Abstract
Our understanding of the mechanisms by which ducts and lobules develop is derived from model organisms and three-dimensional (3D) cell culture models wherein mammalian epithelial cells undergo morphogenesis to form multicellular spheres with a hollow central lumen. However, the mechanophysical properties associated with epithelial morphogenesis are poorly understood. We performed multidimensional live-cell imaging analysis to track the morphogenetic process starting from a single cell to the development of a multicellular, spherical structure composed of polarized epithelial cells surrounding a hollow lumen. We report that in addition to actively maintaining apicobasal polarity, the structures underwent rotational motions at rates of 15–20 μm/h and the structures rotated 360° every 4 h during the early phase of morphogenesis. Rotational motion was independent of the cell cycle, but was blocked by loss of the epithelial polarity proteins Scribble or Pard3, or by inhibition of dynein-based microtubule motors. Interestingly, none of the structures derived from human cancer underwent rotational motion. We found a direct relationship between rotational motion and assembly of endogenous basement membrane matrix around the 3D structures, and that structures that failed to rotate were defective in weaving exogenous laminin matrix. Dissolution of basement membrane around mature, nonrotating acini restored rotational movement and the ability to assemble exogenous laminin. Thus, coordinated rotational movement is a unique mechanophysical process observed during normal 3D morphogenesis that regulates laminin matrix assembly and is lost in cancer-derived epithelial cells.
Keywords: extracellular matrix, tubulogenesis, plasticity
Tissues are composed of a community of cells that act in a coordinated manner to achieve and maintain normal architecture and perform their function. The mechanisms by which cells coordinate their behavior during tissue morphogenesis is poorly understood in part due to technical bottlenecks associated with studying the process in vivo. Three-dimensional (3D) culture systems provide an environment in which normal tissue morphogenesis can be recapitulated and thus is a powerful tool for investigating the molecular signals that specify epithelial tissue architecture (1–3). Unlike monolayer cultures, epithelial cells grown in 3D recapitulate numerous features seen in vivo, including the formation of acini-like spheroids with a hollow lumen, apicobasal polarization of cells comprising these acini, and the deposition of basement membrane components collagen IV and laminin-332 (2).
The initial stages of 3D morphogenesis are characterized by the ability of epithelial cells to detect surfaces with which they come in contact. Cells use integrins and dystroglycans to contact the extracellular matrix (ECM), and cadherins and desmosomes to contact their neighbors (3). These initial contacts trigger a series of cell remodeling events, which result in polarization of cells to create a basolateral surface and an apical surface with asymmetric distribution of membrane proteins. Whereas development of membrane protein asymmetry can occur in monolayer cultures, formation of multicellular structures of finite size and with a hollow central lumen is unique to 3D cultures. Epithelial cells immediately surrounding the lumen point their apical pole toward the lumen. This ability of cells to polarize is needed for morphogenesis and inhibiting polarization affects lumen formation and laminin assembly around 3D cysts (4). Exogenous laminin-1 rescues polarity and lumen formation, suggesting laminin assembly is an important step that initiates and promotes morphogenesis (4). The mechanisms by which cells establish polarization and assemble matrix around them are poorly understood.
To better understand 3D morphogenesis, we engineered Michigan Cancer Foundation (MCF)-10A human mammary epithelial cells to express fluorescent protein reporters and followed them by high-resolution four-dimensional imaging. We report a surprising observation that adds a unique insight into the morphogenetic process. We find that the 3D structures undergo a coordinated rotational movement during the early stages of morphogenesis and that this process is required for assembly of laminins and collagen around the 3D structures. Both 3D structures derived from MCF-10A cells lacking expression of polarity proteins and cancer-derived cell lines fail to undergo rotational motion and are defective in their ability to assemble laminin matrix.
Results
MCF-10A Cells Maintain Cell Polarity and Display Rotational Motion During 3D Morphogenesis.
To investigate the events that occur during in vitro 3D morphogenesis of human mammary epithelial cells, we created an MCF-10A reporter cell line stably coexpressing fluorescent histone 2B (H2B)–Venus and Golgin A2-mCherry (5) fusion proteins to mark the nucleus and the Golgi apparatus, respectively. Using these cells grown fully embedded in a Matrigel matrix, we performed live-cell imaging of acinar morphogenesis over a period of 10 d with a time resolution of 2 h, starting with single cells and culminating in multicellular acini. Live-cell imaging data were processed using Imaris software (Bitplane). Three-dimensional intensity thresholding and tracking analysis functions allowed us to identify and track nuclei and Golgi movements over time. Golgi orientation was used as a surrogate marker of the apical pole of a cell undergoing morphogenesis. We discovered that cell polarity, as monitored by the presence of the apical pole, is actively maintained during multiple rounds of cell division, despite marked cell movement and reorganization (Fig. 1A and Movie S1). Because MCF-10A cells do not develop tight junctions, a hallmark of apicobasal membrane polarity, it is likely that development and maintenance of the apical pole is independent of the cell’s ability to establish apicobasal membrane polarity.
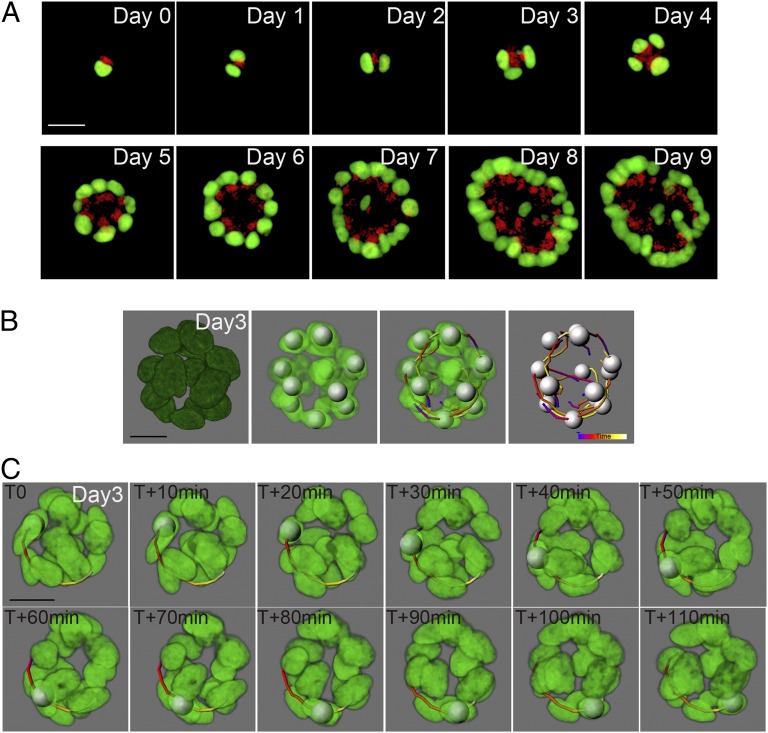
Fig. 1.
MCF-10A acini maintain cell polarity and display a coordinated rotational motion during morphogenesis. (A) Selected equatorial views of an MCF-10A human mammary epithelial cell acinus expressing a nucleus marker (H2B–Venus, green) and a Golgi apparatus marker (GOLGA2–mCherry, red). Images were taken at 2-h intervals for 10 d (Movie S1). (B) Data were processed in Imaris software allowing detection of nuclei (shown as silver spheres) and their movement over time (shown here as colored tracks). Blue end indicating t0, progressing to purple, red, yellow, and ending with white indicating t+2h. In most structures, the white end is masked by the silver sphere. (C) One nucleus was tracked within an MCF-10A acinus, at day 3. The nucleus was followed every 10 min for a period of 2 h. (Scale bars, 20 μm.)
To better understand the morphogenetic process from a single cell to multicellular acini, we increased the time resolution, scanning 3D structures every 10 min for 2 h every day from day 1 to day 9. Surprisingly, we found that cells moved following rotational movements along the periphery and diagonal movements across the lumen during morphogenesis (Fig. 1B). The structure rotated 360° every 4 h (Fig. 1C). These observations highlight the dynamic nature of the morphogenetic program.
Rotational Motion Occurs During Early Stages of 3D Morphogenesis and Depends on Microtubule Networks.
Detailed analysis of coordinated rotational motion from day 0 to day 8 showed that, whereas on day 0 single MCF-10A cells did not rotate (Fig. S1A), beginning from day 1 the cells displayed rotational motion for the first 4 d of morphogenesis (Fig. 2A), which stopped by day 5, despite the fact that the structures continued to grow. The cells, however, continued to show random movement until day 7 and all types of movement stopped in mature (day 8 or older) acini. Quantifications showed that 90–100% of days 2–3 acini showed rotational motion and more than 70% of structures underwent rotational movement on day 4 of 3D morphogenesis (Table S1) with speeds ranging from 13 µm⋅h−1 to 17 µm⋅h−1 (Fig. 2B). No preference for left- or right-hand orientation was detected, and few structures changed the direction of rotation during the course of morphogenesis. The random movement we observed after day 5 is similar to the noninvasive movement previously reported in MCF-10A acini (6), and the meandering movement of epithelial cells in salivary bud (7); however, in both cases, no movement was observed in mature acini, consistent with our observations. These observations demonstrate that rotational and random movements are observed during early stages of morphogenesis and that random movements can occur in the absence of rotational movements, suggesting that rotational and random movements may be regulated by distinct mechanisms.
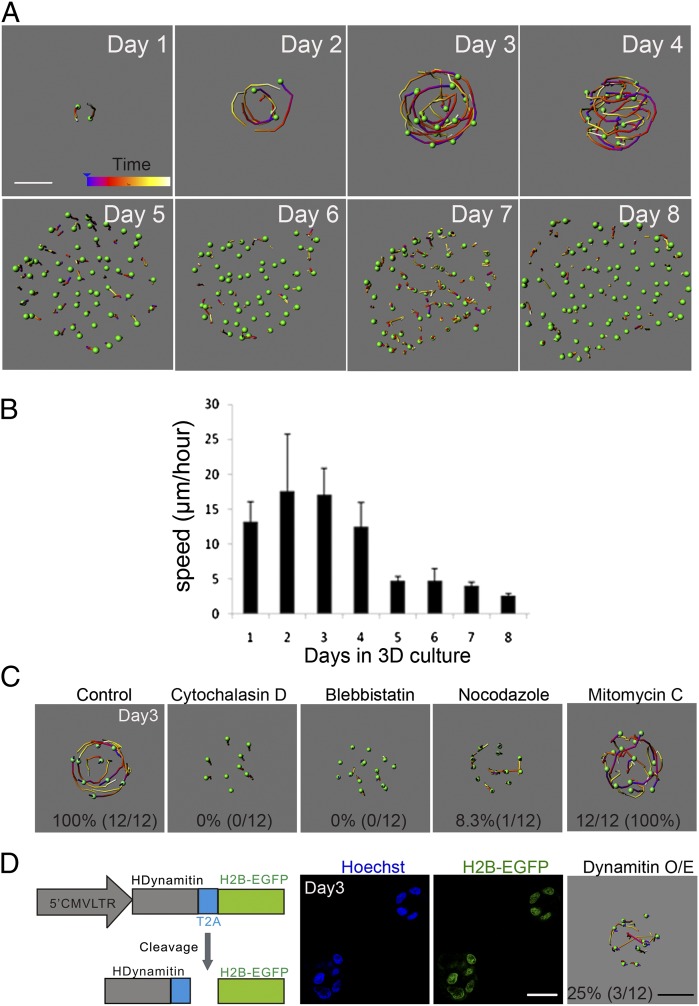
Fig. 2.
Coordinated rotational motion occurs during early stages of MCF-10A 3D morphogenesis and is dependent on microtubule dynamics. (A) Analysis of cell movement in MCF-10A acini. Z stacks of MCF-10A acini expressing H2B–Venus were acquired every 10 min for 2 h during 8 d of morphogenesis. (B) MCF-10A speed analysis. Average nuclei speed was calculated using data from three independent experiments. (C) MCF-10A acini cell movement analysis after treatment with the indicated inhibitors on day 3 of morphogenesis. Percentages represent the number of structures that show normal rotational motion. (D) Schematic representation of the pMSCV–hDynamitin–T2A–H2B–EGFP construct. Day 3 acini from MCF-10A cells infected with retrovirus expressing hDynamitin–T2A–H2B–EGFP. (White scale bars, 20 μm; black scale bar, 15 μm.)
The actin cytoskeleton plays a critical role during morphogenesis and actomyosin contractility is required for noninvasive movement (9). We first investigated the role of actin fibers during rotational motion. To monitor changes in actin dynamics during morphogenesis, we generated MCF-10A cells expressing red fluorescent protein-tagged Lifeact (Lifeact-TagRFP), a 17-amino-acid peptide that labels filamentous actin (F-actin) (Fig. S1B). Interference of F-actin with cytochalasin D or inhibition of myosin II with blebbistatin is known to block stress fiber formation. Treatment of day 3 acini with 5.0 μM cytochalasin D or 50.0 μM blebbistatin disrupted cortical actin (Fig. S1B) and strongly inhibited both rotational and random movements (Fig. 2C). To test whether increased actomyosin contractility can induce rotational motion, the acini were treated with lysophosphatidic acid (LPA), an extracellular signaling phospholipid known to activate myosin II by stimulating phosphorylation of myosin light chain (MLC) (10, 11). However, we did not observe accelerated rotational motion in 3D acini treated with LPA (Fig. S1E). Thus, inhibiting actomyosin contractility reduces both rotational and random cell movements in 3D acini.
To test the relationship between cell cycle progression and rotational motion, we treated day 3 MCF-10A acini with mitomycin C, a DNA cross-linking agent, for 7 h before imaging (Fig. S1C). Rotational motion was still detectable in acini treated with mitomycin C (Fig. 2C) and no significant changes in movement speed were found in these structures (Fig. S1E). This is consistent with previous observations that hyperproliferation is not sufficient to induce noninvasive movement (9) and that mitotic inhibition does not block epithelial cell movement during submandibular gland morphogenesis (12).
Microtubules have been shown to mediate directional cell migration and Golgi positioning in cells (13). To test whether the microtubule cytoskeleton is involved in rotational motion, we disrupted the microtubule network using the microtubule depolymerizing drug, nocodazole (Fig. S1D). This drug blocked rotational movement of day 3 acini where only 8.3% rotated in treated groups compared with 100% of rotating structures in the control groups, without completely inhibiting random movements (Fig. 2C and Fig. S1E), demonstrating a selective role for microtubule dynamics during rotational movement. Lifeact-TagRFP images did not show a difference in the organization of cortical actin in nocodazole-treated cells (Fig. S1B), suggesting that the nocodazole effect was not due to indirect effects on actin. Dynein motors play important roles during microtubule-mediated processes. To genetically interfere with microtubule function we ovexpressed dynamitin and disrupted dynein function (Fig. S2A) (14, 15). We observed a 75% decrease in the number of structures that rotated and a 50% decrease in the speed of movement (Fig. 2D and Fig. S2B). However, dynamitin overexpression did not block the random movement of cells within acini (Fig. 2D). Consistent with previous reports (14), overexpression of dynamitin disrupted Golgi organization and orientation (Fig. S2C), suggesting that organalle localization may play an important role during rotational motion. Thus, microtubule polymerization and dynein motors are required for coordinated rotational motion but are dispensable for random migration.
Rotational Motion Depends on Cell Polarity and Is Impaired in 3D Structures Generated from Cancer Cell Lines.
Because loss of cell polarity proteins is known to interfere with morphogenesis (8), we investigated the effect of depleting expression of polarity proteins Scribble and Pard3 on rotational movement. We down-regulated expression of Scribble and Pard3 using RNA interference (Fig. S2 D and E). To confirm that loss of these polarity proteins affects cell polarization in 3D acini, we monitored the orientation of the Golgi apparatus, which marks the apical pole of polarized epithelial cells. Whereas the Golgi was oriented toward the lumen in control acini (Fig. 3A), this orientation was disrupted in both Scribble knockdown (ScribKD) and Pard3 knockdown (Pard3KD) cells, with Pard3KD having a stronger effect than ScribKD (Fig. 3 B and C). The ScribKD and Pard3KD day 4 acini failed to display any rotational motions (Fig. 3 B and C); however, the cells retained the ability to undergo random movements. These observations demonstrate that the cell polarity proteins Scribble and Pard3 are required for rotational motion, whereas they are dispensable for the random movement in 3D acini.
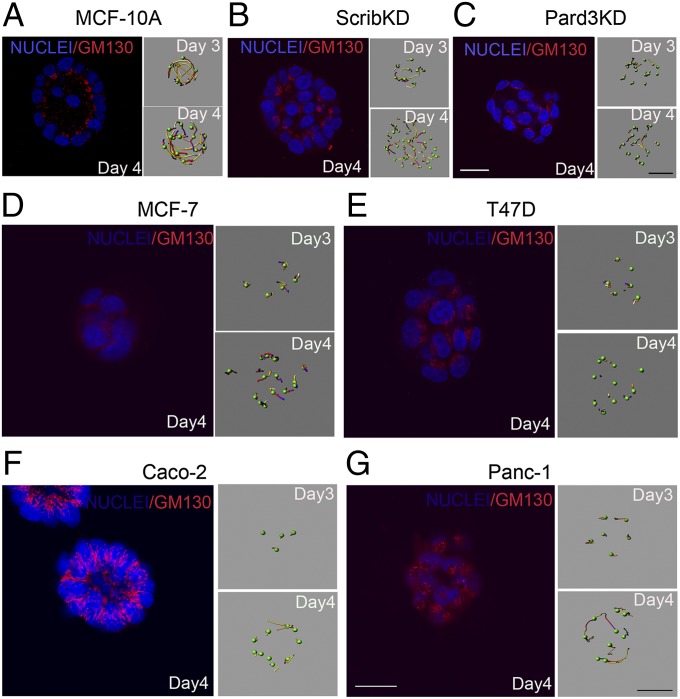
Fig. 3.
Coordinated rotational motion, loss of apicobasal polarity, and cancer cells. (A–C) Days 3 and 4 wild-type (MCF-10A), Scribble shRNA knockdown (ScribKD) and Pard3 shRNA knockdown (Pard3KD) acini were immunostained for GM130 and counterstained with Hoechst to show nuclei (blue). Right shows analysis of cell movement. MCF-7 (D), T47D (E), Caco-2 (F), and Panc-1 (G) cancer-derived cell lines expressing H2B–Venus were grown inside Matrigel and their movement was monitored on days 3 and 4. Day 4 structures were immunostained for GM130. (White scale bars, 20 μm; black scale bar, 15 μm.)
Considering that morphogenetic programs are frequently altered in tumor-derived epithelial cells, we analyzed several human cancer-derived cell lines: breast cancer-derived MCF-7 and T47D, colon cancer-derived Caco-2, and pancreatic cancer-derived Panc-1 cells. We chose these cells to represent diverse epithelial duct-containing organs and due to their ability to grow as spheres when embedded within a 3D matrix, thus allowing for a direct comparison with MCF-10A acini. Whereas the spheres formed by MCF-7, T47D, and Panc-1 cells did not have a lumen (Fig. 3 D, E, and G), Caco-2 cells formed cyst-like structures with a single epithelial cell layer surrounding a central lumen (Fig. 3F), consistent with previous reports (16). Strikingly, none of the spheres formed by cancer-derived cell lines displayed rotational motion. The inability to undergo rotational movement was not due to differences in growth rates because these cancer cell lines developed into structures of similar or larger sizes and with similar cell numbers per structure, compared with MCF-10A acini. The lack of rotational motion was also not due to differences in lumen formation because structures developed from Caco-2 cells had a proper lumen (Fig. 3F). Interestingly, the orientation and/or organization of the Golgi apparatus was disrupted in all cancer cell-derived structures (Fig. 3 D–G), indicating that, like ScribKD and Pard3KD MCF-10A cells (Fig. 3 A and B), cancer-derived cells lack the ability to orient their apical pole within 3D structures. We note that, whereas Caco2 cells retain the ability to establish apicobasal membrane polarity (as defined by tight junctions and apical membrane proteins) (16), they fail to establish an apical pole (as defined by Golgi orientation), suggesting that establishment of apicobasal membrane polarity and establishment of the apical pole are regulated by distinct mechanisms. Thus, we demonstrate that rotational motion is lost in cancer cells and identify a direct relationship between ability of cells to orient their apical pole (as defined by Golgi localization) toward the lumen in 3D cysts and rotational motion.
Consistent with the role for microtubules during rotation of MCF-10A acini, microtubule organization was aberrant in all cancer cell lines analyzed compared with that observed in MCF-10A cells (Fig. S3A). Microtubule networks play critical roles during vesicle transport. To investigate whether there were gross differences in vesicle transport between MCF-10A and cancer cell lines, we monitored overall organization of early endosomes using a reporter that marks early endosomes using Rab5a targeting sequence. We notice that MCF-10A cells show the expected concentrated localization of endosomes in a region above the nucleus, whereas the cancer cell lines show a diffused signal throughout the cells, suggesting a defect in endosomal trafficking process (Fig. S3B). Consistent with the possible differences in trafficking processes, alpha6 integrin, failed to show basal localization in cancer cells compared with MCF-10A cells (Fig. S3C), suggesting that localization of matrix receptors may contribute to the defect in the ability of cancer cells to rotate.
Rotational Motion and Assembly of Laminins 111 and 332 and Collagen IV.
Basement membranes are specialized layers of extracellular matrix proteins that underlie the basal side of all epithelia and play a critical role during morphogenesis (17, 18). We tested if the rotational motion observed in MCF-10A acini directs assembly of basement membrane components, like collagen IV and laminins, that are secreted by MCF-10A cells during morphogenesis.
Using antibodies specific to human laminin beta 1 and gamma 2 chains, and human collagens IV, we monitored basement membrane organization around 3D acini. Laminin-332 (visualized by laminin gamma 2 chain immunostaining) was primarily cytosolic on day 1 of morphogenesis; however, on day 2 we observed increased deposition of laminin-332 at the periphery of developing acini (Fig. 4A). By day 3, laminin-332 was primarily localized to the basal surface, and by day 4 the acini were surrounded by a thin, compact, continuous layer of laminin and collagen IV (Fig. 4 A and C). Acini derived from ScribKD had disrupted organization of both laminin-332 and laminin-111 (visualized by laminin beta 1 chain immunostaining), showing abnormal patches and a multilayered laminin matrix (Fig. S4 A and B), and collagen IV deposition was either diffuse or discontinuous (Fig. S4C). Loss of Pard3 had a more dramatic effect in that laminins or collagen were primarily present in the cytosol (Fig. 4 B and C and Fig. S4B). Dynamitin overexpression showed a delay in assembly for both types of matrix proteins that was apparent at days 2 and 2.5 (Fig. S5). Interestingly, day 4 structures from all of the cancer cell lines showed clear defects in laminin-111 and laminin-332 matrix organization compared with that observed around MCF-10A acini (Fig. 4B and Fig. S4 A and B). In addition, structures derived from T47D, Panc-1, MCF-7, and Caco-2 cells did not form collagen matrix and collagen was primarily detected within the cytosol (Fig. 4C and Fig. S4C).
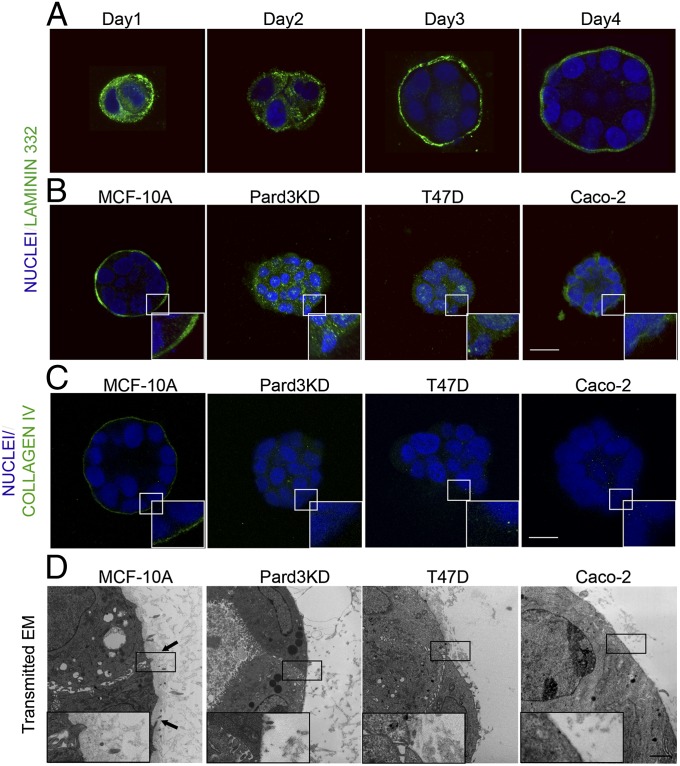
Fig. 4.
Structures with impaired rotational motion do not assemble a proper basement membrane. (A) Assembly of laminin-332 during 3D morphogenesis. MCF-10A cells grown inside Matrigel were immunostained for laminin-332 (green) at indicated days after plating. Confocal images through the equator of the structures are shown. (B and C) Day 4 structures of wild-type and Pard3KD MCF-10A, T47D, and Caco-2 cells were immunostained for laminin-332 or collagen IV. Lower Right Inset shows a magnification of the indicated white rectangular region. (Scale bar, 20 μm.) (D) Transmission electron microscopy images of basement membrane organization in MCF-10A, Pard3KD MCF-10A, T47D, and Caco-2 structures. Arrows indicate regions of densely organized basement membrane immediately adjacent to the basal cell surface of wild-type MCF-10A acinus. Lower Left Inset shows the digital magnification of the indicated black rectangular region. (Scale bar, 2.0 μm.)
Basement membrane structure and organization can also be evaluated by transmission electron microscopy (TEM) (19). The ultrastructure of wild-type MCF-10A acini basement membrane showed a dense, sheet-like organization (Fig. 4D, arrows). However, basement membranes of structures that did not rotate were nonexistent or loosely organized and discontinuous (Fig. 4D). These results demonstrate a correlation between the ability of 3D structures to undergo rotational motion and assemble the basement membrane.
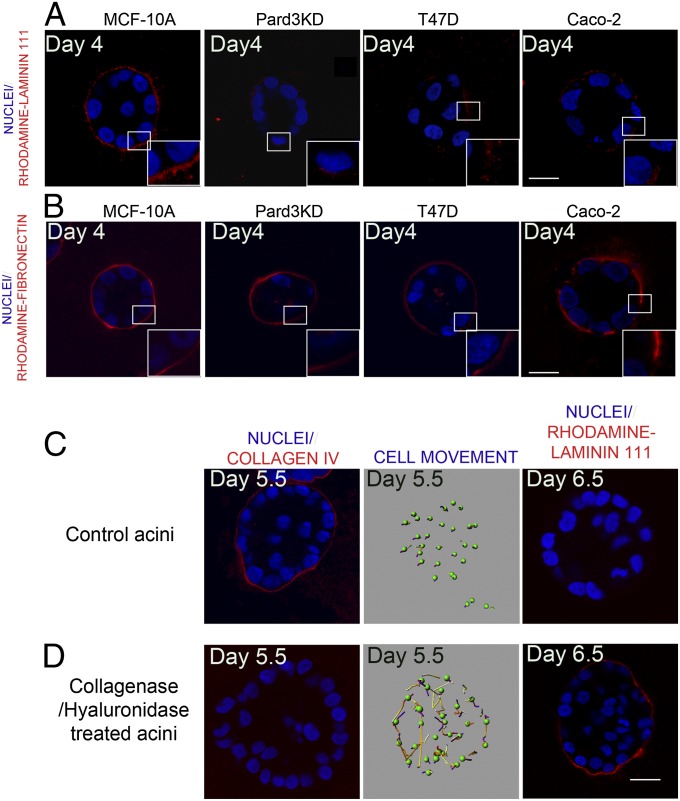
Next we investigated the relationship between rotational motion and basement membrane assembly. To overcome any caveats associated defective secretion of laminin, we examined the assembly of exogenous laminin by adding rhodamine-labeled mouse laminin-111 to the medium. Similar to endogenously produced laminins, MCF-10A acini assembled a continuous, compact layer of rhodamine–laminin-111 (Fig. 5A) surrounding the acinus. However, rhodamine–laminin-111 assembly around acini from MCF-10A dynamitin overexpressing, Pard3KD and ScribKD cells appeared less uniform and was diffusely localized to the basal cell surface (Fig. 5A and Fig. S6A). All cancer cell-derived structures showed an incomplete and less condensed rhodamine–laminin-111 assembly (Fig. 5A and Fig. S6A). These observations demonstrate a direct relationship between the ability of 3D structures to rotate and their ability to assemble exogenous laminin.
Fig. 5.
Relationship between rotational motion and laminin-111 assembly. MCF-10A, Pard3KD MCF-10A, T47D, and Caco-2 cells were grown for 3 d in Matrigel, and then 50 μg/mL of rhodamine-labeled mouse laminin-111 (A) or fibronectin (B) was added into the culture medium. After 24 h (laminin) or 8 h (fibronectin), the structures were fixed and rhodamine-labeled matrix proteins were directly visualized in the 568-nm channel (red). Normal MCF-10A acini (C) or acini treated with collagenase/hyalurondiase treatment (D) were immunostained for collagen IV (Left), monitored for rotational movement (Center) or incubated with rhodamine–laminin-111 (Right). (Scale bars, 20 μm.)
To determine if rotation is required for assembly of all matrix proteins or specific to components of basement membrane, we examined the assembly of rhodamine-labeled fibronectin, an ECM protein that is not a basement membrane component (22). Three-dimensional structures from all cell lines tested assembled rhodamine–fibronectin in a relatively continuous, uniform ring-shape organization regardless of their ability to undergo rotational motion (Fig. 5B and Fig. S6B).
To strengthen the relationship between acini rotation and basement membrane assembly, we tested if development of basement membrane was contributing to the cessation of rotational motion observed after day 4 during MCF-10A morphogenesis. Strikingly, rotational movements were restored in day 5.5 structures if the basement membrane was digested by a collagenase/hyaluronidase treatment (Fig. 5 C and D). Furthermore, whereas day 6.5 MCF-10A acini lacks the ability to assemble exogenous rhodamine–laminin-111, collagenase/hyaluronidase treated day 6.5 regained the ability to assemble exogenous laminin (Fig. 5 C and D), suggesting that restoring rotational motion is sufficient to assemble exogenous laminin by MCF-10A acini. Taken together our findings indicate that coordinated rotational motion during 3D morphogenesis of epithelial mammary acini is a unique regulator of the basement membrane assembly process.
Discussion
We report that 3D acini undergo rotational motion during the first 4 d of morphogenesis. Whereas our observation is consistent with recent studies on rotational motions (23, 24), we find an unexpected role for rotational motion during assembly of basement membrane matrix. Interestingly, assembly of fibronectin matrix did not require the rotational motion. Thus, we report a unique mechanophysical property associated with basement membrane assemble during 3D acinar morphogenesis.
In addition, we find that both cell polarity and microtubule dynamics are required for the rotational motion. Whereas inhibition of actomyosin contractility blocked both rotational and random cell movement, disruption of microtubules inhibited rotational motion without affecting random movement. We are intrigued by the specificity of the role played by microtubules because of the intimate relationship between the microtubule cytoskeleton and polarized cellular processes such as migration and vesicle trafficking (25). In polarized epithelia, microtubules align along the lateral membrane with minus ends facing the apical and plus ends facing the basal domains (26). Consistent with this notion, we find that the overall organization of microtubules was disrupted in 3D structures derived from cancer cell lines that failed to rotate (Fig. S3A). Whereas we observe defects in organization of Rab5a early endosome and basal localization of alpha6 integrin (Fig. S3 B and C), the mechanisms may also involve orientation of centrosomes, or Golgi or directed vesicle transport. It is also possible to establish polarity by regulating orientation of the apical pole by positioning organelles such as the Golgi or centrosomes along the axis of polarity. Golgi elements directly interact with microtubule motors, dynein and kinesin. The interaction of the Golgi with the dynein1 complex is critical for both compaction of the Golgi and maintenance of Golgi stacks (27). Disruption of directional Golgi orientation by loss of golgins, golgin-160 and GMAP210, impairs directional cell migration and wound healing, demonstrating a direct relationship between polarization of Golgi stacks and directional migration (28). It is likely that the relationship between Golgi orientation (apical pole) and rotational motion involves similar mechanisms, highlighting the need to understand how cells establish and coordinate directionality during rotation.
We document an unexpected relationship between rotational motion and assembly of basement membrane. We observed loss of basement membrane by electron microscopy and loss of laminin-111, laminin-332, and collagen IV components of basement membrane. In addition to collagen IV and laminin, basement membranes contain nidogens, perlecan, and agrin. Whereas the process of basement membrane assembly is not well understood, it is thought that interaction of laminin with cell surface receptors integrin and dystroglycans initiate the process of basement membrane assembly. The laminin matrix is linked to the collagen IV by the action of the linking proteins, nidogen, perlecan, and agrin during the maturation of the basement membrane lattice (29). The role rotational motion plays during this process is not known. Consistent with our observations, a recent study identifies a role for tissue revolutions playing a role during ECM assembly in developing Drosophila eggs (30). Together these studies suggest a hitherto unexpected, evolutionarily conserved role for rotational motion during assembly of basement membrane.
Properly assembled basement membrane plays a critical role during establishment of cell polarity and epithelial morphogenesis (31). Defects in ECM assembly will also affect the ability of cancer cells to invade during metastasis (32). Thus, our observations identify a unique aspect of morphogenesis that can impact both normal morphogenesis and carcinoma. Our results also highlight the need to place more emphasis on understanding the mechanisms that regulate plasticity and dynamics of cellular interactions where cell–cell and cell–matrix interactions are in a state of constant flux.
Materials and Methods
Cell Culture and 3D Morphogenesis Assay.
The culture condition of MCF-10A, Pard3KD, and ScribKD cells was described previously (33). T47D cells were cultured in RPMI medium 1640 supplemented with 10% (vol/vol) FBS and 10 µg/mL insulin. MCF-7 cells were cultured in Eagle's Minimum Essential Medium (ATCC 30-2003) supplemented with 10% (vol/vol) FBS, 10 µg/mL insulin; Panc-1 cells were cultured in DMEM supplemented with 10% FBS. Caco-2 cells were cultured in EMEM medium supplemented with 10% FBS and 2 mM glutamine. The details of 3D morphogenesis assay were described previously (33).
Electron Microscopy.
Three-dimensional acini were fixed for transmission electron microscopy with 2% glutaraldehyde in 100 mM Na-Cacodylate buffer (pH 7.4) for 1–2 h and processed subsequently by the University of Toronto EM facility (www.medresearch.utoronto.ca/mil_home.html).
Confocal Microscopy and Image Analysis.
Day 3 and day 4 acini were fixed and processed for immunohistochemistry as previously described (33). The following primary antibodies were used. Anti-GM130 for visualizing the Golgi complex (1/100; BD), antilaminin-gama2 for laminin-332 (1/100; Millipore), anticollagen IV (1/100; DAKO), antilaminin β1 subunit for laminin-111 (1/100; Millipore), anti-Ki67 (1/100; Zymed), anti-α-tubulin (1/100; Sigma), and anti-β1-integrin (1/100; DSHB). After overnight incubation at 4 °C, secondary antibodies conjugated with Alexa 488 or 568 (Molecular Probes) were applied for 2 h at room temperature. Slides were incubated for 15 min with Hoechst 33342 (Molecular Probes) to label nuclei and examined using an Olympus LV1000 confocal microscope.
Detailed information on reagents used, molecular biology, production of stable cell lines, 4D confocal microscopy, immunoblotting, and statistical analysis can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the S.K.M. laboratory for helpful discussions, Laurie Seifried for technical assistance, and Jennifer Hayes for assistance with manuscript preparation. This work was supported by Grants CA098830 and CA105388 from National Cancer Institute; Era of Hope Scholar Award BC075024 from Department of Defense Breast Cancer Research Program; Lee K. Margaret Lau Chair for Breast Cancer Research; and Campbell Family Institute For Breast Cancer Research (S.K.M.). This work was also funded in part by the Ontario Ministry of Health and Long Term Care. The views expressed do not necessarily reflect those of the OMOHLTC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201141110/-/DCSupplemental.
References
- 1.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1(1):46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien LE, Zegers MMP, Mostov KE. Opinion: Building epithelial architecture: Insights from three-dimensional culture models. Nat Rev Mol Cell Biol. 2002;3(7):531–537. doi: 10.1038/nrm859. [DOI] [PubMed] [Google Scholar]
- 4.O’Brien LE, et al. Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nat Cell Biol. 2001;3(9):831–838. doi: 10.1038/ncb0901-831. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura N, et al. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131(6 Pt 2):1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson GW, Hunter T. Real-time imaging reveals that noninvasive mammary epithelial acini can contain motile cells. J Cell Biol. 2007;179(7):1555–1567. doi: 10.1083/jcb.200706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci. 2006;119(Pt 16):3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- 8.Huang L, Muthuswamy SK. Polarity protein alterations in carcinoma: A focus on emerging roles for polarity regulators. Curr Opin Genet Dev. 2010;20(1):41–50. doi: 10.1016/j.gde.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson GW, Hunter T. Real-time imaging reveals that noninvasive mammary epithelial acini can contain motile cells. J Cell Biol. 2007;179(7):1555–1567. doi: 10.1083/jcb.200706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70(3):389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 11.Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 12.Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci. 2006;119(Pt 16):3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- 13.Etienne-Manneville S. From signaling pathways to microtubule dynamics: The key players. Curr Opin Cell Biol. 2010;22(1):104–111. doi: 10.1016/j.ceb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139(2):469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquot G, Maidou-Peindara P, Benichou S. Molecular and functional basis for the scaffolding role of the p50/dynamitin subunit of the microtubule-associated dynactin complex. J Biol Chem. 2010;285(30):23019–23031. doi: 10.1074/jbc.M110.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J Cell Biol. 2008;183(4):625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timpl R. Structure and biological activity of basement membrane proteins. Eur J Biochem. 1989;180(3):487–502. doi: 10.1111/j.1432-1033.1989.tb14673.x. [DOI] [PubMed] [Google Scholar]
- 18.Yurchenco PD, Schittny JC. Molecular architecture of basement membranes. FASEB J. 1990;4(6):1577–1590. doi: 10.1096/fasebj.4.6.2180767. [DOI] [PubMed] [Google Scholar]
- 19.Vracko R. Basal lamina scaffold-anatomy and significance for maintenance of orderly tissue structure. Am J Pathol. 1974;77(2):314–346. [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki T, et al. Deficiency of beta 1 integrins in teratoma interferes with basement membrane assembly and laminin-1 expression. Exp Cell Res. 1998;238(1):70–81. doi: 10.1006/excr.1997.3837. [DOI] [PubMed] [Google Scholar]
- 21.Aumailley M, Pesch M, Tunggal L, Gaill F, Fässler R. Altered synthesis of laminin 1 and absence of basement membrane component deposition in (beta)1 integrin-deficient embryoid bodies. J Cell Sci. 2000;113(Pt 2):259–268. doi: 10.1242/jcs.113.2.259. [DOI] [PubMed] [Google Scholar]
- 22.Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Curr Pharm Des. 2009;15(12):1277–1294. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanner K, Mori H, Mroue R, Bruni-Cardoso A, Bissell MJ. Coherent angular motion in the establishment of multicellular architecture of glandular tissues. Proc Natl Acad Sci USA. 2012;109(6):1973–1978. doi: 10.1073/pnas.1119578109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmaras A, et al. A mathematical method for the 3D analysis of rotating deformable systems applied on lumen-forming MDCK cell aggregates. Cytoskeleton (Hoboken) 2010;67(4):224–240. doi: 10.1002/cm.20438. [DOI] [PubMed] [Google Scholar]
- 25.Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol. 2008;9(11):833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119(Pt 20):4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 27.Sengupta D, Linstedt AD. Control of organelle size: The Golgi complex. Annu Rev Cell Dev Biol. 2011;27:57–77. doi: 10.1146/annurev-cellbio-100109-104003. [DOI] [PubMed] [Google Scholar]
- 28.Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20(6):1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yurchenco PD. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3(2):1–27. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haigo SL, Bilder D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science. 2011;331(6020):1071–1074. doi: 10.1126/science.1199424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu W, et al. Beta1-integrin orients epithelial polarity via Rac1 and laminin. Mol Biol Cell. 2005;16(2):433–445. doi: 10.1091/mbc.E04-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: Architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12(5):308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93(21):11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.