Abstract
Neuropathic pain is a chronic condition that is often refractory to treatment with available therapies and thus an unmet medical need. We have previously shown that the voltage-gated sodium channel Nav1.3 is upregulated in peripheral and central nervous system (CNS) of rats following nerve injury, and that it contributes to nociceptive neuron hyperexcitability in neuropathic conditions. To evaluate the therapeutic potential of peripheral Nav1.3 knockdown at a specific segmental level, we constructed adeno-associated viral (AAV) vector expressing small hairpin RNA against rat Nav1.3 and injected it into lumbar dorsal root ganglion (DRG) of rats with spared nerve injury (SNI). Our data show that direct DRG injection provides a model that can be used for proof-of-principle studies in chronic pain with respect to peripheral delivery route of gene transfer constructs, high transduction efficiency, flexibility in terms of segmental localization, and limited behavioral effects of the surgical procedure. We show that knockdown of Nav1.3 in lumbar 4 (L4) DRG results in an attenuation of nerve injury-induced mechanical allodynia in the SNI model. Taken together, our studies support the contribution of peripheral Nav1.3 to pain in adult rats with neuropathic pain, validate Nav1.3 as a target, and provide validation for this approach of AAV-mediated peripheral gene therapy.
Introduction
Neuropathic pain—pain initiated or caused by a lesion or disease of the somatosensory nervous system1—is a chronic condition that is often refractory to treatment with currently available therapies and thus remains a major unmet medical need.2 Neuropathic pain, including pain associated with diabetic neuropathy, post herpetic neuralgia (herpes zoster), and traumatic injuries, affects up to 17.9% of the general population3 (Institute of Medicine report, Relieving Pain in America, 2011). The lack of clear understanding of the molecular bases and mechanisms of neuropathic pain has hampered the development of new approaches and therapeutics for effective and safe treatment of chronic pain.4
Under normal conditions, nociceptive pain is adaptive, proportional to the stimulus strength and ceases after the latter is withdrawn.5 However, under pathological conditions, for example following peripheral nerve injury, pain can be triggered by innocuous stimuli (allodynia), noxious stimuli are amplified (hyperalgesia), and in many cases pain is spontaneous.5,6 These hallmarks of chronic pain can persist long after the initial injury, spread to neighboring uninjured areas (secondary hyperalgesia), and can have a substantial impact on the patient's quality of life, and retarding recovery and rehabilitation after injury.
A common motif of various neuropathic pain conditions is hyperexcitability and spontaneous firing by pain-signaling primary sensory neurons, whose cell bodies are located in dorsal root ganglia (DRG), even in the absence of a painful sensory stimulus. It is now well established that voltage-gated sodium channels are essential for this ectopic activity and are thus potential targets for pain therapy.7 Clinical use of existing sodium channel blockers, while showing some efficacy, is hampered by their nonselectivity, causing off-target cardiac and central nervous system (CNS) side-effects.8 Studies from our laboratory and others have shed light on the contribution of various sodium channels (Nav1.3, Nav1.7, Nav1.8, and Nav1.9) to DRG neuron hyperexcitability using in vitro neuronal cultures and in vivo animal models of pain, and in heritable human pain disorders.7,9 In particular, Nav1.3 is upregulated in DRG and dorsal spinal cord and thalamic neurons of adult rats with peripheral nervous system or CNS injuries,10,11,12,13 and accumulates at the axonal tips within peripheral human and animal neuromas which form after axotomy.14,15 The biophysical properties of Nav1.3 make it well-suited to support high firing frequencies16,17 and it is thus a potential target for development of novel pain therapies.
In the absence of isoform-selective, effective and safe small molecule sodium channel blockers for treatment of chronic pain, gene therapy in the nervous system offers several potential advantages: (i) increased specificity (targeting one specific sodium channel isoform), (ii) reduced side-effects, (iii) prolonged therapeutic effect.18 Recent studies have shown significant success using nonvirulent adeno-associated virus (AAV), with high affinity for sensory neurons to deliver genes (gain of function) and RNA interference molecules (short hairpin RNA (shRNA) for gene knockdown) in rodents.19,20,21,22 In this study, we assessed the therapeutic potential of Nav1.3 knockdown via highly specific second-generation gene-silencing shRNAmir23 technology, directly delivered to the DRG via an AAV vector, to assess the therapeutic potential of Nav1.3 knockdown in a rat model of neuropathic pain.
Results
Design, selection, and validation of siRNA sequences against Nav1.3
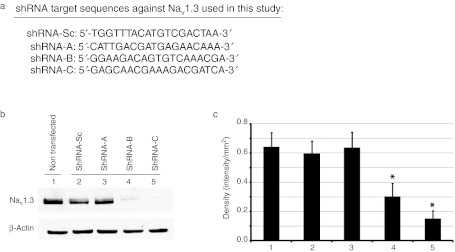
Small interfering RNA (siRNA) mediates gene-specific silencing primarily via recognizing and inducing the degradation of mRNA of targeted genes, thus significantly reducing the gene product. siRNA molecules targeting different sites in a transcript can manifest different potencies, and only a fraction of them are highly effective and can be successfully used in vivo. Sequence selection in the design of the siRNA based on stringent criteria is therefore crucial (reviewed in ref. 24). To this end, we used a recently developed computational algorithm (http://jura.wi.mit.edu/bioc/siRNA) to narrow down a working list of siRNA sequences for Nav1.3.24 This bioinformatic analysis of Nav1.3 produced a short list of ten Nav1.3-specific and potentially effective molecules, as well as a scrambled control sequence, that were advanced for experimental validation.
The selected RNA interference molecules were cloned into expression vectors as shRNA driven by hU6 promoter. The vector also coexpressed a CMV promoter-driven enhanced green fluorescent protein (GFP) to mark infected neurons. shRNA sequences were tested individually for knockdown efficiency in cultured HEK293 cells that stably express rat Nav1.3 (HEK-Nav1.3).25 Among the ten shRNA sequences, two molecules, shRNA-B and shRNA-C (Figure 1a), showed significant decrease in Nav1.3 mRNA levels as measured by quantitative reverse transcription-PCR (data not shown) when transfected into HEK-Nav1.3 cells. The remaining sequences, e.g., shRNA-A (Figure 1a), did not significantly affect Nav1.3 expression. Nav1.3 protein levels were then examined by western blots and normalized to β-actin. Figure 1b and c shows that shRNA treatment significantly reduced Nav1.3 protein levels by 53 and 76% when cells were transfected with shRNA-B and shRNA-C, respectively, but not when transfected by shRNA-A. Importantly, Nav1.3 protein level was not significantly altered by transfection of the HEK-Nav1.3 cell line with control scrambled sequence (shRNA-Sc), indicating that the observed decrease in Nav1.3 mRNA and protein levels was specific to the selected shRNA sequences (Figure 1b, c).
Figure 1.
shRNA-B and shRNA-C knockdown Nav1.3 protein levels in HEK-Nav1.3 cells. (a) List of the short hairpin RNA (shRNA) target sequences used in this study against Nav1.3. (b) Western blot analysis using pan-sodium channel antibody on HEK-Nav1.3 cells lysates treated as indicated. β-Actin antibody was used to normalize the signal. (c) Graph depicting the quantification of the Nav1.3 protein in the different treatments. Note that shRNA-B and shRNA-C, but not shRNA-A or shRNA-Sc (scrambled control), significantly reduce Nav1.3 protein (P < 0.05). Data are presented as means ± SEM.
Selected siRNA sequences significantly reduce Nav1.3 current in cultured cells
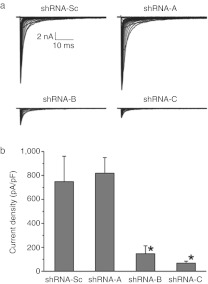
To examine whether knockdown of Nav1.3 transcripts reduces functional Nav1.3 channels in plasma membrane, whole-cell voltage-clamp recording was performed to measure Nav1.3 currents in HEK-Nav1.3 cells transiently transfected with plasmids producing shRNA-A, shRNA-B, shRNA-C, or shRNA-Sc. Figure 2a shows the representative sodium currents recorded from HEK-Nav1.3 cells. HEK-Nav1.3 cells transfected with shRNA-Sc produced large sodium currents (749 ± 212 pA/pF, n = 8). Consistent with the western blot results (Figure 1b, c), shRNA-B and shRNA-C reduced sodium currents by several fold (shRNA-B: 147 ± 66 pA/pF, n = 10, P < 0.01 versus shRNA-Sc; shRNA-C: 68 ± 16 pA/pF, n = 11, P < 0.01 versus shRNA-Sc), while shRNA-A had no effect on current density (819 ± 131, n = 10, P > 0.05 versus shRNA-Sc) (Figure 2b). Together, these results show that shRNA-B and shRNA-C effectively knockdown Nav1.3 mRNA, protein, and currents in HEK-Nav1.3 cells.
Figure 2.
Introducing short hairpin RNA (shRNA)-B and shRNA-C into HEK-Nav1.3 cells effectively reduce voltage-gated sodium currents. (a) Representative sodium currents generated from HEK-Nav1.3 cells transfected with plasmids producing shRNA-Sc, shRNA-A, shRNA-B, or shRNA-C. (b) Average sodium current densities of HEK-Nav1.3 cells transfected with plasmids producing shRNA-Sc, shRNA-A, shRNA-B, or shRNA-C. shRNA-A had no effect on sodium current density, while shRNA-B and shRNA-C significantly reduced current densities in HEK-Nav1.3 cells. Data were presented as means ± SEM.
Efficient AAV-mediated gene delivery to L4 DRG neurons
With effective and experimentally validated shRNA molecules against Nav1.3 in hand, the next step was to test the efficacy of these sequences in vivo and assess the therapeutic potential of Nav1.3 knockdown in the DRG of rats with chronic pain. To achieve a persistent expression of shRNA after a single injection, we chose to use AAV vector as a delivery vehicle. Recently, AAV2 serotype 5 was shown to efficiently and persistently infect DRG.21 For increased transduction efficiency and tissue specificity, AAV was injected directly into DRG of adult rats. This delivery method has been recently adapted to viral vectors.21,26 Here, we extended the applicability of DRG injection to test the therapeutic potential of shRNAmir-Nav1.3 in a chronic pain model.
Direct AAV injection into rodent DRG results in robust infection rate
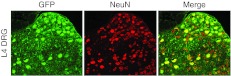
To confirm transduction efficiency following DRG injection in adult rats, we first used a control AAV2/5 virus-expressing GFP. Since pain threshold testing is conducted on the plantar area of the rat hindpaw, we injected the lumbar 4 (L4) DRG which innervates this area of the foot. For each operated animal, one L4 DRG was injected. Based on results from previous time-course studies of GFP expression after DRG injection of AAV2/5,21,22,26 we collected DRGs for GFP imaging at 3 weeks after injection. AAV2/5 significantly transduced the injected (ipsilateral) DRG (Figure 3), but not the contralateral DRG (data not shown). Furthermore, we used NeuN/GFP double staining to determine the percentage of GFP+ neuronal profiles. On average, direct L4 DRG injection in adult rats resulted in infection of 45 ± 2% of total neuronal profiles within sections showing the entire ganglion.
Figure 3.
Direct AAV-GFP injection into rat lumbar 4 (L4) dorsal root ganglion (DRG) results in robust transduction rate. Double immunostaining of green fluorescent protein (GFP) (green) and NeuN neuronal marker (red) on a representative transverse section of L4 DRG collected from an adult rat 3 weeks after AAV-GFP injection. Transduction rate (45 ± 2%) was determined by counting neuronal profiles (NeuN-positive) that are also GFP-positive (n = 3 rats). AAV, adeno-associated virus.
Direct DRG injection produces transient peripheral sensitization
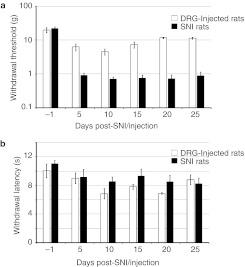
To determine the effect of shRNA-mediated knockdown of Nav1.3 on pain thresholds after peripheral nerve injury, it was critical to first assess the pain behavioral consequences of DRG injections per se. We therefore tested the mechanical pain thresholds and thermal latencies of rats injected with the AAV2/5-GFP (vector) over a time course of 25 days after injection. Five and ten days after DRG injection, rats exhibited a transient sensitization, with mechanical thresholds falling to 32 and 23% of baseline (from 19.8 ± 3.6 g to 6.3 ± 1.5 g and 4.5 ± 0.9 g, respectively) (Figure 4a). This sensitization, however, gradually resolved albeit partially at 15 days postinjection and persisted until the end of the experiment (P < 0.05). Thus, rats showed a two- to threefold recovery of their mechanical pain thresholds (up to 59% of baseline, 11.6 ± 0.6 g) at the end of the experiment compared to their day 10 thresholds (Figure 4a). Together, our results agree with previous reports.26 Importantly, DRG injections had a relatively small effect at day 20 and thereafter on mechanical allodynia when compared to the permanent reduction caused by spared nerve injury (SNI) model used in this study: a 1.7-fold versus 30-fold reduction in pain thresholds compared to presurgical baseline (DRG injection, from 19.8 ± 3.6 g to 11.6 ± 0.6 g; SNI, from 22.1 ± 2.4 g to 0.7 ± 0.2 g) (Figure 4a).27
Figure 4.
Dorsal root ganglion (DRG) injection induces a smaller transient peripheral sensitization than that induced by spared nerve injury (SNI). (a) Von Frey measurements of mechanical thresholds from hindpaws of SNI rats (n = 5) versus rats that are DRG-injected with AAV-GFP (n = 5) over the examined time course. Both DRG injection and SNI significantly reduced mechanical thresholds (P < 0.05). However, sensitization due to DRG injection was significantly smaller than that due to SNI (P < 0.05). (b) Measurements of heat hyperalgesia using the Hargreaves apparatus for SNI rats (n = 5) versus rats that are DRG-injected with AAV-GFP (n = 5) over the examined time course. No significant change was observed. Data are presented as means ± SEM. AAV, adeno-associated virus; GFP, green fluorescent protein.
The thermal latency after exposure of ipsilateral hindpaws of injected rats to radiant heat did not show a significant reduction over the same time course, despite a trend for a mild transient decrease 10–20 days after injection (Figure 4b). Similarly, as previously observed,27 SNI did not result in significant radiant heat sensitization over the time-course analysis. Taken together, these results show that direct DRG injection of AAV5/2 results in robust transduction of the targeted DRG, and the requisite surgery for viral delivery to L4 DRG is accompanied by small transient mechanical and thermal sensitization. Taken together, these results demonstrate that direct DRG infection by AAV is suitable for gene and shRNA transfer for in vivo studies in chronic pain.
In vivo downregulation of Nav1.3 expression by AAV-shRNAmir
SNI is an established animal model for peripheral neuropathic pain.27 A sharp decrease in mechanical thresholds develops as early as 3 days after the surgery and lasts for at least several months (Figure 3).27 We and others have previously shown that peripheral nerve injury results in an upregulation of Nav1.3 in DRG (see Introduction section). Our in vitro studies showed that two independent shRNA sequences, shRNA-B and shRNA-C, directed against different regions in Nav1.3 cDNA resulted in robust reduction of Nav1.3 expression (Figures 1 and 2). Thus, we chose those two sequences to study the contribution of Nav1.3 in DRG neurons in chronic pain.
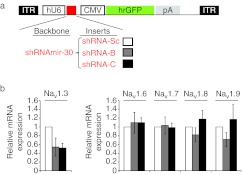
In order to determine whether shRNA-B and -C delivered directly to L4 DRG of rats can achieve Nav1.3 knockdown in vivo, we conducted two experiments, in two groups of rats, each receiving: (i) shRNA-B versus shRNA-Sc (scrambled control shRNA sequence), or (ii) shRNA-C versus shRNA-Sc (Figure 5a). Injected DRG were collected 25 days postinjection and total RNA was extracted for relative quantification of Nav1.3 mRNA. In agreement with the results obtained in vitro, quantitative reverse transcription-PCR analysis showed that Nav1.3 mRNA decreased by 46 and 48% in DRG from SNI rats injected with shRNA-B and shRNA-C, respectively, when compared to DRG injected with the shRNA-Sc control (Figure 5b). It is important to note that in this analysis, cDNA was prepared from L4 DRG including both infected and noninfected cells. Therefore, the observed downregulation should be considered an underestimation of the effect of the shRNA on a per-cell basis.28
Figure 5.
Short hairpin RNA shRNA-B and shRNA-C specifically knockdown Nav1.3 in vivo. (a) A schematic depicting the AAV-shRNA vector used in vivo. (b,c) Graphs showing the relative expression of the indicated Nav channels in lumbar 4 (L4) dorsal root ganglion (DRG) extracts from animals treated with shRNA-Sc (n = 7 animals) versus shRNA-B (n = 3 animals) or shRNA-C (n = 5 animals). GAPDH was used for normalization. Data are presented as means ± SEM. AAV, adeno-associated virus; CMV, cytomegalovirus promoter; GFP, green fluorescent protein; hrGFP, humanized Renilla green fluorescent protein; hU6, human U6 promoter; ITR, inverted terminal repeat; pA; SV40 poly adenylation signal.
Since Nav1.3 is a member of the voltage-gated sodium channel family, we used several approaches to achieve and confirm isoform-selectivity. We explored the potential crossover effect of shRNA-B and shRNA-C on other sodium channel isoforms. A BLAST sequence analysis detected 6–18 mismatches (out of 19 nucleotides) when the sequences of selected shRNA were compared to those of cDNA of other sodium channel isoforms that are expressed in DRG neurons (Nav1.6, Nav1.7, Nav1.8, and Nav1.9).29 Importantly, mismatches were found at the 5′ end of the guide shRNA sequences, minimizing potential off-target effects (data not shown). To empirically confirm the isoform-specificity of shRNA-B and shRNA-C in vivo, we conducted quantitative reverse transcription-PCR analysis for the expression of Nav1.6, Nav1.7, Nav1.8, and Nav1.9 in DRG of rats treated with shRNA-Sc versus shRNA-B and shRNA-C. In agreement with the bioinformatic analysis, we did not detect off-target effects of shRNA-B and shRNA-C against these channels (Figure 5c).
Taken together, these results clearly show that both shRNA-B and shRNA-C are able to specifically knockdown Nav1.3 transcript levels in vivo, in comparison to a nontargeting scrambled control shRNAmir.
Partial attenuation of SNI-induced mechanical allodynia after Nav1.3 knockdown in L4 DRG neurons
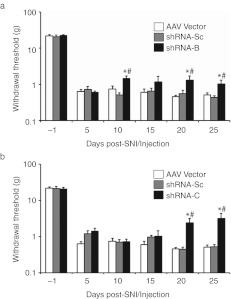
To determine whether knockdown of peripheral nerve injury-induced Nav1.3 upregulation can attenuate mechanical allodynia in the SNI model, we first injected two groups of rats with shRNA-B and shRNA-Sc, respectively on the day of the SNI surgery (day 0). We then measured their mechanical thresholds over a time course of 25 days (Figure 6a). Rats injected with shRNA-Sc exhibited a significant drop in their mechanical thresholds starting from 5 days and lasting throughout the testing period (Figure 6a,b; P < 0.05), similar to animals which underwent SNI without further treatment (Figure 4). Animals injected with shRNA-B exhibited mechanical allodynia at 5 days after injection/SNI similar to shRNA-Sc treated animals. However, starting on day 10, the mechanical thresholds of animals injected with shRNA-B exhibited a 180% increase in mechanical pain threshold when compared to those of animals injected with shRNA-Sc (1.5 ± 0.3 g versus 0.5 ± 0.1 g, P < 0.05). This recovery of mechanical threshold persisted throughout the rest of the period of assessment (Figure 6a, P < 0.05). At day 25, the shRNA-B treated animals displayed a 2.3-fold recovery of their mechanical thresholds compared to control (1.0 ± 0.3 g versus 0.40 ± 0.04 g, P < 0.05). Importantly, treatment with the other shRNA sequence targeting Nav1.3, shRNA-C, also resulted in a significant attenuation of mechanical allodynia when compared to treatment with shRNA-Sc control over the investigated time course (Figure 6b, P < 0.05). The restoration of mechanical thresholds in animals injected with shRNA-C started at day 20 and increased until the experimental endpoint of the time course at 25 days where the animals showed a sixfold recovery of mechanical threshold; pain threshold changed from 2.5 to 15.2% of baseline when compared to control shRNA-Sc (P < 0.05).
Figure 6.
Nav1.3 knockdown in lumbar 4 (L4) dorsal root ganglion (DRG) neurons partially attenuates spared nerve injury (SNI)-induced mechanical allodynia. (a,b) Von Frey measurements of mechanical thresholds from hindpaws of SNI rats injected with (a) short hairpin RNA shRNA-Sc (n = 5) versus shRNA-B (n = 5), or (b) shRNA-Sc (n = 5) versus shRNA-C (n = 6) over the examined time course. SNI rats injected with AAV-GFP vector control (n = 5) are also shown (a,b). A partial but significant attenuation of mechanical allodynia over the indicated time course is observed when animals injected with AAV-GFP or shRNA-Sc are compared to those injected with (a) shRNA-B (P < 0.05), or (b) shRNA-C (P < 0.05). No difference was noted between animals injected with AAV-GFP versus animals injected with shRNA-Sc (P > 0.05). Animals injected with shRNA-B showed significant restoration of mechanical threshold at D10, D20, and D25 when compared to those injected with shRNA-Sc (*P < 0.05), or AAV-GFP (#P < 0.05). Animals injected with shRNA-C showed significant restoration of mechanical threshold at D20, and D25 when compared to those injected with shRNA-Sc (*P < 0.05), or AAV-GFP (#P < 0.05). AAV, adeno-associated virus; GFP, green fluorescent protein.
To control for potential effects of the AAV vector on the mechanical thresholds, we injected a separate group of rats with AAV-GFP vector on the day of the SNI surgery as was done for shRNA-Sc, shRNA-B, and shRNA-C injected animals. The animals developed mechanical allodynia over the entire studied time course similar to shRNA-Sc injected animals (Figure 6, P > 0.05). Importantly, animals treated with shRNA-B and shRNA-C also showed significant recovery when compared to AAV-GFP vector-treated animals (P < 0.05). In summary, our behavioral data show that knockdown of Nav1.3 in a single DRG by two independent shRNA sequences directed against Nav1.3 partially but significantly attenuates SNI-induced mechanical allodynia.
Discussion
Blockade of aberrantly active sodium channel isoforms peripherally while sparing normal neuronal activity peripherally and centrally is considered a promising approach for pain therapy but, to date, most investigations have focused on small molecule channel blockers. Direct DRG injection offers a potential therapeutic approach that is unique in its ability to target specific spinal segments for gene therapy. In the present study, we sought to confirm the contribution of Nav1.3 to injury-induced mechanical allodynia, and to explore the use of DRG-targeted AAV-mediated knockdown of Nav1.3 channel by shRNA as a proof-of-principle for pain therapy. Several potential shRNA molecules were identified by in silico analysis, and two sequences (shRNA-B and shRNA-C) were selected for functional assays in vivo after verification of a robust knockdown of Nav1.3 at the protein and current levels in a stable cell line. We show that delivery of two independent shRNAs targeting Nav1.3 in vivo to a single sensory ganglion using an AAV2/5 virus platform ameliorates injury-induced mechanical allodynia.
Within DRG, Nav1.3 is predominantly expressed in embryonic and neonatal neurons, is not detectable in adult neurons, but is upregulated after nerve injury,10 and accumulates at injured axonal tips within neuromas,14,15 a site which is known to produce spontaneous firing.30 Several independent lines of evidence have correlated nerve injury-induced Nav1.3 upregulation with neuropathic pain in rats: (i) Nav1.3 drives a fast-recovering TTX-sensitive current capable of supporting high frequency firing of neurons,16,31 (ii) TTX concentrations that do not affect TTX-R channels or block action potential conduction reduce pain in injured animals,32,33 (iii) Nav1.3 upregulation, enhanced repriming of TTX-S currents in DRG neurons, and neuropathic pain behavior are reversed by administration of glial cell line-derived neurotrophic factor (GDNF) to rats,34 and (iv) intrathecal AS-ODN treatment is reported to transiently abrogate Nav1.3 upregulation in dorsal spinal cord and alleviate mechanical allodynia for 3 days in rats after chronic constriction injury.12 It should be noted that one report using another AS-ODN sequence against Nav1.3 in DRG of neuropathic rats did not show significant restoration of pain thresholds despite a reported 50% reduction of Nav1.3 levels.13 This discrepancy could be due to the varying efficacies of ODN sequences. Additionally, ODN exhibit varying degrees of stability and in vivo uptake by cells.35 Thus, the utility of Nav1.3 knockdown for pain therapy has remained in question. To address this issue directly, we used second-generation shRNAmir driven by a U6 promoter from an AAV vector delivered directly to the DRG which innervates the area of the foot where pain thresholds are assessed. The combination of direct DRG injection and AAV-shRNA delivery platform offers long-lasting production of shRNA sequences with greater potency and less off-target effects (Figures 5 and 6).23
The rationale behind injecting only one of the lumbar DRG was to address the specific contribution of peripheral Nav1.3 to nerve injury-induced pain in a proof-of-principle effort, while minimizing the effects of the surgical procedure. Previous studies have successfully injected multiple DRG but the pain thresholds following the surgery were not assessed.21 We show here that direct DRG injection of AAV2/5 achieves a 45% neuronal transduction rate in rats, similar to a recently published report.22 Interestingly, although the necessary surgical procedure transiently decreases pain thresholds of operated animals, this sensitization is minimal compared to that caused by SNI nerve injury, and therefore it is unlikely to impede studies on chronic pain using the SNI model (Figure 4). Using the above described tools, we show that Nav1.3 expression in DRG neurons from injured rats is reduced by ~50% when treated with either of two independent shRNA sequences against Nav1.3 (shRNA-B and shRNA-C), thus increasing confidence that behavioral effects are not caused by off-target effects of treatment. Importantly, this knockdown of Nav1.3 in a single DRG resulted in significant attenuation of nerve injury-induced mechanical allodynia. Taken together, our studies lend support to a contribution of peripheral Nav1.3 to pain in adult neuropathic rats, and to the concept that an isoform-specific therapeutic agent engaging a single target may be sufficient for pain relief.
Specific treatment of nerve-injured rats with shRNA targeting Nav1.3 in L4 DRG resulted in an up to sixfold recovery of mechanical threshold (Figure 6b), offering a proof-of-principle that reducing peripheral Nav1.3 could be a potential target for neuropathic pain treatment. Despite a drastic reduction in Nav1.3 currents by shRNA-B and shRNA-C sequences in a cell culture assay (Figure 2), the use of these sequences in vivo resulted in 50% knockdown of total Nav1.3 expression in treated DRG of neuropathic animal (Figure 5). This partial knockdown does not necessarily reflect reduced efficacy of shRNA in vivo, but may reflect the 45% DRG transduction efficiency of AAV2/5 virions obtained in this study. Thus, the observed incomplete resolution of mechanical allodynia could be due in part to the hyperexcitability of L4 DRG neurons which were not infected by the AAV2/5 virus, as well as neurons in other lumbar DRG innervating the ipsilateral hindpaw. Finally, we have previously reported that after peripheral nerve injury, Nav1.3 is also upregulated in CNS pain circuitry centers (dorsal spinal cord and thalamus) where it can also contribute to neuropathic pain.12,36 Taken together, it is not surprising that AAV-shRNA injection into a single ganglion yielded significant but partial recovery of pain thresholds.
We have used two shRNA sequences directed against separate regions in the Nav1.3 cDNA and showed, in two independent sets of animals with independent, contemporaneous controls, that each of these shRNAs can attenuate nerve injury-induced mechanical allodynia. While demonstrating the effect of selective Nav1.3 knockdown in DRG in our pain model, the two sequences (shRNA-B and shRNA-C) showed differences in the time-of-onset and extent of therapeutic effect. Treatment with shRNA-B increased mechanical thresholds by twofold starting from day 10, whereas shRNA-C resulted in a sixfold increase in thresholds starting on day 20. This apparent difference could reflect experimental variability in shRNA efficacy in vivo.37 Our proof-of-principle study did not address this question. Future time-course studies correlating Nav1.3 downregulation and the behavioral outcome might shed light on the dynamics of U6 promoter-driven shRNA expression and effect in vivo, and inform the design of potential gene therapy treatment regimens.
A species-specific effect of Nav1.3 on neuropathic pain following injury has also been reported. Global and sensory neuron-specific knockouts of Nav1.3 in mice did not show a neuropathic pain phenotype.38 The discrepancy between the results of Nassar et al., and Hains et al., and our present study may be due to differences between rats and mice. An apparent species-specific contribution of Nav1.8 to chronic pain has also been reported.39,40,41 Additional confounding factors might be related to developmental and perinatal genetic compensation for the loss of Nav1.3 in mice.4 Understanding the apparent species-specific difference requires future studies.
Pain gene therapy approaches have also used another viral vector, herpes simplex virus, to deliver various molecules to suppress pain.42,43 Herpes simplex virus-expressing human preproenkephalin and delivered to the sensory nervous system via subcutaneous injection has now gone through a human phase I clinical trial for pain and showed promising results.44 Human clinical trials have also suggested safety and tolerability of recombinant AAV in gene therapy for several CNS disorders.45,46 Given AAV2 serotype 5′s high affinity for and efficient transduction within peripheral neurons, its ability to drive long-term expression of transgenes,47 and its apparent lack of immunogenicity,19 it may prove to be an effective alternative vector for pain gene therapy.
In summary, our data provide molecular and behavioral evidence for a contribution of Nav1.3 to SNI-induced neuropathic pain in rat, and demonstrate the efficacy of a gene therapy approach as a potential therapeutic option. AAV-mediated shRNAmir knockdown offers the possibility of long-term tissue-specific delivery of a therapeutic molecule that could be effective for pain treatment, with limited side-effects that are usually associated with nonselective sodium channel blockers.19 More generally, we enhance and validate a platform that is poised to provide a novel degree of flexibility to the (i) conduction of proof-of-principle studies of pharmacogenetic therapies for chronic pain, and (ii) gene function studies in the adult peripheral nervous system at selected segmental levels.
Materials and Methods
Animals. Adult male Sprague–Dawley rats weighing 200–225 g (Harlan, IN) were used in this study. Animals were housed in groups of two per cage and were kept in a room at a constant temperature (22 °C) and relative humidity (60%) with alternating 12 hours light-dark cycle. All experiments were conducted during the light cycle. Food and water were supplied ad libitum. All experiments were in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals, and the protocols were approved by the VA Connecticut Healthcare System Institutional Animal Use Committee.
siRNA design, cloning, and viral vector use. siRNA sequences against Nav1.3 were designed using an algorithm developed at MIT (http://jura.wi.mit.edu/bioc/siRNA). For the experiments in cell culture, the indicated sequences were cloned as shRNA downstream of a U6 promoter in pSicoR vector (Addgene, Cambridge, MA). For in vivo experiments, shRNAmir backbone23 was used and the indicated sequences were cloned downstream of a human U6 promoter in a pAAV2-CMV-GFP vector provided by the Gene Transfer Vector Core (Iowa University). Recombinant virus was produced using a Bacoluvirus system48 by the Gene Transfer Vector Core (Iowa University) and the provided titers were as follows: shRNA-Sc, 7.27 × 1013 vg/ml; shRNA-B, 1.35 × 1013 vg/ml; shRNA-C, 7.05 × 1013 vg/ml. Before their use in the animals, viral solutions were desalted using Slide-A-Lyser mini dialysis (Thermo Scientific, Rockford, IL) for 1 hour at 4 °C according to the manufacturer's protocol.
Cell culture. HEK293 cells stably expressing Nav1.3 were generated as previously described,25 and maintained in Dulbecco's modified Eagle's medium/F-12 (1:1) (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (Hyclone, Rockford, IL), penicillin (100 units/ml), and streptomycin (100 µg/ml) (Invitrogen).
Immunodetection methods. Western blots (Figure 1), as well as immunohistochemistry on cryosections (Figure 3) were done as previously described.49 The following primary antibodies were used: pan-sodium channel primary antibody was used at 1/10,000 dilution, rabbit anti-GFP (Invitrogen; 1/1,000), and mouse anti-NeuN (Millipore, Billerica, CA; 1/1,000).
Voltage-clamp recordings. HEK293 cells stably expressing wild-type NaV1.3 channels were transfected with plasmids expressing shRNA against NaV1.3 or scrambled sequence using Lipofectamine 2000 (Invitrogen). Cells were replated and seeded at low density on 12-mm coverslips 24 hour after transfection. Whole-cell voltage-clamp recordings were obtained 48 hr after transfection with an Axopatch 200B amplifier (Axon Instruments, Foster City, CA) at room temperature (20–22 °C). Fire-polished electrodes (0.6–1.3 MΩ) were fabricated from 1.6-mm outer diameter borosilicate glass micropipettes (World Precision Instruments, Sarasota, FL). The pipette potential was adjusted to zero before seal formation, and liquid junction potential was not corrected. Capacity transients were cancelled and voltage errors were minimized with 80–90% series resistance compensation. Currents were acquired with Clampex 9.2, 5 min after establishing whole-cell configuration, sampled at a rate of 100 kHz, and filtered at 5 kHz.
The pipette solution contained (in mmol/l): 140 CsF, 10 NaCl, 1 EGTA, 10 dextrose, and 10 HEPES, pH 7.32 (adjusted with CsOH), and the osmolarity was adjusted to 308 mOsmol/l with sucrose. The extracellular bath solution for voltage-clamp contained (in mmol/l): 140 NaCl, 3 KCl, 1 MgCl2, 1 CaCl2, 10 dextrose, 10 HEPES, pH 7.35 (adjusted with NaOH), and the osmolarity was adjusted to 315 mOsmol/l with sucrose.
To measure NaV1.3 currents, cells were held at –120 mV and stepped to a range of potentials (–80 to +60 mV in 5 mV increments) for 100 ms. Current density was obtained by normalizing maximal peak inward currents with cell capacitance. Data were analyzed using Clampfit 9.2 (Molecular Devices, Sunnyvale, CA) and OriginPro 8 (Microcal Software, Northampton, MA), and presented as means ± SE. One-way ANOVA followed by Tukey post hoc test for multigroup analysis was used for statistical significance.
Surgical procedures
Direct DRG injection: Direct DRG injections were adapted from recent studies.21,26 Rats were anesthetized by exposure to isoflurane (1–3%) administered by a calibrated vaporizer. Each animal was then prepared for aseptic surgery by shaving the skin on the back around the lumbar area using an electric clipper. It was then placed on a sterile, disposable absorbent pad and the prepared area draped with fenestrated gauze pad. A 2–3 cm incision was made in the skin along the dorsal midline. The superficial muscular fascia was incised and the paraspinal muscles were separated by sharp and blunt dissection, exposing the lateral aspect of the left fourth (L4) lumbar vertebra, and the dorsal aspect of the medial portion of its transverse process. To expose the DRG, a partial laminectomy whereby the lateral process of the vertebra was clipped using a fine Rongeur (Fine Science Tools). To inject solutions into the DRG, we used a custom-designed sharp glass micropipette (20–30 µm tip diameter) mounted on a 10-µl Hamilton needle. The exposed epineurium or DRG capsule was then punctured and the solution injected via a pump (Harvard Apparatus) at a rate of 0.4 µl/min. Once the volume was dispensed, the micropipette was left in place for an additional 1–2 minutes to prevent backflow. We injected 2 × 1 µl in two different sites for each DRG. The overlying muscle and skin were then closed in two layers using 4.0 monofilament nylon sutures.
SNI. The SNI model is a well-establish model of neuropathic pain in rats.27 Briefly, animals were anesthetized by exposure to isoflurane (1–3%). The surgical field (left leg) was shaved and disinfected. An incision (~2 cm) was made on the lateral mid-thigh and the underlying muscles separated to expose the sciatic nerve. The three branches of the sciatic (tibial, common peroneal, and sural) were then carefully separated. Efforts were made to minimize any contact with the sural branch. The tibial and common peroneal were then individually ligated with 6.0 sutures and cut distally 2–3 mm of each of tibial and common peroneal branches were removed distal of the ligation. The muscle and skin were closed in two layers using 4.0 monofilament nylon sutures. In Figure 6, SNI and direct DRG injections were performed on the same day (SNI first, followed immediately by direct DRG injection).
Pain behavioral assays. The experimenter was blinded to the treatment groups of the animals. All rats were acclimatized to the behavior room and testing apparatus in three to four habituation sessions. Baselines were then recorded one to two days prior to the surgery (considered as day 0), and then at defined intervals afterwards as indicated. For mechanical thresholds measurement, the rats were placed on an elevated wire grid and the lateral plantar surface of the left paw (ipsilateral to the surgery/injection) of each animal was presented with a series of calibrated Von Frey hairs (Stoeling, Wood Dale, IL). The 50% withdrawal threshold was determined using the “up-down” method.50 For thermal sensitivity, the plantar paw surface of animals was exposed to radiant heat pain using a Hargreaves apparatus (IITC, Woodland Hills, CA) according to the Hargreaves method.51 Paw withdrawal latency was then measured (beam intensity was adjusted to result in a latency of 9–11 seconds for control baselines). The heat stimulation was repeated three times (at an interval of 5 minutes for each rat) and the average calculated. The heat source was automatically cut-off at 20.5 seconds to prevent tissue damage.
Quantitative real-time reverse transcription-PCR. L4 dorsal root ganglia from rats injected with virus were freshly dissected and immediately processed individually for RNA extraction using RNasy Microkit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Three hundred nanogram of total RNA from each rat L4 DRG was used to generate 1st strand cDNA using Superscript III (Invitrogen) according to the manufacturer's protocol. Real-time Taqman PCR assays for rat: Nav1.3 (assay id: Rn01485332_m1), Nav1.6 (assay id: Rn00570506_m1), Nav1.7 (Rn00591020_m1), Nav1.8 (Rn00568393_m1), Nav1.9 (Rn00570487_m1), and rat GAPDH (assay id: Rn01775763_g1) were purchased from Applied Biosystems and used with Universal Taqman PCR master mix (20×) (Applied Biosystems, Carlsbad, CA). One microliter of the cDNA was used as a template in a 20 µl and the reaction was run in duplicates for each animal (number of biological replicates here is the number of animals used per treatment group. shRNA-Sc, n = 7; shRNA-B, n = 3; shRNA-C, n = 5) according to the manufacturer's instructions using the Eppendorf realplex (USA). Normalization and relative expression analysis of Nav1.3 mRNA were done using the 2–δδCt method with GAPDH as the control.28
Statistical analyses. We used t-test to analyze significance of protein knockdown by shRNA sequences in Figure 1 (n = 4 for each of nontransfected cells, shRNA-Sc, shRNA-A, and shRNA-B, shRNA-C). One-way repeated ANOVA followed by Bonferroni correction was used for analyses of mechanical and thermal sensitivity over the studied time course within each treatment group. To compare mechanical and thermal sensitivity between two treatment groups over the indicated time course, two-way repeated ANOVA followed by Bonferroni correction was calculated. Mann–Whitney U-test was used to calculate the significance at specific time points when comparing treatment groups. Log scale was used to plot mechanical threshold. P < 0.05 was considered significant.
Acknowledgments
This work was supported in part by grants from the Medical Research Service and Rehabilitation Research Service, Department of Veterans Affairs, and the Nancy Taylor Foundation for Chronic Diseases. The Center for Neuroscience and Regeneration Research is a Collaboration of the Paralyzed Veterans of America and Yale University.
REFERENCES
- Jensen TS, Baron R, Haanpaa M, Kalso E, Loeser JD, Rice AS.et al. (2011A new definition of neuropathic pain Pain 1522204–2205. [DOI] [PubMed] [Google Scholar]
- Mao J, Gold MS., and, Backonja MM. Combination drug therapy for chronic pain: a call for more clinical studies. J Pain. 2011;12:157–166. doi: 10.1016/j.jpain.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth C, Lander J., and, Wiebe S. The prevalence and impact of chronic pain with neuropathic pain symptoms in the general population. Pain Med. 2009;10:918–929. doi: 10.1111/j.1526-4637.2009.00655.x. [DOI] [PubMed] [Google Scholar]
- von Hehn CA, Baron R., and, Woolf CJ. Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron. 2012;73:638–652. doi: 10.1016/j.neuron.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum AI, Bautista DM, Scherrer G., and, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Scholz J., and, Woolf CJ. Neuropathic pain: a maladaptive response of the nervous system to damage. Annu Rev Neurosci. 2009;32:1–32. doi: 10.1146/annurev.neuro.051508.135531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj SD, Cummins TR, Black JA., and, Waxman SG. Sodium channels in normal and pathological pain. Annu Rev Neurosci. 2010;33:325–347. doi: 10.1146/annurev-neuro-060909-153234. [DOI] [PubMed] [Google Scholar]
- Dworkin RH, O'Connor AB, Audette J, Baron R, Gourlay GK, Haanpää ML.et al. (2010Recommendations for the pharmacological management of neuropathic pain: an overview and literature update Mayo Clin Proc 853 SupplS3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., and, Wood JN. The roles of sodium channels in nociception: implications for mechanisms of neuropathic pain. Pain Med. 2011;12 Suppl 3:S93–S99. doi: 10.1111/j.1526-4637.2011.01158.x. [DOI] [PubMed] [Google Scholar]
- Waxman SG, Kocsis JD., and, Black JA. Type III sodium channel mRNA is expressed in embryonic but not adult spinal sensory neurons, and is reexpressed following axotomy. J Neurophysiol. 1994;72:466–470. doi: 10.1152/jn.1994.72.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hains BC, Klein JP, Saab CY, Craner MJ, Black JA., and, Waxman SG. Upregulation of sodium channel Nav1.3 and functional involvement in neuronal hyperexcitability associated with central neuropathic pain after spinal cord injury. J Neurosci. 2003;23:8881–8892. doi: 10.1523/JNEUROSCI.23-26-08881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hains BC, Saab CY, Klein JP, Craner MJ., and, Waxman SG. Altered sodium channel expression in second-order spinal sensory neurons contributes to pain after peripheral nerve injury. J Neurosci. 2004;24:4832–4839. doi: 10.1523/JNEUROSCI.0300-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindia JA, Köhler MG, Martin WJ., and, Abbadie C. Relationship between sodium channel NaV1.3 expression and neuropathic pain behavior in rats. Pain. 2005;117:145–153. doi: 10.1016/j.pain.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Black JA, Cummins TR, Plumpton C, Chen YH, Hormuzdiar W, Clare JJ.et al. (1999Upregulation of a silent sodium channel after peripheral, but not central, nerve injury in DRG neurons J Neurophysiol 822776–2785. [DOI] [PubMed] [Google Scholar]
- Black JA, Nikolajsen L, Kroner K, Jensen TS., and, Waxman SG. Multiple sodium channel isoforms and mitogen-activated protein kinases are present in painful human neuromas. Ann Neurol. 2008;64:644–653. doi: 10.1002/ana.21527. [DOI] [PubMed] [Google Scholar]
- Cummins TR, Aglieco F, Renganathan M, Herzog RI, Dib-Hajj SD., and, Waxman SG. Nav1.3 sodium channels: rapid repriming and slow closed-state inactivation display quantitative differences after expression in a mammalian cell line and in spinal sensory neurons. J Neurosci. 2001;21:5952–5961. doi: 10.1523/JNEUROSCI.21-16-05952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert A, Hains BC., and, Waxman SG. Upregulation of persistent and ramp sodium current in dorsal horn neurons after spinal cord injury. Exp Brain Res. 2006;174:660–666. doi: 10.1007/s00221-006-0511-x. [DOI] [PubMed] [Google Scholar]
- Glorioso JC., and, Fink DJ. Gene therapy for pain: introduction to the special issue. Gene Ther. 2009;16:453–454. doi: 10.1038/gt.2009.18. [DOI] [PubMed] [Google Scholar]
- Beutler AS. AAV provides an alternative for gene therapy of the peripheral sensory nervous system. Mol Ther. 2010;18:670–673. doi: 10.1038/mt.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques SJ, Ahmed Z, Forbes A, Douglas MR, Vigenswara V, Berry M.et al. (2012AAV8(gfp) preferentially targets large diameter dorsal root ganglion neurones after both intra-dorsal root ganglion and intrathecal injection Mol Cell Neurosci 49464–474. [DOI] [PubMed] [Google Scholar]
- Mason MR, Ehlert EM, Eggers R, Pool CW, Hermening S, Huseinovic A.et al. (2010Comparison of AAV serotypes for gene delivery to dorsal root ganglion neurons Mol Ther 18715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Chou B, Fitzsimmons B, Miyanohara A, Shubayev V, Santucci C.et al. (2012In vivo gene knockdown in rat dorsal root ganglia mediated by self-complementary adeno-associated virus serotype 5 following intrathecal delivery PLoS ONE 7e32581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ.et al. (2005Second-generation shRNA libraries covering the mouse and human genomes Nat Genet 371281–1288. [DOI] [PubMed] [Google Scholar]
- Pei Y., and, Tuschl T. On the art of identifying effective and specific siRNAs. Nat Methods. 2006;3:670–676. doi: 10.1038/nmeth911. [DOI] [PubMed] [Google Scholar]
- Shah BS, Rush AM, Liu S, Tyrrell L, Black JA, Dib-Hajj SD.et al. (2004Contactin associates with sodium channel Nav1.3 in native tissues and increases channel density at the cell surface J Neurosci 247387–7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Kostic S, Nakai H, Park F, Sapunar D, Yu H.et al. (2011Direct injection into the dorsal root ganglion: technical, behavioral, and histological observations J Neurosci Methods 19943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decosterd I., and, Woolf CJ. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain. 2000;87:149–158. doi: 10.1016/S0304-3959(00)00276-1. [DOI] [PubMed] [Google Scholar]
- Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ.et al. (2010Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels Science 33055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj SD., and, Waxman SG. Isoform-specific and pan-channel partners regulate trafficking and plasma membrane stability; and alter sodium channel gating properties. Neurosci Lett. 2010;486:84–91. doi: 10.1016/j.neulet.2010.08.077. [DOI] [PubMed] [Google Scholar]
- Devor M, Keller CH., and, Ellisman MH. Spontaneous discharge of afferents in a neuroma reflects original receptor tuning. Brain Res. 1990;517:245–250. doi: 10.1016/0006-8993(90)91033-d. [DOI] [PubMed] [Google Scholar]
- Cummins TR., and, Waxman SG. Downregulation of tetrodotoxin-resistant sodium currents and upregulation of a rapidly repriming tetrodotoxin-sensitive sodium current in small spinal sensory neurons after nerve injury. J Neurosci. 1997;17:3503–3514. doi: 10.1523/JNEUROSCI.17-10-03503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omana-Zapata I, Khabbaz MA, Hunter JC, Clarke DE., and, Bley KR. Tetrodotoxin inhibits neuropathic ectopic activity in neuromas, dorsal root ganglia and dorsal horn neurons. Pain. 1997;72:41–49. doi: 10.1016/s0304-3959(97)00012-2. [DOI] [PubMed] [Google Scholar]
- Lyu YS, Park SK, Chung K., and, Chung JM. Low dose of tetrodotoxin reduces neuropathic pain behaviors in an animal model. Brain Res. 2000;871:98–103. doi: 10.1016/s0006-8993(00)02451-3. [DOI] [PubMed] [Google Scholar]
- Boucher TJ, Okuse K, Bennett DL, Munson JB, Wood JN., and, McMahon SB. Potent analgesic effects of GDNF in neuropathic pain states. Science. 2000;290:124–127. doi: 10.1126/science.290.5489.124. [DOI] [PubMed] [Google Scholar]
- Kole R, Krainer AR., and, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Waxman SG., and, Hains BC. Sodium channel expression in the ventral posterolateral nucleus of the thalamus after peripheral nerve injury. Mol Pain. 2006;2:27. doi: 10.1186/1744-8069-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C.et al. (2012A pipeline for the generation of shRNA transgenic mice Nat Protoc 7374–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar MA, Baker MD, Levato A, Ingram R, Mallucci G, McMahon SB.et al. (2006Nerve injury induces robust allodynia and ectopic discharges in Nav1.3 null mutant mice Mol Pain 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Gold MS, Kim CS, Bian D, Ossipov MH, Hunter JC.et al. (2002Inhibition of neuropathic pain by decreased expression of the tetrodotoxin-resistant sodium channel, NaV1.8 Pain 95143–152. [DOI] [PubMed] [Google Scholar]
- Stirling LC, Forlani G, Baker MD, Wood JN, Matthews EA, Dickenson AH.et al. (2005Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice Pain 11327–36. [DOI] [PubMed] [Google Scholar]
- Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP.et al. (2008The cell and molecular basis of mechanical, cold, and inflammatory pain Science 321702–705. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M, Zhou Z, Hao S, Mata M., and, Fink DJ. Reduction of voltage gated sodium channel protein in DRG by vector mediated miRNA reduces pain in rats with painful diabetic neuropathy. Mol Pain. 2012;8:17. doi: 10.1186/1744-8069-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorioso JC., and, Fink DJ. Herpes vector-mediated gene transfer in the treatment of chronic pain. Mol Ther. 2009;17:13–18. doi: 10.1038/mt.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink DJ, Wechuck J, Mata M, Glorioso JC, Goss J, Krisky D.et al. (2011Gene therapy for pain: results of a phase I clinical trial Ann Neurol 70207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplitt MG, Feigin A, Tang C, Fitzsimons HL, Mattis P, Lawlor PA.et al. (2007Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial Lancet 3692097–2105. [DOI] [PubMed] [Google Scholar]
- Janson C, McPhee S, Bilaniuk L, Haselgrove J, Testaiuti M, Freese A.et al. (2002Clinical protocol. Gene therapy of Canavan disease: AAV-2 vector for neurosurgical delivery of aspartoacylase gene (ASPA) to the human brain Hum Gene Ther 131391–1412. [DOI] [PubMed] [Google Scholar]
- Xu Y, Gu Y, Xu GY, Wu P, Li GW., and, Huang LY. Adeno-associated viral transfer of opioid receptor gene to primary sensory neurons: a strategy to increase opioid antinociception. Proc Natl Acad Sci USA. 2003;100:6204–6209. doi: 10.1073/pnas.0930324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urabe M, Ding C., and, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- Persson AK, Gasser A, Black JA., and, Waxman SG. Nav1.7 accumulates and co-localizes with phosphorylated ERK1/2 within transected axons in early experimental neuromas. Exp Neurol. 2011;230:273–279. doi: 10.1016/j.expneurol.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Chaplan SR, Bach FW, Pogrel JW, Chung JM., and, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Hargreaves K, Dubner R, Brown F, Flores C., and, Joris J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32:77–88. doi: 10.1016/0304-3959(88)90026-7. [DOI] [PubMed] [Google Scholar]