Significance
The innate immune system, including natural killer (NK) cells, is responsible for limiting virus spread during the initial phases of an infection. Here, we show in mice that NK cells are recruited to lymph nodes that drain the site of infection with cowpox virus, which is endemic in wild rodents. Full NK-cell recruitment was dependent on their expression of a specific receptor for soluble attractants termed chemokines, a hormone-like substance called IFN-γ, and subcapsular sinus macrophages. Therefore, this study provides a framework for how NK cells are recruited to a site of infection.
Abstract
Natural killer (NK) cells provide in vivo control of orthopoxvirus infections in association with their expansion in the draining lymph node (LN), where they are normally very rare. The mechanism of this expansion is unclear. Herein, we determined that NK-cell depletion results in enhanced infection following footpad inoculation of cowpox virus, a natural pathogen of rodents. Following cowpox virus infection in normal mice, NK cells were greatly expanded in the draining LN, were not replicating, and displayed markers similar to splenic NK cells, suggesting specific recruitment of splenic NK cells rather than in situ proliferation. Moreover, NK-cell expansion was abrogated by prior injection of clodronate-loaded liposomes, indicating a role for subcapsular sinus macrophages. Furthermore, recruitment of transferred splenic NK cells to the draining LN was pertussis toxin-sensitive, suggesting involvement of chemokine receptors. Comprehensive analysis of chemokine mRNA expression in the draining LN following infection suggested the selective involvement of CCR2, CCR5, and/or CXCR3. Mice deficient for CCR2 or CCR5 had normal NK-cell recruitment, whereas CXCR3-deficient mice displayed a major defect, which was NK cell-intrinsic. Interestingly, both induction of transcripts for CXCR3 ligands (Cxcl9 and Cxcl10) and NK-cell recruitment required IFN-γ. These data indicate that NK-cell recruitment is mediated by subcapsular sinus macrophages, IFN-γ, and CXCR3 during orthopoxvirus infection.
Orthopoxvirus infections have caused significant mortality and morbidity, as exemplified by variola virus, the causative agent of smallpox, being highly lethal to humans (1). Cross-protective vaccination efforts with vaccinia virus (VV) by the World Health Organization successfully removed the threat of natural smallpox infection, but concerns remain about its use as a bioterrorism agent. Additionally, other orthopoxviruses cause emerging zoonotic infections in humans, including monkeypox virus, which is mainly limited to Africa but caused an outbreak in the midwestern United States (2, 3). Sporadic but increasing numbers of zoonotic infections with cowpox virus (CPXV) have recently been reported in Europe (4–7,) where CPXV is endemic in wild rodents (8). Inoculation by skin scratch appears to be the predominant means for zoonotic infections with CPXV. Because vaccination for orthopoxviruses is no longer routinely recommended for the general public, in part due to significant side effects, vaccine-unprotected individuals will grow in number, and even among vaccinated subjects, immunity may wane (9, 10). Moreover, immunocompromised patients and those with atopic dermatitis are particularly susceptible to severe infections (11). Therefore, continued study of primary immune responses to orthopoxviruses is of interest to provide additional insight into the host immune response, which may lead to novel clinical advances to protect against emerging orthopoxvirus infections.
Innate host immune responses to orthopoxvirus infection are of particular interest because they occur soon after inoculation. Natural killer (NK) cells play an important role in the innate immune response to tumors and infections by causing lysis of transformed or infected cells and producing cytokines that help shape other immune responses. Activation of NK-cell function is mediated through two distinct mechanisms: integration of signaling through inhibitory and activation receptors (12, 13) and cytokine stimulation (14–16). Down-regulation of MHC class I molecules on a cellular target, which otherwise mediate strong signals through inhibitory receptors, renders the cell sensitive to NK lysis (17). Ligands for activation receptors can either be pathogen-derived or host-derived and up-regulated on cell stress (18, 19). Although stimulation with cytokines, including IL-12, IL-15, and IL-18, has also been shown to be effective at activating NK cells to produce other cytokines, NK-cell effector functions are largely determined locally because cell-to-cell contact is required for NK-cell lysis of infected cells.
To allow productive interactions between NK cells and their targets, NK cells need to be recruited to the relevant site. In noninfectious settings, several studies have implicated the chemokine receptor CXCR3, whose ligands (CXCL9, CXCL10, and CXCL11) are induced by IFN-γ following corneal epithelial injury (20), implantation of tumor cells that endogenously express IFN-γ (21), and LPS-matured dendritic cell footpad injection (22). By contrast, in parasitic and viral infections, NK cells have differential requirements for chemokine receptors, generally not CXCR3. Following intratracheal inoculation of neutropenic mice with Aspergillus fumigatus, CCR2 is required (23), whereas CCR5 is required in Toxoplasma gondii (24) or herpes simplex virus 2 (HSV2) (25) infections, with a higher pathogen burden in CCR5-deficient mice. However, Aspergillus and HSV2 are not natural pathogens of rodents. For protection against murine cytomegalovirus (MCMV), a natural pathogen of mice, recruitment of NK cells to the liver is mediated through CCR2 (26, 27). CXCR3 and CCR5 are also important for NK-cell trafficking from the red pulp to the T-cell zone in the spleen following injection of polyinosinic:polycytidylic acid and MCMV (28). Finally, CXCR6 is required for memory NK cells in the liver, which respond to contact-mediated hypersensitivity or viral-like particles (29). Thus, several chemokine receptors are involved in recruitment of NK cells, although little is known about NK-cell recruitment to the draining lymph node (LN) in the context of natural rodent pathogens.
In terms of orthopoxvirus infections, much has been learned regarding NK cells during infections in mice with VV (30) and ectromelia virus (ECTV) (31, 32), although the endemic hosts for these viruses remain obscure. In the absence of NK cells either genetically or by anti-NK1.1 depletion, VV and ECTV infections were uncontrolled. Moreover, during ECTV infections, the NK-cell receptors, CD94 and NKG2D, are required for protection and NK cells expand in the draining LN, suggesting that local expansion of NK cells is required for viral control (32, 33). However, the basis for NK-cell expansion was not determined.
Because CPXV has coevolved with its rodent hosts, CPXV infection of rodents is appropriate for study of orthopoxvirus–host interactions, but the role of NK cells in CPXV infection has not been investigated. CPXV has the largest genome in the orthopoxvirus genus (34) and contains several ORFs not found in either the VV or ECTV genome. This includes two ORFs, CPXV012 and CPXV203, that potently down-regulate the expression of MHC class I (35–38). Although this might render the infected cell sensitive to NK cell-mediated lysis, CPXV also encodes another ORF, CPXV018, that binds in vitro with high affinity to the NKG2D activation receptor and blocks its recognition of ligands on target cells (39). Additionally, all orthopoxviruses, including CPXV, encode a number of cytokine binding proteins, which could also limit the activation of NK cells, such as one specifically targeting IL-18 (40, 41), a well-known mediator of NK-cell activation. Given the potential for CPXV to encode a larger repertoire of genes for evasion of immune responses, possibly including NK cells, it was of interest to determine whether NK cells could control CPXV infection, if NK cells are recruited to the site of infection, and the basis for this potential recruitment.
In this study, we determined the role of NK cells following footpad inoculation of CPXV infection that mimics natural viral transmission within the rodent population as well as zoonotic spread. We found that NK cells inhibited early viral replication and prevented dissemination of CPXV to other organs. NK cells expanded in the draining LN due to recruitment of splenic NK cells. Full NK-cell recruitment was dependent on subcapsular sinus (SCS) macrophages, mediated by IFN-γ, and required the intrinsic expression of the CXCR3 chemokine receptor on NK cells.
Results
NK Cells Limit Viral Replication Following CPXV Footpad Infection.
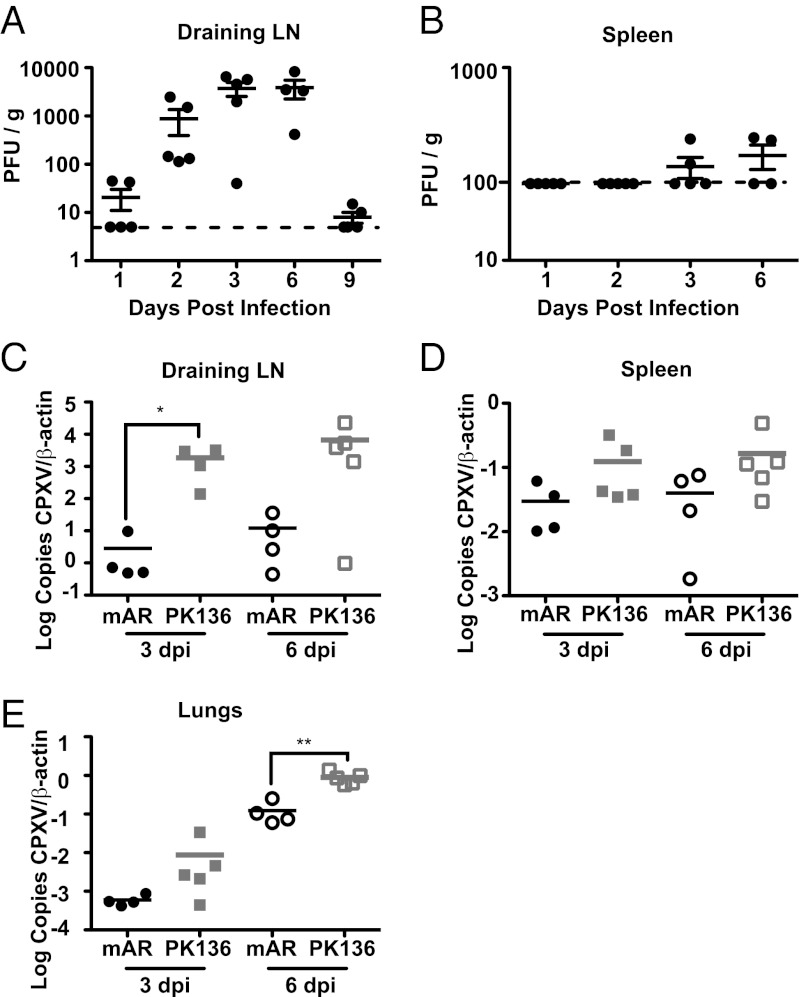
Following footpad inoculation of C57BL/6 mice, we observed that CPXV was predominantly found in the draining popliteal LN, with peak titers occurring between 3 and 6 d postinfection (dpi) (Fig. 1A). Furthermore, at 3 dpi, CPXV was barely detectable in the spleen (Fig. 1B) and could not be detected in the liver by traditional plaque assay. Although plaques were detected at low levels on 3 and 6 dpi in the spleen, the relative absence of active replication contrasts with other orthopoxviruses, such as VV, which resulted in readily detectable plaques in the spleen as early as 2 dpi following footpad inoculation (Fig. S1). When we used quantitative PCR as a sensitive measure of viral genome copies, we were able to detect CPXV at relatively higher levels in the foot and draining LN and at lower levels in spleen, lungs, liver, kidney, and uterus and ovaries at 3 dpi, with an increase in these copy numbers at 6 dpi, except in the spleen (Fig. 1 and Fig. S2). Thus, early following footpad inoculation, CPXV infection is generally limited to the draining LN and the site of inoculation.
Fig. 1.
NK cells limit viral replication and spread following footpad infection. C57BL/6 mice were infected with 1.5 × 106 pfu of CPXV in the footpad; at the indicated time points, organs were removed and viral titers were assessed in the draining LN (A) and spleen (B) by plaque assay. Dotted lines indicate the limit of detection by the assay. Symbols indicate individual mice, and lines indicate the mean. n = 4–5 mice per group were analyzed for each time point. (C–E) C57BL/6 mice were injected i.p. with 200 μg of control antibody (mAR) or PK136 antibody 2 d before infection and 4 dpi. At 3 and 6 dpi, organs were removed and DNA was isolated. Quantitative PCR was used to assess viral burden in the LN (C), spleen (D), and lung (E). CPXV copy number was normalized to β-actin copy number and then multiplied by 1,000. Symbols indicate individual mice, and lines indicate the mean. n = 4–5 mice per group were analyzed for each time point. Data are representative of three independent experiments.
Although NK cells are important for resistance to several orthopoxvirus infections (30–32), their role in controlling CPXV infection was unknown. To address this issue, we used PK136 antibody (anti-NK1.1) to deplete NK cells systemically (31, 42) before infection with CPXV (Fig. S2A). Although systemic depletion of NK cells did not lead to increased mortality, we observed that viral replication was substantially increased in the draining popliteal LN (Fig. 1C). Additionally, there was a trend for increased viral replication in the spleen and the lungs of NK cell-depleted mice at both 3 and 6 dpi, which was statistically significant in the lungs at 6 dpi (Fig. 1 D and E). However, NK-cell depletion did not affect all organs, because an increase in viral replication was not found in the foot, liver, kidney, or uterus and ovaries (Fig. S2 B–E). These data suggest that NK cells limit early viral replication in the draining LN and, in doing so, prevent early dissemination of virus to certain distant sites.
NK-Cell Expansion Following CPXV Infection.
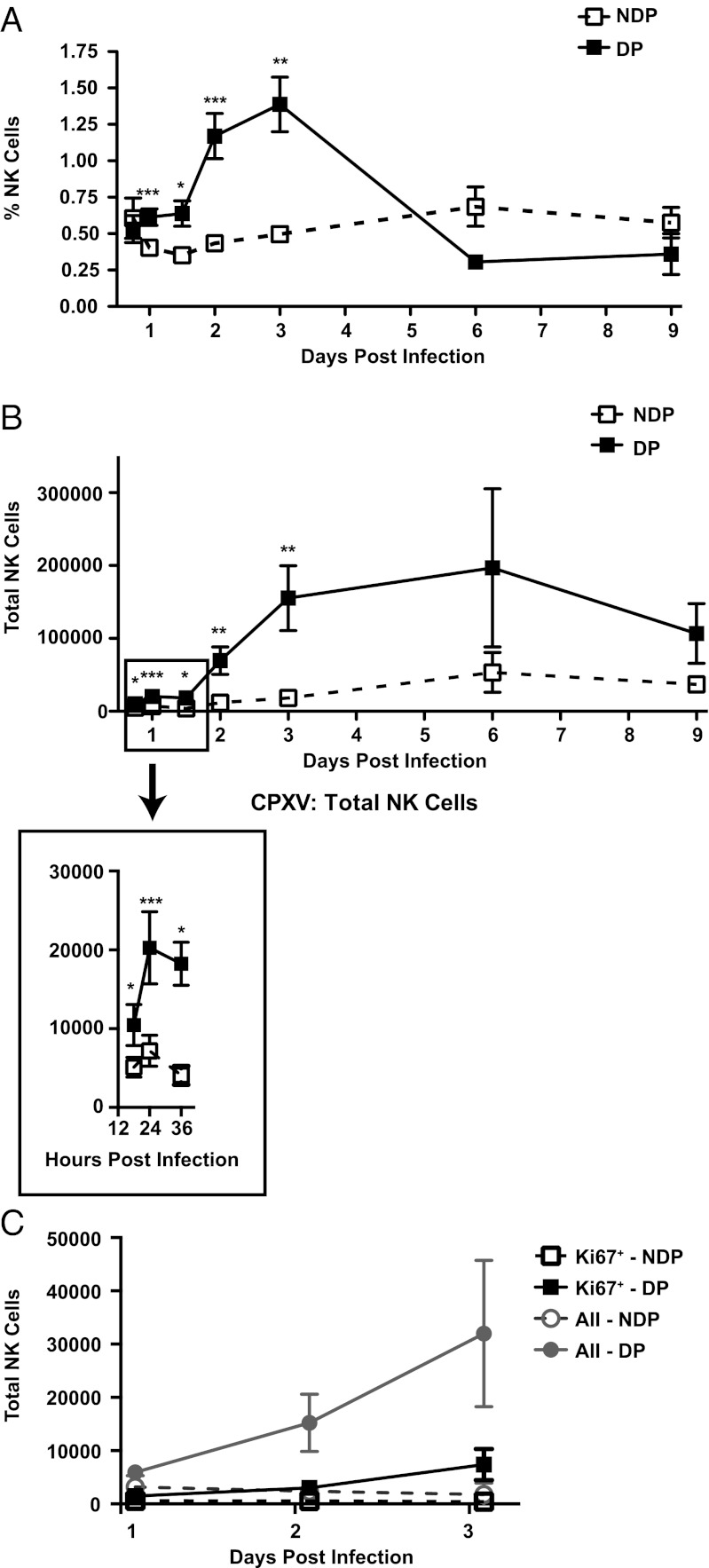
The aforementioned experiments indicated local control of CPXV by NK cells in the draining LN even though NK cells are rare in LNs of naive mice (43). Following CPXV infection, however, there was a statistically significant increase in both the percentage and total cell number of NK cells, detectable as early as 18 h postinfection (hpi) (Fig. 2 A and B, Inset). By 48 hpi, there was a substantial 10-fold increase in total NK cells in the draining LN in comparison to the nondraining LN. The NK-cell population was still increased up to 6 dpi and had contracted at 9 dpi in line with resolution of CPXV titers in the draining LN (Fig. 1A). Few NK cells in the draining LN stained for Ki67, a marker of cell cycle (Fig. 2C). These data suggest that NK-cell expansion was not due to in situ proliferation but to active recruitment.
Fig. 2.
CPXV causes an expansion of NK cells in the draining LN. C57BL/6 mice were infected as before and at the indicated time points, and cells from the draining and nondraining LN were isolated and stained. The percentages of (A) and total (B) NK cells were determined by staining for NK1.1+CD3−CD19− cells. Cell numbers were ascertained using trypan blue exclusion and then multiplied by the percentage of population found by FACS analysis. The graphs depict average NK-cell populations in the nondraining popliteal (NDP) or draining popliteal (DP) LN of n = 6 independent experiments (18 hpi): n = 14 independent experiments (1 dpi), n = 3 independent experiments (36 hpi), n = 8 independent experiments (2 and 3 dpi), and n = 2 independent experiments (6 and 9 dpi). Paired Student's t test: *P < 0.05; **P < 0.005; ***P < 0.0005. (C) Following extracellular staining, cells were fixed, permeabilized, and intracellularly stained for Ki67. Data are representative of three independent experiments.
Because there are several subsets of NK cells that reside in different tissues (44–46), we stained NK cells for cell surface molecules that distinguished between conventionally studied splenic NK cells and alternatively differentiated thymic NK cells (44, 45). NK cells isolated from the draining LN displayed high levels of Mac-1 (CD11b), Ly49 molecules (ADCIFH), L-selectin (CD62L), DX5 (CD49b), NKG2D (CX5), and CD94 (Fig. S3 A–F). The expression levels of these markers were the same on both NK cells isolated from the draining LN and from the spleen. In contrast, NK cells isolated from the thymus had lower expression of these molecules. We also examined a specific thymic NK-cell marker, the IL-7 receptor α-chain (IL-7Rα, CD127) and found that NK cells in the draining LN and spleen had little to no expression of this marker (Fig. S3G). When we examined the surface expression of these markers on NK cells isolated from the nondraining LN, we found that these NK cells expressed an intermediate level of Mac-1, L-selectin, and IL-7Rα. This intermediate expression level could be due to the small numbers of NK cells in the naive LN representing a unique LN subset of NK cells or a mixture of both splenic and thymic NK cells present at steady state. Regardless, NK cells resembling the splenic NK-cell subset were enriched in the draining LN following CPXV infection.
Macrophage Depletion Leads to Diminished NK-Cell Expansion.
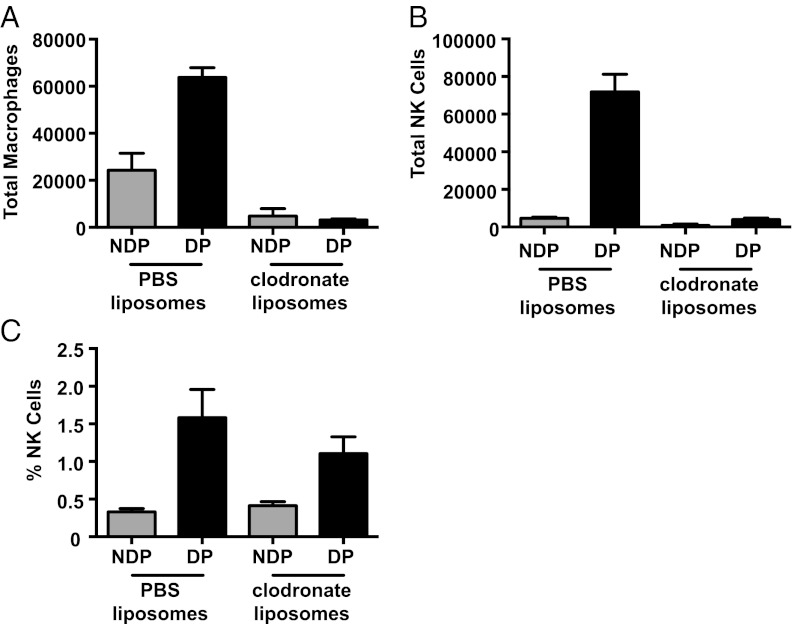
We previously showed that i.p. infection with CPXV, which expresses GFP in the place of CPXV203, showed virus colocalization with SCS macrophages in the draining mediastinal LN, indicating that SCS macrophages are infected in the draining LN following CPXV inoculation (47). Similarly, others have shown that macrophages are infected in the draining LN in cutaneous infection with VV, and, more recently, the colocalization of CD169-expressing SCS macrophages and NK cells has been noted following vesicular stomatitis virus, modified vaccinia Ankara (MVA), T. gondii, and Pseudomonas aeruginosa infection in draining LN (48–52). These data suggested that SCS macrophages may play a role in the recruitment of NK cells following CPXV infection. To investigate this possibility, 5 d before CPXV infection, we used clodronate-loaded liposomes to deplete SCS macrophages selectively, as others have done (48–53), in the draining LN. Following clodronate-loaded liposome treatment, we observed the expected decrease in the macrophage population in the draining LN following infection (Fig. 3A). There was also a corresponding significant decrease in the NK-cell population in the draining LN following CPXV infection (Fig. 3B) but not in percentage of NK cells (Fig. 3C). Thus, SCS macrophages appear to be required for NK-cell expansion following CPXV infection.
Fig. 3.
Macrophage depletion leads to decreased NK-cell recruitment. Mice were injected in both rear footpads with either PBS- or clodronate-loaded liposomes 5 d before infection. The macrophage population (A; F4/80+CD11b+) and the total (B) and percentage of the (C) NK-cell population (NK1.1+CD3−CD19−) were observed by flow cytometry 3 dpi in the nondraining popliteal (NDP) or draining popliteal (DP) LN. Representative results from two independent experiments are shown.
NK-Cell Expansion Can Be Attributed to Recruitment to Draining LN.
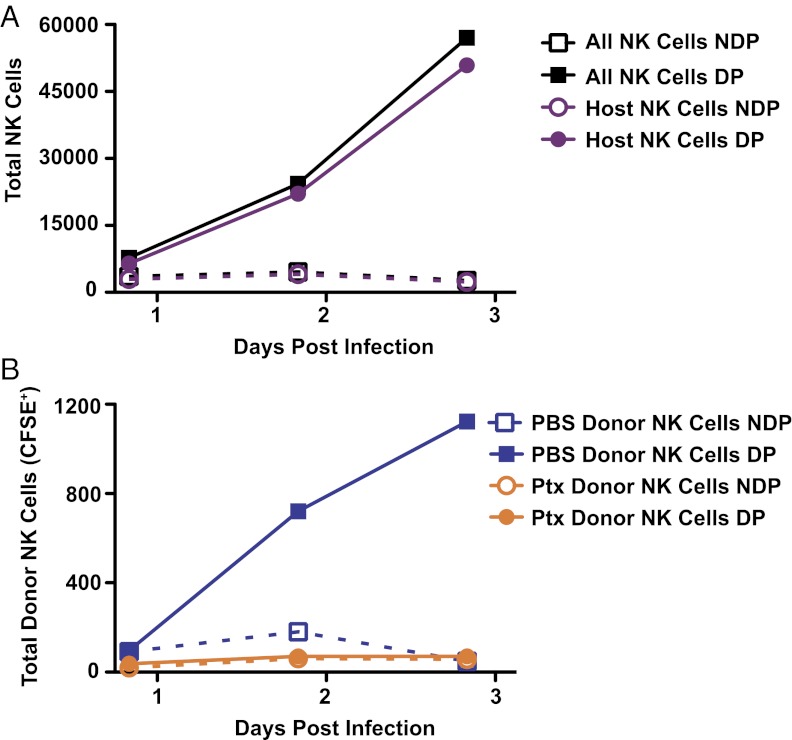
Given the rapid kinetics of the NK-cell population increase, the minimal number of NK cells expressing a specific cell cycle marker, the splenic marker profile of the NK cells isolated from the draining LN, and the dependence on SCS macrophages, we examined the possibility that NK cells were being recruited to the draining LN. We isolated splenocytes from Ly5.1 and Ly5.2 congenic mice and pretreated them in vitro for 2 h with either PBS or pertussis toxin. Following pretreatment, the cells were washed extensively, labeled with carboxyfluorescein succinimidyl ester (CFSE), and mixed together before adoptive transfer into congenic C57BL/6 (Ly5.2) mice. We found that donor splenic NK cells treated with PBS specifically increased in the draining LN following adoptive transfer and CPXV infection (Fig. 4). The increase in the transferred PBS-treated NK cells was similar to the kinetics of host NK-cell increase following CPXV infection, supporting the hypothesis that splenic NK cells are being recruited to the draining LN following CPXV infection.
Fig. 4.
NK expansion in the draining LN is due to pertussis toxin-sensitive recruitment. Splenocytes isolated from congenic CD45.1 and CD45.2 B6 mice were pretreated in vivo with PBS or pertussis toxin for 2 h at 37 °C on 5% CO2 before being washed, CFSE-labeled, mixed, and coinjected i.v. into C56BL/6 mice. Immediately following adoptive transfer, the mice were infected with 1.5 × 106 pfu of CPXV in the footpad. At the indicated time points, LNs were harvested and NK cells (NK1.1+CD3−CD19−) were identified in the nondraining popliteal (NDP) and draining popliteal (DP) LN. Host NK-cell recruitment (A; CFSE−) or donor NK-cell recruitment (B; CFSE+) is shown over time. Data are representative of two independent experiments.
These findings led us to consider chemokine receptor-dependent NK-cell recruitment into the draining LN. As with other G protein-coupled receptors, chemokine receptors signal via guanine nucleotide binding proteins (G proteins); these signals are blocked by pertussis toxin (54, 55). We thus used pertussis toxin to test whether chemokine receptors were important for the NK-cell increase in the draining LN. Pertussis toxin-pretreated splenic NK cells were never found in the draining LN at any time point observed following CPXV infection (Fig. 4B). Although the cells were viable before transfer, as determined by trypan blue exclusion, it is difficult to control fully for off-target effects of pertussis toxin treatment. Despite these concerns, these data further suggested the possibility that splenic NK cells are being recruited to the draining LN following CPXV infection and additionally suggested that NK-cell recruitment occurs via chemokine receptors.
Chemokines Up-Regulated in Draining LN Following CPXV Infection.
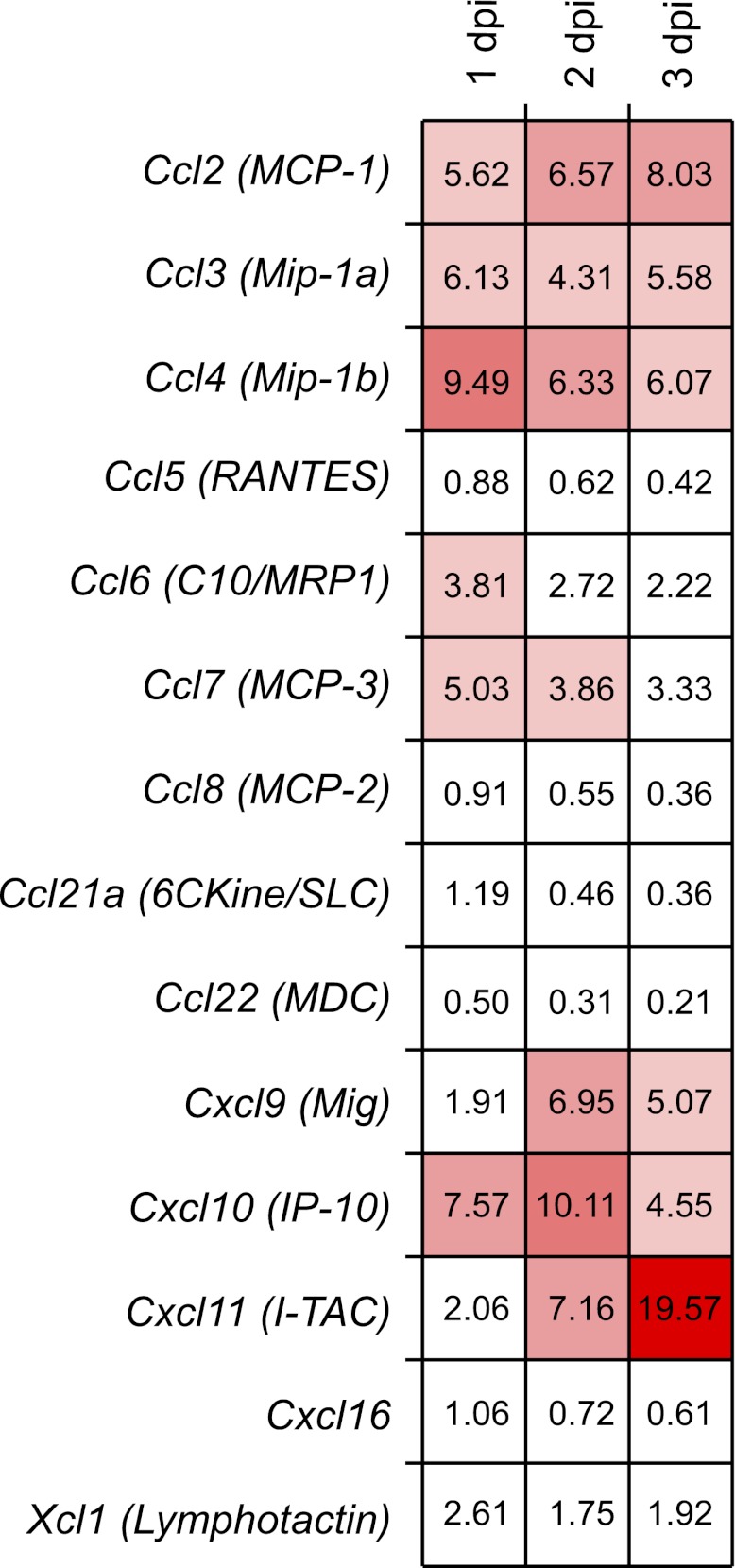
Because the data suggested a role for chemokines and chemokine receptor-mediated recruitment of NK cells following CPXV infection, we determined specifically which of these were required. To examine chemokine expression in the draining LN, we assessed transcript levels over time following CPXV infection by the use of a comprehensive bead-based array. In both draining and nondraining LN over time, we examined 33 transcripts simultaneously: Ccl1, Ccl2, Ccl3, Ccl4, Ccl5, Ccl6, Ccl7, Ccl8, Ccl9, Ccl11, Ccl12, Ccl17, Ccl19, Ccl20, Ccl21a, Ccl22, Ccl24, Ccl25, Ccl27a, Ccl28, Cxcl1, Cxcl2, Cxcl3, Cxcl5, Cxcl9, Cxcl10, Cxcl11, Cxcl12, Cxcl13, Cxcl14, Cxcl15, Cxcl16, and Xcl1 (Fig. S4).
Our analysis of these data was then informed by gene expression microarray data from a previously published study (56) and confirmed by our laboratory, indicating that NK cells express transcripts for Ccr2, Ccr5, Cxcr3, Cxcr4, and Cxcr6. We found that transcripts of ligands for CCR2 and CCR5, including Ccl2, Ccl3, and Ccl7, were up-regulated approximately threefold to ninefold relative to the nondraining LN transcripts (Fig. 5). Additionally, the ligands for CXCR3, namely, IFN-γ–inducible transcripts Cxcl9, Cxcl10, and Cxcl11, were up-regulated from fivefold to 19-fold relative to transcript levels in the nondraining LN. Although transcripts for Cxcl11 were the most highly induced, CXCL11 protein is not expressed in C57BL/6 mice due to a point mutation (57). Finally, we did not find increased transcripts for CXCR4 and CXCR6 ligands (Cxcl12 and Cxcl16, respectively) (Fig. S4). Nonetheless, CPXV infection induces up-regulation of chemokine ligands for chemokine receptors expressed on NK cells.
Fig. 5.
Inflammatory chemokine transcripts are up-regulated following CPXV infection. RNA transcript levels were assessed in C57BL/6 mice infected with 1.5 × 106 pfu of CPXV in the footpad at the indicated time points by multiplex gene expression analysis. LNs from three independently infected mice were isolated. Lysates from either the nondraining or draining LN were generated and run in triplicate, and chemokine transcripts were normalized to the control Hprt1 transcript for each sample. The fold change located in each square depicts the average transcript level in the draining LN relative to the nondraining LN. Here, we show transcript levels for chemokines whose receptors are expressed on NK cells. The results for other chemokines are illustrated in Fig. S4.
Specific Chemokine Receptor-Dependent Recruitment of NK Cells Following CPXV Infection.
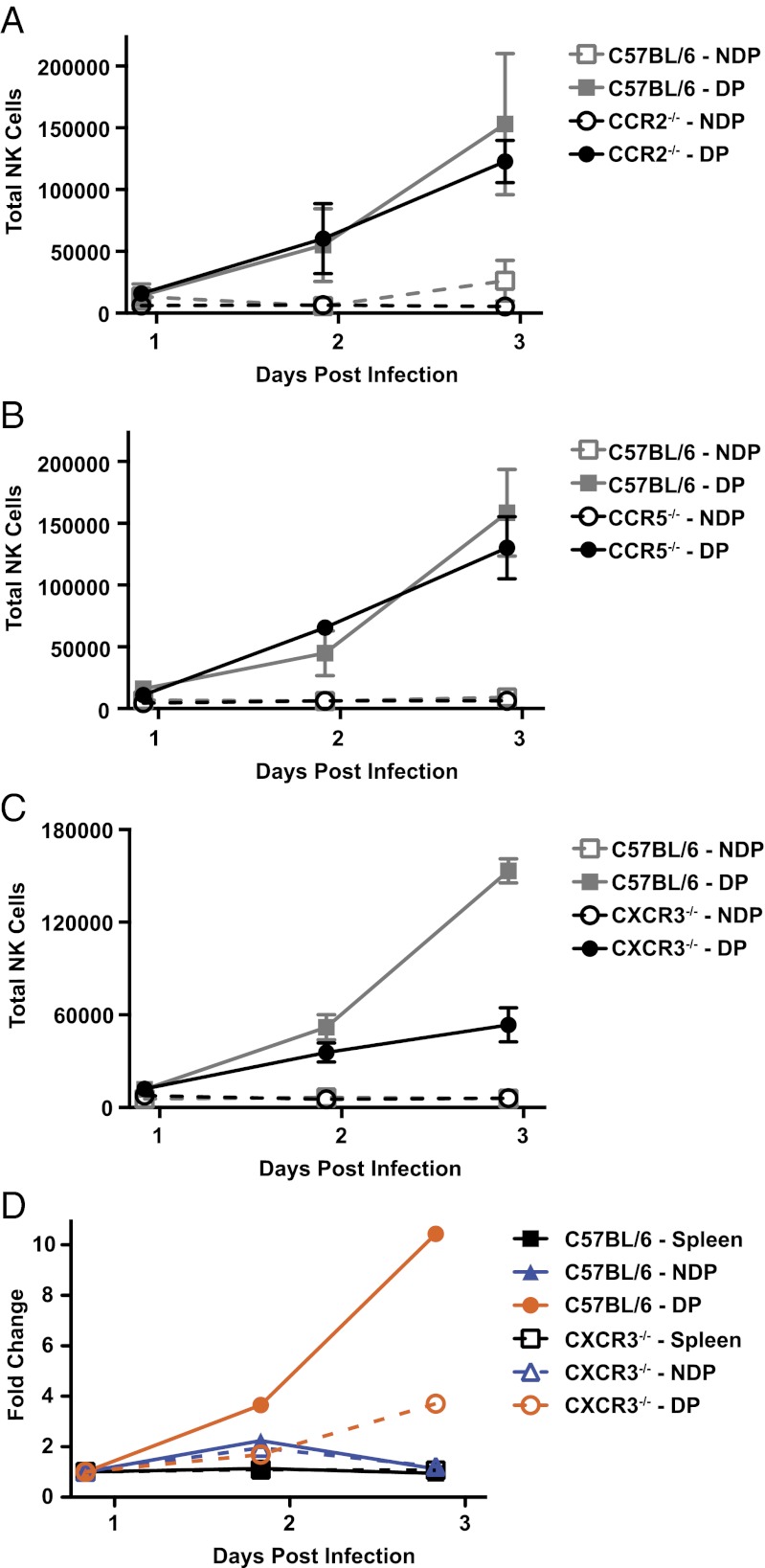
Our analysis of chemokine up-regulation following CPXV infection and NK-cell expression of specific chemokine receptors suggested selective involvement of CCR2, CCR5, or CXCR3 in the recruitment of NK cells. To analyze these possibilities further, we examined NK-cell recruitment in chemokine receptor-deficient mice. Despite up-regulation of transcripts for Ccl2, Ccl3, and Ccl7, there were little to no defects in the recruitment of NK cells in mice lacking either CCR2 or CCR5 (Fig. 6 A and B). Thus, CCR2 and CCR5 individually do not contribute to NK-cell recruitment following CPXV footpad infection.
Fig. 6.
CXCR3, but not CCR2 or CCR5, is required for full NK recruitment. CCR2−/− (A), CCR5−/− (B), or CXCR3−/− (C) mice and age-matched control C57BL/6 mice were infected as before, and NK-cell recruitment was monitored at the indicated time points in the nondraining popliteal (NDP) or draining popliteal (DP) LN. The graph depicts the average of three independent experiments for each chemokine-deficient strain. (D) C57BL/6 (CD45.1) and CXCR3−/− (CD45.2) splenocytes were isolated and enriched by negative selection before being CFSE-labeled, mixed, and cotransferred i.v. into C57BL/6 mice. Eighteen hours after the transfer, mice were infected as before, and NK-cell populations were examined at the indicated time points. To analyze the recruitment in this system, the total number of recruited WT or chemokine KO NK cells at 2 and 3 dpi were compared relative to the initial time point at 1 dpi in the NDP and DP LN. A representative result of three independent experiments is shown.
We next studied NK-cell trafficking to the draining LN in CXCR3-deficient mice. In these mice, we found that there was a substantial defect at 3 dpi, resulting in a marked decrease in trafficking of NK cells following CPXV infection, although NK-cell recruitment at earlier times was relatively unaffected (Fig. 6C). To determine whether these CXCR3-dependent trafficking defects were intrinsic or extrinsic to NK cells, we adoptively transferred a mixture of enriched splenic NK cells from WT and CXCR3-deficient mice into congenic C57BL/6 mice (Fig. 6D). We found that the fold change of recruited donor-derived NK cells for the adoptively transferred CXCR3-deficient NK cells was lower than for the adoptively transferred WT NK cells, especially at later time points, as observed directly in the CXCR3-deficient mice (Fig. 6C). These results indicate that NK cells intrinsically require CXCR3 expression for maximal recruitment.
To determine if CXCR3-dependent recruitment of the NK cells was required to limit viral replication and/or spread, we examined viral replication in C57BL/6 or CXCR3-deficient mice treated with a control or NK cell-depleting antibody (Fig. S5A). Regardless of NK-cell depletion, viral loads in the draining LN in the CXCR3-deficient mice were comparable to those found in C57BL/6 mice (Fig. S5B). Similarly, viral load was unaffected in a distant site, such as the lung (Fig. S5C). All together, these data indicate that CPXV can be controlled in the absence of CXCR3, even though NK-cell recruitment, especially at later time points, is affected.
IFN-γ Is Required for Full NK-Cell Recruitment.
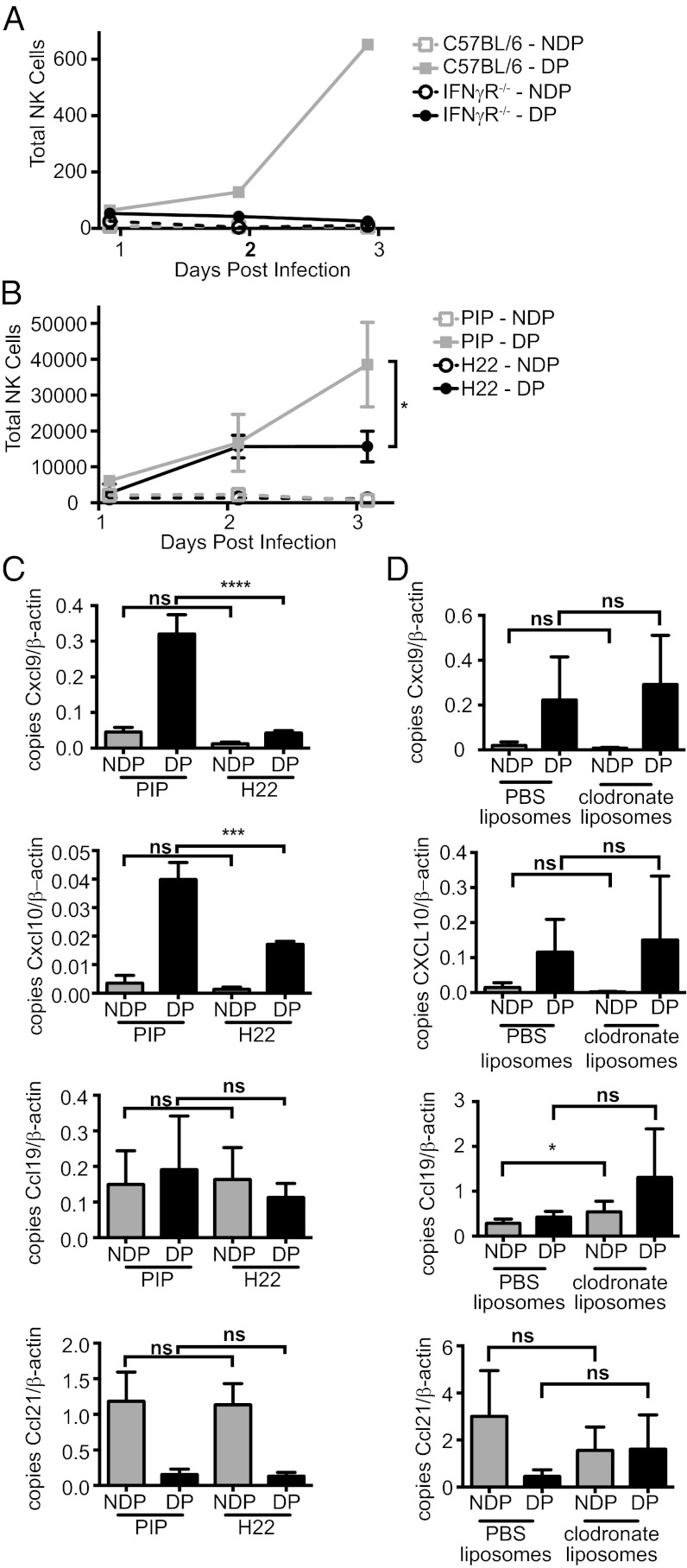
Inasmuch as chemokine ligands for CXCR3 are IFN-γ–inducible, we investigated NK-cell recruitment in mice deficient for the IFN-γ receptor. These mice displayed an impairment of NK-cell recruitment (Fig. 7A). These results were recapitulated in C57BL/6 mice that were pretreated with IFN-γ neutralizing antibody but not with a control antibody (Fig. 7B). When we examined expression of Cxcl9 and Cxcl10 transcripts by quantitative PCR, we found they were not induced in the draining LN following CPXV infection in mice treated with the IFN-γ neutralizing antibody (Fig. 7C). For controls, we examined transcripts for Ccl19 and Ccl21, chemokines responsible for homeostatic trafficking of lymphocytes to secondary lymphoid organs, which were unaffected in mice treated with IFN-γ neutralizing antibody (Fig. 7C). These data indicate that IFN-γ is required for the induction of CXCR3 ligands, and thus full recruitment of NK cells during CPXV infection.
Fig. 7.
Full NK-cell recruitment requires IFN-γ. (A) Mice deficient for IFN-γ receptor or age-matched control C57BL/6 mice were infected as before, and NK-cell recruitment was monitored at the indicated time points in the nondraining popliteal (NDP) or draining popliteal (DP) LN. A representative result of two independent experiments is shown. (B and C) C57BL/6 mice were pretreated with 250 μg per mouse of control (PIP) or IFN-γ neutralizing (H22) antibody 2 d before infection. (B) NK-cell recruitment was monitored as previously described at the indicated time points. (D) C57BL/6 mice were injected in both rear footpads with either PBS- or clodronate-loaded liposomes 5 d before infection. (C and D) Transcript analysis of Cxcl9, Cxcl10, Ccl19, and Ccl21 was analyzed by quantitative PCR. Data were pooled from two independent experiments with two (C) or three (D) mice per group for each experiment. Unpaired Student's t test: *P < 0.05; ***P < 0.0005; ****P < 0.0001; ns, not significant.
Because macrophages are capable of producing many inflammatory chemokines, including CXCL9 and CXCL10 (58, 59), it seemed likely that they might be responsible for the CXCR3-mediated NK-cell recruitment. Surprisingly, however, when we examined chemokine expression in the draining LN of mice treated with either control PBS-loaded liposomes or clodronate-loaded liposomes, we found no decrease in transcripts for Cxcl9 or Cxcl10 (Fig. 7D). As controls for liposome injection, we examined transcripts for Ccl19 and Ccl21 in the nondraining and draining LNs and found little to no change. These data suggest that NK cells are being recruited through two independent mechanisms.
Discussion
Here, we determined that NK cells were required for control of viral replication and spread following CPXV footpad inoculation. However, in contrast to ECTV, there was no increase in mortality or morbidity following systemic NK-cell depletion (31). Some of these differences may be due to the larger repertoire of immune evasion genes in CPXV, including CPXV018, which binds with high affinity to the NK-cell activation receptor NKG2D in vitro (39). Nonetheless, our studies revealed NK-cell expansion in the draining LN, consistent with previous studies showing NK-cell expansion without early BrdU incorporation following ECTV infection (32), providing an opportunity to dissect the mechanisms behind this expansion. Here, we further showed that expansion following CPXV infection requires SCS macrophages and is largely due to recruitment of NK cells from the spleen because the expanded NK cells resemble splenic NK cells. Additionally, transferred splenic NK cells are recruited in a pertussis toxin-dependent manner, chemokine ligands for NK-cell chemokine receptors are expressed in the infected LN, and full recruitment is dependent on NK-cell expression of CXCR3 as well as on IFN-γ.
In the LN, murine NK cells represent a very small population under homeostatic conditions (43). It is thought that this is due to their lack of expression of CCR7, the chemokine receptor that responds to the homeostatic expression of CCL19 and CCL21 by high endothelial venules in secondary lymphoid organs (43). We found that early following footpad infection, the NK-cell population in the draining popliteal LN dramatically expanded. The resolution of the expanded NK-cell population corresponded to the resolution of CPXV titers in the draining LN. In contrast to studies using other pathogens and NK-cell recruitment to other organs (23–27), we found no role for CCR2 or CCR5 individually in recruitment of NK cells to the draining LN following CPXV inoculation. However, it is still possible that they function redundantly in the individually deficient mice, because CCR2 and CCR5 share several of the same ligands (54). To determine formally if CCR2 and CCR5 are required, a mouse lacking both CCR2 and CCR5 would have to be generated and tested. This will likely require the production of a single targeting construct containing deletions of both CCR2 and CCR5 because their genes are located within about 15 kb of each other (www.genome.ucsc.edu), making it unlikely that a double KO can be readily generated by mating the CCR2- and CCR5-deficient mice.
On the other hand, we found that CXCR3 was intrinsically required for full recruitment following CPXV infection. Expression of CXCR3 on NK cells has been shown to be important for the recruitment NK cells to tumor sites (21) and to the draining LN following deliberate LPS-matured dendritic cell footpad injection (22). Ligands for CXCR3 (CXCL9, CXCL10, and CXCL11) can be induced by IFN-γ, as we show here for Cxcl9 and Cxcl10 transcripts, consistent with prior studies on systemic VV infection (60). Expression of Cxcl9 and Cxcl10 transcripts has been correlated with expression of CXCL9 and CXCL10 protein in microglial cells following HSV2 infection, suggesting that protein expression of these chemokines in response to viral infection is regulated at the transcriptional level (61). The kinetics of up-regulation of these inflammatory chemokine transcripts also corresponded to the importance of CXCR3 on NK cells at 3 dpi. In addition, when mice could not respond to IFN-γ, as in the IFN-γ receptor-deficient mice or C57BL/6 mice treated with an IFN-γ neutralizing antibody, NK cells were not recruited and the mice also lacked expression of transcripts for Cxcl9 and Cxcl10. Additionally, although CPXV encodes an IFN-γ binding protein, it does not bind murine IFN-γ, suggesting that it is not playing a role in modulating IFN-γ–induced chemokine expression (62). Thus, CXCR3 and its IFN-γ–induced ligands are required for full recruitment of NK cells to the draining LN during CPXV infection.
During the preparation of this paper, several studies reported the colocalization of NK cells in the SCS macrophage region of the draining LN following infection with several pathogens, including a highly attenuated orthopoxvirus, MVA (48–50). SCS macrophages, which express CD169, are ideally placed to sense infection when viral particles arrive in the LN from draining lymphatics. Indeed, we previously found that when CPXV was injected i.p., GFP-expressing CPXV lacking CPXV203 was found to infect CD169-expressing cells in the draining mediastinal LN (47), consistent with other studies indicating that macrophages are the predominant infected cell type in the LN following s.c. VV infection (63). When administered in the footpad, clodronate-loaded liposomes specifically deplete the macrophages that line the SCS of the LN and partially deplete macrophages in the medulla (64). We showed that clodronate-loaded liposome injection before CPXV infection abrogates NK-cell recruitment. Thus, consistent with and extending recently published reports, we show that SCS macrophages are required for NK-cell recruitment to the draining LN in CPXV infection.
These data suggest a framework model in which SCS macrophages are initially infected in the draining LN by CPXV. An IFN-γ–dependent pathway is set off for inducing chemokine production, allowing NK-cell recruitment by CXCR3. As such, these findings provide unique molecular insight into how NK cells are recruited to the draining LN during infection.
Although there are many details yet to be discovered to fill out this model, such as the source of IFN-γ and chemokines, our data suggest that this framework model is likely to be more complex. The recruitment defect in the CXCR3-deficient mice or in mice unable to respond to IFN-γ is primarily evident at 3 dpi when the largest increase in NK cells normally occurs. Earlier time points are less affected, suggesting that there is at least one other mechanism for NK-cell recruitment that is independent of CXCR3 and IFN-γ. This mechanism may allow early CXCR3-independent recruitment of sufficient numbers of NK cells for viral control. This could be CCR2/CCR5-dependent or due to another chemokine receptor that is up-regulated on NK cells during active infection. We did not evaluate the latter possibility because of the low numbers of NK cells in the draining LN and the procedural concern over FACS sorting of CPXV-infected samples. Regardless, our preliminary data indicate that LN viral titers are not affected by the absence of CXCR3. Taken together, these data suggest that CXCR3 and at least one other chemokine receptor, such as CCR2/CCR5, may function to recruit NK cells to the draining LN and limit viral replication following footpad infection with CPXV.
Another surprising finding suggests additional complexity to our framework model. Despite the smaller NK-cell population in the draining LN following treatment with clodronate-loaded liposomes, there was no reduction in the transcript levels of Cxcl9 or Cxcl10, suggesting that macrophages are not responsible, either directly or indirectly, for induction of these CXCR3 ligands. Furthermore, despite continued Cxcl9 and Cxcl10 transcript expression, NK cells were not recruited in the absence of macrophages, suggesting that expression of these chemokines alone is not sufficient for NK-cell recruitment.
It is possible that the SCS macrophages provide a second signal required for NK-cell recruitment. For example, because SCS macrophages are preferentially infected following VV and CPXV infection, the lack of this population in clodronate-loaded, liposome-treated mice may lead to a lack of pathogen sensing, via pattern recognition receptors or nuclei acid sensors, for example, which would otherwise induce production of other mediators that affect NK-cell recruitment. These mediators could include other chemokines that are initially required, such as the ligands for CCR2/5, or proinflammatory factors, such as IL-12 and IL-18 (65). However, CPXV encodes an IL-18 binding protein that binds with high affinity to murine IL-18, which could potentially inhibit stimulation of SCS macrophages; thus, it is possible that SCS macrophage stimulation may occur through an alternate pathway (41, 65). All together, these results suggest that although macrophages play an important role following both parasitic and viral infection, their precise role in poxvirus infection will require further investigation.
One additional complexity is that viruses encode molecules that affect chemokine–chemokine receptor interactions and functions (66). Viral molecules include chemokine receptors, chemokine mimics, and chemokine binding proteins (CKBPs) that can bind a wide variety of chemokines and modulate inflammation. The poxvirus CKBPs are especially relevant to our studies because CPXV conserves these molecules. CPXV can bind to chemokines through their smallpox virus-encoded chemokine receptor (SECRET) domain localized in CrmB and CrmD (67). The CrmB SECRET domain from variola virus has been shown to bind with high affinity to human CCL20, CCL25, CCL28, CXCL12b, CXCL13, CXCL14, and CXL1. Only transcripts for Ccl20 were found to be up-regulated in our chemokine expression screen, but the expression of its chemokine receptor, CCR6, was not found on murine splenic NK cells. Mucosal NKp44+ and IL-2 or IL-15 in vitro-activated human NK cells do express CCR6, which could affect NK-cell recruitment following zoonotic CPXV infection of certain tissues in humans (46). Additionally, CPXV encodes a protein (vCCI, 35-kDa CKBP) that can bind to the CC family of chemokines, including both CCL2 and CCL3, with high affinity (68), which may also explain why we failed to find a defect in NK-cell recruitment in CCR2- and CCR5-deficient mice. Despite these complexities, our studies suggest a framework for further study of NK-cell recruitment in poxvirus infections.
Finally, our studies are likely to be relevant to human infections. Our footpad inoculation of CPXV mimics contact-mediated spread, the most likely form of viral dissemination for zoonotic infections both within the rodent host reservoir as well as to other mammals, including humans. Indeed, zoonotic CPXV human infection is thought to be mediated through skin abrasions and is commonly manifested as localized pustular lesions at the site of inoculation and lymphadenopathy (4–6), similar to what we observed here after deliberate footpad inoculation of mice. Thus, the mechanisms uncovered in this and future studies may be applicable to humans.
Materials and Methods
Mice.
Mice were maintained according to institutional guidelines under specific pathogen-free conditions. C57BL/6NCr mice and B6-LY5.2/Cr mice were obtained from the National Cancer Institute. CCR2-deficient (B6.129S4-Ccr2tm1Ifc/J), CXCR3-deficient (B6.129P2-Cxcr3tm1Dgen/J), and IFN-γ receptor 1-deficient (Ifngr1tm1Agt) mice were obtained from the Jackson Laboratory and bred in our facility. CCR5 (B6.129P2-Ccr5tm1Kuz N10) KO and C57BL/6 control mice were obtained from Taconic Farms. All mice were on the C57BL/6 genetic background and were used between 6 and 10 wk of age. Animal studies were approved by the Animal Study Committee at Washington University in St. Louis.
Cells and Viruses.
CV-1 and Vero cells were obtained from the American Type Culture Collection (ATCC) and maintained in recommended media. Cowpox strain Brighton Red and VV Western Reserve were plaque-purified three times on CV-1 tissue culture cells from laboratory stocks initially obtained from the ATCC. Viral stocks were amplified in Vero cells and isolated following cell lysis by centrifugation through 36% (g/vol) sucrose solution. Viral titers of these stocks were assessed by standard plaque assays on confluent CV-1 monolayers.
Infection.
Mice under anesthesia were infected in the right rear footpad with 1.5 × 106 pfu in a 25-μL volume of virus diluted in PBS. At 1, 2, 3, 6, 9, or 12 dpi, the LN, spleen, kidneys, lungs, liver, feet, and uterus and ovaries were isolated. Organs that were used to determine viral titers were frozen at −80 °C until processing, and organs that were used to assess viral genome copies were frozen at −20 °C until DNA extraction.
Flow Cytometry.
For samples analyzed by flow cytometry, LNs and spleens were crushed in R10 (RPMI, 10% FCS, L-glutamine, and penicillin/streptomycin) through a 70-mm cell strainer. When macrophages were stained, spleens and LNs were diced and incubated in liberase TL (Roche) for 45 min at 37 °C at 5% CO2 before being crushed. RBCs in splenocytes were lysed before staining for flow cytometry. Before extracellular staining, the cells were stained with cell fixable viability dye (eBioscience).
For flow cytometry analysis, the following antibodies were purchased from BD Pharmingen: NK1.1 (PK136), NKp46 (29A1.4), CD19 (1d3), CD45.1 (Ly5.1, clone A20), CD45.2 (Ly5.2, clone104), and CD62L (MEL-14). The following antibodies were purchased from eBioscience: CD3 (145-2C11), CD11b (M1/70), F4/80 (BM8), CD49b (DX5), Ly49CIFH (14B11), Ly49AD (12A8), NKG2D (CX5), CD94 (18d3), and CD127 (A7R34). Surface staining of cells was performed on ice in staining buffer (3% FBS and 0.1% NaN3 in PBS) with 2.4G2 (anti-FcγRII/III) to prevent nonspecific antibody binding. Ki67 (B56; BD Pharmingen) was intracellularly stained with the Mouse FoxP3 Buffer Set (BD Pharmingen). Flow cytometry data were collected on a FACSCanto using FACSDiva software (both from Becton Dickinson) and analyzed using FlowJo software (TreeStar).
Viral Titers.
Traditional plaque assays were done using organ lysates generated by beat beading three times in 1 mL of RPMI. Serial dilutions were plated in duplicate on confluent CV-1 monolayers grown using minimal FCS. Cells were stained at 2 dpi using a crystal violet solution, and plaques were counted. Quantitative PCR was used as a more sensitive measurement of viral genome copy numbers. DNA was generated from organs and was then amplified using StepOnePlus (Applied Biosystems) in a 10-μL volume containing 2× Universal PCR Master Mix and No AmpErase UNG (Roche). Quantitative PCR DNA probes are nonextendable oligonucleotides labeled with 5′ 6-FAM reporter and 3′ Zen/Iowa Black FQ quencher dyes. Primer/probe sets were obtained from IDT DNA, and sequences are indicated in Table S1. Samples were run in duplicate, genome copy number was normalized to β-actin copy number, and numerical values obtained were multiplied by 1,000.
Depletion.
To deplete NK cells systemically, 200 μg of NK1.1 antibody (PK136) or control antibody (mAR) was injected i.p. 2 d before infection and 4 dpi. For neutralization of IFN-γ, 250 μg per mouse of H22 antibody or control (PIP) antibody, a kind gift from the laboratory of Robert Schreiber (Washington University), was injected 2 d before infection. To deplete macrophages, 50 μL of commercially available liposomes (Encapsula) that encapsulated clodronate or PBS was injected in the footpad 5 d before infection.
Adoptive Transfers.
For adoptive transfer experiments, donor splenocytes were purified from C57BL/6 (CD45.1) or CXCR3 (CD45.2) KO mice and RBC-lysed. For the pertussis toxin or PBS pretreatment, cells were plated at a density of 3 × 107 cells/mL in R10 with either 100 ng/mL pertussis toxin or PBS, incubated at 37 °C in 5% CO2 for 2 h, and then washed four times with PBS. Before adoptive transfer, cells were labeled with 2.5 μM CFSE (CellTrace; Invitrogen), washed, and passed through a 40-μm cell strainer; viability was confirmed by trypan blue exclusion before animals were injected with a 200-μL volume.
Chemokine RNA Expression.
Draining and nondraining LNs were isolated, and RNA was extracted using TRIzol following the manufacturer’s instructions (Invitrogen). cDNA was then generated using 500 ng of the isolated RNA utilizing SuperScript III Reverse Transcriptase according to the manufacturer’s protocol (Invitrogen). Quantitative PCR was then performed as described for viral genome copies, except values were only normalized to β-actin and were otherwise unmanipulated. Primer/probe set sequences are indicated in Table S1.
Multiplex Gene Expression Analysis.
LNs from CPXV-infected mice were isolated, homogenized in QuantiGene sample processing buffer (Affymetrix), and then incubated at 65 °C for 30 min before being stored at −80 °C. Samples were further processed and run using a custom multiplex set of beads (Affymetrix) for controls and all listed chemokines (Fig. S4) by the Genome Technology Access Center (Washington University). Chemokine expression was normalized first to Hprt1 transcripts, and fold change was assessed by normalizing to the appropriate nondraining LN control. Heat maps were generated using Jcolorgrid (University of California, San Francisco) (69).
Supplementary Material
Acknowledgments
We thank Bijal Parikh and Beatrice Plougastel-Douglas for helpful advice on experimental design. This work was supported by the Midwest Center for Regional Excellence in Biodefense and Emerging Infectious Diseases Research (National Institutes of Health Grant U54 AI057160) and by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220456110/-/DCSupplemental.
References
- 1.Fenner F. Smallpox and Its Eradication. Geneva: World Health Organization; 1988. [Google Scholar]
- 2.Kile JC, et al. Transmission of monkeypox among persons exposed to infected prairie dogs in Indiana in 2003. Arch Pediatr Adolesc Med. 2005;159(11):1022–1025. doi: 10.1001/archpedi.159.11.1022. [DOI] [PubMed] [Google Scholar]
- 3.Breman JG, et al. 1980. Human monkeypox, 1970–79. Bulletin of the World Health Organization 58:165.
- 4.Campe H, et al. Cowpox virus transmission from pet rats to humans, Germany. Emerg Infect Dis. 2009;15(5):777–780. doi: 10.3201/eid1505.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel S, et al. The Munich outbreak of cutaneous cowpox infection: Transmission by infected pet rats. Acta Derm Venereol. 2012;92(2):126–131. doi: 10.2340/00015555-1227. [DOI] [PubMed] [Google Scholar]
- 6.Ninove L, et al. Cowpox virus transmission from pet rats to humans, France. Emerg Infect Dis. 2009;15(5):781–784. doi: 10.3201/eid1505.090235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vorou RM, Papavassiliou VG, Pierroutsakos IN. Cowpox virus infection: An emerging health threat. Curr Opin Infect Dis. 2008;21(2):153–156. doi: 10.1097/QCO.0b013e3282f44c74. [DOI] [PubMed] [Google Scholar]
- 8.Chantrey J, et al. Cowpox: Reservoir hosts and geographic range. Epidemiol Infect. 1999;122(3):455–460. doi: 10.1017/s0950268899002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mack TM. Smallpox in Europe, 1950-1971. J Infect Dis. 1972;125(2):161–169. doi: 10.1093/infdis/125.2.161. [DOI] [PubMed] [Google Scholar]
- 10.Kim S-H, Oh M-D. Persistence of cell-mediated immunity to vaccinia virus. J Infect Dis. 2007;196(5):805–806. doi: 10.1086/520519. [DOI] [PubMed] [Google Scholar]
- 11.Pelkonen PM, et al. Cowpox with severe generalized eruption, Finland. Emerg Infect Dis. 2003;9(11):1458–1461. doi: 10.3201/eid0911.020814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonsson AH, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol. 2009;101:27–79. doi: 10.1016/S0065-2776(08)01002-X. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama WM, Plougastel BFM. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3(4):304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 14.Andrews DM, Scalzo AA, Yokoyama WM, Smyth MJ, Degli-Esposti MA. Functional interactions between dendritic cells and NK cells during viral infection. Nat Immunol. 2003;4(2):175–181. doi: 10.1038/ni880. [DOI] [PubMed] [Google Scholar]
- 15.Cooper MA, Fehniger TA, Fuchs A, Colonna M, Caligiuri MA. NK cell and DC interactions. Trends Immunol. 2004;25(1):47–52. doi: 10.1016/j.it.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: Function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 17.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319(6055):675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 18.Gasser S, Raulet DH. Activation and self-tolerance of natural killer cells. Immunol Rev. 2006;214:130–142. doi: 10.1111/j.1600-065X.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- 19.Scalzo AA, Yokoyama WM. Cmv1 and natural killer cell responses to murine cytomegalovirus infection. Curr Top Microbiol Immunol. 2008;321:101–122. doi: 10.1007/978-3-540-75203-5_5. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, Smith CW, Zhang W, Burns AR, Li Z. NK cells modulate the inflammatory response to corneal epithelial abrasion and thereby support wound healing. Am J Pathol. 2012;181(2):452–462. doi: 10.1016/j.ajpath.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendel M, Galani IE, Suri-Payer E, Cerwenka A. Natural killer cell accumulation in tumors is dependent on IFN-gamma and CXCR3 ligands. Cancer Res. 2008;68(20):8437–8445. doi: 10.1158/0008-5472.CAN-08-1440. [DOI] [PubMed] [Google Scholar]
- 22.Martín-Fontecha A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-γ for T(H)1 priming. Nat Immunol. 2004;5(12):1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 23.Morrison BE, Park SJ, Mooney JM, Mehrad B. Chemokine-mediated recruitment of NK cells is a critical host defense mechanism in invasive aspergillosis. J Clin Invest. 2003;112(12):1862–1870. doi: 10.1172/JCI18125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan IA, et al. CCR5 is essential for NK cell trafficking and host survival following Toxoplasma gondii infection. PLoS Pathog. 2006;2(6):e49. doi: 10.1371/journal.ppat.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thapa M, Kuziel WA, Carr DJJ. Susceptibility of CCR5-deficient mice to genital herpes simplex virus type 2 is linked to NK cell mobilization. J Virol. 2007;81(8):3704–3713. doi: 10.1128/JVI.02626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salazar-Mather TP, Lewis CA, Biron CA. Type I interferons regulate inflammatory cell trafficking and macrophage inflammatory protein 1α delivery to the liver. J Clin Invest. 2002;110(3):321–330. doi: 10.1172/JCI15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hokeness KL, Kuziel WA, Biron CA, Salazar-Mather TP. Monocyte chemoattractant protein-1 and CCR2 interactions are required for IFN-alpha/beta-induced inflammatory responses and antiviral defense in liver. J Immunol. 2005;174(3):1549–1556. doi: 10.4049/jimmunol.174.3.1549. [DOI] [PubMed] [Google Scholar]
- 28.Grégoire C, et al. Intrasplenic trafficking of natural killer cells is redirected by chemokines upon inflammation. Eur J Immunol. 2008;38(8):2076–2084. doi: 10.1002/eji.200838550. [DOI] [PubMed] [Google Scholar]
- 29.Paust S, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11(12):1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191(5):771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker AK, Parker S, Yokoyama WM, Corbett JA, Buller RM. Induction of natural killer cell responses by ectromelia virus controls infection. J Virol. 2007;81(8):4070–4079. doi: 10.1128/JVI.02061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang M, Lanier LL, Sigal LJ. A role for NKG2D in NK cell-mediated resistance to poxvirus disease. PLoS Pathog. 2008;4(2):e30. doi: 10.1371/journal.ppat.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang M, et al. CD94 is essential for NK cell-mediated resistance to a lethal viral disease. Immunity. 2011;34(4):579–589. doi: 10.1016/j.immuni.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll DS, et al. Chasing Jenner’s vaccine: Revisiting cowpox virus classification. PLoS ONE. 2011;6(8):e23086. doi: 10.1371/journal.pone.0023086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dasgupta A, Hammarlund E, Slifka MK, Früh K. Cowpox virus evades CTL recognition and inhibits the intracellular transport of MHC class I molecules. J Immunol. 2007;178(3):1654–1661. doi: 10.4049/jimmunol.178.3.1654. [DOI] [PubMed] [Google Scholar]
- 36.Byun M, Wang X, Pak M, Hansen TH, Yokoyama WM. Cowpox virus exploits the endoplasmic reticulum retention pathway to inhibit MHC class I transport to the cell surface. Cell Host Microbe. 2007;2(5):306–315. doi: 10.1016/j.chom.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Byun M, et al. Two mechanistically distinct immune evasion proteins of cowpox virus combine to avoid antiviral CD8 T cells. Cell Host Microbe. 2009;6(5):422–432. doi: 10.1016/j.chom.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alzhanova D, et al. Cowpox virus inhibits the transporter associated with antigen processing to evade T cell recognition. Cell Host Microbe. 2009;6(5):433–445. doi: 10.1016/j.chom.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell JA, Trossman DS, Yokoyama WM, Carayannopoulos LN. Zoonotic orthopoxviruses encode a high-affinity antagonist of NKG2D. J Exp Med. 2007;204(6):1311–1317. doi: 10.1084/jem.20062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reading PC, Smith GL. Vaccinia virus interleukin-18-binding protein promotes virulence by reducing gamma interferon production and natural killer and T-cell activity. J Virol. 2003;77(18):9960–9968. doi: 10.1128/JVI.77.18.9960-9968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith VP, Bryant NA, Alcamí A. Ectromelia, vaccinia and cowpox viruses encode secreted interleukin-18-binding proteins. J Gen Virol. 2000;81(Pt 5):1223–1230. doi: 10.1099/0022-1317-81-5-1223. [DOI] [PubMed] [Google Scholar]
- 42.Seaman WE, Sleisenger M, Eriksson E, Koo GC. Depletion of natural killer cells in mice by monoclonal antibody to NK-1.1. Reduction in host defense against malignancy without loss of cellular or humoral immunity. J Immunol. 1987;138(12):4539–4544. [PubMed] [Google Scholar]
- 43.Grégoire C, et al. The trafficking of natural killer cells. Immunol Rev. 2007;220:169–182. doi: 10.1111/j.1600-065X.2007.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vargas CL, Poursine-Laurent J, Yang L, Yokoyama WM. Development of thymic NK cells from double negative 1 thymocyte precursors. Blood. 2011;118(13):3570–3578. doi: 10.1182/blood-2011-06-359679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vosshenrich CAJ, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7(11):1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 46.Cella M, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457(7230):722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu KM, Pratt JR, Akers WJ, Achilefu SI, Yokoyama WM. Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J Gen Virol. 2009;90(Pt 1):33–43. doi: 10.1099/vir.0.006668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kastenmüller W, Torabi-Parizi P, Subramanian N, Lämmermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell. 2012;150(6):1235–1248. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia Z, et al. Subcapsular sinus macrophages promote NK cell accumulation and activation in response to lymph borne viral particles. Blood. 2012;120:4744–4750. doi: 10.1182/blood-2012-02-408179. [DOI] [PubMed] [Google Scholar]
- 50.Coombes JL, Han S-J, van Rooijen N, Raulet DH, Robey EA. Infection-induced regulation of natural killer cells by macrophages and collagen at the lymph node subcapsular sinus. Cell Rep. 2012;2(1):124–135. doi: 10.1016/j.celrep.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iannacone M, et al. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature. 2010;465(7301):1079–1083. doi: 10.1038/nature09118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hickman HD, et al. Chemokines control naive CD8+ T cell selection of optimal lymph node antigen presenting cells. J Exp Med. 2011;208(12):2511–2524. doi: 10.1084/jem.20102545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Junt T, et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450(7166):110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 54.Allen SJ, Crown SE, Handel TM. Chemokine: Receptor structure, interactions, and antagonism. Annu Rev Immunol. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 55.Deng Q, Barbieri JT. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol. 2008;62:271–288. doi: 10.1146/annurev.micro.62.081307.162848. [DOI] [PubMed] [Google Scholar]
- 56.Fehniger TA, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26(6):798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 57.Groom JR, Luster AD. CXCR3 ligands: Redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89(2):207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melchjorsen J, Sirén J, Julkunen I, Paludan SR, Matikainen S. Induction of cytokine expression by herpes simplex virus in human monocyte-derived macrophages and dendritic cells is dependent on virus replication and is counteracted by ICP27 targeting NF-kappaB and IRF-3. J Gen Virol. 2006;87(Pt 5):1099–1108. doi: 10.1099/vir.0.81541-0. [DOI] [PubMed] [Google Scholar]
- 59.Hardison JL, Wrightsman RA, Carpenter PM, Lane TE, Manning JE. The chemokines CXCL9 and CXCL10 promote a protective immune response but do not contribute to cardiac inflammation following infection with Trypanosoma cruzi. Infect Immun. 2006;74(1):125–134. doi: 10.1128/IAI.74.1.125-134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahalingam S, Karupiah G. Expression of the interferon-inducible chemokines MuMig and Crg-2 following vaccinia virus infection in vivo. Immunol Cell Biol. 2000;78(2):156–160. doi: 10.1046/j.1440-1711.2000.00894.x. [DOI] [PubMed] [Google Scholar]
- 61.Aravalli RN, Hu S, Rowen TN, Palmquist JM, Lokensgard JR. Cutting edge: TLR2-mediated proinflammatory cytokine and chemokine production by microglial cells in response to herpes simplex virus. J Immunol. 2005;175(7):4189–4193. doi: 10.4049/jimmunol.175.7.4189. [DOI] [PubMed] [Google Scholar]
- 62.Alcamí A, Smith GL. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol. 1995;69(8):4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Norbury CC, Malide D, Gibbs JS, Bennink JR, Yewdell JW. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat Immunol. 2002;3(3):265–271. doi: 10.1038/ni762. [DOI] [PubMed] [Google Scholar]
- 64.Delemarre FG, Kors N, Kraal G, van Rooijen N. Repopulation of macrophages in popliteal lymph nodes of mice after liposome-mediated depletion. J Leukoc Biol. 1990;47(3):251–257. doi: 10.1002/jlb.47.3.251. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Chaudhri G, Jackson RJ, Karupiah G. IL-12p40 and IL-18 play pivotal roles in orchestrating the cell-mediated immune response to a poxvirus infection. J Immunol. 2009;183(5):3324–3331. doi: 10.4049/jimmunol.0803985. [DOI] [PubMed] [Google Scholar]
- 66.Alcami A, Lira SA. Modulation of chemokine activity by viruses. Curr Opin Immunol. 2010;22(4):482–487. doi: 10.1016/j.coi.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alejo A, et al. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc Natl Acad Sci USA. 2006;103(15):5995–6000. doi: 10.1073/pnas.0510462103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alcami A, Symons JA, Collins PD, Williams TJ, Smith GL. Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. J Immunol. 1998;160(2):624–633. [PubMed] [Google Scholar]
- 69.Joachimiak MP, Weisman JL, May BCh. JColorGrid: Software for the visualization of biological measurements. BMC Bioinformatics. 2006;7:225. doi: 10.1186/1471-2105-7-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.