Abstract
The MAM domain-containing GPI anchor proteins MDGA1 and MDGA2 are Ig superfamily adhesion molecules composed of six IG domains, a fibronectin III domain, a MAM domain, and a GPI anchor. MDGAs contribute to the radial migration and positioning of a subset of cortical neurons during early neural development. However, MDGAs continue to be expressed in postnatal brain, and their functions during postnatal neural development remain unknown. Here, we demonstrate that MDGAs specifically and with a nanomolar affinity bind to neuroligin-2, a cell-adhesion molecule of inhibitory synapses, but do not bind detectably to neuroligin-1 or neuroligin-3. We observed no cell adhesion between cells expressing neuroligin-2 and MDGA1, suggesting a cis interaction. Importantly, RNAi-mediated knockdown of MDGAs increased the abundance of inhibitory but not excitatory synapses in a neuroligin-2–dependent manner. Conversely, overexpression of MDGA1 decreased the numbers of functional inhibitory synapses. Likewise, coexpression of both MDGA1 and neuroligin-2 reduced the synaptogenic capacity of neuroligin-2 in an artificial synapse-formation assay by abolishing the ability of neuroligin-2 to form an adhesion complex with neurexins. Taken together, our data suggest that MDGAs inhibit the activity of neuroligin-2 in controlling the function of inhibitory synapses and that MDGAs do so by binding to neuroligin-2.
Keywords: inhibitory synapse formation, synaptic cell adhesion, autism, schizophrenia
Recent studies of synapse formation have uncovered a multitude of synaptic adhesion molecules, and human genetic studies have implicated many of these molecules in neuropsychiatric and neurodevelopmental disorders (1–4). However, little is known about the specific pathophysiological mechanisms by which dysfunctions of synaptic adhesion molecules contribute to these complex disorders.
Neurexins and neuroligins (NLs) are arguably the most extensively studied synaptic adhesion molecules (1). They are dispensable for initial synapse establishment but act in an isoform-dependent manner to specify the maturation of either excitatory or inhibitory synapses (5). There are four NL members in rodents (NL1–NL4) that show distinct synaptic localizations and functions (5). NL2, in particular, has received considerable attention because of its unique localization and function at inhibitory synapses (6). For instance, NL2 controls perisomatic inhibitory synapse maturation together with gephyrin and collybistin, which regulate GABA receptor clustering on neurons (7, 8). Moreover, NL2 exhibits differential functions at different types of inhibitory synapses on the same postsynaptic neuron (9). All four NLs likely mediate synapse-promoting activities through direct interactions with presynaptic neurexins, but NLs also perform additional functions in synapse validation that are independent of their binding to neurexins (10).
MAM domain-containing GPI anchor proteins (MDGAs), also termed “GPIMs” or “MAMDCs,” initially were identified in tumor cells (11). The two homologous MDGA proteins, MDGA1 and MDGA2, possess a characteristic domain organization composed of six Ig domains, a fibronectin type III repeat (FNIII), a single meprin/A5 protein/receptor protein tyrosine phosphatase mu (MAM) domain, and a C-terminal GPI anchor (12). Like other Ig-domain superfamily members, including neural cell adhesion molecule (NCAM) and L1 cell adhesion molecule (L1-CAM), MDGA proteins may mediate homophilic cell adhesion (13, 14). MDGAs are highly expressed in the developing brain (12, 15, 16). Knockout of MDGA1 causes a discrete phenotype in neuronal migration and impairs rostral growth of commissural axons, suggesting an important role during the initial development of the brain (17, 18). Interestingly, SNP-based, large-scale human genetic analyses suggested that MDGA1 and MDGA2 may be susceptibility genes for schizophrenia, bipolar disorder, and autism spectrum disorder (19–21). However, despite continued high-level expression of MDGAs in adult brain, their functions in mature neurons remain unknown.
Here, we demonstrated that MDGAs interact directly with NL2, but not with NL1 or NL3. Knockdown of MDGAs using RNAi specifically increased inhibitory synapse numbers in an NL2-dependent manner. Moreover, we observed that MDGA1 decreased the synaptogenic activity of NL2 in artificial synapse-formation assays. These results suggest that MDGAs regulate inhibitory synapses via their direct interaction with NL2.
Results
In preliminary screening experiments, we used cell-surface binding assays with IgC-neuroligin-1 (IgNL1), IgC-neuroligin-2 (IgNL2), IgC (negative control), and a variety of cell-surface proteins to identify potential ligand(s) for NLs (Table S1). Our screening revealed that MDGA1 binds selectively and directly to NL2 (see below). Therefore, we sought to characterize the interaction between MDGAs and NL2 and to investigate the functional importance of this interaction.
Expression of MDGA mRNAs in Adult Rat Brains.
NLs are known to validate initially established synaptic connections during synapse development in an activity-dependent manner (22–24). Thus, to qualify as potential NL ligands, MDGAs should be expressed in early and in adult stages of brain development. Previously, the expression of MDGA mRNAs was examined by Northern blot analysis (12). MDGA1 expression was prominent in the basilar pons, cortices, hippocampus, amygdale, olfactory bulb, and thalamus from embryonic day 15 (E15) to postnatal day 7 (P7) of rat brain. In contrast, MDGA2 was expressed more widely throughout various brain areas (12). To determine whether MDGA expression persists throughout the adult rodent brain, we performed in situ hybridization of rat brain sections at later stages of development and in adults (Fig. S1). As reported previously, we found that MDGA1 mRNAs were highly expressed in the embryonic neocortical neuroepithelium, postnatal neocortical superficial layer, and cerebellar granular layer. Notably, continued robust MDGA1 expression was observed in various regions of adult rat brains (Fig. S1). In contrast to MDGA1, MDGA2 was expressed at relatively higher levels in embryonic brains. NL2 mRNAs were widely distributed in various brain areas throughout embryonic and postnatal stages. Taken together, these data indicate that MDGA mRNAs are widely expressed in adult rat brain.
MDGA1 Binds Selectively to NL2 but Not to NL1 or NL3.
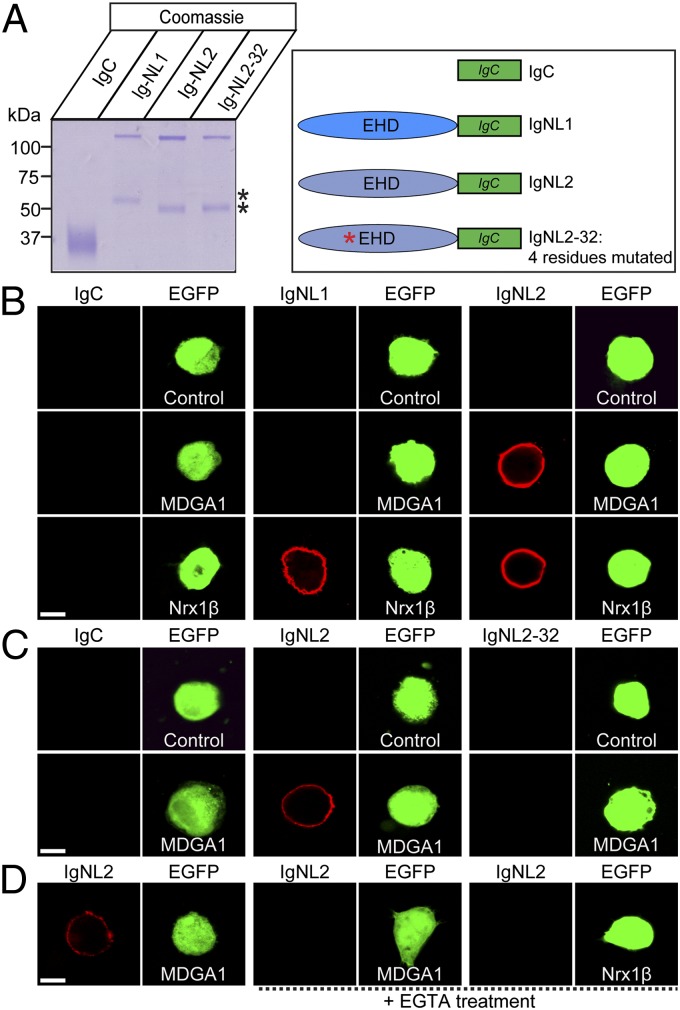
Based on our preliminary screening results (Table S1), we sought to investigate the interaction between NLs and MDGA1 using cell-surface binding assays. We produced recombinant NL-Ig fusion proteins [IgNL1, IgNL2, and IgC-neuroligin-2–32 (IgNL2-32)] in HEK293T cells and purified the expressed proteins (Fig. 1A; 25). Next, we incubated the Ig-fusion proteins with HEK293T cells that had been cotransfected with eGFP and full-length MDGA1 (Fig. 1). We then measured the binding of the Ig-fusion proteins to the transfected HEK293T cells by indirect immunofluorescence. We found that IgNL2, but not IgNL1 or IgC, bound strongly to MDGA1-expressing cells (Fig. 1B). To confirm further the specificity of MDGA1 interaction, we performed the binding assay with an NL2 mutant, NL2-32, that is modeled after the NL1-32 mutant that is unable to bind to neurexins (10, 26). Intriguingly, mutant NL2-32 failed to bind to MDGA1, indicating that MDGA1 interacts with NL2 via a binding mechanism similar to that of neurexins (Fig. 1C). To examine whether the MDGA1–NL2 interaction requires extracellular Ca2+, we performed the cell-surface binding assays in the presence of 10 mM EGTA, a Ca2+ chelator. Interestingly, we observed that the MDGA1–NL2 interaction was abolished by EGTA (Fig. 1D). Taken together, these results indicate that MDGA1 binds specifically to NL2 in a Ca2+-dependent manner at a similar site as neurexins.
Fig. 1.
Analysis of MDGA1–NL interaction using cell-surface binding assays. (A) Coomassie-stained gel of recombinant IgC, IgNL1, IgNL2, and IgNL2-32 used for cell-surface binding assays. The lower band (denoted by asterisk) is likely a degradation product of the full-length NL Ig-fusion proteins. (B–D) Cell-surface binding assays. HEK293T cells expressing full-length MDGA1 (MDGA1), full-length Nrx1β, or eGFP alone (Control) were incubated with IgC (control Ig-fusion protein) or the neuroligin–Ig fusion proteins IgNL1, IgNL2, or IgNL2-32 (a neurexin binding-deficient mutant). Cells were cotransfected with eGFP to allow identification of transfected cells. Cells then were analyzed by immunofluorescence imaging for the Ig-fusion proteins (red) and eGFP (green). All binding reactions were performed in 2 mM CaCl2 and 2 mM MgCl2, except for the reaction marked “+EGTA treatment,” which was performed in the presence of 10 mM EGTA. (Scale bar, 5 μm in all images.)
MDGA1 Binds to NL2 with a Nanomolar Affinity.
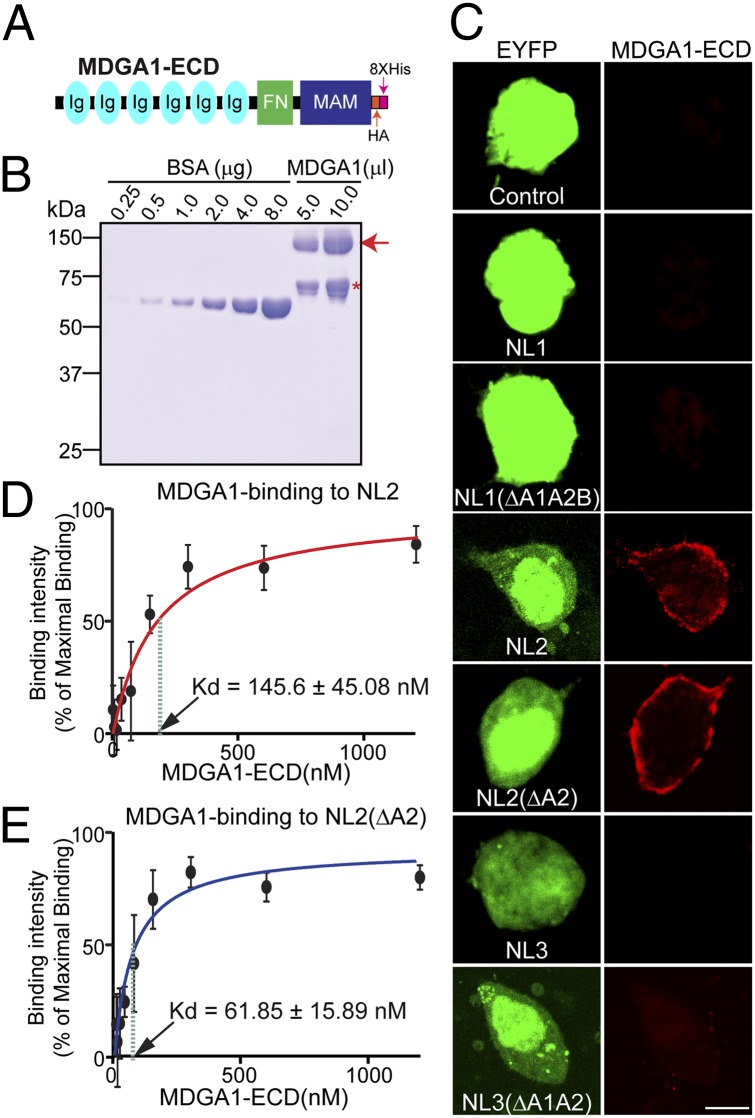
To determine the binding specificity of MDGA1 for NLs, we expressed NL1, NL2, and NL3 containing or lacking various splice-site inserts on the surface of transfected HEK293T cells. We then produced a recombinant MDGA1 protein (MDGA1-ECD-HA-His; Fig. 2 A and B), incubated the transfected cells with recombinant MDGA1 protein, and observed robust binding only to NL2 but not to NL1 or NL3. NLs are alternatively spliced at an identical site (termed “splice site A,” SSA), and alternative splicing of NLs is known to regulate their interactions with neurexins (27–29). Thus, we asked whether the alternative splicing in NLs also regulates their interaction with MDGA1. However, we observed binding of MDGA1 to both splice variants of NL2 but to none of the splice variants of NL1 and NL3 tested (Fig. 2C). We next investigated whether MDGA1 binding to NL2 exhibits an affinity commensurate with a physiological interaction and whether this affinity differed in the two NL2 splice variants. To this end, we used a quantitative cell-surface binding assay allowing calculations of approximate binding affinities between MDGA1 and NL2 variants. We incubated HEK293T cells transfected with the full-length NL2 splice variants with recombinant MDGA1 at concentrations ranging from 3.9 to 1,000 nM (Fig. 2 D and E). We assumed a single, independent NL2 binding site for MDGA1, computed the net binding of the MDGA1 recombinant protein at any given concentration, and analyzed the binding using the Michaelis–Menten equation. The reaction exhibited a dissociation constant (Kd) of 145.6 ± 45.08 nM for MDGA1 for NL2 containing an insert in SSA and 61.85 ± 15.89 nM for NL2 lacking an insert in SSA, indicating a high-affinity interaction of NL2 with MDGA1 that may be modulated by alternative splicing.
Fig. 2.
Analysis of NL2-MDGA1 interaction using quantitative cell-surface binding assays. (A and B) Schematic diagram (A) and purification (B) of recombinant MDGA1 protein (MDGA1-ECD). It has a molecular mass of approximately 130 kDa (red arrow). The asterisk indicates a nonspecific band. (C) Cell-surface binding assay: 20 μg/mL of recombinant MDGA1 protein was overlaid onto HEK293T cells coexpressing indicated NLs and eYFP. (Scale bar, 10 μm in all images.) (D and E) Analysis for Kd calculation. HEK293T cells expressing two different NL2 splice variants (NL2 and NL2ΔA2) were incubated with the recombinant MDGA1 protein with various concentrations, washed, fixed, and probed with anti-HA antibodies and HRP-conjugated secondary antibodies. The amount of antibody bound was determined by calorimetry and plotted as a function of recombinant MDGA1 protein concentration.
MDGA Ig Domains, but Not the FNIII Repeat or the MAM Domain, Are Required for Their Interaction with NL2.
To determine which MDGA domains interact with NL2, we performed cell-surface binding assays using constructs expressing fragments of MDGA1 and MDGA2 composed of their Ig FNIII, or MAM domain (Fig. S2). Individual MDGA fragments were expressed on the surface of transfected HEK293T cells (Fig. S2A). As shown above, IgNL2 bound to HEK293T cells expressing HA-tagged full-length MDGA1 and MDGA2 (Fig. S2). In addition, NL2 bound strongly to the surface of cells expressing the Ig domains of MDGA1 or MDGA2 but did not exhibit significant binding to cells expressing the FNIII repeat or MAM domain (Fig. S2B). We further subdivided the Ig domains of MDGA1 to map the NL2-binding site in greater detail and found that NL2 bound to the surface of HEK293T cells when the three N-terminal Ig domains (Ig1–3) but not the C-terminal Ig-domains (Ig4–6) of MDGA1 were expressed (Fig. S2B). These results indicate that NL2 binds to the three N-terminal Ig domains of MDGA1.
MDGA1 Binding to NL2 Does Not Mediate Cell Adhesion.
To investigate whether MDGA1 binds NL2 in a trans or cis configuration, we examined cell adhesion of cells expressing MDGA1 with cells expressing NL2, using cells expressing Nrx1β as a positive control. HEK293T cells were transfected with (i) mVenus or mCherry alone, (ii) mVenus fusion proteins of NL2 (NL2-mVenus), (iii) mCherry-fusion proteins of Nrx1β splice variants lacking splice site #4, (iv) mCherry-cotransfected with untagged MDGA1, or (v) mCherry-cotransfected with untagged protein tyrosine phosphatase, receptor type t (PTPRT). We found that cells expressing MDGA1 did not form aggregates with cells expressing NL2, whereas cells expressing Nrx1β formed massive aggregates (Fig. S3 A and B). Moreover, when we cocultured cells expressing tagged MDGA1 or NL2, we found that MDGA1 did not cluster with NL2 expressed on an adjacent cell (Fig. S4). Taken together, these data suggest that, unlike neurexins, MDGA1 is unable to form an intercellular junction via a trans interaction with NL2.
Native Rat Brain MDGA1 Can Be Pulled Down with Recombinant NL2.
Next, we conducted pull-down experiments on lysates of HEK293T cells expressing HA-tagged MDGA1, HA-tagged MDGA2, or the cell-surface proteins Slitrk3 or GluA1 using IgNL2 or IgC (Fig. S3C). We found that immobilized IgNL2 effectively captured both MDGA1 and MDGA2 proteins but not Slitrk3 or GluA1. Next, we performed similar experiments using proteins solubilized from rat synaptosomal membrane fractions (Fig. S3D). We found a high degree of MDGA1 enrichment in the IgNL2-bound fraction. No other cell-recognition molecules or receptors examined were enriched in the parallel experiments, suggesting that MDGA1 proteins form specific complexes with NL2 in rat brains. These results support the notion that both MDGA1 and MDGA2 interact with NL2.
MDGA Knockdown Enhances Inhibitory Synapse Formation in an NL2-Dependent Manner in Cultured Neurons.
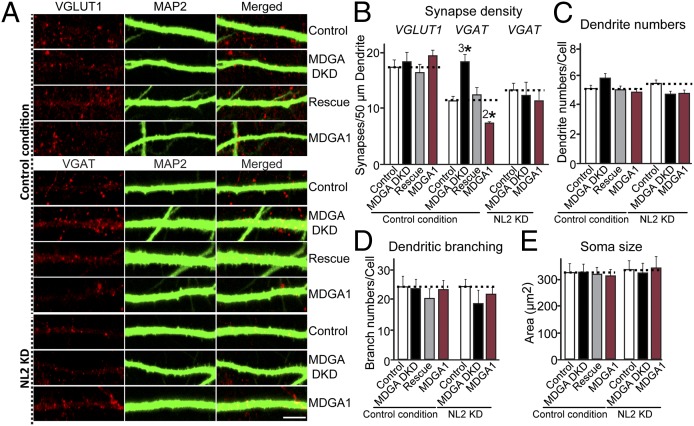
To investigate the functional consequences of the binding of MDGAs to NL2, we knocked down MDGA1 and MDGA2 expression in cultured hippocampal neurons using shRNAs and examined the effect of the knockdowns on synapse density (Fig. 3). For this approach, we designed a series of lentiviral shRNA vectors to knock down MDGA1, MDGA2, or NL2 (Fig. S5A) and quantified the endogenous target mRNA levels by real-time RT-PCR (Fig. S5B). The selected shRNAs efficiently suppressed endogenous mRNAs for MDGA1 (C10), MDGA2 (E5), and NL2 (J90) (Fig. S5B). To avoid a lack of phenotype caused by redundancy between MDGA1 and MDGA2, we next constructed a lentiviral vector that expresses shRNAs to both targets (Fig. S5 A and C). The resulting MDGA double-knockdown (MDGA DKD) was used to suppress expression of both MDGAs simultaneously but not of NLs (Fig. S5C). Cultured hippocampal neurons were infected at day in vitro 3 (DIV3) with lentiviruses expressing empty control vector (Control), MDGA DKD shRNAs (MDGA DKD), MDGA DKD shRNAs plus full-length human MDGA1 (Rescue), or full-length human MDGA1 (MDGA1). At DIV14, infected neurons were immunostained for MAP2 (a somatodendritic marker), and for either VGLUT1 (an excitatory presynaptic marker) or VGAT (an inhibitory presynaptic marker). Knockdown of MDGAs significantly increased inhibitory synapse density but did not affect excitatory synapse density (Fig. 3 A and B). MDGA knockdown did not detectably alter the number of primary dendrites and dendritic branches or the neuronal cell body size (Fig. 3 C–E). Conversely, lentiviral overexpression of MDGA1 significantly decreased inhibitory synapse density (Fig. 3 A and B). Strikingly, when we reduced the level of endogenous NL2 using lentiviral expression of shRNAs targeting NL2, we abolished the increase in inhibitory synapse density produced by the MDGA DKD (Fig. 3 A and B). Collectively, these observations demonstrate that MDGAs act as a “brake” on inhibitory synapses in a manner requiring NL2.
Fig. 3.
MDGA knockdown increases inhibitory synapse density in an NL2-dependent manner in cultured neurons. (A) Representative images of cultured hippocampal neurons infected (i) with control lentiviruses (l-309 empty vector), or NL2 KD lentiviruses [l-309 NL2 vector (J90); see Fig. S5] at DIV2, as indicated; (ii) with a lentiviral vector lacking shRNA expression (Control) at DIV4; with a lentiviral vector expressing shRNAs targeting MDGAs (sh-MDGAs) (MDGA DKD); (iii) with an MDGA DKD vector together with a lentiviral vector expressing full-length human MDGA1 (hMDGA1 vector) (Rescue); or (iv) with an hMDGA1 expression vector (MDGA1). Neurons were analyzed at DIV14 by double immunofluorescence using antibodies against MAP2 and VGLUT1 or VGAT. In the NL2 KD condition NL2 shRNA lentiviruses are infected to reduce endogenous NL2 levels in cultured neurons. (Scale bar, 5 μm in all images.) (B) Summary graph of the effects of MDGA DKD on excitatory and inhibitory synapse density, quantified using VGLUT1 and VGAT immunoreactivity. (C–E) Summary graphs of the effects of MDGA DKD on primary dendrite numbers (C), on dendritic branching (D), and on neuronal cell body size (E). Data shown in B–E are means ± SEM. 2*P < 0.01; 3*P < 0.001; Student’s t test compared with controls.
Overexpression of MDGA1 Decreases Inhibitory Synaptic Transmission in Cultured Neurons.
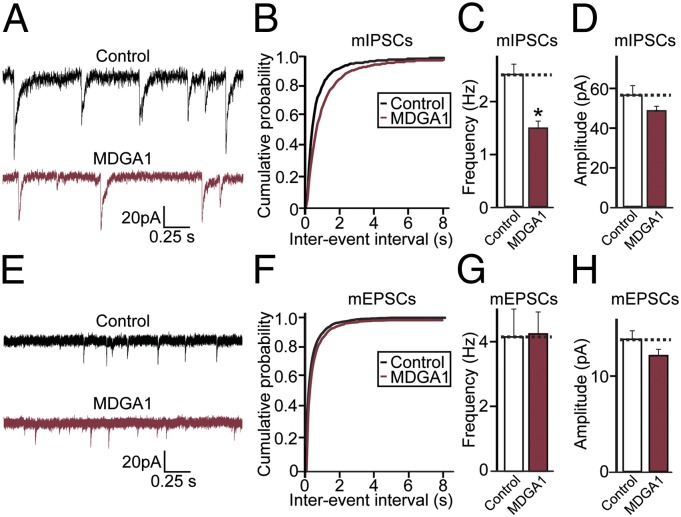
We observed a decrease in the number of inhibitory synapses, as reflected by the number of VGAT puncta, in MDGA1-expressing neurons (Fig. 3B). Therefore, to test whether the effect of MDGA1 on inhibitory synapse numbers influences synaptic transmission, we measured miniature inhibitory postsynaptic currents (mIPSCs) using whole-cell voltage-clamp recordings (Table S2). Expression of MDGA1 led to a significant reduction in mIPSC frequency and to a small, nonsignificant decrease in mIPSC amplitudes compared with neurons expressing eGFP alone (Fig. 4 A–D); this result correlates well with a reduced number of inhibitory synapses (Fig. 3 A and B). Analysis of miniature excitatory postsynaptic currents (mEPSCs) showed no significant differences in the frequency or amplitude of these synaptic events in MDGA1-expressing neurons (Fig. 4 E–H). Taken together, these data support the hypothesis that the MDGA1 level controls the formation and/or maintenance of inhibitory but not excitatory synapse in cultured neurons.
Fig. 4.
MDGA1 overexpression decreases inhibitory, but not excitatory, synaptic transmission in cultured cortical neurons. (A–D) mIPSCs were recorded from DIV13–15 cortical neurons expressing eGFP (Control) or coexpressing eGFP with MDGA1 (MDGA1). Shown are sample traces (A), cumulative plot of mIPSC amplitude (B), and histograms of mIPSC frequency (C) and amplitude (D). (E–H) mEPSCs were recorded from DIV13-15 cortical neurons expressing eGFP (Control) or coexpressing eGFP with MDGA1 (MDGA1). Shown are sample traces (E), cumulative plots of mEPSC amplitude (F), and histograms of mEPSC frequency (G) and amplitude (H).
MDGA1 Inhibits NL2-Mediated Synaptogenic Activity and Disrupts the NL2–Neurexin Interaction.
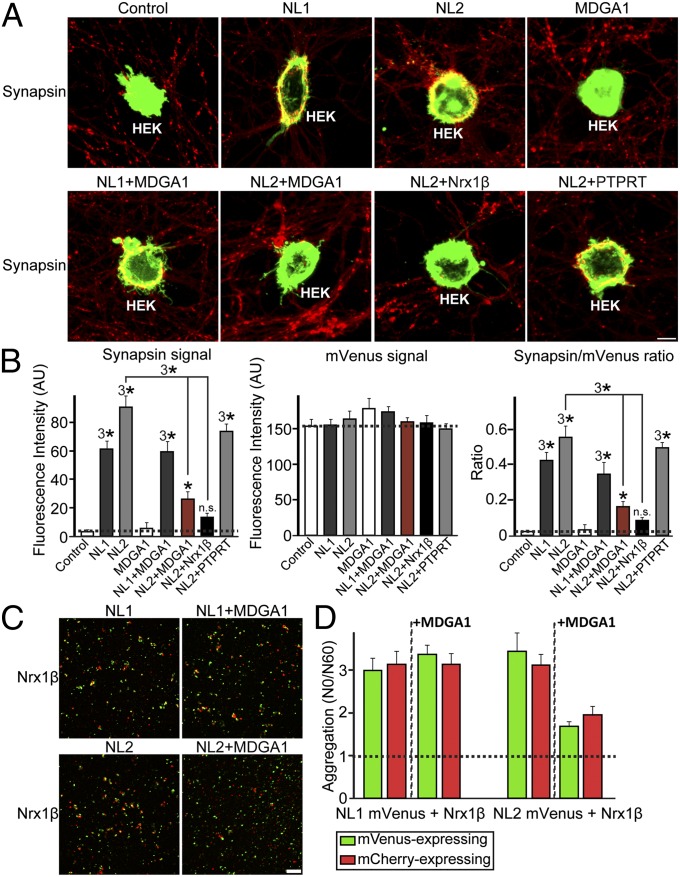
Loss-of-function (the MDGA DKD) and gain-of-function (the MDGA1 overexpression) experiments in cultured hippocampal neurons suggested that MDGAs negatively regulate inhibitory synapses in an NL2-dependent fashion (Fig. 3). To characterize the effect of MDGA1 further, we used artificial synapse-formation assays (Fig. 5). HEK293T cells were transfected with eGFP, NL1, NL2, MDGA1, Nrx1β, or PTPRT alone or together as indicated and were cocultured with hippocampal neurons for 2 d. Formation of synapses onto the transfected HEK293T cells then was analyzed by synapsin staining. NLs strongly induced presynaptic differentiation, visualized as a clustering of synapsin I puncta around the transfected HEK293T cells (Fig. 5 A and B), as previously reported (10, 30, 31). However, MDGA1 did not induce presynaptic clustering, indicating that it lacks synaptogenic activity per se. Coexpression of MDGA1 or of Nrx1β with NL2 in HEK293T cells in a cis configuration strongly impaired the synaptogenic activity of NL2 (Fig. 5 A and B), as reported previously for Nrx1β (32). In contrast, cis expression of MDGA1 with NL1 did not affect the synaptogenic activity of NL1, consistent with the observation that MDGA1 does not interact with NL1 (Figs. 1 and 2). We further tested whether the cell-adhesion properties of NLs are affected by coexpression with MDGA1 (Fig. 5 C and D). HEK293T cells expressing NL1 or NL2 formed aggregates with cells expressing Nrx1β, whereas cells coexpressing NL2 with MDGA1 did not form aggregates with cells expressing Nrx1β (Fig. 5 C and D). Moreover, the interaction between NL2 and Nrx1β in cell-surface binding assays was abolished specifically by treatment with excess recombinant MDGA1 protein (Fig. S6), suggesting that MDGA1 and Nrx1β share an overlapping binding interface for NL2 (also see Fig. 1C). It is conceivable that coexpression of MDGA1 with NL2 impairs surface expression of NL2 in HEK293T cells, thus reducing its synaptogenic activity. To examine this possibility, we compared the localization of recombinant NL2 fused to mVenus (NL2-mVenus) when transfected alone or cotransfected with MDGA1 (Fig. S7). Coexpression of MDGA1 did not interfere significantly with the surface expression of NL2, supporting the notion that cis expression of MDGA1 with NL2 blocks the neurexin–NL interaction, rendering NL2 inactive in artificial synapse-formation assays. Finally, we addressed the possibility that MDGA1 might reduce the synaptogenic activity of NL2 by binding to neurexins (Fig. S8). No binding between MDGA1 and Nrx1β was observed in cell-surface binding assays, indicating that MDGA1 interferes with the action of NL2 by binding directly to NL2 and not to Nrx1β. Taken together, these results strongly suggest that cis expression of MDGA1 blocks the synaptogenic activity of NL2 by inhibiting its trans interaction with presynaptic neurexins and other ligands.
Fig. 5.
MDGA1 cis expression inhibits NL2 activity in artificial synapse-formation assays and cell-adhesion assays. (A) Representative images of artificial synapse-formation assays. Cultured hippocampal neurons were cocultured from DIV8 to DIV10 with HEK293T cells expressing eGFP alone (Control), NL1-mVenus (NL1), NL2-mVenus (NL2), MDGA1 and eGFP (MDGA1), NL1 and MDGA1 (NL1+MDGA1), NL2 and MDGA1 (NL2+MDGA1), or NL2 and PTPRT (NL2+PTPRT). Neurons then were immunostained with antibodies against synapsin I (red) and eGFP (green). (Scale bar, 25 μm in all images.) (B) Quantification of the artificial synapse-formation activity of NLs. Activity was quantified by measuring the average fluorescence intensities of synapsin I (Left), eGFP (Center), and the ratio of synapsin I to eGFP (Right) fluorescence (for absolute red and green fluorescence values). *P < 0.05, 3*P < 0.001, n.s., not significant; Student’s t test compared with controls. (C and D) Representative images (C) and quantitation (D) of cell-adhesion assays. HEK293T cells expressing mVenus alone (Control), NL2-mVenus (NL2), or NL1-mVenus (NL1) or coexpressing NL2-mVenus and MDGA1 (NL2+MDGA1) or NL1-mVenus and MDGA1 (NL1+MDGA1) were mixed with HEK293T cells expressing mCherry alone (Control) or mCherry-neurexin-1β fusion protein (Nrx1β). Cells then were imaged, and free cell numbers were counted immediately after the respective cell populations had been mixed (To) and again after 60 min (T60), indicated in representative images in C. (Scale bar, 100 μm in all images.)
Discussion
Here, we identified MDGAs as NL2-specific ligands that regulate the NL2-dependent inhibitory synapse formation in cultured neurons. We made the following principal observations. (i) MDGA mRNA expression continued at high levels in adult rats (Fig. S1). (ii) MDGA1 interacted directly and specifically with NL2 with a nanomolar affinity (Figs. 1 and 2). (iii) MDGA1 did not interact with NL2 in cell-adhesion assays in trans but did bind when added as a soluble protein to the medium, suggesting that MDGA1 may interact with NL2 in a cis configuration (Figs. S3 A and B and S4). (iv) Knockdown of MDGAs specifically increased inhibitory synapse density, whereas overexpression of MDGA1 decreased inhibitory synapse density and transmission in cultured neurons, indicating that MDGAs negatively regulate inhibitory synapses by binding to NL2 (Figs. 3 and 4). (v) Cis-expressed MDGA1 disrupted NL2-mediated artificial synapse formation and blocked the ability of NL2 to interact with neurexins in trans to mediate cell adhesion (Fig. 5 and Fig. S6), suggesting that cis-binding of MDGA1 to NL2 blocks the NL2–neurexin interaction.
Identification of MDGAs as NL2-Specific Ligands.
Neurexin-NL trans-synaptic interactions constitute a key synaptic adhesion pathway that governs the formation, maturation, and plasticity of neuronal synapses (1, 33). Recently, alternative trans-synaptic interaction proteins have been discovered for neurexins, including leucine-rich repeat transmembrane neuronal proteins (LRRTMs) and cerebellins/GluRδ2 (34–37). Intriguingly, LRRTM1 and LRRTM2 function redundantly with excitatory NL isoforms (NL1 and NL3) in the validation of the structure and function of excitatory synapses (24, 38). In artificial synapse-formation assays, NLs induce presynaptic differentiation by binding to presynaptic neurexins, whereas in cultured neurons NL overexpression increases the density of synapses independently of their neurexin-binding activities (10). These findings suggested that NLs require other extracellular ligands besides neurexins. Two proteins, PTPRT and thrombospondin-1, have been reported to regulate synapse formation via direct interactions with NLs in cultured hippocampal neurons (39, 40). However, because PTPRT is localized specifically to excitatory synapses, and the localization of thrombospondin-1 at synapses remains unclear, these findings cannot account fully for isoform-dependent differential NL activities in synapse validation. Moreover, we did not observe any specific effects of PTPRT when coexpressed with NL2, indicating that PTPRT may function with other NL isoforms, such as NL1 or NL3 (Fig. 5) (39).
Several proteins have been shown previously to decrease the density of mammalian synapses, including Ephexin 5 (RhoA guanine nucleotide exchange factor) and Nogo receptors (41, 42). In addition, regulator of synaptogenesis-1 (RSY-1) has been shown to regulate SYD-2/liprin-α–mediated presynaptic assembly negatively in Caenorhabditis elegans (43). The notion that MDGAs function in an inhibitory capacity as NL2-specific extracellular ligands is significant, in that MDGAs are among the few known negative regulators of synapses. Moreover, MDGAs are unique in that, to the best of our knowledge, they are the only negative regulatory factors yet reported to function specifically at an inhibitory synapse.
Functional Significance of MDGA–NL2 Interactions for Inhibitory Synapses.
Our data suggest that MDGAs suppress inhibitory synapse formation by binding directly to NL2 on the cell surface in a cis configuration (Figs. 3 and 5). An alternative scenario would be that MDGAs inhibit the surface delivery of NL2 by trapping NL2 in the secretory or endocytic pathway. However, recombinant NL2 proteins were well localized to the plasma membrane of HEK293T cells, whether expressed alone or coexpressed with MDGA1, arguing against this possibility (Fig. S7 A and B). It is more likely that MDGAs compete with neurexins or other as yet unidentified NL2 ligands in binding to NL2 at inhibitory synapses, leading to disruption of neurexin–NL interactions, followed by destabilization of inhibitory synapse structure and function. Supporting this idea, cis expression of MDGA1 with NL2 suppressed the adhesion mediated by NL2 and Nrx1β (Fig. 5 C and D). Although the molecular details remain to be determined, the data presented here unequivocally argue that MDGAs are extracellular proteins that specifically bind to NL2.
Because extensive human genetic studies have pinpointed synaptic genes, including those for NLs and neurexins, as causative factors underlying human brain disorders, it is not surprising that MDGAs also have been linked to various neuropsychiatric disorders (1, 19–21). Although the etiologies of these neuropsychiatric disorders are complex, a broad hypothesis that is widely cited is that an imbalance between excitatory and inhibitory synapses is a contributing factor (44). Our data suggest that understanding MDGA functions will provide insight into the pathophysiology of schizophrenia and autistic spectrum disorders, insofar as dysfunctions of MDGAs are likely to influence a subset of NL–neurexin adhesion pathways at inhibitory synapses, possibly increasing the inhibition of neural circuits where MDGAs, NL2, and neurexins function and thereby decreasing the excitation-to-inhibition ratio. Our present study raises a series of new important questions: for example, what is the exact cellular localization of MDGA proteins in vivo? Given that MDGAs interact with NL2 in a cis configuration, are MDGAs localized only to NL2-containing synapses? Do MDGAs have other functions besides binding to NL2, as indicated by the developmental phenotype of MDGA-KO mice that differs from that of NL2-KO mice? The answers to these emerging questions would contribute to our understanding of how the synaptic adhesion molecules shape synaptic connectivity, maturation, and learning-related plasticity. An immediate next step should be to confirm the role of MDGAs in inhibitory synapse development in vivo, a challenging goal given the early developmental function of MDGAs (15, 16) that suggests that conditional KO will be required.
Methods
Artificial synapse-formation assays, cell-adhesion assays, and cell-surface binding assays were performed as previously described (35). Cultured hippocampal neurons were prepared and infected with the indicated lentiviruses at DIV3 and immunostained at DIV13. For a detailed description of methods, see SI Methods.
Supplementary Material
Acknowledgments
We thank Drs. Eunjoon Kim (KAIST, Korea) and Dennis O’Leary (The Salk Institute) for the kind gifts of reagents and Dr. Ji Won Um (Yale University) for the critical comments on the manuscript. This work was supported by a TJ Park Junior Faculty Fellowship from the POSCO TJ Foundation (to J.K.) and by Grant 2011-0028337 from the National Research Foundation of Korea (to J.K.), Grant 177850 from the Simons Foundation (to T.C.S.), grants from the Singapore National Medical Research Council (to H.S.J.) and from the Singapore Ministry of Education (to H.S.J.), and Grant 2012-0005820 from the Korea Science and Engineering Foundation (to H.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219987110/-/DCSupplemental.
References
- 1.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lips ES, et al. International Schizophrenia Consortium Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol Psychiatry. 2012;17(10):996–1006. doi: 10.1038/mp.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011;14(12):1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko J. The leucine-rich repeat superfamily of synaptic adhesion molecules: LRRTMs and Slitrks. Mol Cells. 2012;34(4):335–340. doi: 10.1007/s10059-012-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Missler M, Südhof TC, Biederer T. Synaptic cell adhesion. Cold Spring Harb Perspect Biol. 2012;4(4):a005694. doi: 10.1101/cshperspect.a005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 2004;83(9):449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- 7.Poulopoulos A, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63(5):628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Jedlicka P, et al. Increased dentate gyrus excitability in neuroligin-2-deficient mice in vivo. Cereb Cortex. 2011;21(2):357–367. doi: 10.1093/cercor/bhq100. [DOI] [PubMed] [Google Scholar]
- 9.Gibson JR, Huber KM, Südhof TC. Neuroligin-2 deletion selectively decreases inhibitory synaptic transmission originating from fast-spiking but not from somatostatin-positive interneurons. J Neurosci. 2009;29(44):13883–13897. doi: 10.1523/JNEUROSCI.2457-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko J, et al. Neuroligin-1 performs neurexin-dependent and neurexin-independent functions in synapse validation. EMBO J. 2009a;28(20):3244–3255. doi: 10.1038/emboj.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Juan C, et al. Genomic organization of a novel glycosylphosphatidylinositol MAM gene expressed in human tissues and tumors. Oncogene. 2002;21(19):3089–3094. doi: 10.1038/sj.onc.1205383. [DOI] [PubMed] [Google Scholar]
- 12.Litwack ED, Babey R, Buser R, Gesemann M, O’Leary DD. Identification and characterization of two novel brain-derived immunoglobulin superfamily members with a unique structural organization. Mol Cell Neurosci. 2004;25(2):263–274. doi: 10.1016/j.mcn.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Dityatev A, Bukalo O, Schachner M. Modulation of synaptic transmission and plasticity by cell adhesion and repulsion molecules. Neuron Glia Biol. 2008;4(3):197–209. doi: 10.1017/S1740925X09990111. [DOI] [PubMed] [Google Scholar]
- 14.Katidou M, Vidaki M, Strigini M, Karagogeos D. The immunoglobulin superfamily of neuronal cell adhesion molecules: Lessons from animal models and correlation with human disease. Biotechnol J. 2008;3(12):1564–1580. doi: 10.1002/biot.200800281. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi A, O’Leary DD. Radial migration of superficial layer cortical neurons controlled by novel Ig cell adhesion molecule MDGA1. J Neurosci. 2006;26(17):4460–4464. doi: 10.1523/JNEUROSCI.4935-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi A, Hamasaki T, Litwack ED, O’Leary DD. Novel IgCAM, MDGA1, expressed in unique cortical area- and layer-specific patterns and transiently by distinct forebrain populations of Cajal-Retzius neurons. Cereb Cortex. 2007;17(7):1531–1541. doi: 10.1093/cercor/bhl064. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa T, et al. IgSF molecule MDGA1 is involved in radial migration and positioning of a subset of cortical upper-layer neurons. Dev Dyn. 2011;240(1):96–107. doi: 10.1002/dvdy.22496. [DOI] [PubMed] [Google Scholar]
- 18.Joset P, et al. Rostral growth of commissural axons requires the cell adhesion molecule MDGA2. Neural Dev. 2011;6:22. doi: 10.1186/1749-8104-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahler AK, et al. Association analysis of schizophrenia on 18 genes in involved in neuronal migration: MDGA1 as a new susceptibiligy gene. Am J Med Genet B Neuropsychiatr. 2008;147B(7):1089–1100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- 20.Bucan M, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5(6):e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, et al. The MDGA1 gene confers risk to schizophrenia and bipolar disorder. Schizophr Res. 2011;125(2-3):194–200. doi: 10.1016/j.schres.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Varoqueaux F, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51(6):741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Chubykin AA, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54(6):919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko J, Soler-Llavina GJ, Fuccillo MV, Malenka RC, Südhof TC. Neuroligins/LRRTMs prevent activity- and Ca2+/calmodulin-dependent synapse elimination in cultured neurons. J Cell Biol. 2011;194(2):323–334. doi: 10.1083/jcb.201101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boucard AA, Ko J, Südhof TC. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem. 2012;287(12):9399–9413. doi: 10.1074/jbc.M111.318659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araç D, et al. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56(6):992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Ichtchenko K, Nguyen T, Südhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271(5):2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- 28.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48(2):229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 29.Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51(2):171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101(6):657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 31.Chubykin AA, et al. Dissection of synapse induction by neuroligins: Effect of a neuroligin mutation associated with autism. J Biol Chem. 2005;280(23):22365–22374. doi: 10.1074/jbc.M410723200. [DOI] [PubMed] [Google Scholar]
- 32.Taniguchi H, et al. Silencing of neuroligin function by postsynaptic neurexins. J Neurosci. 2007;27(11):2815–2824. doi: 10.1523/JNEUROSCI.0032-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krueger DD, Tuffy LP, Papadopoulos T, Brose N. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr Opin Neurobiol. 2012;22(3):412–422. doi: 10.1016/j.conb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 34.de Wit J, et al. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64(6):799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko J, Fuccillo MV, Malenka RC, Südhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009b;64(6):791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141(6):1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33(8):1447–1461. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- 38.Soler-Llavina GJ, Fuccillo MV, Ko J, Südhof TC, Malenka RC. The neurexin ligands, neuroligins and leucine-rich repeat transmembrane proteins, perform convergent and divergent synaptic functions in vivo. Proc Natl Acad Sci USA. 2011;108(40):16502–16509. doi: 10.1073/pnas.1114028108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim SH, et al. Synapse formation regulated by protein tyrosine phosphatase receptor T through interaction with cell adhesion molecules and Fyn. EMBO J. 2009;28(22):3564–3578. doi: 10.1038/emboj.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Xiao N, Xia J. Thrombospondin 1 accelerates synaptogenesis in hippocampal neurons through neuroligin 1. Nat Neurosci. 2010;13(1):22–24. doi: 10.1038/nn.2459. [DOI] [PubMed] [Google Scholar]
- 41.Margolis SS, et al. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell. 2010;143(3):442–455. doi: 10.1016/j.cell.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wills ZP, et al. The nogo receptor family restricts synapse number in the developing hippocampus. Neuron. 2012;73(3):466–481. doi: 10.1016/j.neuron.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel MR, Shen K. RSY-1 is a local inhibitor of presynaptic assembly in C. elegans. Science. 2009;323(5920):1500–1503. doi: 10.1126/science.1169025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gogolla N, et al. Common circuit defect of excitatory-inhibitory balance in mouse models of autism. J Neurodev Disord. 2009;1(2):172–181. doi: 10.1007/s11689-009-9023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.