Abstract
Polycations such as polyethylenimine (PEI) are used in many novel nonviral vector designs and there are continuous efforts to increase our mechanistic understanding of their interactions with cells. Even so, the mechanism of polyplex escape from the endosomal/lysosomal pathway after internalization is still elusive. The “proton sponge ” hypothesis remains the most generally accepted mechanism, although it is heavily debated. This hypothesis is associated with the large buffering capacity of PEI and other polycations, which has been interpreted to cause an increase in lysosomal pH even though no conclusive proof has been provided. In the present study, we have used a nanoparticle pH sensor that was developed for pH measurements in the endosomal/lysosomal pathway. We have carried out quantitative measurements of lysosomal pH as a function of PEI content and correlate the results to the “proton sponge ” hypothesis. Our measurements show that PEI does not induce change in lysosomal pH as previously suggested and quantification of PEI concentrations in lysosomes makes it uncertain that the “proton sponge ” effect is the dominant mechanism of polyplex escape.
Introduction
Polyethylenimine (PEI) is one of the most commonly used nonviral vectors for DNA/RNA transfection both in vitro and in vivo.1,2,3,4 One of the important features of PEI and other cationic transfection systems5 is the high concentration of positively charged nitrogen atoms, which makes it suitable for condensing large negatively charged molecules such as DNA resulting in the formation of polyplexes.6 It has been shown by several groups that polyplexes enter cells via endocytosis;7,8,9,10 however, the mechanistic details of intracellular transport from endosomes to the nucleus remains elusive.
Because of its many nitrogen atoms, PEI seems to exhibit considerable buffer capacity also at the low pH values of the lysosomes.11 In 1997 this led Behr12 to propose the “proton-sponge ” hypothesis describing that unprotonated amines of PEI can absorb protons as they are pumped into the lysosome, resulting in more protons being pumped in leading to an increased influx of Cl– ions and water. A combination of the osmotic swelling and a swelling of the PEI because of repulsion between protonated amine groups causes the rupture of the lysosomal membrane with subsequent release of its contents into the cytoplasm.12,13
Different strategies have been employed in order to confirm the “proton sponge ” effect and studies have particularly focused on the pH of lysosomes, as PEI accumulates to a very high degree in these compartments and the buffering capacity of PEI has been interpreted to cause an increase in the pH in PEI containing lysosomes.1,12,14 In order to measure pH of the immediate surroundings of the polyplexes, strategies have been employed where either PEI15,16 or the DNA17 has been labeled with a pH-sensitive and a reference fluorophore for ratiometric pH measurements. These experiments show that the polyplexes experience an initial decrease in pH after internalization, but in most cell lines examined the pH was never below pH 5.5. Godbey et al.18 attempted to measure the pH of lysosomes after treatment with PEI polyplexes with a probe independent of both PEI and DNA, namely LysoSensor Yellow/Blue DND-160. They find no increase in pH after uptake of polyplexes, and therefore conclude that the “proton sponge ” effect must be incorrect. However, as they and others10 also show that LysoTracker (a pH-insensitive version of the LysoSensor) do not colocalize with PEI, it is not certain that they measure pH of the PEI containing vesicles. However, we and others have shown that PEI does reach the lysosomes by colocalization with the lysosome-associated membrane protein-1 (LAMP-1).10 Many of the measurements have been performed by flow cytometry15,17,19 where spatial resolution within cells cannot be resolved; thus, the pH measurement could be an average from polyplexes residing in different cellular compartments.19 The mean pH of ~5.5 could therefore be a consequence of averaging effects caused by polyplexes in e.g., cytoplasm and lysosomes. Furthermore, the majority of the previous measurements have been performed using a sensor system with a pH sensitivity range of 2 pH units from 5.5 to 7.516,17,19 precluding the possibility to measure any pH <5.5.20
Herein, we have used a nanoparticle-based pH sensor (nanosensor) that we recently developed21 to measure pH changes of lysosomes after addition of PEI to the cells. This nanosensor is known to reside in the lysosomes and have a dynamic measurement range from pH 3.2–7.0 covering the whole range of the endosomal system. Investigations of lysosomes with colocalized nanosensor and PEI allowed us to determine the pH in lysosomes as a function of PEI concentration. Hence, we have with high spatial resolution investigated the pH changes induced by PEI in single lysosomes.
Results
Transfection with PEI
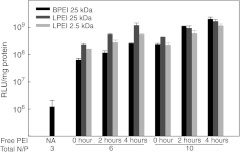
Polyplexes are prepared with a desired N/P ratio (where N is the number of PEI nitrogen atoms and P the number of DNA phosphorus atoms) that has strong influence on polyplex integrity. At N/P ~3 all DNA and PEI is complexed into the polyplex, but at higher N/P ratios the excess PEI chains are free in solution.22,23 The fraction of free PEI is favorable when preparing polyplexes, as particle aggregation is observed when N/P ratios are too low.22 Furthermore, it has been shown that PEI, free in solution or only loosely associated with the polyplexes, enhances the transfection efficiency more than a 1,000-fold.24 In addition, the toxicity of polyplexes is mainly due to the free fraction of PEI.23 From these studies it is clear that free PEI plays an important role in gene transfection. Figure 1 shows the luciferase activity of HeLa cells transfected with polyplexes of DNA condensed by 25 kDa branched PEI (BPEI) at N/P = 3 followed by addition of different free PEI chains (BPEI 25 kDa, linear PEI (LPEI) 25 and 2.5 kDa) simultaneously and 2 or 4 hours after addition of polyplexes. It is clear that addition of free PEI (as N/P = 3 and 7, corresponding to polyplex PEI of N/P = 6 and 10, respectively) greatly enhances the transfection efficiency even if added up to 4 hours after the polyplexes. Furthermore, the short LPEI 2.5 kDa enhances the transfection efficiency of BPEI polyplexes, even though it has poor transfection efficiency on its own.25 This indicates that the free fraction of PEI could be a key factor in polyplex escape from the endosome/lysosome system and thereby highly interesting in relation to the “proton sponge ” hypothesis. We have therefore mainly been focusing on this free fraction of PEI and have performed experiments where cells have been treated with free PEI corresponding to a N/P of 3 and 7, which would be the free fraction in polyplexes with total N/P rations of 6 and 10, respectively, when added in amounts that would give 0.8 µg DNA per well, (as DNA is fully condensed at N/P 3 and excess PEI becomes a “free fraction”).
Figure 1.
In vitro transfection efficiency of polyplexes with free polyethylenimine (PEI) chains. HeLa cells were transfected with branched PEI (BPEI) 25 kDa polyplexes of N/P = 3 in the presence of different free PEI chains (BPEI 25 kDa, LPEI 25, and 2.5 kDa). Free PEI chains of N/P = 3 and 7 (corresponding to polyplexes of N/P = 6 and 10, respectively) were added simultaneously (0), 2, and 4 hours after administration of polyplexes. RLU, relative light units; LPEI, linear PEI. Results presented as mean of triplicate ± SD. Representative of two independent experiments.
Colocalization of nanosensor and PEI with LAMP-1
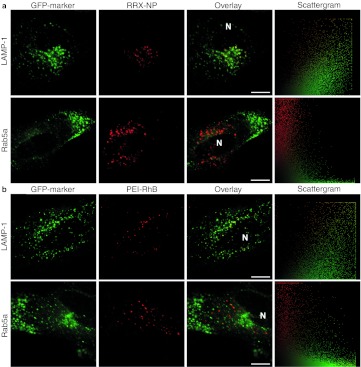
With the aim of measuring pH in lysosomes as a function of PEI concentration, colocalization studies were carried out between the nanoparticle used for the nanosensors and endosomes/lysosomes. Cells were treated with a rhodamine red X (RRX)-labeled nanoparticle and colocalized with the green fluorescent protein (GFP) tagged lysosomal marker LAMP-1 (Figure 2a). The actual nanosensor cannot be used for these colocalization experiments because it contains both fluorescein and Oregon Green which are indistinguishable from GFP. However, the RRX-nanoparticle is prepared from the same nanoparticle batch as the nanosensor and has equal physicochemical properties. Transient expression of GFP-LAMP-1 was obtained in HeLa cells using CellLight reagents and cells were then treated with nanoparticles for 24 hours. Significant colocalization was observed and all nanoparticles have reached a lysosome, whereas colocalization with the early endosomal marker Rab5a fused to GFP showed no colocalization (Figure 2a).
Figure 2.
Colocalization of nanoparticle and polyethylenimine (PEI) with lysosomes. Colocalization of (a) RRX-NP and (b) PEI-RhB with lysosomal marker GFP-LAMP-1 and early endosomal marker GFP-Rab5a. HeLa cells were transduced with plasmids encoding green fluorescent protein (GFP)-tagged marker and incubated with RRX-NP for 24 hours or PEI-RhB for 4 hours. Scattergram: all pixels in the corresponding overlay image presented as red intensity in relation to green intensity. Bar = 10 µm. Representative of three independent experiments. LAMP-1, lysosome-associated membrane protein-1; RRX, rhodamine red X; RhB, rhodamine B; NP, nanoparticle; N, nucleus.
To determine whether PEI reaches the lysosomes, colocalization after 4 hours between rhodamine B (RhB)-labeled BPEI 25 kDa, N/P = 7 (corresponding to polyplexes of N/P = 10) and GFP-LAMP-1 or GFP-Rab5a was performed (Figure 2b). Four hours treatment time was chosen because it has been shown earlier that PEI is efficiently taken up after 4 hours and that effective transfection have been obtained with 4 hours treatment followed by incubation in normal growth medium for expression of reporter gene.19,23 We show significant colocalization with LAMP-1, indicating that most PEI-RhB have reached the lysosomes. We observed no PEI in early endosomes and small quantities of PEI that potentially could have escaped to the cytoplasm are in concentrations that is below the detection limit. This is in agreement with earlier colocalization studies with LAMP-1.10 Unlike Godbey et al.18 we also showed colocalization with LysoSensor Green DND-189 (Supplementary Figure S1), likewise Merdan et al.26 showed colocalization with LysoTracker Blue. Colocalization with LysoSensor indicates that the pH of the lysosomes have not increased to a level where LysoSensor leaves the lysosomes.
Lysosome pH in response to PEI
Determination of the pH in lysosomes in response to PEI can yield important information on the proposed “proton sponge ” effect in lysosomes. We have recently developed and reported a triple-labeled nanosensor with the two pH-sensitive fluorophores Oregon Green and fluorescein and the pH-insensitive fluorophore RhB for ratiometric pH measurements inside living cells by confocal microscopy.20 With two pH-sensitive fluorophores this nanosensor is superior to earlier reported pH sensors with respect to the sensitivity range (pH 3.2–7.0), especially when obtaining measurements in the endosome–lysosome pathway. Furthermore, image acquisition with confocal microscopy and following image analysis provides valuable information on the intracellular distribution of pH.
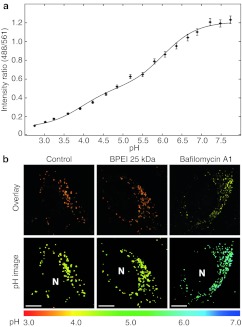
Calibration of the nanosensor was carried out in buffer, as we have previously shown that this is appropriate for this type of nanosensor.20 The calibration curve is presented in Figure 3a fitted to the following equation for a triple-labeled pH sensor:
Figure 3.
Calibration curve of pH nanosensor and pH measurements in cells. (a) In vitro calibration of triple-labeled pH nanosensor performed in buffer. Ratiometric measurements of the nanosensor are related to pH of the buffer and fitted to Equation (1). Mean ± SD are presented. (b) Nanosensor internalized during 24 hours by HeLa cells imaged by confocal microscopy without further treatment or treated with free branched PEI (BPEI) 25 kDa or bafilomycin A1. The ratio of the pH-sensitive and reference fluorophores was converted into pH via the calibration curve and color coded on a common linear scale according to pH. Top panel: Overlay images of the green pH sensitive signal with the red insensitive signal. Bottom panel: pH images, N, nucleus. Bar = 10 µm. Representative of four independent experiments.
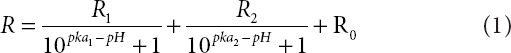 |
where R is the ratio of emission intensities excited at 488 and 561 nm, R0 = Rmin (the ratio for the fully protonated form), (R1+R2+R0) = Rmax (the ratio for the fully deprotonated form), and pKa1 and pKa2 are the specific pKa values of the two pH-sensitive fluorophores in the nanoparticle.
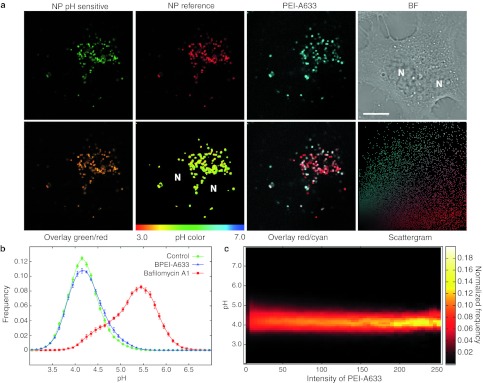
For the intracellular pH measurements cells were treated for 24 hours with nanosensor and were then exposed to free BPEI 25 kDa at N/P = 7 for 4 hours and imaged by confocal microscopy. Figure 3b upper panel shows images of cells as an overlay of the green and red signal and at the bottom the same cells with a new color coding according to a pH color scale. When compared to a control cell (with internalized nanosensor but without PEI) it can be seen that BPEI 25 kDa do not seem to change the pH of the lysosomes. To show that we with this method are indeed able to measure an increase in pH, we have included an image of a cell treated with the Vacuolar-type H+-ATPase (V-ATPase) inhibitor bafilomycin A1, which is known to inhibit the acidification of lysosomes. As the color turns from yellow in the control cell to green/blue in the bafilomycin A1-treated cell it is clear that the pH have increased dramatically. Similar results were obtained with free PEI at N/P = 3 (corresponding to total polyplex of N/P = 6), the LPEI 2.5 and 25 kDa versions and with polyplexes (DNA and BPEI 25 kDa) of N/P = 6 (Supplementary Figure S2). Even though both the nanosensor and PEI have been shown to localize to the lysosomes, it is not assured that the PEI and the nanosensor resides in the same lysosomes. In order to make sure that the measured pH arises from lysosomes that also contain PEI we labeled PEI with Alexa Fluor 633 (A633), which can be distinguished from the fluorophores of the nanosensor, in order to colocalize PEI directly with the nanosensor of which we could read out the pH. Figure 4a shows representative images obtained of cells that were treated with nanosensor for 24 hours to ensure lysosomal localization, and then for 4 hours with 25 kDa BPEI-A633. From the colocalization between the nanosensor reference and PEI-A633 and the corresponding scattergram it can be observed that there is a good degree of colocalization between the nanosensor and PEI; however, nanosensors and PEI that do not colocalize are also observed. Presenting data as a histogram of pH comparing control cells with PEI treated cells reveals no difference in pH distribution (Figure 4b). However, with this method we are able to present the pH as a function of the intensity in the PEI channel, hence as a function of the PEI concentration (Figure 4c). This data presentation, allows direct comparison of measurements in lysosomes without PEI (background intensity of PEI = 0–20) with lysosomes containing high concentrations of PEI, and we observe no change in pH even at the highest PEI concentrations.
Figure 4.
Lysosomal pH in response to polyethylenimine (PEI). (a) HeLa cells with internalized nanosensor for 24 hours were treated with PEI-A633 for 4 hours and imaged by confocal microscopy. Top row: The green pH-sensitive signal and the red reference signal from the nanosensor can be distinguished from the dark red signal of PEI-A633 (here presented as cyan for improved visualization and colocalization). Bright field (BF) image of cells. Bottom row: Overlay image of green and red signal. The ratio of green to red was converted to pH via the calibration curve and color coded on a linear scale according to pH. Overlay red/cyan shows colocalization (white signal) of the nanosensor with PEI-A633 with the corresponding scattergram. Bar = 10 µm. (b) Histogram showing pH distribution of nanosensor containing cells without further treatment (control) or treated with PEI-A633 or bafilomycin A1. Mean ± SEM (n = 9, 13, and 8 images for control, BPEI-A633 and Bafilomycin A1, respectively) are presented. (c) pH of PEI-A633 treated cells plotted as a function of the intensity of PEI-A633 in single lysosomes. The pH measurements of the pH color coded images are presented in relation to the corresponding pixel intensities of PEI-A633. Representative of three independent experiments. BPEI, branched PEI; NP, nanoparticle; A633, Alexa Fluor 633; N, nucleus.
Additionally, we investigated whether higher concentrations of free PEI would have an impact on pH. We have in the above studies utilized concentrations of PEI that would correspond to the addition of 0.8 µg DNA per well, however, others have used higher amounts of DNA (from 0.6–5.0 µg DNA per well)15,18,19,23 and we therefore tested higher concentrations of free PEI. With BPEI 25 kDa we tested: addition of free PEI at what would correspond to N/P = 15, 30, and 45, with 0.8 µg DNA per well. This can also be expressed as addition of free PEI at N/P = 7 at what would correspond to 1.7, 3.4 and 5.1 µg DNA per well. These experiments were also performed with 4-hour treatment of cells, but did not reveal any change in pH compared to control cells. Hence differences in obtained pH profiles between our results and others15,16,19 do not seem to arise because of differences in PEI concentration.
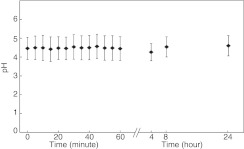
Finally, we tested whether an influence on pH could happen earlier or later in the transfection process compared to the 4 hours treatment time the previous experiments were performed with. We imaged the cells immediately after addition of free BPEI 25 kDa and up to 24 hours after addition. As shown in Figure 5 where the mean ± SD of the frequency distributions are presented these experiments revealed no change in pH compared to control cells.
Figure 5.
Measurements of lysosomal pH in response to polyethylenimine (PEI) over time. HeLa cells with internalized nanosensor for 24 hours were exposed to branched PEI (BPEI) 25 kDa and images were collected about one every minute of different cells for 1 hour. After 4 hours, the PEI was washed off and five images were collected for each time point 4, 8, and 24 hours after addition of PEI. Time point zero was collected just before addition of PEI. Four to five images for each 5 minutes interval were grouped for the analysis of each time point. Presented is mean ± SD of the pH frequency distribution obtained from the image analysis. Representative of three independent experiments.
PEI-buffering capabilities
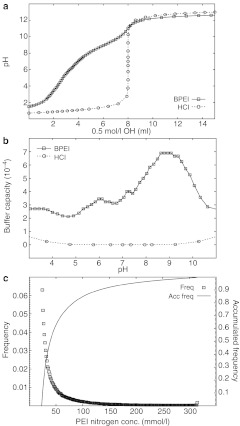
The many amines of PEI are titratable and even though the theoretical pKa values of primary, secondary, and tertiary amines all are >9.0, PEI seem to have a buffering capacity covering the whole physiologically relevant range down to pH 3. From a titration of BPEI 25 kDa with 0.5 mol/l NaOH shown in Figure 6a the buffering capacity of PEI can be calculated β = dn(OH—)/dpH and expressed as a function of pH as presented in Figure 6b. A titration of HCl was carried out as a reference to show that the buffering capacity of PEI is significantly higher than the buffering capacity of water at low pH where a plateau was obtained from pH 3–7 due to multiple pKa values. These pKa values arise because of the close proximity of the amine groups in the molecule, where protonated amines results in electrostatic repulsion of protons, lowering the apparent pKa values of the neighboring amines.27 While the free PEI chains loosely associated with a polyplex will exhibit this type of buffering capability, the PEI directly bound to DNA is more likely to have higher apparent pKa values. This is because the negatively charged DNA will favor positively charged amine groups. The lower pKa values, and therefore buffering capacity at low pH, of the free PEI may explain why excess PEI is an important factor in the enhanced gene transfection efficiency observed when free PEI chains are present, i.e., the endosomal/lysosomal escape in relation to the “proton sponge ” hypothesis, is driven by the buffer capacity and “proton sponge ” effect of free PEI.
Figure 6.
Titration of polyethylenimine (PEI) and lysosomal content of PEI. (a) Titration of an acidified solution of branched PEI (BPEI) 25 kDa with an initial concentration of 232 mmol/l PEI nitrogen atoms and 200 mmol/l HCl. For comparison 200 mmol/l HCl was titrated accordingly. Representative of three independent experiments. (b) The buffer capacity of BPEI 25 kDa was calculated from a, and presented as a function of pH. The buffer capacity of aqueous HCl is presented for comparison. (c) HeLa cells were treated with BPEI-RhB at N/P = 7 for 4 hours and images were collected. Intensity values were then converted to a concentration of PEI nitrogen atoms according to a calibration curve. Presented are the frequency distribution and the corresponding accumulated frequency distribution. Representative of two independent experiments.
Discussion
“Proton sponge ” hypothesis
Our experiments show that PEI reaches lysosomes to a high extent; however, no change in pH in the lysosomes is observed. Even though, we cannot visualize very small amounts of PEI or polyplexes outside the lysosomes, a few polyplexes must have escaped, as transfection is observed. From these experiments we cannot elucidate the mechanism of escape but the measurements of lysosomal pH impacts our understanding of the transfection process in different ways: (i) Even though the majority of PEI reach the lysosomes a small fraction may escape the endosomal pathway before they reach the lysosomes, and it could be this fraction of polyplexes that mediate the transfection of the cell, whereas the polyplexes that reach the lysosomes stays there for degradation or is exocytosed. (ii) Another possibility is that the polyplexes escapes the lysosomes by the “proton sponge ” effect even though the pH does not increase. Many publications state that PEI buffers the lysosomes;8,9,17,26,28 however, the term (buffering) is unspecific. Some publications interpret this as an increase in pH,1,12,15,16,18,29,30 and some have therefore focused on trying to measure this increase in pH in order to prove the “proton sponge ” hypothesis. It is clear that PEI is capable of binding a large amount of protons and hence is buffering the lysosomes, which should lead to an increased proton transport into the lysosomes by ATP driven V-ATPase. This pump is capable of continuous transport of protons and as long as there is sufficient ATP available in the cytosol the V-ATPase will try to secure the proton gradient across the lysosome membrane. It is therefore reasonable to assume that even though PEI is buffering the lysosome, the V-ATPase pump is still able to keep the bulk of the lysosome acidic by increasing the influx of protons. Now, because of the increased influx of protons the “proton sponge ” hypothesis can actually be correct even though no change in pH is observed. The “proton sponge ” hypothesis has been questioned in several publications,10,27 and one issue concerns the concentration of PEI inside the endosomes and lysosomes, as this has an influence on the buffering capacity. Figure 6c shows the accumulated frequency distribution of the concentration of PEI nitrogen atoms inside lysosomes of HeLa cells treated with free 25 kDa BPEI-RhB at N/P = 7 (corresponding to a total PEI polyplex of N/P = 10) (see also Supplementary Figure S3). The concentration can reach >300 mmol/l but <1% of lysosomes reach that concentration and 50% of lysosomes have <40 mmol/l PEI nitrogen. Higher concentrations of PEI leads to higher buffering and therefore influx of Cl– which leads to osmotic pressure differences with influx of water and increased tension on the lysosomal membrane. From the obtained concentrations of PEI in lysosomes and the titration curve of PEI we can approximate the change in osmotic pressure. According to the Young–Laplace equation and using a critical membrane tension of 10 mJ/m2 (tension at which the vesicle will burst),31,32 which corresponds to a simple model membrane with ~20% cholesterol, the critical size of the lysosomes before burst can be calculated in relation to the PEI concentration (for detailed description of these calculations see Supplementary Materials and Methods: Osmotic pressure and critical size of lysosomes). If the calculation based on model liposome membranes with a critical membrane tension of 10 mJ/m2 is a good approximation of lysosomes, then lysosomes with a PEI nitrogen concentration of 300 mmol/l can have a maximum diameter of 260 nm before they burst. However, lysosomes with 100 and 50 mmol/l PEI nitrogen can reach sizes of 775 nm and 1.6 µm, respectively before they burst. These approximations are oversimplified and we expect that higher PEI concentrations are necessary before the lysosomes burst, as a number of factors are not considered in the calculations; The lysosomal membrane is likely to be more stable than accounted for here as it has been shown that it contains ~27 mol% cholesterol33 why the critical membrane tension should be larger than 10 mJ/m2. Also, the permeability of the lysosomal membrane is tightly regulated by several proteins in order to prevent accidental cell death induced by lysosomal proteases that have been shown to trigger apoptosis if released into the cytoplasm.34 Furthermore, the change in pH from pH 7.4–4.2 does not happen in an instance, and in reality there will be time for ion exchange over the lysosomal membrane, potentially leading to equalization of ion gradients during the time it takes for the polyplex to reach the lysosomes. Finally, the pH change might not be as large as 3.2 pH units, since the polyplex is residing very close to the membrane in order to be taken up, and the local pH at the cell membrane will be <7.4 due to the negative charge of the plasma membrane. All these factors lead to larger measured critical diameters of the lysosomes before they burst. It therefore seems uncertain that the “proton sponge ” effect is the dominant effect of polyplex escape from lysosomes, which could be deleterious to the cell and could account for the toxicity of PEI.35 (iii) Another possibility is that the polyplexes actually do escape from the lysosomes through membrane pores/holes as visualized previously by electron microscopy.10 These pores arise due to an interaction between PEI and the membrane, possibly combined with increased membrane tension due to PEI functioning as a “proton sponge”. If these pores close shortly after release of excess content as demonstrated previously for giant unilamellar vesicles36 it is possible that the outward flow of liquid slows down the exchange of protons with the cytoplasm, hence the concentrations of the lysosomal contents (e.g., PEI and H+) will not change considerably even though the volume decreases. If this is the case, little change in pH should be observed, although small amounts of polyplexes or DNA escape to the cytoplasm. Also Merdan et al.26 show release of polyplexes to the cytoplasm due to bursting of the polyplex containing organelles. However, under normal conditions and concentrations of polyplex they find that only one or two bursting events happen per cell. These events allow only small amounts of polyplexes to escape the lysosomes, which is in accordance with the fact that only a fraction of internalized polyplexes actually reaches the nucleus.
We have used a nanoparticle-based pH sensor to measure lysosomal pH in cells in response to treatment with PEI. By colocalization studies we were able to measure the pH in individual lysosomes as a function of PEI concentration. We did not observe any change in pH due to PEI within a timeframe of 0–24 hours. Even though the buffering capacity of PEI has previously been argued to result in lysosomal pH changes in relation to the “proton sponge ” hypothesis, we did not observe this effect. We do believe that PEI functions as a “proton sponge ” but the ATPase pump can overcome this effect and stabilize the pH. Furthermore, with measurements of lysosomal PEI concentration and calculations of the resulting membrane tension we do not believe that the “proton sponge ” effect is the dominant mechanism of polyplex release.
Materials and Methods
Materials. LPEI 2.5 kDa and 25 kDa (Polysciences, Warrington, PA) and BPEI 25 kDa (Sigma-Aldrich, St Louis, MO) was used without further purification. Due to earlier problems with impurities in the commercial LPEI 25 kDa in the form of incomplete deacylation of N-propionyl during synthesis, the polymer was tested by 1H-NMR, which revealed that the LPEI 25 kDa used was 97% deacylated. For polyplex formation plasmid pCMV-LUC (sequence available upon request) was utilized. CellLight Lysosomes-GFP *BacMam 2.0* and CellLight Early Endosomes-GFP *BacMam 2.0* and fluorophore conjugates were purchased from Invitrogen (Paisley, UK). Bafilomycin A1 was purchased from Sigma-Aldrich. 35 mm culture dishes with 10 mm microwell glass bottoms were purchased from MatTek (Ashland, MA). 9 mm round cover glasses and 25 µl stick-on wells were purchased from Thermo Scientific (Waltham, MA). Images were captured by a Leica TCS SP5 AOBS confocal microscope with a 63× water-immersed objective (Leica Microsystems, Wetzlar, Germany). The microscope was equipped with an incubator box and CO2 supply for optimal growth conditions during imaging (Life Imaging Services, Basel, Switzerland).
Cell culture. HeLa cells were maintained in full growth medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 UI/ml penicillin and streptomycin). Cell cultures were incubated in a 5% CO2 humidified incubator at 37 °C. Cells were either imaged in full growth medium without phenol red or imaging medium (Dulbecco's modified Eagle's medium without phenol red and bicarbonate, but supplemented with 30 mmol/l HEPES, 10% fetal bovine serum and 100 UI/ml penicillin and streptomycin) for imaging without CO2 supply.
Preparation of nanosensor, PEI, and polyplexes. Triple-labeled nanosensors were prepared as described earlier,20 where the pH-sensitive dyes Oregon Green and fluorescein was covalently attached to a crosslinked polyacrylamide matrix along with the pH-insensitive dye RhB. In the same way, a corresponding nanoparticle with only the pH-insensitive dye RRX was covalently attached. BPEI 25 kDa was labeled with RhB isothiocyanate or A633 succinate using 1 eq. pr. BPEI polymer and was purified for 4 days in the dark using dialysis tubes with a 3.5 kDa molecular weight cutoff. Polyplexes were prepared as described earlier,24 briefly, polyplexes were prepared by addition of polymer to an equal volume of DNA. Each solution mixture was first vortexed gently for 5 seconds and then incubated for 5 minutes at room temperature. For cell treatments polyplex was added for a final amount of 0.8 µg DNA per well. Solutions of free PEI, PEI-RhB, and PEI-A633 were prepared as the polyplexes, but with pure water instead of DNA at concentrations corresponding to N/P = 3 and 7 (corresponding to polyplexes of N/P = 6 and 10, respectively) and added to cells in amounts that would equal 0.8 µg DNA per well.
Transfection. HeLa cells were seeded in 24-well plates with a density of 50,000 cells per well 24 hours before transfection. The DNA/PEI solution mixture with a desired N/P ratio was further diluted in full growth medium and then added with a final amount of 0.8 µg DNA per well. Cells were incubated with polyplexes for 4 hours in full growth medium where after cells were washed with heparin/phosphate-buffered saline (PBS) (20 U/ml) and medium was exchanged to full growth medium without polyplexes or PEI. In the cases where free PEI was added later than polyplex, the medium exchange was 4 hours after addition of free polymer. After 24 hours, cells were lysed in Reporter Lysis Buffer and luciferase activity was measured using the luciferase assay system (Promega, Madison, WI). The standard assay conditions using a Lumat LB9507 luminometer (Berthold, Bad Wildbad, Germany) involve injection of 100 µl of assay buffer to a tube containing 20 µl cell lysate, with measurement of luminescence for 10 seconds starting 1 second after assay buffer addition. The total lysate protein concentration was measured using a BCA kit (Pierce, Rockford, IL). Luciferase activity is expressed as relative luminescence unit per mg cellular protein.
Colocalization. 2 × 104 HeLa cells per well were seeded in 24-well plates on 9 mm round cover glasses for 24 hours. They were then transduced with CellLight reagents according to the manufacturer. Briefly, cells were incubated with 20 µl of the CellLights solution with baculovirus (containing either GFP-LAMP-1 or GFP-Rap5a plasmids) in full growth medium for 20 hours at normal growth conditions. For colocalization with RRX-labeled nanoparticles the medium additionally contained 10 µg/ml nanoparticle during the 20 hours incubation with virus. For colocalization with PEI-RhB cells were washed once with PBS after the 20 hours incubation with virus and then incubated in full growth medium containing PEI-RhB for 4 hours. Before imaging all cells were washed three times with ice-cold PBS supplemented with heparin, once with PBS and kept in imaging medium for observation by confocal microscopy.
Treatment of cells for pH measurements. 2 × 104 HeLa cells per well were seeded in 24-well plates on 9 mm cover glasses for 24 hours. Cells were then treated with 10 µg/ml nanosensor in full growth medium for 20 hours washed three times with ice cold PBS supplemented with heparin and once with PBS, then kept in imaging medium or treated with PEI, polyplex, or bafilomycin A1. PEI or polyplex treatment was performed in full growth medium for 4 hours where after cells were washed and kept in imaging medium for observation by confocal microscopy. Treatment with 200 nmol/l bafilomycin A1 was performed in full growth medium without phenol red for 45 minutes. For imaging, cells were transferred to imaging medium with 200 nmol/l bafilomycin A1 without prior washing. For continuous pH imaging, 7 × 104 HeLa cells per well were seeded in 35 mm culture dishes with a 10 mm microwell glass bottom for 24 hours. Cells were then treated with 10 µg/ml nanosensor in full growth medium for 24 hours, washed and kept in full growth medium without phenol red. Cells were then imaged before and continuously after addition of free PEI (corresponding to N/P = 7 and 4 µg DNA as a 35 mm culture dish is five times larger than the 24-wells used for the other pH measurements, hence more DNA is added). As a positive control another culture dish was treated with 200 nmol/l bafilomycin A1 for 45 minutes and imaged.
Image acquisition. Cover glasses with growing cells were transferred to microscope slides with stick-on wells of 25 µl. Cells were then covered with 15 µl imaging media and the well was closed with a cover glass. Cells were imaged within 45 minutes. Culture dishes were mounted on the microscope and cells were imaged directly in the dish. Images were collected by sequential line scanning, with excitation at 488 nm (fluorescein and Oregon Green), 561 nm (RhB and RRX) and 633 nm (A633). Emission was collected by photomultiplier tubes in the ranges 498–560, 571–632, and 643–750 nm, respectively, obtained by tunable high-reflectance mirrors. For colocalization studies a differential interference contrast image was also collected, and for pH measurements two bright field images were collected, one for each of the two laser lines 488 nm and 561 nm for correction of laser power.
Calibration. In vitro calibration curves were generated from fluorescence images of the nanosensor at 8 mg/ml in 60 mmol/l buffers (20 mmol/l phosphate/20 mmol/l citric acid/20 mmol/l maleic acid/100 mmol/l NaCl) from pH 2.8–8.2. The microscope was focused in a plane within the solution, and with the same settings as were employed for imaging of corresponding cells with internalized nanosensor. Images were corrected for background by subtraction of an average value for each channel obtained by imaging of pure buffer. The fluorescence images were then corrected for fluctuations in laser power by normalization with corresponding bright field images, and mean ± SD of pixels in an image of nanosensor in buffer was plotted against pH values.
Image analysis. For colocalization studies images were background subtracted, determined by a background region of interest in an area without cells. For pH measurements every image series was subtracted the same background value as the corresponding calibration curve. Images were then normalized according to the bright field images. Background subtraction and normalization was performed with custom-made software which includes further image analysis as described earlier.20 Briefly, image processing was used in order to determine which pixels are actual signal from nanosensors, and the included pixels were then converted to pH via the calibration curve. pH histograms are presented as mean ± SEM for each column.
Titration of BPEI 25 kDa. 200 mg of BPEI 25 kDa was dissolved in 10 ml of a 200 mmol/l NaCl solution and acidified with 1 mol/l HCl to pH 1.5 and adjusted to a final volume of 20 ml with deionized water to final concentrations of 10 mg/ml BPEI (232 mmol/l nitrogen atoms), 100 mmol/l NaCl and 200 mmol/l HCl. This solution was titrated with 0.5 mol/l NaOH at 37 °C. The buffering capacity (β) was calculated according to β = dn(OH—)/dpH. For comparison 200 mmol/l HCl was titrated accordingly.
PEI content of lysosomes. 2 × 104 HeLa cells per well were seeded in 24-well plates on 9 mm round cover glasses for 24 hours. Cells were treated with BPEI-RhB 25 kDa for 4 hours at N/P = 7 (corresponding to polyplexes of N/P = 10), washed and kept in imaging medium for observation by confocal microscopy. With the same settings images were obtained of BPEI-RhB in solution at descending concentrations to obtain a standard curve.
SUPPLEMENTARY MATERIAL Figure S1. Colocalization of RhB-PEI with Lysosensor Green DND-189. Figure S2. Measurements of lysosomal pH. Figure S3. Lysosomal PEI content. Materials and Methods.
Acknowledgments
This work was financially supported by the Danish Cancer Society and the Danish Council for Independent Research [Technology and Production Sciences (FTP) grant no. 274-08-0534 and 274-07-0172]. We would furthermore thank Klaus Bechgaard (Copenhagen University, Denmark) for valuable discussions and Honghao Sun (Technical University of Denmark, Denmark) for providing the nanosensors. The authors declared no conflict of interest.
Supplementary Material
Colocalization of RhB-PEI with Lysosensor Green DND-189.
Measurements of lysosomal pH.
Lysosomal PEI content.
REFERENCES
- Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B.et al. (1995A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine Proc Natl Acad Sci USA 927297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintzer MA., and, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Satterlee A., and, Huang L. In vivo gene delivery by nonviral vectors: overcoming hurdles. Mol Ther. 2012;20:1298–1304. doi: 10.1038/mt.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant RR, Sriraman SK, Navarro G, Biswas S, Dalvi RA., and, Torchilin VP. Polyethyleneimine-lipid conjugate-based pH-sensitive micellar carrier for gene delivery. Biomaterials. 2012;33:3942–3951. doi: 10.1016/j.biomaterials.2011.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito L, Little S, Langer R., and, Amiji M. Poly(beta-amino ester) and cationic phospholipid-based lipopolyplexes for gene delivery and transfection in human aortic endothelial and smooth muscle cells. Biomacromolecules. 2008;9:1179–1187. doi: 10.1021/bm7011373. [DOI] [PubMed] [Google Scholar]
- Dunlap DD, Maggi A, Soria MR., and, Monaco L. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res. 1997;25:3095–3101. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J, Bragonzi A., and, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- von Gersdorff K, Sanders NN, Vandenbroucke R, De Smedt SC, Wagner E., and, Ogris M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol Ther. 2006;14:745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Rémy-Kristensen A, Clamme JP, Vuilleumier C, Kuhry JG., and, Mély Y. Role of endocytosis in the transfection of L929 fibroblasts by polyethylenimine/DNA complexes. Biochim Biophys Acta. 2001;1514:21–32. doi: 10.1016/s0005-2736(01)00359-5. [DOI] [PubMed] [Google Scholar]
- Bieber T, Meissner W, Kostin S, Niemann A., and, Elsasser HP. Intracellular route and transcriptional competence of polyethylenimine-DNA complexes. J Control Release. 2002;82:441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]
- Suh J, Paik H, Hwang BK. Ionization of Poly(ethylenimine) and Poly(allylamine) at various pH's. Bioorg Chem. 1994;22:318–327. [Google Scholar]
- Behr J. The proton sponge: A trick to enter cells the viruses did not exploit. Chimia. 1997;51:34–36. [Google Scholar]
- Nel AE, Mädler L, Velegol D, Xia T, Hoek EM, Somasundaran P.et al. (2009Understanding biophysicochemical interactions at the nano-bio interface Nat Mater 8543–557. [DOI] [PubMed] [Google Scholar]
- Guillem VM., and, Aliño SF. Transfection pathways of nonspecific and targeted PEI-polyplexes. Gene Ther Mol Biol. 2004;8:369–384. [Google Scholar]
- Forrest ML., and, Pack DW. On the kinetics of polyplex endocytic trafficking: implications for gene delivery vector design. Mol Ther. 2002;6:57–66. doi: 10.1006/mthe.2002.0631. [DOI] [PubMed] [Google Scholar]
- Sonawane ND, Szoka FC., Jr, and, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- Akinc A., and, Langer R. Measuring the pH environment of DNA delivered using nonviral vectors: implications for lysosomal trafficking. Biotechnol Bioeng. 2002;78:503–508. doi: 10.1002/bit.20215. [DOI] [PubMed] [Google Scholar]
- Godbey WT, Barry MA, Saggau P, Wu KK., and, Mikos AG. Poly(ethylenimine)-mediated transfection: a new paradigm for gene delivery. J Biomed Mater Res. 2000;51:321–328. doi: 10.1002/1097-4636(20000905)51:3<321::aid-jbm5>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Akinc A, Thomas M, Klibanov AM., and, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- Benjaminsen RV, Sun H, Henriksen JR, Christensen NM, Almdal K., and, Andresen TL. Evaluating nanoparticle sensor design for intracellular pH measurements. ACS Nano. 2011;5:5864–5873. doi: 10.1021/nn201643f. [DOI] [PubMed] [Google Scholar]
- Sun H, Almdal K., and, Andresen TL. Expanding the dynamic measurement range for polymeric nanoparticle pH sensors. Chem Commun (Camb) 2011;47:5268–5270. doi: 10.1039/c1cc10439j. [DOI] [PubMed] [Google Scholar]
- Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E., and, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- Yue Y, Jin F, Deng R, Cai J, Dai Z, Lin MC.et al. (2011Revisit complexation between DNA and polyethylenimine–effect of length of free polycationic chains on gene transfection J Control Release 152143–151. [DOI] [PubMed] [Google Scholar]
- Dai Z, Gjetting T, Mattebjerg MA, Wu C., and, Andresen TL. Elucidating the interplay between DNA-condensing and free polycations in gene transfection through a mechanistic study of linear and branched PEI. Biomaterials. 2011;32:8626–8634. doi: 10.1016/j.biomaterials.2011.07.044. [DOI] [PubMed] [Google Scholar]
- Kircheis R, Wightman L., and, Wagner E. Design and gene delivery activity of modified polyethylenimines. Adv Drug Deliv Rev. 2001;53:341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- Merdan T, Kunath K, Fischer D, Kopecek J., and, Kissel T. Intracellular processing of poly(ethylene imine)/ribozyme complexes can be observed in living cells by using confocal laser scanning microscopy and inhibitor experiments. Pharm Res. 2002;19:140–146. doi: 10.1023/a:1014212630566. [DOI] [PubMed] [Google Scholar]
- Won YY, Sharma R., and, Konieczny SF. Missing pieces in understanding the intracellular trafficking of polycation/DNA complexes. J Control Release. 2009;139:88–93. doi: 10.1016/j.jconrel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K, Oba M, Nakanishi M, Fukushima S, Yamasaki Y, Koyama H.et al. (2008Polyplexes from poly(aspartamide) bearing 1,2-diaminoethane side chains induce pH-selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity J Am Chem Soc 13016287–16294. [DOI] [PubMed] [Google Scholar]
- Funhoff AM, van Nostrum CF, Koning GA, Schuurmans-Nieuwenbroek NM, Crommelin DJ., and, Hennink WE. Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules. 2004;5:32–39. doi: 10.1021/bm034041+. [DOI] [PubMed] [Google Scholar]
- Creusat G, Rinaldi AS, Weiss E, Elbaghdadi R, Remy JS, Mulherkar R.et al. (2010Proton sponge trick for pH-sensitive disassembly of polyethylenimine-based siRNA delivery systems Bioconjug Chem 21994–1002. [DOI] [PubMed] [Google Scholar]
- Needham D., and, Nunn RS. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys J. 1990;58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslov MM., and, Markin VS. A theory of osmotic lysis of lipid vesicles. J Theor Biol. 1984;109:17–39. doi: 10.1016/s0022-5193(84)80108-3. [DOI] [PubMed] [Google Scholar]
- Schoer JK, Gallegos AM, McIntosh AL, Starodub O, Kier AB, Billheimer JT.et al. (2000Lysosomal membrane cholesterol dynamics Biochemistry 397662–7677. [DOI] [PubMed] [Google Scholar]
- Roberg K, Kågedal K., and, Ollinger K. Microinjection of cathepsin d induces caspase-dependent apoptosis in fibroblasts. Am J Pathol. 2002;161:89–96. doi: 10.1016/S0002-9440(10)64160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhamifar L, Larsen AK, Hunter AC, Andresen TL., and, Moghimi SM. Polycation cytotoxicity: a delicate matter for nucleic acid therapy-focus on polyethylenimine. Soft Matter. 2010;6:4001–4009. [Google Scholar]
- Karatekin E, Sandre O., and, Brochard-Wyart, F. Transient pores in vesicles. Polym Int. 2003;52:486–493. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Colocalization of RhB-PEI with Lysosensor Green DND-189.
Measurements of lysosomal pH.
Lysosomal PEI content.