Abstract
In applying the Poisson-Boltzmann (PB) equation for calculating the electrostatic free energies of solute molecules, an open question is how to specify the boundary between the low-dielectric solute and the high-dielectric solvent. Two common specifications of the dielectric boundary, as the molecular surface (MS) or the van der Waals (vdW) surface of the solute, give very different results for the electrostatic free energy of the solute. With the same atomic radii, the solute is more solvent-exposed in the vdW specification. One way to resolve the difference is to use different sets of atomic radii for the two surfaces. The radii for the vdW surface would be larger in order to compensate for the higher solvent exposure. Here we show that radius re-parameterization required for bringing MS-based and vdW-based PB results to agreement is solute-size dependent. The difference in atomic radii for individual amino acids as solutes is only 2–5% but increases to over 20% for proteins with ~200 residues. Therefore two sets of radii that yield identical MS-based and vdW-based PB results for small solutes will give very different PB results for large solutes. This finding raises issues about two common practices. The first is the use of atomic radii, which are parameterized against either experimental solvation data or data obtained from explicit-solvent simulations on small compounds, for PB calculations on proteins. The second is the parameterization of vdW-based generalized Born models against MS-based PB results.
I. Introduction
The Poisson-Boltzmann (PB) equation is widely used for modeling electrostatic effects and solvation of biomolecules.1–30 The calculated electrostatic free energy of a solute molecule depends on the permanent partial charges on the atoms of the solute and the boundary of the low-dielectric solute and the high-dielectric solvent. Even when the radii of the atoms are given, there is still considerable freedom in specifying the dielectric boundary. In particular, two choices widely used in PB calculations are the van der Waals (vdW) surface and the molecular surface (MS) (see Figure 1). The vdW surface consists of the exposed surfaces of the spheres representing the solute atoms. The MS, introduced by Richards,31 relies on a spherical solvent probe. According to the MS, the atomic spheres and all crevices inaccessible to the solvent probe are all treated as part of the solute dielectric (the MS hence has also been referred to as the solvent-exclusion surface). The added crevices reduce the exposure of the solute charges to the solvent. Since solute charges have strong interactions with the solvent, the cumulative effects of the reduced solvent exposure of all the solute atoms can lead to a significant change in the electrostatic solvation energy. As a result, the electrostatic interaction free energy between an oppositely charged protein-protein pair or protein-RNA pair can change from negative to positive when the dielectric boundary is switched from vdW to MS.13, 20, 29, 30, 32 The electrostatic contribution of even a single mutation to the folding stability of a protein or the binding stability of a protein-protein or protein-RNA complex can be predicted very differently by the two choices of the dielectric boundary.8, 10, 13, 20, 32 One possible way to reconcile the differences in calculated electrostatic free energy is to use different sets of atomic radii for the two choices of the dielectric boundary.5, 26 Specifically, to compensate for the higher solvent exposure by the vdW specification, atomic radii would be increased relative to those in the MS specification. We carried out such radius re-parameterization and found that the changes in atomic radii are very dependent on the solute size. The difference in atomic radii for individual amino acids as solutes is only 2–5% but increases to over 20% for proteins with ~200 residues.
Figure 1.
Definitions of (a) the van der Waals surface and (b) the molecular surface. In this two-dimensional illustration, atoms are represented by gray disks. In (a), the exposed boundaries of the disks, shown in dark, constitute the van der Waals surface. In (b), a spherical solvent probe is rolled around the solute molecule. In addition to the van der Waals spheres, small crevices inaccessible to the solvent probe are now part of the solute region. The boundary of this filled-up solute region, shown in dark, is the molecular surface.
There is a widely held perception that, between vdW and MS, the latter is a better choice for the dielectric boundary, though a convincing argument has not been laid out. To the contrary, it has been shown that PB calculations with the vdW choice consistently give better agreement with experimental results for mutational effects on protein folding and binding stability8, 10, 13, 20, 32 and for electrostatic contributions to protein binding rates.29, 30 This paper does not aim to settle the difference between MS and vdW. Rather, the significance of our finding lies in its implications for two common practices in PB calculations. The first is parameterization of atomic radii using either experimental solvation data or explicit-solvent simulations, which are restricted to small solute molecules.19, 23, 26, 33, 34 Our finding would suggest that, on these solute molecules, the values of atomic radii obtained using either vdW or MS as the dielectric boundary differ very little (e.g., < 5%). However, when these radii are then used for PB calculations on proteins, the electrostatic solvation energies will be very different depending whether vdW or MS is specified as the dielectric boundary. The uncertainty on calculated solvation energies for proteins thus diminishes the value of experimental and explicit-solvent data on small solutes for parameterizing the PB model.
The second common practice occurs in developing generalized Born (GB) methods35 as a fast substitute of the PB model. In some GB methods, the MS specification of the dielectric boundary is directly implemented and the GB results are benchmarked against MS-based PB results.36–39 In many other GB methods,40–46 the vdW specification of the dielectric boundary is implemented, and the resulting GB results are then benchmarked against the MS-based PB results through additional parameterization. Our finding suggests that the parameterization required for matching vdW-based GB and MS-based PB is protein dependent, and the use of a uniform set of parameterization introduces a new source of error for individual proteins.
II. Calculation Details
We carried out different sets of PB calculations over 55 test proteins. One set, used as the target, had MS as the dielectric boundary and Bondi radii47 for the protein atoms. All the other sets had vdW as the dielectric boundary and the atomic radii increased by various percentages (denoted as %Δr) from the Bondi values. The aim of the variation in %Δr was to find the value which would lead to agreement in the electrostatic solvation energy, ΔGsolv, between the MS-based and vdW-based calculations for a particular protein. At the end, a collection of 55 “optimized” %Δr values was obtained for the test proteins.
The 55 test proteins have been used in our previous studies to find an empirical dependence of Δ Gsolv on solute and solvent dielectric constants21 and to develop GB methods as substitutes of the linearized and full PB equation.48, 49 These proteins were collected from the Protein Data Bank (http://www.rcsb.org/pdb) using the following criteria: sequence identity less than 10%, resolution better than 1.0 Å, and number of residues less than 250. For protein structures without hydrogen atoms, hydrogen atoms were added with the LEAP module in the AMBER package,50 then energy minimized in vacuum with heavy atoms fixed. The PDB code, total number of atoms (Natom), net charge (Q) for each of the 55 test proteins are listed in Table I.
Table 1.
Number of atoms, net charge, and MS and vdW solvation energy (in kcal/mol) for 55 test proteins
| PDB | Natom | Q | ΔGsolvMS | ΔGsolvvdW(0%Δr) |
|---|---|---|---|---|
| 1a6m | 2435 | 2 | −1893.3 | −2716.3 |
| 1aho | 967 | −2 | −943.3 | −1299.3 |
| 1byi | 3383 | −4 | −2408.9 | −3578.1 |
| 1c75 | 987 | −4 | −1094.4 | −1398.5 |
| 1c7k | 1929 | −5 | −1672.0 | −2439.7 |
| 1cex | 2867 | 1 | −1863.7 | −2873.1 |
| 1eb6 | 2572 | −15 | −4062.5 | −5094.0 |
| 1ejg | 678 | 0 | −356.4 | −574.7 |
| 1etl | 145 | 0 | −213.1 | −288.2 |
| 1exr | 2240 | −25 | −8081.8 | −9253.6 |
| 1f94 | 982 | 1 | −858.6 | −1206.0 |
| 1f9y | 2535 | −5 | −2018.1 | −2915.9 |
| 1g4i | 1842 | −1 | −1659.2 | −2402.0 |
| 1g66 | 2794 | −2 | −1628.6 | −2945.2 |
| 1gqv | 2143 | 7 | −1768.6 | −2561.5 |
| 1hje | 179 | 1 | −221.7 | −275.3 |
| 1iqz | 1171 | −17 | −4149.4 | −4598.3 |
| 1iua | 1207 | −1 | −873.0 | −1289.8 |
| 1j0p | 1597 | 8 | −2242.3 | −2810.4 |
| 1k4i | 3253 | −6 | −2696.4 | −3888.8 |
| 1kth | 894 | 0 | −1104.7 | −1454.4 |
| 1l9l | 1230 | 11 | −2684.4 | −3084.0 |
| 1m1q | 1265 | −7 | −1945.0 | −2379.0 |
| 1nls | 3564 | −7 | −2927.5 | −4680.5 |
| 1nwz | 1912 | −6 | −2015.0 | −2728.9 |
| 1od3 | 1900 | −3 | −1307.3 | −2026.4 |
| 1ok0 | 1076 | −5 | −1153.9 | −1546.3 |
| 1p9g | 529 | 4 | −556.0 | −745.6 |
| 1pq7 | 3065 | 4 | −1484.9 | −2574.2 |
| 1r6j | 1230 | 0 | −972.9 | −1337.4 |
| 1ssx | 2750 | 8 | −1674.4 | −2623.6 |
| 1tg0 | 1029 | −12 | −2815.9 | −3191.5 |
| 1tqg | 1660 | −7 | −2373.2 | −2903.5 |
| 1tt8 | 2676 | 1 | −1655.7 | −2604.9 |
| 1u2h | 1526 | 4 | −1521.1 | −2036.2 |
| 1ucs | 997 | 0 | −705.1 | −1021.8 |
| 1ufy | 1926 | −3 | −1679.0 | −2293.7 |
| 1unq | 1966 | −3 | −2635.0 | −3410.4 |
| 1vb0 | 921 | 3 | −794.7 | −1107.3 |
| 1vbw | 1058 | 8 | −1476.3 | −1805.0 |
| 1w0n | 1756 | −5 | −1685.6 | −2417.1 |
| 1wy3 | 560 | 1 | −600.6 | −768.9 |
| 1x6z | 1741 | 0 | −1511.5 | −2153.4 |
| 1x8q | 2815 | −1 | −2325.5 | −3550.2 |
| 1xmk | 1268 | 1 | −1151.3 | −1589.0 |
| 1yk4 | 770 | −8 | −1578.3 | −1874.2 |
| 1zzk | 1252 | 1 | −1202.8 | −1591.7 |
| 2a6z | 3432 | −3 | −2363.5 | −3636.6 |
| 2bf9 | 560 | −2 | −763.8 | −911.8 |
| 2chh | 1624 | −3 | −1523.6 | −2128.3 |
| 2cws | 3400 | −3 | −1936.4 | −3208.1 |
| 2erl | 573 | −6 | −983.5 | −1167.2 |
| 2fdn | 731 | −8 | −1410.3 | −1702.1 |
| 2fwh | 1830 | −6 | −1629.1 | −2251.1 |
| 3lzt | 1960 | 8 | −1866.9 | −2587.4 |
PB results for ΔGsolv were obtained by using the UHBD program.51 The dielectric boundary was chosen as MS or vdW by the presence or absence of the “nmap 1.4, nsph 500” option in the UHBD input file. By default dielectric smoothing was applied to both choices of the dielectric boundary. UHBD calculations on all the test proteins used a coarse grid with a 1.5-Å spacing followed by a fine grid with a 0.5-Å spacing. The dimensions of the coarse and fine grids were 160 × 160 × 160 and 200 × 200 × 200, respectively. The solute and solvent dielectric constants were set to 1 and 78.5, respectively. No salt was present in the solvent.
For investigating the dependence of optimized %Δr on solute size, we carried out corresponding PB calculations on individual amino acids as solutes. For each of the 20 types of amino acids, 10 conformations were randomly carved out of the 55 test proteins. The UHBD calculations were done on the individual amino acids, with a coarse grid with a 50 × 50 × 50 dimension and a 1.0-Å spacing followed by a fine grid with a 60 × 60 × 60 dimension and a 0.25-Å spacing. For each type of amino acid, the average of optimized %Δr values over the 10 conformations is reported. Results from averaging over 20 conformations for each amino acid were essentially unchanged.
Areas of the dielectric boundary according to the two choices were calculated. For vdW and MS, the respective programs used were Naccess v2.1.1 (http://www.bioinf.manchester.ac.uk/naccess/) with a probe radius of 0 and dms (http://www.cgl.ucsf.edu/chimera/docs/UsersGuide/midas/dms1.html) with a probe radius of 1.4 Å.
III. Results and Discussion
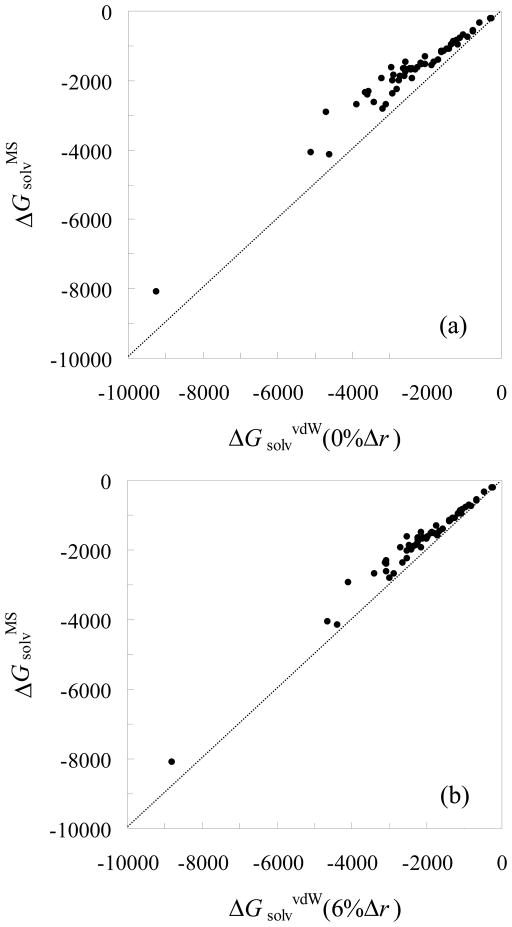
The electrostatic solvation energies of the 55 test proteins, calculated using Bondi radii and either the MS or vdW choice for the dielectric boundary, are listed in Table I and displayed in Figure 2a. It can be seen that the magnitudes of ΔGsolv are consistently larger with the vdW dielectric boundary, due to the resulting higher solvent exposure of solute charges. When the atomic radii are increased in vdW calculations, the magnitudes of ΔGsolv decrease and hence move toward those of the MS results. However, as Figure 2b shows, with a uniform increase of 6% in atomic radii, vdW results still consistently show larger magnitudes than the MS targets.
Figure 2.
Comparison of the electrostatic solvation energies of the 55 test proteins from MS-based and vdW-based PB calculations. For MS-based PB calculations, the Bondi radii are always used. (a) ΔGsolvvdW calculated with Bondi radii. (b) ΔGsolvvdW calculated with atomic radii increased by 6% from the Bondi values.
Figure 3 displays the optimized %Δr values for the 20 types of amino acids as solutes. The increases in atomic radii required to achieve consistency between vdW-based results for ΔGsolv and the MS-based target values are small, falling in the narrow range of 2% to 5%. The small changes in atomic radii are expected. With small solutes, all the atoms are well exposed to the solvent. Hence there are only limited chances that the MS will enclose small crevices outside the vdW surface. Interestingly, even within the narrow range of optimized %Δr values among the 20 types of amino acids, a positive correlation between optimized %Δr and Natom is apparent. Linear regression analysis gave R2 = 0.65.
Figure 3.
The percentage increase in atomic radii from the Bondi values required for ΔGsolvvdW to match with ΔGsolvMS for 20 types of amino acids as solutes.
On the 55 test proteins, the optimized %Δr values increase to at least 10%. As Figure 4a shows, there still seems to be a positive correlation between optimized %Δr and Natom, but the data now exhibits much greater scatter. R2 for linear correlation is now at 0.58. The variations in optimized %Δr within the 20 types of amino acids and within the 55 test proteins as well as between the two collections of solute molecules point to the accumulation of crevices that are outside the vdW surface but inside the MS as the major reason for the increase in optimized %Δr.
Figure 4.
(a) The percent increases in atomic radii, %Δr, for optimal agreement between ΔGsolvvdW and ΔGsolvMS on the 55 test proteins. (b) Percentage of difference in vdW surface area and MS area, 100(SvdW – SMS)/SvdW, against the number of atom.
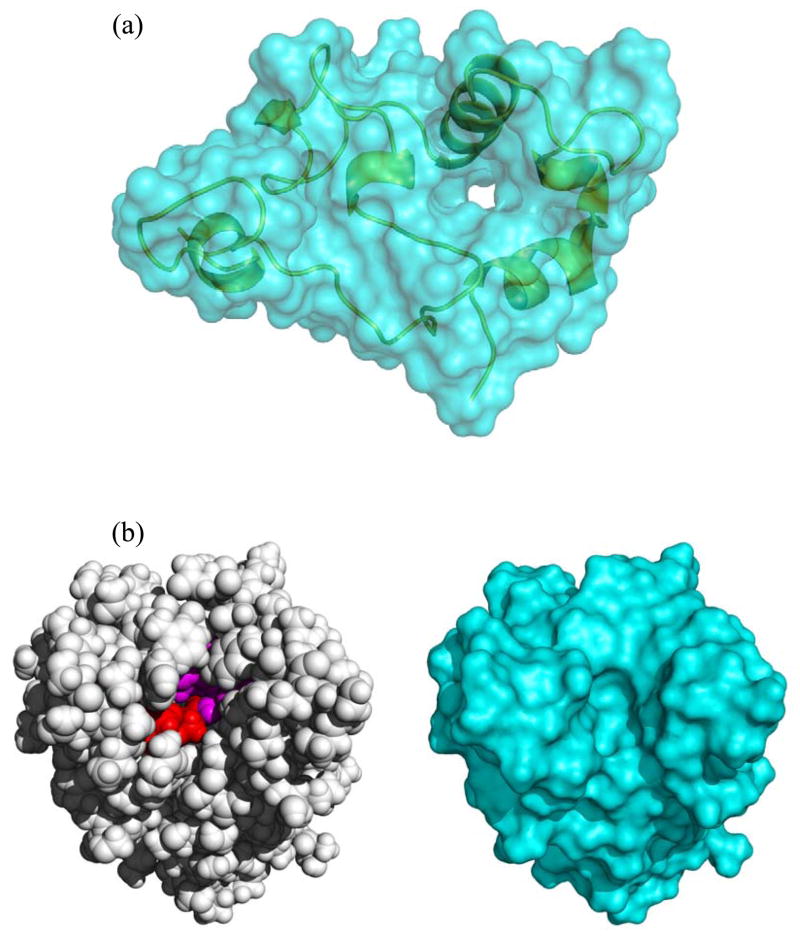
We examined the outliers in the correlation between optimized %Δr and Natom. Some of the low-lying proteins, such as 2bf9, 1m1q, and 1j0p, in the optimized %Δr vs. Natom plot were found to correspond to well-exposed structures (Figure 5a). For these proteins, the difference between the two types of solute surfaces are relatively small, and hence relatively small increases in atomic radii are required to bring vdW-based results for ΔGsolv into agreement with the MS-based target. One way of quantifying the differences between the two types of solute surfaces is by calculating the corresponding surfaces areas. Figure 4b displays %ΔS, the relative differences in MS area and vdW surface area, against Natom. It can be seen that the low-lying proteins, 2bf9, 1m1q, and 1j0p, in the optimized %Δr vs. Natom plot are also below the general trend in the %ΔS vs. Natom plot. However, the correspondence between the two plots is far from being perfect. In particular, a low-lying protein, 1exr, in the %ΔS vs. Natom plot actually occupies a position above the correlation trend line in the optimized %Δr vs. Natom plot, and a high-lying protein, 1etl, in the optimized %Δr vs. Natom plot does not take such a position in the %ΔS vs. Natom plot.
Figure 5.
Comparison of van der Waals and molecular surfaces. (a) A well-exposed protein, 1m1q, which has the shape of a thin disk. The green ribbon representation of the protein is enclosed by the molecular surface in cyan; a hole appears near the center of the disk shape. (b) A protein, 1g66, with a deep channel. In the left panel, the van der Waals surface is presented, and residues lining the wall of the channel are displayed in red (for the catalytic triad) or purple. In the right panel, the molecular surface is presented. The active site now appears as an indent, but there is no channel penetrating into the center of the protein.
We suspected that the high-lying proteins in the optimized %Δr vs. Natom plot correspond to structures with deep channels outside the vdW surface, which become enclosed in the MS and hence are treated as part of the solute dielectric in the MS-based PB calculations. This suspicion did not find support in 1etl, which is the smallest (with Natom = 145) of the 55 test proteins but required a relatively large 17% increase in atomic radii to achieve a match between vdW-based and MS-based results for ΔGsolv. However, a deep channel in the structure of another high-lying protein, acetylxylan esterase with PDB code 1g66, was identified (Figure 5b). Part of the wall of this channel is lined by the catalytic triad; hence this channel is important for access by solvent as well as the substrate. The channel is inaccessible by the 1.4-Å spherical probe used to defined the MS. This example illustrates the artificial nature of using a spherical probe on a static structure to define the boundary between the solute and solvent. Proteins are dynamic, allowing for transient access of water molecules, as seen in NMR experiments52 and molecular dynamics simulations.53 The transient excursions of water molecules into channels and interior positions are accounted for to some extent by choosing the vdW surface as the solute-solvent boundary, which perhaps partly explains the better performance of this choice in reproducing experimental results for electrostatic contributions to protein folding and binding.8, 10, 13, 20, 29, 30, 32
We modeled the trend in the %ΔS vs. Natom plot shown in Figure 4b as a power law,
| (1) |
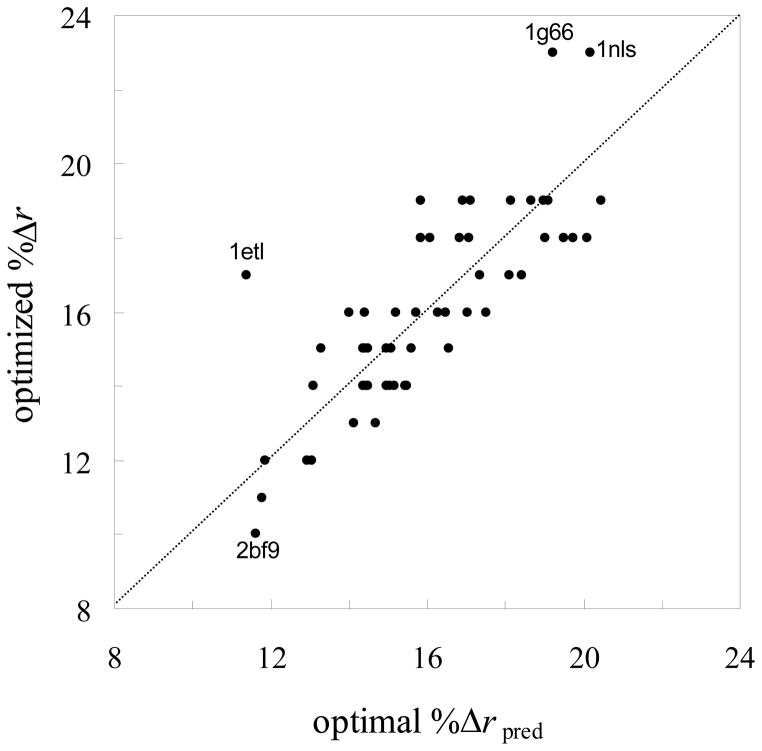
This function, with α = 13.8% and ν = 0.19, fitted the data with R2 = 0.68. Given that the deviations from the trend of eq 1 could explain some of the outliers in Figure 4a, we included the ratio, (%ΔS)/(% ΔSpred), as an independent variable along with Natom in a multi-linear regression to model the variations of optimized %Δr among the 55 test proteins. The inclusion of the new variable led to a modest increase in R2, from 0.58 to 0.65. As Figure 6 shows, there are substantial deviations between actual optimized %Δr values and those predicted from multi-linear regression, especially for 1etl, 2bf9, 1g66, and 1nls. The significant variations in optimized %Δr became apparent after we tested vdW-based and MS-based PB results on a large, diverse collection of proteins. So our study raises caution against using only a small number of test proteins to parameterize the PB model and draw conclusions.
Figure 6.
Comparison of actual optimized %Δr values against those predicted from a multi-linear relation.
Based on the efforts reported here, it seems unlikely that a simple way to predict optimized %Δr values can be found. The chance of bringing MS-based and vdW-based PB results into good agreement for a diverse set of proteins through radius re-parameterization is thus slim. This finding suggests that significant errors are introduced when vdW-based GB methods are parameterized to approximate MS-based PB results. It is interesting to note that, after parameterizing a vdW-based GB method against MS-based PB results for small compounds,40 the deviations of this GB from the MS-based PB were found to increase with increasing sizes of test compounds.54
The overall increase in optimized %Δr with increasing solute size also raises a cautionary note about the use of experimental and explicit-solvent data on small solutes for parameterizing the PB model. Very similar values of atomic radii will be obtained when MS-based and vdW-based PB calculations are benchmarked against the data on small solutes. However, when these radii are then used in respective PB calculations on proteins, the electrostatic solvation energies can differ significantly. Before the issue of the optimal choice for the dielectric boundary is settled, the value of small-solute data seems open to question. This applies not only to MS- and vdW-based PB calculations but also to alternative choices, such as spline-smoothed surfaces, of the dielectric boundary.5, 9, 14, 19, 26, 55–58 A fruitful approach to parameterizing the PB model is to use experimental data obtained on proteins.8, 10, 13, 20, 29, 30, 32
Acknowledgments
This work was supported in part by NIH grant GM058187.
References
- 1.Gilson MK, Sharp KA, Honig B. Calculating the electrostatic potential of molecules in solution: method and error assessment. J Comput Chem. 1987;9:327–335. [Google Scholar]
- 2.Gilson MK, Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4:7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls A, Honig B. A rapid finite difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J Comput Chem. 1991;12:435–445. [Google Scholar]
- 4.Madura JD, Briggs JM, Wade R, Davis ME, Luty BA, Ilin A, Antosiewicz J, Gilson MK, Bagheri B, Scott LR, McCammon JA. Electrostatic and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics program. Comput Phys Commun. 1995;91:57–95. [Google Scholar]
- 5.Nina M, Im W, Roux B. Optimized atomic radii for protein continuum electrostatics solvation forces. Biophys Chem. 1999;78:89–96. doi: 10.1016/s0301-4622(98)00236-1. [DOI] [PubMed] [Google Scholar]
- 6.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rocchia W, Alexov E, Honig B. Extending the applicability of the nonlinear Poisson-Boltzmann equation: multiple dielectric constants and multivalent ions. J Phys Chem B. 2001;105:6507–6514. [Google Scholar]
- 8.Vijayakumar M, Zhou HX. Salt bridges stabilize the folded structure of barnase. J Phys Chem B. 2001;105:7334–7340. [Google Scholar]
- 9.Grant JA, Pickup BT, Nicholls A. A smooth permittivity function for Poisson-Boltzmann solvation methods. J Comput Chem. 2001;22:608–640. [Google Scholar]
- 10.Dong F, Zhou HX. Electrostatic contributions to T4 lysozyme stability: solvent-exposed charges versus semi-buried salt bridges. Biophys J. 2002;83:1341–1347. doi: 10.1016/S0006-3495(02)73904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo R, David L, Gilson MK. Accelerated Poisson-Boltzmann calculations for static and dynamic systems. J Comput Chem. 2002;23:1244–1253. doi: 10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- 12.Zhou HX, Dong F. Electrostatic contributions to the stability of a thermophilic cold shock protein. Biophys J. 2003;84:2216–2222. doi: 10.1016/S0006-3495(03)75027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong F, Vijayakumar M, Zhou HX. Comparison of calculation and experiment implicates significant electrostatic contributions to the binding stability of barnase and barstar. Biophys J. 2003;85:49–60. doi: 10.1016/S0006-3495(03)74453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Q, Luo R. A Poisson-Boltzmann dynamics method with nonperiodic boundary condition. J Chem Phys. 2003;119:11035–11047. [Google Scholar]
- 15.Prabhu NV, Zhu P, Sharp KA. Implementation and testing of stable, fast implicit solvation in molecular dynamics using the smooth-permittivity finite difference Poisson-Boltzmann method. J Comput Chem. 2004;25:2049–2064. doi: 10.1002/jcc.20138. [DOI] [PubMed] [Google Scholar]
- 16.Baker NA. Improving implicit solvent simulations: a Poisson-centric view. Curr Op Struct Biol. 2005;15:137–143. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Dong F, Zhou HX. Electrostatic recognition and induced fit in the κ-PVIIA toxin binding to Shaker potassium channel. J Am Chem Soc. 2005;127:6836–6849. doi: 10.1021/ja042641q. [DOI] [PubMed] [Google Scholar]
- 18.Swanson JMJ, Mongan J, McCammon JA. Limitations of atom-centered dielectric functions in implicit solvation models. J Phys Chem B. 2005;109:14769–14772. doi: 10.1021/jp052883s. [DOI] [PubMed] [Google Scholar]
- 19.Swanson JMJ, Adcock SA, McCammon JA. Optimized radii for Poisson-Boltzmann calculations with the AMBER force field. J Chem Theo Comp. 2005;1:484–493. doi: 10.1021/ct049834o. [DOI] [PubMed] [Google Scholar]
- 20.Dong F, Zhou HX. Electrostatic contribution to the binding stability of protein-protein complexes. Proteins. 2006;65:87–102. doi: 10.1002/prot.21070. [DOI] [PubMed] [Google Scholar]
- 21.Tjong H, Zhou HX. The dependence of electrostatic solvation energy on dielectric constants in Poisson-Boltzmann calculations. J Chem Phys. 2006;125:206101. doi: 10.1063/1.2393243. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, Schlick T. Stereochemistry and position-dependent effects of carcinogens on TATA/TBP binding. Biophys J. 2006;90:1865–1877. doi: 10.1529/biophysj.105.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan C, Yang L, Luo R. How well does Poisson-Boltzmann implicit solvent agree with explicit solvent? A quantitative analysis. J Phys Chem B. 2006;110:18680–18687. doi: 10.1021/jp063479b. [DOI] [PubMed] [Google Scholar]
- 24.Lwin TZ, Zhou R, Luo R. Is Poisson-Boltzmann theory insufficient for protein folding simulations? J Chem Phys. 2006;124:034902. doi: 10.1063/1.2161202. [DOI] [PubMed] [Google Scholar]
- 25.Schnieders MJ, Baker NA, Ren P, Ponder JW. Polarizable atomic multipole solutes in a Poisson-Boltzmann continuum. J Chem Phys. 2007;126:124114. doi: 10.1063/1.2714528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson JMJ, Wagoner JA, Baker NA, McCammon JA. Optimizing the Poisson dielectric boundary with explicit solvent forces and energies: Lessons learned with atom-centered dielectric functions. J Chem Theo Comp. 2007;3:170–183. doi: 10.1021/ct600216k. [DOI] [PubMed] [Google Scholar]
- 27.Qin SB, Zhou HX. Do electrostatic interactions destabilize protein-nucleic acid binding? Biopolymers. 2007;86:112–118. doi: 10.1002/bip.20708. [DOI] [PubMed] [Google Scholar]
- 28.Alsallaq R, Zhou HX. Prediction of protein-protein association rates from a transition-state theory. Structure. 2007;15:215–224. doi: 10.1016/j.str.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Alsallaq R, Zhou HX. Electrostatic rate enhancement and transient complex of protein-protein association. Proteins. 2007 doi: 10.1002/prot.21679. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin SB, Zhou HX. Prediction of salt and mutational effects on the association rate of U1A protein and U1 small nuclear RNA stem/loop II. J Phys Chem B. 2007 doi: 10.1021/jp075919k. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards FM. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 32.Qin SB, Zhou HX. Do electrostatic interactions destabilize protein-nucleic acid binding? Biopolymers. 2007;86:112–118. doi: 10.1002/bip.20708. [DOI] [PubMed] [Google Scholar]
- 33.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
- 34.Nina M, Beglov D, Roux B. Atomic Born radii for continuum electrostatic calculations based on molecular dynamics free energy simulations. J Phys Chem B. 1997;101:5239–5248. [Google Scholar]
- 35.Still A, Tempczyk WC, Hawley RC, Hendrikson R. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 36.Ghosh A, Rapp CS, Friesner RA. Generalized Born model based on a surface integral formulation. J Phys Chem B. 1998;102:10983–10990. [Google Scholar]
- 37.Lee MS, Salsbury FR, Jr, Brooks CL., 3rd Novel generalized Born methods. J Chem Phys. 2002;116:10606–10614. [Google Scholar]
- 38.Lee MS, Feig M, Salsbury FR, Jr, Brooks CL., 3rd New analytic approximation to the standard molecular volume definition its application to generalized Born calculations. J Comput Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 39.Mongan J, Simmerling C, McCammon JA, Case DA, Onufriev A. Generalized Born model with a simple, robust molecular volume correction. J Chem Theo Comp. 2007;3:156–169. doi: 10.1021/ct600085e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu D, Shenkin PS, Hollinger FP, Still WC. The GB/SA continuum model for solvation. A fast analytical method for the calculation of approximate Born radii. J Phys Chem A. 1997;101:3005–3014. [Google Scholar]
- 41.Jayaram B, Sprous D, Beveridge DL. Solvation free energy of biomacromolecules: Parameters for a modified generalized Born model consistent with the AMBER force field. J Phys Chem B. 1998;102:9571–9576. [Google Scholar]
- 42.Onufriev A, Bashford D, Case DA. Modification of the generalized Born model suitable for macromolecules. J Phys Chem B. 2000;104:3712–3720. [Google Scholar]
- 43.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 44.Tsui V, Case DA. Molecular dynamics simulations of nucleic acids with a generalized Born solvation model. J Am Chem Soc. 2000;122:2489–2498. [Google Scholar]
- 45.Onufriev A, Case DA, Bashford D. Effective Born radii in the generalized Born approximation: The importance of being perfect. J Comput Chem. 2002;23:1297–1304. doi: 10.1002/jcc.10126. [DOI] [PubMed] [Google Scholar]
- 46.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 47.Bondi A. van der Waals volumes and radii. J Phys Chem. 1964;68:441–451. [Google Scholar]
- 48.Tjong H, Zhou HX. GBr6: a parameterization-free, accurate, analytical generalized Born method. J Phys Chem B. 2007;111:3055–3061. doi: 10.1021/jp066284c. [DOI] [PubMed] [Google Scholar]
- 49.Tjong H, Zhou HX. GBr6NL: a generalized Born method for accurately reproducing solvation energy of the nonlinear Poisson-Boltzmann equation. J Chem Phys. 2007;126:195102. doi: 10.1063/1.2735322. [DOI] [PubMed] [Google Scholar]
- 50.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER. Vol. 8. University of California; San Fransisco: 2004. [Google Scholar]
- 51.Madura JD, Briggs JM, Wade R, Davis ME, Lutty BA, Ilin A, Antosiewicz J, Gilson MK, Bagheri B, Scott LR, McCammon JA. Electrostatic and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics program. Comput Phys Commun. 1995;91:57–95. [Google Scholar]
- 52.Ernst JA, Clubb RT, Zhou HX, Gronenborn AM, Clore GM. Demonstration of positionally disordered water within a protein hydrophobic cavity by NMR. Science. 1995;267:1813–1817. doi: 10.1126/science.7892604. [DOI] [PubMed] [Google Scholar]
- 53.Damjanovic A, Garcia-Moreno B, Lattman EE, Garcia AE. Molecular dynamics study of water penetration in staphylococcal nuclease. Proteins. 2005;60:433–449. doi: 10.1002/prot.20486. [DOI] [PubMed] [Google Scholar]
- 54.Edinger SR, Cortis C, Shenkin PS, Friesner RA. Solvation free energies of peptides: Comparison of approximate continuum solvation models with accurate solution of the Poisson-Boltzmann equation. J Phys Chem B. 1997;101:1190–1197. [Google Scholar]
- 55.Schaefer M, Karplus M. A comprehensive analytical treatment of continuum electrostatics. J Phys Chem. 1996;100:1578–1599. [Google Scholar]
- 56.Im W, Lee MS, Brooks CL., 3rd Generalized born model with a simple smoothing function. J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 57.Yu Z, Jacobson MP, Friesner RA. What role do surfaces play in GB models? A new-generation of surface-generalized born model based on a novel gaussian surface for biomolecules. J Comput Chem. 2006;27:72–89. doi: 10.1002/jcc.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grant JA, Pickup BT, Sykes MJ, Kitchen CA, Nicholls A. The Gaussian generalized Born model: application to small molecules. Phys Chem Chem Phys. 2007;9:4913–4922. doi: 10.1039/b707574j. [DOI] [PubMed] [Google Scholar]