Significance
Myosin II from the soil amoeba Acanthamoeba castellanii is a member of the largest of the 35 classes of the superfamily of molecular motors that, together with actin filaments, convert the energy of hydrolysis of ATP into force or motion that drives numerous cellular and intracellular processes. In this paper we show that the actin-activated ATPase of Acanthamoeba myosin II is regulated by phosphorylation of a specific serine in a region of the myosin motor domain that is known to be at the myosin–actin interface. No other myosin has been shown to be regulated in this way.
Abstract
It had been proposed previously that only filamentous forms of Acanthamoeba myosin II have actin-activated MgATPase activity and that this activity is inhibited by phosphorylation of up to four serine residues in a repeating sequence in the C-terminal nonhelical tailpiece of the two heavy chains. We have reinvestigated these issues using recombinant WT and mutant myosins. Contrary to the earlier proposal, we show that two nonfilamentous forms of Acanthamoeba myosin II, heavy meromyosin and myosin subfragment 1, have actin-activated MgATPase that is down-regulated by phosphorylation. By mass spectroscopy, we identified five serines in the heavy chains that can be phosphorylated by a partially purified kinase preparation in vitro and also are phosphorylated in endogenous myosin isolated from the amoebae: four serines in the nonhelical tailpiece and Ser639 in loop 2 of the motor domain. S639A mutants of both subfragment 1 and full-length myosin had actin-activated MgATPase that was not inhibited by phosphorylation of the serines in the nonhelical tailpiece or their mutation to glutamic acid or aspartic acid. Conversely, S639D mutants of both subfragment 1 and full-length myosin were inactive, irrespective of the phosphorylation state of the serines in the nonhelical tailpiece. To our knowledge, this is the first example of regulation of the actin-activated MgATPase activity of any myosin by modification of surface loop 2.
Structurally, Acanthamoeba myosin II (AMII) is a typical class II myosin with a pair of identical 1,509-residue heavy chains (1) and two pairs of light chains: a 154-residue light chain 1 (LC1) (2) and a 145-residue light chain 3 (LC2) (this paper). The first 787 amino acids of the heavy chains comprise the globular motor domain which binds F-actin and has actin-activated MgATPase activity, and the next 58 residues (the IQ domain) contain the binding sites for LC1 and LC2 (3). Heptad repeats predicted to form an α-helical coiled-coil of the two heavy chains begin after Pro847 [(1) but see Rimm et al. (4) for an alternative start-site of the coiled-coil helix]. The predicted coiled-coil tail continues, interrupted by a “hinge” region around Pro1244, to residue 1482 (1) followed by a C-terminal nonhelical tailpiece of 27 residues beginning with Pro1483 (1). In the presence of divalent cations at low ionic strength, monomeric myosin molecules assemble through their tail domains forming bipolar minifilaments of 8–16 monomers with a 90-nm bare zone and a 15-nm stagger between heads at both ends (5, 6).
Although the structure of AMII is similar to that of other class II myosins, regulation of the actin-activated ATPase activity of AMII differs from the known regulatory mechanisms of other myosin IIs (7). Striated (skeletal and cardiac) muscle myosin IIs are activated by Ca2+-binding to the tropomyosin/troponin complex associated with the actin filament; vertebrate smooth muscle and nonmuscle myosin IIs and Dictyostelium myosin II filaments are activated by Ca2+-activated kinase phosphorylation of the regulatory light chain; and molluscan muscle myosin II is activated by Ca2+-binding to the essential light chain. None of these regulatory mechanisms is applicable to AMII, whose actin-activated ATPase activity is regulated by phosphorylation of its heavy chains.
AMII isolated from Acanthamoeba castellanii (8, 9) has an average of 1.5 phosphates per heavy chain (10) or 3 P per myosin molecule. The very low actin-activated ATPase activity of AMII increases when the heavy chains are dephosphorylated in vitro by phosphatase to less than 1 phosphates per heavy chain (10). Conversely, the actin-activated ATPase activity of dephosphorylated AMII is down-regulated in vitro by phosphorylation of one or more serines per heavy chain by a partially purified kinase (11). The 27-residue C-terminal nonhelical tailpiece of each heavy chain of AMII, 1483PSSRGGSTRGASARGASVRAGSARAEE1509, contains four serines (residues 1489, 1494, 1499, and 1504) within a sequence RXXSXR. Serines 1489 and 1494 were shown to be phosphorylated in vivo and serines 1489, 1494, and 1499 were found to be phosphorylated in vitro by a partially purified kinase (11–13).
In a series of papers (12–15), E.D.K. inferred from these and other experimental data that phosphorylation of one or more of the serines 1489, 1494, and 1499, and possibly serine 1504, in the nonhelical tailpiece was responsible for inactivation of AMII. Furthermore, because the actin-activated ATPase activity of copolymers of dephosphorylated and phosphorylated AMII was less than the activity of equivalent mixtures of homopolymers, it was suggested that regulation of ATPase activity occurred at the filament level (16, 17). From these and other (18–20) data, it was inferred that only filamentous AMII has phosphorylation-regulated actin-activated ATPase activity and that the activity of each molecule in a filament is a function not of its own level of phosphorylation but of the phosphorylation level of the filament as a whole. Thus, regulation of the actin-activated ATPase of AMII would involve a conformational change in the myosin filament. However, detailed studies by the Pollard laboratory (5, 6, 21) showed no differences in the assembly pathway or structure of AMII with phosphorylation levels of 0.4–2.5 phosphates per heavy chain.
All previous studies of AMII were done with phosphorylated myosin purified from the amoebae and after partial dephosphorylation and rephosphorylation in vitro. The phosphorylation sites in the heavy chains were identified by 2D electrophoresis and chromatography of tryptic and chymotryptic peptides and amino acid sequencing of isolated tryptic peptides by automated Edman degradation (11–13). To determine if the earlier results were affected by unidentified phosphorylation sites or other posttranslational modifications in the endogenous myosin, we studied the effects of phosphorylation on the actin-activated ATPase activity [and filament structure; see accompanying paper (22)] of AMII constructs consisting of recombinant full-length WT, truncated, and mutant heavy chains coexpressed with the two WT light chains in SF-9 cells. We quantified phosphorylation of the recombinant myosins by partially purified AMII heavy-chain kinase, identified the sites of phosphorylation by MS, and assessed the effect of phosphorylation of the myosins on actin-activated ATPase activity and affinity for F-actin.
Contrary to previous conclusions, we found that nonfilamentous heavy meromyosin (HMM) and myosin subfragment 1 (S1) had actin-activated ATPase activity that was down-regulated by phosphorylation. Most importantly, all the recombinant AMII constructs were inactivated by phosphorylation of Ser639 in loop 2 of the motor domain or by its mutation to aspartic acid but not by its mutation to alanine. Phosphorylation, deletion, or mutation of the four serines in the nonhelical tailpiece had no effect on the actin-activated ATPase activity of filamentous AMII. Furthermore, we found that, in addition to the previously identified phosphorylated serines in the nonhelical tailpiece, Ser639 also is phosphorylated in enzymatically inactive endogenous AMII purified from amoebae, and pSer639 is dephosphorylated in phosphatase-treated, enzymatically active AMII. To our knowledge, this is the first example of the regulation of any myosin by covalent modification of any amino acid in loop 2 (7, 23). The effects of phosphorylation of Ser639 and serines in the nonhelical tailpiece on the structure of AMII filaments are described in the accompanying paper (22).
Results
Phosphorylation and Actin-Activated ATPase Activity of AMII Constructs.
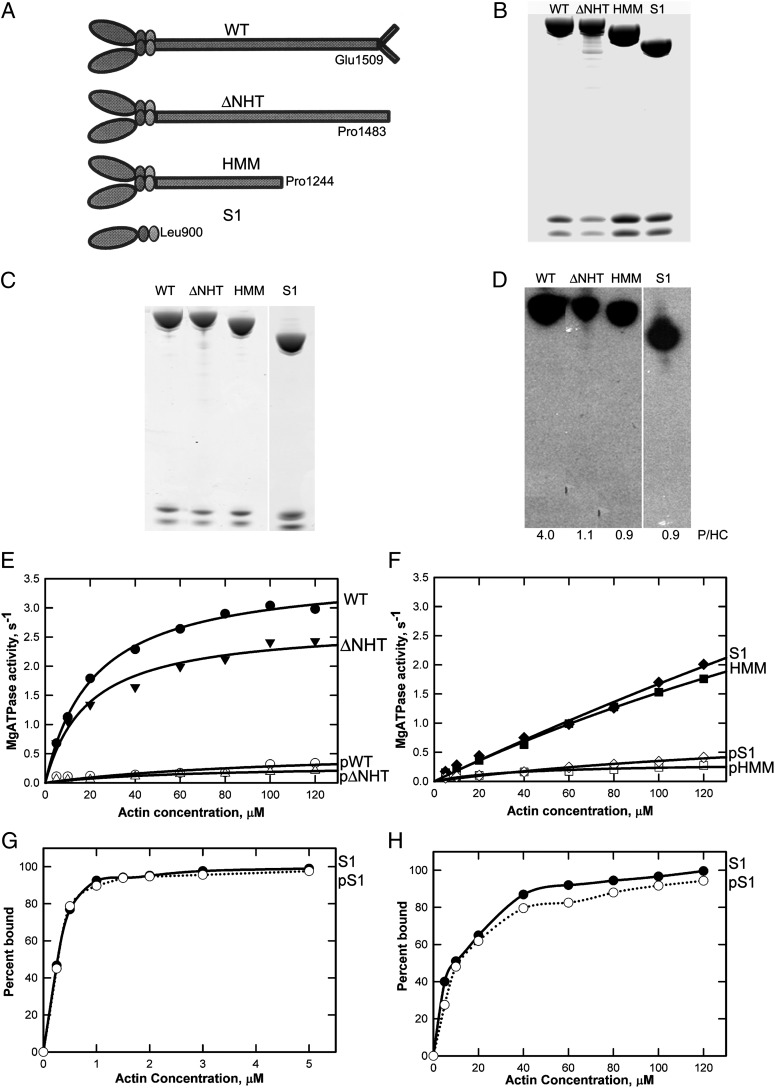
Initially, we expressed the two light chains with full-length heavy chain (WT), heavy chain truncated at Pro1483 to remove the nonhelical tailpiece (ΔNHT), heavy chain truncated at Pro1244 (HMM), and heavy chain truncated at Leu900 (S1) (Fig. 1A). All constructs were expressed at a high level in Baculovirus-infected SF-9 cells (∼2 mg myosin/5 g of cells), and the purified proteins were electrophoretically homogeneous (Fig. 1B). In previous studies, an average of 2.5 phosphates per heavy chain were incorporated when partially dephosphorylated endogenous AMII was phosphorylated in vitro (11, 13) for a total of ∼3 phosphates per heavy chain, consistent with the identification of phosphorylated peptides containing serines 1489, 1494, and 1499 and with the possibility that Ser1504 might also be phosphorylated. Consistent with the previous results, we now have found that 32P was incorporated only into the heavy chain (4 phosphates per heavy chain) when recombinant WT was incubated with AMII heavy-chain kinase and [γ-32P]ATP (Fig. 1 C and D). Unexpectedly, however, 1 phosphates per heavy chain was incorporated into recombinant ΔNHT, HMM, and S1 (Fig. 1D), none of which contained the nonhelical tailpiece.
Fig. 1.
Phosphorylation and actin-activated ATPase activity of AMII constructs. (A) Schematic illustration of WT and recombinant myosins ΔNHT, HMM, and S1. (B) Coomassie blue-stained SDS/PAGE of purified recombinant myosins showing the heavy chain and two light chains. Samples were analyzed on a 1.0-mm 10% NuPAGE gel (Life Technologies). (C and D) Coomassie blue-stained SDS/PAGE (C) and autoradiogram (D) of recombinant myosins phosphorylated in vitro by the kinase preparation. The incorporated phosphates per heavy chain (P/HC; average of two to four experiments) is given under the lanes in D. (E and F) Actin dependence of Mg-ATPase activity of unphosphorylated and phosphorylated WT and ΔNHT (E) and HMM and S1 (F). Data are the averages of two experiments. (G and H) Binding of S1 and pS1 to F-actin in 2.5 mM KCl and 3 mM MgCl2 without ATP (G) or in 2.5 mM KCl and 3 mM MgCl2 with 1 mM ATP (H). In the absence of ATP, F-actin was stabilized by phalloidin at a 1:1 molar ratio to actin.
Furthermore, the maximal actin-activated ATPase activities of nonphosphorylated HMM and S1 were very similar to that of WT (Fig. 1 E and F), although HMM and S1 required a higher actin concentration to have activity similar to WT. Also, as in WT, the enzymatic activities of ΔNHT, HMM, and S1 were down-regulated by phosphorylation (Fig. 1 E and F). Phosphorylation had no effect on Ca-ATPase activity (Table 1), and the affinities of S1 and phosphorylated S1 (pS1) to F-actin were indistinguishable in both the absence and presence of ATP (Fig. 1 G and H). These results are inconsistent with the previous conclusions that nonfilamentous AMII is enzymatically inactive and that actin-activated ATPase activity is regulated by phosphorylation of serines in the nonhelical tailpiece.
Table 1.
Ca-ATPase activity of unphosphorylated and phosphorylated recombinant AMII constructs
| Construct | Ca-ATPase activity, s−1 |
|
| Unphosphorylated | Phosphorylated | |
| WT | 4.1 | 3.9 |
| ΔNHT | 3.1 | 3.4 |
| HMM | 3.4 | 3.9 |
| S1 | 3.5 | 3.1 |
| S639A | 3.4 | 4.1 |
| S639D | 3.8 | 3.6 |
| 4S/E | 3.6 | 3.4 |
Activity was determined in the absence of Mg2+ and F-actin as described in Materials and Methods.
Identification of the Phosphorylation Sites in Recombinant AMII Heavy Chain.
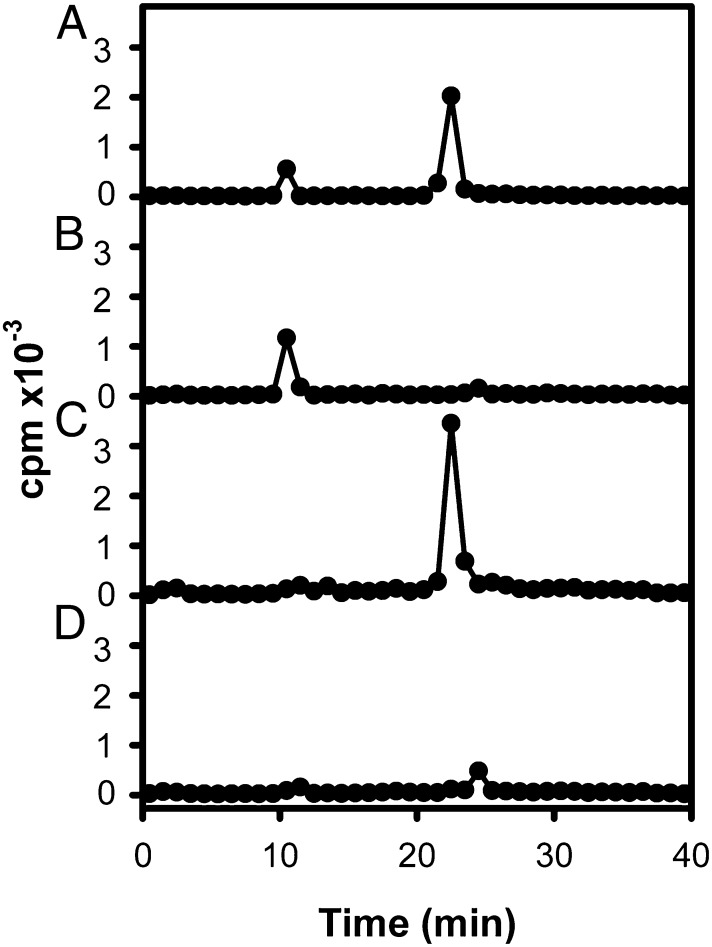
32P-labeled peptides from a LysC digest of 32P-labeled recombinant full-length myosin were separated by reverse-phase chromatography. The eluate had two radioactive peaks (Fig. 2A). LysC digests from unphosphorylated and phosphorylated (nonradioactive) recombinant myosins then were mapped by HPLC-MS. Comparison of the observed masses in the HPLC of the LysC digests of the unphosphorylated and phosphorylated samples revealed two peptide pairs whose mass differences were consistent with phosphorylation and whose elution times, ∼10 and ∼23 min, were the same as the elution times of the radioactive peaks in the chromatography of the 32P-labeled peptides. No other potentially phosphorylated peptides were detected.
Fig. 2.
HPLC chromatograms of LysC digests of 32P-labeled recombinant full-length and mutant myosins. (A) WT. (B) S1489, 1494, 1499, 1504E (4S/E). (C) S639A. (D) S639, 1489, 1494, 1499, 1504A (5S/A). For all samples, LysC digests of 10 µg of protein were analyzed.
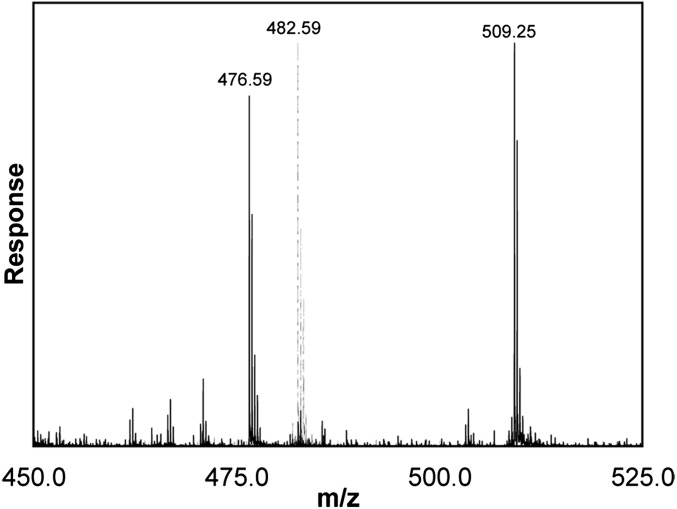
The LysC peptides eluting at 9–10 min had observed masses of 1,444.76 and 1,524.73 for the unphosphorylated and phosphorylated proteins, respectively. These observed masses match the calculated values of 1,444.76 and 1,524.73 for the unphosphorylated and phosphorylated forms of a LysC peptide comprising heavy-chain residues 630–644, AAAGGSRNRSTGRGK. The mass spectrum (Fig. 3) of the triply charged phosphorylated peptide (m/z = 509.248) showed a prominent fragment with m/z = 476.586, corresponding to a decrease in mass of the parent peptide of 97.98 caused by neutral loss of phosphate. This fragmentation confirmed that the peptide was phosphorylated, and MS/MS sequencing identified the phosphorylated residue as Ser639 (Table 2), i.e., both y7-H3PO4 and y6-H3PO4 ions were observed. The absence of a y5-H3PO4 ion and the presence of the y5 ion showed that Thr640 was not phosphorylated, and, similarly, there was no evidence for phosphorylation of Ser635.
Fig. 3.
Mass spectrum of LysC peptide 630–644 from LysC digests of unphosphorylated (dashed line) and phosphorylated (solid line) recombinant AMII. The measured masses of the peptides were 1,444.76 and 1,524.73, respectively, a difference of 79.97 caused by phosphorylation. The parent peaks have m/z values of 482.59 and 509.25 because the ions were triply charged. The peak at 476.58 in the phosphorylated peptide sample resulted from neutral loss of phosphate, establishing that the peptide is phosphorylated. The fragmentor voltage was increased from the usual 235 V to 275 V to enhance the neutral loss of H3PO4.
Table 2.
Tandem mass spectrum of LysC phosphopeptide 630–644 from phosphorylated recombinant myosin
| Residue | a and b ions |
y ions |
||||
| Ion | Calculated | Observed | Ion | Calculated | Observed | |
| Ala630 | a1 | 44.050 | 44.049 | — | — | — |
| Ala631 | a2 | 115.087 | 115.090 | y14 | 1,454.697 | ND |
| b2 | 143.082 | 143.085 | ND | |||
| Ala632 | b3 | 214.119 | 214.116 | y13 | 1,383.660 | ND |
| Gly633 | b4 | 271.141 | 271.143 | y12 | 1,312.623 | ND |
| Gly634 | b5 | 328.162 | 328.154 | y11 | 1,255.602 | ND |
| Ser635 | b6 | 415.194 | ND | y10 | 1,198.580 | ND |
| Arg636 | b7 | 571.295 | 571.301 | y9 | 1,111.548 | ND |
| Asn647 | b8 | 685.338 | 685.359 | y8 – NH3 - H3PO4 | 840.433 | 840.458 |
| Arg638 | b9 | 841.439 | 841.439 | y7 - H3PO4 | 743.417 | 743.416 |
| pSer639 | b10 - H3PO4 | 910.461 | 910.459 | y6 - H3PO4 | 587.316 | 587.326 |
| b10 | 1,008.438 | ND | y6 | 685.303 | ND | |
| Thr640 | b11 | 1,109.485 | ND | y5 | 518.305 | 518.309 |
| Gly641 | b12 | 1,166.507 | ND | y4 | 417.257 | 417.270 |
| Arg642 | b13 | 1,322.608 | ND | y3 | 360.235 | 360.248 |
| Gly643 | b14 | 1,379.629 | ND | y2 | 204.134 | 204.141 |
| Lys644 | — | — | — | y1 | 147.113 | 147.112 |
The measured mass of the peptide was 1,524.72, and the calculated mass for the monophosphorylated peptide is 1,524.73. The doubly charged ion with m/z 763.371 was sequenced. The dominant product ion in the spectrum was at 714.382 and was produced by neutral loss of H3PO4 from the peptide. However, there was sufficient abundance of other peaks to identify Ser639 unambiguously as the phosphorylated residue. Fragmentation of the peptide generated ions whose mass allowed sequencing in both the amino-to-carboxyl direction (a or b ions) and the carboxyl-to-amino direction (y ions). The observed mass of the y5 ion established that Thr640 is not phosphorylated, whereas the masses of the y6 ions with neutral loss of phosphate established that Ser639 was phosphorylated. ND, the ion was not detected.
Tryptic cleavage of phosphorylated recombinant myosin generated nonphosphorylated peptide 630–636 (observed mass, 588.299; calculated mass, 588.2980) and phosphorylated peptide P639-642 (observed mass, 499.179; calculated mass, 499.1792), confirming the LysC results. In addition, we incubated a synthetic peptide with the sequence of residues 629–647, KAAAGGSRNRSTGRGKGGA, with the kinase preparation. By MS analysis, this peptide was fully phosphorylated on the residue corresponding to Ser639 of AMII heavy chain, and no phosphorylation of residues corresponding to Ser635 or Thr640 was detected.
The LysC peptides of unphosphorylated and phosphorylated myosins eluting at ∼23 min also differed in mass in a manner consistent with phosphorylation. The unphosphorylated peptide had an observed mass of 3,770.86, matching the calculated value of 3,770.8644 for the C-terminal peptide 1472–1509, IAQLQDEIDGTPSSRGGSTRGASARGASVRAGSARAEE. Serines of this peptide had been shown previously to be phosphorylated in vivo and in vitro (see Introduction). The LysC map of the phosphorylated myosin had no detectable unmodified peptide 1472–1509, but did have masses matching peptides with two (6%), three (38%), and four (56%) phosphorylations. Tryptic mapping of phosphorylated recombinant AMII identified four peptides from the C terminus that were phosphorylated and exhibited neutral loss of phosphate. MS/MS sequencing (Table 3) identified the following sites of phosphorylation: tryptic peptide 1487–1491, GGSTR, phosphorylated on Ser1489; tryptic peptide 1492–1496, GASAR, phosphorylated on Ser1494; tryptic peptide 1497–1501, GASVR, phosphorylated on Ser1499; and tryptic peptide 1502–1506, AGSAR, phosphorylated on Ser1504. The first three sites had been shown previously to be phosphorylated, but not Ser1504 (11–13).
Table 3.
Tandem mass spectrum of the tryptic phosphopeptides in the C terminus of endogenous myosin
| Peptide and elution (min) | Residue | b ions |
y ions |
||||
| Ion | Calculated | Observed | Ion | Calculated | Observed | ||
| P1487–1491 (4.04) | Gly1487 | b1 | 58.029 | ND | — | — | — |
| Gly1488 | b2 | 115.051 | 115.082 | y4 | 402.210 | ND | |
| pSer1489 | b3 | 184.072 | 184.067 | y3 | 345.188 | 345.186 | |
| Thr1490 | b4 | 285.120 | 285.113 | y2 | 276.167 | ND | |
| Arg1491 | — | — | — | y1 | 175.119 | 175.119 | |
| P1492–1496 (4.56) | Gly1492 | b1 | 58.029 | ND | — | — | — |
| Ala1493 | b2 | 129.066 | 129.063 | y4 | 386.215 | ND | |
| pSer1494 | b3 | 198.088 | 198.087 | y3 | 315.178 | 315.187 | |
| Ala1495 | b4 | 269.125 | 269.121 | y2 | 246.156 | 246.150 | |
| Arg1496 | — | — | — | y1 | 175.119 | 175.120 | |
| P1497–1501 (9.02) | Gly1497 | b1 | 58.029 | ND | — | — | — |
| Ala1498 | b2 | 129.066 | 129.064 | y4 | 414.246 | 414.246 | |
| pSer1499 | b3 | 198.088 | 198.084 | y3 | 343.209 | 343.211 | |
| Val1500 | b4 | 297.156 | 297.158 | y2 | 274.187 | 274.186 | |
| Arg1501 | — | — | — | y1 | 175.119 | 175.120 | |
| P1502–1506 (5.42) | Ala1502 | b1 | 72.045 | ND | — | — | — |
| Gly1503 | b2 | 129.066 | 129.067 | y4 | 372.199 | ND | |
| pSer1504 | b3 | 198.088 | 198.088 | y3 | 315.178 | 315.183 | |
| Ala1505 | b4 | 269.125 | 265.129 | y2 | 246.156 | 246.256 | |
| Arg1506 | — | — | — | y1 | 175.119 | 175.121 | |
Sequencing was performed on the singly charged ions of phosphorylated peptides which experienced neutral loss of H3PO4, thus converting phosphoserine to dehydroalanine. The calculated masses of the ions in the table are with the neutral loss. Fragmentation of peptides generates ions whose mass allow sequencing in both the amino-to-carboxyl direction (b ions) and the carboxyl-to-amino direction (y ions). ND, the ion was not detected.
Phosphorylation of Site-Specific Mutants.
Having identified five serine residues (639, 1489, 1494, 1499, and 1504) that can be phosphorylated in vitro, we prepared three mutant myosins: S639A in which the C-terminal serines were unchanged; S1489, 1494, 1499, 1504E (4S/E), in which Ser639 was unchanged; and S639, 1489, 1494, 1499, 1504A (5S/A), in which all five serines were mutated. These constructs were phosphorylated in vitro with [γ32P]ATP. Reverse-phase HPLC of LysC digests (Fig. 2 B, C, and D) were compared with that of the WT LysC map (Fig. 2A). The C-terminal mutant, 4S/E, showed the expected peak at ∼10 min because of phosphorylation of Ser639, and the peak at ∼23 min was almost, but not completely, eliminated (Fig. 2B). The S639A mutant showed the reciprocal change with almost complete elimination of the peak at ∼10 min and retention of the 23-min peak (Fig. 2C). The 5S/A mutant showed very blunted peaks at both ∼10 and ∼24 min (Fig. 2D).
LysC and tryptic peptide maps of the phosphorylated quintuple mutant, 5S/A, then were analyzed by HPLC-MS. The LysC analysis established that peptide 630–644, AAAGGSRNRATGRGK, was partially phosphorylated (0.1 phosphate per peptide) and underwent neutral loss of phosphate but, with this low level of phosphorylation, we could not determine whether Ser635 or Thr640 was minimally phosphorylated. LysC mapping also demonstrated that the C-terminal peptide 1472–1509, IAQLQDEIDGTPSSRGGETRGAEARGAEVRAGSARAEE, was partially phosphorylated and experienced neutral loss of phosphate. Tryptic peptide mapping showed that peptide 1472–1486, IAQLQDEIDGTPSSR, eluted at 22.5 min. This peptide was partially phosphorylated (0.23 phosphate per peptide) and the phosphopeptide exhibited neutral loss of phosphate. Tryptic peptide1472–1486 has two serine residues but no threonines. De novo sequencing established that Ser1484 was phosphorylated; no phosphorylation of Ser1485 was detected. We conclude that the five major sites of phosphorylation of AMII are heavy-chain serines 639, 1489, 1494, 1499, and 1504. The two minor sites of phosphorylation, Ser635 (or Thr640) and Ser1484, lack an arginine at the −3 position that is present in all five major phosphorylation sites.
Actin-Activated ATPase Activity of Serine Mutants.
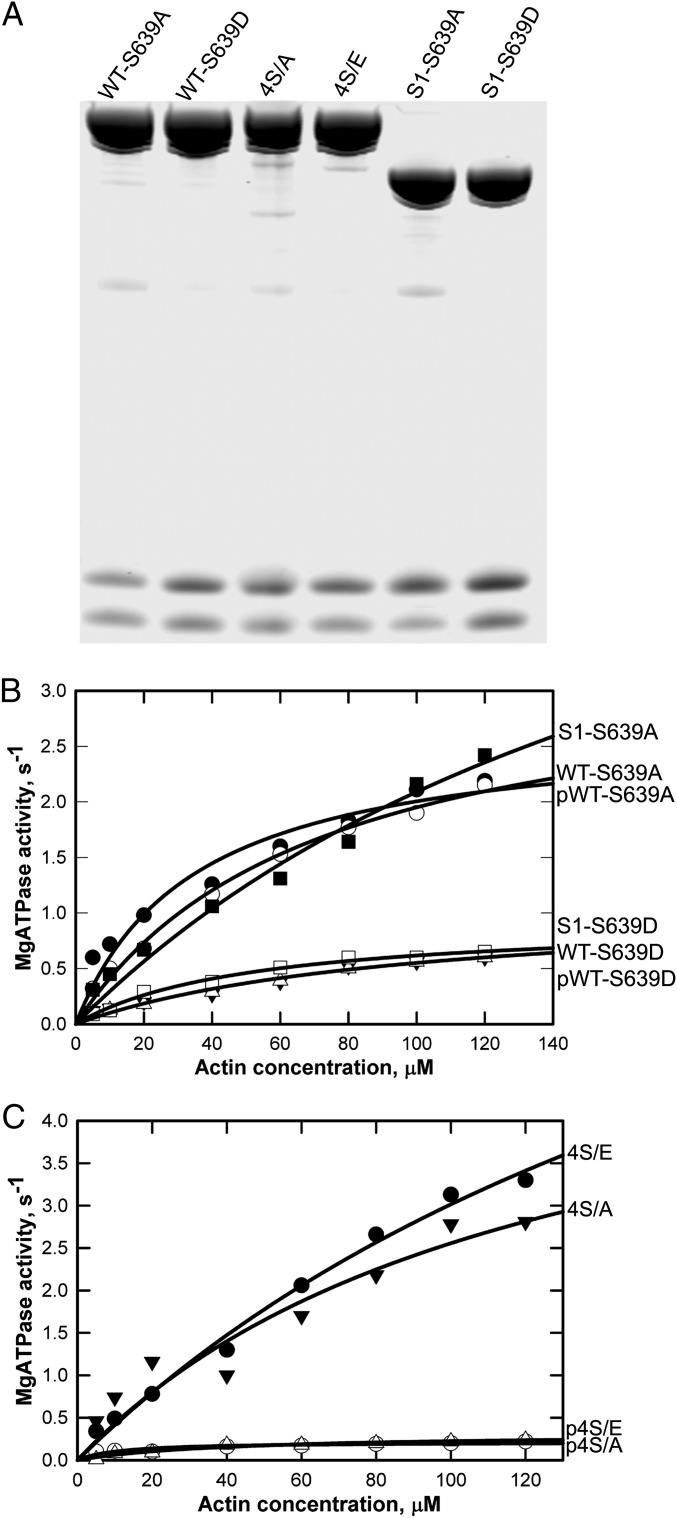
The fact that phosphorylation inhibited the activity of ΔNHT, HMM, and S1, none of which contains the nonhelical tailpiece, made it likely that actin-activated ATPase activity of AMII is regulated solely by phosphorylation of Ser639. To confirm this conclusion, we determined the effect of phosphorylation on the enzymatic activity of WT with Ser639 mutated to alanine or aspartic acid (WT-S639A and WT-S639D; Fig. 4A), WT with the four nonhelical tailpiece serines mutated to alanine or glutamic acid (4S/A and 4S/E; Fig. 4A), and S1 with S639 mutated to alanine or aspartic acid (S1-S639A and S1-S639D; Fig. 4A). The data in Fig. 4B show that the phosphomimetic mutation S639D inactivated both S1 and WT, whereas the S639A mutation had no effect on ATPase activity but did protect WT from inactivation by phosphorylation. Consistent with these results, phosphorylation inhibited the activity of both 4S/E and 4S/A (Fig. 4C). None of the mutations affected Ca-ATPase activity (Table 1).
Fig. 4.
Actin-activated MgATPase of unphosphorylated, phosphorylated, and phosphomimetic site-specific mutant myosins. (A) Coomassie blue-stained gel of SDS/PAGE of unphosphorylated site-specific mutants WT-S639A, WT-S639D, 4S/A, 4S/E, S1-S639A, and S1-S639D. (B and C) Actin-activated ATPase activities of recombinant myosins shown in A. The data are the averages of two experiments for each construct.
Phosphorylated Amino Acids in Endogenous AMII.
To determine if the phosphorylation sites identified in recombinant AMII phosphorylated in vitro also were phosphorylated in vivo, we isolated endogenous AMII from the amoebae and dephosphorylated one portion with phosphatase for comparison with the as-isolated phosphorylated sample. The actin-activated MgATPase activities of the isolated myosin and dephosphorylated myosins were 0.34 s−1 and 2.27 s−1, respectively, at 100 µM F-actin, similar to the activities of phosphorylated and unphosphorylated recombinant full-length myosins (Fig. 1E). MS analysis of both LysC and tryptic digests of endogenous AMII established that the sites of phosphorylation in endogenous myosin are identical to those found in recombinant myosin phosphorylated in vitro, including Ser639 (Fig. 5). However, although recombinant myosin phosphorylated in vitro had no detectable unphosphorylated LysC peptide 630–644 or 1472–1509 (see above), 31% of LysC peptide 630–644 and 44% of LysC peptide 1472–1509 were unphosphorylated in the endogenous AMII (Table 4). Also, the peptides of endogenous myosin that were phosphorylated were incompletely phosphorylated, with an average of 0.8 phosphates on peptide 630–644, which has one phosphorylatable serine, and an average of 0.9 phosphates on peptide 1472–1509, which has four phosphorylatable serines (Table 2). This total of 1.7 phosphates per heavy chain of endogenous AMII determined by MS agrees with the value of 1.5 phosphates per heavy chain previously determined by chemical analysis (10, 11). If phosphorylation were completely random, 96% of endogenous AMII molecules (two heavy chains) would be phosphorylated on at least one Ser639, and 99% would be phosphorylated on at least one of the serines in the nonhelical tailpiece. However, if phosphorylation and dephosphorylation (14) occur at the filament level (22), the phosphorylation levels of Ser639 and the nonhelical tail serines of endogenous filaments could vary with the functional activity and location of the filaments in the cell.
Fig. 5.
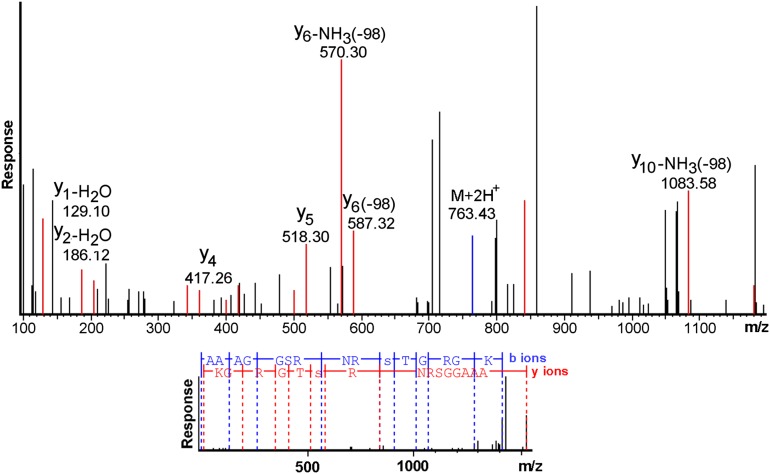
MS/MS sequencing of LysC peptide 630–644 from endogenous myosin. (Upper) The plot has been magnified to allow easier visualization of the peaks that established that Ser639 was phosphorylated. Fragmentation of the peptide generated ions whose masses allow sequencing in both the amino-to-carboxyl direction (b ions) and carboxyl-to-amino direction (y ions). The y ions are shown in red, the b ions in blue, and unassigned ions in black. For clarity, only y ions are labeled. The observed mass of the y5 ion establishes that Thr640 is not phosphorylated, whereas the masses of the y6 ions with neutral loss of phosphate establish that Ser639 is phosphorylated. (Lower) The peptide fragments that were assigned from the b and y ions. Phosphoserine is denoted by a lowercase letter “s.” The spectrum was sequenced de novo by the PEAKS program.
Table 4.
Phosphorylation level of LysC peptides of the heavy chain of endogenous AMII
| Number of phosphorylated residues | Fraction of total peptide |
|
| 630–644 | 1472–1509 | |
| 0 | 0.31 | 0.44 |
| 1 | 0.62 | 0.27 |
| 2 | 0.07 | 0.23 |
| 3 | 0 | 0.06 |
| 4 | 0 | 0 |
| Average P per peptide | 0.8 | 0.9 |
The distributions of phosphorylated peptides were calculated from the area of each peptide in the mass spectrum. The data are the average of results from two digests prepared and analyzed on different days.
Discussion
The results in this paper clearly establish that nonfilamentous forms of AMII, i.e., HMM and S1, have actin-activated ATPase activity with maximal activity similar to that of full-length myosin but with a significantly higher actin concentration required for half-maximal activity. The difference in actin dependence of the ATPase activities of filamentous and nonfilamentous myosins may result from cooperative binding of actin by the clustered heads of bipolar filaments. A possible explanation of the previous conclusion that chymotrypsin-cleaved AMII, which forms parallel dimers but not filaments (18), did not have actin-activated ATPase activity is that 12 µM F-actin was used in those experiments (18); at that concentration, the activities of HMM and S1 in the present experiments are only ∼20% of the activity of full-length recombinant myosin (Fig. 1 E and F).
Importantly, the data reported in this paper establish unequivocally that the actin-activated MgATPase activity of AMII is down-regulated by phosphorylation of Ser639 and not by phosphorylation of serine residues in the nonhelical tailpiece, as was thought previously. Phosphorylation of Ser639 may have been missed previously either because the small phosphorylated peptide pSTGR was not recovered in the 2D electrophoresis chromatography of the tryptic lysate (the more likely explanation) or because the peptide comigrated with one of the phosphorylated C-terminal tryptic peptides and was not detected by Edman sequencing.
Comparison of the sequence of AMII heavy chain with the heavy chain of other myosins (e.g., 24–26) shows that Ser639 lies within loop 2 of AMII, 613FDEDLKPSFKAAPAEEEKAAAGGSRNRSTGRGKGGAQFITVG653, the proteolysis-susceptible, unstructured surface loop that separates the central 50-kDa and the C-terminal 20-kDa domains of S1 (Fig. 6) (3, 28). Ser639 lies within a positively charged pocket near the C-terminal end that is present in loop 2 of all myosins (residues R636–K644 in AMII). The partial phosphorylation of Ser639 in endogenous AMII could explain the earlier finding (29) that the adjacent residue, Arg638, is less susceptible to cleavage by endoprotease Arg-C in purified endogenous AMII than in dephosphorylated AMII. Whether the same or different kinases phosphorylate Ser639 in the motor domain and the four serines in the nonhelical tailpiece is an open question that can be answered only by purification of the crude kinase. However, it is worth noting the sequence similarity of an arginine three residues before all five phosphorylated serine residues.
Fig. 6.
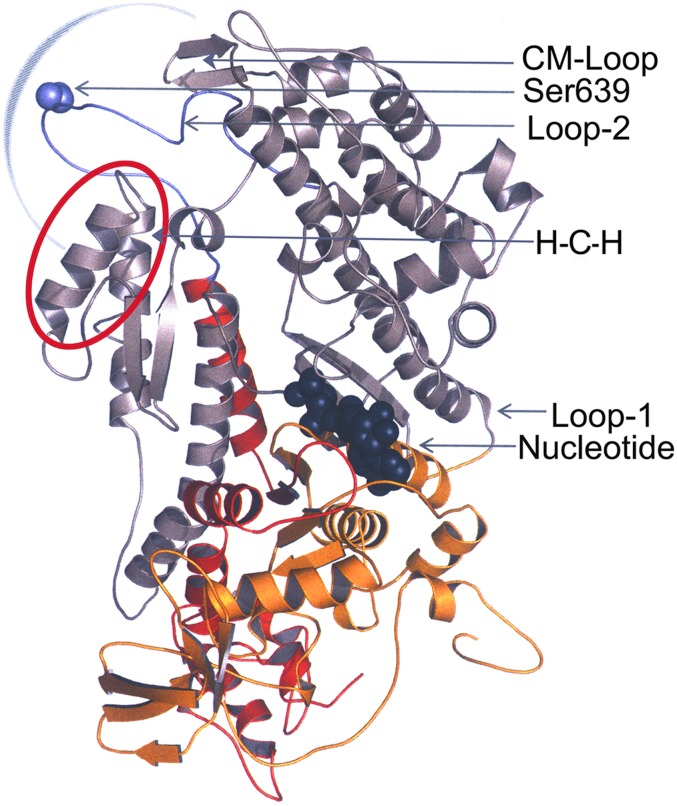
Homology model of the motor domain of Acanthamoeba myosin II. The motor domain shown in ribbon representation is based on the X-ray structure of Dictyostelium myosin II (Protein Data Bank ID, 2XEL) encompassing Met1 to Gln788.The N-terminal 25-kDa domain (red), central 50-kDa domain (gray), and C-terminal 20-kDa domain (yellow) are highlighted. Loop 1 connects the 25-kDa and 50-kDa domains, and loop 2 (purple) connects the 50-kDa and 20-kDa domains. The positions of Ser639 (purple) in loop 2 and the nucleotide-binding site, the helix–loop–helix motif (H-C-H, red circle), and the cleft in the 50-kDa domain that closes when myosin binds to F-actin (blue semicircle) are indicated. The model was generated using Modeller (27) and visualized using PyMol Version 1.4.1, Schrodinger, LLC).
There is considerable evidence that loop 2 interacts with actin (30, 31), probably through residues in its basic pocket interacting with negatively charged amino acids in actin (32, 33). Furthermore, there is considerable evidence that alterations of loop 2 in vitro affect the enzymatic activities of myosins, but there is no consensus regarding the step(s) in the actomyosin ATPase cycle that is (are) affected. The actin-activated ATPase activities of chimeras of Dictyostelium myosin II with its loop 2 replaced by loop 2 of vertebrate smooth and striated muscle (25, 34) and nonmuscle (35) myosin IIs correlated with the activity of the donor myosin, indicating that loop 2 has a role in the affinity of Dictyostelium myosin for actin and the conformational change following release of inorganic phosphate (Pi) from the actomyosin ADP·Pi complex (34, 35). In other experiments, mutation of the basic pocket of Dictyostelium myosin II from a net charge of +2 to +6 or higher increased its affinity for actin in the absence of ATP, the Kactin (actin concentration at half-maximal ATPase activity) for actin-activated ATPase activity, and coupling between the actin- and ATP-binding sites (36). In contrast to the results for Dictyostelium myosin II, changes in the net positive charge of the basic pocket of chicken myosin V variable region had no effect on the maximal velocity of its actin-activated ATPase, although it did have a similar effect on Kactin of myosin V (37). Mutations of chicken smooth muscle myosin HMM showed a role for loop 2 in the conformational change in the actomyosin ATPase cycle that is required for Pi release (38), myosin’s affinity for F-actin (38, 39), and the final step in the ATPase cycle, release of ADP (39).
Although one should not put too much emphasis on the location of an unstructured loop in homology models, it is interesting that loop 2 is near loop 4, the cardiomyopathy loop (CM loop), and the helix-coil-helix motif (Fig. 6), all of which are involved in the interaction of myosin with F-actin (26, 40). Behrmann et al. (41) have suggested that the helix-coil-helix motif may interact with and stabilize loop 2 and also pointed out that loop 2 is in the center of the actin–myosin interface and the cleft in the 50-kDa domain that closes when myosin binds to actin (Fig. 6, semicircle). The CM loop, so-named because mutations in this loop are associated with hypertrophic cardiomyopathy (42), contains the TEDS site (Glu404 in AMII), which has an essential role in actomyosin ATPase activity (43).
Interestingly, Ser846 in loop 2 of Limulus myosin III, which is homologous to Ser639 in AMII loop 2, is autophosphorylated in vitro by its N-terminal kinase domain and also by protein kinase A (44). However, Limulus myosin III does not have ATPase activity and the effect of phosphorylation on its very high actin affinity was not determined. Smooth (but not striated) muscle myosin II also has a serine, Ser647, at the position corresponding to AMII Ser639/Thr640 (45), but there is no evidence for its phosphorylation.
In summary, at this time, AMII is the only myosin known to be phosphorylated or to have any modification on loop 2 in vivo. Phosphorylation of Ser639 in loop 2, which decreases the net charge of the basic pocket from +4 to +2, substantially decreases the maximal velocity of actin-activated MgATPase activity and probably increases the actin concentration required to reach half-maximal activity, but we found no effect of phosphorylation of Ser639 on the affinity of AMII for F-actin in the presence or absence of ATP or on the CaATPase activity of AMII in the absence of F-actin. At this time, it seems most likely that phosphorylation of Ser639 of AMII interferes with communication between the actin-binding site and the catalytic site, inhibiting the conformational change required for the release of Pi from the actomyosin ADP·Pi complex; i.e., loop 2 regulates the rate-limiting step in the ATPase cycle. Because phosphorylation of Ser639 down-regulates single-headed, nonfilamentous S1, the regulatory mechanism does not involve either intramolecular (7) or intermolecular interaction of myosin heads.
Materials and Methods
Cloning of cDNAs.
AMII heavy-chain cDNA was the kind gift of T. D. Pollard (Department of Cell Biology, Yale University, New Haven, CT). LC1 cDNA was cloned based on its published amino acid sequence (2). The cloned cDNA (GenBank accession no. JX532101) has a deduced amino acid sequence of 154 amino acids (Fig. 7), identical to the sequence previously determined by protein sequencing (2). To clone LC2 cDNA, we separated LC2 by SDS/PAGE of AMII purified from Acanthamoeba, performed in-gel trypsin digestion, and designed degenerate primers based on peptide sequences determined by MS. Cloned LC2 cDNA (GenBank accession no. JX532102) has a deduced amino acid sequence of 145 residues (Fig. 7). AMII heavy-chain cDNA was mutated by standard molecular biological protocols. S1 was truncated at Leu900, HMM was truncated at Pro1244, and mutant ∆NHT was truncated at Pro1483 to delete the nonhelical tailpiece. For some experiments, Ser639 was replaced by alanine or aspartic acid, and serines 1489, 1494, 1499 and 1504 were replaced by alanine or glutamic acid. A FLAG peptide was added to the N terminus of each heavy chain to facilitate purification of the recombinant myosins.
Fig. 7.
Deduced amino acid sequences of Acanthamoeba myosin II light-chain cDNAs. The deduced sequence of light chain 1 (accession number JX532101) is identical to the protein sequence determined by Kobayashi et al. (2) (accession number 1810397A). The deduced sequence of light chain 2 (accession number JX532102) is consistent with the sequence of tryptic peptides from in-gel digests of the light chain.
Expression and Purification of Recombinant AMII and Mutant Myosins.
The two light-chain cDNAs and WT or mutant heavy-chain cDNAs were subcloned onto pFastBac4.5 vector (Invitrogen). Light-chain and heavy-chain viruses were coinfected into SF-9 cells grown in the presence of serum according to instructions provided by Invitrogen. Infected cells were harvested 72 h postinfection, and 1 g of cells was mixed with 10 mL of extraction buffer containing 200 mM NaCl, 10 mM Tris (pH 7.5), 1 mM PMSF, and proteinase inhibitor [one proteinase inhibitor tablet (Roche)/80 mL] and ruptured in a French press. The cell lysate was centrifuged in a Beckman centrifuge at 125,000 × g for 1 h, and the supernatant was mixed with pre-equilibrated FLAG-affinity resin (Sigma) and incubated for 1 h on a shaker in a cold room. The resin was packed in a small column and washed with extraction buffer. WT, ∆NHT, and the site-specific mutants were eluted with 0.1 mg/mL FLAG peptide in extraction buffer, dialyzed (in tubing with nominal cutoff of 75,000 Mr) overnight against buffer containing 10 mM imidazole (pH 7.0) and 12 mM MgCl2, and centrifuged at 15,000 × g for 5 min; then the pellets were dissolved in 300 mM NaCl, 10 mM imidazole (pH 7.0), 50% (wt/wt) glycerol, and 1 mM DTT. After elution from FLAG resin, S1, HMM, and the site-specific mutants of S1 were dialyzed against 10 mM imidazole (pH 7.0), 50% (wt/wt) glycerol, and 25 mM KCl. After the final dialysis, aliquots of 60 µL or less of all myosins were transferred to Eppendorf tubes, rapidly frozen in liquid N2, and kept in liquid N2 until use.
Preparation of AMII Heavy-Chain Kinase and Endogenous AMII.
The kinase was partially purified according to the previously published method (11) with several modifications. Acanthamoeba (200 g) was homogenized in 400 mL extraction buffer containing 10% (wt/wt) sucrose, 20 mM imidazole (pH 7.0), 2 mM EGTA, 1 mM ATP, 100 mM KCl, 1 mM sodium pyrophosphate, 1 mM PMSF, and one proteinase inhibitor tablet (Roche)/50 mL. The lysate was centrifuged in a Beckman centrifuge at 125,000 × g at 4 °C for 60 min. The supernatant was mixed with 45 g of pre-equilibrated P11 (Whatman) on a shaker in a cold room for 60 min and then centrifuged at 100,000 × g for 5 min. The supernatant was decanted, and a small amount of extraction buffer was added to resuspend and transfer the resin to a 25 × 40 cm column. The column was washed with 200 mL of extraction buffer but without proteinase inhibitors. The kinase was eluted with a linear gradient of KCl formed by 60 mL of 10% (wt/wt) sucrose, 20 mM imidazole (pH 7.5), 2 mM EGTA, 100 mM KCl, and 60 mL of the same buffer with a KCl concentration of 400 mM. Kinase activity was detected by autoradiography using 10 µL of each fraction and 7 µg of purified recombinant AMII as substrate. The fractions that contained kinase activity were pooled and stored in liquid N2. Pooled kinase (6 mL) was purified further by chromatography on a 25 × 60 cm Sephacryl S-200 column (GE Healthcare) with buffer containing 10 mM imidazole (pH 7.0), 2 mM EGTA, 0.1 M KCl, 10% (wt/wt) sucrose, and 1 mM sodium pyrophosphate. Pooled kinase fractions were concentrated, and aliquots were stored in liquid N2 until use. Endogenous AMII was purified as described previously (46) with the addition of a final step of chromatography on a 26 × 60 cm column of Sephacryl S-200 (GE Healthcare).
Phosphorylation and Dephosphorylation of Myosins.
About 1 mg of recombinant myosin in 200 mM KCl was bound to 1.5 mL of FLAG-resin (bed volume) and washed with 10 mL of 10 mM imidazole (pH 7.0), containing 12 mM MgCl2, 1 mM sodium pyrophosphate, 1 mM EGTA, 1 mM DTT, and 2.5 mM ATP. The resin was transferred to a 15-mL test tube with 1 mL of wash buffer, and 0.25 mL of kinase (4.5 mg/mL) was added. Because KCl inhibits AMII heavy-chain kinase activity, phosphorylation was performed in the absence of KCl. The mixture was incubated at 30 °C for 40 min with occasional stirring and was kept on ice overnight. The resin was washed in a column with 50 mL of 200 mM NaCl in 10 mM Tris (pH 7.5). The phosphorylated myosin was eluted with 0.1 mg/mL of FLAG-peptide in the wash buffer and then was concentrated by Amicon Ultracentrifugal filtration (Ultracell-100K), dialyzed overnight against 10 mM imidazole (pH 7.0) and 25 mM KCl, and kept on ice for use within 3 d. For dephosphorylation, a 40-µL aliquot of AMII (0.5 mg/mL) in 10 mM imidazole (pH 7.0) and 25 mM KCl was mixed with 5 µL of 10× concentrated phosphatase buffer [20 mM DTT, 01.% Brij 35, 500 mM Tris-HCl (pH 7.0), 1.0 mM EGTA, 1 M NaCl; New England BioLabs] and 5 µL of 10× concentrated MnCl2 (10 mM MnCl2, New England BioLabs) and was incubated with 1 µL of lambda protein phosphatase (New England BioLabs) at room temperature for 10 min.
The synthetic peptide, KAAAGGSRNRSTGRGKGGA (American Peptide Co.) was dissolved at 1 mg/mL in 15 mM imidazole (pH 7.0) containing 15 mM MgCl2, 1.5 mM sodium pyrophosphate, 1.5 mM EGTA, and 2 mM DTT. A 20-µL aliquot was mixed with 5 µL of kinase and 5 µL of 12 mM ATP and incubated for 1 h at 30 °C.
Actin-Activated Mg ATPase and CaATPase Activities.
Myosins were dialyzed against 10 mM imidazole (pH 7.0) and 25 mM KCl overnight. Steady-state actin-activated MgATPase activity was assayed in 10 mM imidazole (pH 7.0), 2.5 mM KCl, 1 mM [γ-32P]ATP, 120 nM myosin, 3 mM MgCl2, and F-actin at the indicated concentrations. The production of 32Pi was determined as described (46) after incubation at 30 °C for 12 min, during which period the reaction rates were constant. CaATPase activity was determined in a buffer of 10 mM imidazole (pH 7.0), 10 mM CaCl2, 50 mM KCl, and 1 mM [γ-32P]ATP at 30 °C for 8 min with 32Pi determined as described (12).
Binding Assay.
S1 and pS1were dialyzed against 10 mM imidazole (pH 7.0) and 25 mM KCl and then were mixed with F-actin and diluted in the same buffer to a final concentration of 120 nM S1 and pS1 and the concentrations of F-actin with either no ATP or 1 mM ATP, as indicated in Fig. 1 G and H. The mixtures were centrifuged for 40 min at 100,000 rpm in a Beckman TL-100 centrifuge (rotor TLA 100.3), and the unbound S1 and pS1 in the supernatant were determined by CaATPase activity.
Other Biochemical Assays.
SDS/PAGE was carried out according to Laemmli (48). Protein concentrations were determined by the Bradford method (49) with BSA as a standard.
HPLC, MS, and Sequence Analysis.
The peptides in tryptic (Promega) and LysC (Wako) digests were separated and analyzed by reverse-phase chromatography/MS as described (50, 51), except that the reverse-phase column was a Zorbax 300SB-C18, 1.0 × 50 mm, 3.5 µm (particle diameter), and the mass spectrometer was an Agilent model 6520 QTOF capable of MS/MS sequencing of peptides. In some experiments, the fragmentor voltage was increased from 235 V to 275 V to increase neutral loss of phosphate from phosphopeptides. Peptide masses were extracted from the chromatograms with the Find by Molecular Feature function of Agilent Masshunter version 4. MS/MS spectra were sequenced de novo by PEAKS version 6 (Bioinformatics Solutions) and by matching to those calculated by GPMAW version 9 (Lighthouse Data). The measured masses have an error ≤5 ppm.
When peptides were labeled with 32P, they were separated with the same gradient but on a larger-diameter column, Zorbax 300SB-C18, 2.1 × 50 mm, and with a solvent flow rate of 200 µL/min. One-minute fractions were collected, and radioactivity was determined by liquid scintillation counting (TriCarb 2900TR; PerkinElmer).
Acknowledgments
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH). The homology model of the motor domain of Acanthamoeba myosin II (Fig. 6) was prepared by Dr. Sarah Heissler, Laboratory of Molecular Physiology, NHLBI, NIH.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hammer JA, 3rd, Bowers B, Paterson BM, Korn ED. Complete nucleotide sequence and deduced polypeptide sequence of a nonmuscle myosin heavy chain gene from Acanthamoeba: Evidence of a hinge in the rodlike tail. J Cell Biol. 1987;105(2):913–925. doi: 10.1083/jcb.105.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi T, Zot HG, Pollard TD, Collins JH. Functional implications of the unusual amino acid sequence of the regulatory light chain of Acanthamoeba castellanii myosin-II. J Muscle Res Cell Motil. 1991;12(6):553–559. doi: 10.1007/BF01738443. [DOI] [PubMed] [Google Scholar]
- 3.Sellers JR. 1999. Myosins. Protein Profile 2nd Ed, (Oxford Univ Press, Oxford, UK)
- 4.Rimm DL, Sinard JH, Pollard TD. Location of the head-tail junction of myosin. J Cell Biol. 1989;108(5):1783–1789. doi: 10.1083/jcb.108.5.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollard TD. Structure and polymerization of Acanthamoeba myosin-II filaments. J Cell Biol. 1982;95(3):816–825. doi: 10.1083/jcb.95.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinard JH, Stafford WF, Pollard TD. The mechanism of assembly of Acanthamoeba myosin-II minifilaments: Minifilaments assemble by three successive dimerization steps. J Cell Biol. 1989;109(4 Pt 1):1537–1547. doi: 10.1083/jcb.109.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowey S, Trybus KM. Common structural motifs for the regulation of divergent class II myosins. J Biol Chem. 2010;285(22):16403–16407. doi: 10.1074/jbc.R109.025551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maruta H, Korn ED. Acanthamoeba myosin II. J Biol Chem. 1977;252(18):6501–6509. [PubMed] [Google Scholar]
- 9.Pollard TD, Stafford WF, Porter ME. Characterization of a second myosin from Acanthamoeba castellanii. J Biol Chem. 1978;253(13):4798–4808. [PubMed] [Google Scholar]
- 10.Collins JH, Korn ED. Actin activation of Ca2+-sensitive Mg2+-ATPase activity of Acanthamoeba myosin II is enhanced by dephosphorylation of its heavy chains. J Biol Chem. 1980;255(17):8011–8014. [PubMed] [Google Scholar]
- 11.Côté GP, Collins JH, Korn ED. Identification of three phosphorylation sites on each heavy chain of Acanthamoeba myosin II. J Biol Chem. 1981;256(24):12811–12816. [PubMed] [Google Scholar]
- 12.Collins JH, Côté GP, Korn ED. Localization of the three phosphorylation sites on each heavy chain of Acanthamoeba myosin II to a segment at the end of the tail. J Biol Chem. 1982;257(8):4529–4534. [PubMed] [Google Scholar]
- 13.Côté GP, Robinson EA, Appella E, Korn ED. Amino acid sequence of a segment of the Acanthamoeba myosin II heavy chain containing all three regulatory phosphorylation sites. J Biol Chem. 1984;259(20):12781–12787. [PubMed] [Google Scholar]
- 14.McClure JA, Korn ED. Purification of a protein phosphatase from Acanthamoeba that dephosphorylates and activates myosin II. J Biol Chem. 1983;258(23):14570–14575. [PubMed] [Google Scholar]
- 15.Sathyamoorthy V, Atkinson MAL, Bowers B, Korn ED. Functional consequences of the proteolytic removal of regulatory serines from the nonhelical tailpiece of Acanthamoeba myosin II. Biochemistry. 1990;29(15):3793–3797. doi: 10.1021/bi00467a028. [DOI] [PubMed] [Google Scholar]
- 16.Kuznicki J, Albanesi JP, Côté GP, Korn ED. Supramolecular regulation of the actin-activated ATPase activity of filaments of Acanthamoeba Myosin II. J Biol Chem. 1983;258(10):6011–6014. [PubMed] [Google Scholar]
- 17.Ganguly C, et al. Regulation of the actin-activated ATPase activity of Acanthamoeba myosin II by copolymerization with phosphorylated and dephosphorylated peptides derived from the carboxyl-terminal end of the heavy chain. J Biol Chem. 1990;265(17):9993–9998. [PubMed] [Google Scholar]
- 18.Kuznicki J, Côté GP, Bowers B, Korn ED. Filament formation and actin-activated ATPase activity are abolished by proteolytic removal of a small peptide from the tip of the tail of the heavy chain of Acanthamoeba myosin II. J Biol Chem. 1985;260(3):1967–1972. [PubMed] [Google Scholar]
- 19.Atkinson MAL, Appella E, Corigliano-Murphy MA, Korn ED. Enzymatic activity and filament assembly of Acanthamoeba myosin II are regulated by adjacent domains at the end of the tail. FEBS Lett. 1988;234(2):435–438. doi: 10.1016/0014-5793(88)80132-7. [DOI] [PubMed] [Google Scholar]
- 20.Atkinson MAL, Lambooy PK, Korn ED. Cooperative dependence of the actin-activated Mg2+-ATPase activity of Acanthamoeba myosin II on the extent of filament phosphorylation. J Biol Chem. 1989;264(7):4127–4132. [PubMed] [Google Scholar]
- 21.Sinard JH, Pollard TD. The effect of heavy chain phosphorylation and solution conditions on the assembly of Acanthamoeba myosin-II. J Cell Biol. 1989;109(4 Pt 1):1529–1535. doi: 10.1083/jcb.109.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, et al. Phosphorylation of serines in the non-helical tailpiece of Acanthamoeba myosin II alters filament structure. Proc Natl Acad Sci USA. 110:E33–E40. doi: 10.1073/pnas.1219727110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brzeska H, Korn ED. Regulation of class I and class II myosins by heavy chain phosphorylation. J Biol Chem. 1996;271(29):16983–16986. doi: 10.1074/jbc.271.29.16983. [DOI] [PubMed] [Google Scholar]
- 24.Warrick HM, Spudich JA. Myosin structure and function in cell motility. Annu Rev Cell Biol. 1987;3:379–421. doi: 10.1146/annurev.cb.03.110187.002115. [DOI] [PubMed] [Google Scholar]
- 25.Uyeda TQP, Ruppel KM, Spudich JA. Enzymatic activities correlate with chimaeric substitutions at the actin-binding face of myosin. Nature. 1994;368(6471):567–569. doi: 10.1038/368567a0. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz M, Holmes KC. The actin-myosin interface. Proc Natl Acad Sci USA. 2010;107(28):12529–12534. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with Modeller. Meth Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 28.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 29.Ganguly C, Martin B, Bubb M, Korn ED. Limited proteolysis reveals a structural difference in the globular head domains of dephosphorylated and phosphorylated Acanthamoeba myosin II. J Biol Chem. 1992;267(29):20905–20908. [PubMed] [Google Scholar]
- 30.Rayment I, et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261(5117):58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 31.Mornet D, Pantel P, Audemard E, Kassab R. The limited tryptic cleavage of chymotryptic S-1: An approach to the characterization of the actin site in myosin heads. Biochem Biophys Res Commun. 1979;89(3):925–932. doi: 10.1016/0006-291x(79)91867-9. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto K. Binding manner of actin to the lysine-rich sequence of myosin subfragment 1 in the presence and absence of ATP. Biochemistry. 1989;28(13):5573–5577. doi: 10.1021/bi00439a035. [DOI] [PubMed] [Google Scholar]
- 33.Pliszka B, Martin BM, Karczewska E. Ionic interaction of myosin loop 2 with residues located beyond the N-terminal part of actin probed by chemical cross-linking. Biochim Biophys Acta. 2008;1784(2):285–291. doi: 10.1016/j.bbapap.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Murphy CT, Spudich JA. The sequence of the myosin 50-20K loop affects Myosin’s affinity for actin throughout the actin-myosin ATPase cycle and its maximum ATPase activity. Biochemistry. 1999;38(12):3785–3792. doi: 10.1021/bi9826815. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi M, et al. Functional characterization of vertebrate nonmuscle myosin IIB isoforms using Dictyostelium chimeric myosin II. J Biol Chem. 2001;276(2):1034–1040. doi: 10.1074/jbc.M005370200. [DOI] [PubMed] [Google Scholar]
- 36.Furch M, Geeves MA, Manstein DJ. Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry. 1998;37(18):6317–6326. doi: 10.1021/bi972851y. [DOI] [PubMed] [Google Scholar]
- 37.Yengo CM, Sweeney HL. Functional role of loop 2 in myosin V. Biochemistry. 2004;43(9):2605–2612. doi: 10.1021/bi035510v. [DOI] [PubMed] [Google Scholar]
- 38.Joel PB, Trybus KM, Sweeney HL. Two conserved lysines at the 50/20-kDa junction of myosin are necessary for triggering actin activation. J Biol Chem. 2001;276(5):2998–3003. doi: 10.1074/jbc.M006930200. [DOI] [PubMed] [Google Scholar]
- 39.Joel PB, Sweeney HL, Trybus KM. Addition of lysines to the 50/20 kDa junction of myosin strengthens weak binding to actin without affecting the maximum ATPase activity. Biochemistry. 2003;42(30):9160–9166. doi: 10.1021/bi034415j. [DOI] [PubMed] [Google Scholar]
- 40.Milligan RA. Protein-protein interactions in the rigor actomyosin complex. Proc Natl Acad Sci USA. 1996;93(1):21–26. doi: 10.1073/pnas.93.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behrmann E, et al. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150(2):327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geisterfer-Lowrance AA, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62(5):999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 43.Bement WM, Mooseker MS. TEDS rule: A molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil Cytoskeleton. 1995;31(2):87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- 44.Kempler K, et al. Loop 2 of limulus myosin III is phosphorylated by protein kinase A and autophosphorylation. Biochemistry. 2007;46(14):4280–4293. doi: 10.1021/bi062112u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodson HV, Warrick HM, Spudich JA. Specialized conservation of surface loops of myosin: Evidence that loops are involved in determining functional characteristics. J Mol Biol. 1999;287(1):173–185. doi: 10.1006/jmbi.1999.2565. [DOI] [PubMed] [Google Scholar]
- 46.Collins JH, Korn ED. Purification and characterization of actin-activatable, Ca2+-sensitive myosin II from Acanthamoeba. J Biol Chem. 1981;256(5):2586–2595. [PubMed] [Google Scholar]
- 47.Pollard TD, Korn ED. Acanthamoeba myosin. I. Isolation from Acanthamoeba castellanii of an enzyme similar to muscle myosin. J Biol Chem. 1973;248(13):4682–4690. [PubMed] [Google Scholar]
- 48.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 49.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 50.Paone G, et al. ADP-ribosyltransferase-specific modification of human neutrophil peptide-1. J Biol Chem. 2006;281(25):17054–17060. doi: 10.1074/jbc.M603042200. [DOI] [PubMed] [Google Scholar]
- 51.Paone G, et al. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci USA. 2002;99(12):8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]