Abstract
Initiation is the primary target of translational control for all organisms. Regulation of eukaryotic translation is traditionally thought to occur through initiation factors and RNA structures. Here, we characterize a transcript-specific translation initiation mechanism that is mediated by the ribosome. By studying vesicular stomatitis virus (VSV), we identify the large ribosomal subunit protein rpL40 as requisite for VSV cap-dependent translation but not bulk cellular or internal ribosome entry site–driven translation. This requirement is conserved among members of the order Mononegavirales, including measles virus and rabies virus. Polysome analyses and in vitro reconstitution of initiation demonstrate that rpL40 is required for 80S formation on VSV mRNAs through a cis-regulatory element. Using deep sequencing, we further uncover a subset of cellular transcripts that are selectively sensitive to rpL40 depletion, suggesting VSV may have usurped an endogenous translation pathway. Together, these findings demonstrate that the ribosome acts as a translational regulator outside of its catalytic role during protein synthesis.
Keywords: ribosome code, rhabdovirus, alternative translation, uba52, paramyxovirus
Translation initiation in eukaryotes proceeds generally by a cap-dependent scanning mechanism. The rate-limiting step in this process is recognition of the 5′ m7GpppN mRNA cap structure by eukaryotic translation initiation factor 4E (eIF4E) (1). The cap-binding complex recruits the small 40S subunit in complex with initiation factors, which then scans along the mRNA until it reaches the initiation codon AUG. Start codon recognition causes the release of the initiation factors, and the large 60S subunit joins, forming an elongation competent 80S complex (2, 3).
The study of viral gene expression has uncovered several exceptions to this general mechanism of translation initiation. Poliovirus mRNA contains an ∼750-nt highly structured 5′ UTR that directly recruits the ribosome to an internal RNA site independently of the cap-recognition complex (4). Such internal ribosome entry sites (IRESs) are present in many RNA viruses, and those used by members of the Dicistroviridae family, including Plautia stali intestine virus and cricket paralysis virus, remarkably do not require any initiation factors or the initiating methionine tRNA (5–7). Investigation of the mechanism by which capped but nonpolyadenylated viral mRNAs are translated has also demonstrated that structures or viral proteins can negate the need for poly(A) binding protein during mRNA translation (8, 9). Such studies on translation initiation in viruses have invariably led to subsequent identification of cellular RNAs that are translated by similar mechanisms (10, 11).
Replication of negative-strand RNA viruses in the order Mononegavirales induces profound host shutoff, and inhibition of cellular translation effectively suppresses the host immune response and antiviral immunity (12). However, the mechanistic basis of selective viral translation by these viruses during host shutoff has remained enigmatic, as the viral mRNA transcripts are capped, methylated, and polyadenylated and are structurally indistinguishable from cellular mRNAs (13). For vesicular stomatitis virus (VSV), a prototype negative-strand RNA virus, inhibition of host protein synthesis is achieved in part by induction of the hypophosphorylation of eIF4E-binding protein 1 (4E-BP1), causing sequestration of eIF4E and halting formation of the eIF4F complex (14, 15). VSV infection also interferes with the processing of the 45S precursor rRNA to 28S and 18S ribosomal RNA, thus diminishing the pool of ribosomal RNA (16). The viral matrix protein impedes export of ribosomal RNA from the nucleus by inhibition of the Rae1 messenger ribonucleoprotein (mRNP) export pathway and by blocking transcription of ribosomal RNA (17, 18). Despite such extensive manipulation of the host translational apparatus, efficient synthesis of VSV proteins persists in a mechanism that is not understood.
Measurements of translational efficiency demonstrate that specific translation of VSV mRNAs is not caused by overabundance during infection (19, 20). Furthermore, exogenous proteins expressed by a recombinant VSV in the context of viral 5′ and 3′ UTRs are synthesized during infection, suggesting that cis-acting RNA elements outside the coding region bestow resistance of VSV translation from host shutoff (21). Although viral mRNA translation requires a methylated cap structure, VSV protein synthesis is unaffected by eIF4E sequestration, rapamycin treatment, hypoxic conditions, and eIF4G cleavage (22–25). These observations suggest that, despite their structural similarity to cellular transcripts, VSV mRNA translation proceeds via a distinct mechanism.
While viral infection often inhibits initiation factor function, the ribosome itself cannot be compromised because it is the catalytic machinery for peptide bond formation. Although studied primarily for cap-independent translation, prior studies suggest direct interactions with ribosomal proteins may be a method for alternative translation (26, 27). For instance, rps25 directly interacts with the Dicistroviridae IRES to facilitate translation (28–31). To determine whether specific ribosomal proteins facilitate cap-dependent translation of VSV mRNAs during host shutoff, we performed an siRNA screen of ribosomal proteins. We show that VSV mRNA translation depends specifically on a 60S ribosomal protein, rpL40. We demonstrate that rpL40 is not essential for bulk cellular or cap-independent translation but is necessary for replication of VSV and other viruses within the order Mononegavirales. By recapitulating this pathway in vitro using yeast extracts, we further determine that viral translation requires rpL40 for 80S formation, and rpL40 function in this pathway occurs as part of the ribosome. In vitro and sequencing analyses of polysome-associated mRNAs in yeast depleted of rpL40 indicate this pathway is conserved with select cellular mRNAs. These findings uncover a cellular alternative translation mechanism that negative-sense RNA viruses have usurped for transcript-specific initiation.
Results
siRNA Screen Reveals a Differential Sensitivity of VSV Replication and Cell Viability to Knockdown of Ribosomal Proteins.
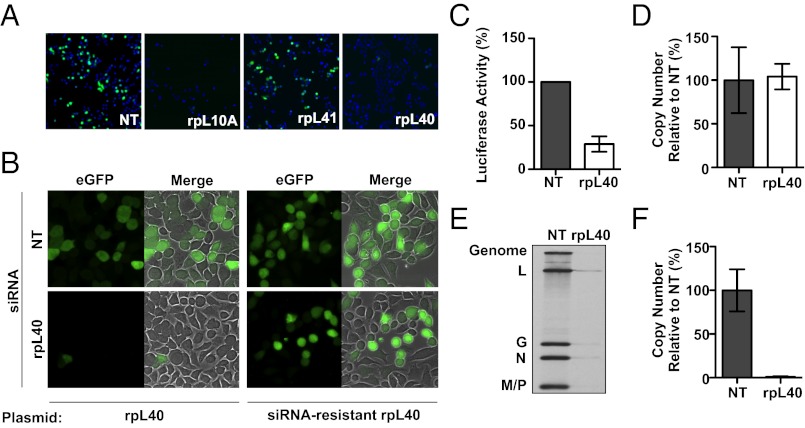
To identify ribosomal proteins required for VSV replication, we performed a targeted siRNA screen. HeLa cells were transfected with pools of four siRNAs against each of the individual ribosomal protein genes. Following 48 h of incubation, cells were infected with a reporter VSV (rVSV-EGFP) that expresses EGFP as a marker of infection, and 7 h later, the percent of infected cells was determined by fluorescence microscopy (Fig. 1A). siRNA targeting of some ribosomal proteins, such as rpL10A, caused cell death, whereas others, including rpL41, had negligible effects on cell proliferation or VSV infection (Fig. 1C; Table S1). Depletion of a subset of ribosomal proteins, such as rpL40, caused inhibition of viral replication by more than 10%, but left cell proliferation unaffected by more than 90% (Fig. 1A; Table S1). To obtain mechanistic insights on the role of ribosomal proteins in VSV replication, we chose to focus on the single candidate protein, rpL40. We selected rpL40 for further study because its knockdown had a profound difference in effects on the virus compared with the host, and its specific role in translation was uncharacterized. RpL40 is a eukaryote-specific ribosomal protein that is encoded as a C-terminal extension of ubiquitin. The ubiquitin is rapidly cleaved off and is not necessary for ribosomal function, but it contributes to the pool of free ubiquitin in the cell (32, 33).
Fig. 1.
RpL40 is required for VSV gene expression. (A) Fluorescence microscopy of cells transfected with a nontargeting (NT) siRNA or indicated ribosomal protein-targeting siRNA and infected with VSV-eGFP. Nuclei are Hochst stained (blue). (B) Rescue of VSV replication by exogenous expression of rpL40. HeLa cells were cotransfected with pcDNA3.1-rpL40, encoding a wild-type or siRNA-resistant form of the gene, and either NT or rpL40 targeting siRNA. Cells were infected and examined by epifluorescence microscopy. (C) Expression of luciferase from purified rVSV-Luc ribonucleoprotein cores transfected into cells treated with an siRNA targeting rpL40. Luciferase activity was normalized to activity from cells treated with a NT siRNA. The results are given as the mean ± SD of three independent experiments performed in triplicate. (D) VSV primary transcription in rpL40 siRNA-transfected cells. Abundance of VSV N mRNA was measured by quantitative RT-PCR. The results are given as the mean ± SD from a single representative quantitative RT-PCR experiment performed in duplicate. (E) Viral RNA synthesis in infected cells treated with rpL40 siRNA. Total [3H]-uridine–labeled cellular RNA was analyzed by electrophoresis on an acid-agarose gel. (F) Quantitative RT-PCR of total viral RNA synthesis. Results were analyzed as in D.
RpL40 Is Required for VSV mRNA Translation.
To ensure that specific silencing of rpL40 was responsible for the reduction in VSV gene expression, we identified individual siRNAs that block infection. Two siRNAs targeting distinct regions of the rpL40 transcript inhibited infection by >90% (Fig. S1A) and reduced rpL40 gene expression at the mRNA (Fig. S1B) and protein (Fig. S1C) levels. Furthermore, knockdown of rpL40 correlated with a ∼2 log10 decrease in virus output (Fig. S1D). The defect in VSV gene expression is caused by a specific loss of rpL40, because infection was restored by transfection of cells with an siRNA-resistant rpL40 variant but not the wild-type gene (Fig. 1B).
We next conducted a systematic examination of each step of viral replication to determine how rpL40 knockdown affects VSV infection. To demonstrate rpL40 is not required for the translation of a cellular factor essential for viral entry, we bypassed the clathrin-dependent entry pathway of VSV by direct transfection of the purified RNP core of a recombinant VSV that expresses firefly luciferase (rVSV-Luc). Circumventing entry did not restore luciferase expression, thus showing that loss of rpL40 specifically impairs a step in viral gene expression (Fig. 1C). VSV gene expression initiates with primary transcription of mRNAs, and translation of these transcripts is essential to provide the proteins necessary for genome replication and subsequent secondary transcription. By measuring VSV mRNA levels through quantitative RT-PCR in cells exposed to the protein synthesis inhibitor cycloheximide, we show that primary transcription is unaffected by rpL40 depletion (Fig. 1D). To measure the translation-dependent steps of genome replication and secondary mRNA transcription directly, we monitored viral RNA levels by metabolic incorporation of [3H]-uridine and also by quantitative RT-PCR. Viral genomic RNA and mRNA levels were reduced >90% by rpL40 depletion (Fig. 1 E and F). These experiments demonstrate that rpL40 is not required for viral entry or transcription, thus specifically implicating rpL40 in translation of viral mRNAs.
RpL40-Dependent Translation Is Transcript-Specific.
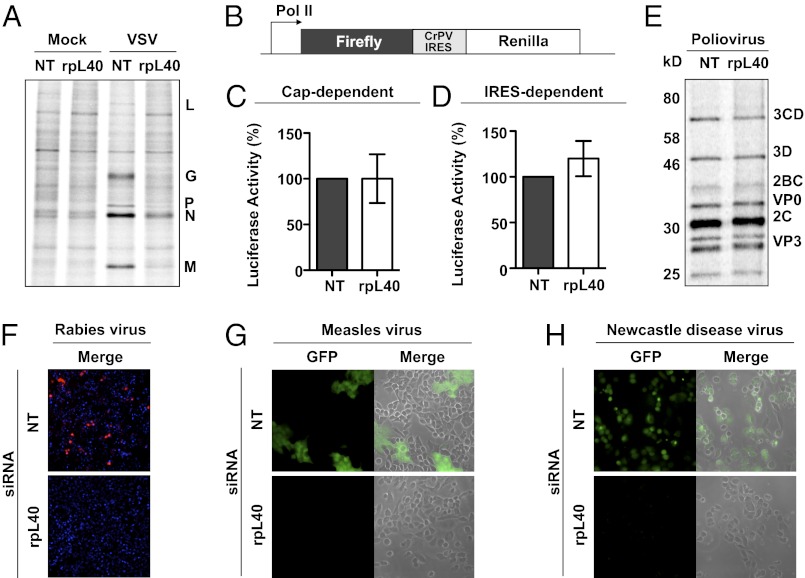
We next determined whether VSV protein synthesis is specifically inhibited by rpL40 depletion. Monitoring metabolic incorporation of [35S]-methionine-cysteine in rpL40-depleted cells revealed an 85% reduction in viral protein synthesis, whereas bulk cellular translation was virtually unaffected (Fig. 2A). We then examined the consequences of rpL40 depletion on transcripts with altered initiation mechanisms. The IRES element of cricket paralysis virus (CrPV) drives translation by direct 80S ribosome formation on the initiation codon, circumventing any requirement for initiation factors, scanning, or the methionine initiator tRNA. In cells transfected with a plasmid encoding a bicistronic luciferase vector separated by the CrPV IRES sequence (pFR-CrPV) (Fig. 2B), neither cap-dependent nor IRES-dependent translation was diminished by depletion of rpL40 (Fig. 2 C and D). Poliovirus infection, which depends on an eIF4E-independent IRES, is similarly unaffected by rpL40 depletion (Fig. 2E). These IRES experiments demonstrate that critical interfaces on the 60S subunit necessary for direct interactions with the small subunit are not altered after knockdown of rpL40. Together, these results confirm that depletion of rpL40 permits the formation of functional ribosomes that mediate efficient cap-dependent and IRES-dependent translation.
Fig. 2.
RpL40-dependent translation is transcript-specific. (A) VSV protein synthesis. Total [35S]-methionine-cysteine–labeled cytoplasmic proteins were analyzed by SDS/PAGE and detected by phosphorimager. Transfection of cells with nontargeting (NT) or rpL40-targeting siRNA is indicated. (B) Schematic diagram of bicistronic CrPV IRES construct. Translation of firefly luciferase is cap-dependent, whereas translation of renilla luciferase is driven by the CrPV IRES. (C) Firefly luciferase protein synthesis driven from pFR-CrPV. Firefly luciferase was measured from pFR-CrPV transfected lysates at 12 h after transfection and normalized to luciferase units from NT-treated cells. The results are given as the mean ± SD of three independent experiments each performed in triplicate. (D) Renilla luciferase protein synthesis driven from pFR-CrPV. Luciferase levels were measured and normalized as in C. (E) Poliovirus protein synthesis. Cells were infected and total cytoplasmic proteins were analyzed as in A. (F) Microscopy of cells infected with rabies virus-mCherry. (G) Microscopy of cells infected with measles virus-GFP. (H) Microscopy of cells infected with Newcastle disease virus-GFP.
We next sought to determine whether viruses more closely related to VSV were dependent on rpL40. VSV is a member of the order Mononegavirales, consisting of nonsegmented negative-sense (NNS) RNA viruses. The transcripts of NNS viruses are capped and polyadenylated, and within each virus, the 5′ ends begin with a conserved gene start site. The replication of multiple NNS viruses, including rabies virus (Fig. 2F), measles virus (Fig. 2G), and Newcastle disease virus (Fig. 2H), is sensitive to rpL40 depletion. These experiments demonstrate that rpL40-dependent translation is transcript-specific, and this pathway is required by an array of NNS viruses.
Ribosome Biogenesis and Maturation Are Not Compromised by rpL40 Depletion.
To determine whether there are defects in ribosome processing following rpL40 depletion, we labeled total cellular RNA by metabolic incorporation of [3H]-uridine and monitored the accumulation of mature rRNA. As in prior studies, rpL40 depletion modestly decreased the total levels of 28S and 18S RNAs (4% and 17%, respectively) but did not abolish processing of mature ribosomal RNAs (Fig. S2A) (34). This result confirms that rpL40 depletion does not inhibit VSV mRNA translation by altering ribosomal RNA processing.
During infection, the VSV matrix protein blocks export of preribosomal RNAs from the nucleus via the Rae1 export pathway, suggesting VSV mRNAs might be dependent on rpL40 as a result of an additive reduction in pools of ribosomes during knockdown and infection. However, a recombinant VSV that does not block nuclear export (rVSV-M51R) (Fig. S2B) remains sensitive to rpL40 depletion (Fig. S2C) (18). Furthermore, rpL22 depletion does not inhibit viral gene expression (Fig. S2 D and E), indicating rpL40 is a specific ribosomal protein requisite for VSV translation. These experiments also show that viral translation is not simply affected by a reduction in the pool of cytoplasmic ribosomes.
RpL40 Is Required for Translation Initiation on VSV mRNAs as a Constituent of the Large Subunit.
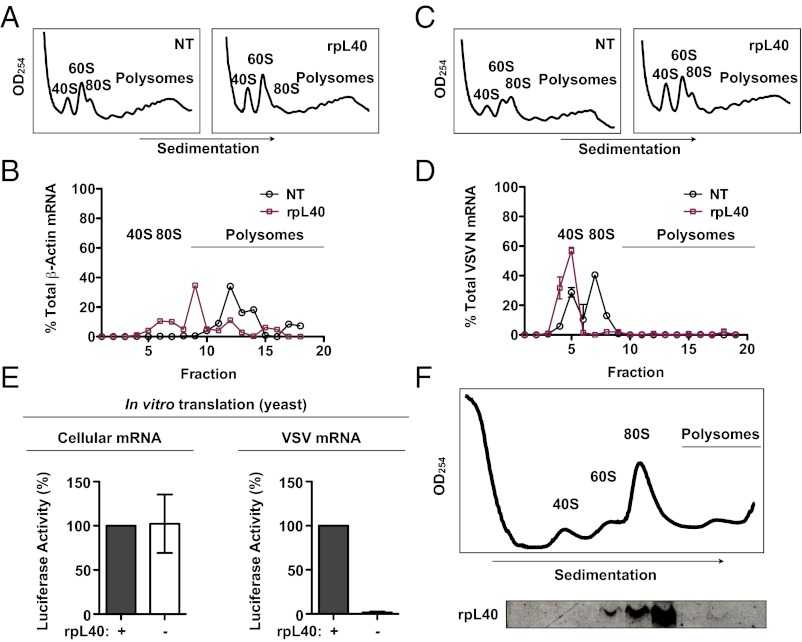
To determine the mechanism of translational control by rpL40, we compared the formation of ribosomal complexes on VSV mRNA with the cellular transcript β-actin. We specifically examined the VSV nucleocapsid (N) mRNA, as this is the most abundant viral mRNA produced during infection. Cells depleted of rpL40 were mock infected or infected with VSV, and lysates were resolved on a 10–50% linear sucrose gradient. Following fractionation, the association of ribosomal subunits with specific mRNA was determined by quantitative RT-PCR. Absorbance monitoring at 254 nm revealed polysome formation, further confirming that bulk translation was not compromised by rpL40 depletion (Fig. 3 A and C). The cellular transcript β-actin remained distributed in 80S ribosomes and polyribosomes following rpL40 depletion, although there was a shift to lighter fractions (Fig. 3B). In contrast, formation of elongation-competent ribosomes on VSV N mRNA was completely abolished by rpL40 depletion (Fig. 3D). We note that the localization of VSV mRNAs to lighter fractions of polysome profiles has been observed previously (35, 36). These experiments establish that rpL40 is required for translation initiation on VSV mRNAs but not canonical cellular transcripts.
Fig. 3.
RpL40 is required for translation initiation on VSV mRNAs as a constituent of the large subunit. (A) Sedimentation profile of mock-infected rpL40-targeting or nontargeting (NT) siRNA-transfected cells. (B) Distribution of β-actin mRNA with ribosomal complexes in cells depleted of rpL40. Transcript number, as determined by quantitative RT-PCR, is expressed as the percentage of total β-actin transcripts recovered and plotted against fraction number. The results are given as the mean ± SD from a representative quantitative RT-PCR experiment performed in duplicate. (C) Sedimentation profile of VSV-infected siRNA-transfected cells. (D) Distribution of VSV N mRNA with ribosomal complexes in cells depleted of rpL40. Lysates were resolved and fractionated, and mRNA distribution was determined as in B. (E) Translation of cellular or VSV-derived luciferase mRNA in yeast extracts expressing or lacking rpL40. Cytoplasmic mRNA was isolated from cells transfected with a luciferase expression plasmid (pRL-CMV) or cells infected with rVSV-Luc and used to program yeast extracts. The results are given as the mean ± SD of three independent experiments performed in triplicate. (F) Distribution of rpL40 with ribosomal complexes in VSV-infected cells.
The polysome data show that rpL40 is crucial for translation initiation on VSV mRNAs. However, as these experiments are performed in the context of viral infection, it is unknown whether rpL40 is required due to alternations to the translation machinery during infection or through aspects of the mRNA itself. Thus, to investigate the initiation defect independent of viral infection, we developed an in vitro translation assay using cytoplasmic extracts from a yeast strain, GAL-RPL40A-∆Ubq (34). Yeast express two rpL40 paralogs, rpL40A and rpL40B, that encode the ribosomal protein rpL40 with an N-terminal ubiquitin moiety (32). The yeast strain GAL-RPL40A-∆Ubq has both forms of rpL40 deleted, and instead, rpL40A lacking its ubiquitin tail is ectopically expressed from a galactose-inducible promoter. In yeast extracts lacking rpL40, translation of a cellular reporter renilla luciferase mRNA was unaffected, whereas translation of rVSV-Luc mRNA was abolished (Fig. 3E). These findings demonstrate that the requirement for rpL40 in VSV translation is dictated by a cis-acting determinant in the mRNA and does not require other components of viral infection. Furthermore, these results indicate that the rpL40-dependent translation strategy is broadly conserved between yeast and mammalian cells. Importantly, the use of an ubiquitin-lacking rpL40 conditional knockout demonstrates that knockdown of rpL40 affects VSV mRNA translation directly rather than through alterations to the cellular ubiquitin pool.
RpL40 could regulate 80S formation as an extraribosomal protein or as part of the ribosome. To distinguish between these possibilities, we monitored the polysomal distribution of rpL40. VSV-infected HeLa cell lysates were resolved on a 5–30% linear sucrose gradient, and the fractions were probed by immunoblot. RpL40 was only found in 60S, 80S, and polysome fractions, with no detection as an extraribosomal population (Fig. 3F). Furthermore, rpL40 has only been found with 60S and 80S fractions of polysome profiles performed in Drosophila and yeast (32, 37). These results indicate that rpL40 regulates translation as a component of the ribosomal large subunit.
RpL40-Dependent Translation Is Used by Select Cellular mRNAs.
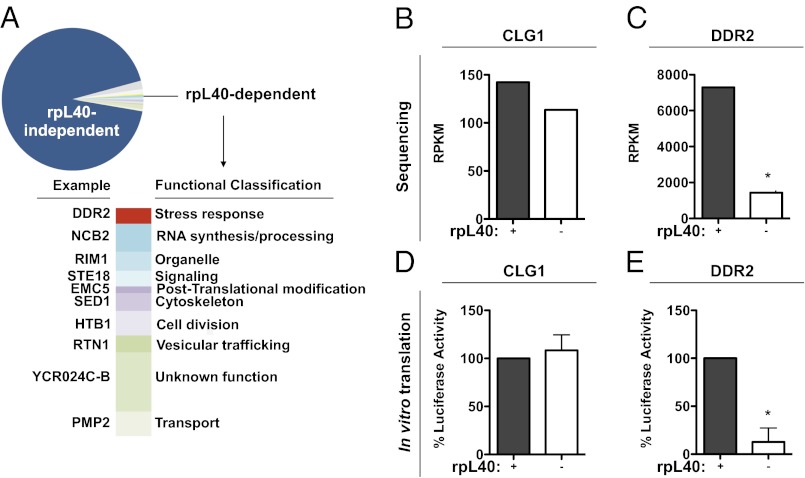
As many alternative translation pathways used by cellular transcripts, including cap-independent translation, were initially characterized with viruses, we next hypothesized that the rpL40-dependent translation pathway used by VSV is shared with a subset of cellular mRNAs (4, 38). To identify such transcripts, we sequenced polysome-associated mRNAs from GAL-RPL40A yeast cells grown in media containing glucose or galactose (Fig. S3). On depletion of rpL40, ∼93% of mRNAs (fold reduction ≤ 3) remained polysome associated, confirming that bulk cellular translation does not depend on rpL40 (Figs. 2A and 4A). The list of candidate cellular mRNAs that require rpL40 for translation included a number of stress response transcripts (Table S2). Intriguingly, VSV infection is resistant to inhibition by stresses, including heat shock and hypertonicity, suggesting the rpL40-dependent translation pathway is available during stress responses (39, 40). Thus, we examined in vitro translation of a representative candidate mRNA that is up-regulated by heat shock and DNA damage, DDR2 (Fig. 4C) (41). In vitro translation of DDR2, but not the control transcript CLG1, was decreased by ∼87% in the absence of rpL40 (Fig. 4 B, D, and E). These results thus suggest that VSV has usurped an endogenous pathway that is shared by select cellular transcripts.
Fig. 4.
RpL40-dependent translation is used by select cellular mRNAs. (A) Functional classification of mRNAs whose polysome association is altered by rpL40 depletion. The gene in each category whose association with polysomes was most decreased by knockdown of rpL40 is listed. (B) Levels of polysome-associated CLG1 mRNA upon rpL40 depletion identified by sequencing analysis. (C) Levels of polysome-associated DDR2 mRNA identified by sequencing analysis. Average reads per kilobase of exon per million mapped reads (RPKM) from biological replicates is graphed in B and C. (D) In vitro translation of CLG1 mRNA. (E) In vitro translation of DDR2 mRNA. The results of D and E are given as the mean ± SD of three independent experiments, performed in triplicate. Conditions that are statistically significant from the +rpL40 conditions are indicated with an asterisk (P < 0.0001).
Discussion
We made the principal finding of a transcript-specific translation initiation strategy that is dependent on rpL40, a protein constituent of the large ribosomal subunit, and is required for replication of multiple NNS viruses. Use of this mechanism is designated by a cis-acting RNA determinant and is conserved among eukaryotes. Our results identify the step of translation at which rpL40 is required and also reveal that select transcripts are translated through an rpL40-dependent mechanism. Together, this work reveals a previously uncharacterized pathway of translational specialization, thus providing evidence that the ribosome controls translation separately of its catalytic function.
Although the ribosome has traditionally been thought to function only as the catalytic machinery for translation elongation, our work substantiates the idea that the ribosome itself can act as an initiation regulator. Before this study, the requirement for ribosomal proteins during viral replication has only been evaluated for cap-independent translation strategies. For example, cap-independent translation of CrPV RNA and hepatitis virus C virus (HCV) RNA requires specific 40S subunit proteins for binding to the IRES elements (30, 42). Here, we demonstrated a requirement for a specific ribosomal protein during cap-dependent viral mRNA translation.
Previous work suggested VSV cap-dependent translation occurs through unconventional mechanisms. Infection alters the cap-binding complex by inducing dephosphorylation of the 4E-BP1 translation repressor, which subsequently sequesters eIF4E, causing translation of host mRNAs to be inhibited while VSV protein production continues (14, 15). These findings suggest that VSV mRNA translation is not limited by eIF4F availability. We have demonstrated in vitro and in mammalian cells that a specific ribosomal protein, rpL40, has a critical role for 80S formation on VSV mRNAs. Although uncommon, there are examples of transcripts with specific requirement for large subunit proteins during translation initiation. For example, during initiation on the CrPV IRES, the RNA must associate with rpL1 for subunit joining of the 60S to the 40S and correct ribosome positioning (43, 44). Furthermore, by studying mice with a short tail phenotype, rpL38 was found to be required for 80S formation on Homeobox mRNAs and thus correct tissue patterning during development (45). RpL40 is localized on the surface of the ribosome, and its amino and carboxy termini are unblocked, providing potential sites to interact with mRNAs or proteins (Fig. S4) (46, 47). It will be important to determine whether structural or sequence elements in the viral UTRs bind to rpL40 directly or through other protein factors. Alternatively, rpL40 may be required for the 60S subunit to undergo correct conformational changes needed for VSV mRNA translation, although its surface localization makes this less likely.
Because viral translation must occur in the face of translation-compromising cellular defense mechanisms and in competition with abundant host transcripts, many viruses have found alternative mechanisms of translation (12). However, viruses must also ensure that these translation pathways are conserved among their host range. HCV only replicates in mammalian hosts, and its genomic RNA cannot be translated in yeast extracts. The inability of HCV RNA to be translated in yeast is postulated to be a result of the high divergence between yeast and mammals for the small subunit proteins that interact directly with the IRES (42). In contrast, VSV has an extremely broad host range, including ruminants, insects, and humans (13). In addition, VSV has a distinct ability to express proteins in almost all vertebrate and insect cells, along with crustacean and Caenorhabditis elegans cells (48, 49). By studying VSV protein synthesis, we demonstrated that mammalian and yeast cells share the rpL40-dependent translation pathway. In agreement, rpL40 is highly conserved in archaea and eukaryotes (Fig. S5). Our data, along with the inspection of rpL40 homologs, indicate this translation strategy is likely present throughout all Eukarya and suggest that ribosome specialization may have an evolutionary impact on host range.
As an extension to our viral studies, we identified select cellular mRNAs that are dependent on rpL40 through sequencing of polysome-associated mRNAs and verified this by examining in vitro translation of DDR2. DDR2 is a stress-response protein that is up-regulated in yeast cells by heat shock and by treatment with the DNA mutagens 4-nitroquinoline-1-oxide and N-methyl-N'-nitro-N-nitrosoguanidine (41). VSV protein synthesis itself is resistant to inhibition by certain stresses, suggesting the viral rpL40-dependent translation pathway is available and used during stress responses. Exposure of cells to hypertonic or heat shock conditions that cause near complete inhibition of host protein synthesis has little effect on VSV mRNA translation (39, 40). The ability of VSV mRNAs to be translated during cell stress is in agreement with the discovery that VSV and DDR2 share an rpL40-dependent translation pathway.
Through studies of VSV mRNA translation, we uncovered an unconventional mechanism of translation initiation that requires rpL40. Our results support a ribosome code dictated by specific ribosomal proteins, thus identifying a new regulatory element during translation that is shared between viruses translated by cap-dependent and cap-independent mechanisms. Further molecular studies will develop our understanding of ribosome-mediated translational control of viral mRNAs and provide new insights into alternative cellular translation.
Methods
Sucrose Gradient Analysis (Polysome Assay).
One 75-cm2 flask of HeLa cells was mock infected or infected at a multiplicity of infection of 1 with VSV for 4 h. Cells were incubated with 100 µg/mL cycloheximide for 5 min at 37 °C and then put on ice. Cells were washed twice with cold PBS and 0.1 mg/mL cycloheximide and collected by cell scraping. Cells were resuspended in 1 mL of RSB-PEB (500 mM Tris, pH 7.5, 2 M NaCl, 15 mM MgCl2, 1% (vol/vol) Triton-X, 2% (vol/vol) Tween-20, 1% (vol/vol) sodium deoxycholate), vortexed briefly, and incubated on ice for 10 min. Lysate was spun for 10 min at 4 °C at 10,000 × g, and supernatant was resolved on a 10–50% (wt/vol) or 5–30% (wt/vol) sucrose gradient, as indicated, by centrifugation at 35,000 rpm at 4 °C for 3 h in a Beckman SW41 Ti rotor. Fractions were collected from the top of the gradient using a BioLogic LP (BioRad) with a Brandel tube piercer. RNA was phenol/chloroform extracted, ethanol-precipitated, and resuspended in dH2O for quantitative RT-PCR analysis. Protein was trichloroacetic acid (TCA) precipitated and analyzed by immunoblot.
In Vitro Translation.
The yeast strain GAL-RPL40A-∆Ubq (a kind gift from P. Milkereit, University of Regensburg, Germany) was grown in galactose-containing medium (YPG) at 30 °C overnight to a final optical density (OD)600 of ∼1.5. Yeast were centrifuged and resuspended in YPG or glucose-containing medium, and a 2-L culture was grown for 6 h to a final OD of 1.5. Saccharomyces cerevisiae cell-free translation extracts were prepared using a modified previously described procedure (50). Small molecules were removed from S30 lysates using a Zeba Desalt Spin Column (Pierce) preequilibrated with Buffer A with 8.5% manniotol and 0.5 mM PMSF (51). The resulting lysate was stored at −80 °C in aliquots. Before in vitro translation, aliquots were treated with 0.8 mM CaCl2 and 800 U/mL micrococcal nuclease (New England Biolabs) for 10 min at 25 °C, and the reaction was halted by addition of 1.2 mM EGTA. For VSV mRNA translation, each sample was 50% lysate, up to 3.94 µL RNA, and buffer to make the final reaction have 0.84 mM ATP, 0.21 mM GTP, 21 mM creatine phosphate, 45 U/mL creatine phosphokinase, 10 mM Hepes, pH 7.6, 2 mM DTT, 2.5 mM magnesium acetate, 100 mM potassium acetate, 8 µM amino acids, 255 µM spermidine, and 9 U murine RNase inhibitor (New England Biolabs). For cellular mRNA translation, the same parameters were used, except that the final reaction had 2 mM magnesium acetate and 200 mM potassium acetate. The magnesium and potassium concentrations were identified by titration to obtain maximum translation. Translation reactions were incubated for 2 h at 25 °C, and luciferase activity was assayed.
Sequencing Library Preparation.
The yeast strain GAL-RPL40A-∆Ubq was grown in YPG at 30 °C overnight to a final OD600 of ∼1.5. Yeast were pelleted and resuspended in YPG or glucose-containing medium, and a 500-mL culture was grown for 4 h to a final OD600 of 0.8. Yeast were incubated with 5 mL of 10 mg/mL cycloheximide for 3 min before being pelleted and washed once with ice cold polysome lysis buffer (20 mM Hepes, pH 7.4, 2 mM magnesium acetate, 100 mM potassium acetate, 3 mM DTT, 0.1 mg/mL cycloheximide). Pellets were resuspended in polysome lysis buffer, and cells were broken by glass bead lysis. Supernatant was clarified by centrifugation at 9,500 × g for 20 min at 4 °C, and aliquots were flash frozen in liquid nitrogen.
Twenty A260 units of sample were loaded on sucrose density gradients (10–50% sucrose in polysome lysis buffer) and spun for 3 h at 35,000 rpm at 4 °C in a Beckman SW41 rotor. Gradients were fractionated, fractions representing the polysomes were pooled, and RNA was isolated using guanidine hydrochloride. Deep sequencing libraries were prepared from total RNA samples using the TruSeq RNA Sample Preparation Kit (Illumina), and libraries were analyzed by sequencing on an Illumina GAII.
Supplementary Material
Acknowledgments
We thank M. J. Moore, S. Ghosh, A. Jacobson, and P. J. Kranzusch for troubleshooting conversations. R. Kerr assisted with performing the initial siRNA screen. This work was supported by National Institutes of Health Grants AI059371 and AI057159. S.P.J.W. is a recipient of a Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award. A.S.-Y.L. is supported by the Department of Defense through the National Defense Science and Engineering Graduate Fellowship Program and the National Science Foundation through the Graduate Research Fellowship Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216454109/-/DCSupplemental.
References
- 1.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA. 1995;1(10):985–1000. [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73(2):1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci USA. 2000;97(4):1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102(4):511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 8.Ito T, Lai MM. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology. 1999;254(2):288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 9.Poncet D, Aponte C, Cohen J. Rotavirus protein NSP3 (NS34) is bound to the 3′ end consensus sequence of viral mRNAs in infected cells. J Virol. 1993;67(6):3159–3165. doi: 10.1128/jvi.67.6.3159-3165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4(12):1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallie DR, Lewis NJ, Marzluff WF. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 1996;24(10):1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh D, Mohr I. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol. 2011;9(12):860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields BN, Knipe DM, Howley PM. 2007. Fields Virology (Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia), 5th Ed, pp 2, v, xix, 3091.
- 14.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433(7025):477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 15.Connor JH, Lyles DS. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J Virol. 2002;76(20):10177–10187. doi: 10.1128/JVI.76.20.10177-10187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zan M, Evans P, Lucas-Lenard J. The inhibition of mouse L-cell 45 S ribosomal RNA processing is a highly uv-resistant property of vesicular stomatitis virus. Virology. 1990;177(1):75–84. doi: 10.1016/0042-6822(90)90461-y. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed M, Lyles DS. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J Virol. 1998;72(10):8413–8419. doi: 10.1128/jvi.72.10.8413-8419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faria PA, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17(1):93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 19.Whitlow ZW, Connor JH, Lyles DS. Preferential translation of vesicular stomatitis virus mRNAs is conferred by transcription from the viral genome. J Virol. 2006;80(23):11733–11742. doi: 10.1128/JVI.00971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnitzlein WM, O’Banion MK, Poirot MK, Reichmann ME. Effect of intracellular vesicular stomatitis virus mRNA concentration on the inhibition of host cell protein synthesis. J Virol. 1983;45(1):206–214. doi: 10.1128/jvi.45.1.206-214.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnell MJ, Buonocore L, Whitt MA, Rose JK. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70(4):2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose JK, Lodish HF. Translation in vitro of vesicular stomatitis virus mRNA lacking 5′-terminal 7-methylguanosine. Nature. 1976;262(5563):32–37. doi: 10.1038/262032a0. [DOI] [PubMed] [Google Scholar]
- 23.Welnowska E, Castelló A, Moral P, Carrasco L. Translation of mRNAs from vesicular stomatitis virus and vaccinia virus is differentially blocked in cells with depletion of eIF4GI and/or eIF4GII. J Mol Biol. 2009;394(3):506–521. doi: 10.1016/j.jmb.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 24.Muthukrishnan S, Morgan M, Banerjee AK, Shatkin AJ. Influence of 5′-terminal m7G and 2′-O-methylated residues on messenger ribonucleic acid binding to ribosomes. Biochemistry. 1976;15(26):5761–5768. doi: 10.1021/bi00671a012. [DOI] [PubMed] [Google Scholar]
- 25.Connor JH, Naczki C, Koumenis C, Lyles DS. Replication and cytopathic effect of oncolytic vesicular stomatitis virus in hypoxic tumor cells in vitro and in vivo. J Virol. 2004;78(17):8960–8970. doi: 10.1128/JVI.78.17.8960-8970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6(18):2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci USA. 2002;99(19):12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35(5):1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schüler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13(12):1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 30.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23(23):2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhs M, et al. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011;39(12):5264–5275. doi: 10.1093/nar/gkr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338(6214):394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 33.Monia BP, et al. Gene synthesis, expression, and processing of human ubiquitin carboxyl extension proteins. J Biol Chem. 1989;264(7):4093–4103. [PubMed] [Google Scholar]
- 34.Pöll G, et al. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS ONE. 2009;4(12):e8249. doi: 10.1371/journal.pone.0008249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David AE. Size distributions of vesicular stomatitis virus-specific polysomes. J Gen Virol. 1978;39(1):149–160. doi: 10.1099/0022-1317-39-1-149. [DOI] [PubMed] [Google Scholar]
- 36.Huang AS, Baltimore D, Stampfer M. Ribonucleic acid synthesis of vesicular stomatitis virus. 3. Multiple complementary messenger RNA molecules. Virology. 1970;42(4):946–957. doi: 10.1016/0042-6822(70)90343-0. [DOI] [PubMed] [Google Scholar]
- 37.Redman KL. The smaller protein formed as a ubiquitin fusion in Drosophila is processed from ubiquitin and found on the 60S ribosomal subunit. Insect Biochem Mol Biol. 1994;24(2):191–201. doi: 10.1016/0965-1748(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 38.Nanbru C, et al. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272(51):32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 39.Nuss DL, Koch G. Differential inhibition of vesicular stomatitis virus polypeptide synthesis by hypertonic initiation block. J Virol. 1975;17(1):283–286. doi: 10.1128/jvi.17.1.283-286.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott MP, Pardue ML. Translational control in lysates of Drosophila melanogaster cells. Proc Natl Acad Sci USA. 1981;78(6):3353–3357. doi: 10.1073/pnas.78.6.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClanahan T, McEntee K. DNA damage and heat shock dually regulate genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6(1):90–96. doi: 10.1128/mcb.6.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8(7):913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spahn CM, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell. 2004;118(4):465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Jang CJ, Lo MC, Jan E. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J Mol Biol. 2009;387(1):42–58. doi: 10.1016/j.jmb.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 45.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145(3):383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334(6058):941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 47.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 48.Makarow M, Nevalainen LT, Kääriäinen L. Expression of the RNA genome of an animal virus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83(21):8117–8121. doi: 10.1073/pnas.83.21.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102(51):18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachs MS, et al. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods. 2002;26(2):105–114. doi: 10.1016/S1046-2023(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 51.Wu C, Amrani N, Jacobson A, Sachs MS. The use of fungal in vitro systems for studying translational regulation. Methods Enzymol. 2007;429:203–225. doi: 10.1016/S0076-6879(07)29010-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.