Abstract
Despite the therapeutic potential of nucleic acid drugs, their clinical application has been limited in part by a lack of appropriate delivery systems. Exosomes or microvesicles are small endosomally derived vesicles that are secreted by a variety of cell types and tissues. Here, we show that exosomes can efficiently deliver microRNA (miRNA) to epidermal growth factor receptor (EGFR)-expressing breast cancer cells. Targeting was achieved by engineering the donor cells to express the transmembrane domain of platelet-derived growth factor receptor fused to the GE11 peptide. Intravenously injected exosomes delivered let-7a miRNA to EGFR-expressing xenograft breast cancer tissue in RAG2–/– mice. Our results suggest that exosomes can be used therapeutically to target EGFR-expressing cancerous tissues with nucleic acid drugs.
Introduction
MicroRNAs (miRNAs) are small (20–22 nucleotides) noncoding RNA molecules that bind to partially complementary mRNA sequences, resulting in target degradation or translation inhibition.1 A growing pool of evidence suggests that miRNA-related gain- or loss-of-function mutations can cause the development and/or progression of cancer.2 For example, let-7a is thought to be a tumor suppressor that inhibits the malignant growth of cancer cells by reducing RAS and HMGA2 expression. Reduced expression levels of let-7 have been observed in colon, lung, ovary, and breast cancer cells.3 Therefore, miRNA replacement therapies have emerged as promising treatment strategies for malignant neoplasms. Yet although miRNA-based modalities may eventually prove effective, their clinical application has been hampered by a lack of appropriate delivery systems.
Exosomes or microvesicles are small vesicles (50–100 nm in diameter) that are secreted by a variety of cell types and tissues.4 Of clinical interest, tumor cells have been shown to release exosomes containing miRNA5 and miRNAs secreted from donor cells can be taken up and function in recipient cells.6,7 These data indicate that exosomes are natural carriers of miRNA that could be exploited as an RNA drug delivery system. For instance, Alvarez-Erviti et al. recently used exosomes with modified membranes containing a neuron-specific peptide to deliver small-interfering RNA (siRNA) to mouse brain tissue.8 Nevertheless, the utility of exosomes as carriers of cancer therapies remains largely unknown.
A number of human tumors of epithelial origin display elevated epidermal growth factor receptor (EGFR) expression, suggesting that EGFR could serve as a receptor target in cancer drug delivery systems.9 Because the EGFR ligand epidermal growth factor (EGF) is strongly mitogenic and neoangiogenic, however, an alternative ligand is needed for clinical applications.
The GE11 peptide (amino-acid sequence YHWYGYTPQNVI) binds specifically to EGFR, but is markedly less mitogenic than EGF.10 Additionally, GE11-conjugated polyethylenimine vectors and polyethylene glycol–conjugated liposomes have been shown to be less mitogenic, and can efficiently transfect genes into cells expressing high levels of EGFR or tumor xenografts.10,11,12,13 These studies indicated that the GE11 peptide is likely superior to EGF for clinically targeting EGFR-expressing tumors.
In this study, we examined exosomes as drug delivery carriers in a model of cancer. Modified exosomes with the GE11 peptide or EGF on their surfaces delivered miRNA to EGFR-expressing cancer tissues; intravenously injected exosomes targeting EGFR delivered let-7a specifically to xenograft breast cancer cells in RAG2–/– mice. These data indicate that exosomes targeted to EGFR-expressing cells may provide a platform for miRNA replacement therapies in the treatment of various cancers.
Results
GE11- and EGF-positive exosomes
GE11 peptide specifically binds to EGFR, but is less mitogenic than EGF.10 To generate GE11- or EGF-positive exosomes, sequence encoding GE11 or EGF was cloned into the pDisplay vector. This vector promotes the expression of proteins on plasma membranes using the transmembrane domain of platelet-derived growth factor receptor (Figure 1a). We transfected human embryonic kidney cell line 293 (HEK293) cells with pDisplay encoding GE11 or EGF, and then cloned cells that were stably expressing the constructs. Exosomes were purified from culture supernatants using an ultracentrifugation protocol (see Materials and Methods section).
Figure 1.
Epidermal growth factor receptor (EGFR) ligands on the outer surfaces of the exosomes. (a) Diagrams of the modified epidermal growth factor (EGF) and GE11 proteins. Signal peptide, Igκ-chain leader sequence; HA, hemagglutinin epitope tag (YPYDVPDYA); Linker, (GGGGS) 3; Myc, Myc epitope (EEKLISEEDL); platelet-derived growth factor receptor (PDGFR) transmembrane domain, transmembrane domain from platelet-derived growth factor receptor. (b) Western blots of HA-tagged constructs in exosomes obtained from culture supernatants of human embryonic kidney cell line 293 (HEK293) cells that had been transfected with pDisplay encoding EGF or GE11. The quality of each exosome preparation was confirmed by hybridization with antihuman leukocyte antigen (HLA) antibodies. (c) For flow cytometry, exosomes from transfected HEK293 cells were incubated with latex beads and stained with anti-Myc tag antibodies. Tetraspanin CD81 was used as a positive control for the exosomes. (d) Immunoelectron microscopy showed that HA-tagged constructs were present on exosomes purified from the supernatants of cells transfected with pDisplay encoding EGF or GE11. Bars = 100 nm. The percentages of HA-positive exosomes are indicated in the graph. Data are expressed as means ± SD.
We then examined the expression of GE11 or EGF in exosomes using anti-hemagglutinin (HA) antibodies and western blot analysis, which revealed bands of the predicted sizes (Figure 1b). In addition, fluorescence-activated cell sorting (FACS) confirmed the presence of GE11 and EGF on the outer membranes of exosomes bound to latex beads, and these complexes were recognized by anti-Myc-tag antibodies (Figure 1c). CD81 was used as a positive control for exosomes. Myc-tag expression was observed more frequently in EGF-positive (73.8%) and GE11-positive (66.2%) exosomes than in vector control (14.3%) exosomes. These data indicated that GE11 or EGF was present on the exosomal membranes. Additionally, immunogold staining with anti-HA antibodies showed that 15.3% and 21.2% of the exosomes were positive for GE11 and EGF, respectively, and no notable morphologic abnormalities were observed in the modified exosomes (Figure 1d).
EGFR-dependent uptake of modified exosomes in vitro
We next examined whether the GE11- or EGF-positive exosomes derived from HEK293 cells bound to recipient cells in an EGFR-dependent manner. We first evaluated EGFR expression in three human breast cancer cell lines. HCC70 cells showed higher EGFR expression levels than HCC1954 and MCF-7 cells (Figure 2a). To examine whether GE11- or EGF-positive exosomes were taken up by recipient cells, exosomes were labeled with PKH67 dye (green) and added to cultures of HCC70 cells (Figure 2b). EGF- and GE11-positive exosomes more efficiently bound to HCC70 cells than HCC1954 or MCF-7 cells. Binding appeared to reflect EGFR expression levels (Figure 2b). Exosomes did not bind to cell surface membranes when the samples were incubated at 4 °C, suggesting that the cells had to be biologically active (Figure 2b). Confocal laser-scanning microscopy demonstrated that the exosomes were internalized by the cells (Figure 2c). To confirm that EGF- and GE11-positive exosomes were taken up by an EGFR-dependent mechanism, we performed assays using breast cancer cell lines with different levels of EGFR expression. First, we prepared MCF-7 cells expressing high levels of EGFR using a retroviral vector (Figure 2d) and examined uptake of EGF- and GE11-positive exosomes (Figure 2e). EGF- and GE11-positive exosomes showed high affinities for these MCF-7 cells compared with cells infected with empty vector or untreated cells. Next, we prepared HCC70 cells in which EGFR expression was knocked down using siRNA. Three days post-transfection, EGFR expression was markedly reduced, and we examined exosome uptake using fluorescence-activated cell sorting analysis (Figure 2f). Because the cells proliferated following transfection of EGFR siRNA, relatively low levels of siRNA were detected in PKH67-labeled exosomes compared with the experiment shown in Figure 2b. Of note, however, siRNA levels decreased to near background levels in HCC70 cells in which EGFR expression was reduced compared with cells expressing high levels of EGFR or cells transfected with nontarget siRNA.
Figure 2.
Uptake of epidermal growth factor (EGF)- and GE11-positive exosomes by breast cancer cell lines. (a) Flow cytometric analysis of epidermal growth factor receptor (EGFR) expression on HCC70, HCC1954, and MCF-7 breast cancer cells. (b) Uptake of fluorescently labeled exosomes by the breast cancer cell lines was detected using flow cytometry. PKH67-labeled exosomes were incubated with the breast cancer cell lines at 37 °C or 4 °C for 4 hours. The degree of uptake was relatively low at 4 °C. (c) Intracellular PKH67-labeled exosomes were detected in HCC70 cells (arrows) using confocal fluorescence microscopy. (d) Flow cytometric analysis of EGFR expression on MCF-7 cells, which were stably infected with retrovirus expressing EGFR. (e) Uptake of PKH67-labeled EGF- and GE11-positive exosomes was compared using MCF-7 cells expressing high levels of EGFR and control cells. (f) Uptake of PKH67-labeled EGF- and GE11-positive exosomes was compared among EGFRLow HCC70, EGFRHigh, and control cells.
To assess whether EGF- or GE11-positive exosomes affected cell growth in vitro, we performed cell proliferation assays using HCC70 cells. EGF-positive exosomes promoted cell proliferation, whereas no effect was noted with control or GE11-positive exosomes (Supplementary Figure S1). These experiments suggested that, unlike EGF-positive exosomes, GE11-positive exosomes do not stimulate EGFR signaling. Thus, GE11-positive exosomes may provide a more suitable drug delivery system than EGF-positive exosomes.
GE11-positive exosomes are functional in vitro
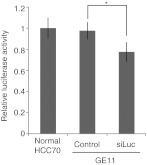
Our results indicated that the modified exosomes were taken up into recipient cells. Next, we investigated exosome-mediated delivery of siRNA or miRNA in vivo, including the effects of the exogenous siRNA or miRNA in the recipient cells. We first transfected HEK293 cells with luciferase-specific siRNA and purified loaded exosomes. Then, we added the exosomes to culture medium containing luciferase-expressing HCC70 cells. After 24 hours, we measured luciferase activities and found that GE11-positive exosomes containing luciferase-specific siRNA reduced luciferase activity in the cells (Figure 3). These exosomes likely contained only a fraction of the transfected siRNA from the exosome-secreting cells. Nevertheless, the encapsulated siRNA significantly inhibited expression of the target luciferase gene.
Figure 3.
The activity of encapsulated small-interfering RNA (siRNA) in luciferase assays. Luciferase-specific siRNA (siLuc) was encapsulated in exosomes, which were incubated for 48 hours with HCC70 cells stably expressing firefly luciferase. Data are expressed as means ± SD. n = 3; *P < 0.05.
GE11-positive exosomes bind tumor cells in vivo
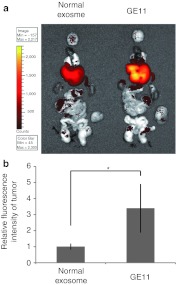
We then examined whether GE11-positive exosomes specifically bind to tumors in vivo. Luciferase-expressing HCC70 cells were transplanted into the mammary fat pads of RAG2–/– mice. GE11-positive and control exosomes were labeled with XenoLight DiR and intravenously injected into tumor-bearing RAG2–/– mice via the tail vein. Twenty-four hours later, the locations of the exosomes were monitored using an in vivo imaging system (IVIS). Signals from the GE11-positive exosomes were detected in the xenograft tumors, whereas little signal was detected in experiments using control exosomes (Figure 4a). We also counted the exosomes and observed that, compared with control exosomes, three times as many GE11-positive exosomes reached the tumor (Figure 4b). In addition, we did not histologically detect any major organ damage in the injected mice (Supplementary Figure S2). These data indicated that GE11-positive exosomes may facilitate the delivery of therapeutic molecules to EGFR-expressing tumors in vivo.
Figure 4.
Migration of GE11-positive exosomes to tumor tissues characterized by high levels of epidermal growth factor receptor (EGFR) expression. (a) Exosomes labeled with XenoLight DiR (near-infrared) were intravenously injected (4 µg of purified exosomes) into mice bearing transplanted HCC70 cells. Brain, heart, spleen, liver, lung, kidney, small intestine, colon, and tumor tissues were harvested 24 hours postinjection for ex vivo imaging. The migration of fluorescently labeled exosomes was detected with an in vivo imaging system (IVIS). (b) The intensity of fluorescent signals from the tumor was measured using an IVIS. Data are expressed as means ± SD. n = 5; *P < 0.05.
GE11-positive exosomes containing let-7 inhibit tumor development in vivo
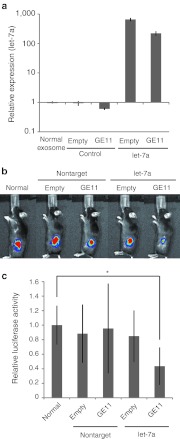
We next investigated the delivery of miRNA to tumors using the GE11-positive exosomes. We used let-7a because elevated let-7 expression in cancer cell lines alters cell cycle progression and reduces cell division, suggesting that let-7 functions as a tumor suppressor.14,15 Let-7a or control miRNA was introduced into GE11-positive or control exosomes using the lipofection method and HEK293 cells, and the amount of loaded miRNA was determined in quantitative real-time reverse transcription-PCRs (Figure 5a). To measure tumor growth in RAG2–/– mice, we prepared tumor-bearing mice with xenograft HCC70 cells that stably expressed luciferase. To analyze tumor growth and development, we injected the tumors with luciferin and monitored signal emission using an IVIS. Exosomes were intravenously injected into tumor-bearing mice via the tail vein. After four injections, tumor growth was measured. Let-7a–containing GE11-positive exosomes markedly suppressed tumor growth (Figure 5b and c; n = 6, P < 0.01). Several studies have reported that let-7a inhibits tumor development by reducing expression levels of HMGA2 or members of the RAS family (K-RAS, H-RAS, N-RAS).16 We examined the expression of these genes in let-7a–transfected HCC70 cells using real-time reverse transcription-PCR analysis, immunoblotting, and immunostaining (Supplementary Figure S3a–c). Furthermore, we immunohistochemically assessed the expression of HMGA2 and RAS family members in xenograft tumors (Supplementary Figure S3d). Let-7a did not affect the expression of HMGA2 or RAS family members in vivo or in vitro. Consistent with previous reports, however, let-7a potently inhibited the expression of HMGA2 mRNA in A549 lung adenocarcinoma cells17 (Supplementary Figure S3a). These data indicated that let-7a inhibits tumor development via previously unidentified or uncharacterized genes in HCC70 breast cancer cells. Taken together, these findings indicated that GE11-positive exosomes are a promising vehicle for delivering drugs to EGFR-expressing tumors.
Figure 5.
Inhibition of breast cancer development in vivo using GE11-positive exosomes containing let-7a. Human embryonic kidney cell line 293 (HEK293) cells expressing GE11 were transfected with synthetic let-7a. Exosomes containing let-7a were purified from culture supernatants and intravenously injected (1 µg of purified exosomes, once per week for 4 weeks) into mice bearing luciferase-expressing HCC70 cells. (a) Let-7a levels in the purified exosomes were measured using quantitative PCRs. Data are expressed as means ± SD. n = 3. (b) Representative images of tumors 4 weeks postinjection are shown. (c) Luciferase signals from the tumors were measured using an in vivo imaging system (IVIS). Data are expressed as means ± SD. n = 5; *P < 0.05.
Discussion
Although miRNA is a promising anticancer therapeutic modality, the clinical use of these RNA molecules has been hampered by a lack of malignant tissue–specific delivery systems. In the present study, we showed that exosomes can be used to efficiently deliver antitumor miRNA to cancer tissues in vivo.
A number of nanocarriers using various materials have been developed for drug delivery systems. Polyethylene glycol–coated liposomes, which are frequently used as carriers for in vivo drug delivery, benefit from easy preparation techniques, acceptable toxicity profiles, and a lack of clearance by the reticuloendothelial system. Liposomes, however, have several drawbacks, including the efficiency of targeting and issues associated with accelerated blood clearance.18 Although exosomes and liposomes have similar phospholipid bilayers, exosomes consist of only biogenic substances. The potential of exosomes as drug delivery carriers, however, is largely unknown. Adding appropriate targeting molecules can cause exosomes to accumulate at sites of disease in vivo (ref. 7 and Figure 4). Thus, the biocompatibility and toxicity profiles of exosomes, which notably are natural carriers of miRNA in vivo, support their application in drug delivery systems.
To use exosomes clinically, however, further studies are needed to resolve several issues. For instance, therapeutic exosomes should not be quickly cleared by the reticuloendothelial system. In this study, when fluorescently labeled exosomes were injected into the mice tail vein, many exosomes accumulated in the liver 24 hours after the injection (Figure 4a). Thus, some modifications are required to avoid normal clearance mechanisms.
This type of delivery strategy requires that the miRNA or siRNA can be efficiently introduced into the exosomes. We transfected donor cells with miRNA and purified exosomes from the culture supernatant. A previous report described electroporation protocols for loading siRNA into exosomes.8 We, however, were unable to use these methods to load our exosomes with miRNA. The differences in the results may have been caused by the different cell types that were used in the two studies.
The composition of exosomes appears to differ depending on the source tissue or cell type. For instance, major histocompatibility complex class II molecules are enriched in exosomes from B lymphocytes, dendritic cells, mast cells, and intestinal epithelial cells, whereas higher levels of growth factors and their receptors are found in exosomes released from cancer cells.19 For allogeneic exosome therapy, the presence of major histocompatibility complex proteins is problematic owing to potential immune responses. Therefore, the appropriate selection of donor cells for exosome production is a key factor for potential clinical applications of exosome-based therapies.
In conclusion, exosomes targeted to tumors may allow systemic administration of miRNA as cancer therapy. Technologic improvements that enhance exosome production and reduce immunogenicity should be explored to further develop this drug delivery approach.
Materials and Methods
Plasmids transfection and retrovirus infection. The pDisplay vector was purchased from Invitrogen (Carlsbad, CA). Sequence encoding GE11 peptide (YHWYGYTPQNVI) with a flexible peptide linker (GGGGS)3 or mature EGF (53 amino acids, GenBank accession number P01133) was directly fused into pDisplay (Figure 1a). HEK293 cells were transfected with pDisplay encoding GE11 or EGF using FuGENE HD transfection reagent (Promega, Madison, WI) and selected with Geneticin (Invitrogen).
EGFR retroviral vector was purchased from Addgene (Cambridge, MA). Viral supernatants were produced by transient transfection of GP2-293T cells with a packaging plasmid (pVSV-G) according to the manufacturer's instructions (Clontech, Mountain View, CA). MCF-7 cells were infected with viral supernatants using polybrene at a final concentration of 8 µg/ml.
Cell culture and small RNA transfection. Breast cancer cell lines (HCC70, HCC1954, and MCF-7) and a HEK293 were purchased from the American Type Culture Collection (Manassas, VA). Cells were cultured according to the manufacturer's instructions. HCC70 cells express firefly luciferase, as previously described.20 miRNA and siRNA used in this study were as follows: has-let-7a sense (5′-UGAGGUAGUAGGUUGUAUAGUU-3′) and antisense (5′-CUAUACAAUCUACUGUCUUUC-3′); nontarget control miRNA sense (5′-AUCCGCGCGAUAGCACGUAUU-3′) and antisense (5′-UACGUACUAUCGCGCGGAUUU-3′); EGFR-specific siRNA sense (5′-GUGAGGUGGUCCUUGGGAATT-3′) and antisense (5′-UUCCC AAGGACCACCUCACTT-3′); luciferase-specific siRNA sense (5′-CUU ACGCUGAGUACUUCGATT-3′) and antisense (5′-UCGAAGUACUCA GCGUAAGTT-3′); and nontarget control siRNA sense (5′-AUCCGCGC GAUAGCACGUATT-3′) and antisense 5′-UACGUACUAUCGCGCGGA UTT-3′). Oligonucleotides were transfected into cells using HiPerFect reagent (final concentration, 50 nmol/l; Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Western blot analysis. Western blot analysis was performed as previously described.21 Exosome samples were lysed in sodium dodecyl sulfate loading buffer. After boiling, equal amounts (4 µg) of the proteins were electrophoresed on 15% sodium dodecyl sulfate-polyacrylamide gels and transferred to Immobilon membranes (Millipore, Bedford, MA) using semidry blotting. Then, using standard techniques, the membranes were probed with antibodies, including anti-HA (HA7) (Sigma-Aldrich, St Louis, MO) and anti-HLA-A/B/C (H-300) (Santa Cruz Biotechnology, Santa Cruz, CA). Labeling was visualized using Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore) and signals were examined on an LAS-3000 mini system (Fujifilm, Tokyo, Japan).
RNA isolation and quantitative real-time reverse transcription-PCRs. RNA was isolated from exosomes using Isogen reagent (Nippon Gene, Osaka, Japan) according to the manufacturer's instructions. miRNA levels were quantified using TaqMan miRNA assays (Applied Biosystems, Carlsbad, CA). Copy numbers were calculated based on a standard curve created using synthetic RNA. miRNA levels were normalized based on has-miR-16 levels. Quantitative PCRs were run on a Stratagene MX3000P thermocycler and analyzed with MxPro (Agilent Technologies, Santa Clara, CA).
Preparation of exosomes. Exosomes were prepared from HEK293 cells that had been cultured for 48 hours in Dulbecco's modified eagle medium supplemented with 1% GlutaMax (Invitrogen). Cell supernatants were subjected to differential centrifugation. To eliminate cellular debris, samples were centrifuged at 2,000g for 20 minutes and 10,000g for 30 minutes. Exosomes were pelleted via ultracentrifugation at 120,000g for 70 minutes and washed once in phosphate-buffered saline. Protein content in the exosomes was measured using a Protein Assay Rapid Kit (Wako Pure Chemicals, Osaka, Japan). The average exosome yield was 69.2 µg from 100 ml (2–5 × 107 cells) of culture supernatant (n = 8).
Flow cytometry. For fluorescence-activated cell sorting, exosomes from HEK293 cells were adsorbed onto 4-µm aldehyde-sulphate latex beads (Interfacial Dynamics, Tualatin, OR), incubated with Alexa Fluor 488–conjugated anti-Myc tag antibodies (Millipore, Temecula, CA) or allophycocyanin-conjugated anti-CD81 antibodies (BD Pharmingen, San Jose, CA), and analyzed on a FACSCalibur system (Becton Dickinson, San Diego, CA).
Immunoelectron microscopy. Purified exosomes from HEK293 cells were fixed in 2% paraformaldehyde and loaded onto Formvar-coated Ni electron microscopy grids. The samples were incubated overnight at 4 °C with anti-HA antibodies (Sigma-Aldrich) followed by 1 hour at room temperature with anti-mouse immunoglobulin G conjugated with 15-nm colloidal gold particles. The samples then were fixed in 2% glutaraldehyde, stained with 1% phosphotungstic acid, air-dried, and analyzed using a Hitachi H-7000 electron microscope (Hitachi High-Technologies, Tokyo, Japan). Exosomes positive or negative for gold particles were counted in 10 grids (~1,000–2,000 exosomes).
Coculture of PKH67-labeled exosomes and breast cancer cell lines. Exosomes were stained with green PKH67 fluorescent dye (Sigma-Aldrich). After staining, exosomes were washed with phosphate-buffered saline and centrifuged at 120,000g for 70 minutes. One microgram of PKH67-labeled exosomes was incubated with 1 × 105 breast cancer cells at 37 °C or 4 °C for 4 hours. The uptake of PKH67-labeled exosomes was analyzed using flow cytometry and confocal fluorescence microscopy.
Administration of let-7a–containing exosomes in a human tumor xenograft model. Luciferase-expressing HCC70 cells (2 × 106) were injected subcutaneously into the mammary fat pads of 5-week-old RAG2–/– mice. Four weeks after transplantation, tumors were sized using an IVIS (Xenogen, Hopkinton, MA). HEK293 cells expressing GE11 were transfected with synthetic let-7a. Let-7a–containing exosomes were purified from culture supernatants and intravenously injected (1 µg of purified exosomes, once per week for 4 weeks) into mice with transplanted luciferase-expressing HCC70 cells. Let-7a levels in the exosome samples were evaluated using TaqMan miRNA assays and real-time PCRs. Mice were handled according to the Ethical Guidelines of our institution. All experiments were approved by the Committee for Animal Research at our institution.
In vivo imaging of fluorescently labeled exosomes. A stock solution of the lipophilic near-infrared dye XenoLight DiR (Caliper Life Sciences, Hopkinton, MA) was prepared in ethanol. A 300-µmol/l working solution was prepared in diluent-C solution (Sigma-Aldrich). Exosomes isolated from culture supernatant–derived HEK293 cells were incubated with 2 µmol/l DiR for 30 minutes. The exosomes were then washed with 10 ml of phosphate-buffered saline, subjected to ultracentrifugation, and injected intravenously into RAG2–/– mice (4 µg of exosomes/mouse). Migration of fluorescently labeled exosomes in murine organs was detected using an IVIS 24 hours postinjection.
In vivo imaging of xenograft tumors. Mice were anesthetized via isoflurane inhalation, and intraperitoneally injected with 100 µl of 7.5 mg/ml luciferin solution (Promega). Bioluminescence imaging was initiated with an IVIS (Xenogen) 10 minutes postinjection. The region of interest was defined manually, and bioluminescence data are expressed as photon flux values (photons/s/cm2/steradian). Background photon flux was defined using an area of the tumor that did not receive an intraperitoneal injection of luciferin. All bioluminescence data were collected and analyzed using an IVIS.
Statistical analysis. Differences were statistically evaluated using one-way analysis of variance followed by the Fisher protected least significant difference test. P values <0.05 were defined as statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Analysis of cell viability based on assays with (4, 5-dimethylthiazol-2-yl) 2,5-diphenyl-tetrazolium bromide. Figure S2. Hematoxylin and eosin staining of lung, liver, spleen, and kidney tissues from mice injected with exosomes. Figure S3. Expression analysis of the let-7 target genes.
Acknowledgments
This work was done in Shinjyu-ku, Tokyo, Japan. This work was supported by the “Private University Strategic Research-Based Support Project: Epigenetics Research Project Aimed at a General Cancer Cure Using Epigenetic Targets” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and in part by a grant-in-aid for scientific research on PriorityAreas (B) and (C) from MEXT (Japan) and the Tokyo Medical University Cancer Research Foundation (Japan). We are also indebted to Roderick J. Turner, Edward F. Barroga, and J. Patrick Barron for their editorial review of the English manuscript. The authors declared no conflict of interest.
Supplementary Material
Analysis of cell viability based on assays with (4, 5-dimethylthiazol-2-yl) 2,5-diphenyl-tetrazolium bromide.
Hematoxylin and eosin staining of lung, liver, spleen, and kidney tissues from mice injected with exosomes.
Expression analysis of the let-7 target genes.
REFERENCES
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Calin GA., and, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Barh D, Malhotra R, Ravi B., and, Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol. 2010;17:70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotvall J., and, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr. 2007;1:156–158. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M.et al. (2008Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers Nat Cell Biol 101470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao Y, Iio A, Itoh T, Noguchi S, Itoh Y, Ohtsuki Y.et al. (2011Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages Mol Ther 19395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y., and, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S., and, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Woodburn JR. The epidermal growth factor receptor and its inhibition in cancer therapy. Pharmacol Ther. 1999;82:241–250. doi: 10.1016/s0163-7258(98)00045-x. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhao R, Wu X, Sun Y, Yao M, Li J.et al. (2005Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics FASEB J 191978–1985. [DOI] [PubMed] [Google Scholar]
- Song S, Liu D, Peng J, Sun Y, Li Z, Gu JR.et al. (2008Peptide ligand-mediated liposome distribution and targeting to EGFR expressing tumor in vivo Int J Pharm 363155–161. [DOI] [PubMed] [Google Scholar]
- Klutz K, Schaffert D, Willhauck MJ, Grünwald GK, Haase R, Wunderlich N.et al. (2011Epidermal growth factor receptor-targeted (131)I-therapy of liver cancer following systemic delivery of the sodium iodide symporter gene Mol Ther 19676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A, Pahnke A, Schaffert D, van Weerden WM, de Ridder CM, Rödl W.et al. (2011Disconnecting the yin and yang relation of epidermal growth factor receptor (EGFR)-mediated delivery: a fully synthetic, EGFR-targeted gene transfer system avoiding receptor activation Hum Gene Ther 221463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H.et al. (2004Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival Cancer Res 643753–3756. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D.et al. (2007The let-7 microRNA represses cell proliferation pathways in human cells Cancer Res 677713–7722. [DOI] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Hau A, Murmann AE., and, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- Lee YS., and, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., and, Kiwada H. Accelerated blood clearance (ABC) phenomenon upon repeated injection of PEGylated liposomes. Int J Pharm. 2008;354:56–62. doi: 10.1016/j.ijpharm.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Simpson RJ, Jensen SS., and, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- Murohashi M, Hinohara K, Kuroda M, Isagawa T, Tsuji S, Kobayashi S.et al. (2010Gene set enrichment analysis provides insight into novel signalling pathways in breast cancer stem cells Br J Cancer 102206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K, Ohbayashi T, Kiyono T, Nishi H, Isaka K, Umezawa A.et al. (2004Expression of a novel human gene, human wings apart-like (hWAPL), is associated with cervical carcinogenesis and tumor progression Cancer Res 643545–3549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of cell viability based on assays with (4, 5-dimethylthiazol-2-yl) 2,5-diphenyl-tetrazolium bromide.
Hematoxylin and eosin staining of lung, liver, spleen, and kidney tissues from mice injected with exosomes.
Expression analysis of the let-7 target genes.