Abstract
The generation of induced pluripotent stem cells (iPSCs) often results in aberrant epigenetic silencing of the imprinted Dlk1-Dio3 gene cluster, which compromises the cells’ ability to generate entirely iPSC-derived adult mice (“all-iPSC mice”). Here, we show that reprogramming in the presence of ascorbic acid attenuates hypermethylation of Dlk1-Dio3 by enabling a chromatin configuration that interferes with binding of the de novo DNA methyltransferase Dnmt3a. This allowed us to generate all-iPSC mice from mature B cells, which have thus far failed to support the development of exclusively iPSC-derived postnatal animals. Our data demonstrate that transcription factor-mediated reprogramming can endow a defined, terminally differentiated cell type with a developmental potential equivalent to that of embryonic stem cells. More generally, these findings indicate that culture conditions during cellular reprogramming can strongly influence the epigenetic and biological properties of resultant iPSCs.
Introduction
Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) by the enforced expression of defined transcription factor combinations, such as Oct4, Klf4, Sox2 and c-Myc (OKSM)1. Since iPSCs can differentiate into virtually any somatic cell type, they provide an invaluable tool for the study of development and disease2. Recent reports have suggested that, compared to blastocyst-derived embryonic stem cells (ESCs), iPSCs harbor genetic and epigenetic abnormalities, including the dysregulation of imprinted genes, gene copy number variations, accumulation of point mutations and aberrant methylation patterns3. To harness the full potential of iPSCs technology, it is important to understand the mechanisms underlying these aberrations and to find ways to prevent them.
We have previously used microarrays to show that RNA expression patterns of ESCs and iPSCs are essentially indistinguishable with the exception of a few maternally-expressed, non-coding transcripts (e.g., Gtl2 and Rian) and miRNAs originating from the imprinted Dlk1-Dio3 gene cluster4, which is silenced in the majority of iPSC lines5. We termed iPSC lines exhibiting aberrant silencing of Dlk1-Dio3 transcripts Gtl2off iPSCs and cell lines with an ESC-like expression Gtl2on iPSCs. In accordance with developmental defects seen in mutants encompassing the Dlk1-Dio3 cluster4,6, Gtl2off iPSCs failed to yield all-iPSC mice upon tetraploid (4n) blastocyst injections5,7, the most stringent assay for developmental potential. Based on these results, we concluded that the stable repression of maternal Dlk1-Dio3 transcripts acts as a roadblock for the establishment of full pluripotency in iPSCs.
In this manuscript, we offer novel insights into the molecular mechanisms of aberrant Dlk1-Dio3 silencing in iPSCs and provide an efficient way to prevent it by supplementing reprogramming cultures with ascorbic acid. We further demonstrate the utility of this approach by generating entirely iPSC-derived mice from terminally differentiated B lymphocytes.
Dlk1-Dio3 hypermethylation occurs late and requires Dnmt3a
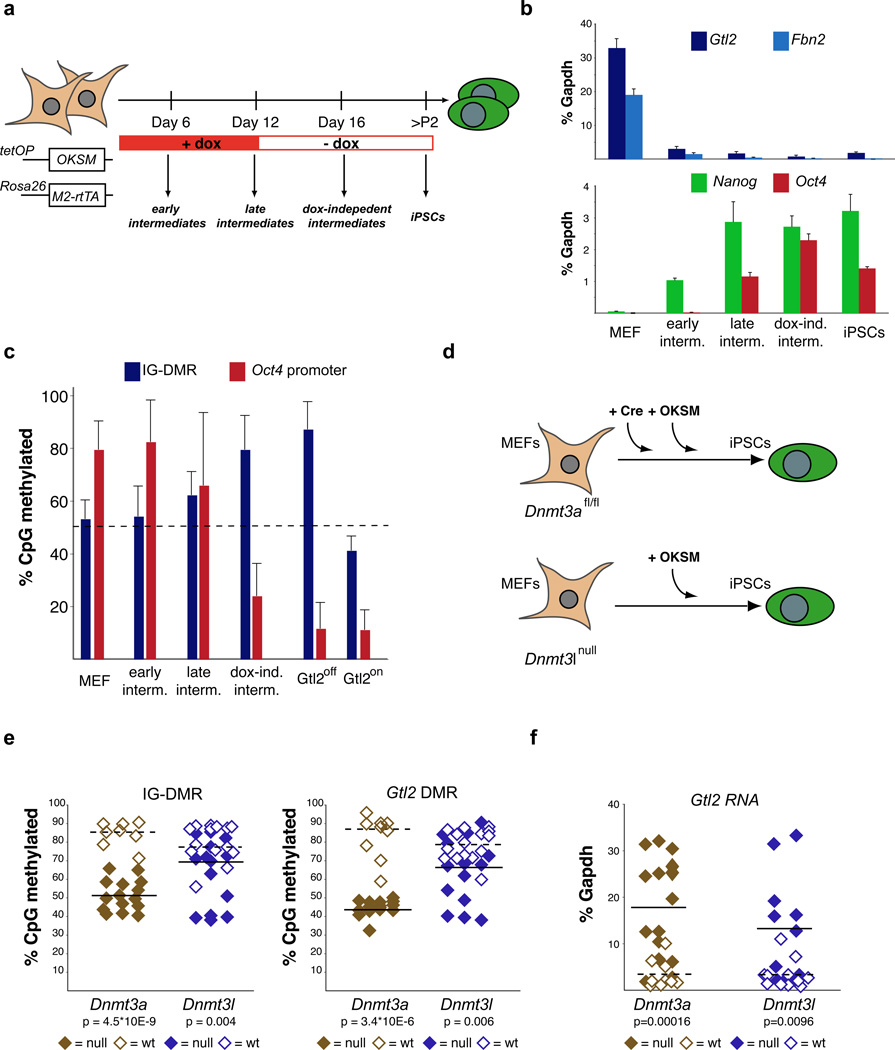
We first determined the kinetics of Gtl2 expression by analyzing defined, purified reprogramming intermediates8 obtained from murine embryonic fibroblasts (MEFs) carrying a transgenic reprogramming system9 (Figure 1a). Analysis of these intermediates showed rapid downregulation of Gtl2 RNA upon OKSM expression, concurrent with the extinction of the fibroblast marker gene Fbn2, but prior to activation of the pluripotency genes Nanog and endogenous Oct4 (also called Pou5f1) (Figure 1b). In contrast to the early downregulation of Gtl2 RNA, abnormal hypermethylation of CpG-dinucleotides within the IG-DMR (intergenic differentially methylated region), which correlates with stable gene silencing of maternally-encoded transcripts4, was only evident at later reprogramming stages. Note that wild type somatic cells and ESCs show methylation levels of ~50% at the IG-DMR, reflecting the silenced and completely methylated paternal copy of Dlk1-Dio3. Interestingly, the timing of IG-DMR hypermethylation coincided with the demethylation of the endogenous Oct4 promoter (Figure 1c), which indicates successful epigenetic reprogramming to pluripotency2. Therefore, repression of maternal Dlk1-Dio3 transcripts appears to occur in two distinct waves, with transcriptional downregulation preceding the acquisition of aberrant DNA methylation and thus stable gene silencing.
Figure 1. Gtl2 hypermethylation occurs late during reprogramming and requires Dnmt3a.
(a) Strategy for isolation and study of reprogramming intermediates using the doxycycline-inducible Collagen-OKSM system. (b) Q-PCR showing the kinetics of Gtl2 repression during reprogramming in relation to the expression of the fibroblast gene Fibrillin-2 (Fbn2) and the pluripotency genes Nanog and Oct4 (Pou5f1). iPSC values were obtained with two Gtl2off clones. (c) DNA methylation analyses for the IG-DMR and Oct4 promoter in MEFs, reprogramming intermediates and established Gtl2off or Gtl2on iPSC clones. Error bars indicate standard deviations (n=28 for IG-DMR and n=5 for Oct4; number of CpGs analyzed). (d) Strategy for the generation of Dnmt3a- and Dnmt3l-deficient (null) iPSCs. Homozygous Dnmt3l null MEFs were transduced with OKSM virus alone, whereas Dnmt3a conditional null MEFs (floxed, fl/fl) were con-transduced with OKSM virus and a Cre-expressing retrovirus. (e) DNA methylation analyses for the IG-DMR and Gtl2 DMR in Dnmt3a null (n=15), Dnmt3a wild-type (wt) (n=8), Dnmt3l null (n=14) and Dnmt3l wt (n=14) iPSC clones. (f) Gtl2 expression levels, as measured by RT-PCR in Dnmt3a null, Dnmt3l null and corresponding wt iPSC clones (see also Suppl. Figure 1). Dashed lines indicate mean values.
During male germ cell development, the IG-DMR is methylated by the de novo DNA methyltransferase Dnmt3a to establish an imprint that is maintained throughout adulthood10. Additionally, the non-enzymatic protein Dnmt3l has been implicated in Dlk1-Dio3 imprinting, although its involvement in this process remains controversial10–12. To genetically test whether Dnmt3a and Dnmt3l are responsible for the hypermethylation observed in iPSCs, we reprogrammed MEFs lacking either Dnmt3a10 or Dnmt3l12 (Figure 1d). The majority of resultant Dnmt3a-deficient iPSC clones exhibited normal (40–50%) methylation levels at both the IG-DMR and a second DMR proximal to the Gtl2 promoter termed Gtl2 DMR4, indicating that Dnmt3a catalyzes the hypermethylation seen in Gtl2off iPSCs (Figure 1e). As expected, Dnmt3a-deficient iPSC clones expressed higher Gtl2 transcript levels compared with control cells (Figure 1f and Supplementary Figure 1a). In contrast, Dnmt3l-deficient iPSCs exhibited, on average, only a small decrease of methylation levels and a less pronounced increase of Gtl2 expression levels (Figure 1e, f and Supplementary Figure 1b). Together, these results provide genetic evidence that Dnmt3a is essential for, while Dnmt3l promotes, Dlk1-Dio3 hypermethylation during the generation of Gtl2off iPSCs. The observation that rare Dnmt3a-deficient iPSCs (2/15) exhibited slightly elevated DNA methylation levels at the IG-DMR (Supplementary Figure 1a) and expressed reduced Gtl2 RNA levels might be explained by delayed Cre-mediated excision of Dnmt3a during reprogramming or by alternative mechanism of gene silencing.
Serum replacement and ascorbic acid prevent Gtl2 silencing
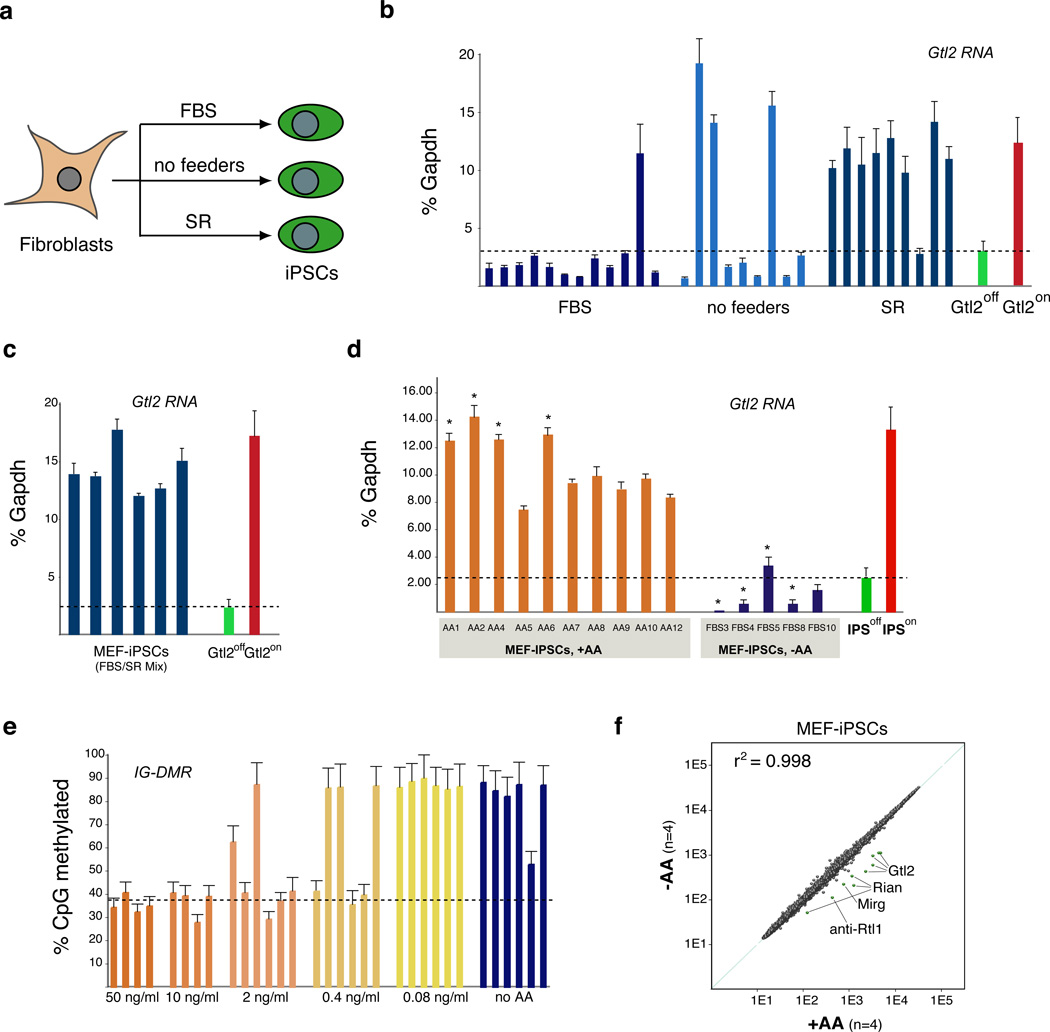
We next sought to identify an efficient and safe approach to prevent Dlk1-Dio3 silencing, that does not depend on genetic interference with Dnmt3a, by testing alternative iPSC derivation protocols. Specifically, we evaluated the effects of omitting feeder cells or using serum replacement (SR)13 instead of FBS during reprogramming (Figure 2a). As shown in Figure 2b, the majority of iPSCs derived in FBS-containing media in the presence or absence of feeders expressed only residual levels of Gtl2 (11/12 and 6/9 of clones, respectively), indicating that feeders do not significantly influence the outcome of Dlk1-Dio3 silencing. Surprisingly, however, 8/9 iPSC clones produced in SR and 6/6 iPSC clones derived in a mix of FBS and SR expressed normal levels of Gtl2, indicating that SR contains a dominant activity that prevents silencing of Dlk1-Dio3 normally incurred by FBS (Figures 2b, c).
Figure 2. Ascorbic acid treatment prevents Gtl2 silencing.
(a) Strategy for the derivation of iPSCs from reprogrammable MEFs in serum-containing media with feeder cells (FBS), serum-containing media without feeder cells (no feeders) and in media containing serum replacement without feeder cells (SR). From the time of doxycycline withdrawal, serum-containing media was used for all cultures. (b) Gtl2 expression levels of iPSC clones generated as outlined in (a). (c) Q-PCR analysis of Gtl2 expression in six independent iPSC clones derived in media containing a 1:1 mix of FBS and SR. (d) Gtl2 expression levels in iPSC clones derived in the presence (+AA) or absence (−AA) of 50 ng/ml ascorbic acid. Upon dox withdrawal, AA was removed from all cultures. Clones marked by asterisks were used for mRNA microarray analysis shown in (f). Error bars in (b)–(d) indicate one standard deviation; n=3. (e) DNA methylation analyses for IG-DMR in iPSC clones generated in medium containing different concentrations of ascorbic acid. Error bars indicate one standard deviation; n=28 (number of CpGs at the IG-DMR). (f) Scatter plot of microarray data comparing iPSC clones derived with ascorbic acid (+AA) or without ascorbic acid (−AA) with differentially expressed genes highlighted in green (two-fold, P<0.05, t-test with Benjamini-Hochberg correction).
SR contains ascorbic acid (also known as vitamin C), which has been shown to influence DNA methylation patterns in human ESCs14 and may thus be responsible for inhibiting aberrant Dlk1-Dio3 silencing. Indeed, addition of ascorbic acid to FBS-containing media during MEF reprogramming abrogated hypermethylation of the IG-DMR and Gtl2 DMR and maintained normal expression levels of Gtl2 in a concentration-dependent manner (Figures 2d, e and Supplementary Figure 2). Consistent with a previous report15, exposure of cells expressing OKSM to ascorbic acid also substantially increased reprogramming efficiencies (from 0.5–1% to about 4%) and speed (five days of OKSM expression were sufficient to generate dox-independent colonies) (Supplementary Figure 3a, b).
Ascorbic acid and alternative reprogramming strategies
Recent reports have documented roles for exogenous c-Myc expression16,17 and reprogramming factor stoichiometry18 in the outcome of Dlk1-Dio3 imprinting in iPSCs, suggesting that ascorbic acid may prevent Gtl2 silencing by modulating these parameters. However, all MEF-iPSC lines (9/9) generated by exogenous expression of only three factors (OKS) in the absence of ascorbic acid exhibited hypermethylation of the IG-DMR and Gtl2 DMR (Supplementary Figure 4). We were further unable to find measurable differences in total or relative protein levels of OKSM between ascorbic acid-treated and untreated reprogrammable MEFs (Supplementary Figure 3c) and hence conclude that ascorbic acid prevents Dlk1-Dio3 silencing independently of factor levels and stoichiometry.
We confirmed the recent observation that reprogramming of MEFs using an alternative polycistronic cassette (OSKM), that produces the four reprogramming factors at different stoichiometries compared with our system (OKSM), gives rise to fewer Gtl2off iPSCs18 (Supplementary Figure 5b). By directly comparing these two transgenic systems, we made additional observations that are worth mentioning here. First, the efficiency of MEF-iPSC formation with the alternative OSKM system was significantly lower (at least one order of magnitude) (Supplementary Figure 5a), raising the possibility that different subsets of cells present in heterogenous fibroblast cultures might have been reprogrammed with the two systems. Second, we noticed that the majority of bone marrow (BM)-derived iPSC clones derived with either transgenic system exhibited hypermethylation of the IG-DMR and Gtl2 DMR (6/8 with OKSM and 15/18 with OSKM), suggesting that factor stoichiometry was either suboptimal in those BM cells that gave rise to iPSCs, or pointing to a cell type-specific effect on Dlk1-Dio3 silencing that is independent of factor stoichiometry (Supplementary Figure 5c, d). Treatment of OSKM BM cells undergoing reprogramming with ascorbic acid also prevented aberrant silencing in the majority of resultant iPSC clones (7/8). Lastly, several MEF-iPSC clones derived with the OSKM transgene (7/18) exhibited hypomethylation (<30% of CpGs methylated) of both the IG-DMR and Gtl2 DMR, irrespective of the presence of ascorbic acid (Supplementary Figure 5b). The biological relevance of this observation remains unclear.
Importantly, seven of seven MEF-iPSC clones generated with four individual lentiviral vectors8 showed IG-DMR hypermethylation in the absence of ascorbic acid, which could be prevented by addition of the compound (Supplementary Figure 6). This indicates that Dlk1-Dio3 silencing is not a peculiarity of the polycistronic construct used in our study but rather a general phenomenon during iPSC derivation in conventional culture conditions.
Specificity of ascorbic acid treatment
MEF-iPSCs generated with or without ascorbic acid showed complete DNA demethylation and acquisition of the activating H3K4me3 histone mark at the Oct4 promoter (Supplementary Figure 7), indicating that epigenetic remodeling of this key pluripotency gene is not affected by ascorbic acid treatment. We also failed to detect differences in the methylation levels of other imprinted genes including the paternally imprinted H19 and Rasgrf1 genes and the maternally imprinted Mest and Peg3 genes (Supplementary Figure 8a, b), demonstrating that ascorbic acid does not globally affect imprinted gene methylation. Following retinoic acid exposure, ascorbic acid-treated iPSC clones upregulated Dlk1 to similar levels as those seen in ESCs (Supplementary Figure 8c), documenting appropriate transcriptional control of the Dlk1-Dio3 locus upon differentiation. Lastly, global gene expression profiling showed that maternal transcripts of the Dlk1-Dio3 locus were the only discernible difference between MEF-iPSCs derived in the presence and absence of ascorbic acid (Figure 2f), respectively, suggesting that its supplementation during reprogramming does not have lasting effects on the expression of other genes in the pluripotent state.
To test whether treatment with ascorbic acid de-represses the normally silenced paternally-inherited allele of Gtl2 and causes a switch of imprinted alleles, we derived iPSCs from M. musculus×M. castaneus hybrid fibroblasts that carry a single nucleotide polymorphism (SNP) on the paternal allele of Gtl2 (Supplementary Figure 9a). All analyzed Gtl2on iPSCs clones (9/9) expressed only the maternal Gtl2 allele (Supplementary Figure 9b,c), indicative of imprint stability at the Dlk1-Dio3 locus.
Ascorbic acid exposure of established Gtl2off iPSCs for 15 passages was insufficient to reverse silencing, as documented by sustained hypermethylation of both the IG-DMR and Gtl2 DMR (Supplementary Figure 10). These observations reveal that ascorbic acid treatment prevents but cannot reverse aberrant DNA methylation of the Dlk1-Dio3 locus.
Ascorbic acid preserves active chromatin marks at Dlk1-Dio3
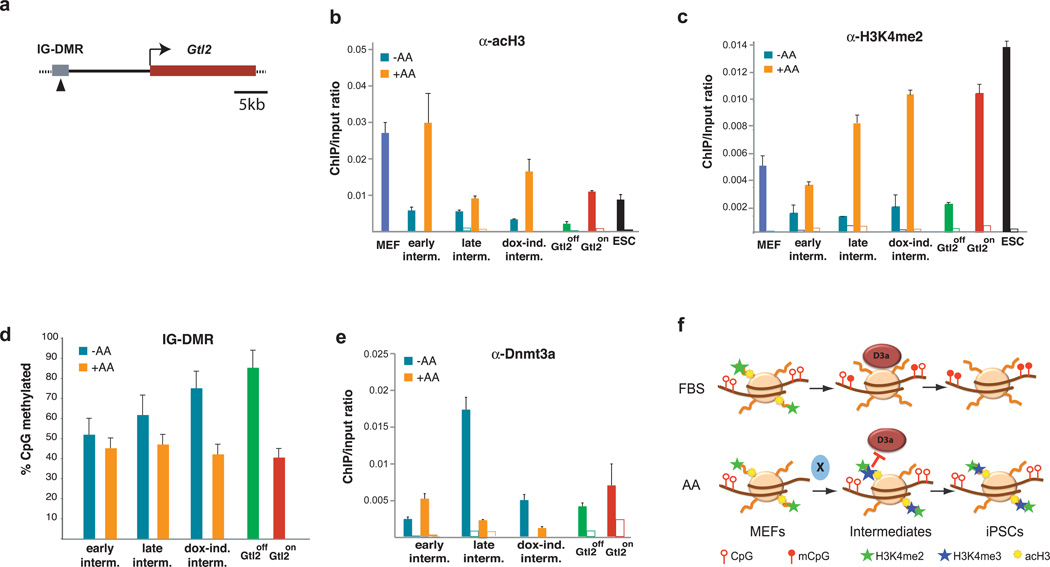
To gain insights into the mechanism by which ascorbic acid might counteract epigenetic silencing of Dlk1-Dio3, we isolated SSEA1+ intermediates from MEFs undergoing reprogramming either in the presence or absence of the compound and analyzed histone modifications at the IG-DMR that have previously been correlated with DNA methylation. As shown in Figure 3, MEFs undergoing reprogramming in FBS alone rapidly lost the activating histone marks acetylated histone H3 (acH3) and H3 lysine 4 dimethylation (H3K4me2) and failed to gain H3 lysine 4 trimethylation (H3K4me3) (Supplementary Figure 11a), a modification seen in ESCs and Gtl2on iPSCs but not in MEFs and Gtl2off iPSCs. In contrast, ascorbic acid-treated intermediates maintained H3K4me2 and acH3 and gained H3K4me3 levels at the IG-DMR during reprogramming, ultimately reaching levels comparable to those seen in ESCs (Figure 3b,c and Supplementary Figure 11a). We did not detect significant differences in the repressive H3 lysine 27 trimethylation (H3K27me3) mark in intermediates or established pluripotent cell lines (Supplementary Figure 11b). Thus, ascorbic acid treatment preserves an active histone configuration at Dlk1-Dio3, which is frequently lost in regular (FBS-containing) reprogramming conditions.
Figure 3. Epigenetic changes induced by ascorbic acid during reprogramming.
(a) Schematic representation of the Gtl2 locus. Arrowhead indicates region within IG-DMR that was used for ChIP and DNA methylation analyses presented here. (b, c) Relative enrichment of the activation marks, acetylated histone H3 (acH3) and dimethylated H3 lysine 4 (H3K4me2), respectively, at the IG-DMR in MEFs, ESCs and reprogramming intermediates isolated from cultures in the absence (−AA) or presence (+AA) of ascorbic acid. Open columns indicate the background levels of IgG antibody control. (d) DNA methylation analyses for the IG-DMR in reprogramming intermediates. (e) ChIP-PCR analysis detecting Dnmt3a binding at the IG-DMR in reprogramming intermediates. (f) Proposed model of how ascorbic acid prevents aberrant methylation of the maternal Gtl2 allele during reprogramming. Factor X depicts as of yet unidentified activity(ies) that act downstream of ascorbic acid to mediate observed epigenetic changes. Error bars in (a)–(e) indicate one standard deviation; n=3.
To correlate the observed changes in histone modifications at Dlk1-Dio3 with DNA methylation patterns, we analyzed methylation levels at the IG-DMR in the same reprogramming intermediates. This showed that untreated fibroblasts undergoing reprogramming become progressively hypermethylated at later reprogramming stages, while ascorbic acid-treated intermediates maintain normal DNA methylation levels throughout the process (Figure 3d, see also Figure 1c). Importantly, aberrant DNA hypermethylation coincided with increased binding of Dnmt3a to the IG-DMR in late reprogramming intermediates in the absence of ascorbic acid (Figure 3e), which is consistent with the previous observation that Dnmt3a can only recognize unmodified histone H3K4 tails19,20. We failed to detect increased expression of DNA methyltransferase genes in untreated intermediates, thus ruling out that hypermethylation is the consequence of transcriptional upregulation of these enzymes (Supplemental Figure 12).
Ascorbic acid has anti-oxidative activity, which may be critical for its inhibitory effect on Dlk1-Dio3 hypermethylation. However, the majority of iPSCs clones derived under reduced oxygen conditions or in the presence of the alternative antioxidant resveratrol still showed IG-DMR hypermethylation (Supplemental Figure 13), thus arguing against the importance of ascorbic acid’s antioxidative properties in preventing Dlk1-Dio3 silencing. It is possible, though, that other antioxidative agents may mirror the effects ascorbic acid treatment on the Dlk1-Dio3 locus. Together, these observations suggest that the rapid loss of activating histone marks in the absence of ascorbic acid facilitates the recruitment of Dnmt3a to the IG-DMR, which catalyzes DNA hypermethylation and leads to stable epigenetic silencing of maternal Dlk1-Dio3 transcripts (see model in Figure 3f).
Gtl2on iPSCs derived from mature B cells
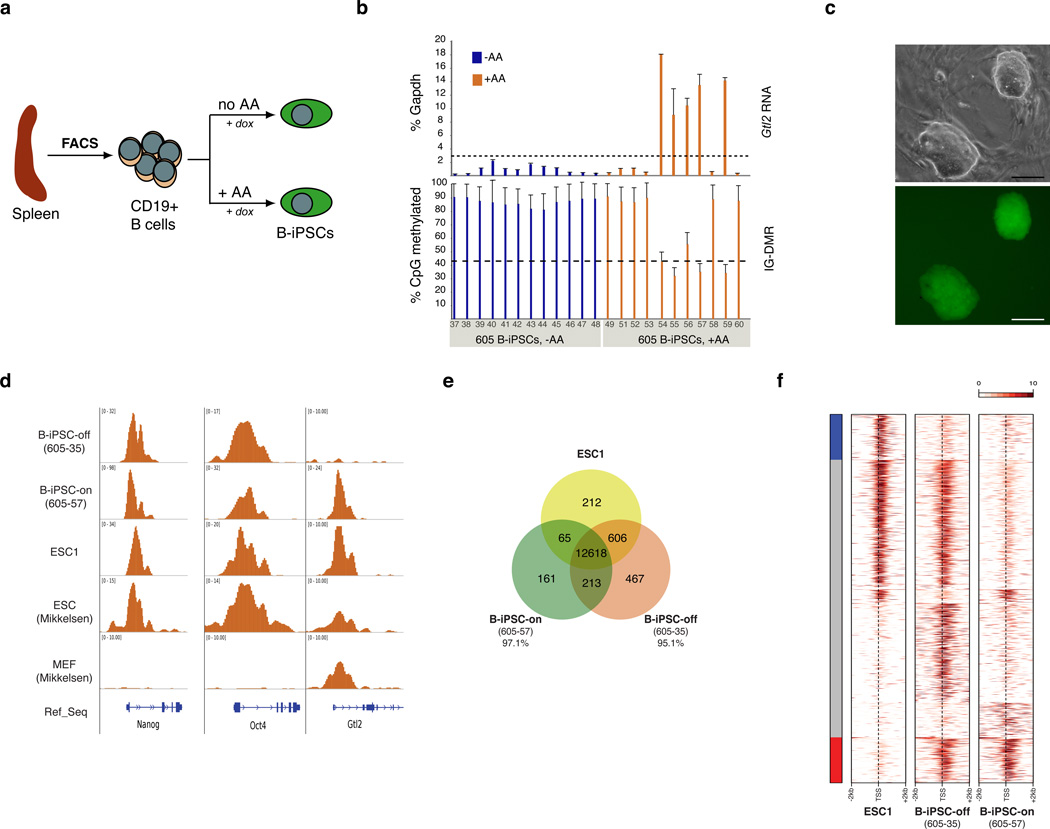
To date, Gtl2on iPSCs have only been obtained from heterogeneous fibroblast cultures while more defined, non-fibroblast populations such as hematopoietic cells and keratinocytes consistently yielded Gtl2off iPSCs with impaired developmental potency as judged by 4n embryo complementation5. To evaluate whether ascorbic acid treatment facilitates the generation of Gtl2on iPSCs from a defined, terminally differentiated cell type, we isolated CD19+ B cells from the spleens of adult reprogrammable mice9 and cultured them in doxycycline-containing media in the presence or absence of ascorbic acid for 14 days until iPSC colonies emerged (Figure 4a and Supplementary Figure 14). The efficiency of iPSC formation from mature B cells (B-iPSC) ranged between 0.02% and 0.06% in the presence of ascorbic acid and between 0.01% and 0.05% in the absence of ascorbic acid (data not shown), representing a less dramatic increase in reprogramming efficiency following ascorbic acid treatment than that seen with fibroblasts (Supplementary Figure 3a). While all B-iPSCs generated without ascorbic acid (36/36) exhibited aberrant hypermethylation of the IG-DMR and Gtl2 DMR and/or diminished Gtl2 transcript levels, 4/17 (24%) iPSC clones established in a mix of FBS and SR and 5/11 (45%) clones derived in the presence of ascorbic acid showed normal DNA methylation patterns and expressed Gtl2 at ESC-like levels (Figure 4b and Supplementary Figure 15). The lower frequency of obtaining Gtl2on B-iPSCs than MEF-iPSCs might be due to the higher baseline methylation levels of the IG-DMR in B lymphocytes and the extremely low expression levels of Gtl2 in these cells (Supplementary Figure 16). Gtl2on B-iPSCs exhibited typical ESC-like morphology, homogenously expressed endogenous pluripotency markers (Figure 4c), showed DNA demethylation and enrichment for H3K4me3 at the Oct4 promoter and had a predominantly normal karyotype (Supplementary Figure 17). Collectively, these data show that ascorbic acid treatment efficiently preserves normal genomic imprinting at Dlk1-Dio3 during the reprogramming of a defined, terminally differentiated non-fibroblast cell type.
Figure 4. Molecular characterization of B-iPSCs generated in the presence of ascorbic acid.
(a) Strategy for the generation of B cell-iPSCs (B-iPSCs) from adult mice. (b) RT-PCR analysis of Gtl2 expression (top graph) and DNA methylation analysis of IG-DMR (bottom graph) in individual B-iPSC clones derived in the absence (−AA) or presence (+AA) of ascorbic acid. The number “605” denotes the mouse used for B cell isolation. Error bars indicate one standard deviation; n=3 PCR reactions (upper panel) and n=28 CpGs covered by pyrosequencing analysis, respectively. (c) Representative image of a B-iPSC clone showing typical morphology (top panel) and expression of Oct4-GFP (bottom panel). Scale bars indicate 100 µm. (d) Reads density profile around the transcription start sites (TSSs) of Nanog, Oct4 and Gtl2 for the H3K4me3 modification following genome-wide ChIP-Seq analysis of the ascorbic acid-derived B-iPSC clones 605–35 (Gtl2off) and 605–57 (Gtl2on) and one ESC line (ESC1). All samples were compared to a published data set19. Gaussian-smoothed read density is normalized by library size. The vertical axis is adjusted to show the whole peak if higher than 10. (e) Venn diagram representing overlap between H3K4me3-enriched genes between ESC1 and the B-iPSC clones. The indicated percentages show overlap with ESC1. Overlapping areas are not proportional to the actual number of genes. (f) Region map of 4kb window around the TSSs of genes enriched for H3K4me3 in either ESC1 or the individual B-iPSC clones. These are the same genes as reported in Figure 4e excluding the 12,618 genes common to all three groups. The side bar marks genes, which are enriched for H3K4me3 in ESC1 only (blue bar, 212 genes) or in both B-iPSC clones only (red bar, 213 genes). The plot reports Gaussian-smoothed read density enrichment over input samples in log2 scale. For each gene, the maximum enrichment across all of the samples is used to compute a scaling factor so that maximum value is equal to 10.
Given that H3K4me3 deposition at the Dlk1-Dio3 locus in reprogramming intermediates was strongly affected by ascorbic acid treatment (see Supplementary Figure 11a), we wondered whether H3K4me3 patterns might be altered on a genome-wide scale in iPSCs by this compound. To this end, we performed ChIP-Seq analysis for the H3K4me3 modification in two B-iPSC clones derived in the presence of ascorbic acid (one Gtl2on and one Gtl2off) and one fully pluripotent ESC line5, that had never been exposed to ascorbic acid. Bioinformatic analysis of sequenced reads from these clones confirmed the absence of the activating H3K4me3 modification at the Gtl2 locus in the Gtl2off clone while pluripotency loci such as Nanog and Oct4 were similarly enriched for H3K4me3 in all pluripotent cell lines (Figure 4d). Both B-iPSC clones exhibited a very high degree of overlap in global H3K4me3 patterns (>95%) compared with the control ESC line (Figure 4e, f). Our ChIP-seq results also showed a large overlap with a previously published dataset on genome-wide H3K4me3 patterns in ESCs21 (Figure 4d and Supplemental Figure 18), further suggesting that ascorbic acid exposure does not cause major global epigenetic aberrations of at least this key epigenetic mark. It remains to be tested whether the few detected differences among the various samples (Figure 4f and Supplementary Table 1) are truly the result of ascorbic acid exposure or rather caused by line-to-line variation as has been seen before among pluripotent cell lines22.
All-iPSC mice generated from mature B cells
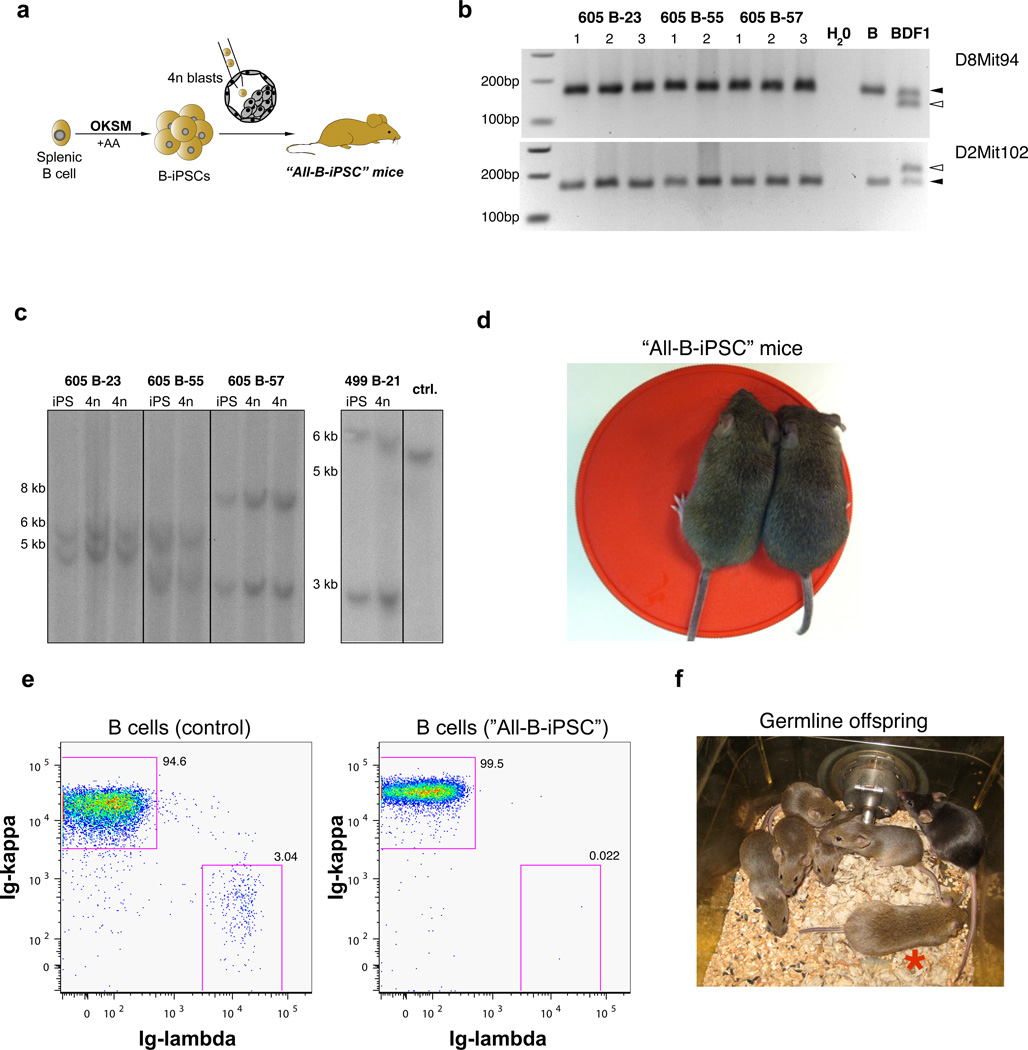
Lastly, we tested the developmental potential of B-iPSCs derived in the presence of ascorbic acid by 4n embryo complementation23,24 (Figure 5a). It should be noted that adult all-iPSC mice have thus far only been produced from fibroblast populations while attempts to produce 4n complementation-competent mice from defined adult cell types such as neural progenitor cells25 or lymphocytes26 have been unsuccessful, likely due to embryonic growth arrest at midgestation25. Remarkably, all Gtl2on B-iPSC clones tested with this rigorous assay (4/4) gave rise to neonatal “all-iPSC” mice whereas two tested Gtl2off clones only yielded late-stage resorptions (Table 1). Given that Gtl2 null animals die pre- or perinatally6, our results do not exclude the possibility that rare Gtl2off all-iPSC mice can develop as far as full-term before they succumb, consistent with a recent report18. We confirmed that viable animals were entirely derived from B-iPSCs by PCR for strain-specific polymorphisms (Figure 5b) and by Southern blot analysis that detects the clonal rearrangements of the immunoglobulin heavy chain locus (Figure 5c). All-iPSC neonates generated from three of the four Gtl2on clones survived at least for several days, and animals derived from two of the clones reached adulthood and exhibited uniform agouti coat color (Table 1 and Figure 5d). Peripheral B cells isolated from an adult all-B-iPSC mouse expressed immunoglobulins composed of essentially only kappa light chain on their surface (Figure 5e) while B cells from control mice produced both kappa and lambda immunoglobulins, further supporting the monoclonal origin of B cells in mice derived from B-iPSCs. We also obtained agouti offspring from all-B-iPSC mice, documenting successful germline transmission (Figure 5f). These data demonstrate that Gtl2on iPSCs produced from terminally differentiated B cells can support the development of entirely iPSC-derived adult monocolonal mice.
Figure 5. Functional characterization of B-iPSC clones generated in the presence of ascorbic acid.
(a) Strategy for the derivation of all-B-iPSC mice by tetraploid embryo complementation. (b) Confirmation of origin of all-B-iPSC mice by PCR for strain-specific polymorphisms. Two different Simple Sequence Polymorphism (SSLP) markers were tested using genomic DNA isolated from tissues of all-BiPSC mice and the parental B cells. Genomic DNA from BDF1 mice served as control for the presence of host blastocyst-derived cells. Triangles indicate the position of strain-specific bands; open triangle = DBA/2 (host blastocyst-specific) and black triangle = C57BL/6 (present in both host blastocysts, parental cells and iPSCs). (c) Southern blot analyses of immunoglobulin heavy chain (IgH) locus rearrangement using genomic DNA isolated from different all-B-iPSC mice and the corresponding four parental B-iPSC clones. DNA isolated from a MEF-iPSC clone was used as a control to visualize unrearranged IgH germline configuration (single band). (d) Image of two agouti all-B-iPSC mice at 5 weeks of age. (e) FACS analysis of B cells isolated from a control (con) mouse and one all-B-iPSC mouse shows expression of only one type of immunoglobulin (Ig) light chain (Ig-kappa or Ig-lambda), thus confirming monoclonality. (f) Image of a three-month-old all-B-iPSC mouse (indicated by red asterisk) and its germline offspring (5 agouti mice).
Table 1.
| Cell line |
Gtl2 expression |
embryos transferred |
pups born |
surviving pups |
Adult mice |
|---|---|---|---|---|---|
| 605B-23 | ON | 138 | 11 (8.0%) | 2 | 2 |
| 499B-21 | ON | 65 | 1 (1.5%) | 0 | 0 |
| 605B-57 | ON | 175 | 19 (10.9%) | 9 | 4 |
| 605B-55 | ON | 131 | 6 (4.6%) | 1 | 0 |
| 605B-35 | OFF | 62 | 15 resorptions | 0 | 0 |
| 499B-22 | OFF | 47 | 12 resorptions | 0 | 0 |
Discussion
Here, we have shown that ascorbic acid treatment efficiently preserves a normal imprinting status at the Dlk1-Dio3 gene cluster during the derivation of mouse iPSCs from different adult cell types and using various means of reprogramming factor expression. Our data suggest that ascorbic acid inhibits Dlk1-Dio3 silencing by interfering with OKSM-induced loss of H3K4 methylation at the maternal IG-DMR by an as yet to be identified mechanism. Sustained H3K4 methylation then prevents the recruitment of Dnmt3a, which is essential for Dlk1-Dio3 DNA hypermethylation. Surprisingly, iPSC lines derived in the presence of ascorbic acid did not exhibit detectable expression changes of transcripts outside the Dlk1-Dio3 cluster, altered DNA methylation patterns at a number of additional imprinted genes or major differences in genome-wide H3K4me3 occupancy, suggesting a remarkable specificity. However, it is possible that ascorbic acid influences other chromatin marks in iPSCs that do not adversely affect their developmental potential as measured by the ability of B-iPSCs to generate adult all-iPSC mice. Given its strong effect on reprogramming speed and efficiency, it is conceivable that ascorbic acid introduces widespread but transient alterations into chromatin that are resolved upon entry into a pluripotent state and hence undetectable in established iPSCs. It is noteworthy that a recent report connected ascorbic acid with the H3K36 demethylase Kdm2b during iPSC formation27. It should thus be informative to test whether Kdm2b may also be involved in preventing Dlk1-Dio3 silencing. Furthermore, the maternally expressed microRNA mir-369 encoded within the Dlk1-Dio3 cluster has recently been suggested to reprogram fibroblasts into iPSCs in combination with other select microRNAs28, raising the possibility that ascorbic acid may in part increase reprogramming efficiency by maintaining normal Dlk1-Dio3 imprinting.
Addition of ascorbic acid to cells undergoing reprogramming allowed us to derive Gtl2on iPSCs from mature B cells, which supported the development of adult all-iPSC mice. To our knowledge, this is the first demonstration that full developmental potential can be reinstated in iPSCs produced from a terminally differentiated adult cell of defined origin. While we were unable to generate Gtl2on B-iPSC clones in the absence of ascorbic acid using our reprogrammable system, we cannot rule out that alternative reprogramming cocktails or constructs may facilitate their derivation in standard conditions. Also, our observation that adult all-iPSC animals could not be obtained from every Gtl2on B-iPSC clone tested indicates that normal expression of Dlk1-Dio3 alone is a valuable indicator but not an absolute predictor for postnatal survival of all-iPSC mice. This finding further implies that other cell line-specific epigenetic and/or genetic abnormalities may be present in iPSCs, which should be identifyable with deep sequencing approaches.
The ability to generate all-iPSC animals from single B lymphocytes might provide an efficient way to produce monoclonal mice with desired antibody specificities for immunological studies. Our observations should also help to streamline the production of mouse iPSCs with increased efficiency and fidelity from different cell types. In this context, it will be important to evaluate if the expression status of Gtl2 correlates with other transcriptional, epigenetic or genetic features of iPSCs, which have escaped detection with the assays employed thus far. More generally, our results make the unexpected observation that alterations of environmental conditions during cellular reprogramming can have profound effects on the epigenetic and biological properties of derivative pluripotent cell lines. Lastly, these data may have ramifications for ongoing comparative studies between ESCs and iPSCs as well as disease modeling approaches.
Experimental procedures
Mice
Derivation, handling and genotyping of Col-OKSM reprogrammable mice was done as described previously9.
Cell culture
ESCs and established iPSCs were cultured on irradiated feeder cells (Global Stem) in KO-DMEM (Invitrogen) supplemented with L-Glutamine, penicillin-streptomycin, non-essential amino acids, β-mercaptoethanol, 1000 U/ml LIF (“ESC media”) and 15% fetal bovine serum (FBS) (HyClone). MEF cultures were established by trypsin-digestion of midgestation (E13.5–15.5) embryos followed by culture in DMEM supplemented with 10% FBS, L-Glutamine, penicillin-streptomycin, non-essential amino acids and β-mercaptoethanol. B cells were defined as CD19+c-kit−Mac-1− cells and isolated from adult spleen using monoclonal antibodies against CD19 (1D3), c-kit (2B8) and Mac-1 (CD11b, M1/70) obtained from eBiosciences. c-kit and Mac-1 were used to ensure the exclusion of progenitor cells and macrophages/monocytes, respectively.
Reprogramming
Reprogramming experiments were routinely conducted in gelatinized plates seeded with a feeder layer of irradiated MEFs, using ESC media supplemented with 1 µg/ml doxycycline (dox) and either 15% FBS or 20% KnockOut Serum Replacement (SR) (Invitrogen). Wherever indicated, ascorbic acid (Sigma A4544) was added to the reprogramming cultures at the same time dox was added. For the reprogramming of B cells, interleukin-4 (80 ng/µl final) and an antibody against CD40 (5 µg/ml final) were added to the cultures. After dox withdrawal, cultures were grown in ESC media with 15% FBS without ascorbic acid. Colonies with ESC morphology were picked four to six days after dox withdrawal. For the isolation of intermediates, Col-OKSM MEFs were seeded on feeders in ESC media containing FBS and dox either in the presence or absence of ascorbic acid. Cells were harvested on day 6 after dox induction (“early intermediates”), on day 12 after dox induction (“late intermediates”) and after another 4–6 days of culture after dox withdrawal on day 12 (“dox-independent intermediates”). Cells were stained with biotinylated anti-SSEA1 (MC-480) and eFluor 450-conjugated anti-CD90.2 (Thy1.2, 53–2.1) antibodies, and the SSEA1+ fraction was either isolated by flow-activated cell sorting (FACS) as described before8 or by magnetic-activated cell sorting (MACS) after incubation with Streptavidin microbeads (Miltenyi Biotec) using the positive selection program on an AutoMACS cell separator according to the manufacturer’s instructions. The purity of all isolated cell fractions was confirmed by flow cytometric analysis using an LSR2 machine (BD). Established iPSCs were isolated by trypsinization followed by 45 minutes of pre-plating to remove feeder cells. DNMT3afloxed/floxed and Dnmt3lnull MEFs were co-transduced with a dox-inducible lentivirus expressing OKSM29, a constitutive lentivirus expressing rtTA8 and, in case of Dnmt3a MEFs, a constitutive retrovirus driving expression of Cre recombinase30. Complete excision of the conditional alleles was confirmed by PCR10. OKS iPSCs were generated using mice carrying a dox-inducible polycistronic cassette encoding for Oct4, Klf4 and Sox2 (E.A. and K.H., unpublished).
Viral vector production
Viral vectors were produced as previously described8. Briefly, 293T cells were co-transfected with vector plasmid and packaging plasmids using Fugene (Roche) transfection reagent. Viral supernatants were harvested between 48 and 72 hours after transfection and concentrated by ultracentrifugation at 20,000 rpm for 1.5 hours at 4°C. Viral concentrates were resuspended in PBS and stored at −80°C. Transduction of MEFs was carried out in MEF media containing 5µg/ml polybrene.
Analysis of DNA methylation
MEFs and B cells were pelleted and then resuspended in lysis buffer, pH 8 (100 mM Tris-HCl, 5 mM EDTA, 0.2% SDS, 200 mM NaCl) with proteinase K. ESCs and dox-independent iPSC clones at passage 2 or higher were seeded on gelatinzed plates without feeders, grown to confluency and then lysed. Genomic DNA was precipitated with 100% isopropanol and reconstituted in TE buffer, pH 7.5 (10 mM Tris-HCl, 1 mM EDTA). DNA was bisulfite-converted using the EpiTect Bisulfite Kit (Qiagen) and analyzed by EpigenDX using the following assays: ADS1452 (IG-DMR), ADS1341 (Gtl2 promoter), ADS438 (ICR1-H19), ADS936 (Rasgrf1) and ASY585 (Oct4 proximal promoter). The precise genomic position of all CpGs assayed is listed in Supplementary Table 2.
SSLP analysis
Genomic DNA was isolated from tail-tip biopsies or primary B cells and PCR reactions set up using primers which detect described polymorphisms in the genome of inbred mouse strains26 (listed in Supplementary Table 3) Reactions were performed with 100 ng of DNA for 30 cycles of 30 seconds at 94° Celcius (C), 30 seconds at 60° C and 60 seconds at 72° C and reaction products analyzed on a 3% agarose gel.
RNA isolation and quantification
MEFs and B cells were pelleted and used for RNA isolation with either the miRNeasy Mini Kit or the RNeasy Mini Kit (Qiagen) without DNase digestion. ESCs and Dox-independent iPSC clones at passage 2 or higher were seeded on gelatinzed plates without feeders and then processed with the kits mentioned above. RNA was eluted from the columns using RNase-free water or TE buffer and quantified using a Nanodrop. cDNA was produced with the Transcriptor First Strand cDNA Synthesis Kit (Roche). Real-time quantitative PCR reactions were set up in triplicate with the Brilliant II SYBR Green QPCR Master Mix (Stratagene) using 5 µl of cDNA (1:100 dilution in water) and previously described primers5,8. Reactions were run on a Mx3000P QPCR System (Stratagene) with 40 cycles of 30 seconds at 95° C, 30 seconds at 58° C and 30 seconds at 72° C. For detection of allele-specific Gtl2 expression in M. musculus×M. castaneus F1 iPSCs, the qPCR amplicon was digested with the endonuclease BstUI and run on a 3% agarose gel.
Western Blot analysis
Cells were harvested and lysed in 1x RIPA buffer. Protein concentration was measured using BCA protein Assay reagent (ThermoScientific). 15 µg of cell extract was loaded and western blot analysis was performed using standard protocols and probed with the following antibodies: anti-Oct4 (sc-5279), anti-Sox2 (ab97959), anti-Klf4 (R&D, AF3158) and anti-cMyc (sc-764).
Gene expression profiling
Total RNA samples (RIN>9) were subjected to transcriptional analyses using Affymetrix Mouse 430_2 mRNA expression microarray similarly as previously described31. Hierarchical clustering was performed using Cluster and Treeview software as well as the GeneSifter server (Geospiza, Seattle).
Southern blot analysis
20 µg of genomic DNA was digested with EcoRI, separated on a 1% agarose gel, blotted onto HybondXL membrane (Amersham Biosciences) and hybridized to a HindIII/EcoRI JH4 fragment probe32. Probe labeling with 32P-α-dCTP was done using the Prime-It® II Random Labeling Kit (Stratagene) following the manufacturer’s instructions.
Chromatin immunoprecipitation
20 million iPSCs, ESCs or MEFs were fixed with 1% formaldeyde for 10 minutes at room temperature (RT) and then lysed in 1ml lysis buffer (50mM Tris-HCl, pH 8.0, 10mM EDTA, 1% SDS, protease inhibitors) for 20 minutes on ice. The lysate was split into three tubes and sonicated using Bioruptor for five times at high intensity, 30 sec. “on” and 30 sec. “off”. After 10 minutes of centrifugation, the supernatant was pre-cleared for 1 hour at 4°C with agarose beads pre-blocked with BSA (1mg BSA for 10ml beads) in IP Buffer (50mM Tris-HCl, pH8, 150mM NaCl, 2mM EDTA, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1% SDS, protease inhibitors). 100ul of pre-cleared chromatin per reaction diluted in 1ml IP Buffer in the presence of 2ug antibody were used for each reaction according to the manufacturer's protocol. The antibodies used were: anti-acetyl histone H3 (Millipore, 17–615), anti-dimethyl K4 of H3 (07–030, Millipore), anti-trimethyl K4 of H3 (Millipore, 17–614), anti-trimethyl K27 of H3 (Millipore, 07–449), anti-Dnmt3a (IMGENEX, IMG-268A) and normal rabbit IgG (Millipore). The precipitate was purified using QIAquick PCR purification Kit and was analyzed by RT-PCR using primers listed in Supplementary Table 3.
ChIP-Sequencing
ChIP libraries were prepared using the NEBNext ChIP-Seq Prep Reagent Set1 of NEB (NEB #E6200) following the manufacturer’s instructions and the Illumina/Solexa Genome Analyzer Primer/Adapter Sequences. The libraries were sequenced using an Illumina GAII Sequencing System.
ChIP-seq data analysis
Illumina sequencing reads were pre-processed with cutadapt tool (http://code.google.com/p/cutadapt/) to trim adapter sequences if present, then aligned to mm9 reference genome with bowtie33, using default alignment parameters but reporting only uniquely aligned reads. Genomic regions with significant enrichment in H3K4me3 ChIP samples over matched input samples were detected using R and spp package procedure for the identification of broad enrichment clusters34. RefSeq transcripts annotations were downloaded from UCSC genome browser database and matched with gene annotations (refGene and refLink tables). For each RefSeq, a window +/−2kb centered at the transcription start site (TSS) is considered. This window is truncated downstream the TSS if the RefSeq transcript is less than 2kb long, and is also truncated upstream of the TSS if another RefSeq transcript end on the same genomic strand is located less than 2kb upstream. Each RefSeq TSS is associated with H3K4me3 enrichment if any overlap exists between the enrichment regions and the selected TSS window. Then these overlaps are summarized at the gene level and each gene is considered as associated to H3K4me3 enrichment if any of its RefSeq transcripts is associated to H3K4me3 enrichment. In Venn diagrams the list of H3K4me3-associated genes are compared across different samples. Venn diagrams were plotted using Vennerable R package. Publicly available data21 were re-analyzed using the same procedure.
Blastocyst injections
2n and 4n blastocyst injections were performed as described before23. Briefly, female BDF1 mice were superovulated by intraperitoneal injection of PMS and hCG and mated to BDF1 stud males. Zygotes were isolated from females with a vaginal plug 24 hour after hCG injection. Zygotes for 2n injections were in vitro cultured for 3 days in KSOM media, blastocysts were identified, injected with ESCs or iPSCs and transferred into pseudopregnant recipient females. For 4n injections, zygotes were cultured overnight until they reached the 2-cell stage, at which point they were electro-fused. One hour later, 1-cell embryos were carefully identified and separated from embryos that had failed to fuse, cultured in KSOM for another 2 days and then injected.
Supplementary Material
Acknowledgements
We are grateful to Bernhard Payer for providing M. musculus×M. castaneus fibroblasts, Duane Wesemann for advice on B cell isolation and immunoglobulin rearrangements, Adlen Foudi for genomic DNA from B cells, Hui Su (Novartis) for technical assistance and Alyssa Riley (MGH) for help with blastocyst injections. We thank past and present members of the Hochedlinger lab for suggestions. M.S. was supported by a postdoctoral fellowship from HHMI, E.A. by a Jane Coffin Childs postdoctoral fellowship, and support to K.H. was from the NIH (grants DP2OD003266 and R01HD058013).
Footnotes
Data access
mRNA microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible under accesion number GSE34761. ChIP-seq data have been submitted to the Sequence Read Archive under accession number SRA048788.
Author Contributions
M.S., E.A. and K.H. conceived the experiments, analyzed the data and wrote the manuscript. M.S. and E.A. performed the majority of experiments. F.F. and P.J.P. analyzed genome-wide histone modification data; T.S. analyzed gene expression data; J.C. and S.Y.K. performed tetraploid embryo complementation experiments; R.M.W. generated Southern blot data; T.C, S.O. and T.B. provided Dnmt3a and Dnmt3l null fibroblasts.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 137:2643–2652. doi: 10.1242/dev.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L, et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 11.Kato Y, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 12.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 13.Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1:245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung TL, et al. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells. 28:1848–1855. doi: 10.1002/stem.493. [DOI] [PubMed] [Google Scholar]
- 15.Esteban MA, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Li W, et al. iPS cells generated without c-Myc have active Dlk1-Dio3 region and are capable of producing full-term mice through tetraploid complementation. Cell research. 2011;21:550–553. doi: 10.1038/cr.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang L, et al. Viable mice produced from three-factor induced pluripotent stem (iPS) cells through tetraploid complementation. Cell research. 2011;21:546–549. doi: 10.1038/cr.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carey BW, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otani J, et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggan K, et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy A, et al. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 25.Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 26.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T, et al. The histone demethylases jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi N, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hock H, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18:2336–2341. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coser KR, et al. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci U S A. 2003;100:13994–13999. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atkinson MJ, Michnick DA, Paige CJ, Wu GE. Ig gene rearrangements on individual alleles of Abelson murine leukemia cell lines from (C57BL/6 × BALB/c) F1 fetal livers. J Immunol. 1991;146:2805–2812. [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.