Abstract
Levels of the phytohormone indole-3-acetic acid (IAA) can be altered by the formation and hydrolysis of IAA conjugates. The isolation and characterization of Arabidopsis thaliana mutants with reduced IAA-conjugate sensitivity and wild-type IAA responses is advancing the understanding of auxin homeostasis by uncovering the factors needed for conjugate metabolism. For example, the discovery that the IAA-Ala-resistant mutant iar1 is defective in a protein in the ZIP family of metal transporters uncovered a link between metal homeostasis and IAA-conjugate sensitivity. To uncover additional factors impacting auxin conjugate metabolism, we conducted a genetic modifier screen and isolated extragenic mutations that restored IAA-amino acid conjugate sensitivity to the iar1 mutant. One of these suppressor mutants is defective in a putative cation diffusion facilitator, MTP5 (At3g12100; formerly known as MTPc2). Loss of MTP5 function restored IAA conjugate sensitivity to iar1 but not to mutants defective in IAA-amino acid conjugate amidohydrolases. Our results are consistent with a model in which MTP5 and IAR1 transport metals in an antagonistic fashion to regulate metal homeostasis within the subcellular compartment in which the IAA-conjugate amidohydrolases reside, and support previous suggestions that the ion composition in this compartment influences hydrolase activity.
Keywords: MTP5, IAR1, auxin conjugates, metal transport, Arabidopsis
Certain trace elements are essential for plant growth and development. For example, Cu2+ and Mn2+ are used in many electron transfer reactions, including those in photosynthesis, Zn2+ ions provide structure to DNA-binding proteins and serve as cofactors for hydrolytic enzymes, and iron is essential for heme proteins such as ferredoxin and catalase [reviewed in (Clemens et al. 2002; Hall and Williams 2003)]. When appropriate levels of any of these ions are not maintained, plants display an array of symptoms, including reduced growth (Marschner 1995).
Numerous cation transporters facilitate ion homeostasis. In Arabidopsis, there are at least 880 putative membrane transporters that fall into 46 families (Mäser et al. 2001). Disruption of some of these transporters leads to measurable metal-related phenotypes in single mutants (Cheng et al. 2003; Delhaize et al. 2007; Li et al. 2008; Kawachi et al. 2009). However, not all mutants defective in putative metal transporters display morphological or ionomic phenotypes due to the complexity and redundancy of the elaborate metal transport system. Establishing function for some of these predicted transporters may require alternative genetic, biochemical, or physiological analyses.
Auxin is a plant hormone that controls a plethora of growth and developmental processes, and auxin metabolism is complex and distributed among several subcellular compartments (Woodward and Bartel 2005; Strader and Bartel 2011). By screening for Arabidopsis mutants with defective indole-3-acetic acid (IAA) conjugate responsiveness, we uncovered several enzymes that hydrolyze conjugates (Bartel and Fink 1995; Davies et al. 1999; Rampey et al. 2004). These IAA-amino acid hydrolases contain predicted N-terminal signal sequences and C-terminal endoplasmic reticulum (ER) retention signals (Bartel and Fink 1995; Davies et al. 1999), and several localize to the ER in organelle proteomics experiments (Dunkley et al. 2006). The observation that the IAA-amino acid hydrolases are metalloenzymes (Bartel and Fink 1995; Davies et al. 1999; Rampey et al. 2004; Bitto et al. 2009) may explain why proteins that are likely to influence metal transport also have emerged from screens for reduced IAA-conjugate responsiveness. Analyses of the IAA-conjugate response mutants iaa-leucine resistant2 (ilr2) (Magidin et al. 2003), ilr3 (Rampey et al. 2006), and iaa-alanine resistant1 (iar1) (Lasswell et al. 2000) reveal a critical role for the metal microenvironment in IAA-conjugate metabolism and indicate that an understanding of subcellular metal homeostasis will be required to fully elucidate mechanisms regulating IAA levels. For example, ilr2 seedlings are resistant to root growth inhibition not only by IAA conjugates but also by Mn2+ and Co2+ (Magidin et al. 2003). ILR2 (At3g18485) is an apparently cytosolic protein that may inhibit an unidentified metal transporter, as ilr2 microsomes have enhanced Mn2+ transport activity compared with wild-type microsomes (Magidin et al. 2003). In addition, the ILR3 (At5g54680) basic helix–loop–helix leucine zipper (bHLH105) transcription factor regulates transcription of metal transporter genes (Rampey et al. 2006) that appear to modulate iron distribution (Gollhofer et al. 2011). A dominant gain-of-function ilr3 allele reduces IAA-Leu and Mn2+ responsiveness, whereas loss of ILR3 heightens responses to both IAA-Leu and Mn2+ (Rampey et al. 2006).
The iar1 mutant has decreased sensitivity to a variety of IAA conjugates (Lasswell et al. 2000) that are in vitro substrates of the Arabidopsis IAA-amino acid hydrolases (LeClere et al. 2002). IAR1 (At1g68100) contains at least seven predicted transmembrane domains and many His-rich regions, consistent with a role in metal binding or transport (Lasswell et al. 2000). IAR1 resembles members of the zinc-regulated transporter iron-regulated transporter-like protein (ZIP) family that transport metals from the vacuole or apoplast into the cytosol (Gaither and Eide 2000). Although there are at least 15 ZIP family members in Arabidopsis (Hall and Williams 2003), IAR1 is the only Arabidopsis member of the LIV-1 zinc transporter (LZT) subfamily (Taylor and Nicholson 2003). Heterologous expression of mouse LZT protein ZIP7/KE4, which is 26% identical to IAR1, complements the Arabidopsis iar1 mutant (Lasswell et al. 2000) and the Saccharomyces cerevisiae yke4 mutant (Kumánovics et al. 2006), suggesting that IAR1, ZIP7, and YKE4 are orthologs that function similarly in plants, animals, and yeast. Although the localization and substrate specificity of Arabidopsis IAR1 have not been determined, mammalian ZIP7/KE4 is a zinc exporter localized in the Golgi membrane (Huang et al. 2005), and yeast Yke4p is a bidirectional zinc transporter localized in the ER membrane (Kumánovics et al. 2006), consistent with the possibility that IAR1 is a zinc transporter in the secretory pathway. Determining the function of IAR1 may provide insight into IAA-conjugate responses and metabolism in plants and contribute to understanding the function of other members of the LZT transporter family.
The observation the IAA-conjugate hydrolases require metal cofactors for activity (Bartel and Fink 1995; Davies et al. 1999; Rampey et al. 2004) underlies the hypothesis that the direct consequences of ilr2, ilr3, and iar1 mutations are alterations in ion homeostasis, and the altered sensitivity to IAA conjugates in these mutants is a secondary effect of this change. Indeed, exogenous manganese suppresses the IAA-Ala resistance of iar1 (Lasswell et al. 2000), and ion level alterations are detected in ilr3 mutants grown on certain media (Rampey et al. 2006). However, ionomic changes have not been identified in either ilr2 (Magidin et al. 2003) or iar1 (Lasswell et al. 2000) plants, and iar1 mutants lack morphological phenotypes expected from a dramatic ion imbalance, suggesting that any such ionomic changes are modest or restricted to specific tissues or subcellular locations.
We have taken a genetic approach to clarify the role of IAR1 in IAA-conjugate sensitivity. We conducted an iar1 suppressor screen to isolate genes that when defective restore wild-type IAA-conjugate sensitivity to iar1 roots. We identified mtp5 as a mutant defective in a cation diffusion facilitator family transporter that suppressed the IAA-conjugate resistance of iar1 but not other IAA-conjugate sensitivity mutants. The isolation of mtp5 as an iar1 suppressor supports the hypothesis that IAR1 functions as a metal transporter and suggests that MTP5 and IAR1 transport ions in an antagonistic fashion to control subcellular metal homeostasis.
Materials and Methods
Plant materials and growth conditions
Plants from the Columbia (Col-0) and Wassilewskija (Ws) accessions were used. For phenotypic assays, seeds were surface-sterilized (Last and Fink 1988) and grown aseptically on plant nutrient medium containing 0.5% (w/v) sucrose [PNS (Haughn and Somerville 1986)] solidified with 0.6% agar. Seedlings were grown in medium alone or medium supplemented with IAA, IAA-l-amino acid conjugates (Sigma-Aldrich) or other hormones (from 0.1, 1, or 100 mM stocks in ethanol), or Basta [glufosinate ammonium, Crescent Chemical, Augsburg, Germany; from a 50 mg/mL stock in 25% (v/v) ethanol]. Media supplemented with metals (from 100 mM MnCl2, 500 mM CoCl2, 2 M CaCl2, 20 mM CdCl2, and 500 mM ZnSO4 stocks in H2O) were prepared without sucrose. Plates were sealed with gas-permeable Leukopor surgical tape (LecTec Corp, Minnetonka, MN) and incubated with constant illumination (25 to 45 µEm-2s-2) at 22° under yellow long-pass filters to slow the breakdown of indolic compounds (Stasinopoulos and Hangarter 1990). Plants transferred to soil (Metromix 200, Scotts, Marysville, OH) were grown at 22 to 25° under continuous illumination by Cool White fluorescent bulbs (Sylvania, Danvers, MA).
Mutant isolation
Approximately 48,000 progeny of iar1-3 (Col-0 accession) mutagenized with ethyl methanesulfonate (Normanly et al. 1997) and iar1-1 (Ws accession) mutagenized with fast-neutron bombardment (Lasswell 2000) were screened for iar1 suppressors by plating ~1000 seeds per 150 mm plate on PNS supplemented with 40 μM IAA-Ala. Seedlings were screened after growing for 8 d in continuous yellow-filtered light at 22°. Seedlings with wild-type sensitivity to IAA-Ala were transferred to PNS and allowed to recover, and seedlings with roots that failed to elongate after transfer were discarded. The remaining mutants were transferred to soil for seed production. Progeny of these plants were tested on 40 μM IAA-Ala and PNS. Homozygous iar1 mutations in the suppressors were confirmed using polymerase chain reaction (PCR) amplification and restriction analyses. The iar1-1 mutation was detected as described previously (Lasswell et al. 2000), and the iar1-3 mutation was detected by PCR amplification with the oligonucleotides 5′-GAACCAGGACAATCATCGTTG-3′ and 5′-CCCAAGCTTGGGATTTCTATATCGGTTAC-3′ followed by digestion of the resulting product with HaeIII to yield a 328-bp product for Col-0 and 258-bp and 70-bp products for iar1-3.
The mtp5-1 mutant was isolated as suppressor of iar1-3, a loss-of-function iar1 allele in the Col-0 accession (Lasswell et al. 2000). To maintain iar1 in the mapping population, the N13 (mtp5-1 iar1-3) isolate was outcrossed to iar1-1 [in the Ws accession (Lasswell et al. 2000)]. The F2 progeny from this outcross were plated on 40 μM IAA-Ala, and seedlings displaying wild-type root lengths were selected for PCR-based recombination mapping. The causative mutation was located to the top of chromosome 3 using the published markers nga172 and nga162 (Bell and Ecker 1994). To further delineate the mapping interval, new PCR-based markers were designed (Table 1).
Table 1. New markers used in the positional cloning of MTP5.
| Markera | Oligonucleotides (5′ to 3′) | Enzyme | Size of Product, bp |
|---|---|---|---|
| F13M14-3+4 | F13M14-3b CTTCTTCTATATTGAGTAGGTAGATTAAAA | BsaBI | Col – 226 |
| F13M14-4 ATTACCTAAAGACTCTGATTTTATACTCTC | Ws – 196, 30 | ||
| T7M13-3+4 | T7M13-3 AGTTAACCAACGATAACAAGCAGATTCGTT | BclI | Col – 130, 20 |
| T7M13-4 CTCCTACAATCCTCTAAGAATCCATTGATC | Ws – 150 | ||
| F24K9-7+8 | F24K9-7 AATTTAAAATTATATGCAAACTAATTAGAT | BglII | Col – 200 + 30 |
| F24K9-8 GTAGCTAAAAAGTTGCTGCAAGCAAGGAAA | Ws – 230 | ||
| F26K24-9+6 | F26K24-6 GATAATAACGAAGAGTATGAAGTAAAAGTA | HpyCHIV | Col – 523 |
| F26K24-9 AGATTCGATTACACTAGGCAATTTGTTATGA | Ws – 128, 395 | ||
| MEC18-7+8 | MEC18-7 TTCGATTCAAGACAAAGTTTAAAGTTACAA | BspHI | Col – 220 |
| MEC18-8b AATACTTTAAGTTTTGGATGTAAGATTCAT | Ws – 190, 30 | ||
| F28J15-3+4 | F28J15-3 AATATCGGCCAACAGTAAGTT | HinP1I | Col – 400, 200 |
| F28J15-4 CATCACGTAACTGAGATTCC | Ws – 600 | ||
| T2E22-9+10 | T2E22-9b AGCCCTATGCACACACATGTAAAAATGGGA | FokI | Col – 213 |
| T2E22-10 ATTCACTGATTTATTTGTTACCTAGCTAAA | Ws – 172, 41 | ||
| MBK21-10+11 | MBK21-10 GATACTCAAGTAGTTATCTGTTACCTTTAG | HincII | Col – 350, 150 |
| MBK21-11 CTAAACCATTATGTGTAATGTGTGAATTAG | Ws – 500 | ||
| MGH6-2+3 | MGH6-2 TAGTTTCTCTGATTACTTGTGTAGATGTGA | MboII | Col – 357 |
| MGH6-3b TATAGCTGTTCCAAGACTATAACACCGGAA | Ws – 327, 30 | ||
| MRP15-3+4 | MRP15-3 GTTAGAATTGGAATTAACAAGTATTACTAG | TaqI | Col – 251, 56 |
| MRP15-4 CTTGATAGCATTGGGAGCAAGCAACGAACC | Ws – 213, 56, 41 |
Col-0, Columbia; dCAPS, derived cleaved amplified polymorphic sequences; PCR, polymerase chain reaction; Ws, Wassilewskija.
Markers reveal polymorphisms between Col-0 and Ws accessions when cut with the indicated restriction enzymes after PCR amplification with the indicated oligonucleotides.
This is a dCAPS oligonucleotide (Michaels and Amasino 1998; Neff et al. 1998); the underlined nucleotide differs from wild-type sequence to create a restriction site in either the Col-0 or Ws PCR product.
A candidate gene within the mapping region, At3g12100, was PCR-amplified and sequenced using DNA from an N13 mapping plant with the following oligonucleotides: T21B14-7 (5′-ATAAAGAATACAACTTTTTCTAGCTTTTAG-3′) and T21B14-9 (5′-TGAATAAAAGTGTCTTCTTGCTTGACTACA-3′), T21B14-13 (5′-GGAAATATGTACACATTCGAGGAACGATT-3′) and T21B14-17 (5′-CTTTGATTTGTTTAATATTTGACATATGTG-3′), T21B14-18 (5′-TATGAGCATCAATTCATACAAGTCTAAACA-3′) and T21B14-19 (5′-ATCGGCAGATGAAGAGACTGTTTCTGCTAA-3′), and T21B14-20 (5′-ATCAGGCTTCTTCCTTGAAGTTGCCATTGCA-3′) and T21B14-21 (5′-TTGGTAACGTAACTGTAAATCTTCTCT-3′). A G-to-A mutation was found in the 3′-splice site preceding exon 8 at nucleotide 2003 (where 1 is the A in the initiator ATG).
The mtp5-2 mutant is a sequence-indexed Arabidopsis T-DNA insertion mutant (GABI_351G01) isolated by the GABI-Kat facility (Rosso et al. 2003). We verified the position of the T-DNA insert in mtp5-2 by using PCR amplification with the oligonucleotides MTPc2-12 (5′-TGTAGTCAAGCAAGAAGACACTTTTATTCA-3′) and MTPc2-13 (5′-TATGCTGCAGCCTACAGAAAAGCAGAAGAT-3′) and the T-DNA specific primer, LB1-GABI (5′-CTTTCTTTTTCTCCATATTGACCATCA-3′). PCR amplification of MTPc2-12 and MTPc2-13 yielded a 309 bp product from wild-type genomic DNA, whereas amplification with MTPc2-13 and LB1-GABI yielded a 250-bp product from mtp5-2 genomic DNA. This product was sequenced, revealing that the T-DNA insertion is located in exon 7 at position 1873 in MTP5 (where 1 is the A in the initiator ATG).
ilr1-5 is a missense mutation the Col-0 accession (Rampey et al. 2006) that was backcrossed to Col-0 five times prior to crossing with mtp5-1. iar3-1 is a missense mutation in the Ws accession (Davies et al. 1999) that was introgressed into Col-0 by three rounds of outcrossing prior to crossing with mtp5-1. mtp5-1 ilr1-5 and mtp5-1 iar3-1 double mutants were isolated from segregating F2 populations using PCR amplification and restriction digest analyses. The ilr1-5 mutation was followed via amplification with 4G12-43 (5′-CAATCATCGCTTCCGCTAC-3′) and 4G12-34 (5′-CCACGCAGCTACACCGCAC-3′) and digestion with RsaI, resulting in 299- and 275-bp products for ILR1 DNA and a 574-bp product for ilr1-5 DNA. Homozygous mtp5-1 and iar3-1 plants were identified by following derived cleaved amplified polymorphic sequences (Michaels and Amasino 1998; Neff et al. 1998). Amplification with MTPc2-1 (5′-CCTAAACATAGGACCTCTGCATTTTCAAGC-3′) and MTPc2-2 (5′-TGCATCAAACTTATATACAGTCAACATGAA-3′) yields a 219-bp product. The altered nucleotide in MTPc2-1 (underlined) creates a HinDIII restriction site in the mtp5-1 product to give 189- and 30-bp products after digestion, whereas the MTP5 product is not cleaved by HinDIII. The iar3-1 mutation was followed by amplification with ILL4-22 (5′-CCTGTGAGTCTAAAGGATCTGCCTCTCGTG-3′) and ILL4-24 (5′-CAAATCAATTGGCATTAGGTCAAGTAAGCT-3′). ILL4-24 creates a HinDIII site (underlined nucleotide) in the iar3-1 PCR product resulting in 174- and 30-bp products after digestion, whereas the 200 bp IAR3 product remains uncut.
MTP5 cDNA isolation
To isolate a MTP5 cDNA, RNA was isolated with RNeasy Mini Kits (QIAGEN, Valencia, CA) from 7-d-old Col-0 seedlings grown on filter paper in 150-mm plates containing PNS at 22° in yellow-filtered light. Reverse transcriptase (RT) reactions used RETROScript (Ambion, Austin, TX) on the Col-0 RNA primed with the oligonucleotide MTPc2-4 (5′-TTGCGGCCGCACCATGATCTCTAGTATACATCC-3′). The resulting cDNA was PCR-amplified using Pfu Turbo DNA polymerase (Stratagene) and the oligonucleotides MTPc2-4 and MTPc2-3 (5′-TCGGATCCTCGACGAAGTTGGAACTTTAAGATC-3′). Underlined base pairs in MTPc2-3 and MTPc2-4 were altered to create a SalI or NotI restriction site, respectively, for subsequent subcloning. The MTP5 RT-PCR product was cloned into the pCR4Blunt-TOPO vector (Invitrogen, Carlsbad, CA) and transformed into TOP10 Escherichia coli (Invitrogen, Carlsbad, CA). Sequence analysis of the TOPO-MTP5 plasmid showed that the MTP5 sequence was mis-spliced (5′ of the mtp5-1 mutation) compared to annotation by The Arabidopsis Information Resource (TAIR; www.arabidopsis.org). Sequence analysis of the RT-PCR product revealed not only the cloned mis-spliced cDNA, designated MTP5-B, but also a second product that matched the predicted MTP5 sequence, designated MTP5-A.
Using this sequence information, MTP5-B was excised from pCR4Blunt-TOPO with EcoRI and ligated into pBluescript KS (+) (Stratagene) cut with EcoRI to obtain pKS-MTP5premut. MTP5-B is oriented in the opposite orientation as LacZ in pKS-MTP5premut. Site-directed oligonucleotide-mutagenesis (Ausubel et al. 1999) was performed using MTPc2-10 (5′-TTTACTCCGTTGATGGAAGTGATGTGTTTTTCGG-3′) and MTPc2-11 (5′-ATCTGCTTTCACTAATGCTCTGTTCCTTATGTTCAT-3′) on pKS-MTP5premut. The underlined base pair in MTPc2-10 was altered to change the A to a G at nucleotide 226 (where 1 is the A in the initiator ATG of the cDNA), which caused an amino acid change from Asn to Ser. MTPc2-11 removed four nucleotides (ACAG) after nucleotide 563 in the cDNA, which changed the splicing pattern into that identified for MTP5-A. The mutagenized cDNA, pKS-MTP5, was identified by PCR amplification and restriction analyses. The MTP5 coding sequence was amplified with T21B14-20 + MTPc2-7. MTP5-B, but not MTP5-A, contains a G at position 226, so the resulting product was digested with TspRI, which cleaves MTP5-B sequence but not MTP5-A. In addition, the PCR product resulting from amplification with T21B14-19 and MTPc2-9 was digested with PvuII, because this site was destroyed when the 4 nucleotides were removed.
MTP5 overexpression in plants
The MTP5 cDNA was excised from pKS-MTP5 with SalI and NotI, restriction sites incorporated at the cDNA ends during the initial RT-PCR amplification, and was ligated into the 35S-pBARN expression vector (LeClere and Bartel 2001) cut with XhoI and NotI. The resultant pBARN-MTP5 plasmid was electroporated (Ausubel et al. 1999) into Agrobacterium GV3101 cells (Koncz et al. 1992) and transformed into Col-0 and mtp5-1 iar1-3 plants using the floral dip method (Clough and Bent 1998). Transformants were selected on PN containing 10 μg/mL Basta, and homozygous lines were selected by following Basta resistance in subsequent generations.
RT-PCR analysis
RT-PCR analysis to identify the mtp5-1 mutant coding sequence was conducted using RNA from leaves of a homozygous mtp5-1 iar1-3 (N13) backcrossed plant as described previously. Sequencing the RT-PCR product with oligonucleotides used for amplification showed that the mtp5-1 mutation shifts the splice site 1 bp 3′ of the original site. This shift causes a frameshift of the coding sequence that results in a stop codon after 33 bp.
To distinguish between MTP5-A and MTP5-B, we reverse transcribed the two transcripts were as described previously by using DNaseI-treated (Amplification Grade, Roche Applied Science, Indianapolis, IN) RNA from Col-0 7-d-old seedlings and the oligonucleotide MTPc2RT-1 (5′-TTCATCTTGTATAAATGCATGAAGAGCTT-3′). cDNAs were amplified with MTPc2RT-2 (5′-GTGTTGTATTCTACAACAGAGCTCTCTAT-3′) and MTPc2RT-1, and the resulting products were digested with PvuII to give a 312-bp product for MTP5-A and 246- and 66-bp products for MTP5-B.
RT-PCR analysis also was completed to determine whether mtp5-2 expresses any intact MTP5 mRNA. RNA was isolated from mtp5-1, mtp5-2, and Col-0 7-d-old seedlings as described previously and RT was performed using MTPc2-RT-4 (5′-CCATCTGAAGCAAGACACCACCAGTGGCTT-3′) and the resulting cDNA was amplified using MTPc2-RT-3 (5′-CGTTTGCTTGCACGTCATATCAGATTCCAT-3′) and MTPc2-RT-4 to yield several splicing products spanning the mtp5 mutations. Products detected were unspliced (404 bp), fully spliced (180 bp), intron 6 present (312 bp), and intron 7 (274 bp).
MTP5 yeast expression
The MTP5-A cDNA was excised from pKS-MTP5 with BamHI and NotI and ligated into BamHI/NotI-cut-pTGPD, a yeast expression vector that contains the glyceraldehyde-3-phosphate dehydrogenase constitutive promoter and the TRP1 biosynthetic gene as a selectable marker (Chang and Lindquist 1994). The resulting plasmid, pTGPD-MTP5, was sequenced with the oligonucleotides MTPc2-3 and T21B14-19 to verify the orientation of the MTP5 cDNA.
To test whether MTP5 could complement yeast metal transport mutants, pTGPD-MTP5 or the empty pTGPD vector were transformed (Gietz and Schiestl 1995) into cm100 (wild type), cm102 (zrc1), cm103 (cot1), and cm104 (zrc1 cot1) yeast strains (Eide et al. 1996). Transformations were plated on selective media (Ausubel et al. 1999) lacking Trp for cm100, lacking Trp and His for cm102, lacking Trp and Ura for cm103, and lacking Trp, His, and Ura for cm104. For metal response analysis, positive transformants were streaked on YPD (Ausubel et al. 1999) plates with no metals or containing ZnSO4 or CoCl2 and incubated at 30° for 3 d.
Results
Isolation of a mutant defective in the cation diffusion facilitator MTP5
The iar1 mutant displays reduced root elongation inhibition on several IAA-amino acid conjugates, including IAA-Ala (Lasswell et al. 2000). To identify genes acting with IAR1 to control conjugate sensitivity, we conducted a genetic modifier screen to isolate extragenic mutations that suppressed the iar1 mutant phenotype. We screened approximately 48,000 progeny of mutagenized iar1 seeds for restored sensitivity to 40 μM IAA-Ala and isolated 61 putative suppressors, of which seven set seeds and retained wild-type IAA-Ala sensitivity in the next generation. One of these suppressors was selected for further analysis.
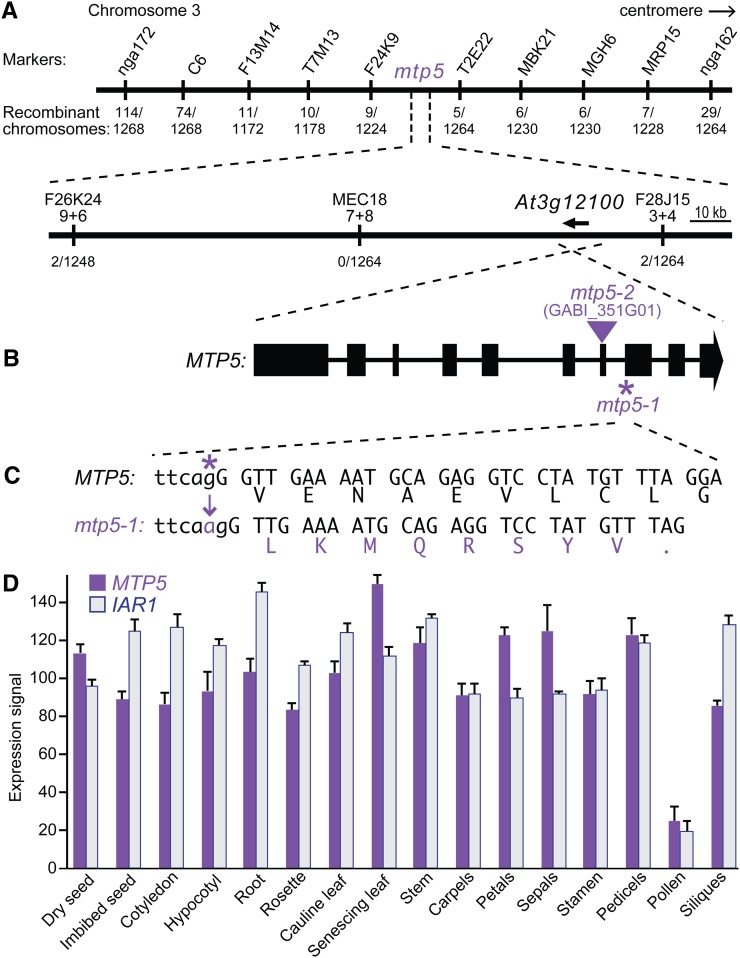
We used PCR-based markers to map the suppressor mutation to the top of chromosome three between nga172 and nga162 (Figure 1A, Bell and Ecker 1994). After identifying new molecular markers within this interval (Table 1), we narrowed the mapping region to ~160 kb between the markers F26K24-9+6 and F28J15-3+4 (Figure 1A). Using a candidate gene sequencing approach, we identified a mutation in the iar1 suppressor mutant in the gene encoding a putative cation diffusion facilitator/metal transport protein, MTP5 (At3g12100; Figure 1B). The mtp5-1 mutation is a G-to-A change at nucleotide 2003 (where 1 is the A in the initiator ATG) in the 3′ splice site preceding exon 8 (Figure 1C). We sequenced MTP5 in our other iar1 suppressors and did not recover any additional mtp5 alleles.
Figure 1 .
Positional cloning of the gene defective in mtp5-1. (A) Recombination mapping of mtp5-1. The lesion suppressing iar1 IAA-Ala resistance was mapped to chromosome 3 between markers F26K24-9+6 and F28J15-3+4 (Table 1), an interval that includes the MTP5 (At3g12100) gene. (B) MTP5 contains 10 exons (boxes) separated by 9 introns (lines). The location of the mtp5-2 T-DNA insertion is indicated by the triangle. (C) The G-to-A mutation in mtp5-1 is located at position 2003 (where 1 is the A in the initiator ATG) and alters the 3′ splice site of the 7th intron (lower-case letters are intronic bases; capital letters are exonic base pairs). RT-PCR analysis revealed that the 3′ splice site in the mtp5-1 mutant occurs 1 bp 3′ of the wild-type site, resulting in a frameshift and a premature termination codon. (D) Relative expression levels of MTP5 and IAR1 mRNAs in selected Arabidopsis tissues. Compiled microarray data were retrieved from the Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/) in October 2012. Tissues queried were dry and imbibed (24 hr) seeds; cotyledons and hypocotyls from 7-d-old seedlings; roots from 17-d-old seedlings; vegetative rosettes from 14-d-old plants; cauline leaves from 21-d-old plants; senescing leaves from 35-d-old plants; stems from 21-d-old plants; carpels, petals, sepals, stamen, and pedicels from stage 15 flowers; mature pollen; and stage 3 siliques. Error bars show SD of mean expression signals scaled to a target intensity value of 100 for each gene.
To determine the molecular consequences of the mtp5-1 mutation, we isolated RNA from mtp5-1 plants and reverse-transcribed and PCR-amplified the mtp5-1 locus. Sequencing the RT-PCR product revealed that the mtp5-1 coding sequence is spliced 1 nucleotide after wild type, creating a frameshift and a premature stop codon nine codons after the splicing mutation (Figure 1C). The mtp5-1 protein, if stable, would thus only encode five of the six conserved transmembrane domains (Figure 2), presumably precluding transport function.
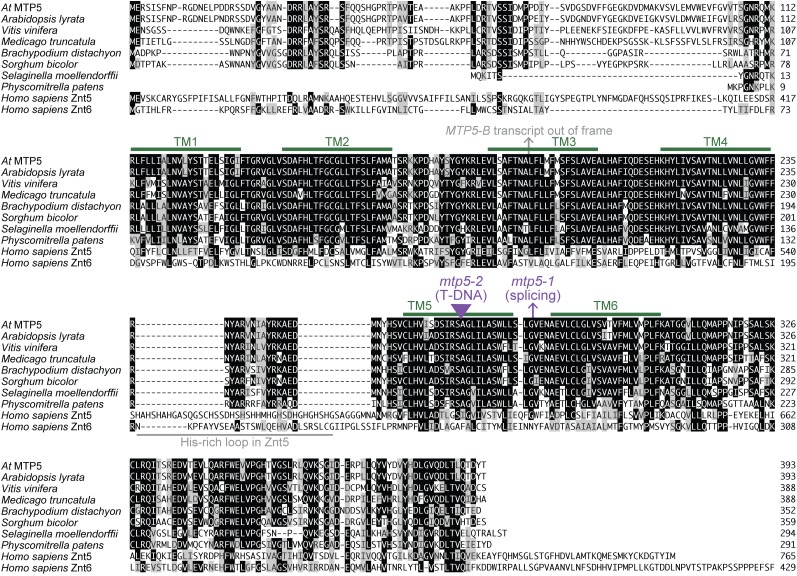
Figure 2 .
Alignment of Arabidopsis MTP5 and related proteins. Arabidopsis thaliana MTP5 (At3g12100; AAT44130.1) was aligned with likely orthologs from other plants (Arabidopsis lyrata XP_002884882.1, Vitis vinifera XP_002279787.1, Medicago truncatula XP_003625268.1, Sorghum bicolor XP_002453105.1, Brachypodium distachyon XP_003570736.1, Selaginella moellendorffii XP_002982633.1, Physcomitrella patens XP_001755969.1) and the human ZnT5 (Homo sapiens NP_075053.2; residues 296-765 of 765 residues) and ZnT6 (Homo sapiens NP_001180442; residues 1-429 of 501 residues) zinc transporters using the MegAlign program (DNAStar) using the Clustal W method. Residues identical in at least five sequences are shaded in black boxes; chemically similar residues in at least five sequences are shaded in gray boxes. Potential transmembrane (TM) domains in Arabidopsis thaliana MTP5 predicted with Aramemnon (Schwacke et al. 2003) are marked with green lines; the His-rich loop found in Znt5 but not MTP5 orthologs is marked by a gray line. The position of the alternative splicing events that would lead to an out-of-frame sequence followed by premature termination codons caused by the mtp5-1 mutation or detected in wild-type mRNA (the MTP5-B transcript) are indicated by arrows. The position of the mtp5-2 T-DNA disruption is indicated with a triangle.
To determine whether MTP5 was expressed in similar tissues as IAR1, we examined publicly available microarray datasets (Winter et al. 2007). We found that MTP5, like IAR1, was widely present at similar levels across many tissues (Figure 1D), consistent with the possibility that the MTP5 and IAR1 proteins might influence similar functions.
mtp5 restores IAA-conjugate sensitivity to iar1
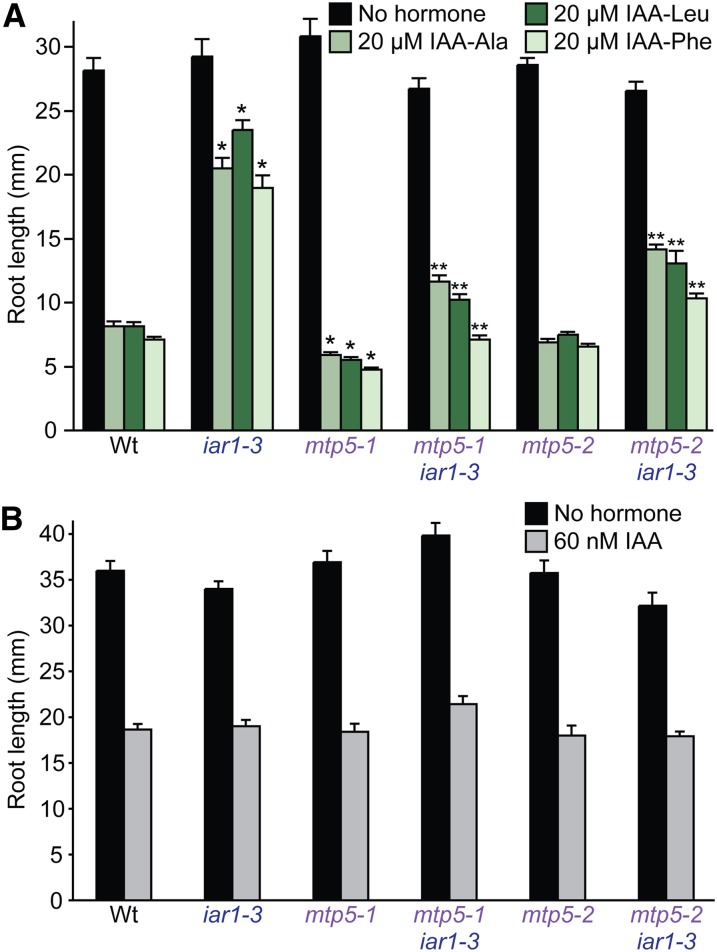
iar1 mutants are resistant to a variety of IAA-amino acid conjugates, including IAA-Ala, IAA-Leu, and IAA-Phe (Lasswell et al. 2000). We found that the mtp5-1 mutation was recessive (data not shown) and restored iar1 root sensitivity not only to IAA-Ala, which we used to isolate the mutant, but also to IAA-Leu and IAA-Phe (Figure 3A). This suppression did not result from generalized auxin resistance because iar1-3 mtp5-1 seedling roots retained wild-type sensitivity to the inhibitory effects of IAA on root elongation (Figure 3B). Beyond restored IAA-amino acid conjugate sensitivity, iar1-3 mtp5-1 seedlings did not exhibit obvious morphological abnormalities and displayed wild-type numbers of lateral roots and normal hypocotyl lengths (data not shown).
Figure 3 .
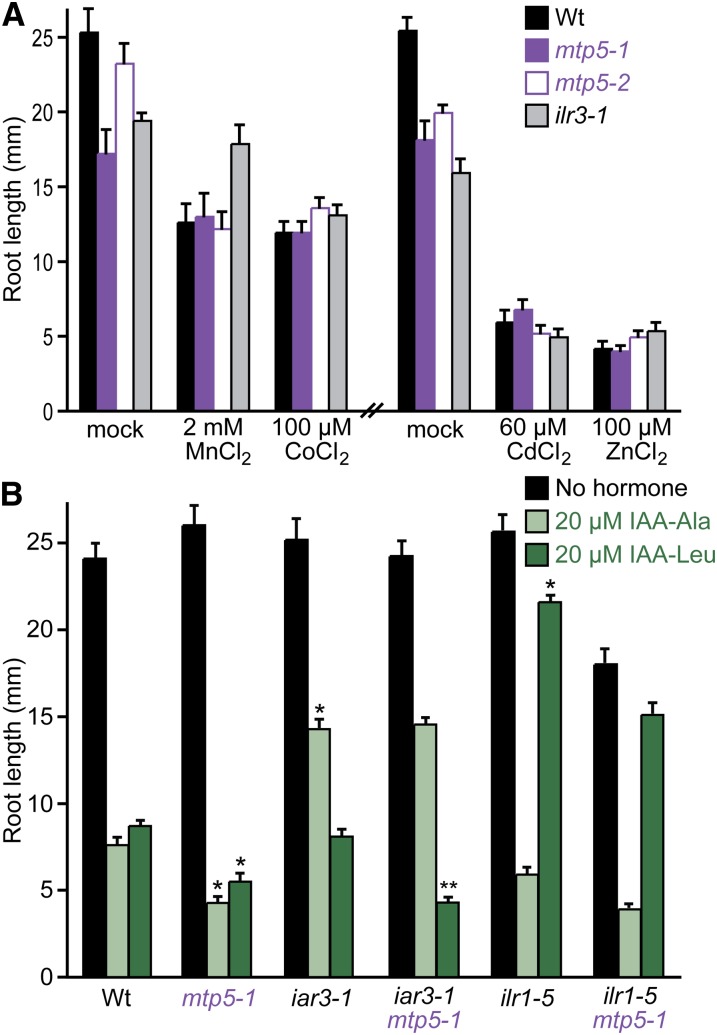
(A) mtp5 mutations restore IAA-conjugate sensitivity to iar1 roots. Col-0 (Wt), iar1-3, mtp5-1, mtp5-2, mtp5-1 iar1-3, and mtp5-2 iar1-3 seedlings were grown on unsupplemented medium or medium containing 20 µM IAA-Ala, IAA-Leu, or IAA-Phe. (B) Like iar1, mtp5, and mtp5 iar1 mutants respond normally to IAA. Seedlings listed in (A) were grown on unsupplemented medium or medium containing 60 nM IAA. Seedlings were grown in constant light under yellow filters for 8 (A) or 9 (B) d at 22°. Error bars indicate standard errors of the mean root lengths (n = 12). Single asterisks indicate single mutant root lengths significantly different from Wt; double asterisks indicate double mutant root lengths significantly different from iar1-3 (two-tailed t-tests; P < 0.001).
Interestingly, when the mtp5-1 mutation was removed from the iar1 mutant background, mtp5-1 IAR1 seedlings were slightly more sensitive to root elongation inhibition by IAA conjugates than wild-type seedlings (Figure 3A). The single mutant was not more sensitive to free IAA (Figure 3B), however, again indicating that MTP5 specifically dampens responsiveness to IAA-amino acid conjugates.
Alternative splicing of MTP5 transcripts
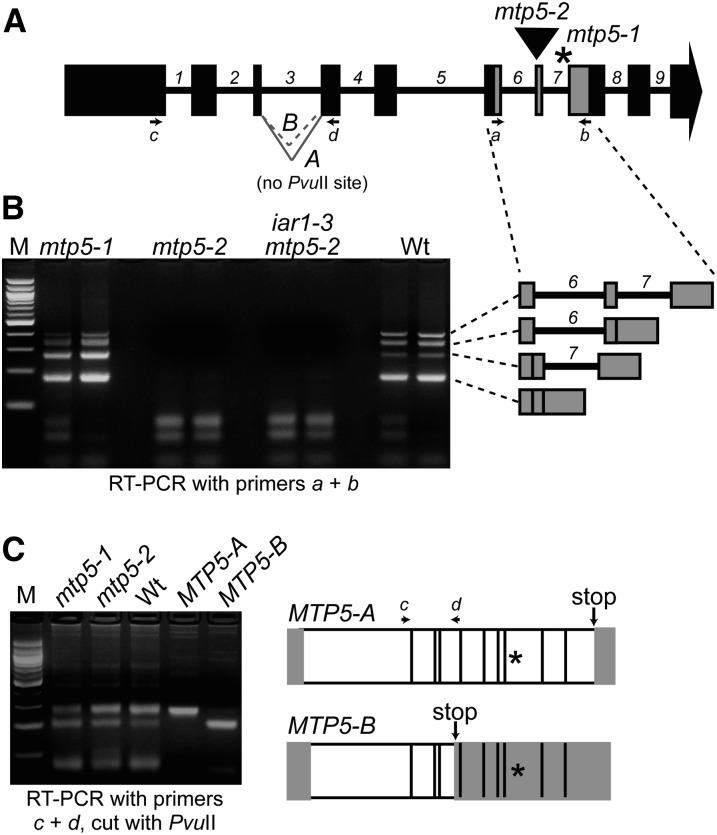
We obtained a second mtp5 allele, designated mtp5-2, from the GABI-Kat collection of sequence-indexed T-DNA insertion mutants (Rosso et al. 2003). Sequencing the insertion site revealed that the T-DNA was inserted in the seventh MTP5 exon (Figure 4A) near the site of the mtp5-1 lesion (Figure 2), and RT-PCR analysis confirmed that the mtp5-2 allele lacked intact MTP5 RNA (Figure 4B).
Figure 4 .
Alternative splicing of MTP5 transcripts. (A) Model of MTP5 gene showing exons (boxes), introns (numbered), positions of mtp5 mutations (asterisk, mtp5-1; triangle, mtp5-2), primers used for RT-PCR amplification (arrows a-d), and the A and B MTP5 splice variants of intron 3. (B) mtp5-2 seedlings lack intact MTP5 transcript, and mtp5-1 and wild-type seedlings inefficiently splice MTP5 introns 6 and 7. MTP5 transcripts were detected via RT-PCR amplification using gene-specific primers (A and B) flanking both mtp5 mutations with RNA isolated from 7-d-old mtp5-1, mtp5-2, iar1-3 mtp5-2, and Col-0 (Wt) seedlings. Adjacent lanes show PCR-amplification products of cDNA from two independent reverse transcription reactions using the same RNA. The identities of the four amplicons from the RT-PCR analysis using primers a and b were determined by sequencing and are shown to the right in gray. (C) mtp5-1 and wild-type seedlings alternatively splice MTP5 intron 3. Using gene-specific primers c and d (A), MTP5 transcripts were amplified from cDNA reverse-transcribed from Col-0 (Wt), mtp5-1, and mtp5-2 RNA. RT-PCR analysis revealed two transcripts in wild type and the two mutant alleles, designated MTP5-A and MTP5-B. Digesting the PCR products with PvuII distinguished MTP5-A, which lacks a PvuII site, from MTP5-B, which includes a PvuII site (agarose gel image in C). Cloned MTP5-A and MTP5-B cDNAs were included as positive controls. The transcripts were differently spliced at the 3′ end of the third intron, resulting in a premature stop codon (arrows) in MTP5-B. Asterisks mark the position of the mtp5-1 mutation. White boxes indicate the open reading frame of the resultant transcripts, and gray boxes show predicted untranslated regions.
In the course of analyzing mtp5-2 RNA, we found that the MTP5 introns 6 and 7 were inefficiently spliced in both wild-type and mtp5-1 seedlings. In addition to fully spliced product, we detected partially processed MTP5 mRNA (Figure 4B). Interestingly, the partially spliced transcript that retained the 7th intron appeared to accumulate more in mtp5-1 than in wild type (Figure 4B), providing evidence that the mtp5-1 splice site mutation at the 3′ end of intron 7 (Figure 1C) affects not only the position, but also the efficiency of MTP5 transcript splicing.
In addition to the inefficient splicing of MTP5 introns 6 and 7, we found that intron 3 was alternatively spliced in both wild type and mtp5 mutants. We detected two fully spliced MTP5 transcripts, MTP5-A and MTP5-B, in 7-d-old seedlings; MTP5-A was more abundant than MTP5-B (Figure 4C). These transcripts used different 3′ splice sites of the third intron (Figure 4C); the MTP5-B transcript would truncate the encoded protein in the third predicted transmembrane domain (Figure 2). Because the stop codon in MTP5-B is at a position prior to the mtp5-1 mutation in intron 7, we concluded that it is the MTP5-A transcript that is defective in mtp5-1.
Characterization of a second mtp5 allele
To confirm that the mutation that we identified in mtp5-1 caused the observed suppression of the IAA-conjugate resistance of iar1, we compared phenotypes of our original mtp5-1 splicing allele and the mtp5-2 T-DNA insertion allele. We crossed mtp5-2 to the iar1-3 mutant to determine whether this allele also restored IAA-conjugate sensitivity to iar1. Indeed, iar1-3 mtp5-2 seedlings were significantly more sensitive to IAA-amino acid conjugates than iar1-3 (Figure 3A), confirming that the mtp5-1 splice-site mutation is a loss-of-function allele and that reducing MTP5 function can compensate for the lack of IAR1 activity in an iar1 mutant. Unlike mtp5-1 (Figure 3A and 5B), mtp5-2 in an otherwise wild-type background was not significantly hypersensitive to IAA conjugates (Figure 3A).
Figure 5 .
mtp5 mutations do not alter metal sensitivity or restore IAA-amino acid sensitivity to IAA-conjugate hydrolase mutants. (A) mtp5 mutant roots respond normally to inhibitory concentrations of various metals. Col-0 (Wt), mtp5-1, mtp5-2, and ilr3-1 seedlings were grown on unsupplemented medium or medium containing 2 mM MnCl2, 100 µM CoCl2, 60 µM CdCl2, or 100 µM ZnSO4 in constant light under yellow filters for 8 d at 22°. Error bars indicate standard errors of the mean root lengths (n ≥ 9). (B) mtp5-1 does not suppress the IAA-conjugate resistance of mutants defective in IAA-amino acid hydrolases. Col-0 (Wt), iar3-1, ilr1-5, mtp5-1, iar3-1 mtp5-1, and mtp5-1 ilr1-5 seedlings were grown on unsupplemented medium or medium containing 20 µM IAA-Ala or IAA-Leu in constant light under yellow filters for 8 d at 22°. Error bars indicate SE of the mean root lengths (n ≥ 9). Single asterisks indicate single mutant root lengths significantly different from Wt; double asterisks indicate iar3-1 mtp5-1 double mutant root lengths significantly different from iar1-3 (two-tailed t-tests; P < 0.001). Significance was not calculated for ilr1-5 mtp5-1 differences because the double mutant roots were shorter than the single mutant roots on unsupplemented medium.
Because MTP5 is a metal transporter homolog, we examined mtp5 sensitivity to various metals in root growth inhibition assays. Unlike the ilr3-1 mutant, which is specifically resistant to Mn2+ (Rampey et al. 2006), both mtp5-1 and mtp5-2 (in the wild-type IAR1 background) displayed responses similar to wild type to all of the metals tested, including Mn2+, Zn2+, Co2+, and Cd2+ (Figure 5A). Moreover, we found that driving expression of the MTP5-A cDNA in wild-type Col-0 seedlings behind the strong CaMV 35S promoter did not alter seedling root lengths on medium supplemented with one of several metals, including Mn2+, Zn2+, Co2+, and Cd2+ (data not shown).
mtp5-1 does not suppress other IAA-conjugate sensitivity mutants
To determine whether mtp5-1 could suppress the IAA-conjugate resistance of mutants in addition to iar1, we constructed double mutants of mtp5-1 with other IAA-conjugate resistant mutants, including the IAA-amino acid conjugate amidohydrolase mutants iar3-1 (Davies et al. 1999) and ilr1-5 (Rampey et al. 2006), which are resistant to the inhibitory effects of IAA conjugates that are substrates of the mutated hydrolases. For example, iar3 is resistant to IAA-Ala and not IAA-Leu and IAR3 cleaves IAA-Ala more efficiently than IAA-Leu in vitro (Davies et al. 1999), whereas ilr1 is resistant to IAA-Leu and not IAA-Ala and ILR1 cleaves IAA-Leu more efficiently than IAA-Ala in vitro (Bartel and Fink 1995). We examined root lengths of each double mutant after growth on IAA-amino acid conjugates. We found that mtp5-1 failed to restore ilr1-5 sensitivity to IAA-Leu or iar3-1 sensitivity to IAA-Ala, and that the heightened sensitivity of mtp5-1 to IAA-Leu was retained in the iar3-1 mtp5-1 double mutant (Figure 5B). This lack of suppression of the amidohydrolase mutants establishes that mtp5-1 is not a general suppressor of all conjugate response mutants. Moreover, the epistatic relationship of these mutants demonstrates that the increased IAA-conjugate sensitivity of the mtp5-1 mutant requires intact conjugate hydrolases with specificity for the indicated IAA-amino acid.
Discussion
Plant cation diffusion facilitator (CDF) transporters
We isolated an extragenic suppressor of the IAA-conjugate response mutant iar1 and found that the defective gene encodes MTP5, a previously uncharacterized member of the CDF transporter family found in bacteria, yeast, animals, and plants. Plant CDF proteins usually have six predicted transmembrane domains (Figure 2) flanked by an N-terminal signature sequence and a C-terminal cation efflux domain (Mäser et al. 2001). There are 12 members of this family in Arabidopsis. Phylogenetic analysis and substrate specificity of characterized members separates the CDF family into three groups: Mn-CDF, Fe/Zn-CDF, and Zn-CDF (Montanini et al. 2007; Gustin et al. 2011). MTP5 is in the Zn-CDF family; all biochemically-characterized members of this subfamily transport at least zinc (Montanini et al. 2007).
Although none are functionally characterized, MTP5 has apparent orthologs in a variety of plants, including Medicago, Brachypodium, Sorghum, Selaginella, and Physcomitrella (Figure 2), suggesting an early emergence of MTP5 during the evolution of land plants. These proteins are between 63 and 73% identical to A. thaliana and A. lyrata MTP5, which are 96% identical to each other. Less similarity is present in the region N-terminal to the first predicted transmembrane domain, which is missing in the MTP5 orthologs from Selaginella and Physcomitrella. The plant MTP5 homologs lack the His-rich loop found between transmembrane domains four and five in some zinc transporters, such as human ZnT5 (Figure 2) and Arabidopsis MTP1 and MTP3 (Kawachi et al. 2012).
Of the 12 Arabidopsis MTP (CDF) proteins, six are in the Zn-CDF subfamily (Montanini et al. 2007; Gustin et al. 2011). This subfamily includes the first characterized plant MTP protein, Arabidopsis ZAT (Van Der Zaal et al. 1999), now designated MTP1 (Mäser et al. 2001). MTP1 transports Zn2+ when heterologously expressed in E. coli proteoliposomes (Bloß et al. 2002), mtp1 mutant roots accumulate less Zn2+ in vacuolar-like organelles (Kawachi et al. 2009), and transgenic plants overexpressing MTP1 hyperaccumulate Zn2+ and have decreased Zn2+ sensitivity (Van Der Zaal et al. 1999). In contrast, we found no alterations in Zn2+ sensitivity when MTP5 was mutated or overexpressed.
Characterized AtMTP1 homologs in other plant species all are proposed to sequester metals in intracellular compartments or catalyze metal efflux from cells. For example, the Thalspi caerulescens (Tc) ZTP1 gene is induced when soil is enriched with Zn2+, Cd2+, or Pb2+ (Assuncao et al. 2001). Heterologous expression of an AtMTP1 homolog from the tropical legume Stylosanthes hamata, ShMTP1, confers Mn2+ tolerance to yeast and Arabidopsis (Delhaize et al. 2003). ShMTP1-GFP fusions localize to plant tonoplasts and the yeast ER (Delhaize et al. 2003). Similarly, heterologous expression of PtdMTP1 from poplar (Populus trichocarpa x P. deltoids) confers Zn2+ resistance to yeast and Arabidopsis, and PtdMTP1-GFP fusions are vacuolar in both yeast and Arabidopsis (Blaudez et al. 2003). Interestingly, the Thalspi goesingense TgMTP1 gene gives rise to three transcript variants, TgMTP1a, TgMTP1b, and TgMTP1c (Kim et al. 2004). Expression of TgMTP1a in yeast confers resistance to Cd2+, Co2+, and Zn2+, whereas TgMTP1b expression confers resistance to Ni2+ (Persans et al. 2001). TgMTP1b localizes to the plasma membrane of Arabidopsis leaf protoplasts and may efflux Zn2+ from cells (Kim et al. 2004). Although we also found evidence for alternative splicing of Arabidopsis MTP5 (Figure 4), only one of these splice products (MTP5-A) is likely to encode an intact transporter.
Several MTP1 proteins have been tested for complementation of the yeast mutant cot1 zrc1, which is defective in vacuolar metal efflux (MacDiarmid et al. 2000). The three TgMTP1 isoforms, TcZTP1, Thaspi arvense MTP1, Thaspi montanum var. fendleri MTP1, Arabidopsis lyrata MTP1, AtMTP1, and PtdMTP1 all confer Zn2+ resistance when expressed in yeast (Blaudez et al. 2003; Kim et al. 2004). However, the authors of a previous study found that AtMTP1 did not complement the cot1 zrc1 mutant (Bloß et al. 2002). Expression of TgMTPb truncations containing the N-terminus, C-terminus, the putative metal-binding His-rich domain, or various combinations of these domains do not complement the cot1 zrc1 mutant, suggesting that the Zn2+ resistance that accompanies TgMTPb overexpression is related to transport and not due to binding excess Zn2+ by the His-rich domain. We transformed the MTP5 cDNA driven by the glyceraldehyde-3-phosphate dehydrogenase constitutive promoter into wild-type and cot1 and zrc1 single and double mutant yeast lines but did not observe changes in relative growth in the presence of Zn2+ or Co2+ (data not shown). It is possible that Arabidopsis MTP5 does not function in yeast membranes. Alternatively, because the localization and identity of metals potentially transported by MTP5 are not known, this lack of complementation may be due to differences in location or metal specificity among MTP5 and the yeast Cot1p and Zrc1p proteins.
An Arabidopsis cation diffusion facilitator protein mutant
Beyond the suppression of the IAA-conjugate resistance of iar1, mtp5-1, and mtp5-2 plants resembled wild type as seedlings and adults (Figures 3 and 5 and data not shown). The mtp5 mutants resembled wild type on media containing metals under all conditions tested (Figure 5A). In contrast, mtp1 mutants have increased Zn2+ sensitivity (Kobae et al. 2004; Kawachi et al. 2009), and mtp11 mutants (defective in a member of the Mn-CDF family) are more sensitive to Mn2+ (Delhaize et al. 2007; Peiter et al. 2007). RNAi lines silencing MTP1 or MTP3 also show Zn2+ hypersensitivity (Desbrosses-Fonrouge et al. 2005; Arrivault et al. 2006). It is possible that other transporters partially compensate for any loss of transport activity in the mtp5 mutants. In this case, the mtp5 mutation may modify subcellular metal concentration(s) adequately to compensate for the presumed transport defect in iar1 seedlings, but not alter levels enough to cause toxic metal accumulation that would increase sensitivity to exogenous metals.
A working model for mtp5 suppression of iar1
In eukaryotes, Zn2+ transport into or out of the cytosol occurs through the opposing action of two protein families, the ZIPs and CDFs (Kambe et al. 2004, 2006). ZIP proteins transport Zn2+ from outside cells and from intracellular compartments into the cytosol. In an opposing manner, CDFs transport Zn2+ out of the cytosol and into extracellular space or intracellular compartments. Homology thus suggests that the MTP5 CDF protein effluxes metals out of cells or into a subcellular compartment, whereas the IAR1 ZIP protein is predicted to move metals into cells or out of a subcellular compartment.
The mutant phenotypes and predicted protein functions suggest that MTP5 and IAR1 transport metals in an antagonistic fashion to regulate metal homeostasis, which in turn influences IAA-amino acid conjugate hydrolysis by compartmentalized enzymes (Figure 6). Based on the membrane topology of IAR1 homologs, one possibility is that IAR1 transports metals inhibitory to IAA-amino acid hydrolase function out of the compartment in which the hydrolases reside (Figure 6A). This compartment is likely to be the ER, because the yeast IAR1 ortholog Yke4p is ER-localized (Kumánovics et al. 2006), the hydrolases contain predicted N-terminal signal sequences and C-terminal ER retention signals (Bartel and Fink 1995; Davies et al. 1999), and both IAR3 and ILR1 have been localized to the ER in proteomics experiments (Dunkley et al. 2006). Metals impact hydrolase activity; for example, the in vitro activities of the IAA-conjugate hydrolases are enhanced by Mn2+, and ILR1 activity is inhibited by Zn2+ (LeClere et al. 2002). Moreover, the crystal structure of the Arabidopsis ILR1-like homolog ILL2 reveals a di-metal binding site in the hydrolase active site that is typical of the M20 metallopeptidases family (Bitto et al. 2009). The IAA-conjugate resistance of iar1 may result from an aberrant accumulation of inhibitory metals in the hydrolase compartment, which decreases hydrolysis of and therefore sensitivity to IAA-amino acid conjugates (Figure 6B). If MTP5 transports inhibitory metals into the hydrolase-containing compartment, an mtp5 iar1 mutant would no longer be IAA-conjugate resistant because inhibitory metals would not accumulate, thus restoring wild-type levels of IAA-conjugate hydrolysis (Figure 6C). This model is consistent with our observation that double mutants defective in both MTP5 and the ILR1 or IAR3 hydrolase genes remain resistant to IAA-Leu or IAA-Ala (Figure 5B), respectively, indicating that MTP5 functions upstream of the hydrolases.
Figure 6 .
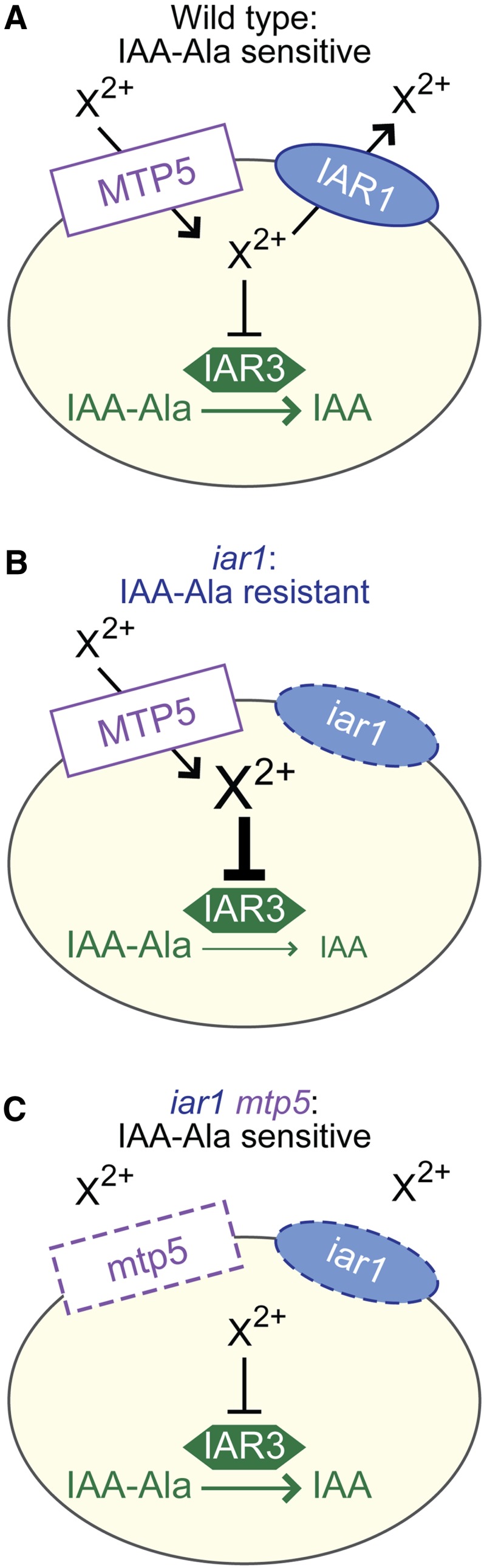
A working model for MTP5 and IAR1 function in IAA-conjugate responses in wild type (A), an iar1 mutant (B), and an iar1 mtp5 double mutant (C). The presence of IAA-amino acid hydrolases in the ER, the localization and function of MTP5 and IAR1 orthologs, and the iar1 and mtp5 mutant phenotypes (see text for details) suggest that MTP5 and IAR1 may transport Zn2+ or other metal(s) that inhibit hydrolase function into and out of, respectively, the ER. Validation or rejection of this model awaits localization of the MTP5 and IAR1 transporters and determination of their substrate specificities.
This compensatory model is consistent with the function of yeast transporters similar to IAR1 and MTP5. When yeast are grown in low zinc medium, the IAR1 ortholog Yke4p moves zinc from the secretory pathway into the cytosol (Kumánovics et al. 2006). Intriguingly, loss of Yke4 can compensate for the loss of the Msc2p (Kumánovics et al. 2006), a CDF protein in the MTP5 subfamily that transports zinc into the ER (Li and Kaplan 2001; Ellis et al. 2005).
Whereas the model depicted in Figure 6 suggests that IAR1 and MTP5 occupy the same membrane, it is also possible that either or both MTP5 and IAR1 are located in a compartment that does not contain the hydrolases. When metal homeostasis is disrupted within this compartment, it may indirectly affect the metal environment of the ER, thus indirectly affecting IAA-conjugate hydrolysis. Distinguishing between these possibilities will be aided by characterization of the subcellular localizations and metal transport specificities of the IAR1 and MTP5 proteins. Regardless of the subcellular localization of IAR1 and MTP5, however, it is clear that genetic studies of IAA-amino acid conjugate responsiveness are exquisitely sensitive to uncovering factors contributing to metal homeostasis. As this screen has not reached saturation, it is possible that continued studies will reveal additional components necessary to control the metal environment in which the hydrolases function.
Interestingly, CDF metal transporters may directly interact with a component from a signaling pathway to modify the binding partner’s activity (Jirakulaporn and Muslin 2004). The C-terminal intracellular portions of mammalian Znt1 and worm CDF-1 bind the amino-terminal regulatory region of Raf-1, a protein kinase that regulates cell proliferation and differentiation (Jirakulaporn and Muslin 2004). Further, Raf-1 activity is dependent on Znt1 function, and in vitro binding does not occur upon addition of Zn2+ (Jirakulaporn and Muslin 2004). Some CDF transporters also form heterodimers, including yeast Msc2p and Zrg17p (Ellis et al. 2005) and the corresponding mammalian homologs ZnT5 and ZnT6 (Ellis et al. 2005; Fukunaka et al. 2009). Identification of Arabidopsis MTP5 binding partners may further elucidate the relationship between metal homeostasis and IAA-conjugate sensitivity.
Acknowledgments
We thank Gretchen Spiess, Andrew Woodward, and Bethany Zolman for comments on the manuscript and the Arabidopsis Biological Resource Center for seeds. This research was funded by the Robert A. Welch Foundation (C-1309) and a Howard Hughes Medical Institute Professors Grant (52005717).
Footnotes
Communicating editor: M. Estelle
Literature Cited
- Arrivault S., Senger T., Kramer U., 2006. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46: 861–879 [DOI] [PubMed] [Google Scholar]
- Assuncao A. G. L., Martins P. D., De Folter S., Vooijs R., Schat H., et al. , 2001. Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 24: 217–226 [Google Scholar]
- Ausubel F., Brent R., Kingston R. E., Moore D. D., Seidman J. G., et al. , 1999. Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York [Google Scholar]
- Bartel B., Fink G. R., 1995. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748 [DOI] [PubMed] [Google Scholar]
- Bell C. J., Ecker J. R., 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Bitto E., Bingman C. A., Bittova L., Houston N. L., Boston R. S., et al. , 2009. X-ray structure of ILL2, an auxin-conjugate amidohydrolase from Arabidopsis thaliana. Proteins 74: 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaudez D., Kohler A., Martin F., Sanders D., Chalot M., 2003. Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 15: 2911–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloß T., Clemens S., Nies D. H., 2002. Characterization of the ZAT1p zinc transporter from Arabidopsis thaliana in microbial model organisms and reconstituted proteoliposomes. Planta 214: 783–791 [DOI] [PubMed] [Google Scholar]
- Chang H. C., Lindquist S., 1994. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J. Biol. Chem. 269: 24983–24988 [PubMed] [Google Scholar]
- Cheng N. H., Pittman J. K., Barkla B. J., Shigaki T., Hirschi K. D., 2003. The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15: 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S., Palmgren M. G., Kramer U., 2002. A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci. 7: 309–315 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davies R. T., Goetz D. H., Lasswell J., Anderson M. N., Bartel B., 1999. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Kataoka T., Hebb D. M., White R. G., Ryan P. R., 2003. Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15: 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Gruber B. D., Pittman J. K., White R. G., Leung H., et al. , 2007. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 51: 198–210 [DOI] [PubMed] [Google Scholar]
- Desbrosses-Fonrouge A. G., Voigt K., Schroder A., Arrivault S., Thomine S., et al. , 2005. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 579: 4165–4174 [DOI] [PubMed] [Google Scholar]
- Dunkley T. P., Hester S., Shadforth I. P., Runions J., Weimar T., et al. , 2006. Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D. J., Broderius M., Fett J., Guerinot M. L., 1996. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C. D., MacDiarmid C. W., Eide D. J., 2005. Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J. Biol. Chem. 280: 28811–28818 [DOI] [PubMed] [Google Scholar]
- Fukunaka A., Suzuki T., Kurokawa Y., Yamazaki T., Fujiwara N., et al. , 2009. Demonstration and characterization of the heterodimerization of ZnT5 and ZnT6 in the early secretory pathway. J. Biol. Chem. 284: 30798–30806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaither L. A., Eide D. J., 2000. Functional expression of the human hZIP2 zinc transporter. J. Biol. Chem. 275: 5560–5564 [DOI] [PubMed] [Google Scholar]
- Gietz R., Schiestl R., 1995. Transforming yeast with DNA. Methods Mol. Cell. Biol. 5: 255–269 [Google Scholar]
- Gollhofer J., Schlawicke C., Jungnick N., Schmidt W., Buckhout T. J., 2011. Members of a small family of nodulin-like genes are regulated under iron deficiency in roots of Arabidopsis thaliana. Plant Physiol. Biochem. 49: 557–564 [DOI] [PubMed] [Google Scholar]
- Gustin J. L., Zanis M. J., Salt D. E., 2011. Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol. Biol. 11: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. L., Williams L. E., 2003. Transition metal transporters in plants. J. Exp. Bot. 54: 2601–2613 [DOI] [PubMed] [Google Scholar]
- Haughn G. W., Somerville C., 1986. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434 [Google Scholar]
- Huang L., Kirschke C. P., Zhang Y., Yu Y. Y., 2005. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J. Biol. Chem. 280: 15456–15463 [DOI] [PubMed] [Google Scholar]
- Jirakulaporn T., Muslin A. J., 2004. Cation diffusion facilitator proteins modulate Raf-1 activity. J. Biol. Chem. 279: 27807–27815 [DOI] [PubMed] [Google Scholar]
- Kambe T., Yamaguchi-Iwai Y., Sasaki R., Nagao M., 2004. Overview of mammalian zinc transporters. Cell. Mol. Life Sci. 61: 49–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambe T., Suzuki T., Nagao M., Yamaguchi-Iwai Y., 2006. Sequence similarity and functional relationship among eukaryotic ZIP and CDF transporters. Genomics Proteomics Bioinformatics 4: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi M., Kobae Y., Mori H., Tomioka R., Lee Y., et al. , 2009. A mutant strain Arabidopsis thaliana that lacks vacuolar membrane zinc transporter MTP1 revealed the latent tolerance to excessive zinc. Plant Cell Physiol. 50: 1156–1170 [DOI] [PubMed] [Google Scholar]
- Kawachi M., Kobae Y., Kogawa S., Mimura T., Kramer U., et al. , 2012. Amino acid screening based on structural modeling identifies critical residues for the function, ion selectivity and structure of Arabidopsis MTP1. FEBS J. 279: 2339–2356 [DOI] [PubMed] [Google Scholar]
- Kim D., Gustin J. L., Lahner B., Persans M. W., Baek D., et al. , 2004. The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. Plant J. 39: 237–251 [DOI] [PubMed] [Google Scholar]
- Kobae Y., Uemura T., Sato M. H., Ohnishi M., Mimura T., et al. , 2004. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in metal homeostasis. Plant Cell Physiol. 45: 1749–1758 [DOI] [PubMed] [Google Scholar]
- Koncz C., Schell J., Rédei G. P., 1992. T-DNA transformation and insertion mutagenesis, pp. 224–273 in Methods in Arabidopsis Research, edited by Koncz C., Chua N.-H., Schell J. World Scientific, Singapore [Google Scholar]
- Kumánovics A., Poruk K. E., Osborn K. A., Ward D. M., Kaplan J., 2006. YKE4 (YIL023C) encodes a bidirectional zinc transporter in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 281: 22566–22574 [DOI] [PubMed] [Google Scholar]
- Lasswell J., 2000. Genetic Analyses of Auxin Metabolism and of the Transition to Flowering in the Model Plant Arabidopsis thaliana Dept. of Biochemistry & Cell Biology, Rice University, Houston, TX [Google Scholar]
- Lasswell J., Rogg L. E., Nelson D. C., Rongey C., Bartel B., 2000. Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. Plant Cell 12: 2395–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last R. L., Fink G. R., 1988. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240: 305–310 [DOI] [PubMed] [Google Scholar]
- LeClere S., Bartel B., 2001. A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol. Biol. 46: 695–703 [DOI] [PubMed] [Google Scholar]
- LeClere S., Tellez R., Rampey R. A., Matsuda S. P. T., Bartel B., 2002. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277: 20446–20452 [DOI] [PubMed] [Google Scholar]
- Li L., Kaplan J., 2001. The yeast gene MSC2, a member of the cation diffusion facilitator family, affects the cellular distribution of zinc. J. Biol. Chem. 276: 5036–5043 [DOI] [PubMed] [Google Scholar]
- Li X., Chanroj S., Wu Z., Romanowsky S. M., Harper J. F., et al. , 2008. A distinct endosomal Ca2+/Mn2+ pump affects root growth through the secretory process. Plant Physiol. 147: 1675–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid C. W., Gaither L. A., Eide D. J., 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19: 2845–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidin M., Pittman J. K., Hirschi K. D., Bartel B., 2003. ILR2, a novel gene regulating IAA conjugate sensitivity and metal transport in Arabidopsis thaliana. Plant J. 35: 523–534 [DOI] [PubMed] [Google Scholar]
- Marschner H., 1995. Mineral Nutrition of Higher Plants. Academic Press, San Diego [Google Scholar]
- Mäser P., Thomine S., Schroeder J. I., Ward J. M., Hirschi K., et al. , 2001. Phylogenetic relationships within cation-transporter families of Arabidopsis thaliana. Plant Physiol. 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M., 1998. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14: 381–385 [DOI] [PubMed] [Google Scholar]
- Montanini B., Blaudez D., Jeandroz S., Sanders D., Chalot M., 2007. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M. M., Neff J. D., Chory J., Pepper A. E., 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Normanly J., Grisafi P., Fink G. R., Bartel B., 1997. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E., Montanini B., Gobert A., Pedas P., Husted S., et al. , 2007. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. USA 104: 8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persans M. W., Nieman K., Salt D. E., 2001. Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc. Natl. Acad. Sci. USA 98: 9995–10000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey R. A., Leclere S., Kowalczyk M., Ljung K., Sandberg G., et al. , 2004. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey R. A., Woodward A. W., Hobbs B. N., Tierney M. P., Lahner B., et al. , 2006. An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174: 1841–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M. G., Li Y., Strizhov N., Reiss B., Dekker K., et al. , 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., Van Der Graaff E., Fischer K., Catoni E., et al. , 2003. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos T. C., Hangarter R. P., 1990. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Bartel B., 2011. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. M., Nicholson R. I., 2003. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim. Biophys. Acta 1611: 16–30 [DOI] [PubMed] [Google Scholar]
- Van Der Zaal B. J., Neuteboom L. W., Pinas J. E., Chardonnens A. N., Schat H., et al. , 1999. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 119: 1047–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., et al. , 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A. W., Bartel B., 2005. Auxin: regulation, action, and interaction. Ann. Bot. (Lond.) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]