Abstract
The irreversible nature of reactions catalysed by P450s makes these enzymes landmarks in the evolution of plant metabolic pathways. Founding members of P450 families are often associated with general (i.e. primary) metabolic pathways, restricted to single copy or very few representatives, indicative of purifying selection. Recruitment of those and subsequent blooms into multi-member gene families generates genetic raw material for functional diversification, which is an inherent characteristic of specialized (i.e. secondary) metabolism. However, a growing number of highly specialized P450s from not only the CYP71 clan indicate substantial contribution of convergent and divergent evolution to the observed general and specialized metabolite diversity. We will discuss examples of how the genetic and functional diversification of plant P450s drives chemical diversity in light of plant evolution. Even though it is difficult to predict the function or substrate of a P450 based on sequence similarity, grouping with a family or subfamily in phylogenetic trees can indicate association with metabolism of particular classes of compounds. Examples will be given that focus on multi-member gene families of P450s involved in the metabolic routes of four classes of specialized metabolites: cyanogenic glucosides, glucosinolates, mono- to triterpenoids and phenylpropanoids.
Keywords: evolution of diversity, general and specialized metabolism, cyanogenic glucosides, glucosinolates, terpenoids, phenylpropanoids and phenolics
1. Introduction: emerging modes of evolution for P450s in specialized metabolism
Plants are characterized by an astonishing diversity of low-molecular-weight natural products that play essential roles in adaptation and defence. These structurally diverse compounds are referred to as plant specialized compounds (i.e. secondary metabolites or natural products). In contrast to general metabolites (i.e. primary), which are prevalent in all plant species, plant specialized compounds are typically found in only a limited number of species, a genus, a single family or a few families. Biochemically, plant specialized compounds can be grouped into three major classes: terpenoids, phenylpropanoids and nitrogen-containing compounds, of which the latter can be further subdivided into alkaloids, cyanogenic glucosides and glucosinolates. Plant specialized compounds are pivotal to the success of a species, as their evolution has provided selective advantages in adaptation and development, and are thus a driving force in speciation. For example, phenylpropanoids show an increasing degree of functional specialization: ranging from UV protectant and radical scavenging, to vascularization, pigmentation, phytoalexins and signalling molecules. In an analogous manner, terpenoids range from structural components of membranes to phytohormones, chemical defence compounds and signalling molecules.
A common feature is that the vast majority of specialized metabolites are oxygenated. It is well documented that cytochromes P450 are often recruited as versatile catalysts in the biosynthesis of plant specialized compounds, and are as such key to the structural diversity and the success of plant specialized compounds [1,2]. It is, therefore, not surprising that plants hold the record in number of P450s per genome. While animal genomes usually contain fewer than 100 P450s, higher land plants typically contain in excess of 250 P450s (e.g. Arabidopsis thaliana 245 genes, Oryza sativa 343 genes) [3]. Non-vascular bryophytes contain a more limited set of P450s (e.g. Physcomitrella patens 71 genes), while the vascular lycopod representative Selaginella moellendorffii contains 227 P450s. These varying numbers suggest that the quantity of P450s in plant genomes has substantially increased through evolution, to a point in angiosperms where they represent approximately 1 per cent of the encoded genes. The advances in next-generation sequencing technologies, combined with progressively decreasing costs, have considerably amplified the number of sequenced plant genomes. Consequently, as more evolutionary diverse aquatic and land species are sequenced, new plant P450 families are being revealed, with the current total standing at 127. Nevertheless, the number of P450 families in angiosperms seems to have plateaued at 59. In the P450 nomenclature system, plants were initially assigned family numbers from 71 to 99. But by 1996, it had already become clear that the diversity of plant P450s was largely underestimated, and new families had to be assigned from 701 to the latest addition of CYP805A (David Nelson's web page, http://drnelson.uthsc.edu/cytochromeP450.html).
Based on the available sequences, land plant P450s can be grouped in 11 phylogenetically distinct clans [4,5]. However, it is to be expected that new clans will emerge as older plant lineages are sequenced, as seen when the genome of the green algae Chlamydomonas was released. Plant clans are named according to the lowest-numbered family member [5]. Originally, the view on plant P450s was more simplified and they were grouped as either A-type or non-A-type [6,7]. The idea behind this division was that the A-type P450s, based on phylogenetic trees, constituted a monophyletic clade, whereas the non-A-type were more diverse and did not form a coherent group in a phylogenetic sense. The A-type P450s are now in the CYP71 clan, whereas the non-A-type P450s are in the remaining 10 clans. Based on the available data at that time, the A-type P450s were involved in ‘plant-specific metabolism’, now commonly referred to as plant specialized metabolism, such as the synthesis of lignin, alkaloids, flavonoids and cyanogenic glucosides. The non-A-types on the other hand were thought to be involved in more basic metabolism such as sterol and lipid oxygenation and hormone metabolism. We know now that this simplified view is incorrect, and that there are several examples of non-A-type P450s that have been recruited for plant specialized metabolism.
Particularly within the A-type/CYP71 clan, there are successive gene duplication events, referred to as blooms [8,9], that have generated often species-specific expanded families. Some of these duplications are reminiscent of ancient whole-genome duplication events in terrestrial plants that have led to duplicated and triplicated polyploid progenitors [10], and opened vast opportunities for neo- and subfunctionalization for the evolution of new chemistries [11,12]. Other duplicated P450s are derived from retroposition events or unequal cross-over events during meiosis. Gene duplication events derived from retroposition are intron-less by nature and blooms of intron-less P450s are prevalent in Arabidopsis CYP86 and CYP96 families in the CYP86 clan; CYP89, CYP81 and CYP98 families in the CYP71 clan; and the CYP710 clan (for a review see [4]). Duplication events from unequal cross-over events result in characteristic tandem arrays of P450s as exemplified by the species-specific bloom of 37 CYP71B genes in Arabidopsis, of which 34 are located in four tandem arrays [13].
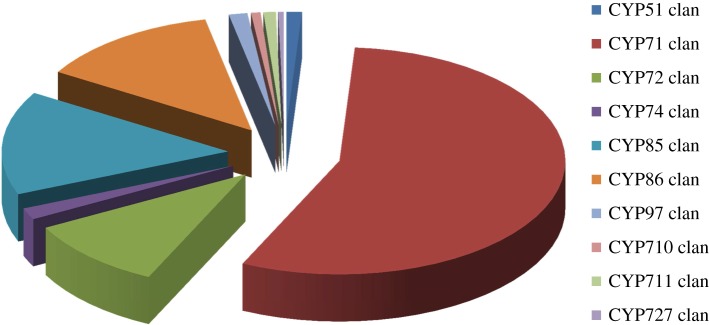
Phylogenetic analyses of plant P450s clearly indicate that the CYP71 clan is the youngest clan [3,4]. Yet, more than half of the P450s found in moss are members of this clan, indicating the CYP71 clan was already dominant in early land plants [3]. When analysing the relative number of angiosperm P450s per clan, it is evident that some clans have expanded more than others (figure 1), and, as a consequence, allowed for their diversified function. For example, CYP51 is the only P450 family in the CYP51 clan and it catalyses the essential sterol 14α-demethylase reaction in sterol biosynthesis in eukaryotes. CYP710A is the only subfamily in the CYP710 clan and it mediates biosynthesis of the phytohormone brassinolide [14,15]. CYP711A is the only family in the CYP711 clan and it is involved in strigolactone signalling and, thereby, the regulation of root–shoot multiplication. Other examples include the CYP74 clan that contains a limited number of subfamilies that are involved in metabolizing oxygenated polyunsaturated C18 fatty acid hydroperoxides to oxylipids in the octadecanoid pathway, generating signalling molecules, such as jasmonates, essential for host immunity and plant development (for a recent review see [16]). Characteristic of these single-family clans is their involvement in essential functions, such as formation of sterols for membranes, and phytohormones for plant development and plant defence. In contrast, the CYP71 clan is highly proliferated and includes P450s involved in metabolism of the majority of plant specialized compounds. Similarly, the CYP85 clan and CYP86 clan are proliferated multi-family clans involved in both general and specialized metabolism. P450s in the CYP85 clan metabolize terpenoid phytohormones and specialized metabolites. Similarly, while P450s of the CYP86 clan are involved in hydroxylation and epoxidation of fatty acids, fatty alcohols or alkanes, a few family members have functionally diversified in specialized metabolism.
Figure 1.
Relative distribution of angiosperm P450s across the clans from a selected taxonomically wide range of angiosperm genomes. The average number of P450s is based on the sequenced and curated (David Nelson) genomes of a wide taxonomic range of angiosperm species: the core eudicotyledons Arabidopsis (Arabidopsis thaliana), papaya (Carica papaya), grape vine (Vitis vinifera), poplar (Populus trichocarpa), castor bean (Ricinus communis), jatropha (Jatropha curcas), soybean (Glycine max), and the monocotyledons rice (Oryza sativa) and Brachypodium (Brachypodium distachyon).
In this review, we will highlight examples of how plant P450s have been continuously recruited for biosynthesis of new specialized compounds, either by divergence from general metabolism after gene duplication, or from pre-existing specialized compound pathways.
2. CYP98A bridges general and specialized phenylpropanoid metabolism
Phenylpropanoids represent one class of metabolites with a key role in the evolution of modern plants, as their diversification led to a plethora of critical functions in development and adaptation. Lignin, built from phenylpropanoid-derived precursors is the second most abundant biopolymer on the planet and has, due to its hydrophobic nature, enabled plants to transport water, and ultimately to grow in height and to colonize the land. Other, equally vital roles of phenylpropanoids include, but are not limited to: protection, as UV pigments, radical scavenging, preformed defence molecules and phytoalexins, as signalling molecules and in reproduction. In all biosynthetic routes, P450s are located at key regulatory positions. For example, three P450 families are involved in biosynthesis of mono-lignols, the building blocks for lignin production: CYP73 (cinnamate 4-hydroxylase), CYP84 (coniferaldehyde and coniferylalcohol 5-hydroxylase) and CYP98 as aromatic meta-hydroxylase (for a review see [17]). The CYP98 family has received substantial attention, as it represents a particularly illustrative example of evolutionary mechanisms driving chemical diversification on the basis of P450s.
The core enzymes of CYP98, which are ubiquitously represented in all land plants at low-gene count, are involved in the general lignin biosynthetic route, where they catalyse hydroxylation of the shikimate and quinate esters of 4-coumarate at the third position of the phenolic ring (meta-hydroxylation) en route to chlorogenic acid and mono-lignols [18]. A mechanistically similar hydroxylation was suggested for the biosynthetic route to the phenylpropanoid specialized metabolite rosmarinic acid, which proceeds via 3-hydroxylation of the 4′-phenyllactic ester of 4-coumarate. Two CYP98A enzymes from species accumulating rosmarinic acid, one from Ocimum basilicum (sweet basil) of the Lamiaceae family, and the other from Lithospermum erythrorhizon of the Boraginaceae family, were suggested to catalyse the required 3-hydroxylation, albeit at a lower rate than with the shikimate ester (O. basilicum) or without comparison to the shikimate ester (L. erythrorhizon) [19,20]. The proof for a truly neofunctionalized CYP98A in rosmarinic acid biosynthesis came in 2009, when Eberle et al. [21] published their study on Coleus blumei (Lamiaceae) CYP98A14. This enzyme efficiently converted, through 3- and 3′-hydroxylation, 4-coumaroyl- and caffeoyl-phenyllactate into rosmarinic acid, without accepting the corresponding shikimate- or quinate-linked phenylpropanoids as alternative substrates. In hexaploid wheat, two of eight detected CYP98A genes were shown to meta-hydroxylate coumaroyl-tyramine to caffeoyl-tyramine, a conjugated phenolic precursor of suberin, and with that demonstrating substrate plasticity and beginning neofunctionalization for members of an expanded CYP98A family [22].
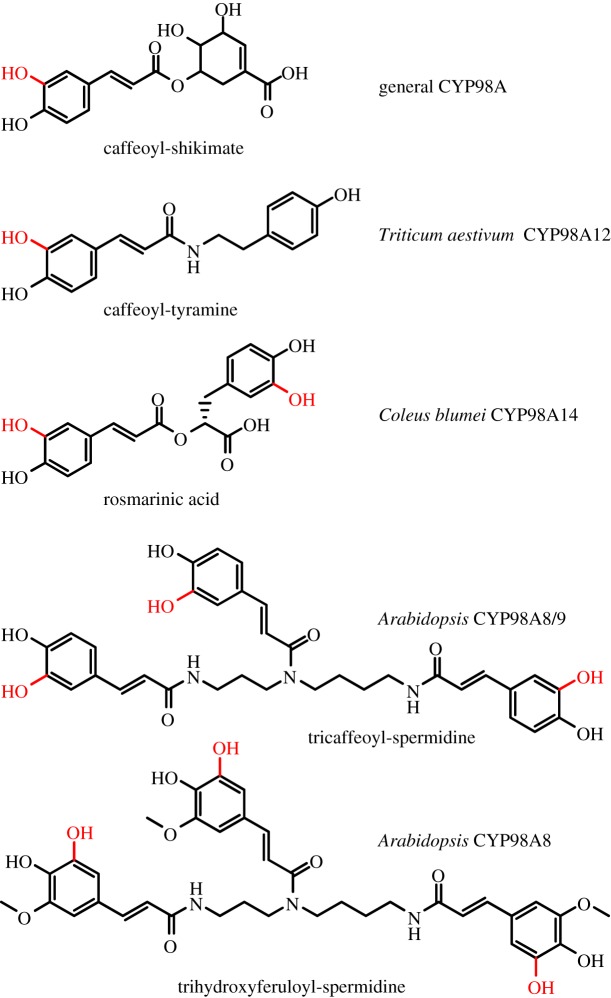
Just 2 years later, members of the same research team followed up on a striking phenomenon detected in Arabidopsis, where gene duplication in the CYP98A family had led to two additional copies that evolved at a highly accelerated rate: CYP98A8 and CYP98A9. Matsuno and co-workers observed that the young members showed, for this family, an unusual lack of intronic sequence and were clustered on chromosome 1. Consequently, it was suggested that the family's expansion was the result of a series of events starting with retrotransposition and tandem duplication, concomitant with neofuntionalization and subsequent further specialization, or broadening of the function [23]. The positions of amino acids under strong positive selection and structure homology modelling indicated enlargement of the substrate access channel and active site cavity in comparison with the ancestral CYP98A. Consistent with the gain of novel regulatory elements upstream of the translational start, which were also partially locally duplicated with the tandem duplication, transcript profiling showed reprogramming of the expression of CYP98A8 and CYP98A9 in male reproductive organs. Metabolite profiling of transgenic plants and characterization of heterologously expressed CYP98A8 and CYP98A9 was used to demonstrate that both enzymes have a redundant activity in the sequential 3-hydroxylation of a pollen-specific triphenylpropanoid, conjugated through a polyamine, but that CYP98A8 may also have an additional 5-hydroxylation activity (figure 2). While it remains unclear whether this specific activity represents the evolution of a novel function in CYP98A8 (broadening), or whether this enzymatic activity was lost in CYP98A9 (specialization), the expansion of the CYP98A subfamily through tandem duplication and the concomitant evolution of the novel phenolic pathway in pollen development represents an illustrative example of pathway elongation (see §8 for further elaboration of this phenomenon).
Figure 2.
Regio- and substrate-specificity of CYP98A family members. While duplicate members of the family have evolved from 4-coumaroyl shikimate to accept and hydroxylate tyramine, 4-coumaroyl 4-hydroxyphenyllactate and polyphenylspermidine-conjugated 4-coumaroyl at the third position, Arabidopsis CYP98A8 has gained in addition the capacity for 5-hydroxylation of polyphenylspermidine-conjugated ferulate.
3. Recurrent recruitments of P450s as catalysts for diversification of mono- to triterpenoids
Terpenoids are the evolutionarily oldest and structurally most diverse class of plant specialized metabolites. Inherently linked with plant development and adaptation, it is not surprising to detect in the most basal land plant lineages, such as the Bryophytes, complex terpenoids that in higher plants have further diversified into vital plant hormones. Chemical diversification of terpenoids has dramatically expanded their role in plant adaptation, protection and, in the broadest sense, interaction of the plant with its biosphere. Derived from C5 carbon building blocks, terpenoids are classified as mono- (C10), sesqui- (C15), di- (C20), tri- (C30) or poly- (n units) terpenoids. The initial biosynthetic step towards the typically cyclic backbone of terpenoids is catalysed by terpene synthases from the ubiquitous precursor molecules geranyl-diphosphate, farnesyl-diphosphate, geranylgeranyl-diphosphate and 2,3-epoxy-squalene (for a recent review see [24,25]). Of the currently more than 43,000 known terpenoids, the majority are oxygenated at one or more carbon positions (table 1). Oxygenation, typically catalysed by P450s, not only modulates physico-chemical properties of the terpenoids (e.g. increase in polarity, decrease in volatility), but also increases the structural diversity and introduces functional groups for auxiliary modification such as glycosylation or acylation.
Table 1.
Currently known mono- to triterpenoids in plants. Dictionary of Natural Products (http://dnp.chemnetbase.com; accessed July 18, 2012; DNP 21.1 Taylor & Francis Group).
| class of terpenoid | number of terpenoids | fraction oxygenated |
|---|---|---|
| C10 monoterpenoid | 3816 | 0.998 |
| C15 sesquiterpenoid | 13 211 | 0.967 |
| C20 diterpenoid | 11 609 | 0.988 |
| C30 triterpenoid | 14 505 | 0.952 |
| combined | 43 141 | 0.970 |
P450s oxidizing terpenoids were among the first P450s identified, characterized and cloned from plants. In addition, they belong to the oldest existing plant P450 families and their role in sterol metabolism or the metabolism of phytohormones represent the most basal known functions, conserved throughout all land plant lineages. Naturally, P450s of general terpenoid metabolism are already distributed over eight of the currently known 11 plant clans, and P450s of the evolutionarily younger terpenoid specialized metabolism have started to proliferate in four of those, indicating that recruitment from existing pathways (i.e. general metabolism) is a key feature in the evolution of new functions for P450s in terpenoid metabolism (table 2).
Table 2.
Distribution of functionally characterized P450s in terpenoid metabolism (see electronic supplementary material, supplementary table for specific biochemical function, species and references).
| clan | subfamily | metabolism | function, class of metabolites |
|---|---|---|---|
| CYP51 | CYP51G | general | triterpenoid sterols |
| CYP51H | specialized | triterpenoid | |
| CYP71 | CYP71A | specialized | monoterpenoid |
| CYP71AR | specialized | monoterpenoid | |
| CYP71AV | specialized | sesquiterpenoid | |
| CYP76B | specialized | iridoid monoterpenoid, xenobiotics detoxification | |
| CYP71BA | specialized | sesquiterpenoid | |
| CYP71BL | specialized | sesquiterpenoid | |
| CYP71D | specialized | mixed, including: monoterpenoid, sesquiterpenoid, diterpenoid, indole alkaloid, flavonoid | |
| CYP71Z | specialized | diterpenoid | |
| CYP76M | specialized | diterpenoid | |
| CYP82G | specialized | terpenoid-derived | |
| CYP93E | specialized | triterpenoid | |
| CYP99A | specialized | diterpenoid | |
| CYP701A | general | diterpenoid phytohormone GA | |
| CYP701A | specialized | diterpenoid | |
| CYP705A | specialized | triterpenoid | |
| CYP706B | specialized | sesquiterpenoid | |
| CYP710 | CYP710A | general | triterpenoid phytohormone brassinolide |
| CYP711 | CYP711A | general | terpenoid signal molecule |
| CYP72 | CYP72A | specialized | monoterpene indole alkaloid, triterpenoid |
| CYP72C | general | triterpenoid phytohormone brassinolide | |
| CYP714A | general | diterpenoid phytohormone GA | |
| CYP714D | general | diterpenoid phytohormone GA | |
| CYP734A | general | triterpenoid phytohormone brassinolide | |
| CYP85 | CYP85A | general | triterpenoid phytohormone brassinolide |
| CYP88A | general | diterpenoid phytohormone GA | |
| CYP88D | specialized | triterpenoid | |
| CYP90A | general | triterpenoid phytohormone brassinolide | |
| CYP90B | general | triterpenoid phytohormone brassinolide | |
| CYP90C | general | triterpenoid phytohormone brassinolide | |
| CYP90D | general | triterpenoid phytohormone brassinolide | |
| CYP707A | general | terpenoid phytohormone ABA | |
| CYP708A | specialized | triterpenoid | |
| CYP716A | specialized | triterpenoid | |
| CYP720B | specialized | diterpenoid | |
| CYP725A | specialized | diterpenoid | |
| CYP86 | CYP724B | general | triterpenoid phytohormone brassinolide |
| CYP97 | CYP97A | general | carotenoids |
| CYP97B | general | carotenoids | |
| CYP97C | general | carotenoids |
The evolution of genes involved in specialized metabolism appears accelerated when compared with general metabolism, with the above-mentioned Arabidopsis CYP98A8/9 versus CYP98A representing a prime example. Hence, duplicated genes and expanded families, for which the evolution of novel specialized terpenoids can be traced back to individual progenitor P450s of general terpenoid metabolism, indicate young inventions. Examples, discussed more in detail below, are found in the CYP51, CYP701 and CYP88 families. Extended families involved in specialized metabolism which are either detected in older land plant lineages, or which are more broadly represented across the plant kingdom, imply an earlier timing of their emergence with the identity of their ancestor remaining uncertain. Examples are CYP725A, CYP720B and CYP716A which are found as distinct families in the CYP85 clan, otherwise consisting largely of families involved in general terpenoid metabolism. Finally, the progenitor(s) for P450 families in specialized metabolism in the CYP71 clan are unknown, and it cannot be excluded that some are derived from CYP71 genes already involved in specialized metabolism followed by further rapid diversification. CYP701, found at the base of this clan, and thereby likely being the oldest existing family in that clan, is the only P450 known to be involved in terpenoid general metabolism (phytohormone gibberellic acid (GA)), whereas 16 of the 17 families with functions in terpenoid metabolism assigned play roles in specialized metabolism.
4. The CYP71 clan: cradle of monoterpenoid and sesquiterpenoid diversity
Monoterpenoids represent the smallest fraction of known terpenoids and P450s involved in their metabolism are, with the exception of Madagascar periwinkle CYP72A1, apparently restricted to the CYP71 clan. CYP72A1 catalyses formation of secologanin, the monoterpenoid moiety of the indole alkaloid strictosidine, through a quite unusual reaction for P450s, cleavage of C–C bonds [26]. Recruitment of CYP72A1 and P450s specific to closely related species for monoterpenoid metabolism may be a curiosity in this family in light of the function of more distantly related members of this subfamily (CYP72A154, Glycyrrhiza plants (liquorice) and CYP72A63, Medicago truncatula) in the biosynthesis of triterpenoids. Consistently, the four other functionally characterized families in the CYP72 clan with involvement in terpenoid metabolism are invariably involved in di- and triterpenoid phytohormone catabolism (see section on triterpenoids for details).
In the CYP71 clan, members of the distantly related subfamilies CYP71A, CYP71D, CYP71AR and CYP76B are involved, or have been suggested to play a role in the oxidation of monoterpenoids. The lack of a shared functionally characterized common ancestor prompts the speculation that this is the result of convergent evolution in these families. Avocado (Persea americana) CYP71A1 was the first plant P450 cloned [27], and is the founding member of the CYP71 family and thus the CYP71 clan. Until this time, P450s with activity towards monoterpenoids had been only identified, isolated and functionally characterized from plant microsomal fractions with activity towards geraniol and nerol from Catharanthus roseus [28] and avocado mesocarp tissue [29]. It was later demonstrated that both the cloned CYP71A1, as well as the P450 from plant microsomes, were capable of oxidizing nerol and geraniol to the corresponding 2,3- and 6,7-epoxides. However, compounds of that class remained undetectable in avocado tissue, hence the function of CYP71A1 in the plant and the physiological substrate are still unclear [30]. CYP71A13, a member of the largest and most diverse family in Arabidopsis, was shown to be involved in the formation of the indole alkaloid specialized metabolite camalexin [31], suggesting that CYP71A family members have acquired highly specific functions across and within individual species. The elusive P450 with in planta geraniol hydroxylase activity was finally discovered in C. roseus, where CYP76B6 was shown to be involved in the route to monoterpenoid indole alkaloids [32]. In both CYP76B and CYP71A families, several reports describe the acceptance of exogenous substrates as well as the capacity for heterologously expressed P450s to degrade xenobiotics [33,34], thus indicating a high degree of functional promiscuity. Both families have also undergone expansions, and at least in Arabidopsis, four CYP76C members appear in a genomic cluster on chromosome 2. For CYP76C1, a function was suggested as geraniol/nerol 10-hydroxylase [35].
Similarly, highly divergent functional specialization is a feature of the CYP71D subfamily, where functionally assigned genes involved in isoprenoid metabolism have been described for monoterpenoids, sesquiterpenoids and diterpenoids in, for example, tobacco and mint species [36–38]. The most recent CYP71D was assigned the name CYP71D353, reflecting the extraordinary size of this subfamily.
Even more limited than for monoterpenoids, P450s involved in sesquiterpenoids metabolism have so far spawned exclusively in five families of the CYP71 clan (table 2). Recent work in the Asteraceae family has yielded highly illustrative insights into the evolution of sesquiterpenoid chemical diversity. Fuelled by the finding that Artemisia annua CYP71AV1 catalyses a three-step oxidation of amorphadiene to artemisinic acid [39,40], a detailed analysis of evolutionarily conserved sesquiterpene lactone biosynthetic routes was performed across a range of Asteraceae [41]. The authors hypothesized that the biosynthesis of commonly occurring germacrene A acid, a transient intermediate in the routes to highly diverse sesquiterpene lactones, represents the evolutionary origin for the biosynthesis of artemisininc acid. Several members of the CYP71AV family from divergent Asteraceae species have consistently been functionally characterized as germacrene A oxidases, but interestingly also accept the non-natural substrate amorphadiene, whereas Artemisia CYP71AV1 displayed no significant activity towards germacrene A [41]. Hence, CYP71AV1 represents an example of biochemical neofunctionalization. The same group of Dae-Kyun Ro identified the subsequent biosynthetic steps in the routes to lettuce (Lactuca sativa) and sunflower (Helianthus annus) sesquiterpene lactones, catalysed by the founding members of the CYP71BL subfamily. It was shown that lettuce CYP71BL1 and its sunflower homologue CYP71BL2 catalyse the distinct 6α - and 8β-hydroxylation of germacrene A acid, and the authors suggest that the sunflower P450 has evolved from an ancestral 6α-hydroxylase in certain Asteraceae lineages [42] (figure 3).
Figure 3.
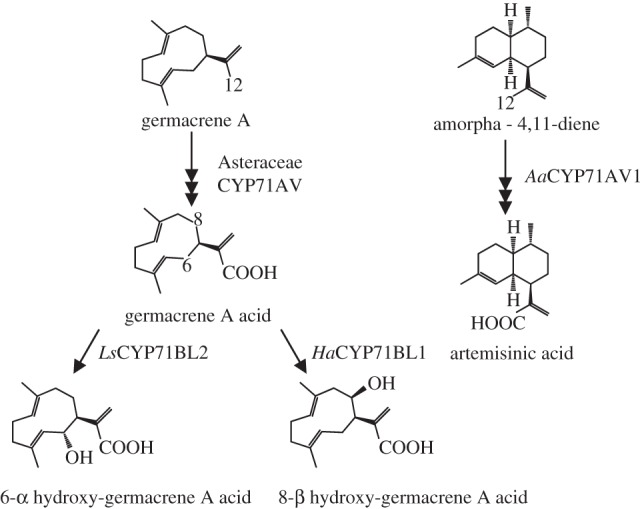
Functional specialization of P450s oxidizing sesquiterpenes. While germacrene A acid formation, catalysed by CYP71AV, is commonly found in Asteraceae, CYP71AV1 in Artemisia annua catalyses conversion of amorpha-diene to artemisinic acid. Similarly, it was suggested that CYP71BL1, taxonomically restricted to sunflower, represents a descendant of CYP71BL2, which catalyses a reaction widely conserved in many lineages of Asteraceae plants. Aa, Artemisia annua; Ls, Lactuca sativa; Ha, Helianthus annus.
5. From phytohormone to specialized metabolites: P450s oxidizing diterpenes are driving chemical diversity
The first P450 function to be detected in plants was described in Echinocystis macrocalpa (Cucurbitaceae), where the successive oxidative conversion of (−)-kaur-16-ene (ent-kaurene) to the corresponding 19-alcohol, 19-aldehyde and 19-acid was observed, which required oxygen and NADPH, and showed inhibition by carbon monoxide [43]. The gene encoding the P450, CYP701A, remains to date the only representative of the general metabolism in the CYP71 clan, and is found ubiquitously in all land plants, typically encoded by a single-copy gene. With the exception of the moss Physcomitrella, which lacks GA phytohormones, the product of CYP701A is further oxidized in higher land plants by a second P450 to GA12. In the genome of rice (O. sativa), five CYP701A subfamily members are found, while only one, CYP701A6, fulfils the vital function of ent-kaurene oxidation en route to GA phytohormones [44]. Additionally, the five P450s are organized in one genomic cluster of 115 kb on chromosome 6. Two of the clustered P450s, which are direct neighbours, are also transcriptionally co-regulated in response to elicitation. Rice contains a broad range of labdane-type diterpenoids with phytoalexin activity against microbial pathogens, and the group of Reuben Peters has pioneered the discovery of the genetic diversity underlying the chemical diversity in rice labdane-type diterpenoids (for a review on the formation of the diterpene backbones see [45]). Expansion of genes typically involved in the formation of ent-kaurene in rice has led to functional divergence and the evolution of the backbone of the phytoalexin diterpenoids. Wang and co-workers demonstrated that one of the extra copies, CYP701A8, has neofunctionalized, and has, in essence, lost the ancestral capacity to oxidize C-19 of ent-kaurene and acquired, with a changed regiospecificity, C-3-hydroxylation activity, not only of ent-kaurene, but also the specialized diterpenoid intermediates ent-sandaracopimaradiene and ent-cassadiene, indicating a broadening of substrate specificity [44]. It remains to be shown whether the observed expansion and clustering of CYP701A in rice has provided the genetic basis for evolution of completely novel routes, or for pathway elongation (see §8 for further elaboration of this phenomenon).
Members of the family CYP88A in the CYP85 clan catalyse the second step in the biosynthetic route to GA phytohormones. In a scenario similar to rice, family expansion in liquorice has been the foundation for evolution of novel functions in triterpenoid metabolism (CYP88D, see below).
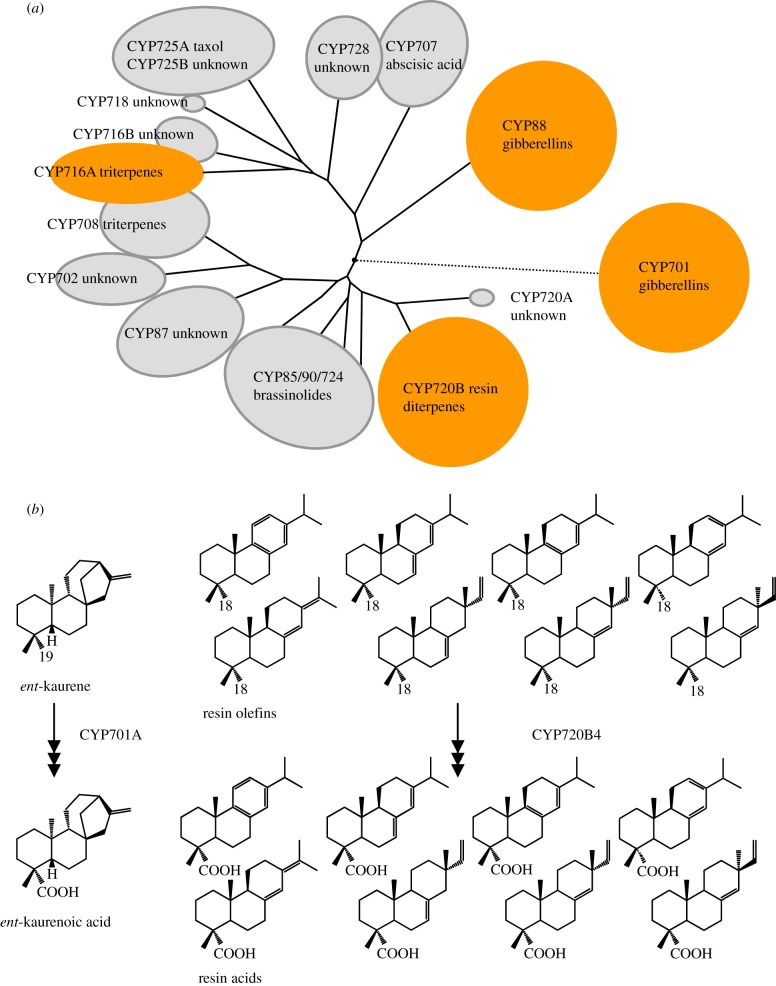
The oldest evolutionary families constituting the core of the CYP85 clan are involved in terpenoid phytohormone metabolism. Here, several families, specific to certain plant lineages have greatly expanded, indicating involvement in specialized metabolism. The specialized metabolism of diterpene resin acids in conifers, constitutive and de novo formed defence compounds, shares striking similarities with the biosynthesis of the GA intermediate ent-kaurenoic acid. Substantial research has addressed the genetic basis underlying the phenomenal diversity of defence-related terpenoids in conifers and the involvement of P450s in their biosynthesis (for a recent review see [46]). Mechanistically, both pathways leading from geranylgeranyl-diphosphate to structurally similar multi-cyclic diterpene acids are the same, catalysed by diterpene synthases and P450s [47,48]. In both pathways, a methyl-group situated on the A-ring (C-18, diterpene resin olefins; C-19, ent-kaurene) of the diterpene backbone is sequentially oxygenated by P450 activity, resulting in the corresponding acid [47,49]. What sets general and conifer specialized diterpenoid metabolism apart is the level of structural diversity of the conifer resin acids. The genetic basis for this diversity is founded through two lines of evolutionary events. Duplication, retention and neofunctionalization of a small family of diterpene synthases, sharing common ancestry with the genes of the GA phytohormone metabolism, leads to a range of diterpene backbones [48,50]. In contrast to this divergent evolution, the emergence of a large family in the CYP85 clan, CYP720B, encoding P450s catalysing the three-step oxidation of diterpene olefins, indicates convergent evolution of this function in the CYP85 and CYP71 clan [47,51]. Additionally, members of this family have acquired a certain degree of promiscuity, accepting with varying affinity a range of structurally related diterpene olefins, alcohols and aldehydes, which is in stark contrast to the known activities of the enzymes of general phytohormone metabolism (figure 4) [47,52]. While the plasticity of individual P450s of the CYP720B family contributes to chemical diversity in the defence-related diterpene resin acids, functional characterization of divergent members of the family encompassing at least a dozen P450s will shed light on the evolutionary advantage of the observed genetic diversity.
Figure 4.
(a) Schematic of the CYP85 clan and the CYP701 family (CYP71 clan), with subfamilies represented with their roles in terpenoid metabolism as known. Highlighted are families discussed in the text. (b) Functional plasticity of diterpene oxidizing P450s in conifer diterpenoid metabolism (adapted from Hamberger et al. [47]; Copyright American Society of Plant Biologists, www.plantphysiol.org).
6. Recruitment of P450s for triterpenoid biosynthesis
Triterpenoids constitute a large and structurally diverse group derived from cyclization of 2,3- oxidosqualene by oxidosqualene cyclases, and to a lesser extent directly from squalene [25,53,54]. The majority of them fall into three classes: sterols, steroids and saponins. The ability to synthesize sterols is vital for eukaryotes as sterols are integrated in eukaryotic membranes and modulate membrane fluidity, and are constituents of lipid rafts. As other eukaryotes that are not sterol heterotrophs, plant genomes contain the orthologous sterol 14α-demethylase CYP51 which catalyses formation of the Δ14–15 double bond in sterols [55]. Saponins can be further divided to triterpenoid saponins and steroidal saponins, depending on if they are derived from 2,3-oxidosqualene directly or via sterols. In sterol biosynthesis, 2,3-oxidosqualene is cyclized to tetracyclic cycloartenol or lanosterol, which is the first committed step in membrane sterol and steroid hormone synthesis. Substantial labelling experiments support that cycloartenol is the major plant sterol precursor, whereas lanosterol biosynthesis has been shown only in a few species. In plants containing triterpenes other than membrane sterols and steroid hormones, 2,3-oxidosqualene is cyclized to typically tetra- or pentacyclic compounds by neofunctionalized cyclases phylogenetically related to the cycloartenol cyclases. Acylic, monocyclic, bicyclic, tricyclic and hexacyclic triterpenes are to a lesser extent also known to occur.
There are a number of conserved P450s involved in the biosynthesis of sterols and steroids. The sterol 14α-demethylase CYP51 catalyses three consecutive oxygen- and NADPH-requiring steps at the C14 position which, via the 14α-alcohol and 14α-aldehyde followed by elimination of formic acid, forms a Δ14–15 double bond [55–57]. Members of the CYP710 family catalyse C-22 saturation [14]. Sterols are precursors of brassinosteroids/brassinolides, which are steroid hormones that promote plant growth and cell division, and of steroidal triterpenoids such as the glycoalkaloids in Solanum species [58]. Four P450 families act downstream of CYP51 and CYP710 in brassinosteroid metabolism: CYP85 catalyses C-6 oxidations, whereas CYP90A, CYP90B, CYP90C and CYP90D subfamily members catalyse C-22 and C-23 hydroxylations [59]. Two P450 families are involved in inactivation of brassinosteroids. CYP734s catalyse C-26 hydroxylations, whereas the precise function of CYP72C1, a subfamily which seems to be restricted to Brassicaceae, is not known [60–63]. The CYP734 family originally belonged to the CYP72B family, but this family has been renamed based on its position in the phylogenetic trees.
Of the P450s listed above, CYP51 belongs to the CYP51 clan and CYP710 to the CYP710 clan, two clans that are characterized by containing only one P450 family. CYP85 and CYP90 belong to the CYP85 clan, and CYP72 and CYP734 fall into the CYP72 clan; these two clans are characterized by containing multiple P450 families and subfamilies [3]. P450s from the CYP51, CYP72 and CYP85 clans are known to be recruited for biosynthesis of triterpenes.
Members of the genus Avena (oat) synthesize and accumulate anti-fungal β-amyrin-derived triterpene saponins known as avenacins in the epidermal cells of young roots [64,65]. This is a novel trait, as monocotyledons generally lack the ability to produce triterpenoid saponins. Based on the structure, avenacins are oxygenated at five positions of β-amyrin, but the genes encoding the P450s responsible for these oxygenations have not been fully documented. However, based on genetic and biochemical analysis of mutants, CYP51H10 acts immediately downstream of β-amyrin [66], and preliminary biochemical analysis of recombinant CYP51H10 confirms that CYP51H10 is a β-amyrin oxidase [67]. In oat, there are three CYP51s known: CYP51G1 which is the orthologous 14α-demethylase, and CYP51H10 and CYP51H11 [66]. CYP51H10 and CYP51H11 are 62 per cent identical at the amino acid level, whereas they are 46–47% identical to CYP51G1. Because the intron position is conserved between CYP51G1, CYP51H10 and CYP51H11, the CYP51H paralogues cannot have arisen via retrotransposition. CYP51H10 transcripts are detected in roots where the avenacins accumulate, whereas CYP51H11 is expressed in flowers. The function of CYP51H11 is currently not known. Interestingly, other monocotyledons such as rice and Brachypodium also contain CYP51H genes although these species are not known to accumulate saponins. Brachypodium has three CYP51H genes and two CYP51H pseudogenes, whereas rice has six CYP51Hs that are likely to be functional and three CYP51H pseudogenes [68], indicating that the CYP51Hs paralogues in these two monocotyledons are not stable. The oat CYP51H10 enzyme is the only example known so far of a CYP51 that has been duplicated and neofunctionalized to produce a specialized compound.
CYP90s and CYP85 in the CYP85 clan act on sterols. It is, therefore, not surprising that this clan has been used for recruitment of P450s for biosynthesis of saponins. Recently, a number of papers have been published on the roles of CYP716As in particular saponin biosynthesis [69–71]. Medicago truncatula CYP716A12 catalyses three sequential oxidation of β-amyrin at the C-28 position, yielding oleanolic acid in biosynthesis of haemolytic, but not soyasaponins, in M. truncatula [69]. Erythrodiol (C-28 hydroxylated β-amyrin) is similarly converted to oleanolic acid by CYP716A12 catalysing sequential hydroxylations from the alcohol to the aldehyde and further on to the carboxylic acid. An elaborate biochemical analysis of M. truncatula CYP716A12 showed, that in addition to β-amyrin, α-amyrin and lupeol, two other 2,3-oxidosqualene cyclized products were also 28-oxidized to the corresponding carboxylic acids, ursolic acid and betulinic acid (figure 5). CYP716A15 and CYP716A17 from grape were included in the same study and were shown to be involved in saponin biosynthesis by oxidizing β-amyrin to oleanolic acid [70].
Figure 5.
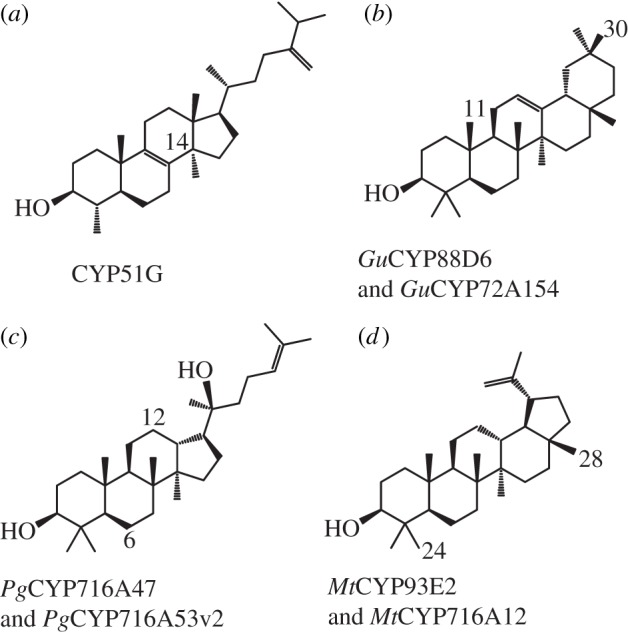
Examples of P450s involved in the oxidation of triterpenes in general and specialized metabolism. The regiospecificity of often multiple reactions is indicated by the number on the triterpenoid backbone structure. (a) Obtusifoliol, (b) β-amyrin, (c) dammarenediol-II and (d) lupeol. Gu, Glycyrrhiza uralensis; Pg, Panax ginseng; Mt, Medicago truncatula.
CYP716 family members are widespread in terrestrial plants. More than 150 are listed on David Nelson's webpage (http://drnelson.uthsc.edu/cytochromeP450.html), and a phylogenetic analysis of 56 CYP716s ranging from moss to Selaginella to gymnosperms and angiosperms shows that CYP716 is an ancient family, consistent with the widespread presence of triterpenes such as oleanolic acid in plants [70]. Interestingly, Panax ginseng CYP716A47 catalyses C-12 hydroxylation of dammarenediol-II to protopanaxadiol, followed by C-6 oxidation to protopanaxatriol by CYP716A53v2 in biosynthesis of ginsenoside saponins, showing that CYP716As are not restricted to C-28 oxidation [71,72].
CYP88As are omnipresent in terrestrial plants, and are, as discussed in the section on diterpenes, involved in biosynthesis of GA. CYP88s have also been recruited for triterpenoid biosynthesis. In biosynthesis of the triterpenoid saponin sweetener glycyrrizin in Glycyrrhiza (liquorice) plants, CYP88D6 catalyses two sequential hydroxylations of β-amyrin to 11-oxo-β-amyrin and to a lesser extent hydroxylation of β-amyrin to 11-α-hydroxy-β-amyrin. CYP88D6 is rather substrate unspecific by catalysing also single oxidations of the β-amyrin derivative 30-hydroxy-β-amyrin to 11α,30-dihydroxy-β-amyrin and 30-hydroxy-11-oxo-β-amyrin in vitro. The functions of the other CYP88 subfamilies are not known, but based on the currently available P450 sequences, CYP88Ds, recruited from CYP88s in gibberellin biosynthesis, are restricted to legumes and catalyse C-11 oxidation of triterpenes.
Originally there were four CYP72 subfamilies, but CYP72Bs have been reclassified as CYP734As. The CYP72A subfamily is highly expanded across higher plants, and often the genes are arranged in tandem arrays as seen with the nine CYP72As in Arabidopsis, which are clustered on chromosome 3. Arabidopsis contains a single CYP72C, which, based on mutant analysis, is involved in deactivation of brassinolides, although the precise biochemical function and substrate is not known [62,73,74]. CYP72C appear to be Brassicaceae-specific, arguing that involvement in brassinolide inactivation is not the original in planta function of CYP72s. Catharanthus roseus CYP72A1 was the first CYP72A to be biochemically fully elucidated, but it catalyses a peculiar reaction en route to monoterpenoid compounds (see section on monoterpenoids for details). In Glycyrrhiza (liquorice), CYP72A154 catalyses three sequential C-30 hydroxylations of 11-oxo-β-amyrin to glycyrrhetic acid and also C-30 hydroxylation of β-amyrin to 30-hydroxy-β-amyrin [75]. Medicago truncatula CYP72A63 catalyses C-30 hydroxylation of β-amyrin to 30-hydroxy-β-amyrin and in contrast to Glycyrrhiza CYP72A154, also catalyses three sequential C-30 hydroxylation of β-amyrin to the corresponding C-30 carboxylic acid 11-deoxoglycyrrhetic acid. Accordingly, CYP72As work in sequence with CYP88Ds in triterpenoid biosynthesis in legumes, and the CYP72 family appears to be a family with a high potential for catalysing a range of specific oxygenations of terpenoids and thus a determinant for the species-specific terpenoid profile as well as in phytohormone homeostasis.
CYP93s belong to the CYP71 clan and are widespread in angiosperms. CYP93As, CYP93Bs, CYP93Cs and CYP93Gs are involved in flavonoid biosynthesis particularly in plant microbe interactions [76–81]. In legumes, CYP93s are involved in isoflavonoid biosynthesis and thus involved in nodulation. But CYP93s have also been recruited for triterpenoid biosynthesis. In legumes, CYP93E1, CYP93E2 and CYP93E3 are involved in biosynthesis of triterpenoid saponins and oxidize β-amyrin at C-24 producing 24-OH-β-amyrin [70,75,82]. Accordingly, CYP93Es appear to be recruited from flavonoid biosynthesis and neofunctionalized for triterpenoid biosynthesis.
7. Recurrent recruitment of CYP79s for amino acid-derived specialized compounds
The CYP79 family is an old angiosperm family present in both monocotyledons and dicotyledons. In phylogenetic trees, CYP79s branch off deep in the CYP71 clan, close to the CYP73 and CYP98 families involved in biosynthesis of lignin and other early phenylpropanoids, and CYP701 in the biosynthesis of gibberellins [3,4,83]. CYP79s are unique P450s as they do not have hydrophobic substrates, but have amino acids as substrates. In all examined cases, CYP79s catalyse the conversion of protein amino acids, or chain elongated amino acids, to their corresponding oximes. They are multi-functional N-hydroxylases that catalyse two consecutive N-hydroxylations, followed by a dehydration and decarboxylation reaction to release the corresponding Z-oxime. Originally, CYP79s were only known to be involved in the biosynthesis of cyanogenic glucosides in angiosperms [83,84]. Cyanogenic glucosides are amino acid-derived β-glycosides of α-hydroxynitriles, and are stored as inactive glucosides in the vacuole. When the tissues they are stored in are damaged, they come into contact with β-glucosidases and release toxic hydrogen cyanide. The sequencing of the Arabidopsis genome and PCR-based strategies revealed CYP79s in species that do not contain cyanogenic glucosides, but instead contain glucosinolates. This was the first indication of how CYP79 homologues in glucosinolate-producing plants show evolutionary conservation of enzymes in conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates [85,86]. While cyanogenic glucosides are prevalent and occur in angiosperms, gymnosperms and pteridophytes (ferns) [83,87], glucosinolates are almost exclusively found in the Brassicales, and to a lesser extent in the evolutionarily distant genus Drypetes in the order Euphorbiales [88]. As no genes have been isolated from the glucosinolate biosynthetic pathway of Drypetes, it cannot be resolved if there has been a parallel evolution of glucosinolate biosynthesis in rosids. Glucosinolates are sulphur-containing amino acid-derived β-thio-glucosides [88] and are, such as cyanogenic glucosides, stored in vacuoles in an inactive form and activated through hydrolysis by thioglucosidases to a range of volatile nitriles and isothiocyanates. The substrates for CYP79s in biosynthesis of cyanogenic glucosides and glucosinolates partly overlap: cyanogenic glucosides are derived from the protein amino acid tyrosine, phenylalanine, isoleucine, leucine and valine, whereas glucosinolates are derived from tyrosine, phenylalanine, tryptophan, alanine, leucine, isoleucine, valine and methionine and from their chain elongated derivatives.
It is now evident that CYP79 homologues have also been recruited for biosynthesis of other specialized classes and subclasses of plant specialized compounds. The indole phytoalexin camalexin is derived from tryptophan. As in indole glucosinolate biosynthesis, a CYP79 catalyses conversion of tryptophan to indole-3-acetaldoxime, which is the branching point between camalexin and indole glucosinolates biosynthesis [89,90]. A number of cyanogenic glucoside-containing plants also contain so-called non-cyanogenic hydroxynitrile glucosides [91]. While cyanogenic glucosides are α-hydroxynitrile glucosides, non-cyanogenic hydroxynitrile glucosides such as the rhodiocyanosides A and D in Lotus japonicus are γ- and β-hydroxynitrile glucosides, respectively. Rhodiocyanosides do not release hydrogen cyanide when hydrolysed by β-glycosidases because they are not α-hydroxynitrile glucosides. The rhodiocyanosides in L. japonicus are derived from isoleucine as is the cyanogenic glucoside lotaustralin. The two pathways branch at the oxime, and share a CYP79 for the first step in their biosynthesis [92,93].
The sequencing of the poplar genome [94] revealed four CYP79 paralogues. The identification of CYP79 sequences was surprising as poplar species were not known to synthesize cyanogenic glucosides. Very recent work from the group of Dr Tobias Köllner (http://www.ice.mpg.de/ext/782.html) has shown that at least two CYP79s in poplar are transcribed and the encoded enzymes produce a mixture of different volatile aldoximes from amino acids (T Köllner and S Irmisch 2012, personal communication). This is in line with the proposal that the cyanogenic glucoside metabolome is dynamic, and may toggle between synthesizing cyanogenic glucosides and oximes by assembly and disassembly of the metabolome in a function-dependent manner [95]. The fact that oximes are the metabolic branch point, and the flexibility of the metabolome to assemble in the above-mentioned pathways, has been used in biotechnological approaches to genetically engineer new glucosinolate profiles in Arabidopsis through the introduction of CYP79s from the cyanogenic glucoside pathway to establish metabolic crosstalk between the two pathways [96,97].
The proposed evolution of glucosinolates, camalexin, non-cyanogenic hydroxynitrile glucosides and volatile oximes from cyanogenic glucosides illustrates how an ancient pathway for cyanogenic glucoside synthesis has given rise to new classes and subclasses of plant specialized compounds for new biological functions (figure 6). The dynamics of CYP79s for evolving novel pathways is reflected in the Arabidopsis and L. japonicus genomes. There are seven CYP79 genes and four pseudogenes in Arabidopsis. The presence of multiple CYP79 genes and a high ratio of pseudogenes, is indicative of recent evolution of the glucosinolate pathway. The involvement of CYP79F1 and CYP79F2 in the biosynthesis of glucosinolates is a typical scenario where two gene copies have both neo- and subfunctionalized to expand the profile of a specialized metabolite. CYP79F1 and CYP79F2 are 88 per cent identical on amino acid level and are positioned in tandem on Arabidopsis chromosome 1 in the same orientation. They are both involved in biosynthesis of homo- to hexahomo-methionine-derived aliphatic glucosinolates, and display overlapping but distinguishable substrate specificity. While CYP79F1 metabolizes homo- to hexahomo-methionine with a preference for short chain glucosinolates, CYP79F2 metabolizes exclusively long-chained penta- and hexahomo-methionine. Their expression patterns overlap slightly but clearly differ both developmentally and tissue-wise, with a preference for CYP79F1 expression in hypocotyledons and roots, and CYP79F2 in rosettes, stems and siliques [98].
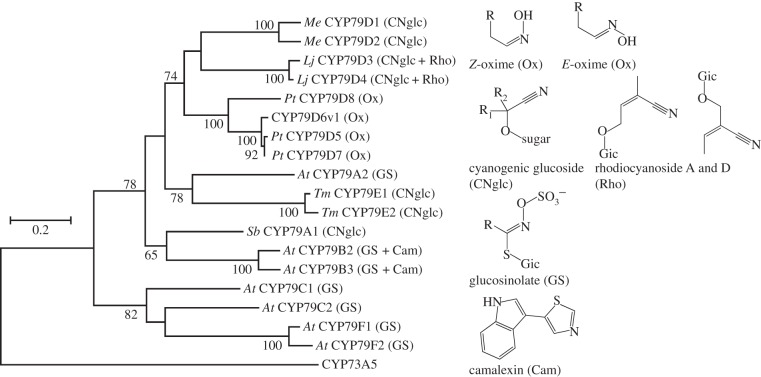
Figure 6.
Maximum-likelihood phylogeny of the CYP79 family. CYP73C5 is included as an out-group. The structures of the products are given and indicated in the abbreviation behind the P450. Bootstrap values greater than 70 are given at the nodes. The scale bar indicates number of changes. Pt, Populus tremuloides; Lj, Lotus japonicus; Me, Manihot esculenta; Tm, Triglochin maritima; Sb, Sorghum bicolor; At, Arabidopsis thaliana.
In the L. japonicus genome, there are two highly similar CYP79D paralogues with a sequence identity of 95 per cent at the amino acid level. Lotus japonicus CYP79D3 and CYP79D4 are not adjacent as the Arabidopsis CYP79F1 and CYP79F2 are, but they are spaced 240 kbp apart, and separated by two genes [99]. In contrast to Arabidopsis CYP79F1 and CYP79F2, they catalyse the same reaction and display the same kinetics, but their promoter sequences have diverged, resulting in differential expression, with transcripts of CYP79D3 accumulating preferentially in aerial tissues and CYP79D4 in the root [92]. This indicates that the two CYP79D paralogues have originated from a gene or whole-genome duplication event and that subsequent subfunctionalization has led to differences in expression patterns, while catalysis has been maintained. The fact that L. japonicus is a paleopolyploid, whereas the most recent autopolyploidy event happened more than 40 Ma [100], is in support of this assumption.
8. Evolution and elongation of pathways by tandem gene duplications
The recent availability of fully sequenced plant genomes has revealed that some pathways for plant specialized metabolites are located in operon-like clusters (review in e.g. [101,102]). Some of these clusters contain multiple P450 paralogues that, after duplication, most likely by unequal cross-over events, diverged to elongate the pathway and to increase metabolic diversity. Even though these represent specific cases, tandem P450 gene duplications may reveal how a pathway can gain diversity and complexity.
In maize, four CYP71Cs are clustered on chromosome four and are part of a functional gene cluster in the grasses for indole-derived benzoxazinoids biosynthesis. They catalyse four consecutive hydroxylations in the pathway of benzoxanoids [103]. Benzoxazinoids are protective and allelophatic metabolites widely found in species of the Poaceae. They are stored as glucosides in the vacuoles and analogous to cyanogenic glucosides and glucosinolates, and are bioactivated through hydrolysis by β-glucosidases stored in separate compartments. In maize the four P450 genes are known as BX2 (CYP71C1) to BX5 (CYP71C4). The availability of other Poaceae genomes has revealed that the pathway has evolved monophyletically in the grasses ([104] and references therein). An evolutionary sequence leading to benzoxazinoids was suggested based on phylogenetic relationship of the P450s involved and by the specific reaction catalysed by each. The ‘founding event’ of the pathway was suggested to be the clustering of duplicated tryptophan synthase α-subunit (TSA) and an ancestral CYP71C gene, followed by neofunctionalization of both into Bx1 and Bx2, respectively, and yielding biosynthetic access to indole and the oxidized indolin-2-one, derived from the primary metabolite indole-3-glyceraldehyde. The elongation of this cluster through duplication of Bx2 (CYP71C1) giving rise to Bx3 (CYP71C2) was followed by two subsequent duplications of Bx3 resulting in Bx4 (CYP71C3) and Bx5 (CYP71C4), and generated the material that, after neofunctionalization, resulted in a series of successive oxidations from indolin-2-one via 3-hydroxy indolin-2-one and 2-hydroxy-1,4(2H)-benzoxazin-3(4H)-one (HBOA) to 2,4-dihydroxy-2H-1,4-benzoxazin-3(4H)-one (DIBOA) [104,105]. Both HBOA and DIBOA are subsequently glycosylated by Bx8/Bx9, further hydroxylated by Bx6 (a 2-oxogluterate-dependent dioxygenase) to 2,4,7-trihydroxy-2H-1,4-benzoxazin-3(4H)-one (TRIBOA)-glc and methoxylated by Bx7 to 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA)-glc ([104] and references therein). A few species of dicotyledons also synthesize benzoxazinoids. Analysis of the first gene in the pathway in grasses and benzoxazinoid-producing dicots revealed an evolution of repeated independent duplications and neofunctionalizations of the TSA gene [106]. Similarly, the UDP-dependent glycosyl transferases (UGTs) have evolved independently [107]. The P450s in benzoxazinoid biosynthesis in dicotyledonous species are not known, and as the CYP71C family appears to be restricted to monocotyledons, the pathway in monocotyledons and dicotyledons apparently evolved convergently.
Recent work has shed light on the assembly and continuous evolution of P450 carrying gene clusters in rice for diterpene biosynthesis [108]. Specifically, four CYP76M paralogues, CYP76M5–8, are situated in a 245 kb gene cluster on chromosome 2, together with three diterpene synthases involved in the biosynthesis of specialized diterpene olefins with ent-configuration. The first of those P450s functionally characterized, CYP76M7, was shown to specifically catalyse C11α hydroxylation of ent-cassadiene [109], the product of two diterpene synthases of that cluster. In contrast, the closest paralogue, CYP76M8, shows very high-substrate promiscuity, hydroxylating several positions of not only diterpene olefin precursors of the ent-, but also the syn-configuration, indicating a function in other pathways. The fact that this P450 showed the capacity to hydroxylate structurally related diterpenes not found in rice could represent latent metabolic capacity for rapid evolution of diterpenoid diversity. The other P450s in this cluster, CYP76M5 and CYP76M6, had partially overlapping and additional specific activities. Thus, the CYP76M5–8 cluster illustrates how subsequent local duplications of P450s involved in one pathway may lead to chemical diversification within the pathway, but also drive diversification of other pathways [108,109].
Arabidopsis produces nearly 40 different glucosinolates derived from methionine, phenylalanine and tryptophan [110]. The core structure of all glucosinolates is synthesized by a similar set of enzymes [88]. Structural diversity arises from subsequent modifications such as oxidation, hydroxylation and methoxylation. Arabidopsis contains four different indole glucosinolates derived from tryptophan. The core indole glucosinolates indol-3-yl-methyl (I3M) is hydroxylated and methoxylated by a set of P450s and methyl-transferases [111]. There are four CYP82s in Arabidopsis: CYP81F3 and CYP81F4 are situated in tandem, spaced from CYP81F1 by genes with unknown function, on chromosome 4, whereas CYP81F2 is positioned on chromosome five. The four CYP81Fs have partly overlapping product specificity: CYP81F1, CYP81F2 and CYP81F3 catalyse hydroxylation of I3M to 4OH-I3M while all four CYP81Fs catalyse the conversion of I3M to 1-OH-I3M, when tested in the transient expression system in Nicotiana benthamiana. Mutant analysis partly confirms the biochemical analysis and, combined with expression analysis based on DNA microarrays, suggests that the CYP81Fs are expressed in an organ- or tissue-specific manner, thus representing both neo- and subfunctionalization [111].
9. Conclusions and perspectives
P450s control key steps in the biosynthetic routes of plant specialized metabolites and are consequently essential drivers in the evolution of chemical diversity. Here, genome and gene duplications leading to family expansion, followed by fixation of the genes in the genome, owing to positive selection of sub- and neofunctionalized paralogues provides the fundamental genetic material for plant adaptation and species radiation. But what are the mechanisms driving this diversification? Arguably the best understood examples may be specialized compounds acting as defence molecules in plant–herbivore and plant–pathogen interactions. Reciprocal evolution of adaptation is, according to macroevolutionary hypotheses for coevolution, followed by surges of speciation giving rise to biodiversity. This progressive diversification is intrinsically connected with diversification of defence metabolites and consistent with coevolution in an arms race.
In this review, we have discussed evolutionary implications of genetic diversity of the P450 multi-gene family, where numerous examples highlight the critical role P450s play in driving chemical diversification. Overall, a common theme appears to be specialization and diversification for production of sophisticated defence molecules in plants, whereas herbivores and pathogens have evolved P450s for detoxification as part of counter-adaptation strategies.
From the biochemist's perspective: the range of reaction mechanisms catalysed by P450s is extremely diverse, but the most commonly found types are oxygenation reactions such as hydroxylation, invariably linked with a gain of bioactivity or ‘activation’ of plant specialized metabolites. At the same time, the catalytic flexibility of P450s makes this class of enzymes ideally suited for the evolution of novel functions. The first examples are emerging, demonstrating that novel P450s involved in specialized metabolism evolve at a much accelerated pace, and taken into account that a considerable fraction of plant genes are P450s, it can be hypothesized that these enzymes will become increasingly instrumental for future plant adaptation and speciation.
From the human perspective: initially evolved as traits linked with an adaptive advantage, plant species accumulating specialized metabolites have been selected and bred by humans for thousands of years. Often considered defence molecules, these particular and often complex traits are what have attracted human interest for nutritional value, flavour and use in traditional medicine, as stimulants, narcotics and poisons. Evidently human selection is linked with an increase in chemical diversity, but in a few species breeding aimed at reduction of specialized metabolites or their chemical complexity. Two cases of this scenario are harmful glycoalkaloids accumulating in wild, but not the modern potato (Solanum commersonii and Solanum tuberosum, respectively), and cyanogenic glucosides determining the bitterness of almond (Prunus dulcis). Further examples with major impacts on human civilization and with key biosynthetic steps catalyzed by diverse P450s include, but are not limited to, flavonoids (grape vine, cocoa (Theobroma cacao), green tea (Camellia sinensis)), terpenes (mint family, Ginkgo biloba) and alkaloids (opium poppy (Papaver somniferum)).
Acknowledgements
We wish to acknowledge David Nelson for helpful discussion and for providing information of P450s family numbers in the different plant species. We thank Allison Maree Maree Heskes for critical reading of the manuscript and the reviewers for helpful comments. B.R.H. acknowledges financial support from the ‘Center for Synthetic Biology’ at the Copenhagen University funded by the UNIK research initiative of the Danish Ministry of Science, Technology and Innovation and the Novo Nordisk Foundation.
References
- 1.Mizutani M, Sato F. 2011. Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 507, 194–203 10.1016/j.abb.2010.09.026 (doi:10.1016/j.abb.2010.09.026) [DOI] [PubMed] [Google Scholar]
- 2.Schuler MA. 2011. P450s in plant–insect interactions. Biochim. Biophys. Acta 1814, 36–45 10.1016/j.bbapap.2010.09.012 (doi:10.1016/j.bbapap.2010.09.012) [DOI] [PubMed] [Google Scholar]
- 3.Nelson D, Werck-Reichhart D. 2011. A P450-centric view of plant evolution. Plant J. 66, 194–211 10.1111/j.1365-313X.2011.04529.x (doi:10.1111/j.1365-313X.2011.04529.x) [DOI] [PubMed] [Google Scholar]
- 4.Bak S, Beisson F, Bishop G, Hamberger B, Hofer R, Paquette S, Werck-Reichhart D. 2011. Cytochromes P450. Arabidopsis Book 9, e0144. 10.1199/tab.0144 (doi:10.1199/tab.0144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S. 2004. Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135, 756–772 10.1104/pp.104.039826 (doi:10.1104/pp.104.039826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durst F, Nelson DR. 1995. Diversity and evolution of plant P450 and P450-reductases. Drug Metab. Drug Interact. 12, 189–206 10.1515/DMDI.1995.12.3-4.189 (doi:10.1515/DMDI.1995.12.3-4.189) [DOI] [PubMed] [Google Scholar]
- 7.Kahn RA, Durst F. 2000. Function and evolution of plant cytochrome P450. In Evolution of metabolic pathways (eds Romeo JT, Ibrahim R, Varin L, de Luca V.). Recent Advances in Phytochemistry, vol 34, pp. 151–189 Rotterdam, The Netherlands: Elsevier [Google Scholar]
- 8.Feyereisen R. 2011. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta 1814, 19–28 10.1016/j.bbapap.2010.06.012 (doi:10.1016/j.bbapap.2010.06.012) [DOI] [PubMed] [Google Scholar]
- 9.Sezutsu H, Le Goff G, Feyereisen R. 2013. Origins of P450 diversity. Phil. Trans. R. Soc. B 368, 20120428. 10.1098/rstb.2012.0428 (doi:10.1098/rstb.2012.0428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proost S, Pattyn P, Gerats T, Van de Peer Y. 2011. Journey through the past, 150 million years of plant genome evolution. Plant J. 66, 58–65 10.1111/j.1365-313X.2011.04521.x (doi:10.1111/j.1365-313X.2011.04521.x) [DOI] [PubMed] [Google Scholar]
- 11.Adams KL, Wendel JF. 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141 10.1016/j.pbi.2005.01.001 (doi:10.1016/j.pbi.2005.01.001) [DOI] [PubMed] [Google Scholar]
- 12.Ober D. 2010. Gene duplications and the time thereafter: examples from plant secondary metabolism. Plant Biol. (Stuttg.) 12, 570–577 [DOI] [PubMed] [Google Scholar]
- 13.Paquette SM, Jensen K, Bak S. 2009. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases (http//www.P450.kvl.dk). Phytochemistry 70, 1940–1947 10.1016/j.phytochem.2009.08.024 (doi:10.1016/j.phytochem.2009.08.024) [DOI] [PubMed] [Google Scholar]
- 14.Morikawa T, Mizutani M, Ohta D. 2006. Cytochrome P450 subfamily CYP710A genes encode sterol C-22 desaturase in plants. Biochem. Soc. Trans. 34, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 15.Morikawa T, Saga H, Hashizume H, Ohta D. 2009. CYP710A genes encoding sterol C22-desaturase in Physcomitrella patens as molecular evidence for the evolutionary conservation of a sterol biosynthetic pathway in plants. Planta 229, 1311–1322 10.1007/s00425-009-0916-4 (doi:10.1007/s00425-009-0916-4) [DOI] [PubMed] [Google Scholar]
- 16.Hughes RK, De Domenico S, Santino A. 2009. Plant cytochrome CYP74 family: biochemical features, endocellular localisation, activation mechanism in plant defence and improvements for industrial applications. ChemBioChem 10, 1122–1133 10.1002/cbic.200800633 (doi:10.1002/cbic.200800633) [DOI] [PubMed] [Google Scholar]
- 17.Ehlting J, Hamberger B, Million-Rousseau R, Werck-Reichhart D. 2006. Cytochromes P450 in phenolic metabolism. Phytochem. Rev. 5, 239–270 10.1007/s11101-006-9025-1 (doi:10.1007/s11101-006-9025-1) [DOI] [Google Scholar]
- 18.Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D. 2001. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 276, 36 566–36 574 10.1074/jbc.M104047200 (doi:10.1074/jbc.M104047200) [DOI] [PubMed] [Google Scholar]
- 19.Gang DR, Beuerle T, Ullmann P, Werck-Reichhart D, Pichersky E. 2002. Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant Physiol. 130, 1536–1544 10.1104/pp.007146 (doi:10.1104/pp.007146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuno M, Nagatsu A, Ogihara Y, Ellis BE, Mizukami H. 2002. CYP98A6 from Lithospermum erythrorhizon encodes 4-coumaroyl-4′-hydroxyphenyllactic acid 3-hydroxylase involved in rosmarinic acid biosynthesis. FEBS Lett. 514, 219–224 10.1016/S0014-5793(02)02368-2 (doi:10.1016/S0014-5793(02)02368-2) [DOI] [PubMed] [Google Scholar]
- 21.Eberle D, Ullmann P, Werck-Reichhart D, Petersen M. 2009. cDNA cloning and functional characterisation of CYP98A14 and NADPH:cytochrome P450 reductase from Coleus blumei involved in rosmarinic acid biosynthesis. Plant Mol. Biol. 69, 239–253 10.1007/s11103-008-9420-7 (doi:10.1007/s11103-008-9420-7) [DOI] [PubMed] [Google Scholar]
- 22.Morant M, Schoch GA, Ullmann P, Ertunc T, Little D, Olsen CE, Petersen M, Negrel J, Werck-Reichhart D. 2007. Catalytic activity, duplication and evolution of the CYP98 cytochrome P450 family in wheat. Plant Mol. Biol. 63, 1–19 10.1007/s11103-006-9028-8 (doi:10.1007/s11103-006-9028-8) [DOI] [PubMed] [Google Scholar]
- 23.Matsuno M, et al. 2009. Evolution of a novel phenolic pathway for pollen development. Science 325, 1688–1692 10.1126/science.1174095 (doi:10.1126/science.1174095) [DOI] [PubMed] [Google Scholar]
- 24.Chen F, Tholl D, Bohlmann J, Pichersky E. 2011. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66, 212–229 10.1111/j.1365-313X.2011.04520.x (doi:10.1111/j.1365-313X.2011.04520.x) [DOI] [PubMed] [Google Scholar]
- 25.Phillips DR, Rasbery JM, Bartel B, Matsuda SP. 2006. Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314 10.1016/j.pbi.2006.03.004 (doi:10.1016/j.pbi.2006.03.004) [DOI] [PubMed] [Google Scholar]
- 26.Irmler S, Schroder G, St-Pierre B, Crouch NP, Hotze M, Schmidt J, Strack D, Matern U, Schroder J. 2000. Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 24, 797–804 10.1046/j.1365-313x.2000.00922.x (doi:10.1046/j.1365-313x.2000.00922.x) [DOI] [PubMed] [Google Scholar]
- 27.Bozak KR, Yu H, Sirevag R, Christoffersen RE. 1990. Sequence analysis of ripening-related cytochrome P-450 cDNAs from avocado fruit. Proc. Natl Acad. Sci. USA 87, 3904–3908 10.1073/pnas.87.10.3904 (doi:10.1073/pnas.87.10.3904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meehan TD, Coscia CJ. 1973. Hydroxylation of geraniol and nerol by a monooxygenase from Vinca rosea. Biochem. Biophys. Res. Commun. 53, 1043–1048 10.1016/0006-291X(73)90570-6 (doi:10.1016/0006-291X(73)90570-6) [DOI] [PubMed] [Google Scholar]
- 29.Hallahan DL, Nugent JH, Hallahan BJ, Dawson GW, Smiley DW, West JM, Wallsgrove RM. 1992. Interactions of avocado (Persea americana) cytochrome P-450 with monoterpenoids. Plant Physiol. 98, 1290–1297 10.1104/pp.98.4.1290 (doi:10.1104/pp.98.4.1290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallahan DL, Lau SM, Harder PA, Smiley DW, Dawson GW, Pickett JA, Christoffersen RE, O'Keefe DP. 1994. Cytochrome P-450-catalysed monoterpenoid oxidation in catmint (Nepeta racemosa) and avocado (Persea americana): evidence for related enzymes with different activities. Biochim. Biophys. Acta 1201, 94–100 10.1016/0304-4165(94)90156-2 (doi:10.1016/0304-4165(94)90156-2) [DOI] [PubMed] [Google Scholar]
- 31.Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, GlazeBrook J. 2007. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19, 2039–2052 10.1105/tpc.107.051383 (doi:10.1105/tpc.107.051383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collu G, Unver N, Peltenburg-Looman AM, van der Heijden R, Verpoorte R, Memelink J. 2001. Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett. 508, 215–220 10.1016/S0014-5793(01)03045-9 (doi:10.1016/S0014-5793(01)03045-9) [DOI] [PubMed] [Google Scholar]
- 33.Bozak KR, O'Keefe DP, Christoffersen RE. 1992. Expression of a ripening-related avocado (Persea americana) cytochrome P450 in yeast. Plant Physiol. 100, 1976–1981 10.1104/pp.100.4.1976 (doi:10.1104/pp.100.4.1976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robineau T, Batard Y, Nedelkina S, Cabello-Hurtado F, LeRet M, Sorokine O, Didierjean L, Werck-Reichhart D. 1998. The chemically inducible plant cytochrome P450 CYP76B1 actively metabolizes phenylureas and other xenobiotics. Plant Physiol. 118, 1049–1056 10.1104/pp.118.3.1049 (doi:10.1104/pp.118.3.1049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizutani M, Ohta D.1998. Plant geraniol/nerol 10-hydroxylase and DNA coding therefor. US Patent number US5753507.
- 36.Ralston L, Kwon ST, Schoenbeck M, Ralston J, Schenk DJ, Coates RM, Chappell J. 2001. Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Arch. Biochem. Biophys. 393, 222–235 10.1006/abbi.2001.2483 (doi:10.1006/abbi.2001.2483) [DOI] [PubMed] [Google Scholar]
- 37.Wang E, Wagner GJ. 2003. Elucidation of the functions of genes central to diterpene metabolism in tobacco trichomes using posttranscriptional gene silencing. Planta 216, 686–691 [DOI] [PubMed] [Google Scholar]
- 38.Wust M, Little DB, Schalk M, Croteau R. 2001. Hydroxylation of limonene enantiomers and analogs y recombinant (−)-limonene 3- and 6-hydroxylases from mint (Mentha) species: evidence for catalysis within sterically constrained active sites. Arch. Biochem. Biophys. 387, 125–136 10.1006/abbi.2000.2248 (doi:10.1006/abbi.2000.2248) [DOI] [PubMed] [Google Scholar]
- 39.Ro DK, et al. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943 10.1038/nature04640 (doi:10.1038/nature04640) [DOI] [PubMed] [Google Scholar]
- 40.Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. 2006. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580, 1411–1416 10.1016/j.febslet.2006.01.065 (doi:10.1016/j.febslet.2006.01.065) [DOI] [PubMed] [Google Scholar]
- 41.Nguyen DT, Göpfert JC, Ikezawa N, MacNevin G, Kathiresan M, Conrad J, Spring O, Ro BK. 2010. Biochemical conservation and evolution of germacrene A oxidase in Asteraceae. J. Biol. Chem. 285, 16 588–16 598 10.1074/jbc.M110.111757 (doi:10.1074/jbc.M110.111757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikezawa N, Göpfert JC, Nguyen DT, Kim S-U, O'Maille PE, Spring O, Ro DK. 2011. Lettuce costunolide synthase (CYP71BL2) and its homolog (CYP71BL1) from sunflower catalyze distinct regio- and stereoselective hydroxylations in sesquiterpene lactone metabolism. J. Biol. Chem. 286, 21 601–21 611 10.1074/jbc.M110.216804 (doi:10.1074/jbc.M110.216804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy PJ, West CA. 1969. The role of mixed function oxidases in kaurene metabolism in Echinocystis macrocarpa Greene endosperm. Arch. Biochem. Biophys. 133, 395–407 10.1016/0003-9861(69)90468-8 (doi:10.1016/0003-9861(69)90468-8) [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Hillwig ML, Wu Y, Peters RJ. 2012. CYP701A8: a rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 158, 1418–1425 10.1104/pp.111.187518 (doi:10.1104/pp.111.187518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters RJ. 2010. Two rings in them all: the labdane-related diterpenoids. Nat. Prod. Rep. 27, 1521–1530 10.1039/c0np00019a (doi:10.1039/c0np00019a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zulak KG, Bohlmann J. 2010. Terpenoid biosynthesis and specialized vascular cells of conifer defense. J. Integr. Plant. Biol. 52, 86–97 10.1111/j.1744-7909.2010.00910.x (doi:10.1111/j.1744-7909.2010.00910.x) [DOI] [PubMed] [Google Scholar]
- 47.Hamberger B, Ohnishi T, Seguin A, Bohlmann J. 2011. Evolution of diterpene metabolism, Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol. 157, 1677–1695 10.1104/pp.111.185843 (doi:10.1104/pp.111.185843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keeling CI, Weisshaar S, Lin RP, Bohlmann J. 2008. Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc. Natl Acad. Sci. USA 105, 1085–1090 10.1073/pnas.0709466105 (doi:10.1073/pnas.0709466105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helliwell CA, Poole A, Peacock WJ, Dennis ES. 1999. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 119, 507–510 10.1104/pp.119.2.507 (doi:10.1104/pp.119.2.507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keeling CI, Madilao LL, Zerbe P, Dullat HK, Bohlmann J. 2011. The primary diterpene synthase products of Picea abies levopimaradiene/abietadiene synthase (PaLAS) are epimers of a thermally unstable diterpenol. J. Biol. Chem. 286, 21 145–21 153 10.1074/jbc.M111.245951 (doi:10.1074/jbc.M111.245951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamberger B, Bohlmann J. 2006. Cytochrome P450 mono-oxygenases in conifer genomes: discovery of members of the terpenoid oxygenase superfamily in spruce and pine. Biochem. Soc. Trans. 34, 1209–1214 10.1042/BST0341209 (doi:10.1042/BST0341209) [DOI] [PubMed] [Google Scholar]
- 52.Ro DK, Arimura G, Lau SY, Piers E, Bohlmann J. 2005. Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc. Natl Acad. Sci. USA 102, 8060–8065 10.1073/pnas.0500825102 (doi:10.1073/pnas.0500825102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaller H. 2010. 1.21—Sterol and steroid biosynthesis and metabolism in plants and microorganisms. In Comprehensive natural products I (eds Lew M, Hung-Wen L.), pp. 755–787 Oxford, UK: Elsevier [Google Scholar]
- 54.Xu R, Fazio GC, Matsuda SP. 2004. On the origins of triterpenoid skeletal diversity. Phytochemistry 65, 261–291 10.1016/j.phytochem.2003.11.014 (doi:10.1016/j.phytochem.2003.11.014) [DOI] [PubMed] [Google Scholar]
- 55.Bak S, Kahn RA, Olsen CE, Halkier BA. 1997. Cloning and expression in Escherichia coli of the obtusifoliol 14 alpha-demethylase of Sorghum bicolor (L.) Moench, a cytochrome P450 orthologous to the sterol 14 alpha-demethylases (CYP51) from fungi and mammals. Plant J. 11, 191–201 10.1046/j.1365-313X.1997.11020191.x (doi:10.1046/j.1365-313X.1997.11020191.x) [DOI] [PubMed] [Google Scholar]
- 56.Cabello-Hurtado F, et al. 1997. Cloning and functional expression in yeast of a cDNA coding for an obtusifoliol 14alpha-demethylase (CYP51) in wheat. Biochem. Biophys. Res. Commun. 230, 381–385 10.1006/bbrc.1996.5873 (doi:10.1006/bbrc.1996.5873) [DOI] [PubMed] [Google Scholar]
- 57.Rahier A, Taton M. 1986. The 14 alpha-demethylation of obtusifoliol by a cytochrome P-450 monooxygenase from higher plants’ microsomes. Biochem. Biophys. Res. Commun. 140, 1064–1072 10.1016/0006-291X(86)90743-6 (doi:10.1016/0006-291X(86)90743-6) [DOI] [PubMed] [Google Scholar]
- 58.Milner SE, Brunton NP, Jones PW, O'Brien NM, Collins SG, Maguire AR. 2011. Bioactivities of glycoalkaloids and their aglycones from Solanum species. J. Agric. Food Chem. 59, 3454–3484 10.1021/jf200439q (doi:10.1021/jf200439q) [DOI] [PubMed] [Google Scholar]
- 59.Bancos S, Nomura T, Sato T, Molnar G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M. 2002. Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130, 504–513 10.1104/pp.005439 (doi:10.1104/pp.005439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohnishi T, Nomura T, Watanabe B, Ohta D, Yokota T, Miyagawa H, Sakata K, Mizutani M. 2006. Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 67, 1895–1906 10.1016/j.phytochem.2006.05.042 (doi:10.1016/j.phytochem.2006.05.042) [DOI] [PubMed] [Google Scholar]
- 61.Sakamoto T, Kawabe A, Tokida-Segawa A, Shimizu B, Takatsuto S, Shimada Y, Fujioka S, Mizutani M. 2011. Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 67, 1–12 10.1111/j.1365-313X.2011.04567.x (doi:10.1111/j.1365-313X.2011.04567.x) [DOI] [PubMed] [Google Scholar]
- 62.Thornton LE, Rupasinghe SG, Peng H, Schuler MA, Neff MM. 2010. Arabidopsis CYP72C1 is an atypical cytochrome P450 that inactivates brassinosteroids. Plant Mol. Biol. 74, 167–181 10.1007/s11103-010-9663-y (doi:10.1007/s11103-010-9663-y) [DOI] [PubMed] [Google Scholar]
- 63.Turk EM, et al. 2005. BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 42, 23–34 10.1111/j.1365-313X.2005.02358.x (doi:10.1111/j.1365-313X.2005.02358.x) [DOI] [PubMed] [Google Scholar]
- 64.Haralampidis K, Bryan G, Qi X, Papadopoulou K, Bakht S, Melton R, Osbourn A. 2001. A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl Acad. Sci. USA 98, 13 431–13 466 10.1073/pnas.231324698 (doi:10.1073/pnas.231324698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner EMC. 1960. The nature of the resistance of oats to the take-all fungus. J. Exp. Bot. 11, 403–412 10.1093/jxb/11.3.403 (doi:10.1093/jxb/11.3.403) [DOI] [Google Scholar]
- 66.Qi X, et al. 2006. A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc. Natl Acad. Sci. USA 103, 18 848–18 853 10.1073/pnas.0607849103 (doi:10.1073/pnas.0607849103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kunii M, Kitahama Y, Fukushima EO, Seki H, Muranaka T, Yoshida Y, Aoyama Y. 2012. Beta-amyrin oxidation by oat CYP51H10 expressed heterologously in yeast cells: the first example of CYP51-dependent metabolism other than the 14-demethylation of sterol precursors. Biol. Pharm. Bull. 35, 801–804 10.1248/bpb.35.801 (doi:10.1248/bpb.35.801) [DOI] [PubMed] [Google Scholar]
- 68.Inagaki YS, Etherington G, Geisler K, Field B, Dokarry M, Ikeda K, Mutsukado Y, Dicks J, Osbourn A. 2011. Investigation of the potential for triterpene synthesis in rice through genome mining and metabolic engineering. New Phytol. 191, 432–448 10.1111/j.1469-8137.2011.03712.x (doi:10.1111/j.1469-8137.2011.03712.x) [DOI] [PubMed] [Google Scholar]
- 69.Carelli M, et al. 2011. Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23, 3070–3081 10.1105/tpc.111.087312 (doi:10.1105/tpc.111.087312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukushima EO, Seki H, Ohyama K, Ono E, Umemoto N, Mizutani M, Saito K, Muranaka T. 2011. CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol. 52, 2050–2061 10.1093/pcp/pcr146 (doi:10.1093/pcp/pcr146) [DOI] [PubMed] [Google Scholar]
- 71.Han JY, Kim HJ, Kwon YS, Choi YE. 2011. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 52, 2062–2073 10.1093/pcp/pcr150 (doi:10.1093/pcp/pcr150) [DOI] [PubMed] [Google Scholar]
- 72.Han JY, Hwang HS, Choi SW, Kim HJ, Choi YE. 2012. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 53, 1535–1545 10.1093/pcp/pcs106 (doi:10.1093/pcp/pcs106) [DOI] [PubMed] [Google Scholar]
- 73.Nakamura M, Satoh T, Tanaka S, Mochizuki N, Yokota T, Nagatani A. 2005. Activation of the cytochrome P450 gene, CYP72C1, reduces the levels of active brassinosteroids in vivo. J. Exp. Bot. 56, 833–840 10.1093/jxb/eri073 (doi:10.1093/jxb/eri073) [DOI] [PubMed] [Google Scholar]
- 74.Takahashi N, et al. 2005. shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels. Plant J. 42, 13–22 10.1111/j.1365-313X.2005.02357.x (doi:10.1111/j.1365-313X.2005.02357.x) [DOI] [PubMed] [Google Scholar]
- 75.Seki H, et al. 2008. Licorice beta-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl Acad. Sci. USA 105, 14 204–14 209 10.1073/pnas.0803876105 (doi:10.1073/pnas.0803876105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akashi T, Aoki T, Ayabe S. 1998. Identification of a cytochrome P450 cDNA encoding -flavanone 2-hydroxylase of licorice (Glycyrrhiza echinata L.; Fabaceae) which represents licodione synthase and flavone synthase II. FEBS Lett. 431, 287–290 10.1016/S0014-5793(98)00781-9 (doi:10.1016/S0014-5793(98)00781-9) [DOI] [PubMed] [Google Scholar]
- 77.Du Y, Chu H, Chu IK, Lo C. 2010. CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol. 154, 324–333 10.1104/pp.110.161042 (doi:10.1104/pp.110.161042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Du Y, Chu H, Wang M, Chu IK, Lo C. 2010. Identification of flavone phytoalexins and a pathogen-inducible flavone synthase II gene (SbFNSII) in sorghum. J. Exp. Bot. 61, 983–994 10.1093/jxb/erp364 (doi:10.1093/jxb/erp364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fliegmann J, Furtwangler K, Malterer G, Cantarello C, Schuler G, Ebel J, Mithofer A. 2010. Flavone synthase II (CYP93B16) from soybean (Glycine max L.). Phytochemistry 71, 508–514 10.1016/j.phytochem.2010.01.007 (doi:10.1016/j.phytochem.2010.01.007) [DOI] [PubMed] [Google Scholar]
- 80.Latunde-Dada AO, Cabello-Hurtado F, Czittrich N, Didierjean L, Schopfer C, Hertkorn N, Werick-Reichhart D, Ebel J. 2001. Flavonoid 6-hydroxylase from soybean (Glycine max L.), a novel plant P-450 monooxygenase. J. Biol. Chem. 276, 1688–1695 [DOI] [PubMed] [Google Scholar]
- 81.Zhang J, Subramanian S, Zhang Y, Yu O. 2007. Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol. 144, 741–751 10.1104/pp.106.095018 (doi:10.1104/pp.106.095018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shibuya M, Hoshino M, Katsube Y, Hayashi H, Kushiro T, Ebizuka Y. 2006. Identification of beta-amyrin and sophoradiol 24-hydroxylase by expressed sequence tag mining and functional expression assay. FEBS J. 273, 948–959 10.1111/j.1742-4658.2006.05120.x (doi:10.1111/j.1742-4658.2006.05120.x) [DOI] [PubMed] [Google Scholar]
- 83.Bak S, et al. 2006. Cyanogenic glycosides: a case study for evolution and application of cytochromes P450. Phytochem. Rev. 5, 309–329 10.1007/s11101-006-9033-1 (doi:10.1007/s11101-006-9033-1) [DOI] [Google Scholar]
- 84.Koch BM, Sibbesen O, Halkier BA, Svendsen I, Moller BL. 1995. The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalyzing the conversion of l-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch. Biochem. Biophys. 323, 177–186 10.1006/abbi.1995.0024 (doi:10.1006/abbi.1995.0024) [DOI] [PubMed] [Google Scholar]
- 85.Bak S, Nielsen HL, Halkier BA. 1998. The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol. Biol. 38, 725–734 10.1023/A:1006064202774 (doi:10.1023/A:1006064202774) [DOI] [PubMed] [Google Scholar]
- 86.Paquette SM, Bak S, Feyereisen R. 2000. Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 19, 307–317 10.1089/10445490050021221 (doi:10.1089/10445490050021221) [DOI] [PubMed] [Google Scholar]
- 87.Seigler D. (ed.) 1991. Cyanide and cyanogenic glucosides. San Diego, CA: Academic Press [Google Scholar]
- 88.Halkier BA, Gershenzon J. 2006. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 10.1146/annurev.arplant.57.032905.105228 (doi:10.1146/annurev.arplant.57.032905.105228) [DOI] [PubMed] [Google Scholar]
- 89.Glawischnig E, Hansen BG, Olsen CE, Halkier BA. 2004. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl Acad. Sci. USA 101, 8245–8250 10.1073/pnas.0305876101 (doi:10.1073/pnas.0305876101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hull AK, Vij R, Celenza JL. 2000. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl Acad. Sci. USA 97, 2379–2384 10.1073/pnas.040569997 (doi:10.1073/pnas.040569997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bjarnholt N, Moller BL. 2008. Hydroxynitrile glucosides. Phytochemistry 69, 1947–1961 10.1016/j.phytochem.2008.04.018 (doi:10.1016/j.phytochem.2008.04.018) [DOI] [PubMed] [Google Scholar]
- 92.Forslund K, Morant M, Jorgensen B, Olsen CE, Asamizu E, Sato S, Tabata S, Bak S. 2004. Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol. 135, 71–84 10.1104/pp.103.038059 (doi:10.1104/pp.103.038059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saito S, Motawia MS, Olsen CE, Moller BL, Bak S. 2012. Biosynthesis of rhodiocyanosides in Lotus japonicus: rhodiocyanoside A is synthesized from (Z)-2-methylbutanaloxime via 2-methyl-2-butenenitrile. Phytochemistry 77, 260–267 10.1016/j.phytochem.2012.01.020 (doi:10.1016/j.phytochem.2012.01.020) [DOI] [PubMed] [Google Scholar]
- 94.Tuskan GA, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 10.1126/science.1128691 (doi:10.1126/science.1128691) [DOI] [PubMed] [Google Scholar]
- 95.Lindberg Møller BL. 2010. Plant science. Dynamic metabolons. Science 330, 1328–1329 10.1126/science.1194971 (doi:10.1126/science.1194971) [DOI] [PubMed] [Google Scholar]
- 96.Bak S, Olsen CE, Petersen BL, Moller BL, Halkier BA. 1999. Metabolic engineering of p-hydroxybenzylglucosinolate in Arabidopsis by expression of the cyanogenic CYP79A1 from Sorghum bicolor. Plant J. 20, 663–671 10.1046/j.1365-313X.1999.00642.x (doi:10.1046/j.1365-313X.1999.00642.x) [DOI] [PubMed] [Google Scholar]
- 97.Mikkelsen MD, Halkier BA. 2003. Metabolic engineering of valine- and isoleucine-derived glucosinolates in Arabidopsis expressing CYP79D2 from cassava. Plant Physiol. 131, 773–779 10.1104/pp.013425 (doi:10.1104/pp.013425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen S, et al. 2003. CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 33, 923–937 10.1046/j.1365-313X.2003.01679.x (doi:10.1046/j.1365-313X.2003.01679.x) [DOI] [PubMed] [Google Scholar]
- 99.Takos AM, et al. 2011. Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J. 68, 273–286 10.1111/j.1365-313X.2011.04685.x (doi:10.1111/j.1365-313X.2011.04685.x) [DOI] [PubMed] [Google Scholar]
- 100.Cannon SB, et al. 2006. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl Acad. Sci. USA 103, 14 959–14 964 10.1073/pnas.0603228103 (doi:10.1073/pnas.0603228103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kliebenstein DJ, Osbourn A. 2012. Making new molecules - evolution of pathways for novel metabolites in plants. Curr. Opin. Plant Biol. 15, 415–423 10.1016/j.pbi.2012.05.005 (doi:10.1016/j.pbi.2012.05.005) [DOI] [PubMed] [Google Scholar]
- 102.Takos AM, Rook F. 2012. Why biosynthetic genes for chemical defense compounds cluster. Trends Plant Sci. 17, 383–388 10.1016/j.tplants.2012.04.004 (doi:10.1016/j.tplants.2012.04.004) [DOI] [PubMed] [Google Scholar]
- 103.Frey M, et al. 1997. Analysis of a chemical plant defense mechanism in grasses. Science 277, 696–699 10.1126/science.277.5326.696 (doi:10.1126/science.277.5326.696) [DOI] [PubMed] [Google Scholar]
- 104.Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. 2009. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70, 1645–1651 10.1016/j.phytochem.2009.05.012 (doi:10.1016/j.phytochem.2009.05.012) [DOI] [PubMed] [Google Scholar]
- 105.Dutartre L, Hilliou F, Feyereisen R. 2012. Phylogenomics of the benzoxazinoid biosynthetic pathway of Poaceae: gene duplications and origin of the Bx cluster. BMC Evol. Biol. 12, 64. 10.1186/1471-2148-12-64 (doi:10.1186/1471-2148-12-64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schullehner K, Dick R, Vitzthum F, Schwab W, Brandt W, Frey M, Gierl A. 2008. Benzoxazinoid biosynthesis in dicot plants. Phytochemistry 69, 2668–2677 10.1016/j.phytochem.2008.08.023 (doi:10.1016/j.phytochem.2008.08.023) [DOI] [PubMed] [Google Scholar]
- 107.Dick R, Rattei T, Haslbeck M, Schwab W, Gierl A, Frey M. 2012. Comparative analysis of benzoxazinoid biosynthesis in monocots and dicots: independent recruitment of stabilization and activation functions. Plant Cell 24, 915–928 10.1105/tpc.112.096461 (doi:10.1105/tpc.112.096461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Q, Hillwig ML, Okada K, Yamazaki K, Wu Y, Swaminathan S, Yamane H, Peters RJ. 2012. Characterization of CYP76M5–8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J. Biol. Chem. 287, 6159–6168 10.1074/jbc.M111.305599 (doi:10.1074/jbc.M111.305599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. 2009. CYP76M7 is an ent-cassadiene C11 alpha-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21, 3315–3325 10.1105/tpc.108.063677 (doi:10.1105/tpc.108.063677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. 2001. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126, 811–825 10.1104/pp.126.2.811 (doi:10.1104/pp.126.2.811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pfalz M, Mikkelsen MD, Bednarek P, Olsen CE, Halkier BA, Kroymann J. 2011. Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23, 716–729 10.1105/tpc.110.081711 (doi:10.1105/tpc.110.081711) [DOI] [PMC free article] [PubMed] [Google Scholar]