Abstract
The idea of targeted therapy, whereby drug or protein molecules are delivered to specific cells, is a compelling approach to treating disease. Immunotoxins are one such targeted therapeutic, consisting of an antibody domain for binding target cells and molecules of a toxin that inhibits the proliferation of the targeted cell. One major hurdle preventing these therapies from reaching the market has been the lack of a suitable production platform that allows the cost-effective production of these highly complex molecules. The chloroplast of the green alga Chlamydomonas reinhardtii has been shown to contain the machinery necessary to fold and assemble complex eukaryotic proteins. However, the translational apparatus of chloroplasts resembles that of a prokaryote, allowing them to accumulate eukaryotic toxins that otherwise would kill a eukaryotic host. Here we show expression and accumulation of monomeric and dimeric immunotoxin proteins in algal chloroplasts. These fusion proteins contain an antibody domain targeting CD22, a B-cell surface epitope, and the enzymatic domain of exotoxin A from Pseudomonas aeruginosa. We demonstrated that algal-produced immunotoxins accumulate as soluble and enzymatically active proteins that bind target B cells and efficiently kill them in vitro. We also show that treatment with either the mono- or dimeric immunotoxins significantly prolongs the survival of mice with implanted human B-cell tumors.
Microscopic eukaryotic green algae play an essential role in converting solar energy into chemical energy through the process of photosynthesis (1), and recently these microorganisms have gained attention as a potential source of renewable fuel (2). In addition to energy production, microalgae can produce a variety of other bioproducts including nutraceuticals and recombinant proteins such as industrial enzymes or therapeutics (3, 4). Using the green algae Chlamydomonas reinhardtii, we have demonstrated that algae are capable of expressing, folding, and accumulating a range of human therapeutic proteins in the chloroplast (5–7). More recently we have shown that recombinant proteins also can be secreted from algae (8). Algae require only trace minerals, fertilizer, and sunlight to be grown at scale, giving them the potential to produce recombinant proteins, including therapeutics, very inexpensively (9). Although cost can be a significant factor in the production of protein-based therapies, producing unique classes of therapeutically relevant proteins is desirable, and algae offer the potential to produce a number of novel proteins because of the unique biochemical environment of the chloroplast (5).
Immunotoxins are antibodies that are either chemically (10) or genetically (11) coupled to eukaryotic toxins. These chimeric proteins are used to deliver the toxin to a specific cancer cell to initiate apoptosis (12). These molecules currently are produced by expressing an antibody in CHO cells and then chemically coupling the purified protein to a toxin (10) or by expressing a chimeric antibody-toxin protein as an insoluble aggregate in bacteria and then denaturing and refolding the protein to produce the functional immunotoxin (11). Eukaryotic expression platforms such as yeast, CHO cells, and insect cells are incapable of producing and accumulating genetically coupled immunotoxins because of the inhibition of host-cell proliferation by the toxin. Therefore the production of genetically coupled immunotoxins is limited to bacterial expression platforms, where the eukaryotic toxin does not inhibit the host’s ability to grow. This restriction limits the complexity of immunotoxins that currently are produced, because bacterial expression platforms lack the ability to fold proteins with multiple domains efficiently or to form disulfide bonds within proteins (13). These limitations reduce immunotoxin production in Escherichia coli to single-chain antibody fragments (scFv) and eukaryotic toxins (14) or disulfide-stabilized variable domains formed ex vivo that are genetically linked to a eukaryotic toxin (15). For chemically linked immunotoxins, antibodies can be expressed in CHO cells and the toxin coupled in vitro, leading to functional complex proteins, but this process results in additional chemical processing steps that lead to more expensive drug conjugates (16). Each of these immunotoxin types has been demonstrated to be a potent and potentially useful tool for the treatment of solid tumor (17).
C. reinhardtii is a eukaryotic alga that contains a single chloroplast that constitutes up to 70% of the cell (18). Chloroplasts contain ribosomes and translation factors that resemble those of photosynthetic prokaryotes (19, 20). However, unlike bacteria, chloroplasts contain a wide range of chaperones (21), protein disulfide isomerases (22), and peptidylprolyl isomerases (PPIases) (23) that allow them to fold the complex proteins of the photosynthetic apparatus. This machinery also allows them to fold complex recombinant proteins, such as full-length human antibodies, which accumulate as soluble and functional molecules within the chloroplast (5).
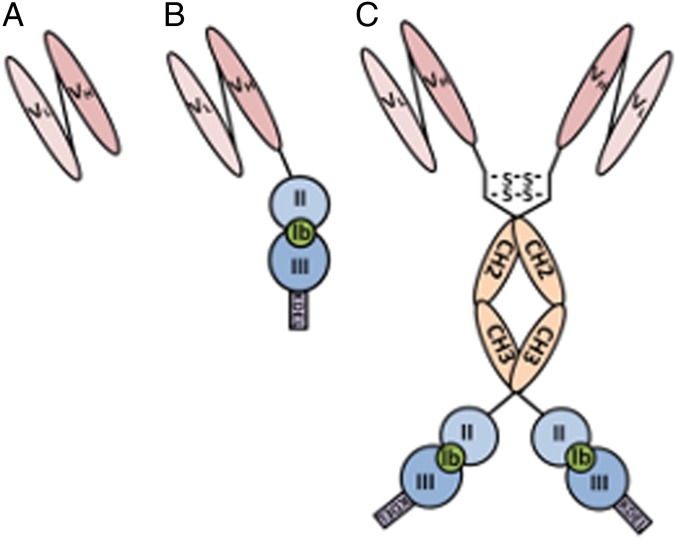
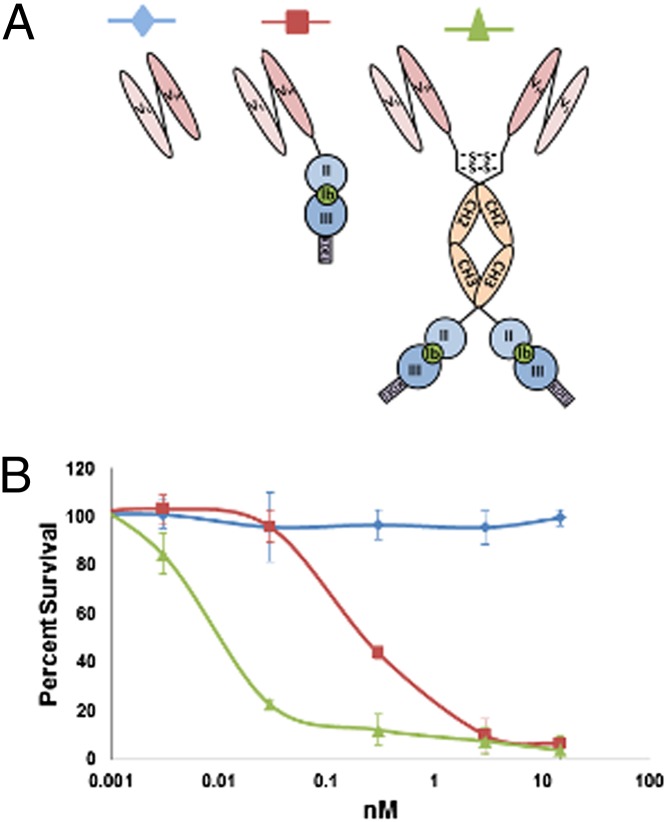
To examine if algae are capable of producing fully functional immunotoxins, we created a recombinant gene encoding a single-chain antibody (scFv) that recognizes CD22, a B-cell surface molecule (Fig. 1A) (24), genetically fused to domains II and III of Exotoxin A (PE40) from Pseudomonas aeruginosa (Fig. 1B). The chimeric gene produced is very similar to one expressed in E. coli called “αCD22PE40” (25). PE40 inhibits the translation of eukaryotic cells by ribosylating eukaryotic elongation factor 2 (eEF2), preventing the elongation of polypeptide chains leading to apoptosis of the targeted cell (26). A significant problem with immunotoxins similar to αCD22PE40 is their short serum half-life resulting from their small size (27). To overcome this potential problem, we also engineered a more complex chimeric immunotoxin gene that contained the hinge and CH2 and CH3 domains of a human IgG1 placed between the αCD22 scFv antibody and PE40, encoding a protein that we have termed “αCD22CH23PE40” (Fig. 1C). This molecule should be capable of forming a dimer through two disulfide bonds in the hinge region, making it significantly larger than αCD22PE40 and doubling the number of CD22-binding domains and PE40 molecules to two.
Fig. 1.
Depiction of algal-expressed immunotoxin proteins. (A) Single-chain antibody (scFv) directed against the CD22 cell-surface antigen made by linking the variable domains of the heavy- and light-chain antibodies with a glycine-serine linker. (B) The CD22-scFv is genetically linked to P. aeruginosa exotoxin A domains 2 and 3. Removal and replacement of domain Ia from exotoxin A with an antibody allows cancer cells to be targeted specifically. (C) The CD22-scFv genetically fused to the hinge and constant domains of an IgG1 and to exotoxin A domains 2 and 3 to create a construct that forms a homodimer through disulfide bonds formed between hinge regions. This fusion allows the molecule to have two binding domains as well as two toxin molecules.
Recently, antibody drug conjugates (ADCs) have garnered a significant amount of attention for their ability to inhibit cancer-cell proliferation (28). ADCs are produced by first purifying an antibody made in a mammalian expression platform, followed by chemical coupling of a toxin molecule to the antibody. This complex production system makes ADCs expensive therapies costing upwards of $100,000 per course of treatment (16). Most ADCs are engineered with special linkers that are cleaved upon acidification in the endosome following antibody binding and endocytosis (29). PE40 provides a biological way to produce the toxin, eliminating the requirement of chemical coupling. PE40 is generated by first removing domain Ia from the full-length exotoxin A, thus making PE40 more than 100-fold less toxic than the native exotoxin A because of its inability to bind to cells (30). Domain II of this protein is cleaved in acidified endosomes by a furin protease, liberating the cytotoxic domain III from the antibody, which is targeted for degradation (31). Additionally, the ability to produce genetically coupled immunotoxins eliminates the possibility of unstable linkers (32) and reduces off-target toxicity. Thus, immunotoxins provide a protein alternative to chemically synthesized drug molecules that currently are being linked to cancer-targeting antibodies.
Here we demonstrate that a eukaryotic cell is capable of accumulating both small monomeric and larger dimeric immunotoxins. These immunotoxins are enzymatically active, bind specifically to cells displaying the CD22 molecule, and are capable of causing those cells to undergo apoptosis. Furthermore, in s.c. tumor cell xenograft mice models, both algal-produced immunotoxins are capable of significantly inhibiting tumor growth.
Results
Protein Engineering and Expression Vector Construction.
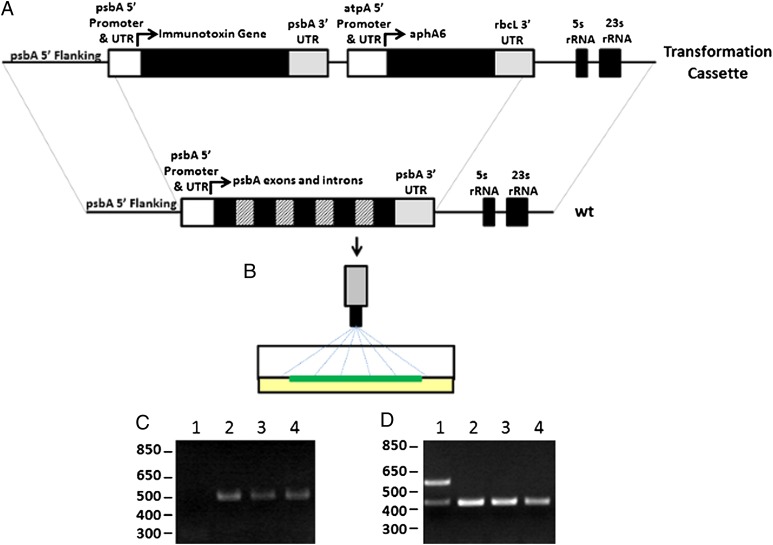
All DNA manipulations were performed by standard methods as previously described (33). The immunotoxin genes and subfragments were synthesized de novo using C. reinhardtii chloroplasts codon bias from www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=3055.chloroplast. The variable domains of a human antibody against the B-cell surface antigen CD22 were separated by a linker consisting of four glycines and a serine repeated four times (4×G4S) to create an scFv that was ligated downstream of a sequence coding for a 1× Flag peptide (DYKDDDDKS) and separated by a sequence that encodes a Tobacco etch virus (TEV) protease cleavage site (ENLYFQG). This gene was termed “αCD22” (Fig. 1A) (Codon adaptation index = 0.722, Nc = 30.5) (34, 35). This scFv was ligated upstream of a 2× G4S linker, the coding sequence for domains II and III of Pseudomonas exotoxin A (PE40), and the sequence coding for a KDEL endoplasmic reticulum localization peptide, which has been shown to increase the activity of exotoxin A-based immunotoxins (36). This molecule was termed “αCD22PE40” (Fig. 1B) (Codon adaptation index = 0.750, Nc = 34.7.) To create the larger dimeric immunotoxin, a sequence encoding the hinge and constant domains 2 and 3 (CH23) of a human IgG1 was ligated between the scFv and PE40, separated on both its amine and carboxyl end by a 2× G4S linker (αCD22HCH23PE40) (Fig. 1C) (Codon adaptation index = 0.759, Nc =35.0). These constructs were placed in a C. reinhardtii chloroplast transformation cassette that contains the psbA promoter and 5′ UTR upstream and the psbA 3′ UTR downstream of the recombinant immunotoxin genes (Fig. 2A). A kanamycin resistance gene, aphA6, was used to select algae that were transformed with the recombinant immunotoxin genes. The aphA6 gene was placed downstream of the atpA promoter and 5′ UTR, which are used to drive its expression, and upstream of the rbcL 3′ UTR (Fig. 2A). Additional chloroplast genome homology is present downstream of the kanamycin gene to facilitate the integration of the immunotoxin gene into the chloroplast genome (6).
Fig. 2.
Integration of genes into the chloroplast genome by homologous recombination. (A) Immunotoxin genes are ligated downstream of the psbA promoter and 5′ UTR and upstream of the psbA 3′ UTR. This construct is placed upstream of an aphA6 gene that confers kanamycin resistance to transformed cells of algae. Regions of chloroplast genome are placed at either end of the transformation vector to allow homologous integration of the entire transformation cassette into the chloroplast genome. (B) Transformation plasmids are precipitated onto gold particles and delivered by particle bombardment into algal chloroplasts, where they recombine into the plastid genome. (C) PCR analysis using primers specific to the αCD22 scFv gene and the psbA 5′ UTR demonstrate that coding sequences for immunotoxins have been integrated into the psbA locus. Lane 1 contains PCR from WT algal cells. Lane 2 contains strains transformed with αCD22. Lane 3 contains strains transformed with αCD22-PE40. Lane 4 contains strains transformed with αCD22-CH23-PE40. (D) PCR analysis is used to confirm homoplasmicity of transformed strains of algae. Primers are used to amplify a control region of the algal chloroplast genome as well the endogenous psbA gene. Loss of the psbA gene (upper band in lane 1) demonstrates homoplasmicity of the transgenic lines.
Analysis of Gene Integration into the Chloroplast Genome.
Transformation vectors were precipitated onto gold particles, transformed into WT C. reinhardtii cells by particle bombardment, and selected on Tris-acetate-phosphate (TAP) plates containing 100 μg/mL of kanamycin (Fig. 2B). Colonies that grew were screened for the presence of heterologous genes. Chloroplasts contain up to 80 copies of their plastid genome, and finding strains in which all 80 copies contain the heterologous gene is essential for identifying strains that stably express the desired gene (37). PCR analysis was used to identify several transgenic lines for each construct in which the αCD22, αCD22PE40, or αCD22HCH23PE40 gene was integrated correctly into the chloroplast genome at the psbA locus (Fig. 2C) (5).
To identify strains homoplasmic for each of these recombinant genes, additional rounds of algae cell cloning and PCR analysis were performed. Primers corresponding to the psbA 5′ UTR and the coding region of the recombinant genes or the native psbA gene were used to amplify DNA from strains homoplasmic for recombinant gene integration, as previously described (5). Control primers for the 16S rRNA region of the chloroplast genome were used for validation that the PCR was successful (5). As shown in Fig. 2D, homoplasmic strains were identified for all three gene constructs.
Analysis of Recombinant Protein Accumulation in Transgenic Algal Strains.
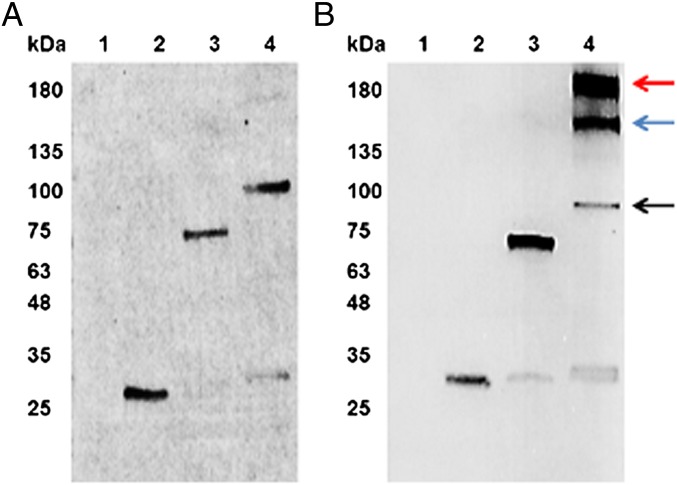
To determine if algal chloroplasts accumulate immunotoxin proteins, Western blot analysis with an anti-Flag antibody conjugated with alkaline phosphatase was used to assay for the presence of αCD22, αCD22PE40, and αCD22HCH23PE40 in each of the respective transgenic strains (Fig. 3A). αCD22, αCD22PE40, and αCD22HCH23PE40 each accumulate as soluble proteins within the respective transgenic alga, and all migrate at the expected size when separated by reducing PAGE. When proteins were separated on a nonreducing PAGE and assayed by Western blot analysis, αCD22HCH23PE40 accumulated as a 190-kDa species, suggesting that chloroplasts assemble this protein into a dimer (Fig. 3B). αCD22 and αCD22PE40 both migrated at the expected mass of the monomer on both the reducing and nonreducing gels. These data suggest that disulfide bonds are formed between the cysteine residues found in the hinge region of a human IgG1, resulting in dimerization of αCD22HCH23PE40 within algal chloroplasts. ELISAs demonstrate that algal chloroplasts express αCD22PE40 at ∼0.3–0.4% and αCD22HCH23PE40 at ∼0.2–0.3%.
Fig. 3.
Western blots demonstrating the accumulation of immunotoxin proteins. (A) Samples (each 20 µg o.d) were separated on a SDS/PAGE gel under reducing conditions, transferred to a nitrocellulose membrane, and probed with an anti-Flag antibody that was conjugated with alkaline phosphatase. The lanes contain the following samples: lane 1, WT total protein; lane 2, αCD22; lane 3, αCD22PE40; lane 4, αCD22CH23PE40. (B) The identical samples were separated on a SDS/PAGE gel under nonreducing conditions to keep disulfide bonds intact. Once separated and transferred to a nitrocellulose membrane, samples were probed with an anti-Flag antibody conjugated with alkaline phosphatase and visualized on the nitrocellulose membrane. The black arrow indicates monomeric αCD22CH23PE40; the red arrow indicates αCD22CH23PE40 that has formed a homodimer which is indicative of an assembled antibody; the blue arrow indicates the formation of an assembled product between αCD22CH23PE40 and a degradation product lacking an scFv binding domain. This result demonstrates that algae produce αCD22CH23PE40 as a dimer, making it a divalent protein containing two exotoxin A molecules.
ADP Ribosyltransferase Assays of Algal-Expressed Immunotoxins.
The enzymatic activity of the exotoxin A protein associated with the immunotoxin is ADP ribosylation of translation elongation factor EF2. An ADP ribosylation assay was used to determine whether algal-expressed αCD22PE40 and αCD22HCH23PE40 molecules were enzymatically active. Biotinylated nicotinamide-adenine dinucleotide (NAD+) (Sigma) the substrate for ribosyltransferases, was incubated with eEF2 and purified, algal-produced immunotoxins. Active PE40 molecules are capable of transferring biotinylated-ADP molecules from biotinylated-NAD+ onto EF2. Successful transfer can be visualized by separating the EF2 protein on a polyacrylamide gel and detecting the biotin on EF2 by Western blot analysis with an anti-biotin alkaline phosphatase-conjugated antibody (Rockland). The detection of a 92-kDa ribosylated eEF2 demonstrates the presence of an enzymatically active PE40 molecule. As shown in Fig. 4, both αCD22PE40 (lane 2) and αCD22HCH23PE40 (lane 3) are capable of ADP ribosylating eEF2, but the control protein αCD22 does not ADP ribosylate eEF2 (lane 1). These data show that algal-expressed immunotoxins containing PE40 are enzymatically active, demonstrating that eukaryotic algal chloroplasts are capable of expressing and accumulating active eukaryotic toxins. This result is somewhat surprising, because even a single molecule of PE40 leaking from the chloroplast into the cytosol would be sufficient to kill the eukaryotic expression host. These data demonstrate that protein translocation in chloroplast is strictly unidirectional and that chloroplast-produced proteins do not appear to transit into the cytoplasm.
Fig. 4.
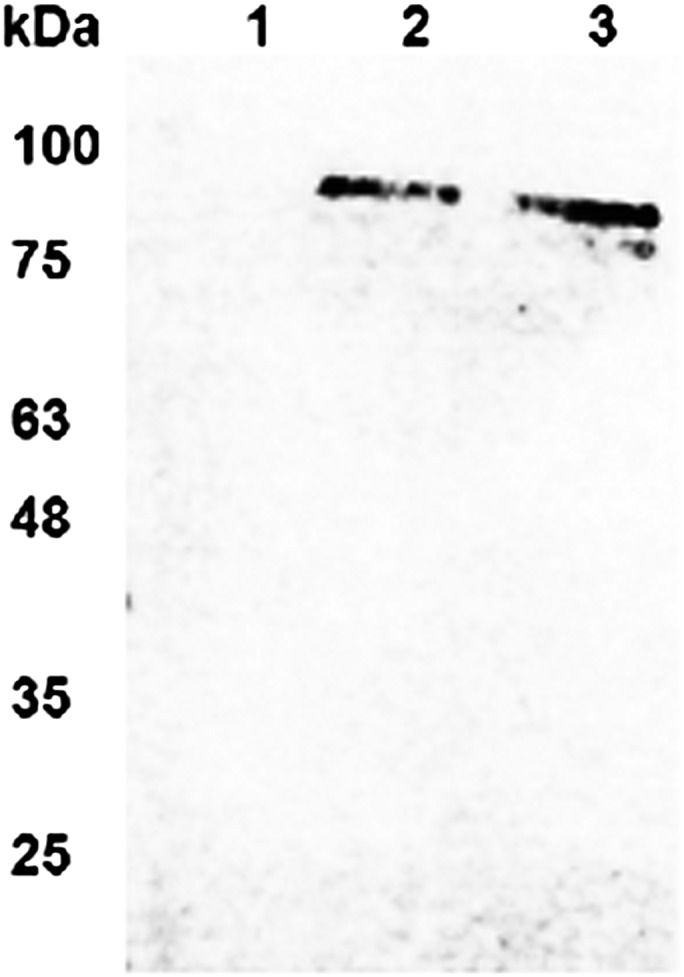
The ADP ribosyltransferase assay demonstrates that algal chloroplasts accumulate enzymatically active immunotoxin proteins. Biotinylated NAD+ was mixed with eEF2 and purified αCD22, αCD22PE40, or αCD22CH23PE40. Biotinylated ADP was transferred to eEF2 by enzymatically active exotoxin A. After reaction completion, samples were separated on SDS/PAGE and blotted onto nitrocellulose membranes. An anti-biotin antibody conjugated with alkaline phosphatase was used to detect eEF2 that was ribosylated with ADP-biotin. Western blot demonstrates that αCD22 does not ribosylate eEF2 (lane 1) but that αCD22PE40 (lane 2) and αCD22CH23PE40 (lane 3) have enzymatically active PE40 and do ribosylate eEF2.
Algal-Produced Immunotoxins Bind Specifically to Target Tumor Cells.
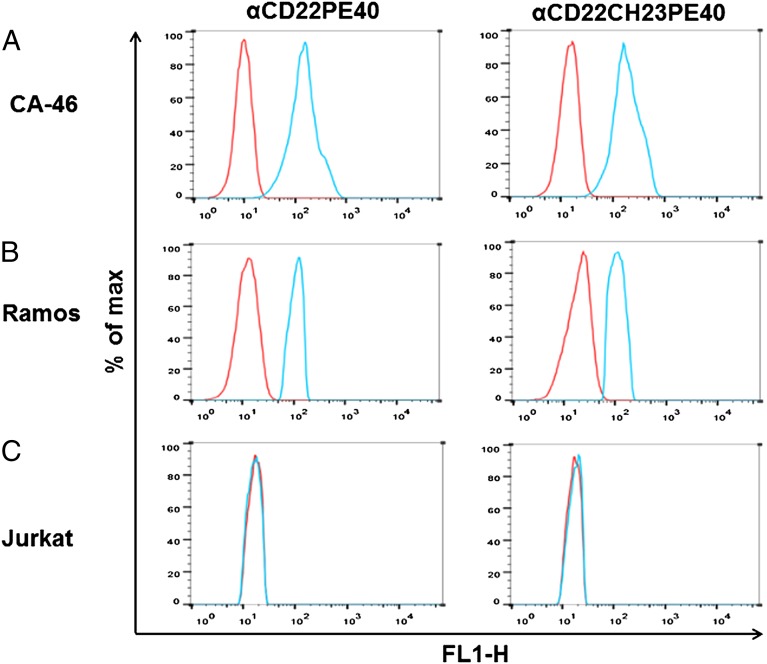
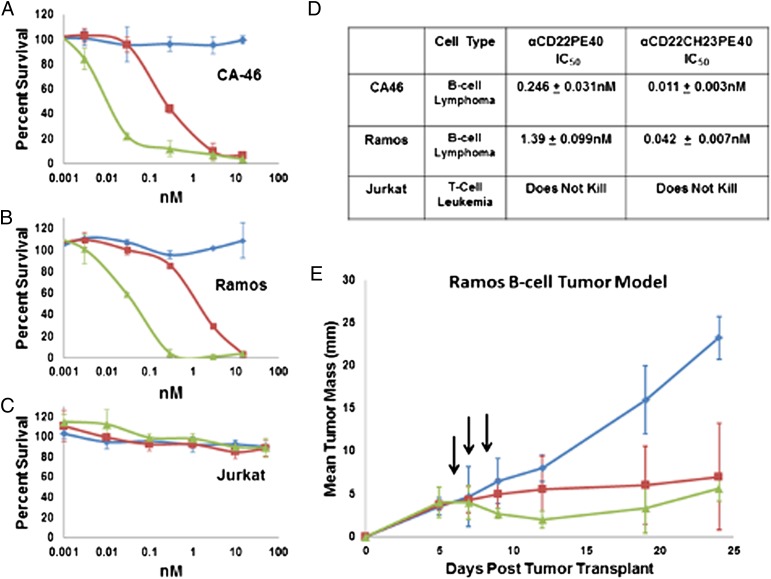
Flow cytometry was used to determine if algal-produced immunotoxins are capable of binding to their target cells. CA-46 B cells, Ramos B cells, and Jurkat T cells were incubated with αCD22PE40 and αCD22HCH23PE40 and subsequently fixed with sodium azide. Following the initial immunotoxin binding, cells were incubated with anti-endothelin receptor A antibody (produced in rabbits) and then with an anti-rabbit DyLight 488-conjugated antibody (Thermo Scientific). The cells then were subjected to FACS analysis on a BD influx (Becton Dickinson). CA-46 B cells treated with either αCD22PE40 or αCD22HCH23PE40 showed a fluorescent shift demonstrating that they were bound by the immunotoxin proteins (Fig. 5A). This shift also was present in Ramos B cells that were incubated with algal-produced immunotoxin proteins (Fig. 5B). Both the algal-produced immunotoxins fail to bind to human Jurkat T cells which do not express the CD22 antigen (Fig. 5C). These data show that algal-produced immunotoxins bind specifically to their target cells that express the antigen and not to other cell types that lack the target antigen, thereby demonstrating the specificity of the antibody component of the immunotoxin proteins.
Fig. 5.
Flow cytometry demonstrates specific binding of algal-produced immunotoxins. αCD22PE40 and αCD22CH23PE40 were incubated with CA-46 B cells, Ramos B cells, or Jurkat T cells. After primary incubation, cells were incubated with anti-exotoxin A produced in rabbit and finally with anti-rabbit DyLight 488. After incubation cells were analyzed by flow cytometry (blue curves). Cells that were not incubated with immunotoxins were used as a baseline of fluorescent intensity (red curves). (A) A shift in the fluorescence spectra demonstrates that αCD22PE40 and αCD22CH23PE40 bind to CA-46 B cells. (B) Fluorescence analysis also demonstrates that αCD22PE40 and αCD22CH23PE40 bind to Ramos B cells. (C) A lack of fluorescence shift demonstrates that algal-produced immunotoxins do not bind nonspecifically to Jurkat T cells.
Algal-Expressed Immunotoxins Are Cytotoxic to B-Cell Lymphoma Cell Lines.
To test the specificity of the cell-killing activity of the immunotoxins, live CA-46, Ramos, and Jurkat cells were treated in triplicate with varying doses of αCD22, αCD22PE40, or αCD22HCH23PE40, and cell survival was measured. Cells treated with PBS plus 0.2% human serum albumin (HSA) were used as a negative control to determine the baseline for 100% survival. Cells also were treated with 10 µg/mL of cycloheximide, which completely inhibits protein synthesis, resulting in 100% cell death, as the positive control. A water-soluble tetrazolium salt (WST-8) assay was used to measure cell viability as previously described (38). The αCD22 scFv lacking the PE40 toxin did not inhibit B cells or T cells from proliferating, but αCD22PE40 and αCD22CH32PE40 inhibited CA-46 B-cell (Fig. 6A) and Ramos B-cell (Fig. 6B) proliferation in a dose-dependent manner. The αCD22 scFv, αCD22PE40, and αCD22CH23PE40 did not inhibit Jurkat T-cell proliferation (Fig. 6C). In cell-based assays the monomeric αCD22PE40 killed CA-46 cells with an IC50 of 0.246 nM and Ramos cells with an IC50 of 1.39 nM (Fig. 6D). The divalent αCD22CH23PE40 killed CA-46 cells with an IC50 of 0.011 nM and Ramos cells with an IC50 of 0.042 nM (Fig. 6D). αCD22CH23PE40 was 22-fold more effective than αCD22PE40 at killing CA-46 cells and was 33-fold more effective at killing Ramos cells.
Fig. 6.
In vitro and in vivo analysis of the effectiveness of algal-expressed immunotoxin against cancer cells. αCD22 (blue traces), αCD22PE40 (red traces), and αCD22CH23PE40 (green traces) were incubated with CA-46 B cells, Ramos B cells, and Jurkat T cells for 72 h in vitro to determine their cytotoxic activity. (A) αCD22PE40 and αCD22CH23PE40 were effective at killing CA-46 B cells, but αCD22 alone was incapable of killing CA-46 cells. (B) Additionally, αCD22PE40 and αCD22CH23PE40 were able to kill Ramos cells, but αCD22 was unable to inhibit Ramos cell proliferation. (C) αCD22, αCD22PE40, and αCD22CH23PE40 were unable to kill Jurkat T cells. (D) The IC50 for each immunotoxin against each cell line was calculated to determine how effective each was at inhibiting cancer-cell proliferation. Both immunotoxins were capable of killing B cells, but dimeric αCD22CH23PE40 was more effective than αCD22PE40 at killing targeted cells in vitro. (E) Ramos cells (3 × 107) were transplanted s.c. into Rag−/− × gc−/− mice until they established tumors with a mean diameter of 4 mm. Mice then were treated each day for 3 d with 240 µg/kg of αCD22, αCD22PE40, or αCD22CH23PE40. Both αCD22PE40 and αCD22CH23PE40 inhibited tumor proliferation more effectively than αCD22 alone.
These data demonstrate that alga-produced immunotoxins are capable of binding to and inhibiting cell proliferation in vitro. Furthermore, in vitro, the divalent immunotoxin αCD22HCH23PE40 is able to cause cell death of target cells at a significantly lower concentration than the monovalent immunotoxin.
Antitumor Efficacy of Monomeric and Dimeric Algal-Produced Immunotoxins Against Established B-Lymphoma Xenografts.
To test the in vivo efficacy of the algal-produced immunotoxins, αCD22PE40 or αCD22HCH23PE40 and the control scFv αCD22 were evaluated for their antitumor activity against established s.c. xenografts in Rag × gc−/− mice. Rag × gc−/− mice were injected with 3 × 107 Ramos cells on day 0. The tumors were grown to an average diameter of 4 mm, which occurred 5 d after transplantation. The mice were treated with three doses (administered every other day by i.p. injection) of αCD22PE40, αCD22HCH23PE40, or αCD22. Mice were treated with 240 µg/kg of each molecule in triplicate. The mice treated with either algal-produced immunotoxin showed a significant inhibition of tumor propagation (Fig. 6E) compared with mice treated with the scFv lacking the PE40 toxin. This result suggests that algal-expressed and purified immunotoxins have a significant effect on tumor progression in animal models.
Discussion
We have shown that chloroplasts of the green algae, C. reinhardtii, are capable of accumulating fully functional immunotoxin proteins that consist of an antibody-binding domain targeting the B-cell surface antigen CD22 and the PE40 toxin domain of exotoxin A. We produced two different types of immunotoxins, single chain and dimeric, and both accumulated as soluble functional proteins within algal chloroplasts. Producing a eukaryotic toxin in a eukaryotic cell was possible because chloroplasts have a prokaryotic-like translational apparatus that is resistant to the toxin and because proteins produced in the chloroplast stay in the chloroplast. A single PE40 molecule escaping the chloroplast should be able to inhibit protein translation in the algal cytosol, resulting in cell death. The survival of algae producing the immunotoxins demonstrates that chloroplasts sequester chloroplast-produced proteins completely within the chloroplast. In addition to sequestering the toxin, allowing the production of immunotoxins in a eukaryotic host, chloroplasts also have the machinery necessary to assemble complex immunotoxins that contain multiple domains, such as αCD22CH23PE40, into larger assembled proteins consisting of two antibody-binding domains and two PE40 molecules. No other expression platform presently is capable of producing such a complex immunotoxin.
Previous studies have produced immunotoxins by expression in E. coli. These proteins generally need to be purified, denatured, and then refolded, because a majority of the protein product accumulates as an insoluble aggregate in E. coli (15). Analysis of immunotoxin proteins produced in algae show that both αCD22PE40 and αCD22CH23PE40 accumulate in algal chloroplasts as soluble, correctly folded molecules that do not require additional chemistry to be functional. This accumulation as a soluble functional molecule should reduce the cost of production significantly, because fewer steps are required to produce the functional therapeutic. Several groups have engineered E. coli to contain complex chaperones (39), protein disulfide isomerases (40), and PPIases (41), but algae already contain this complex protein-folding machinery (21, 42). The ability of chloroplast to assemble complex mammalian proteins was demonstrated previously by using chloroplasts to fold and assemble full-length human antibodies into soluble molecules that bind their target antigen (5). Here we show that C. reinhardtii chloroplasts also can assemble efficiently divalent immunotoxins that contain the hinge and CH2 and CH3 domains of a human IgG1. Two interchain disulfide bonds are formed in the hinge region of αCD22CH23PE40, allowing the protein to form a homodimer (43).
Immunotoxins are multifunctional and require that each individual part of the protein be operational. FACS analysis demonstrates that the antibody portions of αCD22PE40 and αCD22CH23PE40 bind specifically to target cells that express the CD22 antigen. The antibody domain of an immunotoxin is used to direct the toxin molecule to a specific cell type so that cancer cells expressing high levels of CD22 are targeted. An ADP ribosyltransferase assay was used to demonstrate that the PE40 component of both αCD22PE40 and αCD22HCH23PE40 was functional also.
Although enzymatically active immunotoxins that bind to their target cell are crucial for functional therapies, they also must be capable of delivering the catalytic domain of PE40 into the cytosol of the target cell to inhibit its proliferation. Both CD22PE40 and αCD22CH23PE40 were tested in a cell-viability assay to determine how effective they were at inhibiting target cell proliferation. Both algal-produced immunotoxins showed significant cytotoxicity toward two Burkitt lymphoma cell lines (Ramos and CA46). The 190-kDa αCD22HCH23PE40 immunotoxin is 22-fold more effective at killing CA-46 cells and 33-fold more effective at killing Ramos cells than the monomeric αCD22PE40. The increase in cytotoxicity can be attributed to two factors. First, divalent antibodies with multiple binding domains have been shown previously to have a greater binding avidity than monovalent scFv antibodies. The second reason for this increased potency can be attributed to the delivery of two PE40 molecules from αCD22HCH23PE40 as opposed to one from the monomeric αCD22PE40. These two factors appear to be responsible for the increased cytotoxicity of αCD22HCH23PE40 in cell-based in vitro assays.
Importantly, both immunotoxin molecules impact tumor growth in animal models, resulting in significant inhibition of tumor growth and significantly prolonging mouse survival in a tumor-challenge assay. Although αCD22HCH23PE40 may be slightly better at inhibiting tumor growth, the increased effectiveness of αCD22HCH23PE40 appears to be far less in vivo than in vitro. Previously, it was demonstrated that increasing the valence and half-life of an immunotoxin does not always lead to an increased in vivo effectiveness for certain types of cancers (44), but in some instances increased valence and prolonged half-life appear to have a dramatic effect (45). The effectiveness of larger immunotoxins with increased valence in vivo appears to be cancer-type specific.
Recently algae have garnered much attention for their potential use as a source of biofuels, but algae also appear have a value in the production of next-generation protein therapeutics. The ability to fold, assemble, and accumulate multiple domain proteins as soluble molecules is a significant advantage. However, the attributes that truly distinguish algae from other recombinant expression platforms are the presence of chloroplasts and the ability to produce and accumulate immunotoxin proteins in these compartments. Chloroplasts of higher plants such as tobacco show a high degree of conservation of algal chloroplasts and also could be a viable option for expressing immunotoxins (46). However, algae such as C. reinhardtii can be grown in closed bioreactors, thus avoiding problems that might arise through cross-contamination with native species (7, 47). Additionally, the time required to generate transgenic strains of algae and the ability to scale quickly are advantages that algae offer over land plants (7). No other recombinant protein production system has been shown to be capable of accumulating these complex eukaryotic toxin molecules as soluble and enzymatically active proteins. These traits set algae apart from other expression platforms. The potential of immunotoxins as potent and specific anticancer therapeutics is enormous. The use of antibody drug conjugates, using small-molecule drugs, to target and kill cancer cells and minimize the exposure of healthy cells is already a reality (48), and many of these therapies are in late-stage clinical trials (49, 50) or already are approved by the US Food and Drug Administration (51). Protein toxins also have been shown to be highly effective in inhibiting cancer-cell proliferation, but their production is limited to bacterial expression platforms that require the protein to be denatured and subsequently refolded (52), processes that add to both the time and the cost of developing these drugs. Algae provide another avenue for the production of these immunotoxins and add the ability to create more complex molecules than presently can be produced in bacterial systems. Although additional work needs to be done to determine whether larger or smaller immunotoxins are more effective for specific cancers, the αCD22PE40 and αCD22HCH23PE40 we have produced demonstrate that C. reinhardtii chloroplasts are capable of producing complex immunotoxins and provide a potentially significant avenue to produce these next-generation therapeutics.
Methods
Construct Design.
All DNA and RNA manipulations were performed as previously described (53). DNA encoding the variable regions of the heavy-chain and light-chain genes of the antibody RFB4 (10) were synthesized in C. reinhardtii chloroplast codon bias (www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=3055.chloroplast) and fused genetically with a linker sequence coding for 4× (G4S). This sequence was ligated downstream of a sequence coding for a 1× Flag peptide and a TEV protease cleavage site, yielding a 834-bp scFv gene, αCD22 (Codon adaptation index = 0.722, Nc = 30.5). The αCD22 gene was ligated upstream of a C. reinhardtii chloroplast codon optimized sequence coding for a 2× (G4S) linker and domains 2 and 3 of P. aeruginosa exotoxin A, yielding a 1,947-bp gene, αCD22PE40 (Codon adaptation index = 0.750, Nc = 34.7). To generate a dimeric immunotoxin, a C. reinhardtii chloroplast codon optimized gene coding for the hinge and constant domains 2 and 3 of a human IgG1 (HCH23) was ligated between the coding regions of αCD22 and PE40, again separated by a 2× (G4S) linker on both sides of the gene, yielding a 2,664-bp gene, αCD22HCH23PE40 (Codon adaptation index = 0.759, Nc =35.0). All three genes were ligated into the psbA transformation cassette.
Algal Transformations and Selection of Homoplasmic Strains.
DNA transformation plasmids were precipitated onto gold particles (Seashell Technology), and particle bombardment was used to transform chloroplasts of algal strain CC-125 (Duke University) as previously described (6). Transformed cells were selected on TAP plates containing 100 µg/mL of kanamycin sulfate. Subsequently, transformed colonies were patched onto a TAP plate containing 150 µg/mL of kanamycin sulfate to drive the transformants to homoplasmy. PCR analysis was performed as previously described (5) to determine strains that contained the gene of interest. In short, a forward primer was made against the psbA 5′ UTR (5′-gtgctaggtaactaacgtttgattttt-3′), and a reverse primer was made against the αCD22 gene (5′-tggaggtggaggtagtggtggtgg-3′); that sequence is present in all the transformation constructs, and the production of amplicons of 500 bp suggests that genes were integrated into the psbA locus. To identify strains that are homoplasmic, forward primers (5′-ggaaggggaggacgtaggtacataaa-3′) and reverse primers (5′-ttagaacgtgttttgttcccaat-3′) were designed against the psbA gene. A control primer set with forward (5′-ccgaactgaggttgggttta-3′) and reverse (5′-gggggagcgaataggattag-3′) primers was designed against the genomic region coding for the 16S rRNA to be used in the homoplasmic screen. This control primer set ensures that the apparent loss of the psbA gene is not merely a failed PCR. Homoplasmic strains were identified for each recombinant gene.
Accumulation Analysis, Purification, and Characterization of Algal-Produced Immunotoxins.
Accumulation of immunotoxins from transgenic strains of C. reinhardtii was determined by Western blot analysis using anti-Flag antibodies as described previously (6). Transgenic C. reinhardtii cultures were inoculated at 2 × 105 cells/mL and grown to a density of 2 × 106 cells/mL in dim light (200 lux). The 250 mL of culture was used to inoculate a 20-L carboy (VWR) at a density of 2 ×104 cells/mL and grown in light (10,000 lux) and mixed using bubbled air for 96 h before harvesting. Cells were lysed by sonication in lysis buffer [50 mM Tris⋅HCL (pH 8.0), 500 mM NaCl, 0.5% Tween 20 containing complete protease inhibitors (Roche)]. The soluble and insoluble proteins were separated using high-speed centrifugation at 20,000 × g. The soluble protein was applied to an anti-Flag M2 affinity gel (Sigma-Aldrich) and eluted with a Flag-elution buffer [100 mM glycine⋅HCL (pH 3.5) and 500 mM NaCl]. Proteins were purified further using size-exclusion chromatography to remove any degradation products. To visualize accumulation of the protein, 20 μg of total soluble proteins was loaded into each well. Immunotoxin and scFv protein were identified using a mouse anti-Flag alkaline phosphatase-conjugate antibody (Sigma). All procedures were carried out as described in ref. 54. Western blot analysis also was done in reducing conditions to visualize the assembly of dimeric immunotoxins. Proteins then were concentrated and buffered exchanged into PBS (PBS, 3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, pH7.4) using a concentrating column (GE Healthcare). Purified αCD22PE40 or αCD22HCH23PE40 proteins were titrated into crude algal lysate and compared with total soluble lysates made from strains expressing αCD22PE40 or αCD22HCH23PE40 to determine the percentage of total soluble protein that accumulates in C. reinhardtii chloroplasts. Then 20 μg of each lysate was coated on MaxiSorp plates (Nunc) and incubated overnight to allow proteins to coat wells. Wells then were blocked with ELISA blocking buffer (Pierce) for 1 h. Following blocking, samples were washed and probed with an anti-Flag antibody conjugated with HRP. After subsequent binding and washing steps, samples were developed with a 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Pierce) for 30 min. The reaction was stopped with 2 M sulfuric acid, and absorbance measured at 450 nm. Total soluble lysates of strains expressing αCD22PE40 or αCD22HCH23PE40 were compared with total soluble lysate from a WT strain with purified αCD22PE40 or αCD22HCH23PE40 titrated into the lysate.
ADP Ribosyltransferase Assay.
Determination of the enzymatic functionality of the algal-produced PE40 proteins was determined by ADP ribosyltransferase assay. Then 12.5 µM biotinylated-NAD+ (Sigma) was placed in a reaction with 50 mM Tris⋅HCl (pH 7.8), 400 ng of purified eEF2 (gift from Karen Browning of the University of Texas), 1 mM DTT, 1 mM EDTA, and 500 ng of purified αCD22, αCD22PE40, or αCD22HCH23PE40 protein in a 20-μL reaction volume. Each reaction was incubated at 25 °C for 30 min. Samples then were separated by PAGE and blotted onto a nitrocellulose membrane. Once blotted, an anti-biotin alkaline phosphatase-conjugated antibody (Rockfield) was used to identify the presence of biotin molecules on the 9-kDa eEF2 protein, indicating the presence of ribosylated eEF2.
Flow Cytometry Cell-Binding Assay.
CA-46 B cells, Ramos B cells, and Jurkat T cells were incubated in the presence of algal-produced αCD22PE40 or αCD22HCH23PE40 in PBS plus 0.01% sodium azide for 1 h at 4 °C. After primary binding, cells were incubated with an anti-exotoxin A antibody produced in rabbit (Sigma) and diluted 1:20,000 in PBS (Sigma) for 1 h. After secondary binding, cells were incubated with an anti-rabbit DyLight 488-conjugated antibody and analyzed by flow cytometry using a BD influx (Becton Dickinson). Data were analyzed using FlowJo software.
Cytotoxic Cell-Viability Assay.
One hundred microliters of CA-46 B cells, Ramos B cells, and Jurkat T cells (5 ×104 cells/mL) were seeded into each well of a 96-well tissue culture plate (Corning) for 24 h in a humidified incubator at 37 °C and 10% CO2. After incubation, increasing concentrations of αCD22, αCD22PE40, or αCD22HCH23PE40, diluted in PBS containing 0.2% HSA, were added to each well. PBS plus 0.2% HSA was used as a negative control for cell death and represented 100% cell survival. Cycloheximide at a concentration of 10 µM was used as a positive control for cell death and represented 0% cell survival. Cells were incubated with algal-produced αCD22, αCD22PE40, αCD22HCH23PE40, or control reagents for 72 h. After incubation with immunotoxins and controls, 10 μL of WST-8 reagent (Dojindo) was added to each well. The assay was allowed to develop for 4 h, and absorbance was read at 450 nm on a plate reader (Tecan). The IC50 was calculated using Grafit software (Erithacus).
Antitumor Efficacy of Algal-Expressed Immunotoxins Against Established B-Lymphoma Xenografts.
Female RAG2−/− × gc−/− mice (Taconic Farms), which lack adaptive immunity and natural killer cells, were used for the establishment of human lymphoma xenografts. Ramos cells (3 × 107) were transplanted s.c. into mice. When the tumors reached a mean diameter of 5 mm (typically 4 d after transplantation), mice were injected with 240 μg/kg of αCD22, αCD22PE40, or αCD22CH23PE40. Tumors were measured every day for up to 25 d to determine the tumor size and survival of the treated mice. The results shown are representative of three independent experiments. Animal experiments and care were done according to Institutional Animal Care and Use Committee (IACUC) approved animal protocol #S-06201. Any animal experiencing pain and discomfort was euthanized by carbon dioxide anesthesia followed by cervical dislocation by UCSD IACUC- and AVMA approved methods.
Acknowledgments
This work was supported by Grant CBET-1160184 from the National Science Foundation (to S.P.M.). M.T. was supported by a Skaggs Family Foundation predoctoral fellowship. C.V. was supported by a California Department of Labor Edge Internship.
Footnotes
Conflict of interest statement: S.P.M. is a founder of Sapphire Energy and has a financial interest in that company. Sapphire Energy has rights to this technology.
See Author Summary on page 14 (volume 110, number 1).
This article is a PNAS Direct Submission.
References
- 1.Raghavendra AS. Photosynthesis: A Comprehensive Treatise. Cambridge, UK: Cambridge Univ Press; 1998. [Google Scholar]
- 2.Hannon M, Gimpel J, Tran M, Rasala B, Mayfield S. Biofuels from algae: Challenges and potential. Biofuels. 2010;1(5):763–784. doi: 10.4155/bfs.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasala BA, Mayfield SP. The microalga Chlamydomonas reinhardtii as a platform for the production of human protein therapeutics. Bioeng Bugs. 2011;2(1):50–54. doi: 10.4161/bbug.2.1.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregory JA, et al. Algae-produced Pfs25 elicits antibodies that inhibit malaria transmission. PLoS ONE. 2012;7(5):e37179. doi: 10.1371/journal.pone.0037179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran M, Zhou B, Pettersson PL, Gonzalez MJ, Mayfield SP. Synthesis and assembly of a full-length human monoclonal antibody in algal chloroplasts. Biotechnol Bioeng. 2009;104(4):663–673. doi: 10.1002/bit.22446. [DOI] [PubMed] [Google Scholar]
- 6.Rasala BA, et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol J. 2010;8(6):719–733. doi: 10.1111/j.1467-7652.2010.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Specht E, Miyake-Stoner S, Mayfield S. Micro-algae come of age as a platform for recombinant protein production. Biotechnol Lett. 2010;32(10):1373–1383. doi: 10.1007/s10529-010-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasala BA, et al. Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS ONE. 2012;7(8):e43349. doi: 10.1371/journal.pone.0043349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franklin SE, Mayfield SP. Prospects for molecular farming in the green alga Chlamydomonas. Curr Opin Plant Biol. 2004;7(2):159–165. doi: 10.1016/j.pbi.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Shen GL, et al. Evaluation of four CD22 antibodies as ricin A chain-containing immunotoxins for the in vivo therapy of human B-cell leukemias and lymphomas. Int J Cancer. 1988;42(5):792–797. doi: 10.1002/ijc.2910420527. [DOI] [PubMed] [Google Scholar]
- 11.Mansfield E, Amlot P, Pastan I, FitzGerald DJ. Recombinant RFB4 immunotoxins exhibit potent cytotoxic activity for CD22-bearing cells and tumors. Blood. 1997;90(5):2020–2026. [PubMed] [Google Scholar]
- 12.Bogner C, et al. Immunotoxin BL22 induces apoptosis in mantle cell lymphoma (MCL) cells dependent on Bcl-2 expression. Br J Haematol. 2010;148(1):99–109. doi: 10.1111/j.1365-2141.2009.07939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin J, Li G, Ren X, Herrler G. Select what you need: A comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J Biotechnol. 2007;127(3):335–347. doi: 10.1016/j.jbiotec.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhary VK, et al. A recombinant immunotoxin consisting of two antibody variable domains fused to Pseudomonas exotoxin. Nature. 1989;339(6223):394–397. doi: 10.1038/339394a0. [DOI] [PubMed] [Google Scholar]
- 15.Brinkmann U, Reiter Y, Jung SH, Lee B, Pastan I. A recombinant immunotoxin containing a disulfide-stabilized Fv fragment. Proc Natl Acad Sci USA. 1993;90(16):7538–7542. doi: 10.1073/pnas.90.16.7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selyukh A. Seattle genetics cancer drug may top $100,000. Washington, DC: Reuters; 2011. Available at www.reuters.com/article/2011/08/22/us-seattlegenetics-idUSTRE77L5EB20110822. [Google Scholar]
- 17.Cao Y, et al. Single-chain antibody-based immunotoxins targeting Her2/neu: Design optimization and impact of affinity on antitumor efficacy and off-target toxicity. Mol Cancer Ther. 2012;11(1):143–153. doi: 10.1158/1535-7163.MCT-11-0519. [DOI] [PubMed] [Google Scholar]
- 18.Harris EH. Chlamydomonas Sourcebook Introduction to Chlamydomonas and its Laboratory Uses. New York: Academic; 2009. [Google Scholar]
- 19.Beligni MV, Yamaguchi K, Mayfield SP. Chloroplast elongation factor ts pro-protein is an evolutionarily conserved fusion with the s1 domain-containing plastid-specific ribosomal protein-7. Plant Cell. 2004;16(12):3357–3369. doi: 10.1105/tpc.104.026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manuell AL, Quispe J, Mayfield SP. Structure of the chloroplast ribosome: Novel domains for translation regulation. PLoS Biol. 2007;5(8):e209. doi: 10.1371/journal.pbio.0050209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroda M. The Chlamydomonas genome reveals its secrets: Chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth Res. 2004;82(3):221–240. doi: 10.1007/s11120-004-2216-y. [DOI] [PubMed] [Google Scholar]
- 22.Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994;266(5191):1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- 23.Breiman A, Fawcett TW, Ghirardi ML, Mattoo AK. Plant organelles contain distinct peptidylprolyl cis,trans-isomerases. J Biol Chem. 1992;267(30):21293–21296. [PubMed] [Google Scholar]
- 24.Mansfield E, Pastan I, FitzGerald DJ. Characterization of RFB4-Pseudomonas exotoxin A immunotoxins targeted to CD22 on B-cell malignancies. Bioconjug Chem. 1996;7(5):557–563. doi: 10.1021/bc960043y. [DOI] [PubMed] [Google Scholar]
- 25.Mansfield E, Chiron MF, Amlot P, Pastan I, FitzGerald DJ. Recombinant RFB4 single-chain immunotoxin that is cytotoxic towards CD22-positive cells. Biochem Soc Trans. 1997;25(2):709–714. doi: 10.1042/bst0250709. [DOI] [PubMed] [Google Scholar]
- 26.Kondo T, FitzGerald D, Chaudhary VK, Adhya S, Pastan I. Activity of immunotoxins constructed with modified Pseudomonas exotoxin A lacking the cell recognition domain. J Biol Chem. 1988;263(19):9470–9475. [PubMed] [Google Scholar]
- 27.Kreitman RJ, Wang QC, FitzGerald DJ, Pastan I. Complete regression of human B-cell lymphoma xenografts in mice treated with recombinant anti-CD22 immunotoxin RFB4(dsFv)-PE38 at doses tolerated by cynomolgus monkeys. Int J Cancer. 1999;81(1):148–155. doi: 10.1002/(sici)1097-0215(19990331)81:1<148::aid-ijc24>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 28.Bertholjotti I. [Antibody-drug conjugate—a new age for personalized cancer treatment] Chimia (Aarau) 2011;65(9):746–748. doi: 10.2533/chimia.2011.746. [DOI] [PubMed] [Google Scholar]
- 29.Minich SS. Brentuximab vedotin: A new age in the treatment of Hodgkin lymphoma and anaplastic large cell lymphoma. Ann Pharmacother. 2012;46(3):377–383. doi: 10.1345/aph.1Q680. [DOI] [PubMed] [Google Scholar]
- 30. Frankel AD, ed (1992) Genetically Engineered Toxins (Mercel Dekker, New York), pp 439–445.
- 31.Gordon VM, Klimpel KR, Arora N, Henderson MA, Leppla SH. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect Immun. 1995;63(1):82–87. doi: 10.1128/iai.63.1.82-87.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie H, Audette C, Hoffee M, Lambert JM, Blättler WA. Pharmacokinetics and biodistribution of the antitumor immunoconjugate, cantuzumab mertansine (huC242-DM1), and its two components in mice. J Pharmacol Exp Ther. 2004;308(3):1073–1082. doi: 10.1124/jpet.103.060533. [DOI] [PubMed] [Google Scholar]
- 33.Manuell AL, et al. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant Biotechnol J. 2007;5(3):402–412. doi: 10.1111/j.1467-7652.2007.00249.x. [DOI] [PubMed] [Google Scholar]
- 34.Sharp PM, Li WH. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puigbò P, Bravo IG, Garcia-Vallve S. CAIcal: A combined set of tools to assess codon usage adaptation. Biol Direct. 2008;3:38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreitman RJ, Pastan I. Importance of the glutamate residue of KDEL in increasing the cytotoxicity of Pseudomonas exotoxin derivatives and for increased binding to the KDEL receptor. Biochem J. 1995;307(Pt 1):29–37. doi: 10.1042/bj3070029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayfield SP, Schultz J. Development of a luciferase reporter gene, luxCt, for Chlamydomonas reinhardtii chloroplast. Plant J. 2004;37(3):449–458. doi: 10.1046/j.1365-313x.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- 38.Du X, Beers R, Fitzgerald DJ, Pastan I. Differential cellular internalization of anti-CD19 and -CD22 immunotoxins results in different cytotoxic activity. Cancer Res. 2008;68(15):6300–6305. doi: 10.1158/0008-5472.CAN-08-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haacke A, Fendrich G, Ramage P, Geiser M. Chaperone over-expression in Escherichia coli: Apparent increased yields of soluble recombinant protein kinases are due mainly to soluble aggregates. Protein Expr Purif. 2009;64(2):185–193. doi: 10.1016/j.pep.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 40.Frand AR, Cuozzo JW, Kaiser CA. Pathways for protein disulphide bond formation. Trends Cell Biol. 2000;10(5):203–210. doi: 10.1016/s0962-8924(00)01745-1. [DOI] [PubMed] [Google Scholar]
- 41.Outchkourov NS, et al. Correctly folded Pfs48/45 protein of Plasmodium falciparum elicits malaria transmission-blocking immunity in mice. Proc Natl Acad Sci USA. 2008;105(11):4301–4305. doi: 10.1073/pnas.0800459105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Mayfield SP. Protein disulfide isomerase as a regulator of chloroplast translational activation. Science. 1997;278(5345):1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- 43.Bloom JW, Madanat MS, Marriott D, Wong T, Chan SY. Intrachain disulfide bond in the core hinge region of human IgG4. Protein Sci. 1997;6(2):407–415. doi: 10.1002/pro.5560060217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bera TK, Williams-Gould J, Beers R, Chowdhury P, Pastan I. Bivalent disulfide-stabilized fragment variable immunotoxin directed against mesotheliomas and ovarian cancer. Mol Cancer Ther. 2001;1(2):79–84. [PubMed] [Google Scholar]
- 45.Ribbert T, et al. Recombinant, ETA’-based CD64 immunotoxins: Improved efficacy by increased valency, both in vitro and in vivo in a chronic cutaneous inflammation model in human CD64 transgenic mice. Br J Dermatol. 2010;163(2):279–286. doi: 10.1111/j.1365-2133.2010.09824.x. [DOI] [PubMed] [Google Scholar]
- 46.Maliga P. Plastid transformation in higher plants. Annu Rev Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- 47.Mayfield SP, et al. Chlamydomonas reinhardtii chloroplasts as protein factories. Curr Opin Biotechnol. 2007;18(2):126–133. doi: 10.1016/j.copbio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Alley SC, Okeley NM, Senter PD. Antibody-drug conjugates: Targeted drug delivery for cancer. Curr Opin Chem Biol. 2010;14(4):529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 49.Beck A, et al. The next generation of antibody-drug conjugates comes of age. Discov Med. 2010;10(53):329–339. [PubMed] [Google Scholar]
- 50.Lash A. Make the case for antibody-drug conjugates. Pharmaceuticals (Ott) 2010;28(11):32–38. [Google Scholar]
- 51.Younes A, Yasothan U, Kirkpatrick P. Brentuximab vedotin. Nat Rev Drug Discov. 2012;11(1):19–20. doi: 10.1038/nrd3629. [DOI] [PubMed] [Google Scholar]
- 52.Pastan I, Hassan R, FitzGerald DJ, Kreitman RJ. Immunotoxin treatment of cancer. Annu Rev Med. 2007;58:221–237. doi: 10.1146/annurev.med.58.070605.115320. [DOI] [PubMed] [Google Scholar]
- 53.Kontermann R, Dubel S, editors. Expression of Full Length Monoclonal Antibodies (mAb) in Algal Chloroplast. New York: Springer Lab Manuals; 2010. pp. 503–516. [Google Scholar]
- 54.Sambrook J, Fritsch EF, Maniatas T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]