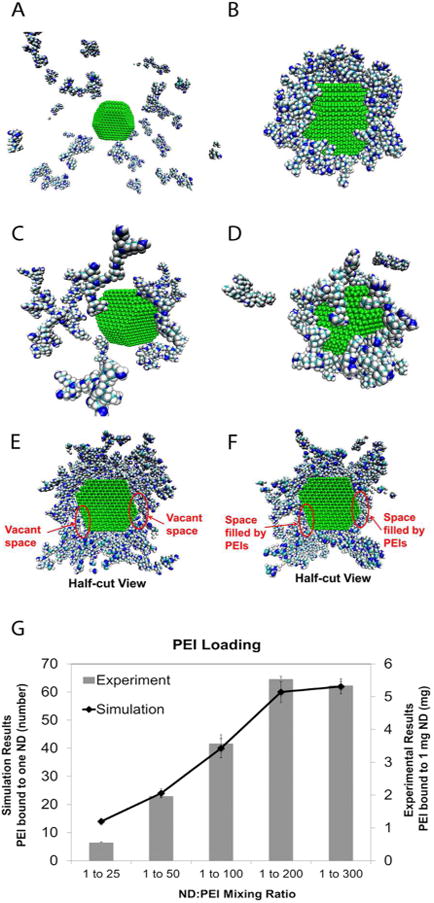
The importance of the ionization of surface functional groups towards mediating PEI loading is demonstrated in parts A and B. A and B show the final structure of a non-functionalized ND and functionalized ND, respectively, with 24 PEIs surrounding the particle after 8 ns of interaction time. The non-functionalized ND is unable to bind with any PEI because the screening of surface electrostatics by the water solvent is too strong to overcome. On the other hand, the functionalized ND is able to load all 24 PEIs successfully. The binding of PEIs to a functionalized ND at two different mixing ratios is shown in parts C-F and the comparison between the loading results of experiments and simulations is shown in part G. Part C shows the initial structure of a functionalized ND with 15 PEIs surrounding the particle, corresponding to a mixing ratio of 1:25 (ND:PEI, by weight). Part D shows the final structure of part C after 8 ns of interaction time, with most PEI's binding to the ND surface. Similarly, parts E and F show the initial and final configuration of a half cut-away functionalized ND with 120 PEIs after 8 ns interaction, corresponding to 1:200 (ND:PEI, by weight) mixing ratio. Part F shows PEI filling in the vacant space found near the ND surface shown in part E. Part G compares loading trends between the experimental and simulation results as a function of mixing ratio. The simulations were able to predict a saturation in PEI loading at a 1:200 mixing ratio, demonstrated by the lack of increasing PEI loading above the 1:200 mixing ratio.
MD simulations and experimental studies were performed to examine the effect of varying mixing ratios on PEI loading. Mixing ratio (1:25, 1:50, 1:100, 1:200, 1:300) refers to the relative concentrations by weight of ND and PEI that are initially mixed to form ND-PEI vectors. Taking advantage of the absorbance properties of PEI, the loading was experimentally quantified by measuring the unbound PEI concentration present in the supernatant (see Characterization of PEI loading onto NDs section). The simulations were constructed to match these ratios so that PEI loading trends could be compared (Figure 2C-F). In a 10 nm cubic simulation domain, one ND was placed at the center followed by the appropriate number of randomly dispersed PEI molecules in a water environment.
For example, a 1:25 mixing ratio (ND 1mg/ml, PEI 25 mg/ml) corresponded to approximately 15 PEI molecules (Figure 2C) when scaled down to the simulation volume. Figure 2D displays the final structure of a ND-PEI complex after 8 ns; here 13 PEIs were bound to the ND (Supporting Information video S3). Since the placement of PEI in the simulation box was randomized, 10 trials for each mixing ratio were performed and averaged. Figure 2E shows the initial structure of ND-PEI for a 1:200 mixing ratio, where 120 PEIs were included. Figure 2F reveals that nearly the entire ND surface was occupied by PEIs after 8 ns, indicating the proximity to the PEI saturation limit (Supporting Information video S4). Table S1 contains both the average and standard deviation of the number of PEIs bound to a ND for each mixing ratio.
The model predicted that the PEI loading would increase with increasing mixing ratio up to a saturation point of approximately 1:200. Figure 2G shows that the result was in excellent agreement with the experimental measurements. Previous experiments have used ND-PEI synthesized by adding PEI in excess, and subsequently removing the unbound PEI.15-16 Using our modeling approach, the optimal mixing ratios were determined using the appropriate input parameters, such as ND surface charge and ionization. Simulation-derived properties such as mixing ratio can assist in maximizing PEI loading, which can be crucial towards improving siRNA condensation and delivery.
siRNA mediates the RNA interference pathway, a post-transcriptional gene-silencing mechanism. In biological systems, such as the human body, the effective delivery of siRNA has the potential to provide precise control of gene expression.24, 47 For example, the C-myc gene normally regulates cellular proliferation, but when overexpressed, can promote cancer. Consequently, the administration of C-myc siRNA has been shown to reduce cancer cell growth both in vitro and in vivo.48 To optimize ND-PEI as a therapeutically-relevant vector, simulations were used to study the loading of C-myc siRNA onto the previously-derived ND-PEI structure.
The interaction between siRNA and both ND-PEI and bare NDs were compared to investigate the condensation mechanism responsible for siRNA loading. One siRNA molecule was placed 5 Å from a [111] facet (Figure 3A). After equilibration, the siRNA was observed to drift away from the surface due to electrostatic repulsion since they were both anionic (Figure 3B) Subsequently, 1 siRNA molecule was placed 5 Å from a ND-PEI complex (1 ND, 57 PEIs as obtained simulations at the saturation mixing ratio of 1:200). The key interaction leading to a stable structure is the electrostatic attraction between protonated amines on PEI and the phosphate groups on siRNA, confirming the critical role of PEI in facilitating siRNA loading.
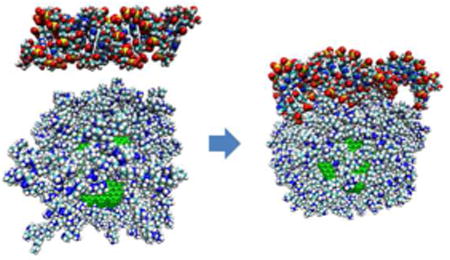
Fig. 3.
The importance of PEI functionalization towards mediating siRNA loading is demonstrated in parts AD. Part A shows the initial structure of a ND (with surface functionalization, but no PEI) with 1 siRNA placed nearest to its [111] facet. Part B shows the final structure of part A after 8 ns of interaction time. This demonstrates that a ND is unable to bind with siRNA without the presence of PEI because the ND surface is not cationic enough to bind with the negatively charged phosphate backbone of the siRNA. Part C shows the initial structure of a ND-PEI (1 ND loaded with 57 PEIs, 1:200 mixing ratio) with 4 siRNA's surrounding the particle, corresponding to a mixing ratio of 1:0.10 (ND:siRNA, by weight). Part D shows the final structure of part C after 8 ns of interaction time, with all siRNA binding to the ND-PEI surface. Part E shows the similar percentage of loading between the experimental and simulation results as a function of mixing ratio. The percentage of siRNA loading decreases above the 1:0.10 ratio while the absolute siRNA loading still increases. The simulations were able to predict the dropoff in efficiency of siRNA loading above the 1:0.10 mixing ratio.
We examined siRNA loading onto ND-PEI platforms by simulating conditions for different mixing ratios. ND-PEI (1:200 ratio) was placed in the middle of a 140 Å cubic box, with multiple siRNAs and an appropriate number of water molecules and counterions. 5 different ND-PEI-siRNA systems were generated corresponding to experimentally-relevant mixing ratios ranging from 1:0.05 to 1:0.2 (ND:siRNA). The number of siRNAs were varied and randomly arranged in the simulation domain. Detailed description of the 5 different systems and simulation results is presented in Table S2. Figure 3C,D shows the initial and final arrangement of a system with a 1:0.10 mixing ratio after 8 ns (Supporting Information video S5). The simulation predicted that almost all siRNA strands would bind to ND-PEI complexes when the siRNA:ND mixing ratio was 1:0.1 or lower. There is a dropoff in siRNA loading when the mixing ratio was increased above 1:0.1, as seen in Figure 3E, due to the repulsion between adjacent siRNAs. Consistent with these results, the experimental values also revealed diminishing siRNA loading above the 1:0.1 ratio. However, a saturation limit was not determined for siRNA loading because testing higher mixing ratios was experimentally infeasible owing to measurement limits, cost, etc. In previous siRNA transfection studies, higher mixing ratios were able to promote increased levels of gene knockdown 15, which suggested that siRNA loading increases at higher mixing ratio. However, our experimental and simulations results demonstrated that, while performance may be improved, there is a tradeoff with loading efficiency.
We have used one of the most complete ND models to date to simulate the formation of ND/PEI complexes and the subsequent loading of siRNA. Our ND model implicitly accounted for the effects of surface functionalization by determining surface charges that resulted from the ionization of acidic sites. pKa values matching experimental titration behavior and a functionalization density consistent with earlier estimates for other drug loading simulations were used as model parameters. The ND/PEI simulations of various mixing ratios, as well as siRNA loading behavior, were consistent with the experimental results. These simulations contributed basic insight on ND behavior, such as the importance of surface functionalization on mediating effective PEI loading. Furthermore, simulations provided important results to guide the optimization of ND-based vectors such as PEI and siRNA saturation limits. The computational framework defined by this model can be extended to other combinations of nanoparticles, polymers and nucleic acids used in drug delivery. In future work, we will examine critical model parameters, such as the nature and extent of surface functionalization, protonation of PEI induced by environmental pH, and their effect on ND-PEI formation and siRNA loading. Further efforts in simulating ND systems will include the modeling of larger scale effects that impact gene and drug delivery, including interparticle interactions, effects of fluid flow, and mixing. In addition, as modeling capabilities improve, detailed simulations of biological processes such as cellular internalization may become realistic. This will provide the ability to not only understand platform synthesis, but also its biological functionality. The effective utilization of multiscale modeling has the potential to enhance the development of NDs as a therapeutic tool.
Quantum Mechanical Calculations
The ND surface charge was calculated using self-consistent-charge density functional based tight-binding (SCC-DFTB) method as implemented in the DFTB+ program package [http://www.dftb-plus.info/] developed by Frauenheim, Seifert, Elstner, and co-workers.49-50 (See SI Materials and Methods for details)
MD Simulations
The simulations were performed at 300 K with an NPT ensemble (constant number of particles, pressure, and temperature) using the LAMMPS software for a total simulation time of 8 ns (http://lammps.sandia.gov).51 A Lennard-Jones potential was used to describe Van der Waals interactions and a Coulombic potential for short range electrostatic interactions. The cutoff for Van der Waals interactions and electrostatic pairwise calculations was set at 10 Å. Particle-particle particle-mesh (pppm)52 was applied for long range electrostatic interactions. The ND structure was modeled as rigid during simulations since the ND structure was already optimized through DFTB calculations. The simulation box was solvated with TIP3P water, and appropriate numbers of counterions (Na+ or Cl−) were added to neutralize the system. All bonds containing hydrogen atoms were constrained through the SHAKE algorithm, which allowed 2 fs time steps for all simulations. For more details of siRNA, see SI Materials and Methods.
Preparation of ND-PEI Complexes
In the experiments, the NDs were dispersed and diluted to a working concentration of 5 mg/ml. NDs were combined with PEI (800 Da, Sigma Aldrich) at various mixing ratios to a final volume of 1 ml and ND concentration of 1 mg/ml. After 1 min of vortexing, samples were incubated (15 min, room temperature) to allow for complexing. Samples underwent centrifugation (1 h, 14000 rpm) to pellet complexed ND-PEI, leaving unbound PEI in the supernatant. After excess PEI was removed, the pellet of ND-PEI was resuspended in water using probe sonication (Fisher Scientific) yielding a clear solution. Three wash steps were performed to remove all excess PEI. ND-PEI complexes were produced and remained stable for >1 month (4° C).
Characterization of PEI loading onto NDs
To determine PEI loading onto NDs, the supernatant of unbound PEI was collected at each of the three wash steps for the various mixing ratios. The supernatant was reacted with CuSO4 to quantify PEI loading colorimetrically (absorbance at 285 nm). For more details, see SI Materials and Methods.
Characterization of siRNA loading onto ND-PEI
0.0875 mg of ND-PEI (ND weight, 1mg/ml ND:PEI) was complexed with siRNA, at final mixing volume to 225 μl. Human C-Myc siRNA (10 μM) (Santa Cruz Biotechnology) was complexed with ND-PEI at various mixing ratios (ND:siRNA w/w). After gentle vortexing, the ND-PEI was incubated with siRNA (15 min, room temp) before centrifugation (30 min, 14000 rpm). Once ND-PEI-siRNA particles were pelleted, the supernatant was collected and quantified using a Quant-iT RNA Assay Kit (Life Technologies) to determine unbound siRNA. For more details, see SI Materials and Methods.
Statistical Analysis
Experiments were performed in at least triplicate and all results were presented as mean ± SE (standard error).
Supplementary Material
Acknowledgments
D.H. gratefully acknowledges support from the National Science Foundation CAREER Award (CMMI-0846323), Center for Scalable and Integrated NanoManufacturing (DMI-0327077), DMR-1105060, V Foundation for Cancer Research Scholars Award, Wallace H. Coulter Foundation Translational Research Award, American Chemical Society Petroleum Research Fund Grant 47121-G10, and National Cancer Institute grant U54CA151880 (The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health, and European Commission funding program FP7-KBBE-2009-3. D.H., G.C.S., B. S., W.K.L., A.M.K., and H.K. acknowledge the support of CMMI-0856492. W.K.L., A.M. K., and H.K. acknowledge support from CMMI-0856333. This research used resources of the QUEST cluster at Northwestern University and the Argonne Leadership Computing Facility at Argonne National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under contract DE-AC02-06CH11357. W.K.L. acknowledges the support of the World Class University Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (R33-10079).
Footnotes
Supporting Information Available: Included are additional materials and methods on simulation techniques and ND characterization and preparation. Also included are methods on the calculation of the pKa of NDs and the derivation of Equation 1. Figure S1: Titration of a ND solution using 0.01M NaOH to determine equivalence point and pKa of NDs. Figure S2: Atomic model of branched PEI-800 structure. Figure S3: Chemical formula of branched PEI-800. Figure S4: Charge assignment on atoms of PEI-800. Figure S5: Initial configuration of ND-PEI binding simulations, corresponding to figure 3A and 3B, showing 24 PEIs surrounding 1 ND. Figure S6: Atomic model of C-myc siRNA structure. Table S1: The detailed results of PEI loading for various mixing ratios (1:25 to 1:300, ND:PEI ratio by weight) for both simulations and experiments. Table S2: The detailed results of siRNA loading for various mixing ratios (1:0.05 to 1:0.20, ND:siRNA ratio by weight) for both simulations and experiments. Video S1: Simulation video of the interaction between 24 PEIs and 1 non-functionalized ND. Video S2: Simulation video of the interaction between 24 PEIs and 1 functionalized ND. Video S3: Simulation video of the interaction between 15 PEIs and 1 functionalized ND. Video S4: Simulation video of the interaction between 120 PEIs and 1 functionalized ND. Video S5: Simulation video of the interaction between 4 C-myc siRNA strands and 1 ND-PEI complex (1:200 weight ratio). This material is available free of charge online at http://pubs.acs.org.
References
- 1.Chow EK, Zhang XQ, Chen M, Lam R, Robinson E, Huang H, Schaffer D, Osawa E, Goga A, Ho D. Nanodiamond Therapeutic Delivery Agents Mediate Enhanced Chemoresistant Tumor Treatment. Sci Transl Med. 2011;3:73ra21. doi: 10.1126/scitranslmed.3001713. [DOI] [PubMed] [Google Scholar]
- 2.Liu WK, Adnan A, Kopacz AM, Hallikainen M, Ho D, Lam R, Lee J, Belytschko T, Schatz G, Tzeng YT, et al. Design of Nanodiamond Based Drug Delivery Patch for Cancer Therapeutics and Imaging Applications. In: Ho D, editor. Nanodiamonds: Applications in Biology and Nanoscale Medicine. Springer Science; 2010. pp. 249–284. [Google Scholar]
- 3.Yan JJ, Guo Y, Altawashi A, Moosa B, Lecommandoux S, Khashab NM. Experimental and Theoretical Evaluation of Nanodiamonds as Ph Triggered Drug Carriers. New J Chem. 2012;36:1479–1484. [Google Scholar]
- 4.Zhang X, Hu W, Li J, Tao L, Wei Y. A Comparative Study of Cellular Uptake and Cytotoxicity of Multi-Walled Carbon Nanotubes, Graphene Oxide, and Nanodiamond. Toxicol Res. 2012;1:62–68. [Google Scholar]
- 5.Bradac C, Gaebel T, Naidoo N, Sellars MJ, Twamley J, Brown LJ, Barnard AS, Plakhotnik T, Zvyagin AV, Rabeau JR. Observation and Control of Blinking Nitrogen-Vacancy Centres in Discrete Nanodiamonds. Nat Nanotechnol. 2010;5:345–349. doi: 10.1038/nnano.2010.56. [DOI] [PubMed] [Google Scholar]
- 6.Fu CC, Lee HY, Chen K, Lim TS, Wu HY, Lin PK, Wei PK, Tsao PH, Chang HC, Fann W. Characterization and Application of Single Fluorescent Nanodiamonds as Cellular Biomarkers. Proc Natl Acad Sci USA. 2007;104:727–732. doi: 10.1073/pnas.0605409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang LCL, Chang HC. Adsorption and Immobilization of Cytochrome C on Nanodiamonds. Langmuir. 2004;20:5879–5884. [PubMed] [Google Scholar]
- 8.Krueger A, Stegk J, Liang Y, Lu L, Jarre G. Biotinylated Nanodiamond: Simple and Efficient Functionalization of Detonation Diamond. Langmuir. 2008;24:4200–4204. doi: 10.1021/la703482v. [DOI] [PubMed] [Google Scholar]
- 9.Rabeau JR, Reichart P, Tamanyan G, Jamieson DN, Prawer S, Jelezko F, Gaebel T, Popa I, Domhan M, Wrachtrup J. Implantation of Labelled Single Nitrogen Vacancy Centers in Diamond Using N-15. Appl Phys Lett. 2006;88 [Google Scholar]
- 10.Ushizawa K, Sato Y, Mitsumori T, Machinami T, Ueda T, Ando T. Covalent Immobilization of DNA on Diamond and Its Verification by Diffuse Reflectance Infrared Spectroscopy. Chem Phys Lett. 2002;351:105–108. [Google Scholar]
- 11.Yu SJ, Kang MW, Chang HC, Chen KM, Yu YC. Bright Fluorescent Nanodiamonds: No Photobleaching and Low Cytotoxicity. J Am Chem Soc. 2005;127:17604–17605. doi: 10.1021/ja0567081. [DOI] [PubMed] [Google Scholar]
- 12.Manus LM, Mastarone DJ, Waters EA, Zhang XQ, Schultz-Sikma EA, MacRenaris KW, Ho D, Meade TJ. Gd(Iii)-Nanodiamond Conjugates for MRI Contrast Enhancement. Nano Lett. 2009;10:484–489. doi: 10.1021/nl903264h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Pierstorff ED, Lam R, Li SY, Huang H, Osawa E, Ho D. Nanodiamond-Mediated Delivery of Water-Insoluble Therapeutics. Acs Nano. 2009;3:2016–2022. doi: 10.1021/nn900480m. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Pierstorff E, Osawa E, Ho D. Active Nanodiamond Hydrogels for Chemotherapeutic Delivery. Nano Lett. 2007;7:3305–3314. doi: 10.1021/nl071521o. [DOI] [PubMed] [Google Scholar]
- 15.Chen M, Zhang XQ, Man HB, Lam R, Chow EK, Ho D. Nanodiamond Vectors Functionalized with Polyethylenimine for SiRNA Delivery. J Phys Chem Lett. 2010;1:3167–3171. [Google Scholar]
- 16.Zhang XQ, Chen M, Lam R, Xu XY, Osawa E, Ho D. Polymer-Functionalized Nanodiamond Platforms as Vehicles for Gene Delivery. Acs Nano. 2009;3:2609–2616. doi: 10.1021/nn900865g. [DOI] [PubMed] [Google Scholar]
- 17.Alhaddad A, Adam MP, Botsoa J, Dantelle G, Perruchas S, Gacoin T, Mansuy C, Lavielle S, Malvy C, Treussart F, et al. Nanodiamond as a Vector for SiRNA Delivery to Ewing Sarcoma Cells. Small. 2011;7:3087–3095. doi: 10.1002/smll.201101193. [DOI] [PubMed] [Google Scholar]
- 18.Wang YH, Liu S, Zhang G, Zhou CQ, Zhu HX, Zhou XB, Quan LP, Bai JF, Xu NZ. Knockdown of C-Myc Expression by RNAi Inhibits Mcf-7 Breast Tumor Cells Growth in Vitro and in Vivo. Breast Cancer Res. 2005;7:R220–R228. doi: 10.1186/bcr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 20.Dorsett Y, Tuschl T. SiRNAs: Applications in Functional Genomics and Potential as Therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 21.Turner JJ, Jones SW, Moschos SA, Lindsay MA, Gait MJ. Maldi-Tof Mass Spectral Analysis of SiRNA Degradation in Serum Confirms an RNAse a-Like Activity. Mol Biosyst. 2007;3:43–50. doi: 10.1039/b611612d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas CE, Ehrhardt A, Kay MA. Progress and Problems with the Use of Viral Vectors for Gene Therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 23.Akhtar S, Benter IF. Nonviral Delivery of Synthetic SiRNAs in Vivo. J Clin Invest. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in Humans from Systemically Administered SiRNA Via Targeted Nanoparticles. Nature. 2010;464:1067–U140. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan N, Chen CS, Hsieh HH, Wu YC, Chang HC. In Vivo Imaging and Toxicity Assessments of Fluorescent Nanodiamonds in Caenorhabditis Elegans. Nano Lett. 2010;10:3692–3699. doi: 10.1021/nl1021909. [DOI] [PubMed] [Google Scholar]
- 26.Yuan Y, Chen Y, Lui JH, Wang H, Liu Y. Biodistribution and Fate of Nanodiamonds in Vivo. Diam Relat Mater. 2009;18:95–100. [Google Scholar]
- 27.Schrand AM, Huang H, Carlson C, Schlager JJ, Ōsawa E, Hussain SM, Dai L. Are Diamond Nanoparticles Cytotoxic? J Phys Chem B. 2007;111:2–7. doi: 10.1021/jp066387v. [DOI] [PubMed] [Google Scholar]
- 28.Ziebarth J, Wang YM. Molecular Dynamics Simulations of DNA-Polycation Complex Formation. Biophys J. 2009;97:1971–1983. doi: 10.1016/j.bpj.2009.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun CB, T'Ang T, Uludag H. Molecular Dynamics Simulations of PEI Mediated DNA Aggregation. Biomacromolecules. 2011;12:3698–3707. doi: 10.1021/bm2009476. [DOI] [PubMed] [Google Scholar]
- 30.Ouyang DF, Zhang H, Parekh HS, Smith SC. Structure and Dynamics of Multiple Cationic Vectors-SiRNA Complexation by All-Atomic Molecular Dynamics Simulations. J Phys Chem B. 2010;114:9231–9237. doi: 10.1021/jp911913c. [DOI] [PubMed] [Google Scholar]
- 31.Adnan A, Lam R, Chen H, Lee J, Schaffer DJ, Barnard AS, Schatz GC, Ho D, Liu WK. Atomistic Simulation and Measurement of Ph Dependent Cancer Therapeutic Interactions with Nanodiamond Carrier. Mol Pharm. 2010;8:368–374. doi: 10.1021/mp1002398. [DOI] [PubMed] [Google Scholar]
- 32.Barnard AS, Sternberg M. Crystallinity and Surface Electrostatics of Diamond Nanocrystals. J Mater Chem. 2007;17:4811–4819. [Google Scholar]
- 33.Lai L, Barnard AS. Interparticle Interactions and Self-Assembly of Functionalized Nanodiamonds. J Phys Chem Lett. 2012;3:896–901. doi: 10.1021/jz300066j. [DOI] [PubMed] [Google Scholar]
- 34.Barnard AS. Self-Assembly in Nanodiamond Agglutinates. J Mater Chem. 2008;18:4038–4041. [Google Scholar]
- 35.Barnard AS, Sternberg M. Substitutional Nitrogen in Nanodiamond and Bucky-Diamond Particles. J Phys Chem B. 2005;109:17107–17112. doi: 10.1021/jp0524126. [DOI] [PubMed] [Google Scholar]
- 36.Chang LY, Osawa E, Barnard AS. Confirmation of the Electrostatic Self-Assembly of Nanodiamonds. Nanoscale. 2011;3:958–962. doi: 10.1039/c0nr00883d. [DOI] [PubMed] [Google Scholar]
- 37.Barnard AS, Sternberg M. Can We Predict the Location of Impurities in Diamond Nanoparticles? Diam Relat Mater. 2007;16:2078–2082. [Google Scholar]
- 38.Barnard AS, Vlasov II, Ralchenko VG. Predicting the Distribution and Stability of Photoactive Defect Centers in Nanodiamond Biomarkers. J Mater Chem. 2009;19:360–365. [Google Scholar]
- 39.Schmidlin L, Pichot V, Comet M, Josset S, Rabu P, Spitzer D. Identification, Quantification and Modification of Detonation Nanodiamond Functional Groups. Diam Relat Mater. 2012;22:113–117. [Google Scholar]
- 40.Potter MJ, Gilson MK, McCammon JA. Small Molecule Pka Prediction with Continuum Electrostatics Calculations. J Am Chem Soc. 1994;116:10298–10299. [Google Scholar]
- 41.Sun H, Mumby SJ, Maple JR, Hagler AT. An Ab-Initio Cff93 All-Atom Force-Field for Polycarbonates. J Am Chem Soc. 1994;116:2978–2987. [Google Scholar]
- 42.Ziebarth JD, Wang YM. Understanding the Protonation Behavior of Linear Polyethylenimine in Solutions through Monte Carlo Simulations. Biomacromolecules. 2010;11:29–38. doi: 10.1021/bm900842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun C, Tang T, Uludağ H, Cuervo Javier E. Molecular Dynamics Simulations of DNA/PEI Complexes: Effect of PEI Branching and Protonation State. Biophys J. 2011;100:2754–2763. doi: 10.1016/j.bpj.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suh J, Paik HJ, Hwang BK. Ionization of Poly(Ethylenimine) and Poly(Allylamine) at Various pHs. Bioorg Chem. 1994;22:318–327. [Google Scholar]
- 45.Nagaya J, Homma M, Tanioka A, Minakata A. Relationship between Protonation and Ion Condensation for Branched Poly(Ethylenimine) Biophys Chem. 1996;60:45–51. [Google Scholar]
- 46.Ōsawa E, Ho D, Huang H, Korobov MV, Rozhkova NN. Consequences of Strong and Diverse Electrostatic Potential Fields on the Surface of Detonation Nanodiamond Particles. Diam Relat Mater. 2009;18:904–909. [Google Scholar]
- 47.Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA Interference Therapy for Cancer. Gene Ther. 2005;13:464–477. doi: 10.1038/sj.gt.3302694. [DOI] [PubMed] [Google Scholar]
- 48.Vita M, Henriksson M. The Myc Oncoprotein as a Therapeutic Target for Human Cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 49.Elstner M, Porezag D, Jungnickel G, Elsner J, Haugk M, Frauenheim T, Suhai S, Seifert G. Self-Consistent-Charge Density-Functional Tight-Binding Method for Simulations of Complex Materials Properties. Phys Rev B. 1998;58:7260–7268. [Google Scholar]
- 50.Porezag D, Frauenheim T, Kohler T, Seifert G, Kaschner R. Construction of Tight-Binding-Like Potentials on the Basis of Density-Functional Theory - Application to Carbon. Phys Rev B. 1995;51:12947–12957. doi: 10.1103/physrevb.51.12947. [DOI] [PubMed] [Google Scholar]
- 51.Plimpton S. Fast Parallel Algorithms for Short-Range Molecular-Dynamics. J Comput Phys. 1995;117:1–19. [Google Scholar]
- 52.Hockney RW, Eastwood JW. Computer Simulation Using Particles. Adam Hilge; New York: 1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.