Abstract
Biosynthetic pathways of secondary metabolites from fungi are currently subject to an intense effort to elucidate the genetic basis for these compounds due to their large potential within pharmaceutics and synthetic biochemistry. The preferred method is methodical gene deletions to identify supporting enzymes for key synthases one cluster at a time. In this study, we design and apply a DNA expression array for Aspergillus nidulans in combination with legacy data to form a comprehensive gene expression compendium. We apply a guilt-by-association–based analysis to predict the extent of the biosynthetic clusters for the 58 synthases active in our set of experimental conditions. A comparison with legacy data shows the method to be accurate in 13 of 16 known clusters and nearly accurate for the remaining 3 clusters. Furthermore, we apply a data clustering approach, which identifies cross-chemistry between physically separate gene clusters (superclusters), and validate this both with legacy data and experimentally by prediction and verification of a supercluster consisting of the synthase AN1242 and the prenyltransferase AN11080, as well as identification of the product compound nidulanin A. We have used A. nidulans for our method development and validation due to the wealth of available biochemical data, but the method can be applied to any fungus with a sequenced and assembled genome, thus supporting further secondary metabolite pathway elucidation in the fungal kingdom.
Keywords: aspergilli, natural products, secondary metabolism, polyketide synthases
No other group of biochemical compounds holds as much promise for drug development as the secondary (nongrowth associated) metabolites (SMs). A review from 2012 (1) found that for small-molecule pharmaceuticals, 68% of the anticancer agents and 52% of the antiinfective agents are natural products, or derived from natural products. The fact that SMs are often synthesized as polymer backbones that are subsequently diversified greatly via the actions of tailoring enzymes sets the stage for combinatorial biochemistry (2), because their biosynthesis is modular.
Major groups of SMs include polyketides (PKs) consisting of -CH2-(C = O)- units, ribosomal and nonribosomomal peptides (NRPs), and terpenoids made from C5 isoprene units. These polymer backbones are, with the exception of ribosomal peptides, made by synthases or synthetases and are modified by a plethora of tailoring enzymes, including (de)hydratases, oxygenases, hydrolases, methylases, and others.
In fungi, these biosynthetic genes of secondary metabolism are organized in discrete clusters around the synthase genes. Although quite accurate algorithms are available for identification of possible SM biosynthetic genes, particularly PK synthases (PKSs), NRP synthetases (NRPSs), and dimethylallyl tryptophan synthases (DMATSs) (3, 4), the assignment and prediction of the members of the individual clusters solely from the genome sequence have not been accurate. Relevant protein domains can be predicted for some of the genes (e.g., cytochrome P450 genes) (5); however, genes in identified clusters often have unknown functions, which makes predicting their inclusion impossible. Furthermore, SM gene clusters often colocalize on the chromosomes (6), which makes separation of clusters solely based on gene function predictions difficult.
The efficient elucidation of the biosynthetic genes for each SM cluster has thus so far been based on laborious single gene deletion of each of the putative members and chemical profiling of the SMs of the deletion strains. This effort has been especially noticeable in the model fungus Aspergillus nidulans, which is presently the fungal species with the largest number (n = 25) of characterized SM synthases/synthetases, due to a massive effort by several groups (7–30). In recent studies, this fungus has also been shown to have cross-chemistry between gene clusters on separate chromosomes (8, 30). Although these reactions are highly interesting for combinatorial chemistry, the identification of gene clusters involved in cross-chemistry is cumbersome because it involves combinatorial deletion of SM synthetic genes, thus greatly increasing the potential number of candidates.
In this study, we propose a general “omics”-based method for the accurate determination of fungal SM gene cluster members. The method is based on an annotated genome sequence and a catalog of gene expression, a set of information that is readily available for many fungal species and can easily be generated for more. To develop, benchmark, and validate this algorithm, we have used A. nidulans as a model organism, which is especially well-suited for this purpose due to the above-stated wealth of information. The algorithm is proven to be very powerful in identifying gene cluster members. We furthermore report an extension of the algorithm, which is proven to be successful in identifying cross-chemistry between gene clusters.
Results
Analysis of SMs A. nidulans on Complex Solid Medium Identifies 42 Compounds.
Initially, we evaluated the production of SMs on four different solid media [oatmeal agar (OTA), yeast extract sucrose (YES), Czapek yeast autolysate (CYA), and CYA with 50 g/L NaCl sucrose (CYAS); Materials and Methods] at 4, 8, and 10 d. The object of this was to identify a selection of media that (i) gave as many produced SMs as possible, (ii) showed one or more SMs unique to each medium, and (iii) had SMs that were only produced on two of the selected media.
These characteristics should allow us to have as many active gene clusters as possible, as well as ensuring unique production profiles for as many SM gene clusters as possible.
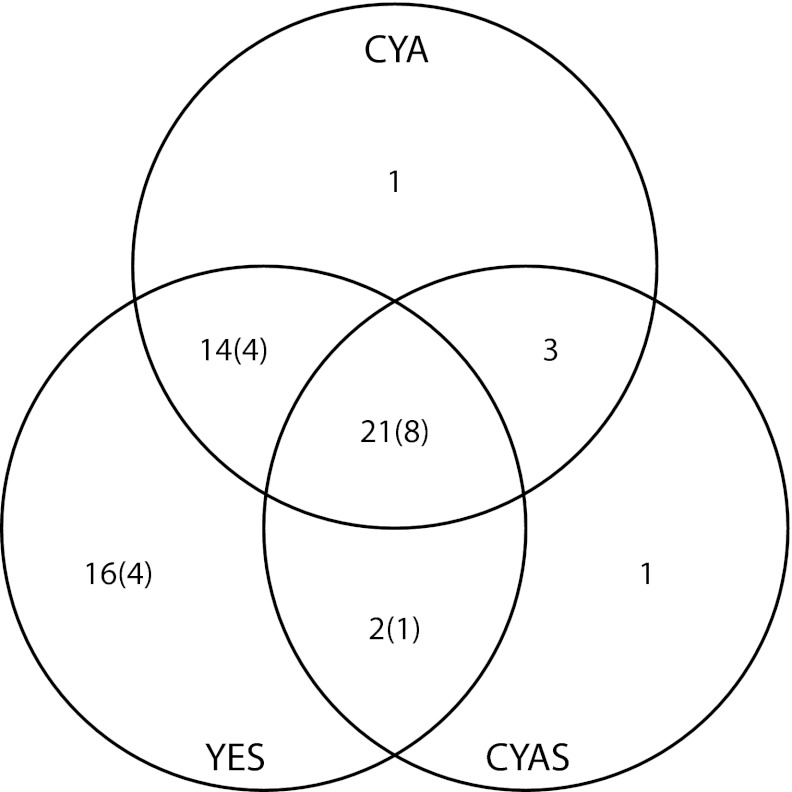
From this initial analysis, we selected the YES, CYA, and CYAS media for transcriptional profiling. On these media, we were able to separate and detect 59 unique SMs, of which we could name 42 by comparison with our extensive in-house library of microbial metabolites (31) and the AntiBase 2010 natural products database. The production profile of the compounds satisfied the three criteria listed above (Fig. 1, Fig. S1, and Dataset S1).
Fig. 1.
Venn diagram of SMs found on three different solid media. The number of different metabolites is sorted according to which media the metabolites have been identified on. The number of metabolites unable to be confidently identified are noted in parentheses. Details can be found in Dataset S1, and the chemical structures are illustrated in Fig. S1.
Generation of a Diverse Gene Expression Compendium for A. nidulans.
Samples were taken for transcriptional profiling from plates cultivated in parallel to those of the SM profiling above. RNA was purified, prepared for labeling, and hybridized to custom-designed Agilent Technologies arrays based on version 5 of the A. nidulans annotation (32).
The produced data were combined with previously published microarray data from A. nidulans bioreactor cultivations (33, 34) to form a microarray compendium spanning a diverse set of conditions, comprising 44 samples in total. The set includes four strains of A. nidulans. Four different growth media are included: three complex media (see above) and one minimal medium. Medium variations include five different defined carbon sources (ethanol, glycerol, xylose, glucose, and sucrose), as well as yeast extract. The combined compendium of expression data is available in Dataset S2.
Correlation-Based Identification of Gene Clusters.
To identify gene clusters efficiently around SM synthases, we developed a gene clustering score (CS) based on the Pearson product-moment correlation coefficient. Our CS gives a numerical value for correlation of the expression profile of a given gene with the expression profiles of the three immediate neighbor genes on either side. Only positive correlation is considered. Values for the CS are available in Dataset S2.
Statistical simulation of the distribution of CS on the given dataset showed that CS values ≥2.13 corresponded to a false-positive rate of 0.05 (Fig. S2). Therefore, CS ≥ 2.13 was used as a guideline for identifying the extent of gene clusters.
Prediction of the Extent of 51 Gene Clusters.
Evaluation of the size of the clusters around SM genes was performed using a precomputed list of 66 putative PKSs, NRPSs, and DMATSs from the secondary metabolite unique regions finder (SMURF) algorithm (3) based on the A. nidulans FGSC A4 gene set (35). In addition to these 66 genes, we added one prenyltransferase gene found in the primary literature (30) and three diterpene synthase (DTS) genes predicted by Bromann et al. (25), resulting in 70 putative biosynthetic genes. All 25 experimentally verified PKSs, NRPSs, DTSs, and prenyltransferases were found to be included in this list (Tables 1–3).
Table 1.
Prediction of PKS gene clusters
| Cluster size |
||||||
| GeneID | Gene | Compound (if known) | Predicted | Known | Medium | Ref(s). |
| AN0150 | mdpG | Monodictyphenone/emodin | 12 | 12 | Solid | (7–10) |
| AN7903 | Violaceol I and II | 12 | ? | Solid | (11) | |
| AN6448 | pkbA | 8 | ? | Solid | (24) | |
| AN7084 | 8 | Solid | ||||
| AN8209 | wA | Green conidial pigment | 6 | ? | Solid | |
| AN7909 | orsA | Orsellinic acid/F9975/violaceols | 5 | 5 | Solid | (11–14) |
| AN1784 | 4 | Solid | ||||
| AN9005 | 4 | Solid | ||||
| AN6000 | aptA | Asperthecin | 3 | 3 | Solid | (15) |
| AN6431 | 3 | Solid | ||||
| AN11191 | 2 | Solid | ||||
| AN7489 | 1 | Solid | ||||
| AN3273 | 1 | Solid | ||||
| AN2547 | easB | Emericellamide | 4 | 4 | Both | (16) |
| AN3230 | pkfA | Orsellinaldehydes | 6 | ? | Both | (24) |
| AN7071 | pkgA | Alternariol/isocoumarins | 7 | ? | Both | (24) |
| AN7825 | stcA | Sterigmatocystin | 24* | 25 | Both | (17–19) |
| AN7815 | stcJ | Sterigmatocystin | 24* | 25 | Both | (17–19) |
| AN8383 | ausA | Austinol | 7 | 4 | Both | (11, 24, 30) |
| AN2032 | pkhA | Unknown | 10 | ? | Liquid | (24) |
| AN2035 | pkhB | Unknown | 10 | ? | Liquid | (24) |
| AN8412 | adpA | Aspyridone | 7† | 8 | Liquid | (20) |
| AN6791 | 1 | Liquid | ||||
| AN8910 | 1 | Liquid | ||||
This table contains predicted PKSs as well as PKS-like genes (AN7489 and AN7815) and a PKS/hybrid gene (AN8412). The medium column describes under which type of medium (liquid, solid, or both) the cluster is expressed. For gene clusters with identified functions and gene members, the number of identified cluster members is given as well as references to the original papers. Further details on the cluster members and the expression profiles of the individual clusters may be found in Dataset S2 and Fig. S4. Chemical structures of all compounds may be found in Fig. S1.
*Difference seemingly due to the current gene calling diverging from the original paper from 1996 (17).
†Algorithm was not able to predict the inclusion of apdG, the outmost gene hypothesized to be a part of the cluster (20). The expression profile of apdG diverges from the rest of the cluster.
Table 3.
Prediction of gene clusters around prenyltransferases and diterpene synthases
| Cluster size |
|||||||
| GeneID | Type | Gene | Compound (if known) | Predicted | Known | Medium | Source |
| AN11194 | DMATS | 18 | Solid | ||||
| AN11202 | DMATS | 18 | Solid | ||||
| AN9259 | DMATS | 12 | 10 | Both | (30) | ||
| AN8514 | DMATS | tdiB | Terrequinone A | 3* | 5 | Solid | (21, 22) |
| AN11080 | DMATS | nptA | Nidulanin A | 1 | Both | This study | |
| AN10289 | DMATS | 1 | Solid | ||||
| AN6784 | DMATS | xptA | Variecoxanthone A | 1 | 1 | Solid | (8–10) |
| AN1594 | DTS | Ent-pimara-8(14),15-diene | 9 | 9 | Solid | (25) | |
| AN3252 | DTS | 7 | Solid | ||||
| AN9314 | DTS | 2 | Solid | ||||
This table contains predicted DMATSs, functionally prenyltransferases, and three DTSs predicted by Bromann et al. (25). The medium column describes on which type of medium (liquid, solid, or both) the cluster is expressed. For gene clusters with identified functions and gene members, the number of identified cluster members is given as well as references to the original papers. Further details on the cluster members and the expression profiles of the individual clusters may be found in Dataset S2, and Fig. S4. Chemical structures of all compounds may be found in Fig. S1.
*Extent of the gene cluster is predicted correctly. The difference is due to the absence of two of the genes on the legacy microarray data, which removes them from the prediction.
Table 2.
Prediction of NRPS gene clusters
| Cluster size |
||||||
| GeneID | Gene | Compound (if known) | Predicted | Known | Medium | Source |
| AN9226 | 18 | Solid | ||||
| AN6444 | 8 | Solid | ||||
| AN4827 | 7 | Solid | ||||
| AN8105 | 8 | Solid | ||||
| AN8513 | tdiA | Terrequinone A | 3* | 5 | Solid | (21, 22) |
| AN1242 | nlsA | Nidulanin A | 3 | Solid | This study | |
| AN6961 | 2 | Solid | ||||
| AN0016 | 1 | Solid | ||||
| AN10486 | 1 | Solid | ||||
| AN7884 | 14 | Both | ||||
| AN3495 | inpA | Unknown | 7 | 7 | Both | (25, 39) |
| AN3496 | inpB | Unknown | 7 | 7 | Both | (25, 39) |
| AN2545 | easA | Emericellamide | 4 | 4 | Both | (16) |
| AN2621 | acvA/pcbAB | Penicillin G | 3 | 3 | Both | (25, 27, 28) |
| AN3396 | mica | Microperfuranone | 3 | 3† | Both | (29) |
| AN2924 | 2 | Both | ||||
| AN10576 | ivoA | N-acetyl-6-hydroxytryptophan | 2 | 2 | Both | (23, 26) |
| AN0607 | sidC | Siderophores | 1 | 1 | Both | (55) |
| AN10297 | 1 | Both | ||||
| AN5318 | 1 | Both | ||||
| AN1680 | 1 | Liquid | ||||
| AN2064 | 1 | Liquid | ||||
| AN9129 | 1 | Liquid | ||||
| AN9291 | 1 | Liquid | ||||
This table contains predicted NRPSs as well as NRPS-like genes (AN3396, AN5318, and AN9291). The medium column describes under which type of medium (liquid, solid, or both) the cluster is expressed. For gene clusters with identified functions and gene members, the number of identified cluster members is given as well as references to the original papers. Further details on the cluster members and the expression profiles of the individual clusters may be found in Dataset S2, and Fig. S4. Chemical structures of all compounds may be found in Fig. S1.
*Extent of the gene cluster is predicted correctly. The difference is due to the absence of two of the genes on the legacy microarray data, which removes them from the prediction.
†Yeh et al. (29), who examined this cluster, found increased transcription of the two extra genes we predict, but they found them to be nonessential for microperfuranone production.
For each of the 70 biosynthetic genes, we examined the genes nearby for high CS values and inspected the expression profiles of the genes manually for additional validation and refinement. Apart from 12 genes that were silent under the conditions tested (Table S1), this allowed prediction of the sizes of gene clusters around 58 biosynthetic genes organized in 51 clusters and counting of a total of 254 genes included in the clusters (an example is shown in Fig. 2). The fact that we can map expression for 58 of the 70 biosynthetic genes (a large proportion of the gene clusters) is surprising, considering that many, or even the majority, of the gene clusters are reported to be silent under standard laboratory conditions (13, 14, 20, 36–38). An example of a cluster previously described as silent but identified here is the inpAB cluster (39). However, those cultivation experiments were conducted on liquid minimal medium and not on solid complex media, where we find that the expression from most of these genes is most pronounced. We therefore see the large number of active clusters as a confirmation of adequate diversity of the cultivation conditions in our microarray compendium.
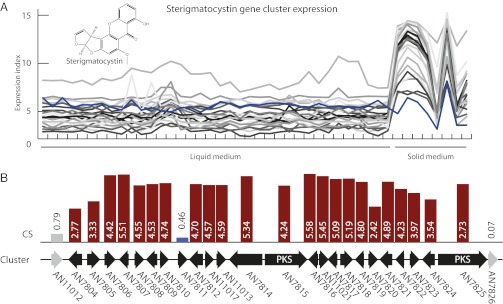
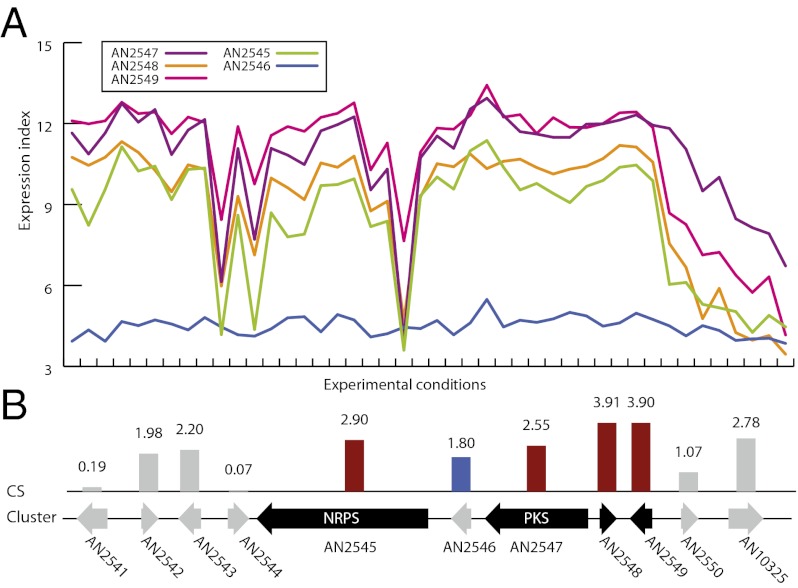
Fig. 2.
Identification of the sterigmatocystin biosynthetic cluster. (A) Gene expression profiles across 44 experiments for the 24 genes (marked in black in B) predicted to be in the sterigmatocystin biosynthetic cluster (liquid and solid cultures are marked for reference). The expression profile of AN7811(stcO) is marked in blue. (B) Illustration of the values of the gene CS for the 24 genes and the two immediate neighbors. Genes included in the predicted cluster are marked in black. AN7811(stcO) did not have a CS above the used cutoff of 2.13 denoting clustering but was added due to the similarity of the expression profile, as shown in blue. The predicted extent of the cluster corresponds with the cluster as originally described by Brown et al. (17), when correcting for the fact that the gene models have changed since then. Full data for all predicted clusters may be found in Dataset S2.
Next, we investigated how our cluster predictions matched those published in the literature. This comparison demonstrated that our algorithm generally predicts gene clusters with excellent accuracy. Specifically, we accurately predict the extent of 11 of the 16 known gene clusters (Tables 1–3). In two of the remaining 5 gene clusters, the difference is due to artifacts. For the gene sterigmatocystin cluster (Fig. 2), the difference of 24 genes relative to 25 genes is caused by differences in the current gene annotation compared with the original paper from 1996 (17). Changes in gene calling are also the reason for discrepancy in the terrequinone cluster, where our legacy microarray data only contain data for 3 of the 5 genes, thus impairing the prediction. For the three remaining cases, the 2 gene clusters involved in meroterpenoid (austinol and dehydroaustinol) biosynthesis and the aspyridone cluster, the divergence seems to be biological. For the austinol/dehydroaustinol double-cluster system, we predict 3 extra genes in one cluster (around AN8383) and 2 extra genes in the other cluster (around AN9259) in addition to genes identified by Lo et al. (30). We individually deleted the 3 extra genes (AN8375, AN8376, and AN8380) in the AN8383 cluster; however, apart from differences in the austinol/dehydroaustinol ratio, we could only confirm the results of Lo et al. (30) of these genes not being essential for austinol/dehydroaustinol biosynthesis (Fig. S3). Because the size of most of the clusters was accurately predicted by our algorithm, we speculate that some or all of the extra genes are involved in biosynthesis of derivatives of austinol/dehydroaustinol. In agreement with this scenario, it is not uncommon that newly detected compounds are linked to known PKS pathways. For example, shamixanthones and arugusins were recently discovered to be products derived from the monodictyphenone cluster (8, 11), and this cluster has been redefined several times (9, 10). For the remaining case, the apdG gene of the aspyridone cluster (20), misprediction of the cluster members is due to a complete divergence between the transcription profiles of apdG and the remainder of the gene cluster. In general, we conclude that the use of CS values in combination with inspection of the expression profiles is a very effective tool to predict the extent of gene clusters, because the borders of 13 of 16 clusters were accurately predicted (when predictions were adjusted to compensate for the two artifacts discussed above) and there was near-accurate prediction of all 16 clusters.
Diverse Gene Expression Compendium Is Important for Accurate Prediction.
To evaluate the compendium size needed for accurate predictions, we used principal component analysis (PCA) on our matrix of expression values (Dataset S2). Greater than 95% of the variation within the set can be described in the first three principal components. This suggests that a theoretical lower limit for this type of analysis would be three arrays if one could select conditions with a near-perfect difference in expression levels, ideally high, medium, and low expression for all genes, and with a maximum difference between all clusters and their surrounding genes. This would be nearly impossible to achieve for all clusters. However, if one is only interested in a single or a few gene clusters of interest, and has the appropriate prior knowledge, it should be possible to select three to five conditions and achieve accurate predictions. Very informative studies have been performed with two conditions, but the boundaries of the cluster can be difficult to determine (e.g., ref. 25).
To test how much it was possible to reduce our dataset, we used an unsupervised PCA-based analysis for incremental reduction of the dataset. In this, we found (unsurprisingly) that our biological replicate samples contain the smallest amount of unique information. Ten of 44 samples can be removed with only an approximately 10% loss in the data variation, and 25 of 44 samples (all replicates) can be removed with less than a 35% loss in data variation. The time sample series on a solid medium presented in this study were not reduced from the set until all biological replicates were reduced. We conclude that in selection of samples for cluster elucidation, one should sample as diversely as possible. Biological replicates are not cost-effective unless already available from prior studies.
Clustering of Synthase Expression Profiles Identifies Superclusters.
Recent work has identified two cases of cross-chemistry between clusters located on separate chromosomes. The production of austinol and derived compounds (the meroterpenoid pathway) has been shown to be dependent on two separate clusters (11, 30), and the biosynthesis of prenyl xanthones is dependent on three separate clusters (8). We were interested in seeing whether this is a general phenomenon and whether such cross-chromosomal “superclusters” could be detected using our expression data.
A full gene-to-gene comparison of expression profiles between all predicted NRPSs, PKSs, DTSs, and prenyl transferases found in the array data was conducted, and the genes were clustered (Fig. 3). This clustering is not based directly on the expression profiles, because expression index variation from silent conditions distorts clustering. Instead, we clustered on the basis of a Spearman-based score of similarity to the expression profiles of the other synthases, which effectively eliminates noise.
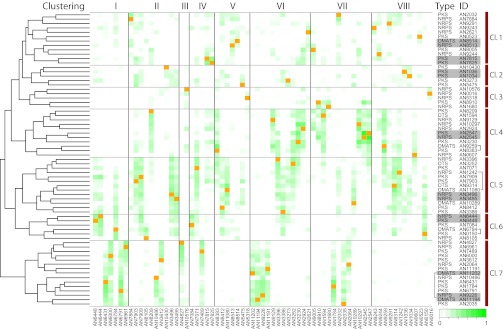
Fig. 3.
Cross-chromosomal clustering. Matrix diagram of the correlation between 67 predicted and known biosynthetic genes. Each square in the matrix shows the compounded squared Spearman correlation coefficient for comparison of the expression profile of the genes color-coded from 0 (white) to 1 (green). Genes are sorted horizontally according to their location on the chromosomes (marked in orange) and vertically according to their scores (Left, marked with a dendrogram). (Right) Genes located in the same clusters are highlighted with a gray box, which is connected with a gray bracket in one case. Genes with known cross-chemistry are marked with a black bracket. An example of cross-chemistry found in this study is marked with a red bracket. Seven putative superclusters are marked. Further details of the clusters may be found in Fig. S4.
The method is efficient for clustering the synthases and transferases according to shared products. Seven of eight sets of genes predicted to be in the same biosynthetic clusters by the method above are found to cluster together in this representation. The exception is AN2032 and AN2035, which do not cocluster due to very low signals from the AN2032 probes on the microarray. Furthermore, the clustering is accurate in terms of cross-chemistry. In examining the two examples of cross-chemistry between gene clusters, it is found that these are predicted correctly. The meroterpenoid pathway includes the PKS AN8383 and the DMATS AN9259, which are illustrated to colocate in Fig. 3. The other example is the prenylxanthone biosynthetic pathway, which includes the PKS AN0150 and the DMATS AN6784. These two genes are also found close to each other in Fig. 3.
We further use the maximum separation distance of two genes in the same biosynthetic cluster in the heat map of Fig. 3 as a cutoff distance for cross-chemistry. This allowed the genes to be sorted into seven larger superclusters. Details on the expression profiles of the individual clusters in each supercluster can be found in Fig. S4. Although we cannot directly separate tight coregulation from cross-chemistry with this method, the presence of these superclusters consisting of individual clusters with similar expression profiles suggests a larger extent of cross-chemistry in A. nidulans than what has been reported to date. To test the predictive power of this clustering further, we performed a gene deletion study within supercluster 5, which contains clusters located on six of the eight chromosomes.
Identification of the Chemical Structure of Nidulanin A Confirms Prediction of Cross-Chemistry Between NRPS AN1242 (NlsA) and Prenyltransferase AN11080 (NptA).
To test the hypothesis of superclusters and whether the analysis above could be used to elucidate cross-chemistry, we constructed a deletion mutant of the NRPS AN1242 and evaluated the SMs found in the mutant relative to a reference strain. Four related compounds (compounds 1–4) were found to be absent in the ΔAN1242 strain (Fig. S5). MS isotope patterns as well as tandem MS (MS/MS) analysis showed compound 1 to have the molecular formula C34H45N5O5, with compounds 2 and 3 likely being oxygenated forms with one and two extra oxygen molecules, respectively. Compounds 1–3 all seem to be prenylated, as shown by spontaneous loss of a prenyl-like fragment, C5H8, in a small fraction of the ions during MS analysis. Compound 4 has a molecular formula of (1)-C5H8, suggesting it to be the unprenylated precursor of compound 1.
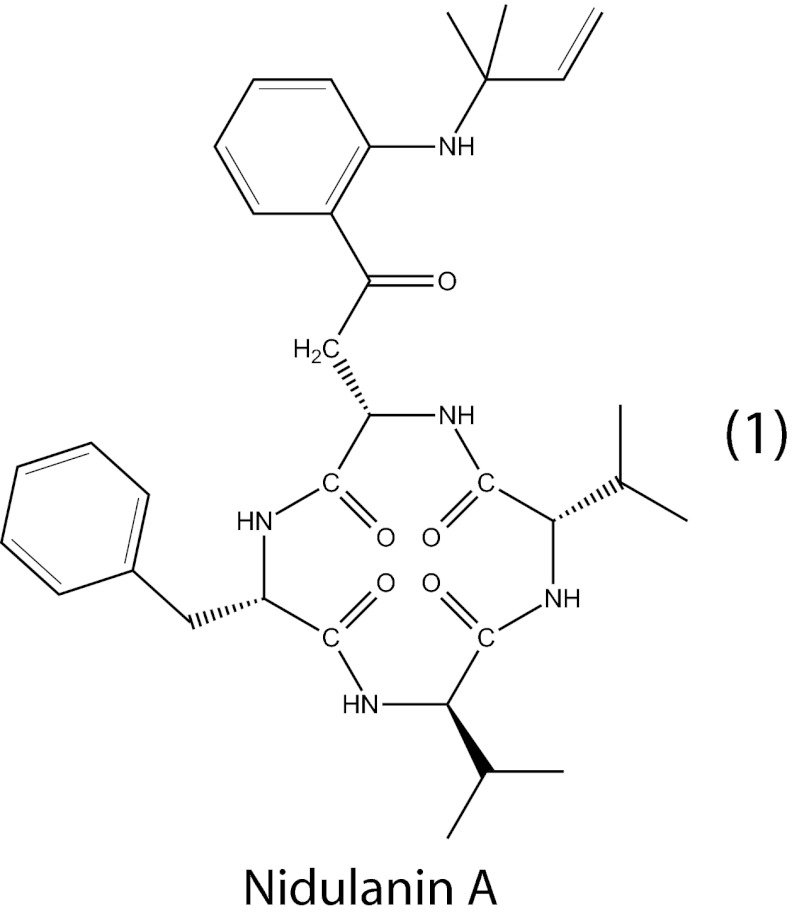
We thus isolated and elucidated the structure of compound 1, henceforth called nidulanin A, based on NMR spectroscopy. The stereochemistry of compound 1 was examined using Marfey’s method (40) and was supported by bioinformatic analysis of the protein domains of AN1242 (SI Text). Altogether, nidulanin A is proposed to be a tetracyclopeptide with the sequence -L-Phe-L-Kyn-L-Val-D-Val- and an isoprene unit N-linked to the amino group of L-kynurenine (Fig. 4).
Fig. 4.
Proposed absolute structure of nidulanin A. Details on the structural elucidation are available in SI Text.
Because no prenyltransferase genes are found near AN1242, cross-chemistry catalyzed by an N-prenylating DMATS is a likely assumption. Examination of supercluster 5 in Fig. 3, where the NRPS AN1242 is found, shows AN11080 to be the DMATS with the expression profile most similar to AN1242. Gene deletion of AN11080 and subsequent ultra-high-performance liquid chromatography (UHPLC) high-resolution MS (HRMS) analysis of the ΔAN11080 strain show that the deprenylated compound 4, but none of the three prenylated forms, is present, thus confirming that nidulanin A and the two oxygenated forms (compounds 3 and 4) are synthesized by cross-chemistry between AN1242 (now NlsA) on chromosome VIII and AN11080 (now NptA) on chromosome V (Fig. S5).
Furthermore, we note that the masses corresponding to compound 3 (nidulanin A + O) and compound 4 (nidulanin A + O2) are not found in the reference strain or in the ΔAN11080 strain. This suggests that compounds 3 and 4 are oxidized after the prenylation step.
Discussion
In this study, we present a method for fungal SM cluster estimation based on similarity of expression profiles for neighboring genes. For the given organism A. nidulans, comparison with legacy data has verified the method to be highly accurate and effective for a large proportion of the gene clusters.
It is clear from our results that the composition of the gene expression compendium has a significant effect on cluster predictions. We show here that it is important with a diverse set of samples, including both liquid and agar cultures as well as minimal medium and complex medium. This is in accordance with previous observations (11, 13, 14, 20, 36) stating that at a given set of conditions, only a fraction of the clusters are active. A reduction analysis of our own data has further shown that the inclusion of biological replicates in the dataset does not improve the analysis as much as inclusion of more unique samples. A diverse set of conditions should remedy regulation at the transcriptional level as well as chromatin-level regulation, which has been shown to have significant effects in fungi (13, 41). Another factor of importance is the quality of genome annotation. Erroneous gene calls inside clusters decrease the value of the CS for genes within a distance of three genes. Furthermore, problems with gene calls can affect expression profiling if a nontranscribed region is included in the gene cluster. However, neither of these seems to be a problem in the data presented here. Including the expression profiles of seven genes in the calculation of the CS also increases the robustness of the method toward erroneous gene calls.
The stated robustness of the CS has the disadvantage that the CS alone performs poorly for clusters with four or fewer genes, because the maximum value of CS for n genes is n − 1. However, in the cases of small clusters, the clustering can still be predicted from the transcription profiles, as shown in this study.
In some cases, we also see that cluster calling based on expression profiles outperforms the combination of gene KO and metabolomics. If a given detected metabolite is not the end product of the biosynthetic pathway, gene deletions will only identify a part of an SM cluster as being relevant for that metabolite, thus missing genes. An example of this is seen in the emodin/monodictyphenone cluster (PKS AN0150), where a subset of the genes is only required for some of the metabolites, resulting in a two-step elucidation of the gene cluster (7, 8). The CS method correctly calls the full cluster.
One aspect of the method is the ability to identify gene clusters simply from identifying groups of genes with high CS values, and not using a seeding set of synthases as was done in this case. This allows the unbiased identification of gene clusters throughout the entire genome. Although we see a surprising amount of these clusters (Dataset S2) not limited to the predicted SM synthases, we have not evaluated these in this study, because data for appropriate benchmarking is not available. However, we believe that there is great potential for biological discoveries to be made here, both in terms of promoter and chromatin-based transcriptional regulation.
The final extension of the algorithm is its ability to identify biosynthetic superclusters scattered across different chromosomes. Although this is a recently reported phenomenon (8), we believe that this is a common phenomenon, at least in A. nidulans and possibly in fungi in general. It is important to note that our method does not allow one to discriminate between tight coregulation and cross-chemistry between two distant clusters. It is therefore most efficient in cases in which it is evident that a given gene cluster does not hold all enzymatic activities required to synthesize the associated compound. In those cases, the use of a diverse transcription catalog, such as the one applied here, is a powerful strategy for identifying cross-chemistry, as shown for the NRPS AN1242 and the assisting prenyltransferase AN11080 in the synthesis of nidulanin A and derived compounds.
In summary, this study provides (i) an updated gene expression DNA array for A. nidulans, (ii) a wealth of information advancing the cluster elucidation in the model fungus A. nidulans, (iii) a powerful tool for prediction of SM cluster gene members in fungi, (iv) a proven methodology for prediction of SM gene cluster cross-chemistry, and (v) a proposed structure for the compound nidulanin A.
Materials and Methods
Strains.
A. nidulans FGSC A4 was used for all transcriptomic experiments in this study. Furthermore, legacy data using the FGSC A4, A. nidulans AR16msaGP74 (expressing the msaS gene from Penicillium griseofulvum) (34), A. nidulans AR1phk6msaGP74 (expressing the msaS gene from P. griseofulvum and overexpressing the A. nidulans xpkA) (34), and A. nidulans AR1phkGP74 (overexpressing the A. nidulans xpkA) (33), were applied.
The A. nidulans FGSC A4 stock culture was maintained on CYA agar at 4 °C. A. nidulans strain IBT 29539 (veA1, argB2, pyrG89, and nkuAΔ) was used for all gene deletions. Gene deletion strains (see below) are available from the IBT fungal collection as A. nidulans IBT 32029, (AN1242Δ::AfpyrG, veA1, argB2, pyrG89, and nkuAΔ) and A. nidulans IBT 32030, (AN11080Δ::AfpyrG, veA1, argB2, pyrG89, and nkuAΔ). For chemical analyses, A. nidulans IBT 28738 (veA1, argB2, pyrG89, and nkuA-trS::AfpyrG) was used as reference strain.
Metabolite Profiling Analysis.
A. nidulans strains were inoculated on CYA agar, OTA, YES agar, and CYAS agar (42). All strains were three-point inoculated on these media and incubated at 32 °C in darkness for 4, 8, or 10 d, after which three to five plugs (6-mm diameter) along the diameter of the fungal colony were cut out and extracted (43).
Samples were subsequently analyzed by UHPLC-UV/vis diode array detector (DAD)-HRMS on a maXis G3 quadrupole time-of-flight mass spectrometer (Bruker Daltonics) equipped with an electrospray injection (ESI) source. The mass spectrometer was connected to an Ultimate 3000 UHPLC system (Dionex). Separation of 1-μL samples was performed at 40 °C on a 100-mm × 2.1-mm inner diameter (ID), 2.6-μm Kinetex C18 column (Phenomenex) using a linear water-acetonitrile gradient (both buffered with 20 mM formic acid) at a flow rate of 0.4 mL/min starting from 10% (vol/vol) acetonitrile and increased to 100% acetonitrile in 10 min, keeping this for 3 min. HRMS was performed in ESI+ with a data acquisition range of 10 scans per second at m/z 100–1,000. The mass spectrometer was calibrated using sodium formate automatically infused before each analytical run, providing a mass accuracy better than 1.5 ppm. Compounds were detected as their [M + H]+ ion ± 0.002 Da, often with their [M + NH4]+ and/or [M + Na]+ ion used as a qualifier ion with the same narrow mass range. SMs with a peak areas >10,000 counts (random noise peaks of approximately 300 counts) were integrated and identified by comparison with approximately 900 authentic standards available from previous studies (31, 44) and dereplicated against the approximately 18,000 fungal metabolites listed in AntiBase 2010 by ultraviolet-visible (UV/Vis) spectra, retention time, adduct pattern, and high-resolution data (<1.5 ppm mass accuracy and isotope fit better than 40 using SigmaFit; Bruker Daltonics) (31, 45).
Array Design.
Initial probe design was done using OligoWiz 2.0 software (46) from the coding sequences of predicted genes from the genome sequence of A. nidulans FGSC A4 (35), using version 5 of the A. nidulans gene annotation, downloaded from the Aspergillus Genome Database (32).
For each gene, a maximum of three nonoverlapping, perfect-match 60-mer probes was calculated using the OligoWiz standard scoring of cross-hybridization, melting temperature, folding, position preference, and low complexity. A position preference for the probes was included in the computations. Pruning of the probe sequences was done by removing duplicate probe sequences.
Also included on the chip were 1,407 standard controls designed by Agilent Technologies. Details of the array are available from the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GPL15899).
Microarray Gene Expression Profiling.
Mycelium harvest and RNA purification.
Whole colonies from three-stab agar plates were sampled for transcriptional analysis by scraping the mycelium off the agar with a scalpel and transferring the agar directly into a 50-mL Falcon tube containing approximately 15 mL of liquid nitrogen. Care was taken to transfer a minimum of agar to the Falcon tube. The liquid nitrogen was allowed to evaporate before capping the lid and recooling the tube in liquid nitrogen before storing the tube at −80 °C until use for RNA purification.
For RNA purification, 40–50 mg of frozen mycelium was placed in a 2-mL microcentrifuge tube precooled in liquid nitrogen containing three steel balls (two balls with a diameter of 2 mm and one ball with a diameter of 5 mm). The tubes were then shaken in a Retsch Mixer Mill at 5 °C for 10 min until the mycelium was ground to a powder. Total RNA was isolated from the powder using the Qiagen RNeasy Mini Kit according to the protocol for isolation of total RNA from plant and fungi, including the optional use of the QiaShredder column. Quality of the purified RNA was verified using a NanoDrop ND-1000 spectrophotometer and an Agilent 2100 Bioanalyzer (Agilent Technologies).
Microarray hybridization.
A total of 150 ng in 1.5-μL total RNA was labeled according to the One Color Labeling for Expression Analysis, Quick Amp Low Input (QALI) manual, version 6.5, from Agilent Technologies. Yield and specific activity were determined on the ND-1000 spectrophotometer and verified on a Qubit 2.0 fluorometer (Invitrogen). A total of 1.65 μg of labeled cRNA was fragmented at 60 °C on a heating block, and the cRNA was prepared for hybridization according to the QALI protocol. A 100-μL sample was loaded on a 4 × 44 Agilent Gasket Slide situated in a hybridization chamber (both from Agilent Technologies). The 4 × 44 array was placed on top of the Gasket Slide. The array was hybridized at 65 °C for 17 h in an Agilent Technologies hybridization oven. The array was washed following the QALI protocol and scanned in a G2505C Agilent Technologies Micro Array Scanner.
Analysis of transcriptome data.
The raw array signal was processed by first removing the background noise using the normexp method, and signals between arrays were made comparable using the quantiles normalization method as implemented in the Limma package (47). Multiple probe signals per gene were summarized into a gene-level expression index using Tukey’s medianpolish, as performed in the last step of the robust multiarray average (RMA) processing method (48). The data are available from the Gene Expression Omnibus database (accession no. GSE39993).
The generated data from the Agilent Technologies arrays were combined with legacy Affymetrix data (accession nos. GSE12859 and GSE7295) using the qspline normalization method (46) to combine the two normalized sets of data to one microarray catalog with expression indices in comparable ranges.
Calculation of the Gene CS.
The CS is calculated for each individual gene along the chromosomes according to the following equation:
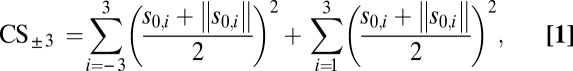 |
where s0,i is the Spearman coefficient for the expression indices of the gene in question and the gene located i genes away in a positive or negative direction relative to the chromosomal coordinate of the gene. The absolute term is added to set inverse correlations to 0. The CS assigned to a specific gene is the average of the CS for the liquid cultures and the CS for the solid cultures to adjust for background expression levels. Genes located less than four genes away from the ends of the supercontigs are assigned a CS of 0. All calculations were performed in the R software suite v. 2.14.0 (49), using the Bioconductor package (50, 51) for handling of array data. An adaptable R script for calculation of the CS is available on request.
Generation of Random Values for Evaluation of CS Significance.
To estimate significance levels of the CS, a random set of scores was generated by selecting six genes at random as simulated neighbors for each of the 10,411 genes in the dataset. Examining this random distribution showed 95% of the population to have a CS <2.13 (Fig. S2). This value was used to have a false discovery rate of 0.05. All calculations were performed in R (49).
Identification of Gene Clusters.
Gene clusters were defined around each NRPS, PKS, and DMATS by examination of the transcription profile of all surrounding genes with a CS ≥2.13 as well as three flanking genes in either direction. All genes with similar expression profiles were included in the cluster.
PCA-Based Analysis of Dataset Variation.
PCA analysis was performed on the data of Dataset S2 using the prcomp-function of R (49). For stepwise reduction of the dataset, all principal components were calculated in each iteration and a sample was eliminated based on the one that had the largest contribution to the last principal component (i.e., with the smallest amount of unique information).
Generation of A. nidulans Gene Deletion Mutants.
The genetic transformation experiments were performed with A. nidulans strain IBT 29539 [veA1, argB2, pyrG89, and nkuAΔ as described by Nielsen et al. (52)]. Fusion PCR-based bipartite gene targeting of substrates using the AFpyrG marker for selection and deletion of AN1242 was performed as described by Nielsen et al. (52), with the exception that all PCR assays were performed with the PfuX7 DNA polymerase (53). The deletion construct for AN11080 was assembled by uracil-specific excision reagent (USER) cloning. Specifically, sequences upstream and downstream of the gene to be deleted were amplified by PCR using primers containing a uracil residue (Table S2). The two PCR fragments were simultaneously inserted into the PacI/Nt.BbvCI USER cassette of pU20002A by USER cloning (54, 55). As a result, AFpyrG is now flanked by the two PCR fragments to complete the gene targeting substrate. The gene targeting substrate was released from the resulting vector pU20002A-AN11080 by digestion with SwaI. All restriction enzymes are from New England Biolabs. Primer sequences for deletion of the targeted genes and verification of strains are listed in Table S2. In addition, internal AFpyrG primers were used in combination with the check primers listed in Table S2 for confirmation of correct integration of DNA substrates (52). Transformants and AFpyrG pop-out recombinant strains were rigorously tested for correct insertions as well as for the presence of heterokaryons by touchdown spore-PCR analysis on conidia with an initial denaturation at 98 °C for 20 min.
MS/MS-Based Characterization of Compounds 1–4.
Analysis was performed as stated above for the UHPLC-DAD-HRMS but in MS/MS mode, where analysis of the target mass and 6 m/z units up (to maintain isotopic pattern) was performed both via a targeted MS/MS list for the target compounds of interest and by the data-dependent MS/MS mode with an exclusion list, such that the same compound was selected several times. MS/MS fragmentation energy was varied from 18 to 55 eV.
Isolation and Structural Elucidation of Nidulanin A.
Two hundred plates of minimal medium were inoculated with A. nidulans, from which SMs were extracted and nidulanin A was isolated in pure form. One-dimensional and 2D NMR spectra were recorded on a Bruker Daltonics Avance 800-MHz spectrometer with a 5-mm TCI Cryoprobe at the Danish Instrument Centre for NMR Spectroscopy of Biological Macromolecules at Carlsberg Laboratory. Stereoisometry of the amino acids was elucidated using Marfey’s method (40). Details are provided in SI Text, Table S3, and Figs. S6–S8.
NRPS protein domains were predicted to identify adenylation domains and epimerase domains (56). Adenylation-domain specificities were predicted using NRPSpredictor (57). Details are provided in SI Text.
Supplementary Material
Acknowledgments
We thank Peter Dmitrov, who treated the raw microarray data. We acknowledge Laurent Gautier for good scientific discussion of experimental design for microarray experiments, Marie-Louise Klejnstrup for assistance in retrieving MS data, and Dorte Koefoed Holm and Francesca Ambri for analysis of the austinol gene deletion mutants. We also thank the Danish Instrument Center for NMR Spectroscopy of Biological Macromolecules for NMR time. This work was supported by the Danish Research Agency for Technology and Production Grants 09-064967 and FI 2136-08-0023.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The gene expression data, gene expression microarray data description, and legacy gene expression data reported in this paper are available from the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE39993, GPL15899, GSE12859 and GSE7295).
See Author Summary on page 24 (volume 110, number 1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205532110/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu T, Chiang YM, Somoza AD, Oakley BR, Wang CC. Engineering of an “unnatural” natural product by swapping polyketide synthase domains in Aspergillus nidulans. J Am Chem Soc. 2011;133(34):13314–13316. doi: 10.1021/ja205780g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khaldi N, et al. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47(9):736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medema MH, et al. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(Web server issue):W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly DE, Krasevec N, Mullins J, Nelson DR. The CYPome (Cytochrome P450 complement) of Aspergillus nidulans. Fungal Genet Biol. 2009;46(Suppl 1):S53–S61. doi: 10.1016/j.fgb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Palmer JM, Keller NP. Secondary metabolism in fungi: Does chromosomal location matter? Curr Opin Microbiol. 2010;13(4):431–436. doi: 10.1016/j.mib.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang YM, et al. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol. 2010;76(7):2067–2074. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez JF, et al. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J Am Chem Soc. 2011;133(11):4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson TJ. Genetic and biosynthetic studies of the fungal prenylated xanthone shamixanthone and related metabolites in Aspergillus spp. revisited. ChemBioChem. 2012;13(11):1680–1688. doi: 10.1002/cbic.201200014. [DOI] [PubMed] [Google Scholar]
- 10.Schätzle MA, Husain SM, Ferlaino S, Müller M. Tautomers of anthrahydroquinones: Enzymatic reduction and implications for chrysophanol, monodictyphenone, and related xanthone biosyntheses. J Am Chem Soc. 2012;134(36):14742–14745. doi: 10.1021/ja307151x. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen ML, et al. A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans. FEMS Microbiol Lett. 2011;321(2):157–166. doi: 10.1111/j.1574-6968.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez JF, et al. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst. 2010;6(3):587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bok JW, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5(7):462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeckh V, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106(34):14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szewczyk E, et al. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol. 2008;74(24):7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang YM, et al. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15(6):527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown DW, et al. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93(4):1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelkar HS, Keller NP, Adams TH. Aspergillus nidulans stcP encodes an O-methyltransferase that is required for sterigmatocystin biosynthesis. Appl Environ Microbiol. 1996;62(11):4296–4298. doi: 10.1128/aem.62.11.4296-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller NP, Watanabe CM, Kelkar HS, Adams TH, Townsend CA. Requirement of monooxygenase-mediated steps for sterigmatocystin biosynthesis by Aspergillus nidulans. Appl Environ Microbiol. 2000;66(1):359–362. doi: 10.1128/aem.66.1.359-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergmann S, et al. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3(4):213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 21.Bouhired S, Weber M, Kempf-Sontag A, Keller NP, Hoffmeister D. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet Biol. 2007;44(11):1134–1145. doi: 10.1016/j.fgb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Schneider P, Weber M, Hoffmeister D. The Aspergillus nidulans enzyme TdiB catalyzes prenyltransfer to the precursor of bioactive asterriquinones. Fungal Genet Biol. 2008;45(3):302–309. doi: 10.1016/j.fgb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Clutterbuck AJ. A mutational analysis of conidial development in Aspergillus nidulans. Genetics. 1969;63(2):317–327. doi: 10.1093/genetics/63.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahuja M, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134(19):8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bromann K, et al. Identification and characterization of a novel diterpene gene cluster in Aspergillus nidulans. PLoS ONE. 2012;7(4):e35450. doi: 10.1371/journal.pone.0035450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birse CE, Clutterbuck AJ. N-acetyl-6-hydroxytryptophan oxidase, a developmentally controlled phenol oxidase from Aspergillus nidulans. J Gen Microbiol. 1990;136(9):1725–1730. doi: 10.1099/00221287-136-9-1725. [DOI] [PubMed] [Google Scholar]
- 27.MacCabe AP, et al. Delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase from Aspergillus nidulans. Molecular characterization of the acvA gene encoding the first enzyme of the penicillin biosynthetic pathway. J Biol Chem. 1991;266(19):12646–12654. [PubMed] [Google Scholar]
- 28.Martin JF. Clusters of genes for the biosynthesis of antibiotics: regulatory genes and overproduction of pharmaceuticals. J Ind Microbiol. 1992;9(2):73–90. doi: 10.1007/BF01569737. [DOI] [PubMed] [Google Scholar]
- 29.Yeh HH, et al. Molecular genetic analysis reveals that a nonribosomal peptide synthetase-like (NRPS-like) gene in Aspergillus nidulans is responsible for microperfuranone biosynthesis. Appl Microbiol Biotechnol. 2012;96(3):739–748. doi: 10.1007/s00253-012-4098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo H-C, et al. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J Am Chem Soc. 2012;134(10):4709–4720. doi: 10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen KF, Månsson M, Rank C, Frisvad JC, Larsen TO. Dereplication of microbial natural products by LC-DAD-TOFMS. J Nat Prod. 2011;74(11):2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- 32.Arnaud MB, et al. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2010;38(Database issue):D420–D427. doi: 10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panagiotou G, et al. Systems analysis unfolds the relationship between the phosphoketolase pathway and growth in Aspergillus nidulans. PLoS ONE. 2008;3(12):e3847. doi: 10.1371/journal.pone.0003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panagiotou G, et al. Studies of the production of fungal polyketides in Aspergillus nidulans by using systems biology tools. Appl Environ Microbiol. 2009;75(7):2212–2220. doi: 10.1128/AEM.01461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galagan JE, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438(7071):1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 36.Brakhage AA, et al. Activation of fungal silent gene clusters: A new avenue to drug discovery. Prog Drug Res. 2008;66(1):3–12. doi: 10.1007/978-3-7643-8595-8_1. [DOI] [PubMed] [Google Scholar]
- 37.Bok JW, et al. Genomic mining for Aspergillus natural products. Chem Biol. 2006;13(1):31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Cullen D. The genome of an industrial workhorse. Nat Biotechnol. 2007;25(2):189–190. doi: 10.1038/nbt0207-189. [DOI] [PubMed] [Google Scholar]
- 39.Bergmann S, et al. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol. 2010;76(24):8143–8149. doi: 10.1128/AEM.00683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marfey P. Determination of D- amino acids. II. Use of a bifunctional reagent, 1,5-di-fluoro-2,4-dinitrobenzene. Carlsberg Res Commun. 1984;49(6):591–596. [Google Scholar]
- 41.Nützmann HW, et al. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc Natl Acad Sci USA. 2011;108(34):14282–14287. doi: 10.1073/pnas.1103523108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frisvad JC, Samson R. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of the food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol. 2004;49:1–173. [Google Scholar]
- 43.Smedsgaard J. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J Chromatogr A. 1997;760(2):264–270. doi: 10.1016/s0021-9673(96)00803-5. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen KF, Smedsgaard J. Fungal metabolite screening: Database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography-UV-mass spectrometry methodology. J Chromatogr A. 2003;1002(1-2):111–136. doi: 10.1016/s0021-9673(03)00490-4. [DOI] [PubMed] [Google Scholar]
- 45.Månsson M, et al. Explorative solid-phase extraction (E-SPE) for accelerated microbial natural product discovery, dereplication, and purification. J Nat Prod. 2010;73(6):1126–1132. doi: 10.1021/np100151y. [DOI] [PubMed] [Google Scholar]
- 46.Workman C, et al. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3(9):research0048. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smyth GK. Limma: Linear models for microarray data. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 48.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2007. Available at www.R-project.org. [Google Scholar]
- 50.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet Biol. 2006;43(1):54–64. doi: 10.1016/j.fgb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen JB, Nielsen ML, Mortensen UH. Transient disruption of non-homologous end-joining facilitates targeted genome manipulations in the filamentous fungus Aspergillus nidulans. Fungal Genet Biol. 2008;45(3):165–170. doi: 10.1016/j.fgb.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Nørholm MH. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 2010;10:21. doi: 10.1186/1472-6750-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen BG, et al. Versatile enzyme expression and characterization system for Aspergillus nidulans, with the Penicillium brevicompactum polyketide synthase gene from the mycophenolic acid gene cluster as a test case. Appl Environ Microbiol. 2011;77(9):3044–3051. doi: 10.1128/AEM.01768-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisendle M, Oberegger H, Zadra I, Haas H. The siderophore system is essential for viability of Aspergillus nidulans: Functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC) Mol Microbiol. 2003;49(2):359–375. doi: 10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- 56.Bachmann BO, Ravel J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 57.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33(18):5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]