Abstract
Of all the myosin filaments in muscle, the most important in terms of human health, and so far the least studied, are those in the human heart. Here we report a 3D single-particle analysis of electron micrograph images of negatively stained myosin filaments isolated from human cardiac muscle in the normal (undiseased) relaxed state. The resulting 28-Å resolution 3D reconstruction shows axial and azimuthal (no radial) myosin head perturbations within the 429-Å axial repeat, with rotations between successive 132 Å-, 148 Å-, and 149 Å-spaced crowns of heads close to 60°, 35°, and 25° (all would be 40° in an unperturbed three-stranded helix). We have defined the myosin head atomic arrangements within the three crown levels and have modeled the organization of myosin subfragment 2 and the possible locations of the 39 Å-spaced domains of titin and the cardiac isoform of myosin-binding protein-C on the surface of the myosin filament backbone. Best fits were obtained with head conformations on all crowns close to the structure of the two-headed myosin molecule of vertebrate chicken smooth muscle in the dephosphorylated relaxed state. Individual crowns show differences in head-pair tilts and subfragment 2 orientations, which, together with the observed perturbations, result in different intercrown head interactions, including one not reported before. Analysis of the interactions between the myosin heads, the cardiac isoform of myosin-binding protein-C, and titin will aid in understanding of the structural effects of mutations in these proteins known to be associated with human cardiomyopathies.
Keywords: thick filaments 3D structure, human heart muscle, electron microscopy, image processing
Muscle contraction involves the interaction between actin and myosin filaments that leads to force production mediated by the hydrolysis of ATP. Myosin filaments are assemblies of myosin molecules and accessory proteins. Myosin molecules consist of two heavy chains arranged to form two globular domains [two subfragment-1s (S1s)] at the N-terminal end and a long rod-like α-helical coiled-coil domain (the myosin tail). Each of the globular domains with its associated essential and regulatory light chains forms a myosin head containing the ATPase activity necessary for contraction (1). This is linked to the N-terminal part of the tail known as the subfragment-2 (S2) region. Myosin filaments are generally bipolar, with the myosin tails packing together in an almost cylindrical backbone and the heads projecting out laterally in a helical or quasihelical fashion at intervals of ∼143 Å. In the myosin filaments of vertebrate striated muscles, the myosin heads are arranged in a three-stranded quasihelical array on the filament surface (2) with the accessory proteins titin (3, 4), myosin-binding protein-C (MyBP-C) (5), and possibly the MyBP-C analog, X protein (6), located on the surface of the backbone. MyBP-C is located within the C zones of the myosin filament, which are regions ∼3,500 Å long centrally located in the two halves of the bipolar filament. Titin is an unusually large elongated protein (3.8 MDa) that runs parallel to the actin and myosin filaments and spans half the length of a sarcomere. It is composed of a linear array of immunoglobullin (Ig) and fibronectin 3 (Fn3) domains connected by short linkers. Within the C zone, the titin domains are distributed in a pattern corresponding to an 11-domain superrepeat every 429 Å. The cardiac isoform of MyBP-C (cMyBP-C) has a mass of ∼140 kDa and is made up of a linear array of 11 Ig and Fn3 domains.
Myosin filament structure has been investigated both by electron microscopy and by X-ray fiber diffraction to explore the different organizations and properties of myosin filaments from a variety of organisms and tissues (7, 8). The most detailed structural description of a myosin filament (to date) derives from helical 3D reconstruction of cryoelectron microscope images of tarantula myosin filaments at a resolution of ∼25 Å (9, 10). In the tarantula myosin filament, pairs of myosin heads were identified forming a similar interaction to that found in the off state of vertebrate smooth muscle myosin (11). Vertebrate skeletal and cardiac muscle myosin filaments differ from those in invertebrates such as tarantula in that the myosin heads depart from an exact helical arrangement. Hence, for vertebrate skeletal and cardiac myosin filaments, helical reconstruction is not a valid approach for the analysis of electron microscope data. Working on a variety of myosin filaments, we have developed an alternative method to determine their 3D structures from electron microscope data based on single-particle analysis (12). This approach is well-suited to the analysis of quasihelical myosin filaments. We have applied single-particle analysis to myosin filaments from a variety of muscle types including vertebrate skeletal and cardiac muscles (13, 14) and invertebrate striated muscle (15). A similar approach has been used by others in the analysis of mouse cardiac myosin filament structure (16), where myosin heads were also found to adopt a paired conformation similar to that in tarantula myosin filaments.
Mutations in human cardiac muscle myosin and its associated proteins, cMyBP-C and titin, are known to cause a number of human cardiomyopathies including familial hypertrophic cardiomyopathy and dilated cardiomyopathy (17, 18). A detailed knowledge of the structure of human cardiac myosin filaments in the normal (undiseased) relaxed state is likely to be important in understanding how the mutations give rise to these cardiomyopathies. To address this issue, we have successfully developed a laboratory method to isolate myosin filaments from human cardiac muscle that preserves the highly ordered pseudohelical structure of the relaxed filaments, thus making them amenable to analysis by electron microscopy and single-particle image analysis. From such samples, we have produced a 3D reconstruction of the C zone of the myosin filament at ∼28-Å resolution, which allows the detailed docking of myosin heads and the modeling of the constituent domains of titin and cMyBP-C.
Results
Three-Dimensional Structure of the Human Cardiac Myosin Filament.
Electron microscope images were recorded from negatively stained preparations of myosin filaments isolated from human ventricular muscle. From these images, well-preserved examples of myosin filaments were selected in which the C zone could be clearly identified (Fig. S1 A and B). From 237 myosin filament C zones, a total of 1,559 segments were extracted, each segment corresponding to two 429-Å repeats. An initial reference 3D structure was calculated using C3 symmetry (threefold rotational symmetry) from a single class average of the segments. Starting from this reference, a refined 3D structure was calculated from the individual segments by successive rounds of forward projection, multireference alignment, and 3D reconstruction. During the refinement, the number of segments was reduced to 285 by selecting only those that agreed visually most strongly with forward projections of the 3D structure. Fourier analysis of the selected segments (Fig. S2A) shows good recovery of detail, with the characteristic layer-line pattern arising from the quasihelical myosin head distribution together with meridional reflections extending to ∼36 Å (the 12th order of the 429-Å repeat). Furthermore, these segments provide an even distribution of projection angles (Fig. S3). The final 3D reconstruction (Fig. 1), which has an estimated resolution of 28 Å (Fig. S4), is characterized by a wealth of detail, and Fourier analysis of reprojections (Fig. S2B) shows good agreement with the original segments. The visual selection is supported by quantitative comparison (Fig. S5) and Fourier analysis (Figs. S2A and S6A) of the selected and rejected segments. Moreover, 3D reconstructions of the rejected segments are characterized by significantly lower levels of detail (Fig. S7) and correspondingly lower resolution (Figs. S6 and S8). Within the 429-Å repeating unit of the final 3D reconstruction, there are three distinct sets of densities on the outer surface of the filament (Fig. 1 A and B) that correspond to the three crowns or levels of myosin heads. The location of the crowns within the C zone matches that described in earlier studies of vertebrate skeletal and cardiac muscle (13, 14), and the crowns are labeled 1, 2, and 3 following the same convention. Here crown 1 is the most massive, with its extra mass arising from cMyBP-C [n.b. this is a different numbering scheme from the unconventional one used by Zoghbi et al. (16) in their analysis of mouse cardiac myosin filament, where the equivalent crowns are labeled 1, 3, and 2, respectively]. Each crown is made up of three pairs of myosin heads. Direct inspection of the density associated with each crown allows the recognition of myosin head pairs that form triangular motifs (Fig. 1 C and D) very similar in appearance to the conformation of myosin head pairs in the off state of vertebrate smooth muscle myosin (Fig. 1E) (11). Similar shapes were seen in the tarantula myosin filament reconstruction (9), where the base of each triangle oriented towards the M band corresponds to the two motor domains and the two sides correspond to the lever arms.
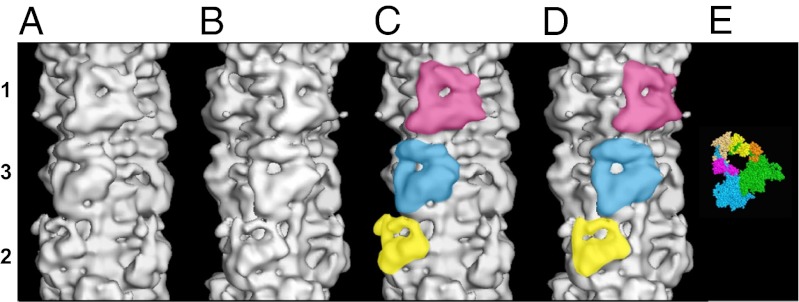
Fig. 1.
(A–D) Surface views of the final 3D reconstruction of the human cardiac myosin filament obtained by single-particle EM analysis and displayed using PyMOL showing a length of a full 429-Å repeat. The reconstruction is shown in two views (A and C; B and D) related by a 25° rotation about the filament axis, such that in A and C, a myosin head pair on level 1 is facing the viewer, and in B and D, a myosin head pair on level 3 is facing the viewer. Note that the two views in A and C and in B and D are quite distinct because of the different perturbations in the crowns. The triangular-shaped configurations for the three head pairs on each crown level are shown in yellow, blue, and pink for levels 2, 3, and 1, respectively, in C and D. The bare zone is at the bottom of the map in all of the four views. (E) Crystal structure for the head pair (11). The blocked and free heads are color-coded as in refs. 9–11. Blocked head: motor domain, green; essential light chain, orange; regulatory light chain, yellow. Free head: motor domain, cyan; essential light chain, pink; regulatory light chain, beige.
Fitting the Crown Densities with Myosin Molecular Models.
Because we identified motifs in each of the crowns with close resemblance to the myosin head pairs in tarantula myosin filaments, we proceeded to dock the corresponding atomic model into the crown densities. For this purpose, we used Protein Data Bank (PDB) ID code 3DTP (10) [a modified version of the off state of vertebrate smooth muscle myosin (11), which also includes part of the coiled-coil S2 structure]. We found that the best fit within the density map could be obtained by uncoupling and separately docking the two S1 domains, resulting in minor changes in the mutual interaction of the motor domains and minor reorientations of the lever arms. Nevertheless, in each case, the overall structure of the docked head pair is not very different from the original structure (11) and had similar intramolecular interactions between the motor domains of the blocked and free heads as described previously (11). We also found it necessary to reorient the S2. Fig. 2 A and B shows a similar view of the map shown in Fig. 1B along with our best-fit structure within the map. The map is also shown as a movie (Movie S1). Overall, the fitted coordinates correspond closely to the density while revealing characteristic features of the individual crowns. In crown 3, there is little density unaccounted for within the head-pair motif, and almost all of the coordinates are contained within the density. However, in the case of crown 1 there are minor excursions associated with the regulatory light chain (RLC) of the blocked head and the essential light chain (ELC) of the free head, whereas for both crowns 1 and 3, the N-terminal SH3 domains (residues 33–78) of both the blocked and free heads extend outside the density. This latter effect may be associated with the conformational variability observed in this region of myosin (19). On the other hand, in the case of crown 2, although the overall shape of the head pair conforms quite well to the fitted coordinates, there are regions where the coordinates lie outside the density more substantially, particularly at the N-terminal SH3 domains of the two heads and in the regions of the lever arms. The least density is recovered for the head domains in crown 2. This may arise because the heads in crown 2 are less well ordered than in crowns 1 and 3.
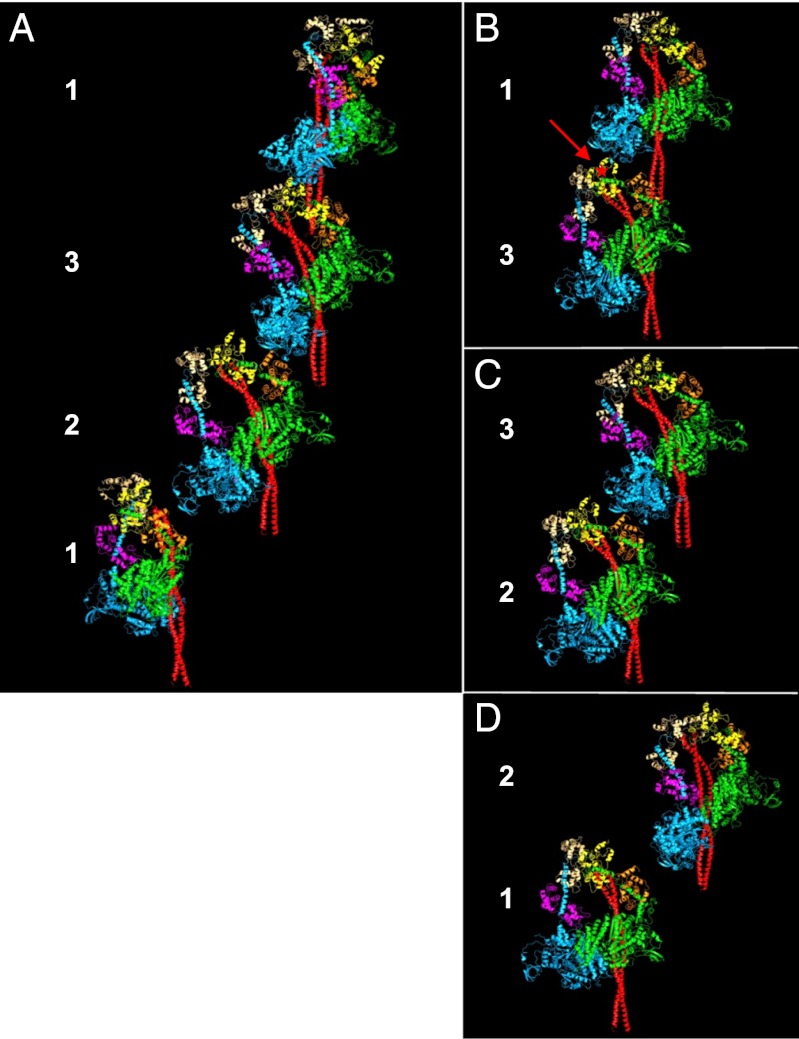
Fig. 2.
(A) Three-dimensional map of the view in Fig. 1B showing a full 429-Å repeat together with fitting of the atomic model of myosin heads in the off state to the head motifs in crowns 2, 3, and 1 after splitting and fitting separately the two heads from Alamo et al. (10). The majority of the atomic structures for the head pairs is well-contained within the density map on crowns 3 and 1 but less on crown 2, especially for the two lever-arm parts. The S2 is also fitted, as described in the text. (B) Same view as in A, but shown without the back part of the map to illustrate the fitting of the heads and S2 within the densities on levels 2, 3, and 1. S2 is shown in red and the bare-zone direction is toward the bottom.
Myosin Head Arrangement in Crowns 1, 2, and 3.
The fitted positions of the myosin heads within the cardiac myosin filament (Fig. 2) can be used to characterize the pseudohelical head arrangement of the filament as summarized in Fig. S9 and Table S1. This gives the axial and angular displacements between successive crowns measured both between the centers of each head pair and between the ends of the lever arm at the C-terminal position of the S1 domains, where they connect to S2. The azimuthal displacement represents the rotation about the filament axis between head pairs on successive crowns (Fig. S9B). Comparison of these parameters with the corresponding values for a regular three-start helix with a repeat of 429 Å allows the perturbations from an exact helical structure to be accurately defined. For the axial displacements (Fig. S9A), the values are very similar whether measured between the centers of the head pairs or between the ends of the lever arms, and it is apparent that crowns 1 and 2 are closer together (axial displacement ∼133 Å) than either crowns 2 and 3 or crowns 3 and 1 (axial displacement ∼148 Å). In comparison, an unperturbed helix would have a uniform axial displacement of 143 Å. On the other hand, the angular displacement between crowns 1 and 2 (60° measured between the centers of the head pairs, and 58° between the ends of the lever arms) is very much greater than the equivalent measurements between crowns 2 and 3 (35° and 30°, respectively) and between crowns 3 and 1 (25° and 32°, respectively). These values can be compared with an unperturbed helix, which would have a constant azimuthal displacement of 40°. Hence, the head arrangement in the cardiac myosin filament is characterized by a sequence of three crowns (2,3,1) with similar axial displacements (∼148 Å) and approximately similar angular displacements (∼30°). This is followed by a discontinuity in which the transition to the next crown has a reduced axial displacement (∼133 Å) but a substantially increased angular separation (∼60°). This gives rise to the phenomenon in which the head pairs in the 2,3,1 crown sequence are quite closely connected along a pseudohelical track, but due to the angular discontinuity, the connection with the next 2,3,1 sequence is not maintained. In addition to the variations in axial and angular displacements between adjacent crowns, the head pairs also exhibit a variation in their tilt angles. This can be most readily appreciated by comparing the orientation (or tilt angle) of the extended density arising from the two motor domains of each head pair (Fig. S9C). Here it is apparent that crowns 2 and 3 have very similar orientations, whereas crown 1 is rotated by 19° in a clockwise direction as viewed from outside the filament. Despite these perturbations in axial displacement, azimuthal displacement, and tilt, the radial perturbation of myosin heads is minor in the human cardiac myosin filament, in agreement with previous observations for both fish and rabbit myosin filaments (13, 14).
Interactions Between the Myosin Heads.
Independent docking of the heads within the head pairs in each crown leads to a similar intramolecular arrangement for the head pairs in which the intramolecular interactions observed in previous studies of smooth muscle myosin (11) and tarantula muscle (9, 10) are substantially conserved. However, intermolecular interactions between the head pairs on adjacent crowns vary much more substantially (Fig. 3A). As noted above, the head pairs in the 2,3,1 crown sequence are quite closely connected, whereas there is a distinct gap between crowns 1 and 2, where the head pairs appear not to be connected (Fig. 3D). In contrast, the head pairs in crowns 2 and 3 are connected through an interaction between the ELC of the blocked head on crown 2 and the motor domain of the free head on crown 3 (Fig. 3C). A similar interaction was observed between heads in successive crowns in the purely helical tarantula myosin filament (9, 10). This structural similarity can be understood because the head pairs on crowns 2 and 3 have the same tilt angles (Table S1 and Fig. S9C). Moreover, the angular rise between crowns 2 and 3 (35°) in the human cardiac myosin filament is quite similar to the angular separation of 30° in tarantula, and the axial spacing of 148 Å is also similar to that in tarantula (145 Å). A notably different type of intermolecular connection is observed between crowns 3 and 1. This is mediated by the interaction between the RLC of the blocked head on crown 3 and the motor domain of the free head on crown 1 (Fig. 3B). The different mode of interaction compared with that between crowns 2 and 3 can be considered as the consequence of the clockwise 19° rotation of the head pair on crown 1 and the reduced angular rise between crowns 3 and 1 (25°), both of which serve to shift the motor domain of the free head on crown 1 from the ELC to the RLC of the blocked head on crown 3 (Fig. 3B). Upon searching the literature, we could not find a record of this type of interaction between myosin head pairs in adjacent crowns. Although in their studies of mouse cardiac myosin filament, Zoghbi et al. (16) presented a docking of myosin head pairs on crowns 3 and 1 (they call them crowns 2 and 1) in which the motor domain of the free head on crown 1 can be seen to make contact with the RLC of the blocked head on crown 3, this was not mentioned by the authors.
Fig. 3.
(A) Fitted crystal structures showing head pairs and S2 on successive crowns along one strand of the myosin filament. Head pairs in the 2,3,1 crown sequence are quite closely connected, whereas there is a distinct gap between crowns 1 and 2. (B) Head pairs on crowns 3 and 1 are connected through an interaction between the RLC of the blocked head on crown 3 and the motor domain of the free head on crown 1. The red arrowed star indicates the predicted position of serine 15 in human myosin RLC. (C) A different type of intermolecular connection is observed between crowns 3 and 2 that is mediated by an interaction between the ELC of the blocked head on crown 2 and the motor domain of the free head on crown 3. (D) No intermolecular interaction is seen between the head pairs in crowns 1 and 2. S2 is shown in red, and the bare zone is oriented vertically down in all views.
Location of S2.
S2 was docked into the reconstruction separately from the myosin head pairs starting from the configuration observed in tarantula myosin filaments (9, 10) (Fig. 2). Furthermore, the variation in tilt angle of the head pairs on different crowns required S2 to be further subdivided into the segment running from the end of the lever arms to the blocked motor domain, the proximal region of S2 (residues 854–927 of S2), and the remaining C-terminal segment, the distal region of S2 (residues 928–972 of S2). Although not fully resolved from the rest of the density of the head pair, the proximal region of S2 can be accommodated within crown 3, where it is integrated with the density of the lever arms of the blocked heads. A similar location with respect to the myosin heads for the proximal region of S2 was obtained in the case of crown 1, although in this case it is not fully accommodated in the density. The less-complete recovery of myosin head-pair density on crown 2, noted above, also applies to this segment of S2. For consistency, given the fact that the head pairs on level 2 have the same tilt angle as the head pair on level 3, the proximal region of S2 is docked in a similar conformation to that in crown 3 but is not accommodated in the density. The distal regions of S2 in all of the three crowns are accommodated in the density and run approximately parallel to the filament axis close to the surface of the filament backbone. The conformation of S2 on each crown is illustrated in Fig. S10, which shows that on each level, S2 is in a similar position in the region between its junction with the C-terminal region of the heads (i.e., end of the lever arm) and its interaction with the blocked head. This conformation is very similar to that observed in tarantula myosin filaments (9, 10), suggesting that the intramolecular interaction between S2 and the actin-binding domain of the blocked motor domain may be conserved between tarantula and human cardiac myosin filaments. Beyond its interaction with the blocked head, S2 continues in a similar direction on level 1, but on levels 2 and 3 it is hinged to the right. The difference in orientation of S2 in this region serves to accommodate the difference between the tilt angle of the head pair on level 1 and the tilt angle of the head pairs on levels 2 and 3 (19°, as documented in Table S1; see also Fig. S9), while allowing the distal regions of S2 to remain approximately parallel to the filament axis.
Location of Nonmyosin Protein Densities: Titin and cMyBP-C.
Having identified the density in the structure of the human cardiac myosin filament that can be ascribed to the myosin heads and S2, we could now seek to identify the nonmyosin components. Alongside the 2,3,1 crown sequence of myosin heads with strong intermolecular interactions, the myosin filament is characterized by regions with prominent and well-defined surface features with a regular modulation every 39 Å, consistent with the size of the Ig and Fn3 domains of titin and cMyBP-C. Accordingly, we sought to develop models of titin and cMyBP-C to fit these densities.
Titin is thought to extend from the Z band (N terminus) to the M band (C terminus), with six titin strands per half thick filament (20, 21), and it is likely that the six titins are grouped as three strands of titin pairs. Titin Ig and Fn3 domains are arranged in long-range patterns or superrepeats. Within the myosin filament bridge region, two types of titin superrepeats are found. There are 7 consecutive copies of the 7-domain “small” superrepeat in the D zone at the distal edge of the A band. These are followed by 11 copies of the 11-domain “large” superrepeat. The 11-domain repeats each follow the sequence (N)-Ig-Fn3-Fn3-Ig-Fn3-Fn3-Fn3-Ig-Fn3-Fn3-Fn3-(C). The axial distance between each large superrepeat copy is 429 Å, and this series ends at the edge of the bare zone (22–25). The 11 copies of the 11-domain large superrepeat of titin cover both the C zone and the P zone (Fig. S1B). cMyBP-C is a modular structure comprising 11 Fn3 and Ig domains (C0–C10) (26, 27).
We used the crystal structure of the Fn3 tandem A77–A78 domains (22) (PDB ID code 3LPW) from the part of titin in the C-zone region to create models of titin and cMyBP-C (Fig. 4 and Movie S2). Each Fn3 domain is also homologous to Ig domains, and is thus likely to be similar in structure, particularly at the resolution of our current analysis (∼28 Å). The titin model consists of a series of 11 appropriately spaced domains: Two parallel copies are docked into the region alongside the 2,3,1 crown sequence of myosin heads extending 429 Å along the filament. We considered it likely that two strands could be side by side and in contact, and indeed these match the density well (yellow in Fig. 4). We also used the Fn3 structure to fit three further domains that are sandwiched between the titin and the myosin head pair on crown 1 (pink in Fig. 4). It is likely that these are the C-terminal part of cMyBP-C domains C8–C10. With this fit, these domains run parallel to the myosin filament axis in agreement with Squire et al. (28) and not as a collar, as suggested by Moolman-Smook et al. (29). With this arrangement, there is an apparent interaction between the C10 domain of cMyBP-C and the motor domain of the free head of crown 1.
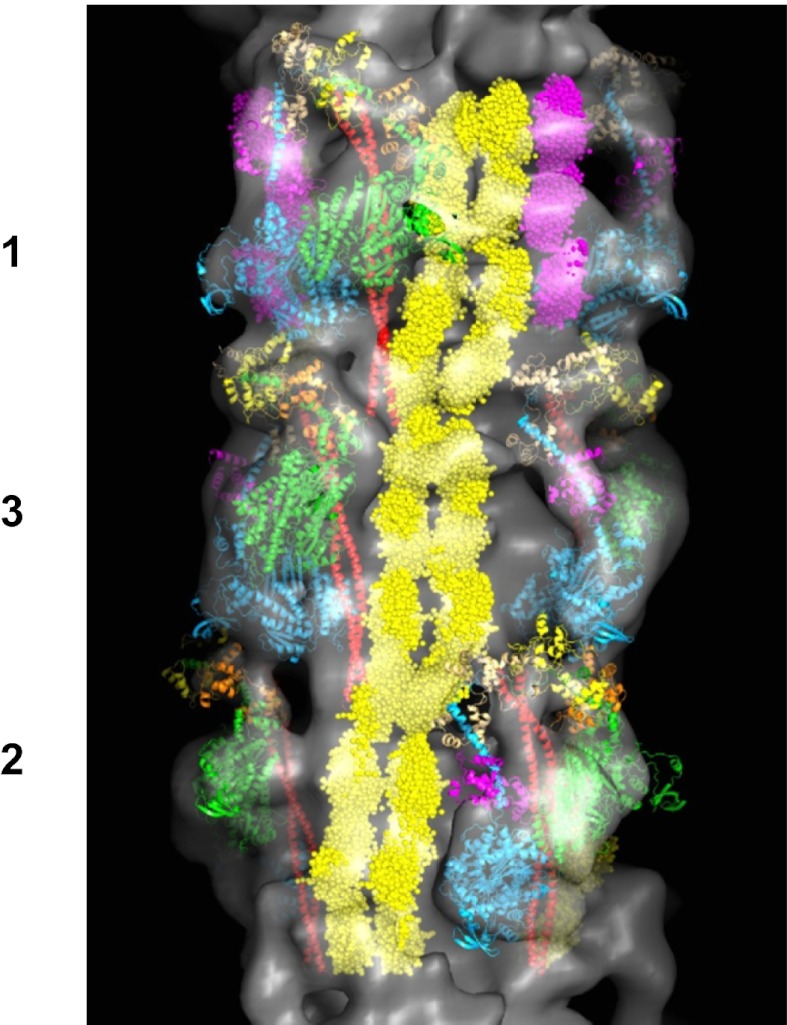
Fig. 4.
Final 3D map (Fig. 1) showing a full 429-Å repeat, shown with myosin head pairs, S2 (red), titin (yellow), and the three C-terminal domains of cMyBP-C (C8–C10) (magenta). The bare-zone direction is toward the bottom.
These modeled locations for titin and cMyBP-C are consistent with the earlier proposals of Zoghbi et al. (16) based on their structural analysis of native and knockout mouse cardiac myosin filaments. However, unlike the previous study, the additional detail achieved in the current analysis allows explicit docking of model structures for the individual domains. The assignment of the three domains to cMyBP-C is also consistent with their location adjacent to the crown 1 myosin heads, whereas the suggestion that these represent C8–C10 is consistent with the location of binding sites to myosin heavy chain (MHC), light meromyosin (LMM), and titin within these domains (30).
Discussion
The structure described here for the human cardiac myosin filament in the normal (undiseased) state is an essential starting point from which to understand the mechanisms of various cardiac diseases, in particular the effects of mutations in myosin, C protein, and titin associated with different cardiomyopathies. The current analysis shows a substantial improvement in the resolution and level of detail compared with previous studies of vertebrate striated muscle myosin filaments and allows detailed modeling of myosin heads and accessory proteins and their interactions. Using a negative-stain approach, we have achieved a degree of resolution and detail that approaches that obtained with frozen-hydrated preparations of highly ordered invertebrate myosin filaments (9, 10). We have been able to exploit the high level of detail to demonstrate that the myosin head pairs in each crown adopt the off-state conformation previously identified in smooth muscle myosin (11) and tarantula myosin filaments (9, 10). These observations are consistent with a previous analysis of mouse cardiac myosin filaments at lower resolution (16) that identified the off-state conformation on crowns 1 and 3. These studies, however, were unsuccessful in recovering head-pair density on crown 2, and it was suggested that this was due to higher mobility. The higher resolution in the current analysis allows us to recognize the off-state head-pair conformation on crown 2 and identify the orientation of the head pair by docking. It is apparent that recovery of density for the head pair is less-complete on level 2, consistent with the earlier suggestion of increased mobility in this region. Increased mobility on crown 2 may be a consequence of the pattern of intermolecular interactions between crowns, because crown 2 lacks the interaction of its free-head motor domain with the lever arm of the adjacent crown 1. The docked locations for titin and cMyBP-C described here are broadly consistent with previous proposals (16). However, the enhanced resolution of the current study makes explicit docking of both the myosin heads and S2 possible and allows the development of a molecular model. The locations we identify for the myosin heads are quite close to the backbone, and the groupings of three crowns show well-defined interactions between myosin head pairs in adjacent crowns. A head conformation recently identified biochemically has been called the superrelaxed (SRX) state, in which the myosin ATPase activity is inhibited (31–33). It was concluded that the myosin heads in the SRX state are strongly stabilized on the backbone of the thick filament and have the J-like motif (11). It could well be this particular structure, with many of the heads making strong (sometimes blocking) interactions with their neighbors, that we have now captured in our current cardiac myosin filament preparation.
In our structure, the interaction between levels 3 and 1 is between the motor domain of the free head on level 1 and the RLC of the blocked head on level 3 below it. By comparing the sequence of the human cardiac RLC with that of tarantula, the origin of the myosin head coordinates used here (10), it is apparent that serine 15, which is the major phosphorylation site in the human RLC, is at the base of an α-helix that forms a prominent contact between the RLC and the free-head motor domain on level 1 (Fig. 3B, red star). Thus, phosphorylation might well act to destabilize this interaction in a similar manner to the phosphorylation effect proposed by Alamo et al. (10) for the tarantula myosin filament. From these considerations, it seems likely that the relaxed myosin filaments studied here are substantially in the dephosphorylated state. Moreover, X-ray diffraction studies of mouse heart muscle (34) have shown an increase in the proximity of the heads to actin and thus a displacement of the heads away from the backbone upon phosphorylation. Conversely, in the dephosphorylated state, the myosin heads would be predicted to be tilting back toward the backbone surface, as observed in our structure, and forming interactions, as proposed by Stewart and colleagues (31–33) for the SRX state.
Materials and Methods
Chemicals.
Type III lyophilized powder elastase from porcine pancreas (code E0127), N-p-tosyl-L-phenylalanine chloromethyl ketone (code T4376), aprotinin from bovine lung (code A1153), pepstatin A from microbial source (code P5318), trypsin inhibitor from Glycine max (soyabean) (code T6522), PMSF (code P7626), and leupeptin hemisulfate salt from microbial source (code L2884) were obtained from Sigma-Aldrich. The elastase obtained from Sigma-Aldrich with code E0127 had an elastase content of 55–85%, elastase enzymatic activity ≥4% unit/mg, and trypsin activity ≤200 unit/mg.
Isolation of Myosin Filaments from Human Heart Muscle.
Myosin filaments were isolated from human ventricular muscle under relaxing conditions (35). To avoid chloride-induced disorder of myosin heads (36), MgCl2 was replaced by MgAc in both the relaxing solution and the rinsing solution for EM grids. Similarly, KCl was replaced by NH4Ac in the EM rinsing solution. Ventricular muscles removed from donor human hearts were frozen and stored in liquid nitrogen until use. Small muscle pieces, about 80 mg, were dissected and thawed for 30 min in relaxing solution containing the components listed in SI Text 1. Samples were cut with a razor blade into small pieces and then teased with a pair of forceps into fine bundles 0.5 mm or less in diameter on a microscope slide and stirred for 1 h in 10 mL of fresh relaxing solution to ensure full relaxation (assessed by light microscope as showing long uncontracted sarcomeres). Myosin filaments were released by incubation for 3 min in 0.22 mg/mL elastase and 0.44 mg/mL trypsin inhibitor in a relaxing solution containing the components listed in SI Text 2. The reaction was stopped by transfer into the same buffer as in SI Text 2, but with 1 mM PMSF and no elastase. After vigorous shaking by hand for 3 min and then gentle pipetting, the muscle broke up, yielding a suspension of separated thick and thin filaments. The suspension was centrifuged for 2 min at 735 × g using an Eppendorf Centrifuge 5415C, and EM grids were made using the supernatant. Donor heart muscle tissue samples were collected by C. dos Remedios, University of Sydney, Australia. Ethical approval was obtained from the Brompton, Harefield, and NHLI Ethics Committee, London, UK and the Australian Red Cross Blood Service and St. Vincent's Hospital, Sydney, Australia. The investigation conformed to the principles outlined in the Declaration of Helsinki.
Negative Staining and Electron Microscopy.
Five microliters of the isolated myosin filament suspension was applied to 400-mesh copper grids with a thin carbon film (5–7 nm thickness) supported by a holey formvar film and left for 10 s. The grids were rinsed sequentially with eight drops of half-strength relaxing solution containing the components listed in SI Text 3 and then five drops of 1% (wt/vol) uranyl acetate and 0.025% glycerol (37, 38). Samples were blotted with filter paper after each drop of uranyl acetate stain except for the last drop of stain, which was left on the grid for 30 s. Most of the stain was then removed with filter paper and the grid was rapidly dried with a hair dryer. Samples were imaged at room temperature with an FEI Tecnai TF20 electron microscope at an accelerating voltage of 200 kV, in low-dose mode with an exposure of ∼100 e−/Å2, a nominal magnification of 29,000×, and an underfocus chosen to place the first minimum in the contrast transfer function at ∼17 Å. Images were recorded using a Tietz F415 (4k × 4k) CCD camera and fields were binned by a factor of 2, resulting in a calibrated sampling of 17 μm per pixel (equivalent to 5.86-Å sampling in the specimen). Only filaments lying over holes were used in the current analysis.
Preparation of Filament Segments and 3D Analysis.
CCD frames were collected and evaluated using MRC (39), IMAGIC (40), and locally developed software. Regions were selected that contained intact half-filaments that were relatively straight and minimally overlapped by actin and other myosin filaments, with readily identifiable bare zones (Fig. S1 A and B). Whole myosin filaments were cut into two half-filaments (each including the whole bare zone) and half-filaments were oriented with their bare zones pointing vertically downward (Fig. S1B). Fourier transforms of the half-filaments were calculated. The spacing of the sixth order of the 429-Å repeat, the 71.5-Å meridional reflection, was used to calibrate the magnification and to adjust the sampling of each half-filament from all of the different micrographs to be exactly 5.94 Å/pixel. This sixth-order meridional reflection was particularly strong in most of the Fourier transforms. From the C zone of these half-filaments, particles/segments were extracted, each segment being centered on a 429-Å repeat and being 950 Å in length (just over 2 × 429 Å repeats). Three-dimensional analysis was conducted using the approach previously described for vertebrate myosin filaments (13, 14). During the later stages of the refinement, we used a brute-force multireference alignment program that performs rotational and translational alignment independently. We also used a locally developed Fourier space 3D reconstruction program (41). Docking of the atomic coordinates into the density was done manually with PyMOL (Version 1.2r3pre; Schrödinger), and then docking was refined computationally using the VEDA (http://mem.ibs.fr/GAEL/index.html) interface to URO (42).
Supplementary Material
Acknowledgments
This work was supported by British Heart Foundation Fellowship FS/07/017/22951 (to H.A.A.-K.). R.W.K. was supported by Minorities Basic Research Support Grant 5SC1HL96017 from the National Institutes of Health (NIH) and, in part, by funding from Research Centers in Minority Institutions Award G12RR-03051 from the National Center for Research Resources, NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The map of the human cardiac muscle myosin filament has been deposited in the EMDataBank (accession code EMD-2240).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212708110/-/DCSupplemental.
References
- 1.Huxley HE. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- 2.Squire JM. General model of myosin filament structure. II. Myosin filaments and cross-bridge interactions in vertebrate striated and insect flight muscles. J Mol Biol. 1972;72(1):125–138. doi: 10.1016/0022-2836(72)90074-5. [DOI] [PubMed] [Google Scholar]
- 3.Labeit S, Kolmerer B. Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270(5234):293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 4.Trinick J. Titin as a scaffold and spring. Cytoskeleton. Curr Biol. 1996;6(3):258–260. doi: 10.1016/s0960-9822(02)00472-4. [DOI] [PubMed] [Google Scholar]
- 5.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J Mol Biol. 1973;74(4):653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 6.Bennett P, Craig R, Starr R, Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J Muscle Res Cell Motil. 1986;7(6):550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 7.Craig R. Isolation, electron microscopy and 3D reconstruction of invertebrate muscle myofilaments. Methods. 2012;56(1):33–43. doi: 10.1016/j.ymeth.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Squire JM. Muscle myosin filaments: Cores, crowns and couplings. Biophys Rev. 2009;1(3):149–160. doi: 10.1007/s12551-009-0017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodhead JL, et al. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436(7054):1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 10.Alamo L, et al. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J Mol Biol. 2008;384(4):780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98(8):4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Khayat HA, Morris EP, Squire JM. Single particle analysis: A new approach to solving the 3D structure of myosin filaments. J Muscle Res Cell Motil. 2004;25(8):635–644. doi: 10.1007/s10974-004-5333-5. [DOI] [PubMed] [Google Scholar]
- 13.Al-Khayat HA, Morris EP, Kensler RW, Squire JM. 3D structure of relaxed fish muscle myosin filaments by single particle analysis. J Struct Biol. 2006;155(2):202–217. doi: 10.1016/j.jsb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Al-Khayat HA, Morris EP, Kensler RW, Squire JM. Myosin filament 3D structure in mammalian cardiac muscle. J Struct Biol. 2008;163(2):117–126. doi: 10.1016/j.jsb.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Khayat HA, Morris EP, Squire JM. The 7-stranded structure of relaxed scallop muscle myosin filaments: Support for a common head configuration in myosin-regulated muscles. J Struct Biol. 2009;166(2):183–194. doi: 10.1016/j.jsb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105(7):2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidman CE, Seidman JG. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: A personal history. Circ Res. 2011;108(6):743–750. doi: 10.1161/CIRCRESAHA.110.223834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marston SB. How do mutations in contractile proteins cause the primary familial cardiomyopathies? J Cardiovasc Transl Res. 2011;4(3):245–255. doi: 10.1007/s12265-011-9266-2. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: Visualization of the pre-power stroke state. Cell. 1998;94(5):559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 20.Liversage AD, Holmes D, Knight PJ, Tskhovrebova L, Trinick J. Titin and the sarcomere symmetry paradox. J Mol Biol. 2001;305(3):401–409. doi: 10.1006/jmbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- 21.Knupp C, Luther PK, Squire JM. Titin organisation and the 3D architecture of the vertebrate-striated muscle I-band. J Mol Biol. 2002;322(4):731–739. doi: 10.1016/s0022-2836(02)00819-7. [DOI] [PubMed] [Google Scholar]
- 22.Bucher RM, Svergun DI, Muhle-Goll C, Mayans O. The structure of the FnIII tandem A77-A78 points to a periodically conserved architecture in the myosin-binding region of titin. J Mol Biol. 2010;401(5):843–853. doi: 10.1016/j.jmb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 23.LeWinter MM, Granzier H. Cardiac titin: A multifunctional giant. Circulation. 2010;121(19):2137–2145. doi: 10.1161/CIRCULATIONAHA.109.860171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krüger M, Linke WA. The giant protein titin: A regulatory node that integrates myocyte signaling pathways. J Biol Chem. 2011;286(12):9905–9912. doi: 10.1074/jbc.R110.173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tskhovrebova L, Trinick J. Roles of titin in the structure and elasticity of the sarcomere. J Biomed Biotechnol. 2010;2010 doi: 10.1155/2010/612482. Article 612482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuda M, Koshida S, Sato N, Obinata T. Complete primary structure of chicken cardiac C-protein (MyBP-C) and its expression in developing striated muscles. J Mol Cell Cardiol. 1995;27(10):2275–2286. doi: 10.1016/s0022-2828(95)91731-4. [DOI] [PubMed] [Google Scholar]
- 27.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: A modulator of cardiac contraction? EMBO J. 1995;14(9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J Mol Biol. 2003;331(3):713–724. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 29.Moolman-Smook J, et al. Identification of novel interactions between domains of myosin binding protein-C that are modulated by hypertrophic cardiomyopathy missense mutations. Circ Res. 2002;91(8):704–711. doi: 10.1161/01.res.0000036750.81083.83. [DOI] [PubMed] [Google Scholar]
- 30.Oakley CE, Chamoun J, Brown LJ, Hambly BD. Myosin binding protein-C: Enigmatic regulator of cardiac contraction. Int J Biochem Cell Biol. 2007;39(12):2161–2166. doi: 10.1016/j.biocel.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci USA. 2010;107(1):430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooijman P, Stewart MA, Cooke R. A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys J. 2011;100(8):1969–1976. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naber N, Cooke R, Pate E. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J Mol Biol. 2011;411(5):943–950. doi: 10.1016/j.jmb.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colson BA, et al. Differential roles of regulatory light chain and myosin binding protein-C phosphorylations in the modulation of cardiac force development. J Physiol. 2010;588(Pt 6):981–993. doi: 10.1113/jphysiol.2009.183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kensler RW, Harris SP. The structure of isolated cardiac myosin thick filaments from cardiac myosin binding protein-C knockout mice. Biophys J. 2008;94(5):1707–1718. doi: 10.1529/biophysj.107.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kensler RW, Levine RJ. An electron microscopic and optical diffraction analysis of the structure of Limulus telson muscle thick filaments. J Cell Biol. 1982;92(2):443–451. doi: 10.1083/jcb.92.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart M, Kensler RW. Arrangement of myosin heads in relaxed thick filaments from frog skeletal muscle. J Mol Biol. 1986;192(4):831–851. doi: 10.1016/0022-2836(86)90032-x. [DOI] [PubMed] [Google Scholar]
- 38.Kensler RW, Stewart M. Frog skeletal muscle thick filaments are three-stranded. J Cell Biol. 1983;96(6):1797–1802. doi: 10.1083/jcb.96.6.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116(1):9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 40.van Heel M, et al. Single-particle electron cryo-microscopy: Towards atomic resolution. Q Rev Biophys. 2000;33(4):307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- 41.da Fonseca PCA, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46(1):54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 42.Navaza J, Lepault J, Rey FA, Alvarez-Rua C, Borge J. On the fitting of model electron densities into EM reconstructions: A reciprocal-space formulation. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 10 Pt 2):1820–1825. doi: 10.1107/s0907444902013707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.