Abstract
Reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) is an epigenetic phenomenon. It has been suggested that iPSC retain some tissue-specific memory whereas little is known about interindividual epigenetic variation. We have reprogrammed mesenchymal stromal cells from human bone marrow (iP-MSC) and compared their DNA methylation profiles with initial MSC and embryonic stem cells (ESCs) using high-density DNA methylation arrays covering more than 450,000 CpG sites. Overall, DNA methylation patterns of iP-MSC and ESC were similar whereas some CpG sites revealed highly significant differences, which were not related to parental MSC. Furthermore, hypermethylation in iP-MSC versus ESC occurred preferentially outside of CpG islands and was enriched in genes involved in epidermal differentiation indicating that these differences are not due to random de novo methylation. Subsequently, we searched for CpG sites with donor-specific variation. These “epigenetic fingerprints” were highly enriched in non-promoter regions and outside of CpG islands–and they were maintained upon reprogramming. In conclusion, iP-MSC clones revealed relatively little intraindividual variation but they maintained donor-derived epigenetic differences. In the absence of isogenic controls, it would therefore be more appropriate to compare iPSC from different donors rather than a high number of different clones from the same patient.
Introduction
Induced pluripotent stem cells (iPSCs) are routinely generated from somatic cells by ectopic expression of a few reprogramming factors and they represent a promising source of differentiated cells for developmental studies, regenerative medicine, and human in vitro disease modeling.1 Similar to embryonic stem cells (ESCs) iPSC have the ability to differentiate into all three embryonic germ layers in vitro, form teratomas, contribute to chimeric mice, and generate viable, fertile animals by tetraploid complementation.2
Most available iPSC lines have so far been generated from dermal fibroblasts which are easily accessible and can be in vitro expanded to large cell numbers. Since the establishment of iPSC technology, a multitude of other cell types, such as keratinocytes, melanocytes, hepatocytes, circulating T lymphocytes, neural stem cells, cord blood-derived endothelial cells,3 and even malignant cells4 have been used to generate iPSC. Reprogramming efficiency is cell type-dependent and more research is needed to determine the best starting population of somatic cells for iPSC generation. In this respect, mesenchymal stromal cells (MSCs) are promising candidates with several advantages: (i) MSC can easily be isolated and culture expanded from various tissues; (ii) they have been tested in many clinical trials and good manufacturing practice -conform cell culture methods are being routinely implemented in many laboratories; and (iii) MSC comprise a multipotent subset of adult stem cells, which may be more prone to reprogramming than terminally differentiated cells. Despite these advantages only relatively few studies have used human MSC for generation of iPSC: these were either derived from adipose,5,6,7 dental,8,9 or synovial tissue,10 and two reports described generation of iPSC from bone marrow-derived MSC.11,12
The process in which a somatic cell acquires a pluripotent state is an epigenetic phenomenon. Several groups provided comprehensive comparisons of DNA methylation profiles between iPSC, ESC, and their parental cells.7,13,14,15 Overall, methylomes of ESC and iPSC are remarkably similar. At early passage iPSC reveal residual DNA methylation signatures, which are characteristic for their tissue of origin. Most of these aberrant tissue-specific epigenetic modifications get erased during long-term culture of iPSC when they become molecularly more similar to ESC.15,16,17,18,19 However, a recent study demonstrated that some residual epigenetic marks of somatic cells still persist after extended culture for many passages of iPSC derived from cord blood or keratinocytes–these residual marks can skew the differentiation potential of the respective iPSC lines.20
In this study, we have generated iPSC from bone marrow-derived MSC (iP-MSC) and analyzed their DNA methylation profiles with a novel high-density DNA methylation array covering more than 450,000 CpG sites in the human genome.21,22 We show that iP-MSC clones from the same donor cluster closely together, whereas iP-MSC from different donors reveal interindividual differences, particularly outside of promoter regions and distant to CpG islands. Thus, iPSC not only retain cell type-specific memory but also donor-specific epigenetic characteristics.
Results
Generation of iPSC from human MSC
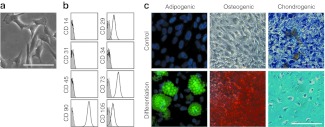
MSCs were isolated from five unrelated donors and culture expanded for two passages in human platelet lysate (HPL) medium. Cell preparations satisfied all criteria for definition of MSC:23 (i) plastic adherent growth with fibroblastoid morphology; (ii) the typical immunophenotype (CD14−, CD29+, CD31−, CD34−, CD45−, CD73+, CD90+, CD105+); and (iii) differentiation toward adipogenic, osteogenic, and chondrogenic lineage (Figure 1).
Figure 1.
Characterization of mesenchymal stromal cells (MSC). (a) MSC from human bone marrow revealed typical plastic adherent cellular morphology, (b) immunophenotypic characteristics, and (c) in vitro differentiation potential toward adipogenic, osteogenic, and chondrogenic lineage. Bars, 100 µm.
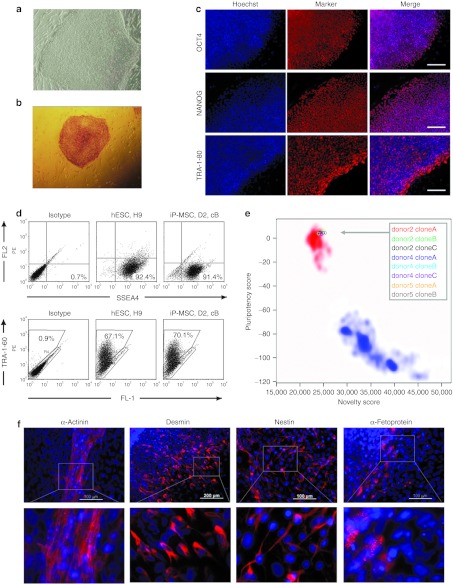
At passage 3, the cells were retrovirally infected in one experiment with three independent biological replica to express OCT-3/4, SOX2, c-MYC, and KLF4. Colonies that exhibited distinct flat and compact ESC-like morphology with well-defined edges were selected 3–4 weeks after transduction and further expanded for validation (Figure 2a). Three different iP-MSC clones were established from each donor-derived MSC line. These iP-MSC clones presented an alkaline phosphatase activity (Figure 2b) and expressed the endogenous pluripotency markers at protein level (Figure 2c,d, Supplementary Figure S1). Upon reprogramming, expression of retrovirally encoded reprogramming factors was silenced whereas expression of endogenous OCT4, NANOG, SOX2, and REX1 was activated (Supplementary Figure S2). Pyrosequencing of the corresponding promoter regions revealed that the DNA methylation level decreased to that present in conventional ESC (Supplementary Figure S3). Robust demethylation of CpG dinucleotides within promoter regions of these pluripotency-associated genes suggests that MSC were stably reprogrammed into iPSC. Furthermore, we generated gene expression profiles of eight iP-MSC clones and PluriTest analysis revealed that all of them were clearly associated with pluripotent cells (Figure 2e).24 We further demonstrated that iP-MSC are able to differentiate in vitro into derivatives of all three embryonic germ layers. Mesoderm differentiation was shown by observation of spontaneously contracting clusters that contained cardiomyocytes exhibiting typical α-actinin–positive sarcomeric structures (Figure 2f, Supplementary Video S1). In addition, pluripotency of iP-MSC was confirmed by the presence of desmin- (mesoderm), α-fetoprotein- (endoderm), and nestin- (ectoderm) positive cells in differentiating embryoid bodies (Figure 2f, Supplementary Figure S4). Microsatellite analyses revealed that all iPSC lines carried the same genotype as parental somatic cells, excluding the possibility of contamination with other ESC or iPSC lines in our laboratories (Supplementary Table S1). We also analyzed the integration sites of the retroviruses using linear amplification-mediated PCR (LAM-PCR; Supplementary Figure S5) followed by next generation sequencing (data not shown). To increase genomic accessibility and to account for possible restriction enzyme biases,25 we used two different enzymes (Tsp509I and HpyCH4IV), and found that insertion sites in each of three clones derived from donors 2 and 4 were different, whereas three clones derived from donor 5 showed identical integration sites. These data prove that the six iP-MSC clones of donors 2 and 4 are derived from different originally transfected cells, whereas the three clones of donor 5 are subclones derived from the same parental cell when cells were split on a feeder layer at day 6 post-transduction. Collectively, these analyses indicate that bone marrow-MSC were successfully reprogrammed into iPSC.
Figure 2.
Characterization of iP-MSC. (a) Human iP-MSC revealed ESC-like morphology. (b) Alkaline phosphatase-positive iP-MSC colony. (c) Immunocytochemical detection of pluripotency markers OCT4, NANOG, and TRA-1-80 in iP-MSC donor 4, clone B. Bars, 200 µm. (d) Expression of pluripotency markers SSEA4 and TRA-1-60 on the surface of human ESC line H9 and iP-MSC donor 2 (D2), clone B (cB) as determined by flow cytometry. (e) Gene expression profiles of eight iP-MSC clones were analyzed by PluriTest to evaluate pluripotency.24 Pluripotency and novelty scores were compared with previously published datasets of pluripotent (red background; 98 reference samples) and non-pluripotent samples (blue background; 1,028 reference samples). (f) Differentiating iP-MSC give rise to beating cardiomyocytes that are positive for α-actinin (mesoderm) and exhibit typical cross-striations (inset) as well as to desmin- (mesoderm), nestin- (ectoderm) and α-fetoprotein- (endoderm) positive cells. hESC, human embryonic stem cell; iP-MSC, induced pluripotent-mesencymal stromal cell.
Global DNA methylation changes upon reprogramming
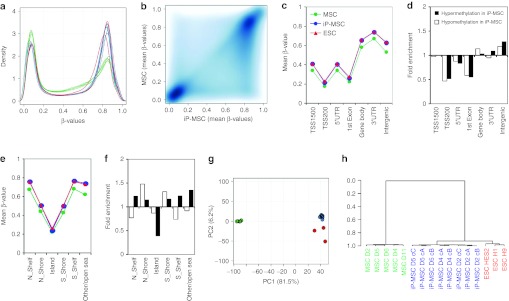
DNA methylation profiles of five MSC samples (passage 2 or 3), eight different iP-MSC clones generated in a single reprogramming experiment from three different donors (passage 9–11) and three established ESC lines (H1, H9, and HES2) were analyzed with the HumanMethylation450 BeadChip. The median methylation level of all CpG sites was 49.4, 70.6, and 70.5% in MSC, iP-MSC, and ESC, respectively, and this is in line with previous reports demonstrating global hypermethylation upon reprogramming.7,18,26 The distribution of β-values over all CpG sites revealed, that reprogramming is associated with the loss of hemimethylated regions and gain of highly methylated CpG sites (Figure 3a); 185,246 CpG sites revealed differential methylation upon reprogramming of MSC into iP-MSC (Figure 3b): 33,941 CpG sites became demethylated and 151,305 CpG sites became hypermethylated in iP-MSC (adjusted P value <0.001).
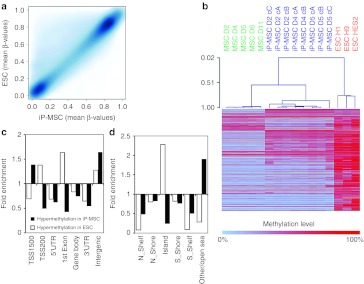
Figure 3.
Global DNA methylation changes upon reprogramming. MSC (green), iP-MSC (blue), and ESC (red) were analyzed with the Infinium HumanMethylation450 BeadChip. (a) Histogram and (b) scatter plot analysis of DNA methylation levels revealed that many hemimethylated CpG sites become highly methylated upon reprogramming. (c) In average, this gain in methylation was observed in all gene regions, whereas both hypermethylation and hypomethylation were significantly enriched in (d) non-promoter and intergenic regions as well as in (e,f) shore regions. (g) Unsupervised principal component analysis demonstrated that iP-MSC clustered closely together and that their methylome was related to ESC. The two principal components PC1 and PC2 showed variances of 81.5 and 6.2%, respectively. (h) Furthermore, hierarchical cluster analysis demonstrated that iP-MSC clones (c) from the same donor (D) clustered always together. ESC, embryonic stem cell; iP-MSC, induced pluripotent-mesencymal stromal cell; UTR, untranslated region.
CpG sites were then classified according to corresponding gene regions: 1,500 bp or 200 bp upstream of the transcription start site (TSS1500 or TSS200), in the 5′-untranslated region, 1st exon, gene body, 3′-untranslated region, and intergenic regions. Overall, methylation levels were lower in promoter regions and in the 1st exon whereas the mean methylation levels increased in all regions upon reprogramming (Figure 3c). Notably, hypermethylated and hypomethylated sites were highly significantly enriched in intergenic regions (P < 10−90; hypergeometric distribution; Figure 3d).
Next, we considered DNA methylation in the context of CpG islands: 2 kb regions upstream or downstream of CpG islands are termed as shore region (“north” and “south”, respectively); 2 kb flanking regions next to these shores were defined as shelf regions; and all other CpG sites were referred to as “open sea”.22 In MSC, the mean DNA methylation level in CpG islands (22.2%) was much lower than in shelf (42.7%), shore (67.5%) or open sea regions (61.8%). Taking into account all CpG sites represented on the platform, the reprogramming-associated DNA methylation changes were preferentially enriched in regions outside of CpG islands (P < 10−64; Figure 3e,f). This supports the notion that non-promoter–associated regions may be relevant for reprogramming.
The relationship between MSC, iP-MSC, and ESC DNA methylation profiles was further analyzed by principal component analysis: the first component clearly separated MSC from pluripotent cells, the second component distinguished between ESC and iP-MSC (Figure 3g). Despite heterogeneity in the MSC starting populations, there was a remarkable similarity between all eight iP-MSC clones. This was further substantiated by unsupervised hierarchical clustering (Pearson correlation). Notably, clones derived from the same MSC donor always clustered together, indicating that there may be donor-derived epigenetic memory (Figure 3h).
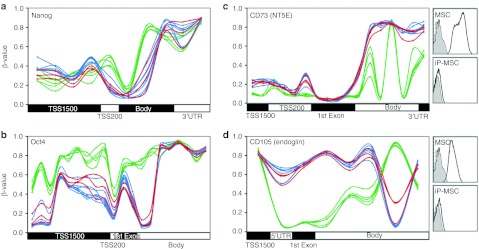
Subsequently, we have exemplarily analyzed DNA methylation changes in specific pluripotency-associated genes. As expected, pluripotency genes such as NANOG, OCT4 as well as DPPA4 and DNMT3b became reproducibly hypomethylated upon reprogramming. Interestingly, significant methylation changes in most of these genes were restricted to relatively small regions in the promoter region and the methylation pattern of individual CpG sites in iP-MSC closely resembled the pattern of ESC. Differential methylation for NANOG and OCT4 is exemplarily demonstrated and the results were validated by pyrosequencing (Figure 4a,b; Supplementary Figure S2). On the other hand, several genes, which are known to be expressed in MSC, such as CD73, endoglin (CD105), VCAM1, and TGFβ-3, became hypermethylated upon reprogramming (Figure 4c,d and data not shown). The remarkable reproducibility of DNA methylation patterns within each of these genes demonstrated that DNA methylation is not just a digital code where genes are either methylated or non-methylated. In fact, the probability of DNA methylation—and, hence, the methylation level within the cell population–is tightly regulated at specific CpG sites upon reprogramming.
Figure 4.
DNA methylation patterns in selected genes. (a,b) β-Values of each CpG site on the HumanMethylation450 BeadChip are exemplarily depicted for the pluripotency genes NANOG and OCT4. (c,d) DNA methylation levels of the MSC surface markers CD73 and CD105. Inserts demonstrate that these proteins are not detected by flow cytometry on iP-MSC. Overall, gene expression is associated with hypomethylation in the promoter regions but the pattern is diverse throughout the genes and remarkably consistent between different cell preparations. Green = MSC; blue = iP-MSC; red = ESC. ESC, embryonic stem cell; iP-MSC, induced pluripotent-mesencymal stromal cell; UTR, untranslated region.
Differences between iP-MSC and ESC
Despite global similarity there are differences in the epigenetic makeup of iPSC and ESC.7,18,20,27 In our data, 3,744 CpG sites revealed highly significant differential methylation between iP-MSC and ESC—3,134 CpG sites were hypomethylated and 610 CpG sites were hypermethylated in iP-MSC as compared with ESC (adjusted P value <0.001; Figure 5a). On first sight, the lower methylation in iP-MSC might be due to incomplete reprogramming but these differences were highly reproducible and often irrespective of the methylation pattern in the parental cells (Figure 5b). Interestingly, CpG sites with higher methylation in ESC were significantly enriched in TSS200, 1st exon, and intergenic regions (P < 10−13), whereas CpG sites with higher methylation in iP-MSC were rather enriched in TSS1500 and intergenic regions (P < 10−6; Figure 5c). In comparison to all CpG sites on the array higher methylation in ESC was 2.3-fold enriched in CpG islands (P < 10−100), whereas higher methylation in iP-MSC was hardly observed in CpG islands (P < 10−45) and was 1.9-fold enriched in open sea regions (P < 10−61; Figure 5d).
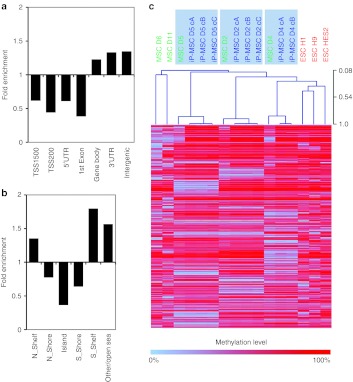
Figure 5.
Differential methylation of iP-MSC and ESC. (a) Scatter plot analysis reveals higher methylation of many CpG sites in ESC as compared with iP-MSC. (b) Heatmap of 3,744 CpG sites with aberrant methylation (3,134 hypermethylated and 610 hypomethylated; adjusted P < 0.001). Many of these sites revealed also stark differences between MSC and iP-MSC indicating that aberrant methylation cannot solely be attributed to epigenetic memory of the parental cells. (c,d) Hypermethylation in ESC was significantly enriched at the transcription start and CpG islands, whereas hypomethylation was particularly enriched at TSS1500, intergenic and open sea regions. ESC, embryonic stem cell; iP-MSC, induced pluripotent-mesencymal stromal cell; UTR, untranslated region.
Gene Ontology (GO) analysis of corresponding genes revealed that the 610 CpG sites, which were hypermethylated in iP-MSC displayed the most significant enrichment in: keratinization (GO:31424; P = 10−14); keratinocyte differentiation (GO:30216; P = 10−11); epidermal cell differentiation (GO:9913; P = 10−10); and epidermis development (GO:8544; P = 10−6.5; Supplementary Table S2). Furthermore, these genes were enriched at chr1p21 (P < 10−14) and chr19p13 (P < 10−6). On the other hand, the 3,134 CpG sites, which showed higher methylation levels in ESC were not significantly over-represented in a functional category (P < 10−4) and they were only significantly enriched in the chromosomal location chr19q13 (P < 10−4). Taken together, epigenetic differences between iP-MSC and ESC are highly reproducible: higher methylation in ESC is primarily observed in the proximal promoter regions and CpG islands whereas higher methylation in iP-MSC is specifically enriched in distal promoter regions, intergenic regions, open sea regions, and in genes which are functionally related to epidermal development.
Donor-specific variation in DNA methylation
Epigenetic profiles in somatic cells are not only cell type-specific but there is also interindividual variation—even in monozygotic twins.28 Yet, the question remains whether donor-specific epigenetic modifications are maintained upon reprogramming. We have selected those CpG sites with a SD of more than 20% methylation differences over the five MSC samples (σ > 0.2); 1,129 CpG sites passed this threshold. These CpG sites corresponded to 565 different genes including several human leukocyte antigens. They were significantly enriched in the GO classification of antigen processing and presentation (GO:19882; P = 10−5.6) and in the chromosomal locations chr13q34 (P = 10−5.5), chr6p21 (P = 10−5.3), chr10q26 (P = 10−3.8), and chr17q25 (P = 10−3.6; Supplementary Table S3). Furthermore, these CpG sites with donor-specific methylation were highly enriched in the gene body, 3′-untranslated region, and intergenic regions whereas promoter regions were rather spared (Figure 6a). Correspondingly, these changes were underrepresented in CpG islands (Figure 6b). The enrichment in non-promoter regions and distant to CpG islands was highly significant with a P value <10−40 for each comparison. The heatmap of the hierarchical cluster analysis revealed, that iP-MSC clustered closely together with their original MSC preparations and retained most of the donor-specific modifications (Figure 6c). Thus, donor-specific epigenetic modifications are hardly affected by reprogramming.
Figure 6.
Interindividual methylation changes. To focus on genomic regions with high variation between individual donor samples, we selected 1,129 CpG sites with a SD of more than 0.2 in the five MSC samples. (a) These CpG sites with interindividual variation were highly significantly enriched in gene body, 3′UTR, and intergenic regions as well as in (b) shelf and open sea regions (P < 10–40). The bars represent the enrichment of differentially methylated CpG sites in comparison to representation of all CpG sites covered by the DNA methylation array. (c) Heatmap analysis of 1,129 CpG sites exhibiting differential methylation between individual donor samples demonstrated that interindividual differences are maintained upon reprogramming—iP-MSC clones from each donor (D) clustered always closely together with their parental MSC preparation. ESC, embryonic stem cell; iP-MSC, induced pluripotent-mesencymal stromal cell; UTR, untranslated region.
Discussion
Overall, reprogramming of somatic cells entails erasure of tissue-specific epigenetic patterns and reestablishment of an embryonic methylome. Despite remarkable similarities in their epigenetic makeup several groups described differences between iPSC and ESC.7,17,18 Furthermore, there is a cell type-dependent variation in iPSC due to “tissue-specific epigenetic memory”, which may result in alteration of the differentiation potential,13,15,20 although most recent studies provided evidence against this notion.27,29 Here, we demonstrate that iP-MSC maintain “donor-specific epigenetic memory” upon reprogramming particularly in non-promoter and intergenic regions.
We show here that MSC from human bone marrow are a suitable source for reprogramming. For clinical application of MSC, xeno-free cell culture conditions are preferred to minimize the risk of transmitting disease or causing immunoreactions towards bovine proteins. HPL has been shown to provide a very effective human alternative to fetal bovine serum.30,31 Therefore, we have used HPL-medium for culture isolation and the cell preparations were thoroughly characterized according to the minimal criteria for MSC.23
iP-MSC generated in this study revealed characteristics of stably reprogrammed cells: they exhibited ESC-like colony morphology, expressed alkaline phosphatase, and pluripotency-associated markers such as cell surface proteins SSEA4, TRA-1-60, TRA-1-80 and transcription factors OCT4 and NANOG, of which the latter was not part of the vectors used for reprogramming. In addition, all established iP-MSC lines strongly silenced retrovirally encoded reprogramming transcripts, upregulated expression of endogenous ESC-specific transcription factors, and exhibited multilineage differentiation potential in vitro. Moreover, analysis of transcriptomes of eight iP-MSC lines with the web-based PluriTest24 revealed that all iP-MSC lines perfectly group together with established PSC lines and are clearly separated from somatic samples. PluriTest has been regarded as a sensitive and highly specific, animal-free alternative to the teratoma assay for assessing pluripotency of human iPSC lines. It uses an algorithm based on genome-wide transcriptional profiles of 223 human ESC and 41 human iPSC lines as well as almost 200 non-pluripotent cells and tissues and can easily differentiate between pluripotent, partially reprogrammed, and non-PSC lines. These data strongly indicate that all of our iP-MSC clones are fully reprogrammed iPSC lines which are transcriptionally highly similar to many other PSC lines established in different laboratories and from different cell types. Acquisition of a pluripotent state in iP-MSC was also confirmed at the epigenetic level by analyzing methylation changes in specific regions of individual genes using the 450k BeadChip, which covers on average 17 CpG sites per gene and facilitates such analyses.21 Many pluripotency genes, such as NANOG, OCT4, DPPA4, and DNMT3b became hypomethylated upon reprogramming and this was especially evident in their promoter regions. This is in line with the fact that these genes become expressed in pluripotent cells. On the other hand, several regions with MSC-specific genes, such as CD73, CD105, CD106, and TGFβ-3 became hypermethylated, which substantiates successful reprogramming.
Reprogramming is associated with global hypermethylation.7,18,26 It has been speculated that this is initially a random process with continuous convergence of DNA methylation profiles towards the embryonic state throughout long-term culture.17,18 We demonstrate that reprogramming-associated changes in the DNA methylation pattern, hypermethylation as well as hypomethylation, are highly reproducible at specific CpGs and particularly involve non-promoter regions and shore regions. This was somewhat unexpected, as previous studies suggested that ESC-associated methylation changes were significantly biased to genes with CpG islands.18,32,33 Apparently, the regions close to these CpG islands are more prone to reprogramming-associated changes than the CpG islands themselves. It has been shown that during development tissue-specific hypomethylation is significantly associated with CpG-poor promoters34,35 as well as regions outside of core promoters33 and conversely, these regions may need to become hypermethylated upon reprogramming.
Several studies indicated highly reproducible epigenetic differences between iPSC and ESC.7,13 Our data demonstrate that this cannot solely be attributed to somatic memory as the DNA methylation pattern at many of these sites varied considerably between iP-MSC and their parental cells. iP-MSC were analyzed at passage 9 or 10 and according to several other studies aberrant methylation changes might diminish with further passaging.18 On the other hand, iP-MSC–specific methylation at early passages appears to be highly reproducible and hypermethylation was enriched in developmental genes. Thus, the detected methylation differences do not seem to be due to incomplete or inaccurate reprogramming—it is rather conceivable that iP-MSC initially take a distinct epigenetic state which assimilates towards the ESC-like state upon long-term in vitro culture. Bock and coworkers proposed that single iPSC lines at passages 15–30 may be indistinguishable from ESC as these states are not well-defined endpoints–due to inherent variability between individual cell lines there is an overlap between iPSC and ESC.17 If there is no unique epigenetic endpoint upon reprogramming the question remains if long-term culture of iPSC is required to fully adopt ESC-like epigenetic profiles. Recent studies actually dismiss this notion by showing that some iPSC lines at higher passages still retain residual epigenetic marks of somatic cells.20 Considering early passages rather than the late ones may be advantageous particularly with regard to possible accumulation of cellular defects and mutations in long-term cultures.36
In this study, we demonstrate that iP-MSC maintain donor-specific differences in DNA methylation. These “epigenetic fingerprints” spared CpG island and promoter regions which is consistent with previous studies demonstrating that the fidelity of CpG methylation patterns in twins and cell culture was higher in promoter regions than in non-promoter regions.28,37 It may be anticipated that DNA methylation in non-promoter regions has relatively little impact on gene expression. However, regulatory regions may also be located in the gene body or intergenic regions and there is recent evidence that they may even govern alternative splicing.38 Therefore, it is expected that at least some of interindividual epigenetic variation contributes to donor-specific functional differences. Recently, it has been shown that 16 individual iPSC lines revealed some variability in differentiation efficiency, which seems to correlate with donor identity and donor gender.27 The iPSC lines in this study were derived from one single reprogramming experiment using MSC of three unrelated donors. In order to account for the potential confounding effect of inter-experimental variations further comparison of many iPSC lines generated from additional donors in more than one reprogramming experiment is required to elucidate such donor-specific differences in iPSC in more detail. Either way, it is remarkable that donor-specific epigenetic patterns are maintained despite global methylation changes upon reprogramming.
It is striking how much the DNA methylation patterns of independent samples are alike—even within individual genes. So far, it is unknown how these precise DNA methylation changes are governed. On the other hand, there is evidence that DNA methylation is complemented by other regulatory mechanisms such as histone modifications.39 Data analysis of the histone code in combination with the methylome and transcriptome may help to elucidate molecular mechanisms responsible for reprogramming of iPSC.
MSC are extremely heterogeneous cell preparations—individual subpopulations reveal distinct morphology,40 proliferation potential,41 growth pattern,31,41 and multilineage differentiation potential.42 To this end, it was unexpected, that the different cell clones derived from the same donor samples revealed very similar epigenetic profiles. On the other hand, the iP-MSC retain donor-specific differences in their DNA methylation pattern—and these had much higher impact than differences between individual clones from the same donor. These results have practical implications: for screening purposes or in vitro disease modeling it may be more relevant to compare different donor samples rather than a high number of different clones from the same patient.
Materials and Methods
Isolation of MSC. MSCs were isolated from human bone marrow from tibia plateau after written consent using guidelines approved by the Ethic Committee on the Use of Human Subjects at the University of Aachen (permit no. EK 128/09). Gender and donor age are provided in Supplementary Table S4. Culture expansion of MSC was performed in standard medium consisting of Dulbecco's modified Eagle's medium (DMEM) (1 g/l glucose; PAA, Cölbe, Germany) supplemented with glutamine (PAA), penicillin/streptomycin (PAA) and 10% HPL. HPL was generated from platelet units by a simple freeze-thaw procedure with addition of heparin.31 Upon 80% confluent growth, cells were harvested by trypsinization and subsequently counted with a Neubauer chamber (Brand, Wertheim, Germany). Re-seeding of cells was performed at a density of 5,000 cells/cm2.
Molecular characterization of MSC. Immunophenotypic surface marker analysis was performed on a FACS Canto II (BD, Heidelberg, Germany) upon staining with the following antibodies as described before:43 CD14-allophycocyanin (APC, clone M5E2; BD), CD29-phycoerythrin (PE, clone MAR4; BD), CD31-PE (clone WM59; BD), CD34-APC (clone 8G12; BD), CD45-APC (clone HI30; BD), CD73-PE (clone AD2; BD), CD90-APC (clone 5E10; BD), CD105-fluorescein isothiocyanate (FITC, clone MEM-226; ImmunoTools, Friesoythe, Germany).
For adipogenic differentiation, MSC were cultured in medium supplemented with 0.5 mmol/l isobutylmethylxanthine, 1 µmol/l dexamethasone, and 10 µg/ml insulin.44 Two to three weeks after induction, cells were stained with the green fluorescent dye BODIPY (4,4-difluoro-1,2,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene) and counterstained with DAPI (4′,6-Diamidino-2-phenylindol; both Molecular Probes, Eugene, OR). Fluorescence microscopic pictures were taken from four randomly chosen areas with a Leica DM IL HC microscope (Leica, Wetzlar, Germany).
Osteogenic differentiation was induced with culture medium supplemented with 10 mmol/l β-glycerophosphate, 0.1 µmol/l dexamethasone, and 0.2 mmol/l ascorbic acid and with medium changes every 3–4 days. After 2–3 weeks, osteogenic differentiation was analyzed by Alizarin Red staining and quantified by acetic acid extraction and neutralization with ammonium hydroxide. Absorbance was measured at 405 nm on a Tecan Infinite M200 plate reader (Tecan, Crailsheim, Germany).
Chondrogenic differentiation was induced in micromass culture for 3 weeks. Subsequently the pellets were fixed with 10% formalin and paraffin embedded; 1 mm sections were stained with Alcian blue in combination with Periodic acid-Schiff in an automated slide stainer and photo-documented.43
Generation of human MSC-derived iPSC. MSC of three different donors at third passage (seeded at a density of 2,000 cells/cm2 in a 6-well plate) were infected in a single experiment with equal amounts of pMXs-based retroviruses (Addgene, Cambridge, MA) encoding the human transcription factors OCT3/4, SOX2, KLF4, and c-MYC as described previously.45 The cells were transduced with freshly produced viruses in the presence of 4 µg/ml polybrene (Sigma, St Louis, MO) for 8 hours per day for 2 days. Transduced cells were cultured for the initial 3 days in α-MEM (Life Technologies/Gibco, Darmstadt, Germany) medium containing 10% HPL. On day 3, medium was changed to a dialyzed fetal bovine serum medium containing high glucose DMEM (Gibco), 20% fetal bovine serum, 1% nonessential amino acids, 1× penicillin/streptomycin, 1× L-glutamine, 0.1 mmol/l β-mercaptoethanol, and 50 ng/ml basic fibroblast growth factor (Peprotech, Hamburg, Germany). On day 6 after the first infection, 500 cells/cm2 were plated on 0.1% gelatin-coated 60 mm dishes containing 5 × 105 irradiated CF1 murine embryonic fibroblasts prepared in our laboratory. From day 7, plates were maintained in dialyzed fetal bovine serum medium supplemented with 20 µg/ml vitamin C (Sigma) and 1 mmol/l valproic acid (Sigma). Human ESC-like colonies were observed between day 21–30 after the first infection. The putative ESC-like colonies were mechanically isolated with 22G needles for expansion and subsequent validation. Established iPSC colonies were maintained on murine embryonic fibroblasts in DMEM/F12 medium supplemented with Glutamax, 20% knockout serum replacer, 1% nonessential amino acids, 0.1 mmol/l β-mercaptoethanol, and 50 ng/ml basic fibroblast growth factor. Cells were passaged by manual dissection of cell clusters every 5–6 days. Human ESC lines H1, H9, and HES-2 (WiCell Research Institute, Madison, WI) were used as controls and cultured under the same conditions as described for established iPSC above. Work with human ESC has been approved by the Robert Koch Institute, Berlin, Germany (permit no. Az 1710-79-1-4-57).
In vitro iP-MSC differentiation. iP-MSC differentiation was induced using the embryoid body differentiation system. Undifferentiated iP-MSC colonies were cut into small clumps and cultivated in suspension for 5 days in the differentiation medium (knockout DMEM supplemented with 20% fetal bovine serum, 1% nonessential amino acids, 0.1 mmol/l β-mercaptoethanol, and 1× L-glutamine). Embryoid bodies were then plated on 0.1% gelatin-coated culture dishes and immunostaining was performed on days 14–19 of differentiation for markers of all three embryonic germ layers.
Alkaline phosphatase staining. iP-MSC were cultured for 5 days, washed twice with phosphate-buffered saline, fixed in 100% methanol for 10 minutes at room temperature, and air-dried. Cells expressing alkaline phosphatase were stained for 15–30 minutes at room temperature with 1:10 dilution of Naphthol AS-MX phosphate (200 g/ml; Sigma) in Fast Red TR salt TM (1 mg/ml; Sigma) that was prepared in Tris-HCl (pH 9.2). The reaction was stopped by rinsing the cells with distilled water and air drying. Colonies were photographed using the phase-contrast microscope equipped with ×4 and ×10 lens (Zeiss, Jena, Germany).
RT-PCR and quantitative RT-PCR. Total RNA was isolated using TRIzol Reagent (Life Technologies) and cDNA was synthesized for semiquantitative or quantitative reverse transcription (RT)-PCR as described previously.31 For semiquantitative RT-PCR, cDNA was amplified using JumpStart RedTaq ReadyMix PCR Reaction Mix (Sigma). For quantitative RT-PCR, the cDNA probes were amplified using SYBR Green PCR Master Mix (Qiagen, Hilden, Germany). GAPDH was used for normalization of expression levels of individual genes. Primers used are listed in the Supplementary Table S5.
Immunocytochemistry. iP-MSC were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 5% fetal bovine serum, and then stained overnight at 4 °C with primary antibodies specific for OCT4 (clone c-10; Santa Cruz Biotechnology, Santa Cruz, CA), SOX2 (Stemgent, Cambridge, MA), Nanog (clone H-155; Stemgent), AlexaFluor 555-conjugated mouse anti-human TRA-1-60 antibodies (BD Biosciences, Heidelberg, Germany), TRA-1-80 (Santa Cruz Biotechnology), AlexaFluor 488-conjugated mouse SSEA4 antibodies (BD Biosciences), desmin (GeneTex, Irvine, CA), nestin (clone 10C2; Miliipore, Billerica, MA) and α-fetoprotein (clone C-19; Santa Cruz Biotechnology). Samples were visualized after staining with fluorescently labeled AlexaFluor secondary antibodies (Molecular Probes) for 1 hour at room temperature. Nuclei were counterstained with Hoechst 33342. Samples were embedded in ProLong Gold antifade reagent (Life Technologies) and evaluated on an Axiovert 200 microscope (Zeiss) equipped with the image processing software Axiovision 4.5.
Flow cytometry. PSC colonies were dissociated by Trypsin/EDTA treatment. Cells were then stained either with isotype control or antigen-specific AlexaFluor 488-conjugated Mouse SSEA4 antibodies (clone MC813-70; BD Biosciences) or with AlexaFluor 555-conjugated mouse anti-human TRA-1-60 antibodies (BD Biosciences), and analyzed after two washing steps on FACScan flow cytometer (BD Biosciences). Propidium iodide was used for dead cell staining.
Microsatellite analysis. Genotype analysis of parental MSCs and MSC-derived iPS cell lines were performed using six highly informative microsatellite markers at D16S2621, D18S976, D1S466, D22S280, D3S1768, and GAAT1A4. Primers for microsatellite analysis are listed in Supplementary Table S6. Fluorescently labeled PCR products were electrophoresed and detected on automated 3730 DNA Analyzers (Applied Biosystems, Foster City, CA). Data were analyzed using Genemapper software version 3.0 (Applied Biosystems).
Bisulfite pyrosequencing. Genomic DNA was isolated with the DNeasy Blood and Tissue Kit (Qiagen) and bisulfite treated using the EpiTect Bisulfite Kit (Qiagen). The methylation status of the promoters of OCT4 and NANOG was analyzed by bisulfite pyrosequencing on a PSQTM 96MA Pyrosequencing System (Biotage, Uppsala, Sweden) with the PyroGold SQA reagent kit (Biotage). Primers for bisulfite pyrosequencing are listed in Supplementary Table S7. Pyro Q-CpG software (Biotage) was used for data analysis.
Pluripotency evaluation via gene expression microarrays. Gene expression profiles of iP-MSC clones were analyzed by Affymetrix GeneChip Human Gene 1.0 ST technology and deposited at NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/; GSE38806). For evaluation of pluripotency with PluriTest,24 we had to project the algorithm from the Illumina Human HT 12 bead array to the Affymetrix GeneChip Human Gene 1.0 ST array by homology matching using the “getLDS” method of the Bioconductor “biomaRt” package.46 The projection was validated by a manually curated reference dataset from publicly available data at GEO—it consists of 98 pluripotent and 1,028 non-pluripotent Affymetrix Human Gene 1.0 ST array samples (Supplementary Table S8). Gene expression profiles of iP-MSC clones were quantile normalized using a target as determined by the reference dataset. Pluripotency and novelty scores were then calculated using the same gene expression patterns (“metagenes”) as described before.24
Clonal analysis of retroviral integration sites. To retrieve retroviral insertion sites, we accomplished LAM-PCR as previously described.47 In brief, 250–1,000 ng of DNA was preamplified by linear PCR. Biotinylated linear PCR products were enriched via magnetic beads, followed by second strand synthesis, restriction digest, ligation of a linker cassette, and two additional exponential amplifications. All samples were analyzed using two different restriction enzymes (Tsp509I, HpyCH4IV) for LAM-PCR to identify insertion sites in separate approaches. LAM-PCR amplicons were purified for downstream sequencing analyses using the QIAquick PCR Purification Kit (Qiagen) followed by pyrosequencing. Bioinformatic analysis of individual LAM-PCR amplicons was performed as described recently.48,49
DNA methylation profiling. Genomic DNA was isolated from 106 cells using the Qiagen DNA Blood Midi Kit. The DNA quality was assessed with a NanoDrop ND-1000 spectrometer (NanoDrop Technologies) and by gel electrophoresis; 600 ng DNA were bisulfite converted using the EZ DNA Methylation Kit (Zymo, Irvine, CA) according to the manufacturer̀s instructions. DNA methylation profiles were analyzed using the novel Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA) according to the manufacturer's instructions. This platform represents 4,82,421 CpG sites, 3,091 non-CpG sites, and 65 random SNPs. It covers 99% of RefSeq genes with multiple probes per gene and 96% of the CpG islands.21 Initial analysis with the Genomestudio 2010.3 (Modul M Version 1.8.5) was performed at the DKFZ Gene Core Facility in Heidelberg, Germany. Data were normalized with internal controls according to Illumina's standard procedures. Methylation level at each locus was calculated with the GenomeStudio Methylation module as β-value (ranging from 0–1). The number of beads per feature varies between chips and β-values were calculated as average of at least three technical replica. The complete information has been deposited at GEO (GSE34688).
Bioinformatics and statistics. For further analysis, we have only considered CpG sites on autosomes. Histograms of DNA methylation level and principal component analysis were calculated with prcomp in R package stats. Unsupervised hierarchical clustering according to Pearson correlation was performed using the MultiExperiment Viewer (MeV, TM4.7.4; http://www.tm4.org/mev/). Differentially methylated regions were selected with limma (R) using a stringent cut-off (adjusted P value <0.001). Genes associated with the differentially methylated CpG sites were classified by GO analysis using GoMiner software (http://discover.nci.nih.gov/gominer/) and enrichment of chromosomal location was analyzed with gene set enrichment analysis (http://www.broadinstitute.org/gsea/index.jsp). Affiliation of CpG sites with specific gene regions or CpG islands were provided in the annotation of the HumanMethylation450 BeadChip and have been described in detail before.21,50 CpG islands were defined as 500 bp with a CG content of more than 50% and a CpG observed/expected ratio of more than 0.6. Statistics for enrichment of differentially methylated CpG sites in specific regions was calculated by hypergeometric distribution.
SUPPLEMENTARY MATERIAL Figure S1. Expression of pluripotency markers in different iP-MSC clones. Figure S2. Expression of endogenous and virally encoded pluripotency factors in different iP-MSC. Figure S3. Methylation status of the promoters of NANOG and OCT4 genes. Figure S4. Immunocytochemical detection of nestin in differentiating iP-MSC derivatives from donor 4, clone B. Figure S5. Linear amplification-mediated PCR analysis of retroviral insertion sites in the genomes of different iP-MSC lines and their subclones. Table S1. Microsatellite markers in three parental MSC preparations and reprogrammed iP-MSC lines. Table S2. Gene Ontology classification of hypermethylated genes in iP-MSC versus ESC. Table S3. Gene Ontology classification of genes with donor-specific variation in DNA methylation. Table S4. MSC used in this study. Table S5. List of primer sequences used for PCR analyses. Table S6. Primers used for microsatellite analysis. Table S7. Genes and primers used for bisulfite pyrosequencing. Table S8. Reference dataset for PluriTest analysis with Affymetrix Human Gene 1.0 ST platform. Video S1. iP-MSC–derived spontaneously contracting cluster on day 28 of differentiation.
Acknowledgments
We are grateful to Matthias Schick (gene core facility at the German Cancer Research Center, Heidelberg, Germany) for array hybridization and initial data analysis. We thank Rebecca Dieterich and Nadin Lange for technical assistance. We also acknowledge Christian Weber and Anna Paruzynski for linear amplification-mediated PCR and bioinformatic analysis. This work was supported by the excellence initiative of the German federal and state governments within the START-Program of the Faculty of Medicine, RWTH Aachen (W.W.); by the state North-Rhine Westphalia within the BioNRW2 project “StemCellFactory” (M.Z., W.W., M.L.) and by the Stem Cell Network of North-Rhine Westphalia (T.Š., W.W.); by the Köln Fortune Program (T.Š.); by the Priority Programme SPP1356 of the German Research Foundation DFG (U.Z.); by the Federal Ministry for Education and Research (T.Š.), and by the Else-Kröner Fresenius Stiftung (T.Š., C.K., W.W.). The authors declared no conflict of interest.
Supplementary Material
Expression of pluripotency markers in different iP-MSC clones.
Expression of endogenous and virally encoded pluripotency factors in different iP-MSC.
Methylation status of the promoters of NANOG and OCT4 genes.
Immunocytochemical detection of nestin in differentiating iP-MSC derivatives from donor 4, clone B.
Linear amplification-mediated PCR analysis of retroviral insertion sites in the genomes of different iP-MSC lines and their subclones.
Microsatellite markers in three parental MSC preparations and reprogrammed iP-MSC lines.
Gene Ontology classification of hypermethylated genes in iP-MSC versus ESC.
Gene Ontology classification of genes with donor-specific variation in DNA methylation.
MSC used in this study.
List of primer sequences used for PCR analyses.
Primers used for microsatellite analysis.
Genes and primers used for bisulfite pyrosequencing.
Reference dataset for PluriTest analysis with Affymetrix Human Gene 1.0 ST platform.
iP-MSC–derived spontaneously contracting cluster on day 28 of differentiation.
REFERENCES
- Zhu H, Lensch MW, Cahan P., and, Daley GQ. Investigating monogenic and complex diseases with pluripotent stem cells. Nat Rev Genet. 2011;12:266–275. doi: 10.1038/nrg2951. [DOI] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T.et al. (2009iPS cells produce viable mice through tetraploid complementation Nature 46186–90. [DOI] [PubMed] [Google Scholar]
- Patel M., and, Yang S. Advances in reprogramming somatic cells to induced pluripotent stem cells. Stem Cell Rev. 2010;6:367–380. doi: 10.1007/s12015-010-9123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD.et al. (2010Generation of iPSCs from cultured human malignant cells Blood 1154039–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F.et al. (2009Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells Proc Natl Acad Sci USA 10615720–15725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugii S, Kida Y, Kawamura T, Suzuki J, Vassena R, Yin YQ.et al. (2010Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells Proc Natl Acad Sci USA 1073558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G.et al. (2011Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells Nature 47168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Qin H, Qu C, Tuan RS, Shi S., and, Huang GT. iPS cells reprogrammed from human mesenchymal-like stem/progenitor cells of dental tissue origin. Stem Cells Dev. 2010;19:469–480. doi: 10.1089/scd.2009.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Yoshimura Y, Ohnishi H, Tadokoro M, Katsube Y, Sasao M.et al. (2010Induction of pluripotent stem cells from human third molar mesenchymal stromal cells J Biol Chem 28529270–29278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Son MJ, Son MY, Seol B, Kim J, Park J.et al. (2011Generation of human induced pluripotent stem cells from osteoarthritis patient-derived synovial cells Arthritis Rheum 633010–3021. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A.et al. (2008Disease-specific induced pluripotent stem cells Cell 134877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi H, Oda Y, Aoki T, Tadokoro M, Katsube Y, Ohgushi H.et al. (2012A comparative study of induced pluripotent stem cells generated from frozen, stocked bone marrow- and adipose tissue-derived mesenchymal stem cells J Tissue Eng Regen Med 6261–271. [DOI] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R.et al. (2009Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts Nat Genet 411350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J.et al. (2009Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming Nat Biotechnol 27353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P.et al. (2010Epigenetic memory in induced pluripotent stem cells Nature 467285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T.et al. (2011Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells Nat Cell Biol 13541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD.et al. (2011Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines Cell 144439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H.et al. (2011DNA methylation dynamics in human induced pluripotent stem cells over time PLoS Genet 7e1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY.et al. (2010Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells Nat Biotechnol 28848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P.et al. (2011Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells Nat Biotechnol 291117–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM.et al. (2011High density DNA methylation array with single CpG site resolution Genomics 98288–295. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M.et al. (2011Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome Epigenetics 6692–702. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D.et al. (2006Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement Cytotherapy 8315–317. [DOI] [PubMed] [Google Scholar]
- Müller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP.et al. (2011A bioinformatic assay for pluripotency in human cells Nat Methods 8315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A.et al. (2009Comprehensive genomic access to vector integration in clinical gene therapy Nat Med 151431–1436. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J.et al. (2009Human DNA methylomes at base resolution show widespread epigenomic differences Nature 462315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ.et al. (2011A functionally characterized test set of human induced pluripotent stem cells Nat Biotechnol 29279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH.et al. (2009DNA methylation profiles in monozygotic and dizygotic twins Nat Genet 41240–245. [DOI] [PubMed] [Google Scholar]
- Tan KY, Eminli S, Hettmer S, Hochedlinger K., and, Wagers AJ. Efficient generation of iPS cells from skeletal muscle stem cells. PLoS ONE. 2011;6:e26406. doi: 10.1371/journal.pone.0026406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallmoser K, Bartmann C, Rohde E, Reinisch A, Kashofer K, Stadelmeyer E.et al. (2007Human platelet lysate can replace fetal bovine serum for clinical-scale expansion of functional mesenchymal stromal cells Transfusion 471436–1446. [DOI] [PubMed] [Google Scholar]
- Cholewa D, Stiehl T, Schellenberg A, Bokermann G, Joussen S, Koch C.et al. (2011Expansion of adipose mesenchymal stromal cells is affected by human platelet lysate and plating density Cell Transplant 201409–1422. [DOI] [PubMed] [Google Scholar]
- Sato S, Yagi S, Arai Y, Hirabayashi K, Hattori N, Iwatani M.et al. (2010Genome-wide DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) residing in mouse pluripotent stem cells Genes Cells 15607–618. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A.et al. (2008Genome-scale DNA methylation maps of pluripotent and differentiated cells Nature 454766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S.et al. (2008An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res 181518–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae G, Isagawa T, Shiraki N, Fujita T, Yamamoto S, Tsutsumi S.et al. (2011Tissue-specific demethylation in CpG-poor promoters during cellular differentiation Hum Mol Genet 202710–2721. [DOI] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT.et al. (2010Identification and classification of chromosomal aberrations in human induced pluripotent stem cells Cell Stem Cell 7521–531. [DOI] [PubMed] [Google Scholar]
- Ushijima T, Watanabe N, Okochi E, Kaneda A, Sugimura T., and, Miyamoto K. Fidelity of the methylation pattern and its variation in the genome. Genome Res. 2003;13:868–874. doi: 10.1101/gr.969603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M.et al. (2011CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing Nature 47974–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L.et al. (2008Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation Cell Stem Cell 2160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colter DC, Sekiya I., and, Prockop DJ. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci USA. 2001;98:7841–7845. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg A, Stiehl T, Horn P, Joussen S, Pallua N, Ho AD.et al. (2012Population dynamics of mesenchymal stromal cells during culture expansion Cytotherapy 14401–411. [DOI] [PubMed] [Google Scholar]
- Russell KC, Phinney DG, Lacey MR, Barrilleaux BL, Meyertholen KE., and, O'Connor KC. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2010;28:788–798. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- Koch CM, Suschek CV, Lin Q, Bork S, Goergens M, Joussen S.et al. (2011Specific age-associated DNA methylation changes in human dermal fibroblasts PLoS ONE 6e16679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R.et al. (2008Replicative senescence of mesenchymal stem cells: a continuous and organized process PLoS ONE 3e2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K.et al. (2007Induction of pluripotent stem cells from adult human fibroblasts by defined factors Cell 131861–872. [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A.et al. (2005BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis Bioinformatics 213439–3440. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Schwarzwaelder K, Bartholomae CC, Glimm H., and, von Kalle C. Detection of retroviral integration sites by linear amplification-mediated PCR and tracking of individual integration clones in different samples. Methods Mol Biol. 2009;506:363–372. doi: 10.1007/978-1-59745-409-4_24. [DOI] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I.et al. (2009Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy Science 326818–823. [DOI] [PubMed] [Google Scholar]
- Arens A, Appelt JU, Bartholomae CC, Gabriel R, Paruzynski A, Gustafson D.et al. (2012Bioinformatic clonality analysis of next-generation sequencing-derived viral vector integration sites Hum Gene Ther Methods 23111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R.et al. (2010CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer Oncogene 296390–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of pluripotency markers in different iP-MSC clones.
Expression of endogenous and virally encoded pluripotency factors in different iP-MSC.
Methylation status of the promoters of NANOG and OCT4 genes.
Immunocytochemical detection of nestin in differentiating iP-MSC derivatives from donor 4, clone B.
Linear amplification-mediated PCR analysis of retroviral insertion sites in the genomes of different iP-MSC lines and their subclones.
Microsatellite markers in three parental MSC preparations and reprogrammed iP-MSC lines.
Gene Ontology classification of hypermethylated genes in iP-MSC versus ESC.
Gene Ontology classification of genes with donor-specific variation in DNA methylation.
MSC used in this study.
List of primer sequences used for PCR analyses.
Primers used for microsatellite analysis.
Genes and primers used for bisulfite pyrosequencing.
Reference dataset for PluriTest analysis with Affymetrix Human Gene 1.0 ST platform.
iP-MSC–derived spontaneously contracting cluster on day 28 of differentiation.