Abstract
Crossing over between homologous chromosomes occurs during the prophase of meiosis I and is critical for chromosome segregation. In baker’s yeast, two heterodimeric complexes, Msh4-Msh5 and Mlh1-Mlh3, act in meiosis to promote interference-dependent crossing over. Mlh1-Mlh3 also plays a role in DNA mismatch repair (MMR) by interacting with Msh2-Msh3 to repair insertion and deletion mutations. Mlh3 contains an ATP-binding domain that is highly conserved among MLH proteins. To explore roles for Mlh3 in meiosis and MMR, we performed a structure−function analysis of eight mlh3 ATPase mutants. In contrast to previous work, our data suggest that ATP hydrolysis by both Mlh1 and Mlh3 is important for both meiotic and MMR functions. In meiotic assays, these mutants showed a roughly linear relationship between spore viability and genetic map distance. To further understand the relationship between crossing over and meiotic viability, we analyzed crossing over on four chromosomes of varying lengths in mlh3Δ mms4Δ strains and observed strong decreases (6- to 17-fold) in crossing over in all intervals. Curiously, mlh3Δ mms4Δ double mutants displayed spore viability levels that were greater than observed in mms4Δ strains that show modest defects in crossing over. The viability in double mutants also appeared greater than would be expected for strains that show such severe defects in crossing over. Together, these observations provide insights for how Mlh1-Mlh3 acts in crossover resolution and MMR and for how chromosome segregation in Meiosis I can occur in the absence of crossing over.
Keywords: DNA mismatch repair, meiotic recombination, Msh4-Msh5, Mlh1-Mlh3, crossing over
During gametogenesis in most eukaryotes, crossing over between homologous chromosomes occurs during prophase of meiosis I and is critical for both chromosome segregation and exchange of genetic information between homologs (Zickler 2006). Meiotic recombination in Saccharomyces cerevisiae is initiated by the induction of approximately 140−170 SPO11-dependent double-strand breaks (DSBs) that are located throughout the genome (Cao et al. 1990; Gilbertson and Stahl 1996; Keeney et al. 1997; Robine et al. 2007; Chen et al. 2008). Roughly 40% of these DSBs are repaired to form crossovers between homologous chromosomes; the rest are repaired as noncrossovers or by using a sister chromatid as template. DSB resection results in 3′ single-strand tails whose repair is directed primarily to the complementary sequence in the other homolog (Schwacha and Kleckner 1995). The 3′ tails are acted upon by strand exchange enzymes to form single-end invasion intermediates (SEIs). SEIs are subsequently converted into double Holliday junctions (dHJs) that are ultimately resolved into crossovers (Hunter and Kleckner 2001).
Two MutS and MutL homolog (MSH and MLH) complexes, Msh4-Msh5 and Mlh1-Mlh3, respectively, promote crossovers that are nonrandomly spaced (interference-dependent crossover pathway). In this pathway the presence of one crossover decreases the likelihood of another nearby (Kleckner et al. 2004; Stahl et al. 2004; Shinohara et al. 2008). A second, interference-independent crossover pathway is mediated by the endonuclease complex Mus81-Mms4 (Clyne et al. 2003; De Los Santos et al. 2003; Argueso et al. 2004; Matos et al. 2011). Little is known about the intermediates in this pathway; however, the Mus81-Mms4 complex is thought to act directly in Holliday junction resolution or by cleaving D-loops and half-HJ intermediates (Kaliraman et al. 2001; Hollingsworth and Brill 2004; Gaskell et al. 2007). Genetic, biochemical, and physical studies have shown that Msh4-Msh5 acts in meiosis to stabilize SEI and dHJ intermediates (Börner et al. 2004; Snowden et al. 2004; Nishant et al. 2010). Mlh3 was found to coimmunoprecipitate with Msh4, suggesting that the Mlh1-Mlh3 heterodimer interacts with the Msh4-Msh5-DNA complex (Santucci-Darmanin et al. 2002). This interaction is thought to reinforce the crossover decision by providing a substrate for a dHJ resolvase(s) during early- to mid-pachytene stages in meiosis (Wang et al. 1999; Santucci-Darmanin et al. 2002; Hoffman and Borts 2004; Whitby 2005; Nishant et al. 2008). Consistent with these observations are cytological observations showing that ∼140 Msh4-Msh5 foci are present per mouse spermatocyte nucleus in zygotene. The number of Msh4 foci decrease to about two to three foci per chromosome in mid-pachytene. At this stage, Mlh1 foci begin to appear. Initially, there is high (95–100%) colocalization between the two foci; however, as pachytene progresses, this colocalization gradually disappears (Kneitz et al. 2000; Santucci-Darmanin et al. 2000; Svetlanov and Cohen 2004). The presence of a large number of Msh4-Msh5 foci in zygotene supports early roles for Msh4-Msh5 in meiosis, perhaps during initial interhomolog interactions (Storlazzi et al. 2010).
Crossover placement in meiosis is carefully regulated through the Msh4-Msh5 interference pathway and the actions of Sgs1 helicase, which may play a role in promoting crossing over, as well as serve as an anticrossover factor by removing aberrant recombination intermediates (Jessop et al. 2006; Oh et al. 2007; De Muyt et al. 2012; Zakharyevich et al. 2012). Crossover levels also are regulated by a homeostasis mechanism that ensures that when DSB levels are reduced crossovers are maintained at the expense of noncrossovers. This mechanism facilitates proper disjunction of homologs (Martini et al. 2006; Zanders and Alani 2009). At least one crossover per homolog, called the obligate crossover, appears necessary for proper homolog disjunction. Steps that ensure the obligate crossover in the interference-dependent pathway are thought to occur during the crossover/noncrossover decision step, just before single-end invasion (Allers and Lichten 2001; Hunter and Kleckner 2001).
During DNA mismatch repair (MMR), the MSH proteins Msh2-Msh6 and Msh2-Msh3 bind to base−base and insertion/deletion mismatches that form primarily as the result of DNA replication errors (Kunkel and Erie 2005). In the baker’s yeast S. cerevisiae Msh2-Msh6 and Msh2-Msh3 interact primarily with a single MLH complex, Mlh1-Pms1, to reinforce the repair decision and activate downstream excision and resynthesis steps. In addition to its role in meiosis outlined previously, Mlh1-Mlh3 performs a minor role in the repair of insertion and deletions, most likely through interactions with Msh2-Msh3 (Flores-Rozas and Kolodner 1998). Mlh3 contains an ATP-binding domain that is highly conserved among MLH proteins. It also contains an endonuclease domain that is detected in specific classes of MLH proteins [Figure 1 (Kadyrov et al. 2006)]. Previous work from our laboratory indicated that the endonuclease domain present near the C-terminus of Mlh3 is critical for its role in MMR and meiotic crossing over (Nishant et al. 2008).
Figure 1 .
The ATPase domain of Mlh3 is highly conserved across eukaryotic species and within the MLH protein family. (A) Location of the mlh3 mutations analyzed in this study with respect to Homo sapiens, S. cerevisiae, and Mus musculus protein sequences. Conserved residues are highlighted in bold. (B) Location of the mlh3 mutations created with respect to the conserved ATPase domains in the Saccharomyces cerevisiae MLH family of proteins (Ban and Yang 1998; Tran and Liskay 2000). ATPase domain IV is not shown.•, locations of mlh3 alleles analyzed in this study.
In this study we investigated the role of Mlh3 in DNA MMR and meiosis by analyzing the phenotype of eight mlh3 ATPase mutants. Our data suggest that ATP hydrolysis by both Mlh1 and Mlh3 is important for both meiotic and MMR functions. In meiotic assays these mutants showed a roughly linear relationship between spore viability and genetic map distance. To further analyze the role of Mlh3 in meiosis, we analyzed crossing over on four chromosomes in mlh3Δ mms4Δ cells and observed a strong decrease in crossing over at all intervals, but higher spore viability than would be expected for strains that show such strong crossover defects. Together these observations provide insights for how Mlh1-Mlh3 acts in crossover resolution and MMR, and for how chromosome segregation in Meiosis I can occur in the absence of crossing over.
Materials and Methods
Media
S. cerevisiae strains were grown at 30° in either yeast extract-peptone, 2% dextrose media, or minimal selective media (SC) containing 2% dextrose, sucrose, or galactose (Rose et al. 1990). When required for selection, geneticin (Invitrogen, San Diego, CA) and nourseothricin (Werner BioAgents, Jena, Germany) were used at recommended concentrations (Wach et al. 1994; Goldstein and McCusker 1999). Sporulation plates and media were prepared as described in Argueso et al. (2004).
Plasmids and strains
Plasmids containing each of the mlh3 alleles were constructed via QuickChange mutagenesis (Stratagene, La Jolla, CA) using the single-step integration vector pEAI254 as a template. pEAI254 contains the SK1 MLH3 gene with a KANMX4 selectable marker inserted 40 bp downstream of the stop codon (Nishant et al. 2008). Mutations created by QuickChange were confirmed by sequencing (Sanger method) the entire MLH3 open reading frame. Primer sequences used to create the mlh3 alleles are available upon request. pEAI254 and mutant derivatives were digested with BamHI and SalI before introduction into yeast by the lithium acetate transformation method (Gietz et al. 1995). Plasmids used for the dominant-negative assay were constructed by QuickChange mutagenesis using pEAE220 (S288C, GAL10-MLH3, 2μ, URA3) as a template (Nishant et al. 2008). The mutated regions created by QuickChange were subcloned into a new pEAE220 backbone to eliminate other possible mutations.
The SK1 mlh3 alleles described in this study were introduced by gene replacement into SK1 congenic and isogenic strain backgrounds (Tables 1 and 2). The effect of the eight alleles on spore viability and crossing over was measured in EAY1108/1112 [SK1 congenic; Figure 2 (Argueso et al. 2004)]. mlh3msh5 double mutants also were constructed in EAY1108/1112. More specifically, mlh3 alleles were introduced by gene replacement into the msh5Δ MATα strain EAY1279, and msh5 alleles were introduced into the mlh3Δ msh5Δ MATa strain EAY3312. The mlh3Δ and mlh3Δ mms4Δ strains analyzed in Figure 2 were derived from the SK1 isogenic NHY942/NHY943 background (De Los Santos et al. 2003).
Table 1. Yeast strains used in this study.
| Strain | Genotype |
|---|---|
| EAY1062 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14 |
| EAY2186 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, MLH3::KANMX4 |
| EAY2037 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3::KANMX4 |
| EAY3117 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3-E31A::KANMX4 |
| EAY3119 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3-N35A::KANMX4 |
| EAY3121 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3-A41F::KANMX4 |
| EAY3123 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3-G63R::KANMX4 |
| EAY3125 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3-K80E::KANMX4 |
| EAY3127 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3-K83A::KANMX4 |
| EAY3129 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3-R96A::KANMX4 |
| EAY3131 | MATa ho::hisG, ura3, leu2::hisG, ade2::LK, his4xB, lys214::insE-A14, mlh3-G97A::KANMX4 |
| EAY1269 | MATa ura3, leu2, trp1, lys2::insE-A14 |
| EAY1366 | MATa leu2, ura3, trp1, his3, lys2::insE-A14 mlh1Δ::KANMX4 |
| EAY3308 | MATa ura3, leu2, trp1, lys2::insE-A14 w/ pEAE220 (GAL10-MLH3, 2μ) |
| EAY3309 | MATa ura3, leu2, trp1, lys2::insE-A14 w/ pEAE374 (GAL10-mlh3-E31A, 2μ) |
| EAY3310 | MATa ura3, leu2, trp1, lys2::insE-A14 w/ pEAE375 (GAL10-mlh3-R96A, 2μ) |
| EAY3311 | MATa ura3, leu2, trp1, lys2::insE-A14 w/ pEAE376 (GAL10-mlh3-G97A, 2μ) |
| EAY1108 | MATa trp1:hisG leu2::hisG ho::hisG ura3 lys2 URA3insertion@CENXV LEU2insertion@chromXV, LYS2 insertion at position 505193 |
| EAY2413 | Same as EAY1108, but mlh3Δ::NATMX4 |
| EAY3007 | Same as EAY1108, but mlh3-E31A |
| EAY3009 | Same as EAY1108, but mlh3-N35A |
| EAY3011 | Same as EAY1108, but mlh3-A41F |
| EAY3013 | Same as EAY1108, but mlh3-G63R |
| EAY3015 | Same as EAY1108, but mlh3-K80E |
| EAY3017 | Same as EAY1108, but mlh3-K83A |
| EAY3019 | Same as EAY1108, but mlh3-R96A |
| EAY3021 | Same as EAY1108, but mlh3-G97A |
| EAY2423 | Same as EAY1108, but msh5-D76A::KANMX4 |
| EAY2439 | Same as EAY1108, but msh5- T423A::KANMX4 |
| EAY2032 | Same as EAY1108, but mlh3Δ::KANMX4, msh5Δ::NATMX4 |
| EAY1281 | Same as EAY1108, but msh5Δ::NATMX4 |
| EAY1847 | Same as EAY1108, but mlh3Δ::KANMX4 |
| EAY1845 | Same as EAY1108, but mms4Δ::NATMX4 |
| EAY2030 | Same as EAY1108, but mlh3Δ::KANMX4, mms4Δ::NATMX4 |
| EAY3312 | Same as EAY1108, but mlh3Δ::HPHMX4, msh5Δ::NATMX4 |
| EAY3313 | Same as EAY1108, but mlh3Δ::HPHMX4, msh5-D76A::KANMX4 |
| EAY3314 | Same as EAY1108, but mlh3Δ::HPHMX4, msh5-T423A::KANMX4 |
| EAY1112 | MATα ura3, trp1::hisG, leu2::hisG, lys2, ho::hisG, ade2::hisG, his3Δ::hisG, TRP1insertion@CENXV |
| EAY1848 | Same as EAY1112, but mlh3Δ::KANMX4 |
| EAY1846 | Same as EAY1112, but mms4Δ::NATMX4 |
| EAY1279 | Same as EAY1112, but msh5Δ::NATMX4 |
| EAY2031 | Same as EAY1112, but mlh3Δ::KANMX4, mms4Δ::NATMX4 |
| EAY2033 | Same as EAY1112, but mlh3Δ::KANMX4, msh5Δ::NATMX4 |
| EAY3315 | Same as EAY1112, but mlh3-R96A::KANMX4, msh5Δ::NATMX4 |
| EAY3316 | Same as EAY1112, but mlh3-G97A::KANMX4, msh5Δ::NATMX4 |
| EAY1425/NHY942 | MATα ho::hisG ade2Δ can1 ura3(ΔSma-Pst) met13-B trp5-S CENVIII::URA3 thr1-A cup1s |
| EAY2904 | Same as EAY1425, but mlh3Δ::KANMX4 |
| EAY3290 | Same as EAY1425, but mms4Δ::KANMX4 |
| EAY3296 | Same as EAY1425, but mlh3Δ::KANMX4 mms4Δ::KANMX4 |
| EAY1426/NHY943 | MATa ho::hisG ade2Δ ura3(ΔSma-Pst) leu2::hisG CENIII::ADE2 lys5-P his4-B cyh2 |
| EAY2906 | Same as EAY1426, but mlh3Δ::KANMX4 |
| EAY3323 | Same as EAY1426, but mms4Δ::NATMX4 |
| EAY3298 | Same as EAY1426, but mlh3Δ::KANMX4 mms4Δ::NATMX4 |
Table 2. Diploids generated by the zero growth mating regime that were analyzed for spore viability and genetic map distance.
| EAY1108/EAY1112 Background (Analyzed in Tables 4, and 5 and Figures 2, 3, and 4) | |
|---|---|
| EAY1108/EAY1112 | wild type |
| EAY1108/EAY1848 | MLH3/mlh3Δ |
| EAY2413/EAY1848 | mlh3Δ/mlh3Δ |
| EAY3007/EAY1848 | mlh3-E31A/mlh3Δ |
| EAY3009/EAY1848 | mlh3-N35A/mlh3Δ |
| EAY3011/EAY1848 | mlh3-A41F/mlh3Δ |
| EAY3013/EAY1848 | mlh3-G63R/mlh3Δ |
| EAY3015/EAY1848 | mlh3-K80E/mlh3Δ |
| EAY3017/EAY1848 | mlh3-K83A/mlh3Δ |
| EAY3019/EAY1848 | mlh3-R96A/mlh3Δ |
| EAY3021/EAY1848 | mlh3-G97A/mlh3Δ |
| EAY1281/EAY1279 | msh5Δ/msh5Δ |
| EAY2032/EAY2033 | msh5Δ mlh3Δ/msh5Δ mlh3Δ |
| EAY2423/EAY1279 | msh5-D76A/msh5Δ |
| EAY2439/EAY1279 | msh5-T423A/msh5Δ |
| EAY3313/EAY3315 | msh5-D76A mlh3G96A/msh5Δ mlh3Δ |
| EAY3313/EAY3316 | msh5-D76A mlh3-G97A/msh5Δ mlh3Δ |
| EAY3314/EAY3315 | msh5-T423A mlh3-R96A/msh5Δ mlh3Δ |
| EAY3314/EAY3316 | msh5-T423A mlh3-G97A/msh5Δ mlh3Δ |
| EAY1845/EAY1846 | mms4Δ/mms4Δ |
| EAY2030/EAY2031 | mlh3Δ mms4Δ/mlh3Δ mms4Δ |
| NHY942/NHY943 background (analyzed in Tables 6, 7, 8, Figure 2) | |
| NHY942/NHY943 | wild type |
| EAY2904/EAY2906 | mlh3Δ/mlh3Δ |
| EAY3290/EAY3323 | mms4Δ/mms4Δ |
| EAY3296/EAY3298 | mlh3Δ mms4Δ/mlh3Δ mms4Δ |
The indicated haploid strains (Table 1, Materials and Methods) were mated and sporulated using the zero growth mating protocol and tetrads were dissected (Argueso et al. 2003).
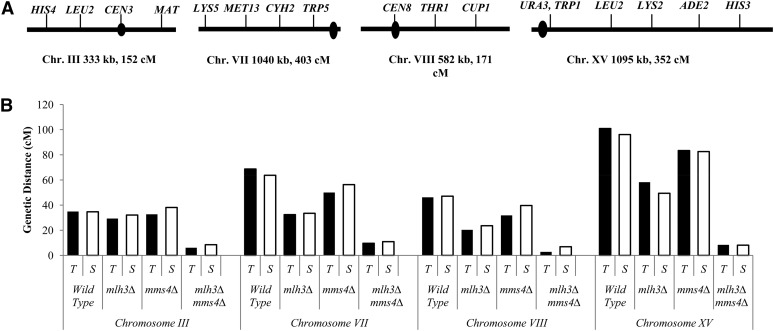
Figure 2 .
Cumulative genetic distances for wild type, mlh3Δ, mms4Δ, and mlh3Δ mms4Δ on four chromosomes. (A) Location of genetic markers used to determine map distances in the NHY942/NHY943 background for chromosomes III, VII, VIII, and the EAY1108/EAY1112 background for chromosome XV. (B) The cumulative genetic distance for each chromosome is shown for both complete tetrad data (black bars) and single spore data (white bars). Raw data are shown in Table 7. Data for wild type for chromosomes III, VII, and VIII are from Zanders and Alani (2009). Data for wild type and mms4Δ for chromosome XV are from Argueso et al. (2004). Data for mlh3Δ and mlh3Δ mms4Δ on chromosome XV are from Nishant et al. (2008). For chromosome III, the physical distances (end of the marker gene to the beginning of the next, in KB) are: HIS4-LEU2, 23; LEU2-CEN3, 22; CEN3-MAT, 90. For chromosome VII, the physical distances are: LYS5-MET13, 56, MET13-CYH2, 36; CYH2-TRP5, 135. For chromosome VIII, the physical distances are: CEN8-THR1, 54; THR1-CUP1, 52. For chromosome XV, the physical distances are: URA3-LEU2, 136; LEU2-LYS2, 43; LYS2-ADE2, 59; ADE2-HIS3, 157.
The isogenic SK1 strain EAY1062 [lys2::InsE-A14 (Nishant et al. 2008)] was used to measure the effect of mlh3 mutations on mutation rate (Table 3). For the dominant-negative assay, pEAE220 (2μ, S288c GAL10-MLH3), and mutant derivatives pEAE374 (GAL10-mlh3-E31A), pEAE375 (GAL10-mlh3-R96A), and pEAE376 (GAL10-mlh3-G97A) were transformed into EAY1269 (S288c, lys::InsE-A14).
Table 3. Reversion of the lys2:InsE-A14 allele in mlh3 strains.
| Genotype | n | Mutation Rate (×10−7) | Relative to WT | Phenotype |
|---|---|---|---|---|
| MLH3 | 110 | 4.71 (3.87–5.11) | 1.0 | + |
| mlh3Δ | 110 | 26.5 (23.5–30.4) | 5.7 | − |
| mlh3-E31A | 15 | 30.5 (16.7–51.6) | 6.5 | − |
| mlh3-N35A | 15 | 31.2 (25.6–44.4) | 6.7 | − |
| mlh3-A41F | 15 | 27.9 (17.1–34.3) | 6.0 | − |
| mlh3-G63R | 15 | 23.8 (18.2–37.1) | 5.1 | − |
| mlh3-K80E | 15 | 16.0 (15.1–27.7) | 3.4 | − |
| mlh3-K83A | 15 | 5.24 (3.49–6.34) | 1.1 | + |
| mlh3-R96A | 15 | 14.8 (6.42–40.6) | 3.2 | − |
| mlh3-G97A | 15 | 16.6 (11.8–26.0) | 3.6 | − |
| MLH3 + empty vector | 11 | 4.42 (1.02-6.05) | 1 | + |
| MLH3 + pGAL10-MLH3 | 11 | 39,100 (15,700-79,900) | 8850 | − |
| MLH3 + pGAL10-mlh3E31A | 11 | 47,800 (28,700-85,900) | 10,800 | − |
| MLH3 + pGAL10-mlh3R96A | 11 | 23,500 (5910-38,400) | 5320 | − |
| MLH3 + pGAL10-mlh3G97A | 11 | 96,000 (45,800-156,000) | 21,700 | − |
| mlh1Δ + empty vector | 11 | 218,000 (121,000-283,000) | 49,300 | − |
The lys2:InsE-A14 SK1 strain EAY1062 and mlh3 derivatives (Table 1) were examined for reversion to Lys+. EAY1269 (lys2:InsE-A14, S288c strain) and an mlh1Δ derivative containing the indicated overexpression plasmids were tested for reversion to Lys+. n, the number of independent cultures tested from at least two independently constructed strains. Median mutation rates are presented with 95% confidence intervals, and relative mutation rates compared with the wild-type strain are shown. WT, wild type.
Genetic map distance analysis
EAY1108/EAY1112 and NHY942/NHY943 background diploids were sporulated using the zero growth mating protocol [Table 2 (Argueso et al. 2003)] and tetrads were dissected. For the EAY1108/EAY1112 background strains, tetrads were dissected and spores were germinated on synthetic complete media. For the NHY942/NHY943 background strains, tetrads were dissected and germinated on yeast extract-peptone, 2% dextrose media supplemented with complete amino acids. Spore clones were incubated 3–4 d at 30° and then replica-plated to various selective media. The replica plates were scored after 1 d of incubation at 30°. Spore clones were analyzed using the recombination analysis software RANA (Argueso et al. 2004), which analyzes map distances. Genetic map distances ± SE were calculated using the Stahl Laboratory Online Tools (http://www.molbio.uoregon.edu/∼fstahl/), which uses the formula of Perkins (1949). Differences in spore formation and viability were analyzed by a χ2 test with P-values < 0.05 considered statistically significant. The genetic intervals measured in this study (illustrated in Figure 2) were: chromosome III-HIS4-LEU2, LEU2-CEN3, CEN3-MAT; chromosome VII-LYS5-MET13, MET13-CYH2, CYH2-TRP5; chromosome VIII-CEN8-THR1, THR1-CUP1; and chromosome XV- URA3-LEU2, LEU2-LYS2, LYS2-ADE2, ADE2-HIS3.
Lys+ reversion assays
The mlh3 allele constructs were transformed into EAY2037 (SK1, mlh3Δ::KANMX4, lys2::InsE-A14), and strains were analyzed for reversion to Lys+ (Tran et al. 1997). At least 15 independent cultures for each allele were analyzed, and experiments were conducted with two independent transformants. Mutation rates were determined as previously described (Drake 1991; Heck et al. 2006). Each median rate was normalized to the wild-type median rate to calculate the fold-increase in mutation rate. 95% confidence intervals were determined as described (Dixon and Massey 1969).
For the dominant-negative assays, EAY1269 bearing pEAE220 and mutant derivatives were grown for 5 d on uracil dropout SC agar plates containing 2% sucrose or 2% sucrose and 2% galactose. Individual colonies were picked and grown overnight in liquid (-agar) versions of the respective media for 26 hr. Appropriate dilutions were made, and cells grown in sucrose only were plated on uracil, lysine dropout SC agar plates containing 2% sucrose, and uracil dropout SC agar plates containing 2% glucose. Cells grown in sucrose and galactose were plated on uracil, lysine dropout SC agar plates containing 2% sucrose and 2% galactose, and uracil dropout SC agar plates containing 2% glucose. Using GAL10-MLH3 and mlh1Δ as controls, we analyzed 11 independent colonies from two independent transformations.
Results and Discussion
ATP hydrolysis by both Mlh1 and Mlh3 is likely to be important for their roles in meiosis and MMR
MLH family proteins each contain an N-terminal ATP binding domain. This domain is thought to regulate asymmetric conformational changes in MLH dimers through cycles of ATP binding and hydrolysis (Ban and Yang 1998; Ban et al. 1999; Tran and Liskay 2000; Hall et al. 2002; Sacho et al. 2008). Previous structure−function studies have shown that the two subunits in yeast Mlh1-Pms1 are functionally asymmetric. For example, the Mlh1 subunit of the yeast Mlh1-Pms1 complex displayed a much greater affinity for ATP compared to the Pms1 subunit, and an ATP hydrolysis mutation in MLH1 (mlh1-E31A) conferred a much greater effect on MMR than the equivalent mutation in PMS1 (pms1-E61A; Tran and Liskay 2000; Hall et al. 2002). Also, in baker’s yeast the Mlh1 subunit has been shown to interact with the downstream MMR factor Exo1 in an ATP-dependent manner. Thus, ATP-dependent and asymmetric conformational changes in MLH proteins are likely to be important to modulate interactions with downstream MMR effector molecules (Pedrazzi et al. 2001; Tran et al. 2001).
Previous genetic and biochemical analyses identified mutations in the ATP-binding domains of yeast MLH proteins that disrupt ATP hydrolysis to a greater extent than ATP binding (e.g., mlh1-E31A). Mutations also were identified that severely disrupt ATP binding [e.g., mlh1-N35A (Hall et al. 2002)]. Other mutations have been made in MLH ATP-binding domains that are predicted to affect ATP binding and/or ATP-dependent conformational changes but have yet to be tested in biochemical assays [Figure 1 (Tran and Liskay 2000; Hall et al. 2002; Ban and Yang 1998; Ban et al. 1999)].
We made mutations in Mlh3 predicted to confer defects in ATP hydrolysis (mlh3-E31A) and binding (mlh3-N35A), and six other mutations that map within or near motifs identified in the GHKL family of ATPases, of which the MLH proteins are members [Figure 1 (Ban and Yang 1998; Ban et al. 1999)]. We tested the effect of these mutations in a MMR repair assay that measures reversion of the lys2::InsE-A14 allele (Tran et al. 1997) and in meiotic assays that measure spore viability and crossing over in four intervals on chromosome XV in EAY1108/1112 SK1 congenic strains [Figure 2 (Argueso et al. 2004)]. Three of the eight mlh3 mutations also were analyzed by Cotton et al. (2010), using similar assays. In the lys2::InsE-A14 reversion assay, mlh3Δ strains display a roughly 6-fold increase in mutation rate compared with wild-type (Harfe et al. 2000; Nishant et al. 2008; this study). We found that all but one of the eight mlh3 alleles conferred MMR defects similar to the null (within 95% confidence intervals), ranging from 3.2 to 6.7-fold greater than wild-type levels. mlh3-K83A strains showed a wild-type phenotype (Table 3). Our results for the mlh3-N35A and mlh3-G97A mutations were similar to those obtained by Cotton et al. (2010). However, for mlh3-E31A, which is thought to disrupt ATP hydrolysis by the Mlh3 subunit, we observed a null MMR phenotype; Cotton et al. (2010) observed a close to wild-type phenotype for this mutant.
To assess Mlh3 expression, we overexpressed mlh3-E31A, mlh3-R96A, and mlh3-G97A in wild-type cells and assessed dominant-negative phenotypes using the lys2::InsE-A14 frameshift reporter, which can detect a roughly four-order of magnitude difference in mutation rate (Tran et al. 1997). This approach was taken because we have been unable to detect single copy levels of Mlh3 in vegetative cells (M. Rogacheva and E. Alani, unpublished observations). We showed previously that overexpressing Mlh3 using the GAL10 promoter conferred a high mutator phenotype in the lys2::InsE-A14, reversion assay with mutation rates more than 1000-fold greater than wild-type. This phenotype was similar to that seen in wild-type strains overexpressing Mlh1 (Shcherbakova and Kunkel 1999; Nishant et al. 2008). Based on these observations, we hypothesized that increased levels of Mlh3 interfered with mismatch repair by outcompeting Pms1 for Mlh1, thus preventing Mlh1-Pms1 from acting in MMR (Wang et al. 1999; Kondo et al. 2001). Consistent with this idea, overexpressing mlh3-E529K, which does not interact with Mlh1, did not confer a dominant-negative phenotype (Nishant et al. 2008). As shown in Table 3, each allele conferred a strong dominant-negative phenotype similar to MLH3, with mutation rates 5000- to 20,000-fold greater than wild-type containing an empty vector. This suggests that an intact Mlh1-mlh3 complex is formed in each of these mutants.
As mentioned previously, mismatch repair rates have been examined in strains bearing mlh1 mutations at positions equivalent to those made in MLH3 (Tran and Liskay 2000; Argueso et al. 2003; Hoffman et al. 2003; Wanat et al. 2007). Consistent with its lesser role in MMR, mlh3 alleles show a lower mutation rate as measured in the lys::InsE-A14 reversion assay compared with equivalent mlh1 alleles; however, they appear to be just as sensitive to mutagenesis. Similar to their equivalent mlh3 mutations, mlh1-K81E, mlh1-R97A, and mlh1-G98A conferred null phenotypes in MMR. mlh1-E31A and mlh1-K84A, however, conferred MMR phenotypes that were different from their equivalent mlh3 mutations, with mlh1-E31A strains appearing more proficient in MMR and mlh1-K84A strains less proficient [Tables 3 and 4 (Tran and Liskay 2000; Hoffman et al. 2003; Wanat et al. 2007; Argueso et al. 2003)]. Thus our work, in conjunction with previous studies, reinforces the hypothesis that the subunits of MLH complexes provide unique contributions to MMR (Tran and Liskay 2000; Hall et al. 2002; Argueso et al. 2003; Hoffman et al. 2003; Wanat et al. 2007; Nishant et al. 2008; Cotton et al. 2010).
Table 4. Spore viabilities, map distances, qualitative MMR phenotypes, and known mlh1 homolog phenotypes for the mlh3 alleles, msh5Δ, and mlh3 msh5 double mutants.
| Strain | Spore Viability, % | cM | MMR | mlh1 allele | MMR |
|---|---|---|---|---|---|
| mlh3 mutant analysis | |||||
| MLH3a | 97.0 | 100.9 (1068) | + | MLH1 | + |
| mlh3Δb | 71.7 | 54.5 (582) | − | mlh1Δ | − |
| mlh3-E31A | 89.2 | 67.0 (330) | − | mlh1-E31Ac,d | +/− |
| mlh3-N35A | 72.7 | 51.5 (229) | − | mlh1-E35A | ND |
| mlh3-A41F | 71.6 | 51.2 (214) | − | mlh1-A41F | ND |
| mlh3-G63R | 74.1 | 51.2 (216) | − | mlh1-G64R | ND |
| mlh3-K80E | 71.8 | 49.8 (221) | − | mlh1-K81Ee | − |
| mlh3-K83A | 94.1 | 100.5 (289) | + | mlh1-K84Ad | +/− |
| mlh3-R96A | 82.4 | 76.4 (177) | − | mlh1-R97Ad | − |
| mlh3-G97A | 81.5 | 61.0 (210) | − | mlh1-G98Ac,f | − |
| msh5 mutant analysis | |||||
| msh5Δa | 36.0 | 37.0 (540) | |||
| msh5Δ mlh3Δ | 31.8 | 38.5 (43) | |||
| msh5-D76Ag | 87.8 | 53.9 (77) | |||
| msh5-T423Ag | 95.2 | 78.3 (101) | |||
| msh5-D76A mlh3 R96A | 57.8 | 45.0 (81) | |||
| msh5-D76A mlh3 G97A | 47.1 | 31.7 (82) | |||
| msh5-T423A mlh3 R96A | 89.6 | 60.9 (160) | |||
| msh5-T423A mlh3 G97A | 78.3 | 54.7 (130) |
Spore viabilities (%) and cumulative genetic map distances from four spore-viable tetrads (number in parentheses) on chromosome XV are shown for wild-type, mlh3, and msh5 strains in the SK1 congenic EAY1108/1112 background (Table 2). The qualitative MMR phenotype of each allele (see Table 3) is shown for comparison. MMR data are also shown for the homologous mlh1 alleles, if known. MMR, mismatch repair; ND, not determined.
Data obtained from Argueso et al. (2004).
Data obtained from Nishant et al. (2008).
Data from Tran and Liskay (2000).
Data from Argueso et al. (2003).
Data from Wanat et al. (2007).
Data from Hoffman et al. (2003).
Data obtained from Nishant et al. (2010).
We tested the effect of mlh3 mutations in meiosis in the EAY1108/1112 SK1 congenic strain background, which is marked to measure map distances over four consecutive genetic intervals on chromosome XV [Materials and Methods; Figure 2 (Argueso et al. 2004)]. In this background, wild-type display 97% spore viability and a cumulative map distance of 100.9 cM over the four intervals, whereas mlh3Δ display 72% spore viability and a cumulative map distance of 54.5 cM (Tables 4 and 5). As shown in Tables 4 and 5, four of eight of the mlh3 mutations (mlh3-N35A, -A41F, G63R, K80E) conferred null phenotypes in the meiotic assays, and one mutation, mlh3-K83A, conferred a wild-type phenotype. Three mutations, mlh3-E31A, mlh3-R96A, and mlh3-G97A, conferred intermediate phenotypes (Tables 4 and 5). Like Cotton et al. (2010), we found that the predicted ATP binding mutation mlh3-N35A conferred a null phenotype in the meiotic assays. However, in contrast to a nearly wild-type phenotype previously seen for mlh3-E31A in both MMR and meiotic assays (Cotton et al. 2010), we found that mlh3-E31A mutants displayed, compared with the wild-type, defects in meiosis (Table 4; 67 cM map distance, 89% spore viability, P < 0.001) and MMR (null phenotype, Table 3). Thus, our analyses are consistent with ATP hydrolysis by Mlh3 being important for its meiotic and MMR functions. We do not have a clear explanation for why our data differ from Cotton et al. (2010). However, one possibility is that the SK1 strain background is more sensitized to defects in MLH3 compared with the Y55 background studied by Cotton et al. (2010). Consistent with this idea, we found that SK1 mlh3Δ strains showed lower spore viability (72%) compared with Y55 mlh3Δ strains [92% (Cotton et al. 2010)].
Table 5. Genetic map distances for chromosome XV from single spores and tetrads with distributions of parental and recombinant progeny.
| Single Spores |
Tetrads |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | n | Par. | Rec | cM | n | PD | TT | NPD | cM |
| URA3-LEU2 | |||||||||
| Wild typea | 4644 | 3635 | 1009 | 21.7 | 1068 | 607 | 456 | 5 | 21.8-23.8 |
| msh5Δa | 5674 | 5352 | 322 | 5.7 | 757 | 643 | 76 | 1 | 5.0-6.4 |
| mlh3Δb | 3023 | 2682 | 341 | 11.3 | 582 | 460 | 114 | 8 | 12.3-15.5 |
| msh5Δ mlh3Δ | 382 | 352 | 30 | 7.9 | 43 | 34 | 8 | 0 | 6.5-12.6 |
| msh5-D76Ac | 351 | 310 | 41 | 11.7 | 77 | 57 | 17 | 0 | 9.0-13.9 |
| msh5-T423Ac | 457 | 378 | 79 | 17.3 | 101 | 62 | 33 | 0 | 14.9-19.8 |
| mlh3- R96A | 840 | 676 | 164 | 19.5 | 177 | 105 | 69 | 0 | 18.0-21.7 |
| mlh3- G97A | 978 | 841 | 137 | 14.0 | 210 | 152 | 55 | 2 | 13.6-18.5 |
| msh5-D76A mlh3 R96A | 462 | 409 | 53 | 11.5 | 81 | 63 | 16 | 0 | 7.9-12.4 |
| msh5-D76A mlh3 G97A | 490 | 455 | 35 | 7.1 | 82 | 71 | 11 | 0 | 4.8-8.6 |
| msh5-T423A mlh3 R96A | 717 | 583 | 134 | 18.7 | 160 | 96 | 64 | 0 | 18.1-21.9 |
| msh5-T423A mlh3 G97A | 622 | 552 | 70 | 11.3 | 130 | 100 | 28 | 1 | 10.3-16.1 |
| LEU2-LYS2 | |||||||||
| Wild typea | 4644 | 3388 | 1256 | 27.0 | 1068 | 496 | 569 | 3 | 26.6-28.4 |
| msh5Δa | 5674 | 5047 | 627 | 11.1 | 757 | 562 | 155 | 3 | 11.0-13.0 |
| mlh3Δb | 3023 | 2610 | 413 | 13.7 | 582 | 424 | 154 | 3 | 12.9-16.6 |
| msh5Δ mlh3Δ | 382 | 338 | 44 | 11.5 | 43 | 31 | 10 | 1 | 11.5-26.6 |
| msh5-D76Ac | 351 | 308 | 43 | 12.3 | 77 | 58 | 16 | 0 | 8.4-13.2 |
| msh5-T423Ac | 457 | 365 | 92 | 20.1 | 101 | 57 | 38 | 0 | 17.5-22.5 |
| mlh3- R96A | 840 | 695 | 145 | 17.3 | 177 | 112 | 62 | 0 | 16.0-19.6 |
| mlh3- G97A | 978 | 825 | 153 | 15.6 | 210 | 140 | 68 | 1 | 15.6-19.8 |
| msh5-D76A mlh3 R96A | 462 | 422 | 40 | 8.7 | 81 | 67 | 12 | 0 | 5.6-9.6 |
| msh5-D76A mlh3 G97A | 490 | 457 | 33 | 6.7 | 82 | 72 | 10 | 0 | 4.3-7.9 |
| msh5-T423A mlh3 R96A | 717 | 606 | 111 | 15.5 | 160 | 111 | 49 | 0 | 13.5-17.1 |
| msh5-T423A mlh3 G97A | 622 | 535 | 87 | 14.0 | 130 | 91 | 37 | 1 | 13.7-19.6 |
| LYS2-ADE2 | |||||||||
| Wild typea | 4644 | 4052 | 592 | 12.7 | 1068 | 803 | 263 | 2 | 12.1-13.7 |
| msh5Δa | 5674 | 5409 | 265 | 4.7 | 757 | 659 | 61 | 0 | 3.7-4.7 |
| mlh3Δb | 3023 | 2822 | 201 | 6.6 | 582 | 501 | 81 | 0 | 6.2-7.7 |
| msh5Δ mlh3Δ | 382 | 363 | 19 | 5.0 | 43 | 39 | 3 | 0 | 1.6-5.6 |
| msh5-D76Ac | 351 | 320 | 31 | 8.8 | 77 | 60 | 14 | 0 | 7.2-11.7 |
| msh5-T423Ac | 457 | 405 | 52 | 11.4 | 101 | 75 | 20 | 0 | 8.4-12.6 |
| mlh3- R96A | 840 | 775 | 65 | 7.7 | 177 | 149 | 25 | 0 | 5.9-8.5 |
| mlh3- G97A | 978 | 898 | 80 | 8.2 | 210 | 173 | 35 | 1 | 7.9-11.7 |
| msh5-D76A mlh3 R96A | 462 | 437 | 25 | 5.4 | 81 | 68 | 11 | 0 | 5.0-8.9 |
| msh5-D76A mlh3 G97A | 490 | 464 | 26 | 5.3 | 82 | 75 | 7 | 0 | 2.7-5.8 |
| msh5-T423A mlh3 R96A | 717 | 669 | 48 | 6.7 | 160 | 141 | 19 | 0 | 4.7-7.2 |
| msh5-T423A mlh3 G97A | 622 | 591 | 31 | 5.0 | 130 | 116 | 13 | 0 | 3.7-6.4 |
| ADE2-HIS3 | |||||||||
| Wild typea | 4644 | 3033 | 1611 | 34.7 | 1068 | 343 | 709 | 16 | 36.5-38.9 |
| msh5Δa | 5674 | 4797 | 877 | 15.5 | 757 | 496 | 215 | 9 | 17.2-20.2 |
| mlh3Δb | 3023 | 2485 | 538 | 17.8 | 582 | 379 | 201 | 2 | 17.1-19.5 |
| msh5Δ mlh3Δ | 382 | 328 | 54 | 14.1 | 43 | 30 | 12 | 0 | 10.8-17.8 |
| msh5-D76Ac | 351 | 277 | 74 | 21.1 | 77 | 43 | 31 | 0 | 18.1-23.8 |
| msh5-T423Ac | 457 | 322 | 135 | 29.5 | 101 | 44 | 49 | 2 | 27.4-36.9 |
| mlh3- R96A | 840 | 600 | 240 | 28.6 | 177 | 74 | 98 | 2 | 28.7-34.5 |
| mlh3- G97A | 978 | 801 | 177 | 18.1 | 210 | 136 | 73 | 0 | 15.8-19.1 |
| msh5-D76A mlh3 R96A | 462 | 395 | 67 | 14.5 | 81 | 57 | 20 | 2 | 14.6-25.9 |
| msh5-D76A mlh3 G97A | 490 | 422 | 68 | 13.9 | 82 | 58 | 24 | 0 | 12.1-17.1 |
| msh5-T423A mlh3 R96A | 717 | 575 | 142 | 19.8 | 160 | 97 | 63 | 0 | 17.8-21.6 |
| msh5-T423A mlh3 G97A | 622 | 507 | 115 | 18.5 | 130 | 83 | 45 | 1 | 16.8-22.8 |
Strains used are isogenic derivatives of the congenic SK1 EAY1108/1112 background (Tables 1 and 2). Single spore data are shown with n, total number of spores, and parental and recombinant data. Map distances (cM) were calculated by recombination frequency (recombinant spores/total spores) × 100. Tetrad data are shown with n, number of complete tetrads. Map distances (cM) were calculated using the Perkins formula (Perkins 1949), and 95% confidence intervals were calculated using the Stahl Laboratory Online Tools website (http://www.molbio.uoregon.edu/∼fstahl/).
Data from Argueso et al. (2004).
Data from Nishant et al. (2008).
Data from Nishant et al. (2010).
It is important to note that five of the eight mlh3 alleles displayed consistent phenotypes in both the MMR and meiosis assays (either wild-type or null in both). However, three mlh3 hypomorph mutants, mlh3-E31A, -R96A, -G97A, displayed null phenotypes in MMR, but intermediate meiotic phenotypes, as measured in meiotic spore viability and crossover assays (Tables 4 and 5). These observations suggest that, as was seen for Mlh1 (Argueso et al. 2003; Hoffman et al. 2003), Mlh3 functions are more easily disrupted for MMR.
mlh3 strains show a roughly linear relationship between crossing over and spore viability
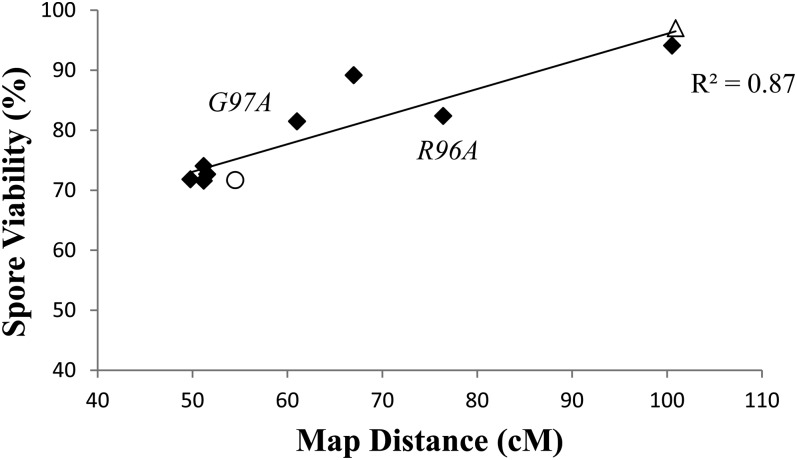
As shown in Figure 3 and Table 4, the mlh3 mutants displayed a relationship where spore viability decreased progressively with map distance (R2 = 0.87). Consistent with this we observed that wild-type spore viability was significantly greater than that seen in mlh3-E31A, -R96A, and -G97A (P ≪ 0.001). This pattern is in contrast to the pattern observed in msh4/5 mutants, where crossing over could be decreased to approximately 50% of wild-type levels (to ∼50 cM across the four intervals in chromosome XV) without an apparent defect in spore viability, after which point spore viability and crossing over decreased linearly (Nishant et al. 2010). Based on this and other observations, Nishant et al. (2010) proposed that crossover designation functions executed by Msh4-Msh5 are prioritized in yeast to maintain the obligate crossover, ensuring that each homolog pair receives at least one disjunction-promoting crossover. The finding that mlh3 mutants show a pattern where spore viability decreased progressively with map distance is consistent with a wealth of data supporting a crossover resolution role for Mlh1-Mlh3 in the interference-dependent crossover pathway (see Introduction). Such a relationship might be expected if Mlh1-Mlh3 acts late in crossover resolution because a decrease in Mlh3 function would be expected to cause a random loss in crossing over, thus not assuring that all obligate crossovers would take place.
Figure 3 .
mlh3 strains show a roughly linear relationship between crossing over and spore viability. Spore viabilities are plotted vs. genetic map distances on chromosome XV for eight mlh3 ATP binding domain mutations, wild type (open triangle), and mlh3Δ (open circle).
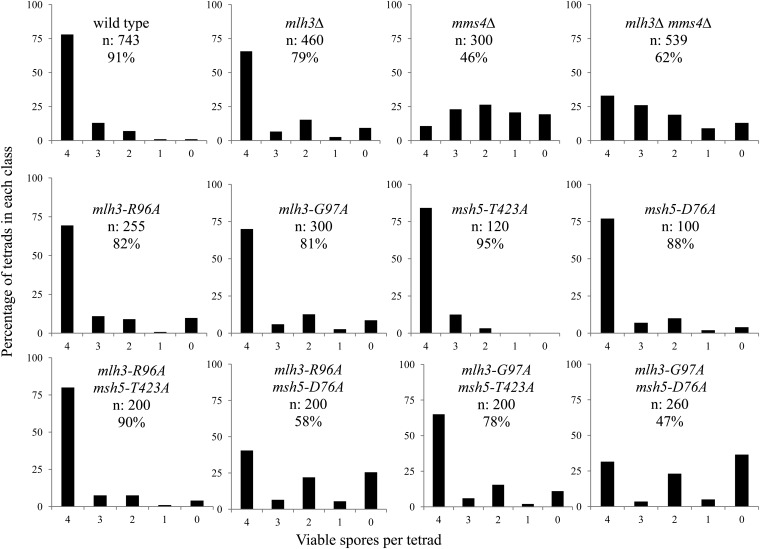
To further test whether the mlh3 spore viability and map distance data support a roughly linear relationship, we more closely examined the phenotype of two mutants, mlh3-G97A and mlh3-R96A. These mutants show a relatively large difference in genetic map distance but a negligible difference in spore viability (P > 0.5). We attempted to detect any difference in phenotype conferred by these mutants by making double mutants with msh5 alleles. When mlh3-R96A was combined with msh5-T423A, very little change in spore viability or map distance was observed compared with single mutants (Table 4; Figure 4). However, when the mlh3-R96A was combined with msh5-D76A, a strong synthetic defect was observed for spore viability in the double mutant; crossing over, however, was only slightly decreased. Similar results were obtained when each of these msh5 alleles was combined with mlh3-G97A, except the results were more extreme. For example, the differences in spore viability between mlh3-G97A msh5-D76A and mlh3-R96A msh5-D76A (P < 0.02) and between mlh3-G97A msh5-D423A and mlh3-R96A msh5-D423A (P < 0.01) were statistically significant. This analysis confirms that mlh3-G97A confers a more severe defect compared with mlh3-R96A, as predicted if the pattern seen for msh4/5 mutants did not hold for the mlh3 mutants. Consistent with these observations, mlh3-G97A conferred a mild nondisjunction phenotype, as measured by an excess of 4, 2, 0 viable spore tetrads compared with 3 and 1 viable tetrads (Ross-Mcdonald and Roeder 1994), but mlh3-G97A msh5-D76A conferred a more extreme nondisjunction pattern (Figure 4).
Figure 4 .
Spore viability profile of wild-type and select mutants. The horizontal axis shows the number of viable spores per tetrad, and the vertical axis shows the percentage of tetrads in each class. n, the total number of tetrads dissected, and percent spore viability are shown. Data for wild-type, mlh3Δ, mms4Δ, and mlh3Δ mms4Δ are from the NHY942/943 background (Tables 6 and 7; the remaining data are from the EAY1108/1112 background (Tables 4 and 5).
mlh3Δ mms4Δ mutants show dramatically decreased crossing over across four different chromosomes but display high spore viability
Our analysis of mlh3 mutants described previously encouraged us to more closely examine mlh3Δ mutants for defects in crossing over. In previous studies authors showed that there are at least two types of crossover pathways in budding yeast: an Msh4-Msh5-Mlh1-Mlh3 pathway and an interference-independent pathway involving Mus81-Mms4 (see Introduction). In addition, three meiotic joint molecule resolvase complexes have been identified: Mus81-Mms4, Yen1, and Slx1-Slx4 (Boddy et al. 2001; Fricke and Brill 2003; Furukawa et al. 2003; Ishikawa et al. 2004; Cromie et al. 2006; Ip et al. 2008; Jessop and Lichten 2008; Oh et al. 2008; Muñoz et al. 2009; Svendsen et al. 2009; Schwartz and Heyer 2011). These resolvases appear to play different roles in different organisms. For example, Mus81-Mms4 plays a major role in fission yeast (Smith et al. 2003), but only a minor role in budding yeast, Arabidopsis, mouse, and Drosophila (De Los Santos et al. 2003; Argueso et al. 2004; Berchowitz et al. 2007; Trowbridge et al. 2007; Higgins et al. 2008; Holloway et al. 2008; Jessop and Lichten 2008; Oh et al. 2008).
Previously we showed that on a large chromosome, mlh1Δ mms4Δ double mutants display significant decreases (∼13- to 15-fold) in crossing over compared with wild type (Argueso et al. 2004). Based on these and other data we suggested that Mus81-Mms4 and Mlh1-Mlh3 act in competing crossover pathways (Argueso et al. 2004), with Mus81-Mms4 dependent crossovers promoting proper chromosome disjunction in the absence of Mlh1-Mlh3. Consistent with this finding, the Hunter lab and Lichten groups recently provided evidence for Msh4-Msh5-Mlh1-Mlh3-Exo1 and Mus81-Mms4 acting independently in crossover resolution (De Muyt et al. 2012;Zakharyevich et al. 2012). The Hunter lab previously showed that mlh3Δ decreases crossover levels without changing joint molecule levels, also suggesting a late role for Mlh3 (Zakharyevich et al. 2010). Using Southern blot analysis at the well-studied HIS4LEU2 hotspot, they showed that compared with the wild-type, exo1 (Exo1 forms a complex with Mlh1-Mlh3) reduced crossing over by 49%, mms4yen1 by 39%, and exo1mms4yen1 by 86%. Strikingly, crossover levels decreased roughly 20-fold in mlh3mms4slx4yen1sgs1 cells (Zakharyevich et al. 2012). The Lichten group (De Muyt et al. 2012) showed that in msh4Δ mms4yen1Δ triple mutants, the bulk of chromosomal DNA fails to segregate. Furthermore, they found that unresolved joint molecules accumulated to similar levels in msh4Δ ndt80Δ, where joint molecule resolution cannot take place, suggesting that the Mus81-Mms4 and Yen1 pathways are responsible for resolving crossover intermediates that are not resolved by the Msh4-Msh5-Mlh1-Mlh3 pathway. Because they found that most joint molecules were resolved in mms4yen1Δ slx1Δ mutants, their data provide evidence that Msh4-Msh5-Mlh1-Mlh3 acts in crossover resolution.
The Hunter and Lichten studies, summarized previously, provide evidence that Exo1-Mlh1-Mlh3 and Mus81-Mms4 are responsible for the majority of crossovers in budding yeast. Although each of the aforementioned studies presented convincing data for the presence of two independent crossover pathways, physical data reported in Zakharyevich et al. (2012) were primarily obtained at a single locus, the HIS4LEU2 hotspot, and genetic data were obtained by Argueso et al. (2004) and Nishant et al. (2008) in only one chromosome arm. To understand the role of Mlh3 in crossing over genome-wide, we analyzed spore viability and crossovers across four chromosomes in mlh3Δ mms4Δ double mutants. A total of 250 cM of map distance was measured, representing ∼6.2% of the yeast genome. mlh3Δ mms4Δ double mutants were chosen for this analysis because they formed viable spores at a reasonable frequency and displayed strong defects in crossing over in one arm of chromosome XV. As shown in Tables 6 and 7 and Figure 2, we found that for all loci examined crossing over was drastically reduced (6- to 17-fold) in mlh3Δ mms4Δ strains compared to wild-type. Interestingly, crossing over was decreased by the smallest amount on chromosome III, a pattern seen in other meiotic mutants (Zanders and Alani 2009). Although mlh3Δ mutants show a characteristic 4:2:0 pattern of viable spores per tetrad indicative of nondisjunction (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Hunter and Borts 1997; Argueso et al. 2003; Nishant et al. 2008; this study), neither mms4Δ nor mlh3Δ mms4Δ showed this pattern (Figure 4). Thus, our analysis provides further support for the hypothesis that Mlh1-Mlh3 and Mus81-Mms4 independently contribute late roles for meiotic crossover formation.
Table 6. Spore viabilities and cumulative genetic map distances for wild type, mlh3Δ, mms4Δ, and mlh3Δ mms4Δ for chromosomes III, VII, VIII, and XV.
| Genotype | Map Distance, cM |
|||||
|---|---|---|---|---|---|---|
| Chromosome | Spore Viability, % | n | III (333 kb) | VII (1040 kb) | VIII (582 kb) | XV (1095 kb) |
| Wild typea | 91.0 | 572 | 34.9 | 68.7 | 46.2 | 96.1b |
| mlh3Δ | 79.0 | 306 | 29.3 | 32.4 | 20.3 | 54.5c |
| mms4Δ | 46.3 | 32 | 32.7 | 50.0 | 31.8 | 83.4b |
| mms4Δd | 45.4 | 272 | 25.2 | 62.1 | 35.3 | |
| mlh3Δ mms4Δ | 61.9 | 170 | 5.7 | 9.6 | 2.8 | 8.4c |
| Fold decrease in mlh3Δ mms4Δ vs. wild type | 6.1 | 7.2 | 16.5 | 11.4 | ||
Spore viabilities (%) and cumulative genetic map distances in cM (number of complete tetrads) on chromosomes III, VII, VIII, and XV are shown for mlh3Δ, msh5Δ, mlh3 alleles, msh5 alleles, and the double mutants (Tables 1 and 2). Sizes of each chromosome are shown below each chromosome number, and the fold decrease in crossing over in mlh3Δ mms4Δ compared with wild type is shown below. Chromosome III, VII, and VIII data are from derivatives of the isogenic SK1 NHY942/943 background. Data for chromosome XV are from derivatives of the congenic SK1 EAY1108/1112 background.
Data from Zanders and Alani (2009).
Data from Argueso et al. (2004).
Data from Nishant et al. (2008).
Data from De Los Santos et al. (2003).
Table 7. Genetic map distances for chromosomes III, VII, and VIII from single spores and tetrads with distributions of recombinant and parental progeny.
| Single Spores |
Tetrads |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | n | Par. | Rec. | cM | n | PD | TT | NPD | cM |
| Chromosome III | |||||||||
| HIS4-LEU2 | |||||||||
| Wild typea | 2711 | 2360 | 351 | 12.9 | 572 | 413 | 141 | 2 | 12.6-15.0 |
| mlh3Δ | 1453 | 1333 | 120 | 8.3 | 306 | 253 | 47 | 1 | 7.4-10.3 |
| mms4Δ | 555 | 508 | 47 | 8.5 | 32 | 21 | 5 | 0 | 5.8-13.5 |
| mlh3Δ mms4Δ | 1336 | 1304 | 32 | 2.4 | 170 | 158 | 2 | 0 | 0.2-1.1 |
| LEU2-CEN3 | |||||||||
| Wild typea | 2711 | 2527 | 184 | 6.8 | 572 | 488 | 68 | 0 | 5.4-6.8 |
| mlh3Δ | 1453 | 1314 | 139 | 9.6 | 306 | 261 | 39 | 1 | 6.1-8.9 |
| mms4Δ | 555 | 482 | 73 | 13.2 | 32 | 22 | 3 | 1 | 5.8-28.8 |
| mlh3Δ mms4Δ | 1336 | 1302 | 34 | 2.5 | 170 | 156 | 4 | 0 | 0.6-1.9 |
| CEN3-MAT | |||||||||
| Wild typea | 2711 | 2309 | 402 | 14.8 | 572 | 395 | 160 | 1 | 13.9-15.9 |
| mlh3Δ | 1453 | 1246 | 207 | 14.2 | 306 | 223 | 78 | 0 | 11.7-14.2 |
| mms4Δ | 555 | 464 | 91 | 16.4 | 32 | 23 | 3 | 0 | 2.6-8.9 |
| mlh3Δ mms4Δ | 1336 | 1288 | 48 | 8.5 | 170 | 153 | 6 | 1 | 1.8-5.8 |
| Chromosome VII | |||||||||
| TRP5-CYH2 | |||||||||
| Wild typea | 2711 | 1803 | 908 | 33.5 | 572 | 197 | 337 | 9 | 34.2-37.8 |
| mlh3Δ | 1453 | 1215 | 238 | 16.4 | 306 | 198 | 100 | 0 | 15.4-18.2 |
| mms4Δ | 555 | 391 | 164 | 29.5 | 32 | 11 | 11 | 0 | 19.7-30.3 |
| mlh3Δ mms4Δ | 1336 | 1289 | 47 | 3.5 | 170 | 151 | 11 | 0 | 2.4-4.4 |
| CYH2-MET1: | |||||||||
| Wild typea | 2711 | 2451 | 260 | 9.6 | 572 | 442 | 101 | 0 | 8.5-10.1 |
| mlh3Δ | 1453 | 1350 | 103 | 7.1 | 306 | 266 | 32 | 0 | 4.5-6.3 |
| mms4Δ | 555 | 500 | 55 | 9.9 | 32 | 18 | 4 | 0 | 5.0-13.2 |
| mlh3Δ mms4Δ | 1336 | 1302 | 34 | 2.5 | 170 | 156 | 6 | 0 | 1.1-3.0 |
| MET13-LYS5: | |||||||||
| Wild typea | 2711 | 2152 | 559 | 20.6 | 572 | 334 | 205 | 4 | 19.6-22.6 |
| mlh3Δ | 1453 | 1307 | 146 | 10.0 | 306 | 242 | 55 | 1 | 8.7-11.7 |
| mms4Δ | 555 | 461 | 94 | 16.9 | 32 | 15 | 7 | 0 | 10.9-20.9 |
| mlh3Δ mms4Δ | 1336 | 1271 | 65 | 4.9 | 170 | 148 | 14 | 0 | 3.2-5.4 |
| Chromosome VIII | |||||||||
| CEN8-THR1: | |||||||||
| Wild typea | 2711 | 2105 | 606 | 22.4 | 572 | 317 | 219 | 2 | 20.2-22.8 |
| mlh3Δ | 1453 | 1305 | 148 | 10.2 | 306 | 251 | 45 | 0 | 6.6-8.6 |
| mms4Δ | 555 | 463 | 92 | 16.6 | 32 | 16 | 6 | 0 | 8.9-18.4 |
| mlh3Δ mms4Δ | 1336 | 1288 | 48 | 3.6 | 170 | 157 | 3 | 0 | 0.4-1.5 |
| THR1-CUP1: | |||||||||
| Wild typea | 2711 | 2043 | 668 | 24.6 | 572 | 277 | 260 | 1 | 23.5-25.9 |
| mlh3Δ | 1453 | 1258 | 195 | 13.4 | 306 | 226 | 69 | 1 | 11.1-14.2 |
| mms4Δ | 555 | 427 | 128 | 23.1 | 32 | 14 | 8 | 0 | 13.1-23.3 |
| mlh3Δ mms4Δ | 1336 | 1292 | 44 | 3.3 | 170 | 154 | 6 | 0 | 1.1-2.6 |
Strains analyzed are isogenic derivatives of the SK1 NHY942/943 background (Tables 1 and 2). Single spore data are shown with n, total number of spores, and parental and recombinant data. Map distances (cM) were calculated by recombination frequency (recombinant spores/total spores) × 100. Tetrad data are shown with n, number of complete tetrads. Map distances (cM) were calculated using the Perkins formula (Perkins 1949), and 95% confidence intervals were calculated using the Stahl Laboratory Online Tools website (http://www.molbio.uoregon.edu/∼fstahl/).
Data from Zanders and Alani (2009).
Previous work showed that mms4Δ strains display low spore efficiency (∼10%) and viability (∼40%) as well as high levels of aberrant recombination events (De Los Santos et al. 2001, 2003). We found that the mlh3Δ mutation can partially suppress the spore viability, sporulation defects, and high frequency of aberrant events observed in mms4Δ strains (Tables 6 and 8). In the SK1 isogenic background NHY942/943, mms4Δ strains displayed low sporulation efficiency (16%) and viability (45%) whereas mlh3Δ displayed greater levels of spore formation (73%, P < 0.001) and viability (79%, P < 0.001). mlh3Δ mms4Δ displayed significantly greater sporulation (43%; P < 0.001) and viability (62%; P < 0.001) compared to mms4Δ. In addition, mlh3Δ mms4Δ mutants showed gene conversion levels that were similar to wild-type but lower than mms4Δ alone (Table 8; aberrant levels for our small mms4Δ data set are similar to those seen in De Los Santos et al. (2003), who analyzed 272 tetrads).
Table 8. Aberrant marker segregation in wild type, mlh3Δ, mms4Δ, and mlh3Δ mms4Δ on chromosomes III, VII, and VIII.
| Chromosome III | Four-spore viable tetrads | HIS4 | LEU2 | ADE2 | MATa | Total |
|---|---|---|---|---|---|---|
| Wild type | 572 | 2.1 | 0.3 | 0.2 | 0.2 | 2.8 |
| mlh3Δ | 306 | 0.7 | 0.7 | 0.3 | 0.0 | 1.7 |
| mms4Δ | 32 | 9.4 | 6.3 | 3.1 | 3.1 | 21.9 |
| mlh3Δ mms4Δ | 170 | 4.1 | 0.6 | 0 | 1.2 | 5.9 |
| Chromosome VII | LYS5 | MET13 | CYH2 | TRP5 | ||
| Wild type | 572 | 1.6 | 2.4 | 0.3 | 0.7 | 5.0 |
| mlh3Δ | 306 | 0.7 | 2.0 | 0.0 | 0.0 | 2.7 |
| mms4Δ | 32 | 9.4 | 0.0 | 6.3 | 0.0 | 15.7 |
| mlh3Δ mms4Δ | 170 | 1.2 | 2.4 | 0.0 | 1.2 | 4.8 |
| Chromosome VIII | URA3 | THR1 | CUP1 | |||
| Wild type | 572 | 0.2 | 5.1 | 0.7 | 6.0 | |
| mlh3Δ | 306 | 0.0 | 3.3 | 0.0 | 3.3 | |
| mms4Δ | 32 | 0.0 | 6.3 | 9.4 | 15.7 | |
| mlh3Δ mms4Δ | 170 | 0.6 | 4.7 | 0.6 | 5.9 |
Aberrant segregation (1:3 or 3:1) of markers is shown. Data are from four-spore viable tetrads analyzed by RANA software (Argueso et al. 2004). Strains analyzed are isogenic derivatives of the SK1 NHY942/943 background (Tables 1 and 2).
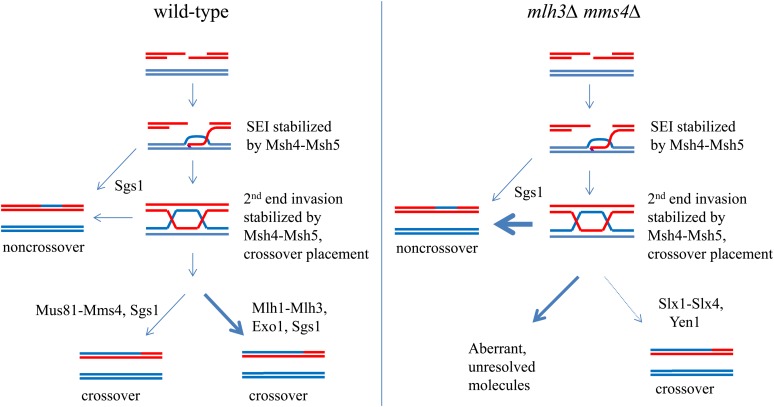
Our measurements of gene conversion in mlh3Δ mms4Δ mutants, coupled with previous analyses of recombination intermediates in crossover resolution mutants, are consistent with meiotically induced DSBs forming at wild-type levels in mlh3Δ mms4Δ strains [Table 8 (Argueso et al. 2004; Nishant et al. 2010; Zakharyevich et al. 2012). Based on this argument, we are left trying to understand how recombination intermediates in mlh3Δ mms4Δ are repaired. Previous genetic and physical studies have identified roles for Sgs1 in resolving aberrant joint molecules that form during meiosis in mutants defective in Mus81-Mms4 and Mlh1-Mlh3 crossover pathways (Van Brabant et al. 2000; Adams et al. 2003; Rockmill et al. 2003; Wu and Hickson 2003; McVey et al. 2004; Bachrati et al. 2006; Jessop et al. 2006; Oh et al. 2007, 2008; Cejka and Kowalczykowski 2010; De Muyt et al. 2012; Zakharyevich et al. 2012). Based on the aforementioned studies we hypothesize that Sgs1 is acting to resolve joint molecules into noncrossovers in mlh3Δ mms4Δ mutants (Figure 5). One explanation for why the spore viability of mms4Δ is lower than that seen in mlh3Δ mms4Δ is that in mms4Δ mutants Mlh1-Mlh3 competes with Sgs1 for joint molecule substrates but is unable to efficiently resolve them. The explanation is consistent with chromosome segregation defects seen in mms4 mutants and the finding that sgs1mms4 mutants accumulate high levels of joint molecules in meiosis (Oh et al. 2008).
Figure 5 .
Model of crossover pathways during meiosis. A summary of the crossover pathways are shown. In wild-type cells (left), DSBs are made and resected, and initial single-end invasion intermediates can be dissolved by Sgs1−dependent mechanisms, leading to noncrossovers. Single-end invasion intermediates that are not resolved as noncrossovers can proceed through the Mus81-Mms4 interference-independent pathway, leading to crossovers, or Msh4-Msh5 can stabilize the SEI in an interference-dependent mechanism. These stabilized joint molecules undergo crossover placement decisions, and are subsequently resolved in an Mlh1-Mlh3-dependent manner. In the absence of Mlh3 and Mms4 (right), initial recombination events occur as in wild type. However, due to the lack of the major Mlh1-Mlh3 and Mus81-Mms4 resolvase functions, other pathways are activated, including Sgs1-dependent resolution to form noncrossovers and other resolution activities (e.g., Slx-Slx4, Yen1), resulting in a larger number of events being resolved into noncrossovers.
Chromosome disjunction appears mostly functional in mlh3Δ mms4Δ despite dramatic genome-wide decreases in crossing over
As indicated previously, spore viability in mlh3Δ mms4Δ is high (62%) despite large reductions (6- to 17-fold) in crossing over. Such reduced levels should yield crossover levels below the obligate number (16) required to segregate all yeast homologs. If we assume that crossover levels decrease to similar extents across the length of a single chromosome, then only chromosome VII would appear to have at least one crossover in mlh3Δ mms4Δ. This calculation is based on high-resolution genotyping of meiotic spore progeny performed by Mancera et al. (2008). They observed in wild-type an average of three, eight, four, and seven crossovers on chromosomes III, VII, VIII, and XV, respectively. Based on these values, multiple chromosomes are unlikely to receive a crossover during meiosis in mlh3Δ mms4Δ.
We offer two explanations for the high spore viability in mlh3Δ mms4Δ, both of which assume achiasmate chromosome disjunction mechanisms. The first suggests that the high spore viability is due to distributive disjunction, which is defined as the process in which “two nonhomologous chromosomes that lack homologs or two homologs that have failed to recombine, disjoin at meiosis I” (Guacci and Kaback 1991). Distributive disjunction has been shown to accurately segregate chromosomes in male Drosophila meiosis and the fourth chromosome in female Drosophila meiosis (Grell 1962, 1976). It also plays a role in budding yeast (Guacci and Kaback 1991; Loidl et al. 1994). However, distributive disjunction in budding yeast acts independently of chromosome homology and chromosome size, at least when only three achiasmate elements are present (Guacci and Kaback 1991; Loidl et al. 1994; Ross et al. 1996). Based on this observation, it is unlikely that such a system would efficiently act to segregate chromosomes in meiosis I if multiple chromosomes lacked chiasma. Indeed, hybrid yeast strains that have severely reduce recombination due to high sequence divergence display low spore viability (∼1%; Hawthorne and Philippsen 1994; Hunter et al. 1996).
A second explanation is that homologous pairing mechanisms are taking place in mlh3Δ mms4Δ that promote disjunction of homologs in the absence of crossing over. We can imagine two ways that this could happen: (1) Chromosome disjunction in mlh3Δ mms4Δ is facilitated by Zip1, a synaptonemal complex protein that promotes homology-independent centromere pairing (Tsubouchi and Roeder 2005; Gladstone et al. 2009; Newnham et al. 2010). Zip1 promotes centromere pairing in both nonhomologous chromosomes and nonexchange homologous chromosomes, providing a mechanism for nonexchange chromosomes to be held together until the first meiotic division, possibly by promoting correct spindle orientation (Newnham et al. 2010; Gladstone et al. 2009). (2) Msh4-Msh5 acts to facilitate disjunction in mlh3Δ mms4Δ by promoting homolog pairing. Consistent with this idea, Msh5 has been shown to act in early steps in homolog pairing in mice and Sordaria (Edelmann et al. 1999; Storlazzi et al. 2010). Experiments aimed at testing these ideas are in progress.
Acknowledgments
We thank members of the Alani laboratory and Sarah Zanders for helpful comments and Rhona Borts for providing information prior to publication of Cotton et al. (2010). M.S.B. was supported by a National Institutes of Health (NIH) Training Grant in Molecular and Cellular Biology. E.L. was supported by a Howard Hughes Medical Institute undergraduate summer research fellowship awarded to Cornell University, and C.C. and E.A. were supported by NIH GM53085. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the NIH.
Footnotes
Communicating editor: M. Johnston
Literature Cited
- Adams M. D., McVey M., Sekelsky J. J., 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267 [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M., 2001. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8: 225–231 [DOI] [PubMed] [Google Scholar]
- Argueso J. L., Kijas A. W., Sarin S., Heck J., Waase M., et al. , 2003. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol. Cell. Biol. 23: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Wanat J., Gemici Z., Alani E., 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168: 1805–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati C. Z., Borts R. H., Hickson I. D., 2006. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 34: 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C., Yang W., 1998. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell 95: 541–552 [DOI] [PubMed] [Google Scholar]
- Ban C., Junop M., Yang W., 1999. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell 97: 85–97 [DOI] [PubMed] [Google Scholar]
- Berchowitz L. E., Francis K. E., Bey A. L., Copenhaver G. P., 2007. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Gaillard P. H., McDonald W. H., Shanahan P., Yates J. R., et al. , 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 [DOI] [PubMed] [Google Scholar]
- Börner G. V., Kleckner N., Hunter N., 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N., 1990. A pathway for generation and processing of double strand breaks during meiotic recombination in S. cerevisiae. Genetics 185: 459–467 [DOI] [PubMed] [Google Scholar]
- Cejka P., Kowalczykowski S. C., 2010. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J. Biol. Chem. 285: 8290–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Y., Tsubouchi T., Rockmill B., Sandler J. S., Richards D. R., et al. , 2008. Global analysis of the meiotic crossover landscape. Dev. Cell 15: 401–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne R. K., Katis V. L., Jessop L., Benjamin K. R., Herskowitz I., et al. , 2003. Polo-like kinase Cdc5 promotes chiasmate formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5: 480–485 [DOI] [PubMed] [Google Scholar]
- Cotton V. E., Hoffman E. R., Borts R. H., 2010. Distinct regulation of Mlh1p heterodimers in meiosis and mitosis in Saccharomyces cerevisiae. Genetics 185: 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie G. A., Hyppa R. W., Taylor A. F., Zakharyevich K., Hunter N., et al. , 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127: 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Santos T., Loidl J., Larkin B., Hollingsworth N. M., 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159: 1511–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Santos T., Hunter N., Lee C., Larkin B., Loidl J., et al. , 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon F. J., Massey W. J., 1969. Introduction to Statistical Analysis, Ed. 3 McGraw-Hill, New York [Google Scholar]
- Drake J. W., 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88: 7160–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W., Cohen P. E., Kneitz B., Winand N., Lia M., et al. , 1999. Mammalian MutS homolog 5 is required for chromosome pairing in meiosis. Nat. Genet. 21: 123–127 [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H., Kolodner R. D., 1998. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. USA 95: 12404–12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W. M., Brill S. J., 2003. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 17: 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Kimura S., Ishibashi T., Mori Y., Hashimoto J., et al. , 2003. OsSEND-1: a new RAD2 nuclease family member in higher plants. Plant Mol. Biol. 51: 59–70 [DOI] [PubMed] [Google Scholar]
- Gaskell L. J., Osman F., Gilbert R. J., Whitby M. C., 2007. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? EMBO J. 26: 1891–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A., 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Gilbertson L. A., Stahl F. W., 1996. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics 144: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladstone M. N., Obeso D., Chuong H., Dawson D. S., 2009. The synaptonemal complex protein Zip1 promotes bi-orientation of centromeres at meiosis I. PLoS Genet. 5: e1000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Grell R. F., 1962. A new hypothesis on the nature and sequence of meiotic events in the female of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 48: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell R. F., 1976. Distributive pairing. Genetics and Biology of Drosophila 1: 425–486 [Google Scholar]
- Guacci V., Kaback D. B., 1991. Distributive disjunction of authentic chromosomes in Saccharomyces cerevisiae. Genetics 127: 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. C., Shcherbakova P. V., Kunkel T. A., 2002. Differential ATP binding and intrinsic ATP hydrolysis by amino-terminal domains of the yeast Mlh1 and Pms1 proteins. J. Biol. Chem. 277: 3673–3679 [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Minesinger B. K., Jinks-Robertson S., 2000. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 10: 145–148 [DOI] [PubMed] [Google Scholar]
- Hawthorne D., Philippsen P., 1994. Genetic and molecular analysis of hybrids in the genus Saccharomyces involving S. cerevisiae, S. uvarum and a new species, S. douglasii. Yeast 10: 1285–1296 [DOI] [PubMed] [Google Scholar]
- Heck J. A., Argueso J. L., Gemici Z., Reeves R. G., Bernard A., et al. , 2006. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc. Natl. Acad. Sci. USA 103: 3256–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. D., Buckling E. F., Franklin F. C., Jones G. H., 2008. Expression and functional analysis of ATMUS81 in Arabidopsis meiosis revelas a role in the second pathway of crossing-over. Plant J. 54: 152–162 [DOI] [PubMed] [Google Scholar]
- Hoffman E. R., Borts R. H., 2004. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet. Genome Res. 197: 232–248 [DOI] [PubMed] [Google Scholar]
- Hoffman E. R., Shcherbakova P. V., Kunkel T. A., Borts R. H., 2003. MLH1 mutations diferentially affect meiotic functions in Saccharomyces cerevisiae. Genetics 163: 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Brill S. J., 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Ponte L., Halsey C., 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739 [DOI] [PubMed] [Google Scholar]
- Holloway J. K., Booth J., Edelmann W., McGowan C. H., Cohen P. E., 2008. MUS81 generates a subset of MLH1–MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4: e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Borts R. H., 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11: 1573–1582 [DOI] [PubMed] [Google Scholar]
- Hunter N., Kleckner N., 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70 [DOI] [PubMed] [Google Scholar]
- Hunter N., Chambers S. R., Louis E. J., Borts R. H., 1996. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15: 1726–1733 [PMC free article] [PubMed] [Google Scholar]
- Ip S. C., Rass U., Blanco M. G., Flynn H. R., Skehel J. M., et al. , 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361 [DOI] [PubMed] [Google Scholar]
- Ishikawa G., Kanai Y., Takata K., Takeuchi R., Shimanouchi K., et al. , 2004. DmGEN, a novel RAD2 family endo-exonuclease from Drosophila melanogaster. Nucleic Acids Res. 32: 6251–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Lichten M., 2008. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell 31: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Rockmill B., Roeder G. S., Lichten M., 2006. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov F. A., Dzantiev L., Constantin N., Modrich P., 2006. Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126: 297–308 [DOI] [PubMed] [Google Scholar]
- Kaliraman V., Mullen J. R., Fricke W. M., Bastin-Shanower S. A., Brill S. J., 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15: 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 357–384 [DOI] [PubMed] [Google Scholar]
- Kleckner N., Zickler D., Jones G. H., Dekker J., Padmore R., et al. , 2004. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA 101: 12592–12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B., Cohen P. E., Avdievich E., Zhu L., Kane M. F., et al. , 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14: 1085–1097 [PMC free article] [PubMed] [Google Scholar]
- Kondo E., Horii A., Fukushige S., 2001. The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res. 29: 1695–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Erie D. A., 2005. DNA mismatch repair. Annu. Rev. Biochem. 74: 681–710 [DOI] [PubMed] [Google Scholar]
- Loidl J., Klein F., Scherthan H., 1994. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J. Cell Biol. 125: 1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E., Diaz R. L., Hunter N., Keeney S., 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., Blanco M. G., Maslen S., Skehel J. M., West S. C., 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Larocque J. R., Adams M. D., Sekelsky J. J., 2004. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. USA 101: 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz I. M., Hain K., Declais A. C., Gardiner M., Toh G. W., et al. , 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell 35: 116–127 [DOI] [PubMed] [Google Scholar]
- Newnham L., Jordan P., Rockmill B., Roeder G. S., Hoffmann E., 2010. The synaptonemal complex protein, Zip1, promotes the segregation of nonexchange chromosomes at meiosis I. Proc. Natl. Acad. Sci. USA 107: 781–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant K. T., Plys A. J., Alani E., 2008. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179: 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant K. T., Chen C., Shinohara M., Shinohara A., Alani E., 2010. Genetic analysis of baker’s yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability. PLoS Genet. 6: e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Lao J. P., Hwang P. Y., Taylor A. F., Smith G. R., et al. , 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Lao J. P., Taylor A. F., Smith G. R., Hunter N., 2008. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell 31: 324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzi G., Perrera C., Blaser H., Kuster P., Marra G., 2001. Direct association of Bloom’s syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 29: 4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., 1949. Biochemical mutants in the smut fungus Ustilago maydis. Genetics 34: 607–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N., Uematsu N., Amiot F., Gidrol X., Barillot E., et al. , 2007. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 275: 1868–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Fung J. C., Branda S. S., Roeder G. S., 2003. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 13: 1954–1962 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Ross L. O., Rankin S., Shuster M. F., Dawson D. S., 1996. Effects of homology, size and exchange of the meiotic segregation of model chromosomes in Saccharomyces cerevisiae. Genetics 142: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P., Roeder G. S., 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080 [DOI] [PubMed] [Google Scholar]
- Sacho E. J., Kadyrov F. A., Modrich P., Kunkel T. A., Erie D. A., 2008. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol. Cell 29: 112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci-Darmanin S., Walpita D., Lespinasse F., Desnuelle C., Ashley T., et al. , 2000. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 14: 1539–1547 [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin S., Neyton S., Lespinasse F., Saunieres A., Gaudray P., et al. , 2002. The DNA mismatch-repair MLH3 protein interacts with MSh4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum. Mol. Genet. 11: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791 [DOI] [PubMed] [Google Scholar]
- Schwartz E. K., Heyer W. D., 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova P. V., Kunkel T. A., 1999. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol. 19: 3177–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M., Oh S. D., Hunter N., Shinohara A., 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40: 299–309 [DOI] [PubMed] [Google Scholar]
- Smith G. R., Boddy M. N., Shanahan P., Russell P., 2003. Fission yeast Mus81-Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165: 2289–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R., 2004. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451 [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Foss H. M., Young L. S., Borts R. H., Abdullah M. F., et al. , 2004. Does crossover interference count in Saccharomyces cerevisiae? Genetics 168: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A., Gargano S., Ruprich-Robert G., Falque M., David M., et al. , 2010. Recombination proteins mediate meiotic spatial organization and pairing. Cell 141: 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen J. M., Smorgorzewska A., Sowa M. E., O’Connell B. C., Gygi S. P., et al. , 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlanov A., Cohen P. E., 2004. Mismatch repair proteins, meiosis, and mice: understanding the complexities of mammalian meiosis. Exp. Cell Res. 296: 71–79 [DOI] [PubMed] [Google Scholar]
- Tran P. T., Liskay R. M., 2000. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha. Mol. Cell. Biol. 20: 6390–6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H. T., Keen J. D., Kricker M., Resnick M. A., Gordenin D. A., 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 17: 2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. T., Simon J. A., Liskay R. M., 2001. Interactions of Exo1p with components of MutLα in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98: 9760–9765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge K., McKim K., Brill S. J., Sekelsky J., 2007. Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics 176: 1993–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T., Roeder G. S., 2005. A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308: 870–873 [DOI] [PubMed] [Google Scholar]
- Van Brabant A. J., Ye T., Sanz M., German J. L., Ellis N. A., et al. , 2000. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39: 14617–14625 [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808 [DOI] [PubMed] [Google Scholar]
- Wanat J. J., Singh N., Alani E., 2007. The effect of genetic background on the function of Saccharomyces cerevisiae mlh1 alleles that correspond to HNPCC missense mutations. Hum. Mol. Genet. 16: 445–452 [DOI] [PubMed] [Google Scholar]
- Wang T. F., Kleckner N., Hunter N., 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes ith distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96: 13914–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M. C., 2005. Making crossovers during meiosis. Biochem. Soc. Trans. 33: 1451–1455 [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I. D., 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Zakharyevich K., Ma Y., Tang S., Hwang P. Y., Boiteux S., et al. , 2010. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday Junctions. Mol. Cell 40: 1001–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S., Alani E., 2009. The pch2Delta mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 5: e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., 2006. From early homologue recognition to synaptonemal complex formation. Chromosoma 115: 158–174 [DOI] [PubMed] [Google Scholar]