Abstract
Two pathways of transcription termination, factor-independent and -dependent, exist in bacteria. The latter pathway operates on nascent transcripts that are not simultaneously translated and requires factors Rho, NusG, and NusA, each of which is essential for viability of WT Escherichia coli. NusG and NusA are also involved in antitermination of transcription at the ribosomal RNA operons, as well as in regulating the rates of transcription elongation of all genes. We have used a bisulfite-sensitivity assay to demonstrate genome-wide increase in the occurrence of RNA-DNA hybrids (R-loops), including from antisense and read-through transcripts, in a nusG missense mutant defective for Rho-dependent termination. Lethality associated with complete deficiency of Rho and NusG (but not NusA) was rescued by ectopic expression of an R-loop-helicase UvsW, especially so on defined growth media. Our results suggest that factor-dependent transcription termination subserves a surveillance function to prevent translation-uncoupled transcription from generating R-loops, which would block replication fork progression and therefore be lethal, and that NusA performs additional essential functions as well in E. coli. Prevention of R-loop–mediated transcription-replication conflicts by cotranscriptional protein engagement of nascent RNA is emerging as a unifying theme among both prokaryotes and eukaryotes.
All bacterial transcription termination occurs by one of two pathways that are referred to as factor-independent (or intrinsic) and factor-dependent (or Rho-dependent), respectively (1). The latter pathway, which requires the action of factors Rho, NusG, and NusA in Escherichia coli, serves to terminate synthesis of transcripts that are not being simultaneously translated, for example, at the ends of various genes and operons (1, 2). The same mechanism is also responsible for the classic phenomenon of nonsense polarity (3), by which a stop codon mutation within the proximal gene of an operon results in absence of transcription of the distal genes; in this manner, Rho-dependent termination provides a back-up to other mechanisms (4, 5) that act to ensure the coupling of transcription with translation in bacteria. NusG and NusA are also involved in transcription antitermination during lytic growth of the lambdoid prophages and at the ribosomal RNA operons, as well as in regulating the rates of transcription elongation of all genes (6, 7).
Rho, NusG, and NusA are each essential for viability of the prototypic WT E. coli strain MG1655. Cardinale et al. (8) have reported that NusG and NusA are dispensable in strain MDS42 [which is an engineered MG1655 derivative with 14% reduced genome content because of deletions of insertion elements and cryptic prophages (9)], based on which they have proposed that the essentiality of Rho-dependent termination stems from its need for silencing of horizontally acquired genes in bacteria. Even so, the Δrho mutation in MDS42, as in MG1655, is lethal (8, 10).
Several phenotypes in nusG, rho, and nusA missense mutants, which are defective for Rho-dependent termination and relieved for nonsense polarity, have earlier also been interpreted as evidence in support of the increased occurrence in them of RNA-DNA hybrids or R-loops, which are generated by 5′-end invasion and reannealing of nascent untranslated transcripts to the upstream DNA (11). These phenotypes include synthetic lethalities with deficiencies of R-loop–removing enzymes RNase HI or RecG, increased copy number of plasmids that are R-loop–dependent for replication, and suppression of RNase E deficiency by postulated R-loop–mediated RNA degradation (12–14). Both R-loops (15) and Rho inhibition (10, 16) are also independently associated with replication fork blockage. A similar model of R-loop formation, by the upstream reannealing of untranslated transcripts, has also been proposed for topA mutants of E. coli (17).
In this article we present two converging lines of evidence that confirm the validity of the R-loop model. An assay using sodium bisulfite (18) was used to demonstrate excessive genome-wide R-loops in a nusG missense mutant defective for Rho-dependent termination. Furthermore, ectopic expression of UvsW, an R-loop helicase of phage T4 (19, 20), restored viability in Δrho derivatives of MDS42 and MG1655, as well as in the ΔnusG derivative of MG1655. Our results therefore establish that the essential role of factor-dependent transcription termination in E. coli is in R-loop prevention.
Results
Bisulfite Strategy for Genome-Wide R-Loop Detection in a nusG Mutant.
Sodium bisulfite targets C residues on the displaced DNA single strand in an R-loop, and the resulting changes can be detected as C-to-T conversions upon subsequent PCR and DNA sequencing (18). In a test of the R-loop model, total nucleic acids from both an MG1655 derivative (designated WT) and its isogenic nusG (-G146D) missense mutant (12) defective for Rho-dependent termination were exposed to bisulfite without prior denaturation and subjected to whole-genome next-generation resequencing. The population of sequence reads (each ∼50 bases long) was mapped to the MG1655 upper (or top) strand reference sequence, and those reads that remained unmapped were then sequentially aligned to two modified versions of this sequence, the first bearing conversions of all C residues to Ts and the second of all Gs to As. It was expected that clustered C-to-T changes on a DNA strand caused by bisulfite would generate reads that fail to align to the native reference sequence, but would now instead map to the C-to-T or G-to-A converted reference sequence (depending upon whether the bisulfite mutagenesis had targeted the upper or lower genomic strand, respectively). As shown in Table 1, the combined aggregate of reads mapping to the two converted reference sequences (expressed as a ratio of those mapping to the native reference sequence) was approximately threefold higher in the nusG mutant (19%) compared with that in the WT strain (7%); these results are indicative of increased bisulfite sensitivity of genomic DNA from the mutant, and thus are in accord with the genetic evidence for increased R-loops in this strain (12, 14).
Table 1.
Read mapping statistics after bisulfite treatment
| Reference genome | Mapped read numbers (× 106) |
||
| WT | nusG | nusG/prnhA+ | |
| Native (A) | 21.53 | 16.12 | 8.50 |
| C-to-T converted (B) | 0.79 | 1.57 | 0.04 |
| G-to-A converted (C) | 0.75 | 1.50 | 0.04 |
| (B) + (C)/(A) | 7.15% | 19.04% | 0.93% |
Gene- and Strand-Wise categorization of Bisulfite Sensitivity.
For the WT and nusG strains, the read populations aligned to each of the three reference genomes (that is, native MG1655 and its C-to-T and G-to-A converted versions) were binned to discrete intervals corresponding to the >4,300 gene and intergene regions of E. coli (Dataset S1). To control for differences in gene length and intensity of sequence coverage, we have expressed the read numbers mapping to the C-to-T and G-to-A converted reference sequences for each gene as percentages of those mapping to the native reference sequence itself. The corresponding normalized values are designated as Nupper and Nlower, respectively (Dataset S2, sheet 1).
Following the binning procedure, it was also noted that in the nusG strain, read numbers mapping to the native MG1655 reference sequence at the rac prophage locus (about 20-kb long, extending from intR to stfR) were markedly lower than the read numbers for genes flanking rac (Dataset S1, sheet 1); these results indicate the existence of a Δrac deletion in >90% of cells in the culture, which was then confirmed by PCR (SI Text and Fig. S1). This finding is in agreement with the findings of Cardinale et al. (8) that rac genes confer reduced fitness when Rho-dependent termination is compromised, and we believe that the Δrac deletion was selected for during the course of routine maintenance of this missense mutant in the laboratory.
Bisulfite Sensitivity of Genes in nusG Mutant Is Biased by Both Transcriptional and Replication Fork Orientations.
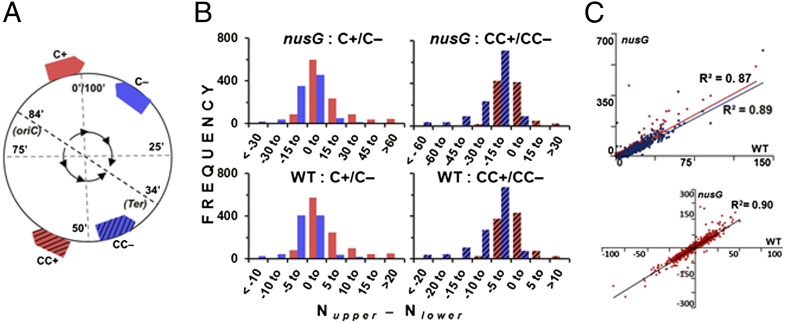
On the 100-min-long circular E. coli chromosome, genes are transcribed in clockwise (+) and counterclockwise (−) orientations, and the replication forks progress bidirectionally on the clockwise (C) and counterclockwise (CC) arms from oriC at around 84 min to Ter at around 34 min. Accordingly, by combining both replication arm disposition and transcriptional orientation, all genes may be placed in four categories: C+, C−, CC+, and CC− (Fig. 1A).
Fig. 1.
Transcription and replication bias in genome-wide bisulfite reactivity. (A) Schematic depiction of the disposition of four categories of genes C+, C−, CC+, and CC− on the E. coli chromosome (outer circle): genes on C and CC replication arms are shown as nonhatched and hatched arrows respectively, and those in + and − transcriptional orientations in red and blue, respectively. The inner circle shows direction of replication fork movement from oriC to Ter on C and CC arms. (B) Frequency distribution of each of the four categotries of genes (symbolized as in A) from nusG mutant and WT strain, in different intervals of [Nupper − Nlower] values as indicated. (C) Correlation between WT and nusG strains for Nupper (red) and Nlower (blue) values of genes in + and − orientations, respectively (Upper), and for [Nupper − Nlower] values (Lower).
Because the gene-specific Nupper and Nlower values are measures, respectively, of bisulfite sensitivity of the upper and lower genomic strands, a large positive value of [Nupper − Nlower] indicates preferential sensitivity of the gene’s upper strand (relative to its lower strand) to bisulfite, and a large negative value the converse. With increasing positive values of [Nupper − Nlower], genes of the nusG strain transcribed in + (relative to those in −) orientation were progressively overrepresented across both the C and CC replication arms of the genome (that is, as one moves from left to right in the top pair of histograms in Fig. 1B), and vice versa. As the upper and lower strands constitute the transcriptional nontemplate strands of, respectively, the + and − oriented genes, our results establish that it is the nontemplate strand of transcription that exhibits preferential bisulfite reactivity across the majority of genes (in accord with the expectations of the R-loop model). At the same time, highly expressed genes, such as those encoding proteins of the transcription-translation apparatus, were not overly bisulfite-sensitive (Dataset S2, sheet 2), suggesting that bisulfite was not simply targeting the unpaired nontemplate strand within an RNA polymerase transcription bubble (16).
After adjustment for transcriptional orientation, an effect of replication fork direction on the read numbers mapping to the converted reference genomes was also detected for the nusG mutant; thus, the values of Nupper and Nlower were respectively higher for genes, in their aggregate, located on the C and CC chromosomal replication arms (Table 2) (compare aggregate Nupper and Nlower values between C and CC pairs of same transcriptional orientation: that is, + or −). Given that the upper and lower strands represent the lagging-strand templates of the C and CC arms, respectively, our results indicate that the single-stranded regions of DNA associated with Okazaki fragment synthesis at the replication forks of asynchronously dividing cells (21) are also targets for bisulfite-induced C-to-T conversions.
Table 2.
Mean Nupper and Nlower values for gene categories aggregated by replication arm and orientation
| Category | Aggregate gene length (kb) | WT |
nusG |
||||||||
| Mapped read nos. (× 105) |
Mean |
Mapped read nos. (× 105) |
Mean |
||||||||
| Native | C-to-T | G-to-A | Nupper | Nlower | Native | C-to-T | G-to-A | Nupper | Nlower | ||
| C+ | 1062.5 | 40.66 | 2.73 | 1.01 | 6.7 | 2.5 | 30.77 | 5.77 | 1.92 | 18.8 | 6.3 |
| CC+ | 894.9 | 42.14 | 1.57 | 1.32 | 3.7 | 3.1 | 31.60 | 3.00 | 2.67 | 9.5 | 8.4 |
| C− | 922.1 | 37.88 | 1.43 | 1.47 | 3.8 | 3.9 | 27.97 | 2.92 | 2.82 | 10.5 | 10.1 |
| CC− | 1146.3 | 51.06 | 1.15 | 2.89 | 2.3 | 5.7 | 37.55 | 2.07 | 5.99 | 5.5 | 15.9 |
Differences Between WT and nusG Strains for Bisulfite Reactivity Are only Quantitative.
Unexpectedly, the patterns of bisulfite reactivity in the WT strain were highly correlated with those in its nusG derivative (Fig. 1 B and C). This finding was true with regard to both the relative sensitivities of, and strand-biasness in, the individual genes; the former was determined by comparisons between the two strains of Nupper values for + and Nlower for − genes, and the latter by comparisons of the [Nupper − Nlower] values (Fig. 1C). Similarly, the contribution of replication fork direction to bisulfite sensitivity was evident also in the WT strain, from the mean Nupper and Nlower values for the four gene categories (Table 2).
Thus, the differences between parent and mutant strains were only quantitative, and no genes were identified that were differentially more or less sensitive to bisulfite in the nusG mutant compared with those in the WT strain. As discussed below, these results are interpreted to signify that R-loops do occur at lower frequencies even in the WT strain. Consistent with this interpretation, the magnitude of global bisulfite sensitivity in a nusG derivative carrying the rnhA gene (encoding RNase HI) on a multicopy plasmid, as assessed by the proportion of reads mapping to the two converted reference sequences versus those to the native reference sequence, was lower even than that in the WT strain (Table 1, last column).
Features of Genes and Regions That Exhibit High Bisulfite Reactivity.
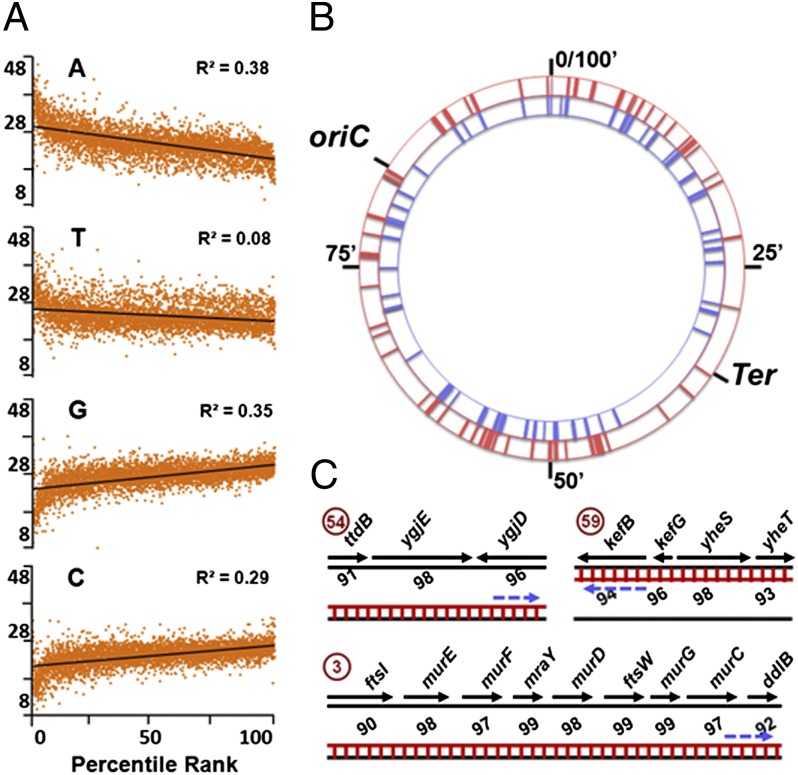
In both the WT and nusG strains, increased bisulfite sensitivity of individual genes was correlated inversely with content of A nucleotides, and directly with those of G and C nucleotides, on the nontemplate strand (Fig. 2A and Dataset S2, sheet 3). By using the set of criteria described in SI Text, around 76 single- and multigene loci or clusters of increased bisulfite sensitivity were identified, each possessing at least one gene that scored above the 97th percentile for Nupper or Nlower values for genes categorized by orientation and replication arm in the two strains (Fig. 2B and Dataset S2, sheet 4). In many clusters, heightened bisulfite sensitivity was confined to one DNA strand but included genes in both orientations or in large operons, reflecting the effects of polarity, antisense transcription, or read-through across normal termination signals, which are the targets of Rho-dependent termination (1–3, 22); examples of three such clusters are shown in Fig. 2C. Some clusters also scored high for bisulfite reactivity on both the strands (Dataset S2, sheet 4), our interpretation for which is that each of these clusters represents two separate cell populations in the culture with R-loop based displacement in them of the upper and lower DNA strands, respectively.
Fig. 2.
Bisulfite-reactive high-ranking clusters and correlation with ATGC content. (A) Correlation between percentile ranks of Nupper for + and Nlower for − genes with percent content of A, T, G, and C nucleotides on nontemplate strand, in both strains (data from Dataset S2, sheet 3). (B) Representation on circular E. coli chromosome of high-ranking clusters for Nupper (red, outer circle) and Nlower (blue, inner circle) values. (C) Examples of three high-ranking clusters (from Dataset S2, sheet 4; numbers encircled) depicting orientation of and percentile ranks for genes in each cluster, DNA and RNA strands as black and red lines respectively (RNA-DNA hybrid as ladder), and direction of RNA polymerase movement by interrupted arrow.
About 50 genes that were above the 90th percentile for bisulfite sensitivity in the present study had also been identified earlier by an approach involving the mapping of RNA polymerase-bound genomic positions in the presence and absence of bicyclomycin (22), to be in very close proximity to putative targets of Rho-dependent termination (Dataset S2, sheet 3). Similarly, several clusters were also in or near the horizontally acquired nonessential gene regions (9) that are silenced by Rho-dependent termination (8) (Dataset S2, sheet 4).
Rescue of Δrho Lethality in Both MDS42 and MG1655 by UvsW Expression.
As another test of the R-loop model, we examined whether ectopic expression of the phage T4-encoded R-loop helicase UvsW (19, 20) would rescue lethality in Δrho mutants. Such ectopic UvsW expression has been shown to suppress synthetic lethality associated with combined deficiency of the R-loop–removing enzymes RNase HI and RecG (19), and we also found in this study that the Ts growth phenotype of a mutant doubly defective for RNases HI and HII (23) can be partially rescued by UvsW (SI Text and Fig. S2).
Because UvsW negatively regulates copy number of many commonly used plasmids (19), its graded expression was achieved from a chromosomal Ptac-UvsW construct [with varying concentrations of isopropyl-β-D-thiogalactopyranoside (IPTG)], and control derivatives similarly expressed a UvsW variant (-K141R) inactive for ATPase and helicase activities (19). Viability of Δrho strains was assessed by the ability of their derivatives carrying a cognate single-copy-number rho+ lacZ+ plasmid to yield spontaneous plasmid-free segregants, which could be distinguished by blue-white screening of colonies on plates supplemented with X-Gal (14).
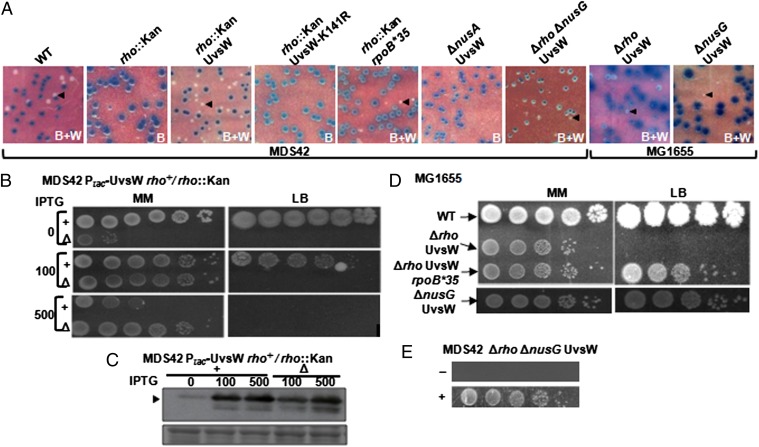
Because ΔnusG and ΔnusA derivatives of MDS42 have been reported to be viable (8), we undertook our initial experiments in MDS42Δlac. With either of two deletion rho alleles, MDS42 derivatives with Ptac-UvsW, but not Ptac-UvsW-K141R, were viable on minimal medium supplemented with ≥ 25 µM IPTG; both strains were inviable on rich medium (LB) at any IPTG concentration (Fig. 3 A and B, and Table S1). The rich-medium sensitivity was elicited also on nutrient agar and on defined medium supplemented with 5% (but not 0.5%) (wt/vol) Casamino acids (Fig. S3), indicating that it is the growth rate and not any specific component of the growth medium that modulates UvsW’s ability to rescue Δrho lethality.
Fig. 3.
Rescue of E. coli ∆rho and ∆nusG lethalities. (A) Blue (B) or mixture of blue and white (B+W) colonies from parental (WT) and deletion derivatives of MDS42 and MG1655 carrying cognate lacZ+-bearing shelter plasmids on X-Gal and IPTG-supplemented minimal medium; representative white colonies are marked by arrowheads. (B) Dilution-spotting of rho+ (+) and rho::Kan (∆) derivatives of MDS42 expressing UvsW on minimal (MM) and rich (LB) media with indicated micromolar IPTG concentrations. (C) Immunoblot (Upper) with anti-GST antibody for (GST-tagged) UvsW levels (arrowhead) in the pair of strains used in B, after growth in minimal medium with indicated micromolar IPTG concentrations; (Lower) representative section of the amido black-stained blot to serve as loading and transfer control. (D) Dilution-spotting of MG1655 (WT) and its indicated derivatives on minimal (MM) and rich (LB) medium with IPTG supplementation at 100 (Upper Left), 10 (Upper Right), 5 (Lower Left), and 2.5 (Lower Right) µM, respectively. (E) Dilution-spotting at 30 °C of MDS42∆rho∆nusG expressing UvsW on minimal medium without (−) or with (+) 20 µM IPTG. In all panels, UvsW and UvsW-K141R expression where shown was from their respective Ptac constructs; all MDS42 and MG1655 derivatives were also ∆lac; and the images shown are reproductions to size, within approximation.
The rho+ Ptac-UvsW derivative was also killed on rich medium with ≥ 50 µM IPTG, indicative presumably of R-loop helicase toxicity through unwinding of the RNA primers for Okazaki fragment synthesis (21); indeed, when equivalent levels of UvsW overexpression were elicited with 0.5 mM IPTG (as assessed by immunoblotting) (Fig. 3C), greater toxicity was observed in the rho+ than Δrho strain, suggestive of a protective titration effect by genome-wide prevalence of R-loops in the latter (Fig. 3B).
In MG1655Δrho as well (Fig. 3 A and D, and Table S1), UvsW expression from Ptac conferred viability, and only so in minimal medium (with the optimal growth rate at 150 µM IPTG of 0.29 h−1; that is, around 48% of that for MG1655). By PCR experiments, it was verified that the resulting strain continued to remain rac+ (Fig. S1), thus establishing that this prophage is not an impediment to viability of Rho-compromised strains (8) as long as R-loop toxicity has been alleviated in them.
Finally, UvsW expression did not alter the nonsense polarity-relief phenotype conferred by a rho missense mutation at two different loci that were tested (Fig. S4A), and even the Δrho strain expressing UvsW was polarity-relieved (Fig. S4B). These data indicate that UvsW was not merely substituting for Rho’s termination function while mediating the rescue of Δrho lethality.
UvsW Expression also Rescues ΔnusG Lethality in MG1655.
We adopted the blue-white screening approach (using an unstable nusG+ lacZ+ shelter plasmid) to test for suppression by UvsW of ΔnusG lethality in MG1655 as well. As with Δrho, the lethality in MG1655 of ΔnusG was rescued by UvsW but not UvsW-K141R (Fig. 3 A and D, and Table S1), this time on both rich and minimal media (with optimal growth rates at 5 µM IPTG of 0.38 h−1 and 0.22 h−1, respectively). Similar results of lethality rescue by UvsW were obtained with three different deletion alleles of nusG that were tested (Table S1), including one encoding loss of only the C-terminal domain after residue 118 so that the N-terminal domain’s function in transcription elongation is retained (24, 25). The double-mutant Δrho ΔnusG derivative of MDS42 but not MG1655 also was viable with UvsW, but only so on minimal medium at 30 °C (Fig. 3 A and E).
ΔnusA Lethality in Neither MDS42 Nor MG1655 Is Rescued by UvsW.
When the similar approach was used with ΔnusA, we observed that it was lethal in MDS42 [contrary to the report of Cardinale et al. (8)], and that UvsW was unable to rescue this lethality (Fig. 3A). Four different deletion alleles were tested in these experiments (Table S1), including the nusA::Cm mutation used earlier (8) and two others that retained their N-terminal domains (residues 1–137 and 1–200, respectively) with postulated distinct functions in transcription elongation and antitermination (26). The nusA deletions were lethal also in MG1655, but our data do provide support to the earlier suggestion (8) that MDS42 requires a considerably lower level of NusA activity than does MG1655 for viability (SI Text and Fig. S5).
Rescue of MDS42Δrho Lethality by RpoB*35.
Rich-medium sensitivity is commonly attributed to the occurrence of lesions that lead to replication fork collapse (27), and our finding that Δrho strains expressing UvsW are rich-medium–sensitive suggested that the residual R-loops in them impede replication fork movement (15). Similarly, we found that the Ts phenotype of a mutant deficient for both RNases HI and HII (23) is manifested only in rich medium (Fig. S2).
RpoB*35 (-H1244Q) is a mutant RNA polymerase that is reported to be less prone to backtracking and to be more readily dislodged from the DNA template; it is hence associated with reduced incidence of transcription-replication conflicts and fork blockage (16, 28, 29). The rpoB*35 mutant is also bicyclomycin-resistant (10, 16), indicating that the mutation confers a reduced requirement for Rho function in the cells. We found that RpoB*35 restored viability to MDS42Δrho in minimal medium with a growth rate of 0.28 h−1, but [as also reported previously (10)] that it was unable to do so on rich media (Fig. 3A, Fig. S3, and Table S1); however, RpoB*35 could rescue neither the Δrho nor ∆nusG derivatives of MG1655 (Table S1). Whereas each by itself (RpoB*35 or UvsW) was only partially effective in suppressing Δrho (with the resulting strains being viable on minimal but not rich medium), the two together conferred viability to Δrho derivatives of both MDS42 and MG1655, even on rich medium (Fig. 3D and Fig. S3); these observations therefore suggest that the effects of the two suppressors are additive.
Suppression of Δrho by UvsW Is Independent of Replication Restart Proteins.
Through its helicase activity, UvsW is able not only to unwind R-loops but also to catalyze the regression of blocked replication forks, both in vivo and in vitro (30, 31). We therefore considered the possibility that suppression by UvsW of lethality associated with compromised Rho-dependent termination is because of an enhanced ability to restart replication following double-strand breaks at blocked forks in these mutants (10, 32). We found, however, that UvsW-mediated rescue of Δrho lethality occurs even in strains that are deficient for RecA, RecB, or PriA (Fig. S6), which are some of the other proteins that are crucially involved in fork restoration and restart (33, 34). Thus, it appears that UvsW is acting more to prevent fork blockage by R-loop unwinding than to resolve the blocked forks through its fork regression activity.
Discussion
Even as NusG and NusA were reported to be dispensable for viability in MDS42, no condition had so far been identified that would suppress Δrho lethality (8, 10). It was therefore suggested that the essential role of factor-dependent termination is to silence the horizontally transferred prophage genes (8), and that Rho performs an additional essential role in E. coli (10, 16).
The results from the present study indicate that lethality of both Δrho and ΔnusG (but not ΔnusA) in MG1655 is rescued by expression of the R-loop helicase UvsW, and that genome-wide prevalence of R-loops is much increased in a nusG mutant compared with that in the WT strain. We conclude that essentiality of Rho-dependent termination is related solely to its function in reducing R-loop occurrence across the genome. Although there is evidence that NusA participates in Rho-dependent termination (8, 10, 13, 35), our findings indicate that it is also needed for other essential functions (6, 7, 26). The increased prevalence of R-loops in Rho-compromised strains may reflect either their reduced removal (16) or, perhaps more likely as explained below, their increased generation by reannealing of nascent untranslated transcripts to upstream DNA (11, 17, 36).
Based on the bisulfite sensitivity data, we propose that there are two categories of untranslated transcripts that can invade and reanneal with DNA to generate R-loops when Rho-dependent termination is compromised. One category comprises mRNAs of protein-coding genes that, for stochastic reasons, are not simultaneously translated; this category would explain the strong transcriptional bias in patterns of bisulfite sensitivity in the strains (Fig. 1B). We suggest that the source of the second category is the unexpectedly pervasive transcription from both genomic strands that has been discovered in E. coli as also in other bacteria (37–40); this category would explain both the pan-genome distribution and long multigene clusters of bisulfite sensitivity. Rho-dependent termination is therefore to be seen as a surveillance mechanism to prevent excessive synthesis of both categories of untranslated transcripts that are otherwise R-loop–prone.
Because our data indicate that the gene-wise patterns of bisulfite sensitivity are identical between the nusG mutant and the WT strain, we suggest that both categories of R-loop–generating untranslated transcripts occur at lower prevalence in the latter. This proposal is supported by earlier findings that strains deficient for either of the R-loop–removing enzymes, RNase HI or RecG, exhibit R-loop–initiated constitutive stable DNA replication [which is transcription-dependent (27)], that the combined deficiency of both enzymes is lethal (19, 27), and that ectopic UvsW expression rescues this synthetic lethality (19). It would therefore appear that accumulation of R-loops to lethal levels in a WT strain is avoided, not by the absence of their generation, but by a balance between their generation and removal.
Our study confirms that strain MDS42 requires less Rho function for its viability than does MG1655 (8). Furthermore, our data for Δrho strains on suppression by RpoB*35 and on rich-medium sensitivity suggest that R-loop lethality is correlated with RNA polymerase backtracking and arrest, leading to replication fork blockage (16, 29, 41, 42). However, these interpretations from the genetic data are not conclusive, and rigorous mechanistic explanations await supporting biochemical and other evidence.
Reminiscent of the bacterial situation in which R-loops occur from nascent transcripts that fail to be simultaneously translated, recent work has established that R-loops also occur in eukaryotic cells in the absence of cotranscriptional splicing, polyadenylation, or export, leading to replication fork blockage and genome instability (reviewed in refs. 43–45). Furthermore, loss in yeast (46) or human (47) cells of Sen-1/senataxin that, like Rho is a 5′-3′ RNA translocase, is associated with both increased R-loops and defective transcription termination. Thus, cotranscriptional engagement of RNA by different proteins appears to be necessary for R-loop prevention in both prokaryotes and eukaryotes.
Methods
Bisulfite Treatment and Whole-Genome Sequencing.
Details of the procedures used for bisulfite treatment of nucleic acid preparations from the different strains and for analysis of DNA sequence reads are given in SI Text. Genomic DNA sequencing was undertaken on the SOLiD 4 System platform (Life Technologies), and the raw sequence read data have been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number SRA059488.
Other Methods.
Other methods, plasmids, and strains are described in SI Text. Isogenic MG1655 derivatives GJ6504 (WT) and GJ6511 (nusG-G146D), described earlier (13), were used in the bisulfite reactivity experiments. Studies with rho, nusG, and nusA deletions were performed in two strain backgrounds, MDS42Δ(argF-lac)U169 and MG1655ΔlacIZYA. The protocols of Datsenko and Wanner (48) and Boyd et al. (49) were used, respectively, for generation of deletions on the chromosome and for chromosomal integration of Ptac-UvsW and Ptac-UvsW-K141R.
Note Added in Proof.
Peters et al. (50) have also shown recently that Rho and NusG suppress pervasive antisense transcription in E. coli.
Supplementary Material
Acknowledgments
We thank the investigators who provided various reagents for this study; Jamshaid Ali for help with determinations of gene-specific nontemplate strand nucleotide composition; Seshadri Gowrishankar for the software programs to bin the sequence reads; and members of the Laboratory of Bacterial Genetics for advice and discussions. The work was supported by a Centre of Excellence in Microbial Biology research grant of the Department of Biotechnology; A.H.S. was the recipient of Junior and Senior Research Fellowships of the Council of Scientific and Industrial Research toward the pursuit of a PhD degree of the Manipal University; and J.G. is recipient of the J. C. Bose Fellowship of the Department of Science and Technology, Government of India.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. SRA059488).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213123110/-/DCSupplemental.
References
- 1.Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: The RNA 3′-end chronicles. J Mol Biol. 2011;412(5):793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabhi M, Rahmouni AR, Boudvillain M. In: RNA Helicases. Jankowsky E, editor. Cambridge, UK: RSC; 2010. pp. 243–271. [Google Scholar]
- 3.Adhya S, Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- 4.Burmann BM, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328(5977):501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 5.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328(5977):504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen R, Challissery J, Muteeb G. Nus factors of Escherichia coli. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. 2008. eds Curtiss R III et al. (ASM, Washington, DC), Module 4.5.3.1, www.ecosal.org. Accessed January 18, 2012.
- 8.Cardinale CJ, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320(5878):935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pósfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312(5776):1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 10.Washburn RS, Gottesman ME. Transcription termination maintains chromosome integrity. Proc Natl Acad Sci USA. 2011;108(2):792–797. doi: 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gowrishankar J, Harinarayanan R. Why is transcription coupled to translation in bacteria? Mol Microbiol. 2004;54(3):598–603. doi: 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 12.Harinarayanan R, Gowrishankar J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J Mol Biol. 2003;332(1):31–46. doi: 10.1016/s0022-2836(03)00753-8. [DOI] [PubMed] [Google Scholar]
- 13.Saxena S, Gowrishankar J. Compromised factor-dependent transcription termination in a nusA mutant of Escherichia coli: Spectrum of termination efficiencies generated by perturbations of Rho, NusG, NusA, and H-NS family proteins. J Bacteriol. 2011;193(15):3842–3850. doi: 10.1128/JB.00221-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anupama K, Leela JK, Gowrishankar J. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol Microbiol. 2011;82(6):1330–1348. doi: 10.1111/j.1365-2958.2011.07895.x. [DOI] [PubMed] [Google Scholar]
- 15.Gan W, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25(19):2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146(4):533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massé E, Drolet M. R-loop–dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J Mol Biol. 1999;294(2):321–332. doi: 10.1006/jmbi.1999.3264. [DOI] [PubMed] [Google Scholar]
- 18.Yu K, Roy D, Huang FT, Lieber MR. Detection and structural analysis of R-loops. Methods Enzymol. 2006;409:316–329. doi: 10.1016/S0076-6879(05)09018-X. [DOI] [PubMed] [Google Scholar]
- 19.Carles-Kinch K, George JW, Kreuzer KN. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 1997;16(13):4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudas KC, Kreuzer KN. UvsW protein regulates bacteriophage T4 origin-dependent replication by unwinding R-loops. Mol Cell Biol. 2001;21(8):2706–2715. doi: 10.1128/MCB.21.8.2706-2715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornberg A, Baker T. DNA Replication. New York: W.H. Freeman; 1992. [Google Scholar]
- 22.Peters JM, et al. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA. 2009;106(36):15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itaya M, et al. Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J Bacteriol. 1999;181(7):2118–2123. doi: 10.1128/jb.181.7.2118-2123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mooney RA, Schweimer K, Rösch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391(2):341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalissery J, et al. Interaction surface of the transcription terminator Rho required to form a complex with the C-terminal domain of the antiterminator NusG. J Mol Biol. 2011;405(1):49–64. doi: 10.1016/j.jmb.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 26.Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Biol. 2010;401(5):708–725. doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogoma T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61(2):212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahdi AA, Buckman C, Harris L, Lloyd RG. Rep and PriA helicase activities prevent RecA from provoking unnecessary recombination during replication fork repair. Genes Dev. 2006;20(15):2135–2147. doi: 10.1101/gad.382306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGlynn P, Savery NJ, Dillingham MS. The conflict between DNA replication and transcription. Mol Microbiol. 2012;85(1):12–20. doi: 10.1111/j.1365-2958.2012.08102.x. [DOI] [PubMed] [Google Scholar]
- 30.Long DT, Kreuzer KN. Fork regression is an active helicase-driven pathway in bacteriophage T4. EMBO Rep. 2009;10(4):394–399. doi: 10.1038/embor.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson SW, Benkovic SJ. Response of the bacteriophage T4 replisome to noncoding lesions and regression of a stalled replication fork. J Mol Biol. 2010;401(5):743–756. doi: 10.1016/j.jmb.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37(11):3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7(12):932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 34.Gabbai CB, Marians KJ. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair (Amst) 2010;9(3):202–209. doi: 10.1016/j.dnarep.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward DF, Gottesman ME. The nus mutations affect transcription termination in Escherichia coli. Nature. 1981;292(5820):212–215. doi: 10.1038/292212a0. [DOI] [PubMed] [Google Scholar]
- 36.Drolet M. Growth inhibition mediated by excess negative supercoiling: The interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006;59(3):723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 37.Selinger DW, et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18(12):1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 38.Dornenburg JE, DeVita AM, Palumbo J, Wade JT. Widespread antisense transcription in Escherichia coli. mBio. 2010;1(1) doi: 10.1128/mBio.00024-10. pii e00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomason MK, Storz G. Bacterial antisense RNAs: How many are there, and what are they doing? Annu Rev Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raghavan R, Sloan DB, Ochman H. Antisense transcription is pervasive but rarely conserved in enteric bacteria. mBio. 2012;3(4) doi: 10.1128/mBio.00156-12. pii e00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149(7):1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merrikh H, Zhang Y, Grossman AD, Wang JD. Replication-transcription conflicts in bacteria. Nat Rev Microbiol. 2012;10(7):449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perales R, Bentley D. “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36(2):178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: Resolving conflicts between replication and transcription. Mol Cell. 2012;45(6):710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Aguilera A, García-Muse T. R loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46(2):115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Mischo HE, et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41(1):21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42(6):794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyd D, Weiss DS, Chen JC, Beckwith J. Towards single-copy gene expression systems making gene cloning physiologically relevant: Lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J Bacteriol. 2000;182(3):842–847. doi: 10.1128/jb.182.3.842-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters , et al. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26(23):2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.