Abstract
One the most intriguing, yet least studied, aspects of the bacterium–host plant interaction is the role of the host ubiquitin/proteasome system (UPS) in the infection process. Increasing evidence indicates that pathogenic bacteria subvert the host UPS to facilitate infection. Although both mammalian and plant bacterial pathogens are known to use the host UPS, the first prokaryotic F-box protein, an essential component of UPS, was identified in Agrobacterium. During its infection, which culminates in genetic modification of the host cell, Agrobacterium transfers its T-DNA—as a complex (T-complex) with the bacterial VirE2 and host VIP1 proteins—into the host cell nucleus. There the T-DNA is uncoated from its protein components before undergoing integration into the host genome. It has been suggested that the host UPS mediates this uncoating process, but there is no evidence indicating that this activity can unmask the T-DNA molecule. Here we provide support for the idea that the plant UPS uncoats synthetic T-complexes via the Skp1/Cullin/F-box protein VBF pathway and exposes the T-DNA molecule to external enzymatic activity.
Keywords: plant genetic transformation, VIP1-binding F-box protein
The pathogen Agrobacterium elicits neoplastic growths on plants, which represent its natural hosts, and also can transform a wide range of other eukaryotes, from fungi (1, 2) to human cells (3). This genetic transformation is achieved by transporting a single-stranded (ss) copy (T-strand) of the bacterial T-DNA from the Ti plasmid into the plant cell nucleus, followed by integration into the host genome by illegitimate recombination (4–6). Two Agrobacterium proteins, VirD2 and VirE2, directly associate with the T-strand, forming a transport (T) complex (7) in which one molecule of VirD2 is covalently attached to the 5′-end of the T-strand, and VirE2, an ssDNA-binding protein, cooperatively coats the rest of the T-strand (4, 7, 8). The WT T-complex can be quite large, reaching up to 9 × 104 kDa and carrying ∼1,200 molecules of VirE2 (9, 10). The complex is imported into the host cell nucleus by VirD2 and VirE2 (11–17); however, the role of VirD2 in this process is not critical, and VirE2 alone is sufficient to transport ssDNA into the nucleus (18). Thus, the VirE2–ssDNA complexes represent the minimal functional T-complex.
T-complex nuclear uptake is facilitated by a cellular protein, VIP1, that binds VirE2 and directs it to the importin α-mediated nuclear import pathway (19, 20). Because VirE2 is associated with the T-strand, VIP1 effectively mediates nuclear import of the entire T-complex. Once inside the nucleus, VIP1 mediates chromatin association of the T-complex by acting as a molecular link between VirE2 and nucleosomes via interactions with the core histones (21, 22).
Whereas VirE2 and VIP1 are critical for nuclear import and chromatin targeting of the T-complex, they become a liability for integration because they physically mask the DNA molecule. Thus, once the T-complex reaches the host chromatin, its proteins must be removed. This process has been proposed to involve the host ubiquitin/proteasome system (UPS) (23–25) based on the observations that challenge of plants by bacteria, including Agrobacterium, induces expression of a defense-related F-box protein, VBF, that recognizes and targets VIP1 and its bound VirE2 for degradation (25). F-box proteins—a large protein family with almost 700 predicted members in the Arabidopsis genome (26)—represent a component of the Skp1/Cullin/F-box protein (SCF) complex (27, 28) that acts as a E3 ubiquitin ligase to polyubiquitinate target proteins and tag them for subsequent degradation by the 26S proteasome. Within the F-box protein molecule, its conserved F-box motif mediates interaction with the rest of the SCF complex via Skp1, whereas other, variable domain(s) mediate interaction with target proteins (29, 30). In the case of VBF, it is presumed to function in the SCFVBF complex and to target VIP1, alone or in association with VirE2, for degradation (25). However, evidence that Agrobacterium can take advantage of the host UPS to uncoat the T-complex and expose its T-DNA molecule has been elusive. Here we provide this evidence by showing that the plant UPS can disassemble synthetic T-complexes, most likely via the SCFVBF pathway, and expose the T-DNA molecule to external enzymatic activity.
Results
VBF-Dependent Proteasomal Degradation of VIP1.
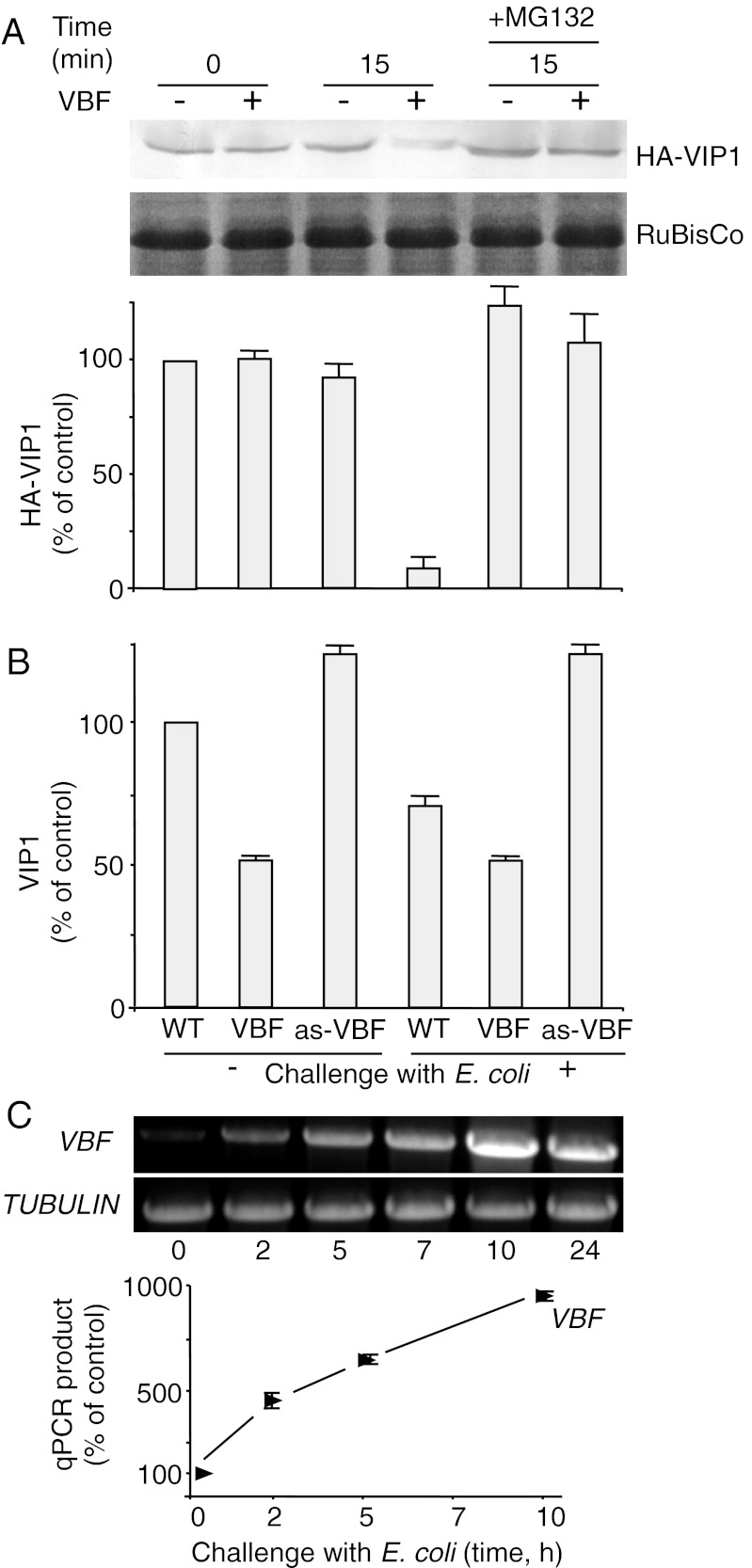
We previously reported that VBF can lead to degradation of VIP1 (25) and, by implication, the immediate substrate. To examine whether VBF can promote removal of VirE2 from ssDNA, we first analyzed the effects of VBF on the amounts of VIP1, the direct interactor of VirE2 (19) and the presumed substrate of VBF (25), using a cell-free proteasomal degradation assay (31). Total cell extracts were prepared from Nicotiana benthamiana plants transiently coexpressing Arabidopsis VBF and/or HA-tagged VIP1 (HA-VIP1), and their HA-VIP1 content was determined by Western blot analysis. Within 15 min, VIP1 amounts declined substantially in the presence of VBF, whereas without VBF, VIP1 remained relatively stable; a small decrease in the VIP1 content in the absence of transient VBF expression most likely was related to low levels of the endogenous tobacco VBF homolog (Fig. 1A). This VBF-mediated destabilization of VIP1 most likely occurred by proteasomal degradation via the SCFVBF pathway, because it was inhibited by MG132 (Fig. 1A), a known selective inhibitor of proteasomal activity (32). Quantification of these data indicate that VIP1 degradation in the presence of VBF was almost complete (≥90 ± 5%), and this effect of VBF was practically blocked by MG132; that, in the presence of MG132, VIP1 accumulated to slightly higher levels than even in the absence of the transient VBF expression in untreated plants may be due to inhibition of the endogenous VBF homolog (Fig. 1A).
Fig. 1.
VBF promotes proteasomal degradation of VIP1. (A) Western blot analysis of VIP1 degradation in the N. benthamiana cell-free system. HA-VIP1 was expressed alone or coexpressed with VBF in N. benthamiana leaves. The resulting protein extracts were incubated for the indicated time periods and analyzed using anti-HA antibodies. The putative RuBisCo large chain was used as a loading control. The quantified Western blot signal was expressed as percent of the signal obtained in the absence of VBF at the start of the incubation period. The data represent average values of three independent experiments with indicated SDs. (B) Western blot analysis of the endogenous VIP1 content in Arabidopsis plants. VBF expression was induced by challenging the plants for 10 h with E. coli. Protein extracts from WT plants, VBF transgenic plants (VBF), or VBF antisense plants (as-VBF) were analyzed with anti-VIP1 antibody, and the resulting signal was quantified and expressed as percent of the signal obtained in WT, unchallenged plants. The data represent average values of three independent experiments with indicated SDs. (C) RT-PCR (Upper) and RT-qPCR analyses (Lower) of VBF gene expression in WT Arabidopsis plants after challenge with E. coli. Constitutively expressed TUBULIN was used as an internal control.
We next examined the effect of VBF on the steady-state levels of the intracellular VIP1 in vivo. Using Western blot analysis, we compared the content of the endogenous VIP1 protein in the WT Arabidopsis, transgenic Arabidopsis plants that constitutively express VBF, and transgenic Arabidopsis plants in which VBF was knocked down by antisense suppression (25). The VBF-expressing transgenic plants efficiently (50 ± 5%) destabilized VIP1, whereas the VBF antisense plants did not support such VIP1 destabilization (Fig. 1B). Furthermore, the VBF antisense plants consistently exhibited ∼25% higher VIP1 levels than the WT plants, suggesting the corresponding contribution of the SCFVBF pathway in determination of the steady-state levels of VIP1 in plant cells.
Previous observations indicate that VBF is an inducible gene, the expression of which is up-regulated by microbial challenge, such as inoculation with Agrobacterium or Escherichia coli (25). Thus, it was interesting to examine whether such microbial challenge can alter the steady-state level of the endogenous VIP1 and, consequently, the cellular potential to uncoat the T-complex. We first sought to gain better understanding of the conditions under which plants can accumulate greater amounts of endogenous VBF. Specifically, we explored the kinetics of the VBF gene induction in Arabidopsis plants. To avoid introducing Agrobacterium-specific virulence effectors into the plant, we chose to use E. coli as the VBF-inducing bacterium. Using RT-PCR, we detected increasingly larger amounts of VBF transcripts in E. coli-inoculated Arabidopsis tissues (Fig. 1C). Quantification of the VBF transcriptional activation by qPCR indicated induction levels as high as 10-fold of the basal amounts of the VBF transcript (Fig. 1C). Equal efficiency of the RT-PCR/qPCR reactions was controlled using transcripts specific for a constitutively expressed TUBULIN gene.
This knowledge allowed us to examine the ability of endogenous VBF, accumulated after the challenge of Arabidopsis with E. coli, to mediate degradation of the endogenous VIP1. Substantial (≥25 ± 3%) and reproducible reduction of the amount of VIP1 was observed in WT Arabidopsis plants that had been challenged for 10 h with E. coli (Fig. 1B). As expected, VIP1 destabilization (50 ± 5%) in the transgenic plants that stably and constitutively expressed VBF was independent of the bacterial challenge. The more efficient VIP1 destabilization by the VBF-expressing transgenic plants compared with the WT plants even after bacterial challenge may be because the high VBF levels in transgenic plants were constitutive, whereas in the challenged plants, the duration of elevated presence of VBF in the cells was relatively short (i.e., 10 h). Importantly, the VBF antisense plants did not support VIP1 degradation even after bacterial challenge (Fig. 1B), suggesting a key role for VBF in regulating VIP1 turnover during plant defense.
VBF-Dependent Proteasomal Uncoating of Synthetic T-Complexes.
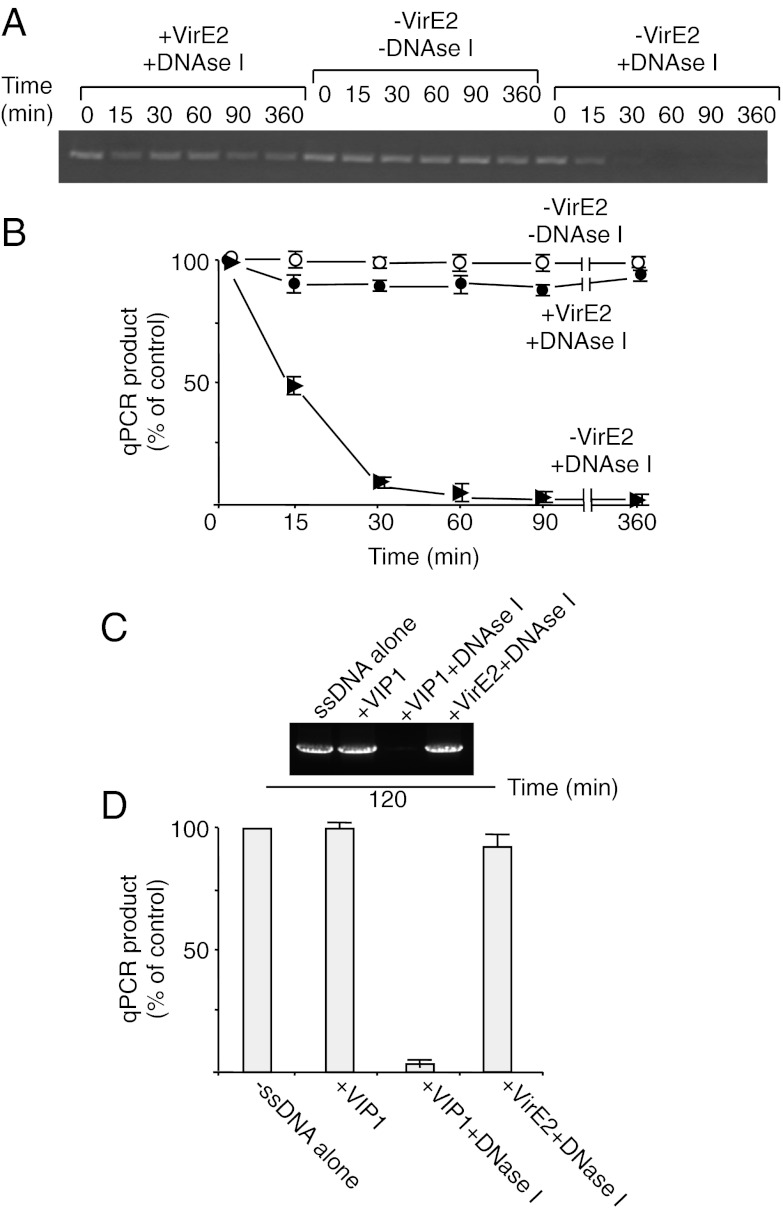
Because VirE2 is the major structural and functional protein component of the T-complex, and because T-DNA is sequence-nonspecific, a simple T-complex can be reconstituted from any ssDNA and VirE2 in vitro. Such synthetic T-complexes have a solenoidal structure (9, 10), within which the ssDNA molecule is tightly packaged by the VirE2 molecules and thus is inaccessible to its environment (33). We monitored this VirE2-mediated shielding of ssDNA by exposing the synthetic T-complexes to exogenous DNase activity and detecting the undigested, protected DNA by PCR. ssDNA, which is stable under our assay conditions for at least 6 h, was rapidly digested by the DNase, becoming almost undetectable already after 30 min of treatment (Fig. 2A). In contrast, ssDNA complexed with VirE2 remained stable for the entire duration of the assay (Fig. 2A). Quantification of these experiments by qPCR demonstrated that the DNase treatment eliminated one-half of the tested ssDNA after 15 min of incubation, and ssDNA hydrolysis was nearly complete after 30 min (Fig. 2B). Under the same conditions, ssDNA–VirE2 complexes were remarkably stable, with only ≤5 ± 2% of the ssDNA susceptible to the DNase. Thus, VirE2 almost completely protected ssDNA from the exogenous DNase activity (Fig. 2B). Unlike VirE2, VIP1 did not affect the stability of free ssDNA or its degradation by DNase I (Fig. 2 C and D).
Fig. 2.
VirE2 in synthetic T-complexes protects ssDNA from nucleolytic attack. Susceptibility to DNase I of ssDNA in the presence or absence of recombinant VirE2 or VIP1 purified as described previously (19, 33) was analyzed by PCR (A and C) and quantified by qPCR (B and D). The quantified data are expressed as percent of the input signal of free ssDNA (control) and represent average values of three independent experiments with indicated SDs.
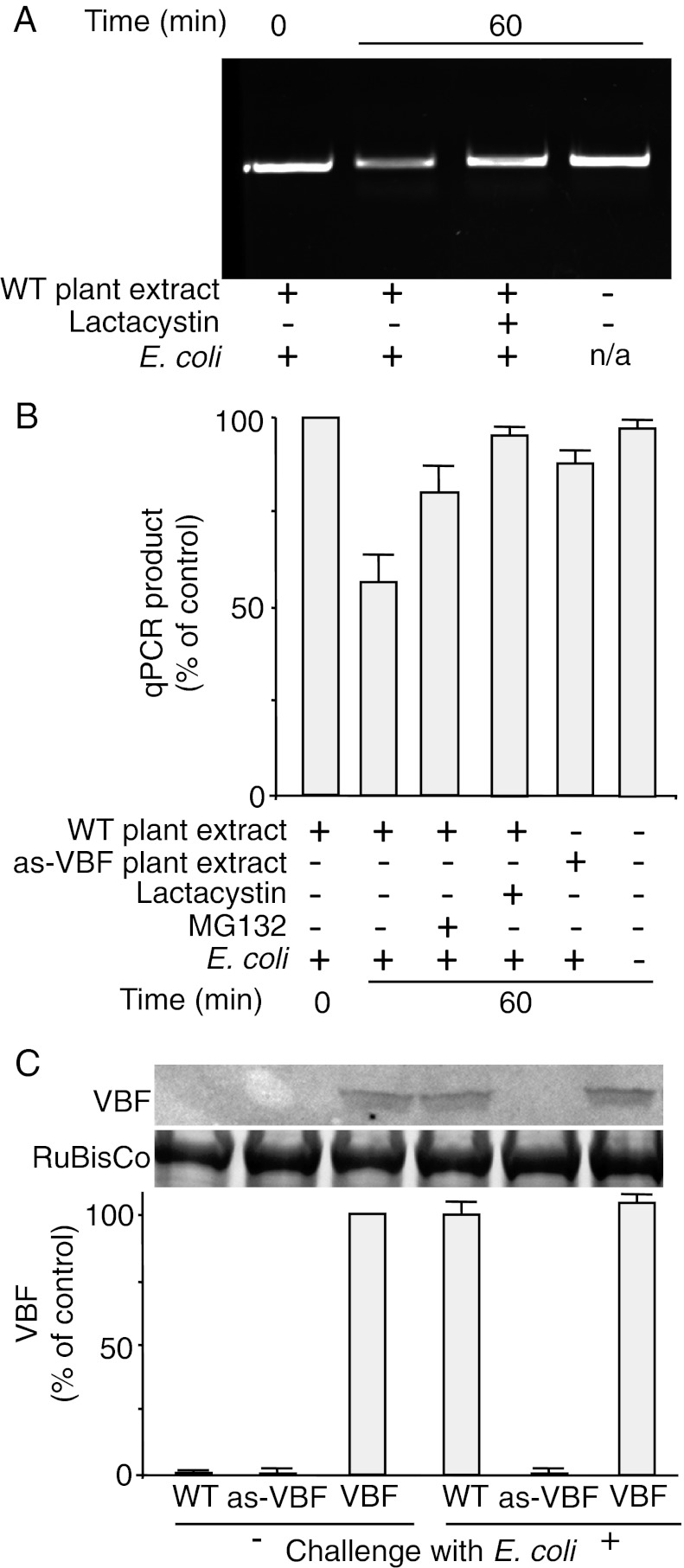
We next combined this experimental system to measure ssDNA accessibility with the cell-free proteasomal degradation assay to evaluate the ability of VBF to “unprotect” ssDNA of the synthetic T-complex in Arabidopsis. To achieve maximal VBF expression (Fig. 1C), we first challenged the WT Arabidopsis plants with E. coli. The addition of cell extracts from these plants to the otherwise stable ssDNA-VirE2 complexes resulted in exposure of the ssDNA to and degradation by the DNase activity (Fig. 3A). This ssDNA deprotection most likely occurred via UPS, because two proteasomal inhibitors, MG132 and lactacystin, largely blocked the ssDNA exposure and stabilized the T-complexes. Quantitative analyses of these effects by qPCR revealed that the ssDNA-deprotecting activity of the Arabidopsis cell extracts destabilized 58 ± 7% of ssDNA (Fig. 3B). This activity was largely inhibited by MG132 and lactacystin (79 ± 6% and 91 ± 1%, respectively). Note that this often-incomplete inhibitory effect of MG132 is a known property of this drug (34). Importantly, cell extracts from the VBF antisense plants lost most of their ability to destabilize the ssDNA-VirE2 complexes, with 85 ± 3% of ssDNA remaining stable (Fig. 3B). Collectively, these data indicate that the VBF-based activity can expose the ssDNA component of the synthetic T-complex, most likely via proteasomal degradation by the SCFVBF pathway.
Fig. 3.
VBF promotes proteasomal uncoating of synthetic T-complexes. (A and B) Protein extracts from WT or VBF antisense plants (as-VBF) challenged for 10 h with E. coli were incubated for the indicated time periods in the presence or absence of MG132 or lactacystin with synthetic T-complexes, and the susceptibility of ssDNA to DNase I was analyzed by PCR (A) and quantified by qPCR (B). The quantified data were expressed as percent of the input signal. (C) Western blot analysis of the endogenous VBF content in Arabidopsis plants. VBF expression was induced by challenging the plants for 24 h with E. coli. (Upper) Protein extracts from WT plants, VBF antisense plants (as-VBF), or VBF transgenic plants (VBF) were analyzed with anti-VBF antibody, with putative RuBisCo large chain used as a loading control. (Lower) The resulting signal was quantified and is expressed as percent of the signal obtained in VBF transgenic, unchallenged plants. All data represent average values of three independent experiments with indicated SDs.
Western blot analysis of the VBF content in different Arabidopsis lines indicated that the amounts of this protein in the WT plants and the VBF antisense plants were below the limit of detection of our anti-VBF antibody, whereas the transgenic VBF-expressing plants accumulated VBF. Challenging these plants with E. coli promoted VBF accumulation only in the WT plants and had no discernible effect on VBF levels in VBF-expressing plants or VBF antisense plants (Fig. 3C). The finding that the VBF antisense plants contained no detectable amounts of VBF yet exhibited residual levels of ssDNA exposure activity suggests the presence of other cellular factors that can compensate for the loss of this VBF function.
VBF-Dependent Proteasomal Uncoating of Synthetic T-Complexes as a Function of the VBF/VIP1 Equilibrium.
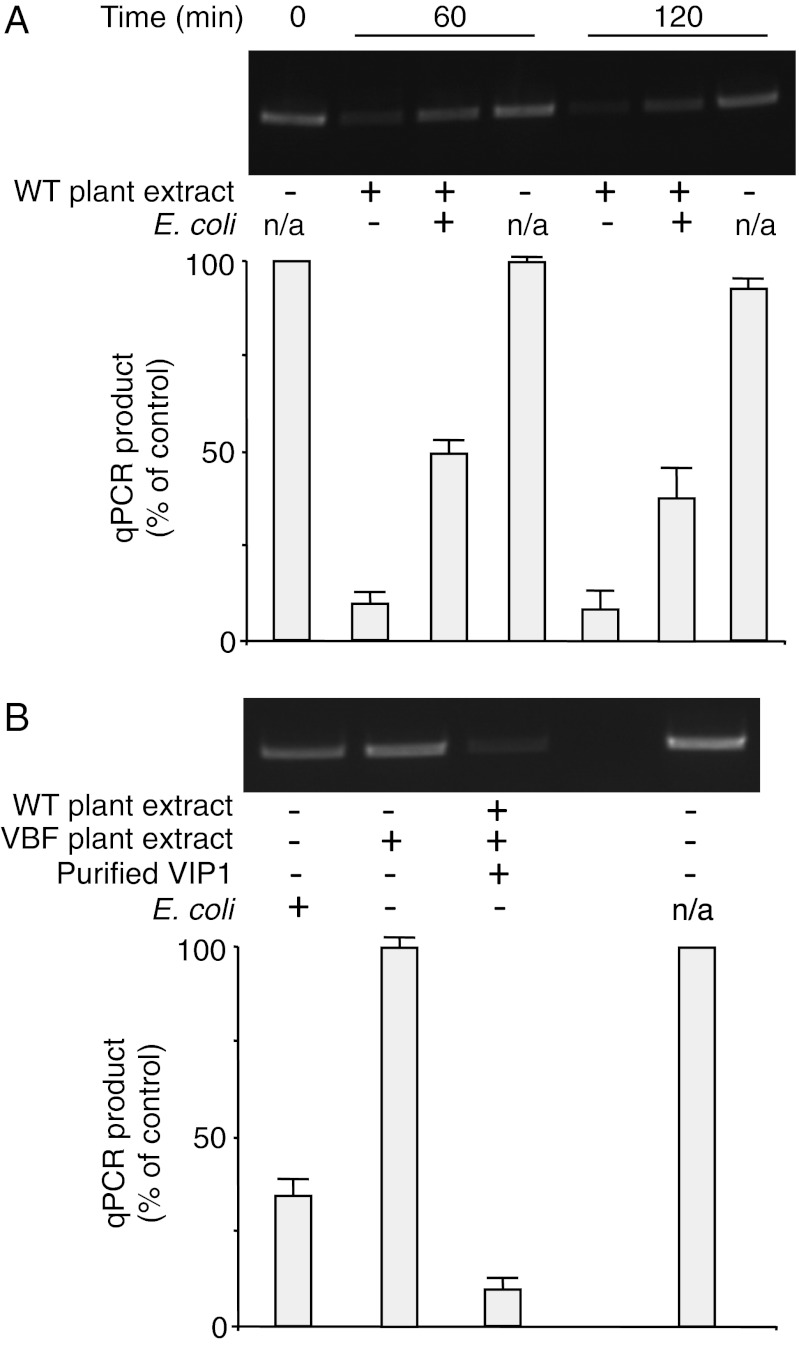
Because VBF and VIP1 are defense-related proteins, their amounts and activities are altered during microbial challenge (25, 35). These changes likely affect the efficiency of VBF-dependent ssDNA uncoating. To test this idea, we altered the endogenous amounts of VBF and/or VIP1 in our ssDNA deprotection assay. We first examined the effect of the intracellular level of VBF by comparing WT plants challenged with E. coli and untreated plants. In an apparent paradox, cell extracts from the E. coli-treated plants promoted ssDNA uncoating to a substantially lesser extent than extracts from the untreated plants (45–50% ± 3–8% vs. 10 ± 3%) (Fig. 4A). As expected, in the absence of plant extracts, the ssDNA remained stable. Increasing the VBF content by an independent approach (i.e., after stable expression in VBF transgenic plants) produced a qualitatively similar effect (i.e., reduction in the ssDNA-deprotecting activity of the plant extract), although quantitatively this inhibitory effect was much more severe (Fig. 4B).
Fig. 4.
VBF and VIP1 levels affect the efficiency of uncoating of synthetic T-complexes. (A) Effect of E. coli challenge. (B) Effects of additional VBF and VIP1. Protein extracts from WT or VBF transgenic plants (VBF) untreated or challenged for 10 h with E. coli were incubated for the indicated time periods in the presence or absence of 1 μg of recombinant VIP1 purified as described previously (19) with synthetic T-complexes, and the susceptibility of ssDNA to DNase I was analyzed by PCR (Upper) and quantified by qPCR (Lower). The quantified data were expressed as percent of the input signal and represent average values of three independent experiments with indicated SDs. n/a, not applicable.
One possible explanation for this effect is that higher and persistent transgenic levels of VBF more efficiently degrade the endogenous VIP1, preventing it from interacting with the ssDNA–VirE2 complex and, by implication, reducing the efficiency of uncoating and/or, during the actual infection, nuclear import of T-complexes, which presumably requires VIP1 (19, 20). Consistent with this idea, our VBF transgenic plants accumulated less VIP1 than the WT plants after challenge with E. coli (Fig. 1B); in addition, in the latter plants, the bacterial challenge could activate the existing VIP1 (35). In this scenario, an increase in VIP1 levels should enhance deprotection. Indeed, Fig. 4B shows that addition of purified VIP1—which by itself does not affect the stability of ssDNA (Fig. 2 C and D)—to the assay system resulted in a substantial increase in ssDNA exposure to the DNase activity. These data suggest that a defined balance between the intracellular amounts of VBF and VIP1 is required for optimal uncoating of the T-complex by the SCFVBF pathway within plant cells, and that this uncoating appears to be remarkably sensitive to VIP1 levels.
Discussion
Conceptually—and, in many aspects, functionally—the T-complex is reminiscent of nonenveloped viruses (36). Both infectious agents travel within the host cell cytoplasm, enter the host cell nucleus, and uncoat their genetic material from its associated proteins. However, whereas proteolysis has long been known to play a role in the uncoating of some viruses (37), the role of proteolytic degradation in T-complex uncoating has emerged much more recently (23, 25, 31). This uncoating has been proposed to occur by proteasomal degradation via the SCF pathway. This hypothesis is based on recognition and proteasomal degradation by the host F-box protein VBF (and its bacterial effector analog VirF) (23, 25) of the host VIP1 protein, which in turn associates with the VirE2 coat protein of the T-complex for its nuclear targeting (19) and chromatin localization (22). Thus, it has been proposed that VBF and VirF promote destabilization of both VIP1 and its associated VirE2, thereby removing the latter from the T-strand molecule and uncoating the T-complex (23, 25).
A critical aspect of this model has remained unproven: whether or not this SCF pathway can expose a DNA molecule packaged within the T-complex. To address this question, we developed a cell-free assay system for proteasomal degradation and ssDNA protection. Using this assay, we demonstrated that plant cell extracts can uncoat the T-complex and expose its DNA component, and that this uncoating involves the VIP1-binding F-box protein VBF. Because F-box proteins represent critical and specific core components of the SCF complex (27, 30), involvement of an F-box protein strongly suggests involvement of SCF. Thus, we propose that the ssDNA is uncoated via the SCFVBF pathway, in which VIP1 acts as a molecular adaptor between SCFVBF and the T-complex by binding to both VBF and VirE2. It is important to note that our synthetic T-complexes does not include the VirD2 component; however, given its very minor contribution to the T-complex structure and composition and its inability to interact with VirE2 or VIP1 (19), we do not envision any substantial role for VirD2 in the uncoating process.
Interestingly VBF, as well as its direct substrate VIP1, represent plant defense-related factors activated by infection (25, 35). Thus, Agrobacterium most likely takes advantage of the very defense pathway that it activates. The notion that Agrobacterium activates the host defense and then subverts it for infection infers that the overall infection efficiency depends on the balance between these two opposing processes. Indeed, our data indicate that in fact maximal induction of VBF expression by bacterial challenge reduces the T-complex uncoating. This interplay possibly may contribute to the well-known, yet poorly understood bimodal expression of T-DNA, in which a small proportion of the T-DNA molecules are integrated and stably expressed, whereas most T-DNA molecules are not integrated and are expressed only transiently (38). Potentially, before the plant defense response is fully mounted, the invading T-complexes are uncoated more rapidly, leading to T-DNA expression before the T-complex can be targeted by VIP1 to the chromatin for integration (22), whereas later the uncoating slows, allowing the T-complex to reach the target chromatin. Furthermore, it is tempting to speculate that, once at the chromatin, the SCFVBF pathway can expose not only the T-DNA by degrading VirE2 attached to VIP1, but also the target host DNA by degrading histone molecules to which VIP1 also attaches (21, 22). Thus, disassembly of the T-complex via the host UPS may represent a more general mechanism for uncoating of DNA molecules within eukaryotic cells; for example, proteasomal degradation of H2A/H2B histones from promoter regions has been shown to promote chromatin disassembly during transcriptional activation (39).
The central role of the SCF pathway in the Agrobacterium infection process is supported by the observation that this microorganism does not rely exclusively on its host to provide the protein machinery for this stage of infection but instead has evolved a “backup” system composed of its own virulence F-box protein, VirF, the first prokaryotic F-box protein discovered (40), which is exported into the host cells and acts as a bacterial functional homolog of VBF, destabilizing VIP1 and VIP1-VirE2 complexes (23). This strategy of Agrobacterium reflects a general ability of pathogenic microorganisms to encode and export protein functions normally provided by the host eukaryotic cell (41).
Materials and Methods
Agroinfiltration and Transgenic Plants.
The Agrobacterium EHA105 strain harboring a binary plasmid expressing HA-VIP1—made by inserting the VIP1 coding sequence into the EcoRV-BglII sites of pSAT6-HA-C1 (42) and cloning the resulting expression cassette into pPZP-RCS2, as described previously (43)—was grown in YEP medium (1% peptone, 1% yeast extract, and 0.5% NaCl) supplemented with 100 mg/L streptomycin and 10 mg/L rifampin overnight at 28 °C. Cells were pelleted, resuspended at OD600 = 0.1 in infiltration buffer [10 mM MgCl2, 10 mM Mes (pH 5.6), 100 μM acetosyringone], incubated for 4 h at room temperature, and infiltrated into the abaxial side of 4- to 6-wk-old N. benthamiana leaves using a 1-mL needleless syringe. Plants were grown for 72 h at 22 °C before harvesting.
Generation of Arabidopsis transgenic plants expressing VBF in the antisense orientation was described previously (25). In parallel experiments, transgenic plants expressing VBF in the sense orientation were produced by cloning the VBF coding sequence into the EcoRI-NdeI sites of pBluescript II (Stratagene), removing it as an EcoRI-SalI fragment and inserting it into the EcoRI-SalI sites of pSAT4-35SP-MCS-35ST (44). The subsequent experimental steps were as described for production of the antisense plants (25).
Cell-Free Degradation Assay and Western Blot Analysis.
Leaves were harvested and ground into fine powder in liquid nitrogen. For treatment with MG132 or lactacystin, leaves were infiltrated with 10 μM MG132 (CalBiochem) or 10 μM lactacystin (Sigma-Aldrich) or mock-treated with 0.1% DMSO or distilled water, respectively, and incubated for 4 h before harvesting. The cell-free degradation assay was performed as described previously (31, 45). In brief, total protein extracts from 12.5 mg of fresh leaf weight, prepared by bead-beating the tissue in 25 μL of degradation/DNase digestion buffer [10 mM Tris-HCl (pH 7.6), 0.5 mM CaCl2, 10 mM MgCl2, 5 mM DTT, 5 mM ATP, and 1× plant protease inhibitor mixture (Thermo Scientific)], were incubated at room temperature for the indicated times in a final reaction volume of 120 μL. Reactions were terminated by boiling in SDS sample buffer. The protein samples were resolved by SDS-acrylamide gel electrophoresis, and HA-VIP1 and VIP1 were detected by anti-HA (Covance) and anti-VIP1 antibodies (22, 46), respectively, followed by goat anti-rabbit antibodies conjugated to alkaline phosphatase (Bio-Rad). A major band near 50 kDa (putative RuBisCo large chains) on Coomassie blue-stained gels served as a loading control. All immunoblots were quantified using ImageJ software.
Synthetic T-Complexes and ssDNA Protection Assay.
M13mp18 ssDNA (0.1. μg) (New England BioLabs) and VirE2 (1.0 μg) purified to near homogeneity as described previously (33) were allowed to form a complex as described previously (19). For uncoating, the complexes (95 μL) were combined with 25 μL of the cell extracts. Then two units of DNase I (New England BioLabs) were added, and incubation continued for the indicated times. ssDNA was then detected by 27 cycles of PCR using ExTaq polymerase (TaKaRa) and 5′ATCAAACAGGATTTTCGCCTGCT3′/5′AACGCCAGGGTTTTCCCAGT3′ primers, which amplify a 461-bp fragment. qPCR was performed with 5′CTCTTGATGAAGGTCAGCCA3′/5′GGAACCGAACTGACCAACTT3′ primers, which amplify a 83-bp fragment, in a Light Cycler 480 Real-Time PCR System (Roche) with SYBR-Green I Master Mix (Roche).
Bacterial Challenge and RT-PCR/qPCR.
Arabidopsis thaliana (Col-0) plants (age 3–4 wk) were inoculated with E. coli (OD600 = 0.1) or mock-inoculated with the bacterial growth medium as described previously (25). At the indicated times after inoculation, total RNA was extracted from tissue samples using Tri-Reagent (Sigma-Aldrich) and treated with RNase-free DNase I (Fermentas). Reverse-transcription was then performed using 0.5 μg of RNA and SuperScript II reverse transcriptase (Stratagene), and the cDNA was PCR-amplified for 29 cycles using VBF-specific primers 5′CATTCGCTCTTCTCGGTTC3′/5′TGGGAAAAGTTTCTACCATCGG3′, which amplify a 522-bp fragment. The absence of contaminating genomic DNA was confirmed using TUBULIN-specific primers flanking an intron sequence to distinguish between PCR products derived from DNA and mRNA templates (47); TUBULIN also served as an internal control of a constitutively expressed gene. For qPCR, the reverse-transcriptase products were amplified in a Light Cycler 480 Real-Time PCR System (Roche) with SYBR-Green I Master Mix (Roche) and 5′TTCAGCTGACTCCGAGAATCTAGA3′/5′CTCGAAAGCTGTTCAAGATCAATA3′ VBF-specific primers, which amplify a 145-bp fragment. The resulting data were normalized to the amount of TUBULIN-specific products, detected using the 5′AGATTCTTCACATCCAGGGTGGTC3′/5′TCTACCGCAACTCGCTTCATTGTA3’ primers in each system.
Western Blot Analyses.
Leaf samples (500 mg) were ground in 1.5 mL of extraction buffer (30 mM NaCl, 0.1% Nonidet P-40, 1 mM PMSF, and 50 mM sodium phosphate buffer; pH 8.0) and clarified by centrifugation at 12,000 × g for 30 min. The resulting extracts (20 µL) were evaluated by Western blot analysis as described previously (25), using anti-HA antibodies (ICL), anti-VIP1 antibodies (46), or polyclonal rabbit anti-VBF antibodies (GenScript) custom-produced against the VBF peptide CVPAETSIKSKNGQI, followed by detection with secondary antibody conjugated to HRP (Pierce) and an ECL kit (Pierce). The Western blot signal was quantified using ImageJ 1.47 software. A major band near 50 kDa (putative RuBisCo large chains) on Coomassie blue-stained gels served as a loading control.
Acknowledgments
The work in our laboratory is supported by grants from the National Institute of Food and Agriculture/US Department of Agriculture (to V.C.), National Institutes of Health (to V.C.), National Science Foundation (to V.C.), and the US-Israel Binational Research and Development Fund (BARD) (to V.C. and Y.G.)
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Piers KL, Heath JD, Liang X, Stephens KM, Nester EW. Agrobacterium tumefaciens-mediated transformation of yeast. Proc Natl Acad Sci USA. 1996;93(4):1613–1618. doi: 10.1073/pnas.93.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Groot MJ, Bundock P, Hooykaas PJJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16(9):839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 3.Kunik T, et al. Genetic transformation of HeLa cells by Agrobacterium. Proc Natl Acad Sci USA. 2001;98(4):1871–1876. doi: 10.1073/pnas.041327598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzfira T, Rhee Y, Chen MH, Kunik T, Citovsky V. Nucleic acid transport in plant–microbe interactions: The molecules that walk through the walls. Annu Rev Microbiol. 2000;54:187–219. doi: 10.1146/annurev.micro.54.1.187. [DOI] [PubMed] [Google Scholar]
- 5.Gelvin SB. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 6.Tzfira T, Li J, Lacroix B, Citovsky V. Agrobacterium T-DNA integration: Molecules and models. Trends Genet. 2004;20(8):375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Zupan J, Zambryski PC. The Agrobacterium DNA transfer complex. Crit Rev Plant Sci. 1997;16:279–295. [Google Scholar]
- 8.Zupan J, Muth TR, Draper O, Zambryski PC. The transfer of DNA from Agrobacterium tumefaciens into plants: A feast of fundamental insights. Plant J. 2000;23(1):11–28. doi: 10.1046/j.1365-313x.2000.00808.x. [DOI] [PubMed] [Google Scholar]
- 9.Citovsky V, Guralnick B, Simon MN, Wall JS. The molecular structure of Agrobacterium VirE2-single stranded DNA complexes involved in nuclear import. J Mol Biol. 1997;271(5):718–727. doi: 10.1006/jmbi.1997.1230. [DOI] [PubMed] [Google Scholar]
- 10.Abu-Arish A, et al. Three-dimensional reconstruction of Agrobacterium VirE2 protein with single-stranded DNA. J Biol Chem. 2004;279(24):25359–25363. doi: 10.1074/jbc.M401804200. [DOI] [PubMed] [Google Scholar]
- 11.Citovsky V, Warnick D, Zambryski PC. Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc Natl Acad Sci USA. 1994;91(8):3210–3214. doi: 10.1073/pnas.91.8.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Citovsky V, Zupan J, Warnick D, Zambryski PC. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992;256(5065):1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 13.Howard EA, Zupan JR, Citovsky V, Zambryski PC. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: Implications for nuclear uptake of DNA in plant cells. Cell. 1992;68(1):109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 14.Koukolíková-Nicola Z, et al. Genetic analysis of the virD operon of Agrobacterium tumefaciens: A search for functions involved in transport of T-DNA into the plant cell nucleus and in T-DNA integration. J Bacteriol. 1993;175(3):723–731. doi: 10.1128/jb.175.3.723-731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mysore KS, et al. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol Plant Microbe Interact. 1998;11(7):668–683. doi: 10.1094/MPMI.1998.11.7.668. [DOI] [PubMed] [Google Scholar]
- 16.Rossi L, Hohn B, Tinland B. The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plant. Mol Gen Genet. 1993;239(3):345–353. doi: 10.1007/BF00276932. [DOI] [PubMed] [Google Scholar]
- 17.Tzfira T, Citovsky V. Comparison between nuclear import of nopaline- and octopine-specific VirE2 protein of Agrobacterium in plant and animal cells. Mol Plant Pathol. 2001;2:171–176. doi: 10.1046/j.1364-3703.2001.00065.x. [DOI] [PubMed] [Google Scholar]
- 18.Zupan JR, Citovsky V, Zambryski PC. Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc Natl Acad Sci USA. 1996;93(6):2392–2397. doi: 10.1073/pnas.93.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20(13):3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis VIP1 gene. Proc Natl Acad Sci USA. 2002;99:10435–10440. doi: 10.1073/pnas.162304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA. 2005;102(16):5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacroix B, Loyter A, Citovsky V. Association of the Agrobacterium T-DNA–protein complex with plant nucleosomes. Proc Natl Acad Sci USA. 2008;105(40):15429–15434. doi: 10.1073/pnas.0805641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431(7004):87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 24.Zaltsman A, Krichevsky A, Kozlovsky SV, Yasmin F, Citovsky V. Plant defense pathways subverted by Agrobacterium for genetic transformation. Plant Signal Behav. 2010;5(10):1245–1248. doi: 10.4161/psb.5.10.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaltsman A, Krichevsky A, Loyter A, Citovsky V. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe. 2010;7(3):197–209. doi: 10.1016/j.chom.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(17):11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9(6):631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 29.Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Genet. 1998;14(6):236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 30.Xiao W, Jang JC. F-box proteins in Arabidopsis. Trends Plant Sci. 2000;5(11):454–457. doi: 10.1016/s1360-1385(00)01769-6. [DOI] [PubMed] [Google Scholar]
- 31.Magori S, Citovsky V. Agrobacterium counteracts host-induced degradation of its F-box protein effector. Sci Signal. 2011;4:ra69. doi: 10.1126/scisignal.2002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang P, et al. Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem. 2004;279(8):6401–6413. doi: 10.1074/jbc.M311977200. [DOI] [PubMed] [Google Scholar]
- 33.Citovsky V, Wong ML, Zambryski PC. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: Implications for the T-DNA transfer process. Proc Natl Acad Sci USA. 1989;86(4):1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchsbaum S, Bercovich B, Ciechanover A. FAT10 is a proteasomal degradation signal that is itself regulated by ubiquitination. Mol Biol Cell. 2012;23(1):225–232. doi: 10.1091/mbc.E11-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science. 2007;318(5849):453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- 36.Howard EA, Citovsky V. The emerging structure of the Agrobacterium T-DNA transfer complex. Bioessays. 1990;12:103–108. [Google Scholar]
- 37.Greber UF, Singh I, Helenius A. Mechanisms of virus uncoating. Trends Microbiol. 1994;2(2):52–56. doi: 10.1016/0966-842x(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 38.Janssen BJ, Gardner RC. Localized transient expression of GUS in leaf discs following cocultivation with Agrobacterium. Plant Mol Biol. 1990;14(1):61–72. doi: 10.1007/BF00015655. [DOI] [PubMed] [Google Scholar]
- 39.Ransom M, et al. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem. 2009;284(35):23461–23471. doi: 10.1074/jbc.M109.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrammeijer B, et al. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol. 2001;11(4):258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 41.Nagai H, Roy CR. Show me the substrates: Modulation of host cell function by type IV secretion systems. Cell Microbiol. 2003;5(6):373–383. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 42.Dafny-Yelin M, Tzfira T. Delivery of multiple transgenes to plant cells. Plant Physiol. 2007;145(4):1118–1128. doi: 10.1104/pp.107.106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzfira T, et al. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57(4):503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 44.Chung SM, Frankman EL, Tzfira T. A versatile vector system for multiple gene expression in plants. Trends Plant Sci. 2005;10(8):357–361. doi: 10.1016/j.tplants.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Más P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426(6966):567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 46.Citovsky V, et al. Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J Biol Chem. 2004;279(28):29528–29533. doi: 10.1074/jbc.M403159200. [DOI] [PubMed] [Google Scholar]
- 47.Zaltsman A, Feder A, Adam Z. Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts: Implications for thylakoid formation and photosystem II maintenance. Plant J. 2005;42(5):609–617. doi: 10.1111/j.1365-313X.2005.02401.x. [DOI] [PubMed] [Google Scholar]