Abstract
The proteins harboring double-stranded RNA binding domains (dsRBDs) play diverse functional roles such as RNA localization, splicing, editing, export, and translation, yet mechanistic basis and functional significance of dsRBDs remain unclear. To unravel this enigma, we investigated transactivation response RNA binding protein (TRBP) consisting of three dsRBDs, which functions in HIV replication, protein kinase R(PKR)–mediated immune response, and RNA silencing. Here we report an ATP-independent diffusion activity of TRBP exclusively on dsRNA in a length-dependent manner. The first two dsRBDs of TRBP are essential for diffusion, whereas the third dsRBD is dispensable. Two homologs of TRBP, PKR activator and R3D1-L, displayed the same diffusion, implying a universality of the diffusion activity among this protein family. Furthermore, a Dicer–TRBP complex on dsRNA exhibited dynamic diffusion, which was correlated with Dicer’s catalytic activity. These results implicate the dsRNA-specific diffusion activity of TRBP that contributes to enhancing siRNA and miRNA processing by Dicer.
Keywords: RNA interference, single molecule FRET
The family of double-stranded RNA (dsRNA) binding proteins (dsRBPs) comprise one or more evolutionarily conserved dsRNA binding domains (dsRBDs) of 65–68 amino acids found in eukaryotes, prokaryotes, and viral-encoded products (1). The dsRBPs interact exclusively with dsRNA in a nonsequence-specific manner (1, 2). The dsRBP family includes Dicer, transactivation response (TAR) RNA binding protein (TRBP), protein activator of protein kinase R (PKR) (PACT), R3D1-L, dsRNA-dependent PKR, adenosine deaminase acting on RNA (ADAR), and nuclear factors associated with dsRNA (NFARs), which are implicated in gene silencing, antiviral immune response, mRNA editing, and transport (3). Such functional diversity is not surprising in light of the majority of cellular and viral RNA species that likely contain extensive dsRNA segments, primarily in the context of 3D RNA structures (4). However, it remains puzzling why most dsRBPs require multiple dsRBDs, as truncation or mutation of one dsRBD greatly diminishes or completely abolishes the protein’s ability to bind dsRNA (5, 6) and their biological function (7–9). Here, we focused our study on the mechanism of the simplest multiple dsRBD-containing proteins, TRBP, and its homologs, PACT and Loquacious (Loqs)–PB [also known as R3D1-L (long)].
TRBP was initially isolated from a HeLa cell expression library using TAR RNA as a binding probe (10). TRBP plays many different roles, including inhibition of PKR in antiviral immune signaling (11), activation of HIV-1 gene expression by binding TAR RNA (12), and regulation of cell growth (13). More recently, TRBP was also identified as an integral component of RNA-induced silencing complexes (RISCs) (14) that include TRBP, Dicer, and Argonaute 2 (Ago2). As an essential partner of Dicer, TRBP is required for optimal gene silencing induced by small interfering RNAs (siRNAs) and microRNAs (miRNAs) (15), although in one study it was dispensable for siRNA function (16). The diverse functions of TRBP are based on its ability to bind dsRNA, mediated by dsRBDs. Therefore, it is crucial to understand how TRBP interacts with dsRNA as a first step toward deciphering its role in higher-order biological processes. Moreover, TRBP consists of three dsRBDs, where the first two N-terminal dsRBDs bind dsRNA and the third C-terminal dsRBD interacts with other partner proteins, but it remains unclear why two dsRBDs are required for interaction with dsRNA. Using single molecule fluorescence detection, we sought to determine the fundamental behavior of TRBP upon dsRNA binding and how it may contribute to the biological function of its partner protein, Dicer, when in a complex. Here, we report an unanticipated dsRNA diffusion behavior of TRBP, which requires two dsRBDs, and its correlation with the role of TRBP in promoting Dicer-induced RNA cleavage. Furthermore, the diffusion activity is conserved in orthologous members of tandem dsRBD-containing proteins, indicating that diffusion may be a general mechanism by which dsRBPs mediate diverse biological processes.
Results
Observation of TRBP Diffusion on dsRNA.
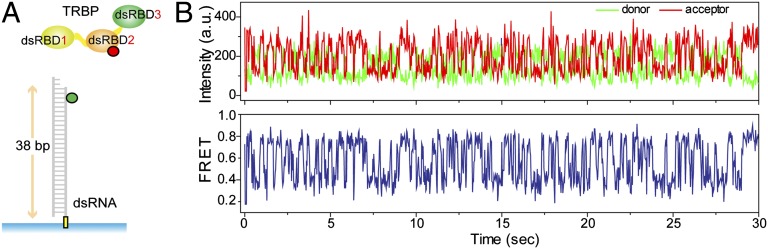
TRBP possesses three dsRBDs, the N-terminal two of which (dsRBD1 and 2) bind dsRNA tightly (17), whereas the third dsRBD (dsRBD3) participates in higher order complex assembly with proteins such as PKR and Dicer (18, 19). To examine the interaction between TRBP and dsRNA by single molecule fluorescence resonance energy transfer (smFRET) (20), we prepared a donor (Cy3, green)-labeled dsRNA and an acceptor (Alexa 647, red)-labeled TRBP. We immobilized the RNA via biotin–neutravidin linkage and added the labeled TRBP to single molecule imaging surface (Fig. 1A). Unexpectedly, we observed rapid FRET fluctuations caused by distance changes between TRBP and one end of the dsRNA (Fig. 1B). The FRET fluctuation doesn’t come from repetitive TRBP binding and dissociation because the FRET fluctuates between 0.3 and 0.8, rather than going down to 0 as expected if dissociation is occurring, as shown in Fig. S1 A and B (Fig. 1A). This repetitive distance change could result either from a diffusion of TRBP on dsRNA or a conformational change of TRBP’s subdomain. To differentiate between the two possibilities, we labeled TRBP site-specifically at its N terminus, which is expected to contact dsRNA tightly. This protein also yielded FRET fluctuations highly analogous to the nonspecifically labeled TRBP shown in Fig. 1B (Fig. S2A). The similarity in FRET fluctuation regardless of a fluorophore labeling position implies that the FRET fluctuations are likely due to the movement of the whole TRBP, rather than its subdomain’s motion relative to the static dsRNA-bound domain. Furthermore, the initial FRET values are heterogeneous due to the binding of TRBP in a nonsequence-specific manner (Fig. S2B).
Fig. 1.
TRBP’s interaction with dsRNA at the single-molecule level. (A) Alexa 647(red)-labeled TRBP was added to an immobilized Cy3(green)-labeled dsRNA, and their interaction was visualized by TIRF microscopy. (B) Repetitive FRET fluctuation was observed at the single molecule level without TRBP dissociation from dsRNA, reflecting a repetitive distance change between TRBP and the end of dsRNA.
To test further whether the observed FRET fluctuation could arise from diffusion of TRBP along dsRNA, we performed one-color protein-induced fluorescence enhancement (PIFE) (21) (Fig. S3 A and B) and three-color FRET (22) assays (Fig. S3 C–E). In the PIFE assay, the intensity fluctuation of Cy3 in the absence of Cy5 indicates that TRBP comes in contact with the fluorophore repeatedly. In three-color FRET assay, the anti-correlated changes between the two acceptors (Cy5 and Cy7) located at both ends of dsRNA indicates that the donor (Cy3)-labeled TRBP is moving across the dsRNA from one end to the other in succession. Again, we rule out the possibility of TRBP association and dissociation because the anticorrelated change between the two acceptor dyes (Cy5 and Cy7) can only result from the continuous and periodic movement of the donor labeled TRBP from one end to the other end of dsRNA axis. Both assays independently support the conclusion that TRBP diffuses on dsRNA. It is noteworthy that this activity was ATP independent, indicating that the diffusion of TRBP does not require an external energy source. Furthermore, this activity likely arises from a single binding event of TRBP based on the observation that the same activity persists even after removal of excess protein by buffer wash. We observed that the fluorophore labeling of TRBP didn’t affect its binding affinity to dsRNA (Fig. S4). Here, we use the term “diffusion” to ascribe 1D diffusion of TRBP on dsRNA (23).
dsRNA Length Dependence of TRBP Diffusion.
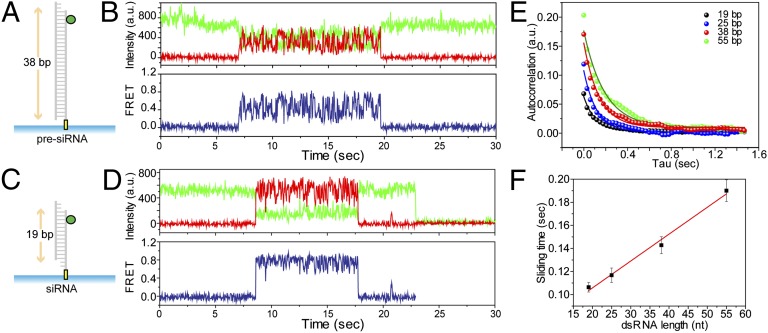
We tested whether the length of the dsRNA could modulate the diffusion activity of TRBP. We prepared dsRNAs of 19, 25, 38, and 55 bp each labeled with a donor at one end. TRBP diffusion on 38 bp (Fig. 2A) results in larger amplitude of FRET change (Fig. 2B) compared with that observed using 19 bp (Fig. 2 C and D), suggesting that TRBP diffuses along the entire length of the dsRNA. To test this hypothesis, we performed an autocorrelation analysis of the FRET fluctuations obtained for all four dsRNAs of different lengths (Fig. 2E). As a way of signal processing, the autocorrelation gives the magnitude of signal change as its initial value and the rate of signal change as its decay time. The initial values of the autocorrelation curve, 0.07, 0.12, 017, and 0.20, were obtained for 19, 25, 38, and 55 bp dsRNA, respectively, indicating that a longer dsRNA displayed larger distance changes as expected. We also confirmed the result by comparing the average FRET histograms obtained from 19 and 55 bp dsRNAs, which showed a shift toward the low FRET state in the case of longer dsRNA (Fig. S5A). Furthermore, the rates calculated from an exponential fitting of each autocorrelation curve, 0.106, 0.117, 0.143, and 0.190 s of diffusion time for 19, 25, 38, and 55 bp dsRNA, respectively, indicate that it takes a longer time to diffuse on the longer dsRNA (Fig. 2F). We shortened the dsRNA length further to determine the minimal length of dsRNA required for TRBP’s diffusion and found that TRBP didn’t bind dsRNA as short as 12 bp and did bind but not diffuse on 15 bp (Fig. S5B). Taken together, our data demonstrate that the observed FRET fluctuation is due to TRBP’s diffusion movement along the entire RNA rather than a conformational change of TRBP’s subdomain.
Fig. 2.
TRBP diffuses on the entire length of dsRNA. (A–F) TRBP diffuses on dsRNA in a length-dependent manner. (A–D) Diagrams of RNA substrates tested (38 bp dsRNA and 19 bp dsRNA with 2 nt 3′ overhang) and their representative FRET traces. Larger FRET changes were observed with a longer dsRNA. (E) Autocorrelation analysis on FRET signal using dsRNAs with four different lengths: 19, 25, 38, and 55 bp. The initial value of the autocorrelation curve (0.07, 0.12, 0.17, and 0.20 for 19, 25, 38, and 55 bp dsRNA, respectively) indicates a larger distance change with a longer dsRNA. More than 100 TRBP diffusion events were analyzed for each dsRNA. (F) Diffusion time (±SD) calculated by an exponential fit to an autocorrelation curve as shown in Fig. 2E displays a longer diffusion time for a longer dsRNA.
TRBP Diffuses Exclusively on dsRNA.
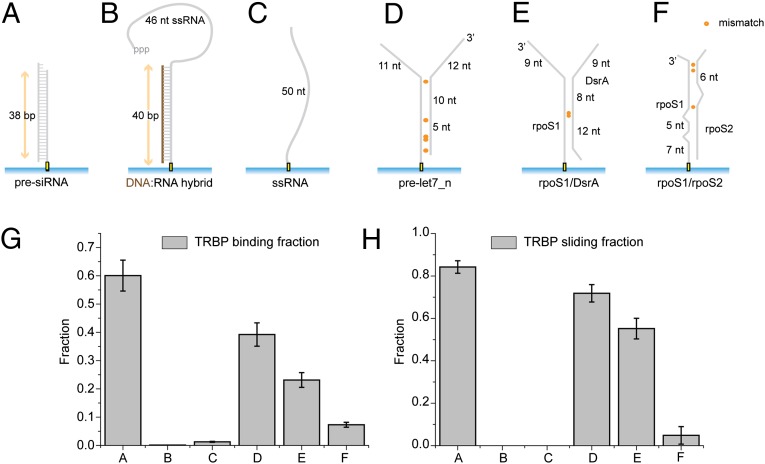
We varied the composition of RNA substrates to investigate the substrate specificity of TRBP diffusion (Fig. 3 A–F). Although TRBP showed ∼60% binding to 38 bp dsRNA (presiRNA), it did not show any binding or movement on a DNA-RNA hybrid or a single-stranded RNA (ssRNA), indicating that both strands of dsRNA are required for TRBP binding (Fig. 3 G and H). It also indicates that A-form structure of nucleic acids is not sufficient for TRBP’s binding because TRBP didn’t bind to DNA-RNA heteroduplex exhibiting A-form duplex structure. Consistent with this possibility, TRBP diffusion behavior was observed only when bound to dsRNA, in which most or all nucleotides are predicted to form canonical Watson–Crick base pairs, and not to dsRNAs with more complex secondary structure such as bulges and loops (Fig. 3H).
Fig. 3.
Substrate specificity of TRBP binding and its diffusion along dsRNA. (A–F) PresiRNA, DNA–RNA heteroduplex, ssRNA, and three dsRNAs with different secondary structure were prepared for testing the substrate specificity. (G) The fraction (±SEM) of the substrate (A–F) bound to TRBP was calculated from ∼5,000 substrate molecules at 10 nM TRBP concentration. TRBP does not bind to DNA–RNA heteroduplex and ssRNA, but binds to all dsRNAs to varying degrees at 10 nM TRBP concentration. (H) The fraction (±SEM) of diffusion among TRBP-bound substrates shows that TRBP diffuses on dsRNAs with several single mismatches but not on dsRNA with more complex secondary structures.
Two dsRBDs Are Responsible for Diffusion Activity.
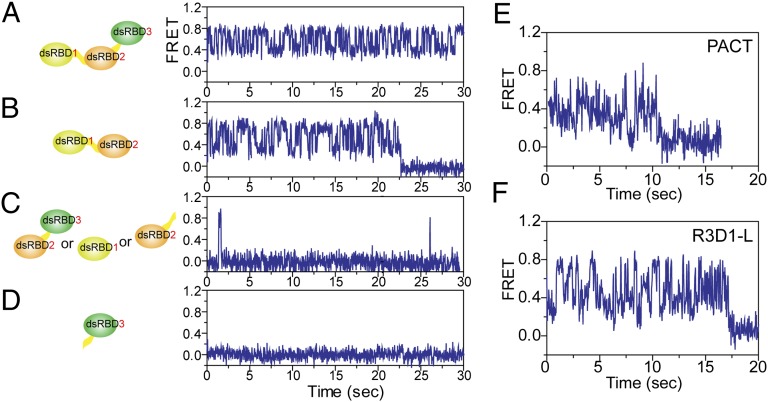
We generated truncation mutants of TRBP to investigate which of three dsRBDs give rise to the diffusion activity. Based on the dissociation constants of individual dsRBD1 (220 nM) and dsRBD2 (113 nM) to dsRNA, which are three orders of magnitude higher than that of dsRBD1+dsRBD2 (0.25 nM) (17), it is clear that both dsRBD1 and dsRBD2 are required for an efficient binding to dsRNA. In addition, both dsRBD1 and -2 play a functionally important role with respect to RNA processing (14, 24, 25), whereas dsRBD3 serves as a connector to Dicer (18, 19, 26). Consistent with these previous findings, we find that the deletion of dsRBD3 did not interfere with the observed diffusion behavior, whereas the deletion of dsRBD1 or dsRBD2 completely abrogated the diffusion activity (Fig. 4 A–D). We note that the short-lived binding observed in Fig. 4C represents an extremely rare event at 10 nM protein that we used for most of our assays, which is likely due to these mutants exhibiting low binding affinity (Fig. S6). Therefore, we conclude that the intact dsRBD1 and dsRBD2 are directly responsible for the diffusion movement of TRBP and that dsRBD3 is dispensable. This result also implies that the diffusion activity requires intact tandem dsRBD1 and -2, and one dsRBD alone is insufficient. We hypothesize that one dsRBD’s binding affinity is not strong enough to show diffusion, whereas the affinity of two dsRBDs is sufficient to support this activity. However, the three orders of magnitude difference in the binding affinity between one dsRBD and two dsRBDs (dsRBD1+2) suggests that it is not due to a simple summation of two independent binding bodies but a new conformational state of two dsRBDs that contributes to nonadditive binding affinity to dsRNA as well as the unique diffusion activity along dsRNA.
Fig. 4.
dsRBD1 and -2 as indispensable components for diffusion. (A) Representative single-molecule trace of wild-type TRBP diffusion on 38 bp dsRNA. (B–D) Several truncation mutants of TRBP show that both dsRBD1 and -2 are required for diffusion. (B) DsRBD1/2 shows a similar FRET trace as those from wild-type TRBP. (C) Short-lived binding but no diffusion was observed with dsRBD2/3, dsRBD1 only, and dsRBD2 only. (D) DsRBD3 does not bind to dsRNA. (E) Representative FRET traces of Alexa 647–labeled PACT with Cy3-labeled 38 bp dsRNA showing its repetitive diffusion motion. (F) R3D1-L also showed a similar diffusion motion on 38 bp dsRNA as seen in the wild-type TRBP.
Conservation of Diffusion Activity in PACT and R3D1-L.
We tested if this diffusion activity is conserved in orthologous proteins with tandem dsRBDs. For this, we purified PACT, another partner protein of human Dicer (27) as well as an activator of PKR, and R3D1-L, a cofactor of Dicer-1 found in Drosophila (28–30). Both of these proteins possess three dsRBDs analogous to those in TRBP. In smFRET experiments similar to those described for TRBP previously, both PACT and R3D1-L exhibited behavior similar to that of TRBP and consistent with diffusion on dsRNA substrates (Fig. 4 E and F). The finding that two additional proteins possess dsRNA diffusion activity strongly suggests that this behavior may be an intrinsic feature of these types of RNA-binding proteins.
TRBP-Driven Diffusion of Dicer–TRBP Complex.
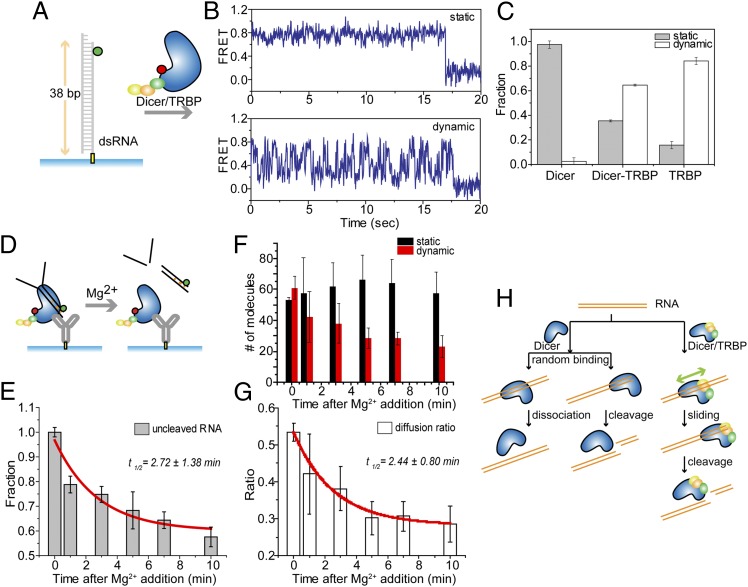
We asked if TRBP diffusion contributes to the function of its partner protein, Dicer. To test whether TRBP diffusion occurs in the context of a Dicer–TRBP complex, we analyzed a reconstituted sample of Dicer–TRBP in smFRET assays in which Dicer–TRBP and dsRNA substrates were labeled as before (Fig. 5A). Upon association with dsRNA, which is detected by the appearance of FRET, some Dicer–TRBP binding events produced a constant FRET signal, whereas others produced a fluctuating FRET signal, consistent with two populations of complexes exhibiting either static binding or diffusion on dsRNA, respectively (Fig. 5B). The molecules exhibiting static binding are not likely due to Dicer alone because Dicer’s binding affinity to dsRNA is at least 10 times lower than Dicer-TRBP as quantified by fluorescence spot numbers (Fig. S7). In these assays, we found that about 60% of the Dicer–TRBP complex also exhibits a smFRET fluctuation pattern that is consistent with diffusion on dsRNA, although almost none (< 5%) of Dicer alone and most (>85%) of TRBP alone does (Fig. 5C), suggesting that TRBP induces the diffusion of Dicer–TRBP complex.
Fig. 5.
Dicer–TRBP diffuses on dsRNA and thereby promotes dsRNA cleavage activity. (A and B) Alexa 647–labeled Dicer–TRBP complex exhibits two populations—static vs. dynamic FRET. Dynamic FRET changes similar to those observed for TRBP only suggest that Dicer–TRBP also diffuses on dsRNA. (C) Fraction (±SEM) of static and dynamic population for Dicer, Dicer–TRBP, and TRBP. Most Dicer showed a static binding, whereas about 60% of Dicer–TRBP and more than 85% of TRBP showed diffusion. (D) Diagram of the cleavage assay by pull-down of Dicer–TRBP using anti-His antibody. The cleavage reaction was initiated by adding Mg2+ on the immobilized Dicer–TRBP–RNA complex. (E) The normalized number of fluorescence spots (±SEM) decreases upon Mg2+ addition, which triggers the cleavage of dsRNA. More than 3,000 molecules were investigated. (F) The number of diffusion molecules (red bar) (±SEM) of Dicer–TRBP decreases over time upon Mg2+ addition, whereas the number of static molecules (black bar) (±SEM) doesn’t change. (G) The calculated ratio of diffusion molecules over both static and diffusion molecules exhibited the similar rate with the RNA cleavage rate by Dicer-TRBP, suggesting the correlation between the RNA cleavage and the diffusion by Dicer–TRBP. (H) The suggested model explains how Dicer–TRBP’s diffusion may help the dsRNA cleavage by positioning Dicer at the correct cleavage site and thereby enhancing the cleavage rate.
TRBP-Mediated Diffusion Correlates with Dicer-Induced RNA Cleavage.
Next, we tested dicing activity of dsRNA by Dicer–TRBP complex. To minimize possible surface effects that may interfere with the access of the Dicer–TRBP complex to dsRNA, we performed a complementary experiment in which the protein complex, rather than the dsRNA, was immobilized (Fig. 5D). The protein complex labeled with red fluorescent dye (Alexa 647) nonspecifically was surface-immobilized using an antibody against the Histidine (His)6-tag (31), and green dye (Cy3)–labeled RNA substrate was added, generating Dicer–TRBP–RNA ternary complex (Materials and Methods). The labeling position in the Dicer–TRBP complex can be either in Dicer or TRBP, but in either case will lead to FRET fluctuations regardless of the labeling position if the protein complex moves along dsRNA. All immobilized molecules are expected to be the Dicer–TRBP complex rather than TRBP or Dicer alone. First, they cannot be TRBP alone because the protein complex was immobilized via the His6-tag tag on Dicer. Second, they cannot be Dicer alone because the binding affinity of Dicer to the RNA substrate is too low to associate with dsRNA at the concentration used here. The Kd for Dicer is ∼1.8 nM, whereas the Kd for Dicer–TRBP is less than 50 pM (32). Fig. S7 also shows the lower RNA binding fraction of Dicer compared with Dicer–TRBP. Dicer itself couldn’t capture as much RNA as Dicer–TRBP, suggesting that the captured RNA signals in Fig. 5D and Fig. S8A come from Dicer–TRBP complexes. They appear as fluorescence spots on the imaging surface (Fig. S8A), and the number is expected to decrease as the RNA is cleaved away. We conducted control measurements to confirm that RNA binding is specific to the Dicer–TRBP complex and not to the surface or the antibody (Fig. S8A).
The RNA cleavage activity was monitored by the loss of fluorescent spots on the surface over time after the addition of Mg2+. The tested cleavable dsRNA contains the sequence of the well-characterized pre-miRNA, pre-let-7a, but with a nick in the middle of its hairpin loop, which is predicted by mFold to retain the same structure (33). The cleavage rate of Dicer–TRBP on this nicked pre-miRNA was consistent with that observed in a bulk cleavage assay with t1/2 of ∼3 min (Fig. 5E) (32). Further analysis of individual single molecule traces revealed both static binding and dynamic diffusion population of molecules. We counted these two classes of molecules over the time interval corresponding to active RNA cleavage after Mg2+ addition.
Surprisingly, our data indicate that the diffusing molecules, but not the static molecules, are selectively lost (Fig. 5F). The calculated ratio of the diffusion molecules over both static and diffusion molecules exhibited the disappearance rate following the same kinetic rate of dsRNA cleavage (Fig. 5G). There are two possible explanations for the two different populations of Dicer–TRBP complex. The tested RNA substrate is asymmetric because only one end of RNA is available for Dicer’s cleavage, whereas the other end is blocked by the imaging surface or flanked. Dicer’s Piwi-Argonaute-Zwille (PAZ) domain requires a terminal dsRNA end to bind, resulting in RNA cleavage at a position 21–23 bp away from it. If Dicer binds in the opposite orientation, Dicer cannot cleave the dsRNA because the PAZ domain cannot position the RNA correctly for cleavage. Molecules positioned in this wrong orientation may explain the static population that didn’t get cleaved over time. Another possibility is that there are two types of associations between Dicer and TRBP, one that inhibits diffusion of the complex and the other that permits it. In this model, the inhibitory association will not allow cleavage unless the binding occurs in the correct position where PAZ domain engages with the open end of the RNA. In contrast, the diffusing molecules can slide along dsRNA to search for the cleavage site. Both scenarios suggest that only the diffusing Dicer–TRBP complex cleaves the RNA substrate, suggesting that TRBP-driven diffusion may play a role in enhancing the cleavage rate of Dicer.
To check for nonspecific cleavage activity, the RNA substrate was incubated for 30 min before the cleavage reaction was triggered by adding magnesium (Mg2+). Experiments carried out in the absence of Mg2+ do not show any change in the number of RNA molecules bound to Dicer–TRBP. When Dicer–TRBP was provided with a noncleavable dsRNA, binding was observed without loss of fluorescent spots during a 1 h period (Fig. S8B). Cumulatively, our finding suggests that TRBP-driven diffusion activity of Dicer–TRBP complex is correlated with its dsRNA cleavage activity.
Discussion
Herein, we present a case of a protein family that diffuses exclusively on dsRNA. Such movement can only be detected by real-time monitoring of individual proteins. It is interesting to note that TRBP, PACT, and R3D1-L that contain tandem dsRBDs possess the intrinsic ability to diffuse on dsRNA, whereas Dicer with only one dsRBD shows transient static binding without diffusion on dsRNA. The diffusion activity seen in three dsRBPs here may represent a general mode of multiple dsRBDs. Diffusion activity tested on TRBP truncation mutants also confirms that one dsRBD is insufficient for its diffusion. The substrate specificity shown in Fig. 3H in which TRBP diffusion was observed in dsRNA with internal mismatches but not with more complex secondary structures may serve as a basis for its RNA scanning function. It is noteworthy that miRNAs also contain internal mismatches on which TRBP can diffuse.
Reduced dimension in diffusion has been suggested to increase the efficiency of protein–nucleic acid interaction. The association and dissociation kinetics of Lac repressor protein showed that it searches for its target not just by a 3D random collision but also by a 1D diffusion (34, 35), which was supported by theoretical approaches (36, 37) Recently, single molecule studies have identified proteins that diffuse on nucleic acids. Rad51 diffuses laterally on dsDNA (38), DNA glycosylase 1 (hOgg1) diffuses on dsDNA by 1D diffusion (23), reverse transcriptase of HIV shuttles back and forth on DNA-RNA hybrid (39), single-stranded DNA binding protein diffuses on single-stranded DNA (40), and DNA repair protein, Msh2–Msh6, also diffuses on dsDNA (41). Herein, we present a case of a protein family that diffuses exclusively on dsRNA. This motion is ATP-independent, unlike RIG-I, which translocates on dsRNA fueled by ATP (42–45). It is interesting and intriguing to note that TRBP, PACT, and R3D1-L, which possess the intrinsic ability to diffuse on dsRNA, are multiple dsRBD-containing proteins as well as cofactors of Dicer in different organisms.
This diffusion activity arising from multiple tandem dsRBDs could at least, in part, unravel the unsolved biological questions regarding the role of dsRBPs. In vitro, TRBP facilitates Dicer’s cleavage of pre-siRNAs or pre-miRNAs (18, 33), but how it contributes to the function of Dicer has remained unknown. Based on the correlation between TRBP-driven diffusion of Dicer–TRBP complex and its cleavage activity, we propose that TRBP-induced diffusion could aid in positioning Dicer at the proper cleavage site (Fig. 5F), resulting in an enhanced cleavage rate of Dicer–TRBP compared with Dicer alone (18, 33). Dicer by itself will find the cleavage site through multiple trials of random binding and dissociation, whereas Dicer–TRBP complex can find it more efficiently through a diffusion mechanism (Fig. 5F). In other words, Dicer–TRBP diffusion can serve to scan pre-siRNA and pre-miRNA substrates and to facilitate Dicer’s catalytic activity by locating the complex at the RNA cleavage site with an improved efficiency and precision (46, 47). It is also likely that TRBP’s diffusion plays another role in RNA-mediated gene silencing, for instance a recruitment of Dicer–TRBP complex to Ago2 (10) or guide strand selection by dsRNA scanning (48, 49).
Furthermore, TRBP and PACT interact with PKR, a major antiviral protein (50, 51), where TRBP inhibits whereas PACT enhances PKR’s activity (11, 52). In light of the diffusion activity exhibited by both TRBP and PACT, it will be interesting to elucidate the distinct mechanisms that lead to disparate regulatory responses in PKR and in the immune response. Our study also provides insight in the role of TRBP in HIV infection as TRBP binds and sequesters the TAR RNA of HIV (53).
Materials and Methods
Protein Purification.
N-terminal His6–Dicer was purified as previously described (54) with several modifications. TRBP, PACT, and their truncations were cloned as cleavable N-terminal His6–Maltose binding protein (MBP) fusions and R3D1-L as a cleavable N-terminal His6–Glutathion S-transferase (GST) fusion. Each was purified separately from bacterial overexpression using methods previously described (54). For details, see SI Materials and Methods.
Dicer–TRBP Reconstitution.
A total of 705 μg (3 nmol) of Dicer and 550 μg (11 nmol) of TRBP were mixed in 250 μL (final volume) of gel filtration buffer. The protein solution was incubated on ice for 1 h and then was applied to a Superose 6 10/30 column (Amersham Pharmacia) equilibrated in gel filtration buffer. Fractions were analyzed by SDS/PAGE, and those fractions containing the complex were pooled and concentrated to 1.5 mg/mL
RNA Constructs Preparation and Protein Labeling.
The sequences of all RNA constructs were displayed in Table S1; 25RNA, 40RNA, and 55RNA were purchased from Dharmacon, and 3′-biotin or 3′-DY547 was incorporated in the process of each RNA synthesis. RNA constructs containing 5′ triphosphate were in vitro transcribed and biotinylated at 3′ end. All other RNA constructs were synthesized and HPLC-purified from Integrated DNA Technology Inc. with proper chemical modifications of each RNA such as an internal C6 amine modifier dT, biotin, and/or Cy3 incorporation at the 5′ and/or 3′ end.
With presiRNA (38RNA), we showed the diffusion activity by TRBP and its truncation mutants by two-color FRET as well as three-color FRET. siRNA, 25RNA, presiRNA, 40RNA, and 55RNA were for investigating the length dependence of TRBP’s diffusion activity, and DNA-RNA, 50ssRNA, DsrA, rpoS1, rpoS2, and prelet-7_n for the substrate structure dependence. For the cleavage assay by Dicer–TRBP complex, nonbiotinylated prelet-7_n was examined.
For smFRET assay, we labeled RNAs and proteins with a fluorophore. Details of RNA and protein labeling can be found in SI Materials and Methods.
Immobilization of RNA and Protein.
For the smFRET assays, RNAs and proteins were immobilized on a polyethylene glycol (PEG)-coated quartz surface through neutravidin–biotin interaction. A total of 0.05 mg/mL of neutravidin was incubated for 5 min and then 30–50 pM biotinylated RNA in T50-BSA buffer (10 mM Tris, pH 8, 50 mM NaCl, and 0.1 mg/mL BSA) was added for RNA immobilization. Between10–100 nM of protein was added to the immobilized RNA for the diffusion assay. For the RNA cleavage assay, N-terminal His6-tag protein was immobilized on a neutravidin-treated PEG surface through biotinylated anti-Penta-His antibody (Qiagen). Between 1–2 μg/mL of the antibody was incubated for 5 min, following the incubation of 0.05 mg/mL neutravidin for 5 min on a PEG surface. Then, 10 nM of His6-tag protein was incubated for another 5 min, and 1–2 nM of RNA was added afterward for the single-molecule assays.
smFRET Assays.
Single-molecule detection of protein dynamics and functional assay was achieved by a homemade wide-field total internal reflection fluorescence (TIRF) microscopy. Details of smFRET assays can be found in SI Materials and Methods.
Data Analysis: Auto-Correlation and Cross-Correlation.
The auto-correlation function was analyzed by MATLAB program, and the equation of auto-correlation used is as below:
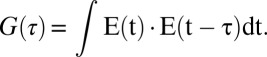 |
E(t) represents FRET, and the FRET auto-correlation, G(τ), gives us an average diffusion rate because FRET changes in our system reflect the diffusion of a protein. More than 30 molecules were analyzed for each auto-correlation curve, and it was fitted with a single exponential decay to obtain an average diffusion rate of the molecules.
In the three-color FRET experiment, the cross-correlation between the fluorescence intensity of Cy5 and the one of Cy7 was analyzed using the following equation:
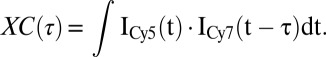 |
Supplementary Material
Acknowledgments
We thank T. Ha, H. Hwang, and Y. Qiu for helpful discussions and R. Zhou, Y. Ishitzuka, Q. Xu, and C. Xu for careful review of the manuscript. This work was funded by the American Cancer Society (Research Scholar Grant RSG-12-066-01-DMC), the Human Frontier Science Program (RGP0007/2012), and the US National Science Foundation Physics Frontiers Center Program (0822613) through the Center for the Physics of Living Cells (to S.M.) and National Institutes of Health Grants AI083025 (to S.M and H.R.K.) and GM073794-06 (to J.A.D. and M.A.K). J.A.D. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.L.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212917110/-/DCSupplemental.
References
- 1.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17(24):7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12(11):5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle M, Jantsch MF. Distinct in vivo roles for double-stranded RNA-binding domains of the Xenopus RNA-editing enzyme ADAR1 in chromosomal targeting. J Cell Biol. 2003;161(2):309–319. doi: 10.1083/jcb.200301034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders LR, Barber GN. The dsRNA binding protein family: Critical roles, diverse cellular functions. FASEB J. 2003;17(9):961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 5.Schmedt C, et al. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR) J Mol Biol. 1995;249(1):29–44. doi: 10.1006/jmbi.1995.0278. [DOI] [PubMed] [Google Scholar]
- 6.Krovat BC, Jantsch MF. Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J Biol Chem. 1996;271(45):28112–28119. doi: 10.1074/jbc.271.45.28112. [DOI] [PubMed] [Google Scholar]
- 7.Micklem DR, Adams J, Grünert S, St Johnston D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19(6):1366–1377. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuldt AJ, et al. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12(12):1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen CP, et al. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 1998;12(12):1837–1846. doi: 10.1101/gad.12.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251(5001):1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 11.Daher A, et al. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J Biol Chem. 2001;276(36):33899–33905. doi: 10.1074/jbc.M103584200. [DOI] [PubMed] [Google Scholar]
- 12.Dorin D, et al. The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit the dsRNA-dependent kinase PKR. J Biol Chem. 2003;278(7):4440–4448. doi: 10.1074/jbc.M208954200. [DOI] [PubMed] [Google Scholar]
- 13.Benkirane M, et al. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16(3):611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Betancur JG, Tomari Y. Dicer is dispensable for asymmetric RISC loading in mammals. RNA. 2012;18(1):24–30. doi: 10.1261/rna.029785.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita S, et al. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Sci. 2011;20(1):118–130. doi: 10.1002/pro.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haase AD, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6(10):961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J Mol Biol. 2008;384(4):967–979. doi: 10.1016/j.jmb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437(7063):1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 21.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108(18):7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87(2):1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103(15):5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daviet L, et al. Analysis of a binding difference between the two dsRNA-binding domains in TRBP reveals the modular function of a KR-helix motif. Eur J Biochem. 2000;267(8):2419–2431. doi: 10.1046/j.1432-1327.2000.01256.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301(5641):1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 26.Gatignol A, Buckler C, Jeang KT. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol Cell Biol. 1993;13(4):2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, et al. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25(3):522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang F, et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19(14):1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Förstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3(7):e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3(7):e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473(7348):484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol. 2010;404(3):392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 35.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor—operator interaction: Kinetic measurements and conclusions. Biochemistry. 1981;20(24):6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 36.Berg OG, Ehrenberg M. Association kinetics with coupled three- and one-dimensional diffusion. Chain-length dependence of the association rate of specific DNA sites. Biophys Chem. 1982;15(1):41–51. doi: 10.1016/0301-4622(82)87015-4. [DOI] [PubMed] [Google Scholar]
- 37.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 38.Granéli A, Yeykal CC, Robertson RB, Greene EC. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc Natl Acad Sci USA. 2006;103(5):1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X. Slide into action: Dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322(5904):1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461(7267):1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17(8):932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323(5917):1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147(2):423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 44.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147(2):409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479(7373):423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311(5758):195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 47.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14(10):934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 48.Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell. 2011;43(1):110–121. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gredell JA, Dittmer MJ, Wu M, Chan C, Walton SP. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry. 2010;49(14):3148–3155. doi: 10.1021/bi902189s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clemens MJ, et al. PKR: Proposed nomenclature for the RNA-dependent protein kinase induced by interferon. J Interferon Res. 1993;13(3):241. doi: 10.1089/jir.1993.13.241. [DOI] [PubMed] [Google Scholar]
- 51.Meurs E, et al. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62(2):379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 52.Patel CV, Handy I, Goldsmith T, Patel RC. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J Biol Chem. 2000;275(48):37993–37998. doi: 10.1074/jbc.M004762200. [DOI] [PubMed] [Google Scholar]
- 53.Bannwarth S, Gatignol A. HIV-1 TAR RNA: The target of molecular interactions between the virus and its host. Curr HIV Res. 2005;3(1):61–71. doi: 10.2174/1570162052772924. [DOI] [PubMed] [Google Scholar]
- 54.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci USA. 2008;105(2):512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.