Abstract
Cytochromes P450 play important roles in biosynthesis of flavonoids and their coloured class of compounds, anthocyanins, both of which are major floral pigments. The number of hydroxyl groups on the B-ring of anthocyanidins (the chromophores and precursors of anthocyanins) impact the anthocyanin colour, the more the bluer. The hydroxylation pattern is determined by two cytochromes P450, flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H) and thus they play a crucial role in the determination of flower colour. F3′H and F3′5′H mostly belong to CYP75B and CYP75A, respectively, except for the F3′5′Hs in Compositae that were derived from gene duplication of CYP75B and neofunctionalization. Roses and carnations lack blue/violet flower colours owing to the deficiency of F3′5′H and therefore lack the B-ring-trihydroxylated anthocyanins based upon delphinidin. Successful redirection of the anthocyanin biosynthesis pathway to delphinidin was achieved by expressing F3′5′H coding regions resulting in carnations and roses with novel blue hues that have been commercialized. Suppression of F3′5′H and F3′H in delphinidin-producing plants reduced the number of hydroxyl groups on the anthocyanidin B-ring resulting in the production of monohydroxylated anthocyanins based on pelargonidin with a shift in flower colour to orange/red. Pelargonidin biosynthesis is enhanced by additional expression of a dihydroflavonol 4-reductase that can use the monohydroxylated dihydrokaempferol (the pelargonidin precursor). Flavone synthase II (FNSII)-catalysing flavone biosynthesis from flavanones is also a P450 (CYP93B) and contributes to flower colour, because flavones act as co-pigments to anthocyanins and can cause blueing and darkening of colour. However, transgenic plants expression of a FNSII gene yielded paler flowers owing to a reduction of anthocyanins because flavanones are precursors of anthocyanins and flavones.
Keywords: anthocyanin; anthocyanidin; flavonoid; flavonoid 3′-hydroxylase; flavonoid 3′,5′-hydroxylase; flavone synthase
1. Introduction
Flower colour is mainly due to three classes of pigment: flavonoids, carotenoids and betalains [1]. Among them, a coloured class of flavonoids, anthocyanins, confer a diverse range of colour from orange to red to violet and blue. Other flavonoids such as aurones and flavones are pale yellow or invisible to human beings. Carotenoids and betalains generally yield yellow or red colours [1].
The final colour of a flower is determined by a combination of various factors: anthocyanin structures, the pH of the vacuole where anthocyanins localize, coexisting flavonoids (co-pigments) and metal ions [2]. Anthocyanidins, aglycones of anthocyanins, are chromophores and precursors of anthocyanins (figure 1). In spite of large numbers of anthocyanins and various tones of flower colour, there are only six major classes of anthocyanidins: pelargonidin, cyanidin, peonidin, delphinidin, petunidin and malvidin. In terms of biosynthesis, peonidin is the methylated derivative of cyanidin and petunidin, and malvidin is the methylated derivatives of delphinidin (figure 1). The number of hydroxyl groups on the B-ring contribute to colour, the more hydroxyl groups the bluer the colour. Blue/violet flowers tend to contain delphinidin-based anthocyanins, magenta/red flowers contain predominantly cyanidin-based anthocyanins and orange/brick red flowers contain pelargonidin-based anthocyanins. Methylation of 3′- or 5′-hydroxyl groups tends to yield slightly redder colours [2]. Modification of anthocyanins with plural aromatic acyl groups (polyacylanthocyanins) tends to shift the colour towards blue as in the cases of Gentiana spp. (gentian) and Clitoria ternatea (butterfly pea) [3]. Vacuolar pH greatly affects anthocyanin colour; anthocyanins are redder and more stable in lower pH and bluer, however more unstable, in higher (neutral) pH. Co-pigments cause bathochromic shifts (bluer and darker colour) of anthocyanins and are typically flavones and flavonols. Ferrous and aluminium ions cause blueing by chelating the hydroxyl groups on the B-ring of anthocyanins. Super-complexes consisting of anthocyanins, flavonoids and metal ions yield blue colour in some plant species [2]. Plants achieve their own colour, especially blue, by a combination of these factors.
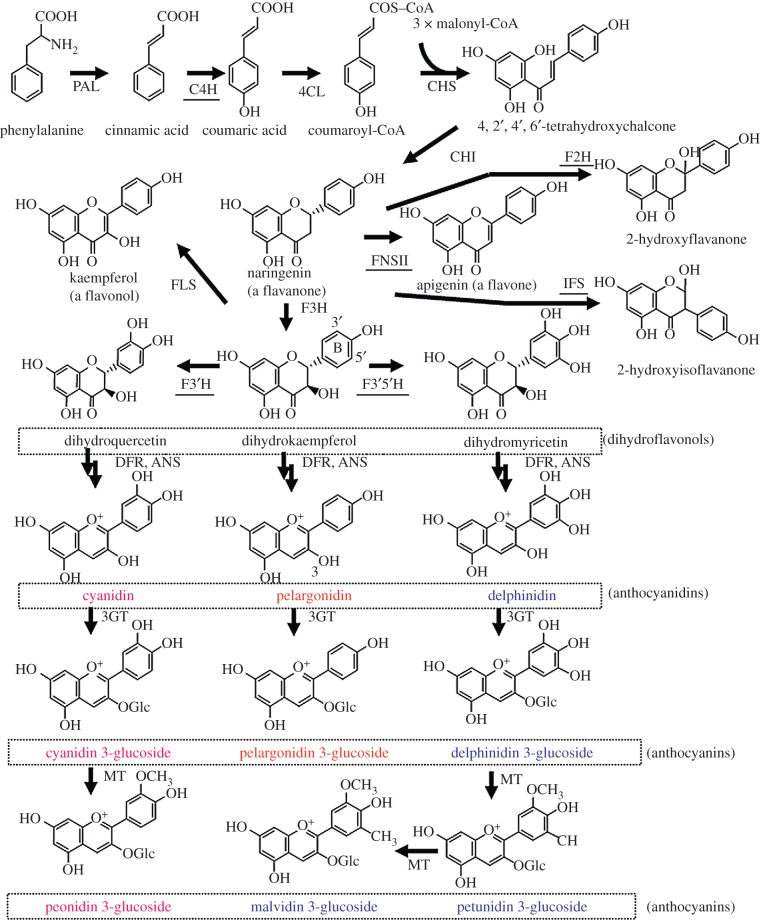
Figure 1.
The flavonoid biosynthetic pathway relevant to flower colour. The pathways leading to anthocyanidin 3-glucosides are conserved in seed plants. Anthocyanidin 3-glucosides are further modified by glycosylation, acylation and methylation in a species-specific manner. P450 enzymes are underlined. In this review, anthocyanins based on peonidin are included as cyanidin-based anthocyanins whilst those based on petunidin and malvidin are included as delphinidin-based anthocyanins. Abbreviations include: PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; CL, 4-coumarate Co-A ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; FNS, flavone synthase; F2H; flavanones 2-hydroxylase; IFS, 2-hydroxyisoflavanone synthase; FLS, flavonol synthase; 3GT, UDP-glucose: anthocyanidin 3-O-glucosyltransferase; MT, S-adenosylmethionine: anthocyanin methyltransferase.
Cytochromes P450 (P450s) play crucial roles in flower colour by determining the number of hydroxyl groups on the B-ring and catalysing flavone biosynthesis [4]. In this review, we will mainly describe biology and biotechnology relevant to these P450s determining flower colour, especially focusing on genetically engineered carnations that have been in market over 10 years and have not yet been scientifically described in detail.
2. Role of cytochromes P450 in flower colour
The flavonoid biosynthetic pathway relevant to flower colour has been well studied, and the pathway leading to anthocyanidin 3-glucosides is generally conserved in flowering plants (figure 1). Two related P450s, flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H), catalyse hydroxylation of the B-ring. They have broad substrate specificity and catalyse hydroxylation of flavanones, dihydroflavonols, flavonols and flavones. Because flavanones and dihydroflavonols are precursors of anthocyanidins and anthocyanins, the two enzymes determine the hydroxylation pattern of the B-ring and thus flower colour. Presence of F3′5′H leads to trihydroxylated delphinidin-based anthocyanins that tend to have violet/blue colours. Many plant species that lack F3′5′H usually lack violet/blue flower colour although cyanidin-based anthocyanins can result in blue colours by forming supermolecular complexes as in the case of cornflower [5] or accumulating polyacyl anthocyanins at alkaline vacuolar pH such as morning glory [6]. Because the top-selling cut flower species (Rosa hybrida (rose), Chrysanthemum morifolium (chrysanthemum), Dianthus caryophyllus (carnation) and Lilium spp. (lily) that occupy more than 50% of cut-flower market) lack delphinidin-based anthocyanins owing to the deficiency of a F3′5′H gene, the gene has attracted considerable attention as a critical molecular tool to engineer blue flower colour in these species using recombinant DNA technology. Because suppression of F3′5′H and/or F3′H genes will redirect the anthocyanidin biosynthetic pathway to pelargonidin and yield intense red colour, the F3′H gene is also a useful molecular tool for flower colour modification. F3′H and F3′5′H are also important in terms of plant and pollinator interaction and therefore have had an impact on plant evolution [7]
F3′H and F3′5′H activities have been detected in the microsomal fractions of mainly petals of many plant species using radio-isotope-labelled flavonoids such as naringenin and NADPH as an electron donor [8]. NADPH cytochrome P450 reductase was shown to catalyse electron transfer from NADPH to F3′5′H in petunia [9]. Petunia has two F3′5′H loci (Hf1 and Hf2) [10], and a cytochrome b5 that specifically transfers electrons to Hf1 F3′5′H that contributes to efficient accumulation of delphinidin-based anthocyanins [11]. Petunia dihydroflavonol 4-reductase (DFR) contributes to efficient accumulation of delphinidin by catalysing dihydromyricetin well but not dihydrokaempferol (DHK) [12].
Most flavonoid biosynthetic enzymes are soluble cytosolic proteins. These enzymes have been proposed to form a multi-enzymatic complex or metabolon anchored to F3′H, F3′5′H and cinnamic acid 4-hydroxylase (CYP73, figure 1) that localize in the endoplasmic reticulum (ER) membrane via protein–protein interaction. Such complexes facilitate metabolic channelling for efficient biosynthesis of flavonoids [13].
(a). Molecular evolution of F3′H and F3′5′H genes
F3′H [14] and F3′5′H [10] genes were first isolated and characterized from petunia and subsequently have been isolated from many plant species. Their biochemical functions are usually identified by measuring their recombinant enzyme expressed in yeast or functional analysis and colour changes in plants. Their isolation from many species revealed that F3′H and F3′5′H were classified into different subfamilies in the same family in P450 nomenclature (CYP75B and CYP75A, respectively; table 1) and that their genes diverged before the speciation of higher plants [15]. CYP75, belonging to CYP71 clan, is found in gymnosperm and angiosperm plants and appeared as early as ferns but is not found in mosses or algae [16].
Table 1.
A list of CYP75A and CYP75B members from cytochrome P450 nomenclature (http://drnelson.uthsc.edu/BiblioD.html). One of each from a subfamily is shown. A plant species often contains plural loci for CYP75A and CYP75B.
| CYP75A | species (common name) | CYP75B | species (common name) |
|---|---|---|---|
| CYP75A1 | Petunia hybrida (petunia) | CYP75B1 | Arabidopsis thaliana |
| CYP75A2 | Solanum melongena (eggplant) | CYP75B2 | Petunia hybrida (petunia) |
| CYP75A3 | Petunia hybrida (petunia) | CYP75B3 | Oryza sativa (rice) |
| CYP75A4 | Gentiana triflora (gentian) | CYP75B4 | Perilla frutescens (an Asiatic mint) |
| CYP75A5 | Eustoma grandiflorum (lisianthus) | CYP75B5 | Callistephus chinensis (aster) |
| CYP75A6 | Campanula medium (bell flowers) | CYP75B6 | Callistephus chinensis (aster) |
| CYP75A7 | Eustoma russellianum (=Eustoma grandiflorum) | CYP75B7 | Matthiola incana (stock) |
| CYP75A8 | Catharanthus roseus (Madagascar periwinkle) | CYP75B8 | Pelargonium hortorum (zonal geranium) |
| CYP75A9 | Lycianthes rantonnei (blue potato bush) | CYP75B9 | Gentiana triflora (gentian) |
| CYP75A10 | Torrenia hybrida (torenia) | CYP75B10 | Torrenia hybrida (torenia) |
| CYP75A11 | Oryza sativa (rice) | CYP75B11 | Oryza sativa (rice) |
| CYP75A12 | Populus trichocarpa (black cottonwood) | CYP75B12 | Populus trichocarpa (black cottonwood) |
| CYP75A13 | Populus trichocarpa (black cottonwood) | CYP75B13 | Pinus taeda (loblolly pine) |
| CYP75A14 | Pinus taeda (loblolly pine) | CYP75B14 | Osteospermum hybrida (African daisy) |
| CYP75A15 | Pinus taeda (loblolly pine) | CYP75B15 | Gerbera hybrida (gerbera) |
| CYP75A16 | Nierembergia sp. (cup flower) | CYP75B16 | Hieracium pilosella (mouseear hawkweed) |
| CYP75A17 | Glycine max (soybean) | CYP75B17 | Osteospermum hybrida (African daisy) |
| CYP75A18 | Solanum tuberosum (potato) | CYP75B18v1 | Pericallis cruenta (=Senecio cruentus, cineraria) |
| CYP75A19v1 | Verbena hybrida (verbena) | CYP75B19 | Ipomoea nil (Japanese morning glory) |
| CYP75A20 | Gossypium hirsutum (cotton) | CYP75B20 | Ipomoea purpurea (common morning glory) |
| CYP75A21 | Delphinium grandiflorum (delphinium) | CYP75B21 | Ipomoea tricolor (morning glory) |
| CYP75A22 | Vinca major (large periwinkle) | CYP75B22 | Ipomoea quamoclit (Cypressvine morning glory) |
| CYP75A23 | Verbena hybrida (verbena) | CYP75B23v1 | Glycine max (soybean) |
| CYP75A24 | Clitoria ternatea (butterfly pea) | CYP75B24 | Allium cepa (onion) |
| CYP75A25 | Cycas rumphii (cycad) | CYP75B25 | Sorghum bicolor (sorghum) |
| CYP75A26 | Camellia sinensis (tea) | CYP75B26 | Verbena hybrida (verbena) |
| CYP75A27 | Gentiana scabra (gentian) | CYP75B27 | Pinus taeda (loblolly pine) |
| CYP75A28 | Vitis vinifera (grapevine) | CYP75B28 | Dianthus caryophyllus (carnation) |
| CYP75A29 | Phalaenopsis hybrid cultivar | CYP75B29 | Antirrhinum majus (snapdragon) |
| CYP75A30 | Carica papaya (papaya) | CYP75B30 | Verbena hybrida (verbena) |
| CYP75A31 | Solanum lycopersicum (tomato) | CYP75B31 | Ageratina adenophora (sticky snakeroot) |
| CYP75A32P | Vitis vinifera (grapevine) | CYP75B32v1 | Vitis vinifera (grapevine) |
| CYP75A33 | Vitis vinifera (grapevine) | CYP75B33 | Trifolium pratense (red clover) |
| CYP75A34 | Vitis vinifera (grapevine) | CYP75B34 | Sorghum bicolor (sorghum) |
| CYP75A35 | Vitis vinifera (grapevine) | CYP75B35v1 | Sorghum bicolor (sorghum) |
| CYP75A36 | Vitis vinifera (grapevine) | CYP75B36 | Sorghum bicolor (sorghum) |
| CYP75A37P | Vitis vinifera (grapevine) | CYP75B37 | Carica papaya (papaya) |
| CYP75A38v1 | Vitis vinifera (grapevine) | CYP75B38 | Vitis vinifera (grapevine) |
| CYP75A39Pv1 | Vitis vinifera (grapevine) | CYP75B39P | Vitis vinifera (grapevine) |
| CYP75A40P | Vitis vinifera (grapevine) | CYP75B40 | Glycine max (soybean) |
| CYP75A41 | Vitis vinifera (grapevine) | CYP75B41 | Glycine max (soybean) |
| CYP75A42 | Vitis vinifera (grapevine) | CYP75B42P | Glycine max (soybean) |
| CYP75A43 | Vitis vinifera (grapevine) | CYP75B43 | Glycine max (soybean) |
| CYP75A44 | Gossypium hirsutum (cotton) | CYP75B44P | Glycine max (soybean) |
| CYP75A45 | Glycine max (soybean) | CYP75B45P | Glycine max (soybean) |
| CYP75A46P | Glycine max (soybean) | CYP75B46 | Glycine max (soybean) |
| CYP75A47 | Antirrhinum kelloggii (climbing snapdragon) | CYP75B47P | Glycine max (soybean) |
| CYP75A48 | Antirrhinum kelloggii (climbing snapdragon) | CYP75B48 | Glycine max (soybean) |
| CYP75A49 | Brachypodium distachyon (purple false brome) | CYP75B49 | Solanum lycopersicum (tomato) |
| CYP75A50 | Lavandula angustifolia (lavender) | CYP75B50 | Antirrhinum kelloggii (climbing snapdragon) |
| CYP75B51 | Brachypodium distachyon (purple false brome) | ||
| CYP75B52 | Brachypodium distachyon (purple false brome) | ||
| CYP75B53 | Brachypodium distachyon (purple false brome) | ||
| CYP75B54 | Brachypodium distachyon (purple false brome) | ||
| CYP75B55 | Calystegia soldanella (beach morning glory) | ||
| CYP75B56 | Calystegia pubescens (Japanese bindweed) | ||
| CYP75B57 | Zea mays (maize) | ||
| CYP75B58 | Pericallis cruenta (=Senecio cruentus, cineraria) | ||
| CYP75B59 | Gynura bicolor (Okinawan spinach) | ||
| CYP75B60 | Cichorium intybus (chicory) |
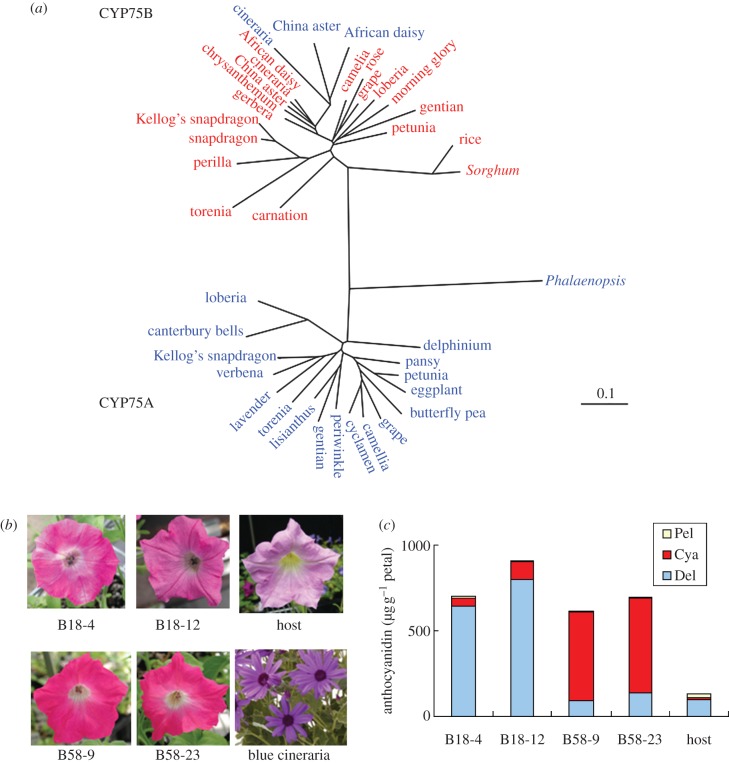
However, convergent evolution of F3′5′H genes was found in some Compositae plants by expressing the isolated CYP75 genes in yeast [17]. The Compositae plants have F3′5′H belonging to CYP75B rather than CYP75A (figure 2; Callistephus chinensis (China aster) (F3′5′H: CYP75B5, F3′H: CYP75B6), Osteospermum hybrida (African daisy) (F3′5′H: CYP75B17, F3′H: CYP75B14) and Senecio cruentus (cineraria) (F3′5′H: CYP75B18, F3′H: CYP75B58)). This suggests that these species lost a CYP75A type F3′5′H gene and then reacquired the F3′5′H gene by duplication and neofunctionalization of a CYP75B gene [17]. The authors also isolated CYP75 members from cineraria and revealed that they are F3′H and F3′5′H by expressing the genes in petunia that was deficient in F3′H and F3′5′H genes (Y. Tanaka and K. Ishiguro 2010, unpublished data; figure 2).
Figure 2.
(a) Phylogenetic tree consisting of CYP75A and CYP75B members from various plant species. F3′5′H and F3′H are shown in blue and red letters, respectively. CYP75A subfamily: Phalaenopsis (CYP75A29, AAZ79451), delphinium (AAX51796), pansy (BAF93855), petunia (CYP75A1, P48418), eggplant (CYP75A2, P37120), butterfly pea (CYP75A24, BAE72870), grape (CYP75A38v4, ABH06585), camellia (AAY23287), cyclamen (ACX37698), periwinkle (CYP75A6, CAA09850), gentian (CYP75A4, BAA12735), lisianthus (CYP75A5, Q96418), torenia (CYP75A10, BAB20076), lavender (CYP75A50, ADA34527), verbena (CYP75A19v2, BAE72871), Kellog's snapdragon (CYP75A48, BAJ16329), canterbury bells (CYP75A6, O04773) and loberia (BAF49321). CYP75B subfamily: carnation (CYP75B28, AAW01411), torenia (CYP75B10, BAB87838), perilla (CYP75B4, BAB59005), snapdragon (CYP75B29, AAW01412), Kellog's snapdragon (CYP75B50, BAB87838), gerbera (CYP75B15, ABA64468), chrysanthemum (AAW01419), China aster F3′H (CYP75B6, AAG49298), cineraria F3′H (CYP75B58), African daisy F3′H (CYP75B14, ABB29899), cineraria F3′5′H (CYP75B18v4), China aster F3′5′H (CYP75B5, AAG49299), African daisy F3′5′H (CYP75B17, ABB43031), camellia (ADZ28515), rose (AAW01418), grape (Q3C211), loberia (BAF49324), morning glory (CYP75B19, BAD00190), gentian (CYP75B9, BAD91808), petunia (CYP75B2, Q9SBQ9), rice (CYP75B3, BAG89180) and Sorghum (CYP75B34, ABG54319). CYP numbers (http://drnelson.uthsc.edu/Nomenclature.html) and/or protein database accession numbers are shown in parenthesis when available. The bar indicates 0.1 substitutions per site. (b) Functional analysis of cDNAs encoding CYP75B18v4 and CYP75B58 derived from blue cineraria petals. Their expression in a petunia line lacking F3′5′H and F3′H activity complemented their deficiency and resulted in colour changes. The two representative flowers expressing the genes (B18-4 and B18-12, B58-9 and B58-23) and the results of their anthocyanidin analysis (c) are shown. Pel, pelargonidin; Cya, sum of cyanidin and peonidin; Del, sum of delphinidin, peonidin and malvidin.
The C-terminal region sequences of F3′H and F3′5′H have been shown to be important in determining F3′H and F3′5′H activity by measuring activities of the chimeric proteins of Gerbera hybrida (gerbera) F3′H and African daisy F3′5′H. Substitution of one or two amino acid residues in the C-terminal region of gerbera F3′H and African daisy F3′5′H changed their ratios of 3′,5′- to 3′-hydroxylation [18]. These results indicate that elucidation of function based on amino acid sequence alone may not be appropriate, and functional analysis in yeast and preferably in plant is essential to identify the function of CYP75 members that are being identified, thanks to advancement of plant molecular biology and genome analysis.
Antirrhinum majus (snapdragon) does not produce delphinidin and lacks blue/violet flowers, whereas Antirrhinum kelloggii (Kellog's snapdragon) does. Functional analysis of CYP75 members of A. kelloggii indicates its CYP75A- and CYP75B-encoded F3′5′H and F3′H, respectively, indicating that A. majus or its ancestor lost the F3′5′H gene after speciation from A. kelloggi or its ancestor [19].
Genome sequencing has enabled a better elucidation of the evolution of F3′H and F3′5′H. The Arabidopsis genome does not contain CYP75A sequences that explain the absence of delphinidin-based anthocyanins in Arabidopsis. The Vitis vinifera (grape) cultivar PN40024 contains 15 copies of F3′5′H genes located on chromosome 6 (seven of which encode full-length amino acid sequence) and one functional and distantly related copy on chromosome 8 (F3′5′Hp). The 15 copies were generated by gene duplication that occurred within this gene family after the Vitaceae lineage separated from other dicot lineages and have different expression profiles from F3′5′Hp. Increase in the copy number might contribute to efficient accumulation of delphinidin-based anthocyanins in grape skin [20].
(b). Mutations of F3′H and F3′5′H genes and their effect on flower colour
Various flower colour mutations have been described in many plant species because such mutations are visible and often add ornamental value to floricultural crops. Interactions with pollinators have driven the evolution of flower colour in many species [21]. Mutations in F3′H or F3′5′H genes result in significant flower colour changes that can impact on pollinators. It has been proposed that the ancestral state of angiosperms is the production of violet/blue flowers containing delphinidin-based anthocyanins [7]. During the evolutionary history of angiosperms, a colour change from blue to red owing to loss of function of F3′5′H or F3′H genes has happened more often than a change from red to blue [21].
Morning glory flowers (Ipomoea spp.) are blue and accumulate anthocyanins derived from cyanidin as mentioned earlier. Flower colour change towards red, and accumulation of pelargonidin-based anthocyanins in Ipomoea nil, I. purpurea and Ipomoea tricolor have been attributed to mutations in their F3′H genes (a nonsense mutation caused by a transition of C to T leading to a truncated protein lacking the haem-binding domain, an insertion of a transposon affecting splicing of the second intron and polyadenylation and single T-insertion in the open reading frame resulting in a truncated protein lacking C-terminal 241 amino acid residues, respectively) [22]. Mutations in the F3′H gene and additional change of substrate specificity of DFR restrict future evolutionary potential to reacquire cyanidin production and blue colour in Ipomoea [23].
Loss of delphinidin production and subsequent flower colour change from blue to pink in two lines of gentian were caused by the insertion of different transposable elements in their F3′5′H genes [24]. In Saintpaulia sp. (African violet), transposon-related insertions or deletions in the promoter region of the F3′5′H gene rather than mutations in its coding region affected transcription level of F3′5′H, flower colour and composition of anthocyanidins (malvidin, peonidin and pelargonidin) [25].
Glycine soja, a wild relative of soybean, has purple flowers. A light-purple flower line was shown to have an amino acid substitution in F3′5′H and contains decreased amounts of anthocyanins derived from delphinidin and an increase in the amount of those derived from cyanidin [26]. A single-base deletion in the F3′H gene in soybean resulted in deletion of the haem-binding domain of the F3′H and colour change of pubescence colour from brown to grey [27].
Transition from light-blue to dark-red flower colour in Phlox drummondii (annual phlox) results from changes of two genes: lower expression of a F3′5H gene and higher expression of a R2R3-Myb transcriptional factor gene regulating the anthocyanin biosynthetic pathway [28].
(c). Flavone synthase II
Flavone biosynthesis from flavanones is catalysed by two types of flavone synthases: flavone synthase I (FNSI, a 2-oxoglutarate-dependent dioxygenase) [29] or flavone synthase (FNSII, a P450 belonging to the CYP93B family). FNSII is more widespread than FNSI. FNSII activity was first demonstrated in snapdragon [30].
In legumes, flavones are synthesized from flavanones by the action of flavanone 2-hydroxylase (F2H; figure 1) and a dehydratase [31]. F2H is a P450 belonging to the CYP93B family (for licorice, CYP93B1). CYP93 also belongs to the CYP71 clan. Homologues of F2H that were isolated from snapdragon and Torenia hybrida (torenia) were shown to encode FNSII that directly catalyse biosynthesis of flavones from flavanones without a dehydratase activity [32]. An FNSII gene was also isolated from gerbera (CYP93B2) [33] and subsequently from many species [4]. More recently, a further pathway for flavone biosynthesis was revealed. In rice, flavone C-glucoside is biosynthesized by F2H (CYP93G2 [34]), UDP-glucose-dependent C-glucosyltransferase [35] and a dehydratase. Engineering of this pathway could be useful to generate flavone C-glucosides that have been shown to act as strong co-pigments to enhance blueing of colour [36,37]. Because flavones also play important roles in plant–micro-organism interaction, engineering flavone biosynthesis might have a wider impact than flower colour modification.
Isoflavones, a subclass of flavonoid mainly found in leguminous plants, have the B-ring at the C-3 position and are phytoalexins and signal molecules to symbiotic soil bacteria. Isoflavone synthase or 2-hydroxyisoflavone synthase (figure 1) that catalyses 2-hydroxyisoflavones synthesis from flavanones is also a P450 belonging to CYP93 [38–40].
3. Efforts to blue
(a). Principal tactics
Successful genetic engineering of plants for commercial production consists of (i) isolation of a useful molecular tool (in this case, F3′H, F3′5′H and FNSII genes), (ii) establishment of efficient transformation systems for target species, because phenotypes of individual transgenic events vary widely and it is often necessary to generate large numbers of transgenic events in order to obtain a few lines with favourable phenotypes, (iii) optimization of transgene expression that can be species-specific as transgene performance has been shown to be impacted by promoter sequences, species of origin of coding regions and terminator sequences and (iv) selection of host cultivars with appropriate genetic backgrounds. A further hurdle to overcome is the regulatory requirements before permission for commercialization of genetically modified plants is granted [41].
Redirecting the flavonoid biosynthetic pathway is feasible by overexpressing exogenous genes. However, it is often necessary to downregulate competing pathways or select mutant lines lacking the competing activities as a host for transformation for efficient redirection and accumulation of a considerable amount of the preferred product. RNAi-induced silencing appears to be the technology of choice for downregulation of an endogenous gene because it is more efficient than antisense or sense suppression [42–44]. Flower colour modification by genetic engineering, that is not limited to engineering P450 genes, has been reviewed recently [41,45–47].
(b). Engineering model plants
Useful knowledge can be obtained by using model plant species that are easily transformable and in which transgene behaviour is predictable. In the case of delphinidin production, petunia and tobacco have been used as model plants.
Expression of petunia F3′5′H cDNAs in a pale pink petunia that was deficient in F3′5′H and F3′H activities resulted in reddish purple transgenic plants that had elevated levels of delphinidin-based anthocyanins in the petals [10]. Various promoters including constitutive and floral-specific promoters have been used to transcribe petunia F3′5′H genes. Commonly used constitutive promoters such as cauliflower mosaic virus 35S promoter (CaMV 35S) or a mannopine synthase promoter derivative [48] have been used to control the expression of flavonoid biosynthetic transgenes in petals to confirm the enzymatic function of the encoded product and analyse resultant phenotypes. Floral-specific promoters of the chalcone synthase (CHS) genes from snapdragon and petunia and F3′5′H Hf1 gene from petunia were successfully used to transcribe F3′5′H cDNAs isolated from a variety of species [49].
Similar flower colour and anthocyanidin changes have been reported by using F3′5′H genes from Eustoma grandiflorum (lisianthus) [50] and Vinca major (periwinkle) [51]. A Campanula medium (canterbury bells) F3′5′H gene in tobacco yielded a higher percentage of delphinidin (up to 99%) than the petunia and lisianthus F3′5′H genes probably owing to a more efficient enzymatic character of the canterbury bells F3′5′H [52]. A butterfly pea F3′5′H gene, and a Viola wittrockiana (pansy) F3′5′H were more effective than a Verbena hybrida (verbena) F3′5′H gene in transgenic verbena plants [53] (J. Togami 2011, unpublished results). These results indicate that the choice of the gene source is important for an efficient change of the pathway.
Suppression of F3′5′H gene in petunia resulted in an increase of cyanidin-based anthocyanins by sacrificing delphinidin-based anthocyanins and a subsequent flower colour change from violet to pink [50]. Similar results were obtained in a commercial petunia line. However, an increase of flavonols was also detected, which indicated competition for dihydroquercetin (DHQ) between FLS and DFR after F3′5′H was downregulated [54].
(c). Carnation
Carnations are one of the most extensively grown cut flowers in the world and are characterized into types based upon flower forms: midi, spray and standard types, with most sales confined to spray and standard types. Classically bred carnations come in white, yellow, pink and red colours. Red and pink carnations generally accumulate flavonols and anthocyanins derived from cyanidin and/or pelargonidin but lack those derived from delphinidin [55,56].
Over the years, a number of strategies have been used to engineer the production of anthocyanins derived from delphinidin using F3′5′H (CYP75A) genes in carnations. Although many of the strategies did lead to the production of delphinidin derivatives, only a few led to commercially viable products.
One of the more successful strategies relied on the identification of white carnation varieties that were specifically deficient in the DFR enzyme [57]. The hydrolysed extracts of white carnation petals were analysed by thin layer or high-performance liquid chromatography (TLC or HPLC). The presence of flavonols indicated that the biochemical pathway leading to dihydroflavonols was intact (figure 1). The absence of quercetin indicated F3′H (CYP75B) activity was lacking. Precursor feeding experiments determined whether DFR activity was lacking and whether the latter part of the anthocyanin pathway was intact as DFR mutants would not accumulate anthocyanins upon incubation with DHK or DHQ but would upon feeding with leucodelphinidin or leucocyanidin (precursors of delphinidin or cyanidin, respectively). Expression of petunia F3′5′H (under the control of a promoter region from the snapdragon chalcone synthase gene) and petunia DFR (under the control of Mac-1 promoter [48]; table 2) in one such DFR mutant (cultivar White Unesco) resulted in an exclusive accumulation of delphinidin derivatives and a significant colour change towards blue [57], FLORIGENE Moondust (figure 3), the first genetically modified floricultural crop to be sold in the world [41,58].
Table 2.
A summary of the purple/blue-coloured transgenic carnations containing introduced F3′5′H genes that have overcome the research and product development hurdles to be launched as commercial products on the market to date. All constructs included a 35S: SurB selectable marker gene giving resistance to chlorsulfuron. WU, White Unesco; U, Unesco; CC, Cream Cinderella; CW, Cerise Westpearl; KC, Kortina Chanel; RHSCC, Royal Horticultural Society Colour Code; %del, percentage of delphinidin out of total anthocyanidins detected in hydrolysed petal extracts; total ac, total amount of anthocyanidins detected in hydrolysed petal extract in mg/g petal (wet weight); P CHS, promoter fragment from the Antirrhinum majus chalcone synthase (CHS) gene; petunia F3′5′H, F3′5′H cDNA clone (Hf1) from Petunia hybrida petals; T D8, terminator region from a phospholipid transfer protein gene (D8) of P. hybrida petals; P Mac 1, hybrid promoter consisting of the promoter from the mannopine synthase (mas) gene of Agrobacterium tumefaciens and a Cauliflower Mosaic Virus 35S (CaMV 35S) enhancer region; petunia DFR, DFR-A cDNA clone from P. hybrida petals; T mas, terminator region from the mas gene of A. tumefaciens; pansy F3′5′H, F3′5′H cDNA clone from Viola (pansy) petals; petunia DFR genomic clone, DFR-A genomic clone from P. hybrida; P 35S, promoter fragment from CaMV 35S; carnation DFR (s/as), sense and antisense fragments of DFR cDNA clone from Dianthus caryophyllus (carnation) petals; T 35S, terminator fragment from CaMV 35S; P ANS, promoter fragment from anthocyanidin synthase (ANS) gene of D. caryophyllus; T ANS, terminator region from ANS gene of D. caryophyllus; petunia cytb5, cytochrome b5 cDNA clone from P. hybrida petals.
| parent |
transgenic petals |
||||||
|---|---|---|---|---|---|---|---|
| product | cultivar | background | type | colour (RHSCC) | %del | total ac (mg g−1) | construct number and transgenes |
| FLORIGENE Moondust | WU | DFR and F3′H mutant | midi | mauve (75A) | 100 | 0.04 |
pCGP1470 P CHS: petunia F3′5′H: T D8 P Mac 1: petunia DFR: T mas |
| FLORIGENE Moonshadow | U | DFR mutant | midi | purple (82A) | 94 | 0.34 |
pCGP1991 P CHS: pansy F3′5′H: T D8 Petunia DFR genomic clone |
| FLORIGENE Moonlite | CC | DFR mutant | standard | light purple (77C) | 75 | 0.09 |
pCGP1470 P CHS: petunia F3′5′H: T D8 P Mac 1: petunia DFR: T mas |
| FLORIGENE Moonvista | CC | DFR mutant | standard | dark purple (77A) | 99 | 1.82 |
pCGP1991 P CHS: pansy F3′5′H: T D8 Petunia DFR genomic clone |
| FLORIGENE Moonshade | CC | DFR mutant | standard | purple (82A) | 94 | 0.47 |
pCGP1470 P CHS: petunia F3′5′H: T D8 P Mac 1: petunia DFR: T mas |
| FLORIGENE Moonaqua | CC | DFR mutant | standard | light purple (84B) | 74 | 0.10 |
pCGP1991 P CHS: pansy F3′5′H: T D8 Petunia DFR genomic clone |
| FLORIGENE Moonpearl | CW | pelargonidin line (F3′H mutant) | spray | light purple (77D) | 61 | 0.27 |
pCGP3366 P 35S: carnation DFR (s/as): T 35S P CHS: pansy F3′5′H: T D8 Petunia DFR genomic clone |
| FLORIGENE Moonberry | CW | pelargonidin line (F3′H mutant) | spray | purple (81B) | 81 | 0.67 |
pCGP3366 P 35S: carnation DFR (s/as): T 35S P CHS: pansy F3′5′H: T D8 Petunia DFR genomic clone |
| FLORIGENE Moonique | KC | cyanidin line (wild-type) | spray | dark purple (82A) | 90 | 2.05 |
pCGP2442 P CHS: salvia F3′5′H: T D8; P ANS: pansy F3′5′H: T ANS; Petunia DFR genomic clone |
| FLORIGENE Moonvelvet | CW | pelargonidin line (F3′H mutant) | spray | dark purple (77A) | 88 | 3.26 |
pCGP2355 P ANS: petunia F3′5′H: T ANS; P CHS: petunia cytb5: T D8 |
Figure 3.
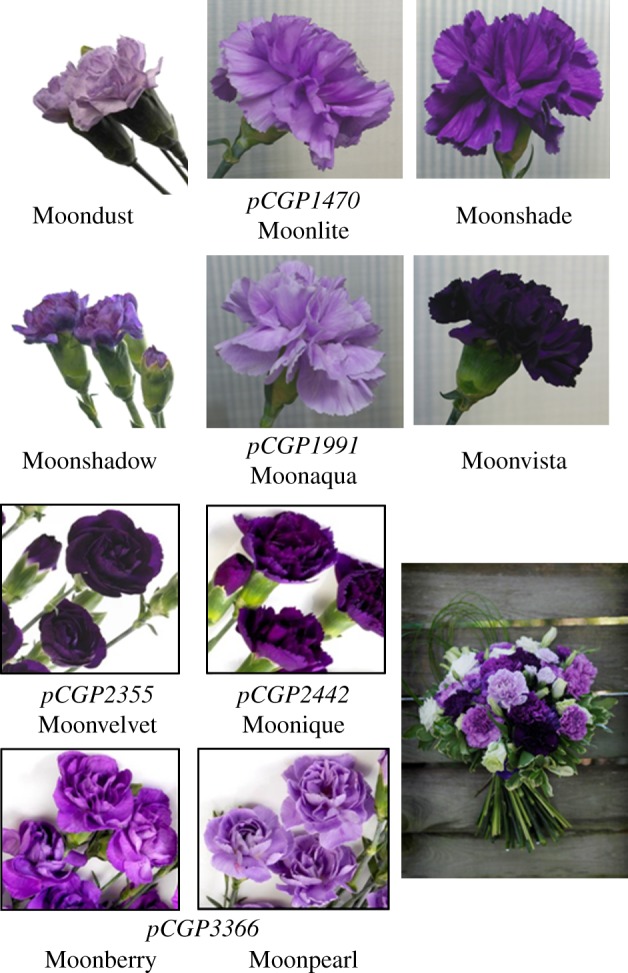
Commercially launched FLORIGENE Moon series transgenic carnations accumulating delphinidin-based anthocyanins by expressing heterologous F3′5′H genes. The flowers have novel blue/violet colours and accumulate various amounts of delphinidin-derived anthocyanins in their petals resulting in differing petal shades. Binary vector numbers (italic) and commercial names are shown (refer to table 2 for description). Phenotypes of these lines are stable. A flower arrangement consisting of the transgenic carnations is also shown.
Experiments testing the efficacy of F3′5′H genes isolated from a variety of sources showed that two F3′5′H genes isolated from pansy resulted in a F3′5′H that was more efficient at producing higher delphinidin levels [59] in carnation and in rose. Therefore, a pansy F3′5′H (under the control of snapdragon CHS promoter) and a petunia DFR-A gene (table 2) were introduced into a carnation DFR mutant of the midi-type (Unesco) and resulted in transgenic flowers with a dark violet colour (FLORIGENE Moonshadow; figure 3) that accumulated delphinidin at higher levels than FLORIGENE Moondust (table 2). The same petunia or pansy gene combinations (table 2) were subsequently introduced into a white standard-type DFR mutant carnation (Cream Cinderella). The colours produced by the resulting transgenic flowers were grouped into nine shades with four shades having novel violet hues. Events from each of the novel shades were selected for field trialling and resulting products (FLORIGENE Moonvista, FLORIGENE Moonshade, FLORIGENE Moonlite and FLORIGENE Moonaqua) have been sold for over 15 years [41] (table 2 and figure 3). Because they are genetically modified plants, it was necessary to obtain appropriate government permission to sell or grow after showing the release of these carnations was most unlikely to affect biodiversity. The regulatory systems and procedures that apply vary extensively between countries and/or regions [41].
The flavonoid composition of FLORIGENE Moonshadow was studied in some detail and found to contain flavone C-glucoside (apigenin 6-C-glucosyl-7-O-glucoside-6′″-malyl ester) which resulted in a strong co-pigment effect to the delphinidin-based anthocyanins producing a bluer colour [37]. The biosynthetic mechanism of flavone C-glucoside in carnation has not yet been clarified. The delphinidin-based anthocyanins in the transgenic carnation (mainly delphinidin 3,5-diglucoside-6″-O-4, 6′″-O-1-cyclic-malyl diester) have the same modification (glucosylation and acylation) of pelargonidin and cyanidin in classically bred carnations.
Although the above ‘DFR-mutant’ strategy was successful in midi and standard-type carnations, it relied on the identification of white lines that were deficient in carnation DFR (and preferably carnation F3′H) with appropriate vacuolar contents to enhance the colour of the delphinidin derivatives once produced. Although DFR mutants of the spray-type were found, none led to commercially viable colours upon the accumulation of anthocyanins derived from delphinidin. Alternative strategies were therefore used using coloured carnations that accumulated pelargonidin or cyanidin-derived pigments.
Expression of a petunia F3′5′H (Hf1) gene along with a petunia cytochrome b5 gene in a carnation cultivar producing cyanidin-based anthocyanins (the cultivar Exquisite) resulted in efficient production of delphinidin-based anthocyanins and subsequent petal colour change [60,61]. The result indicated that enhancement of the electron transfer system to F3′5′H sufficiently out-competed the endogenous F3′H (and DFR) activities. Although the Exquisite transgenic flowers did not pass the product development phase, the strategy was used successfully in a pelargonidin variety (lacking F3′H activity; cultivar Cerise Westpearl) to produce the dark-purple-coloured FLORIGENE Moonvelvet recently launched in the USA (table 2 and figure 3).
The combination of two F3′5′H genes from different species (pansy and Salvia sp.: a blue salvia) along with the petunia DFR gene appeared to be a successful strategy used in a cyanidin-producing carnation line (cultivar Kortina Chanel). The resulting product was a dark-purple-coloured carnation FLORIGENE Moonique (table 2 and figure 3) [62].
Recent improvements in technologies aimed at downregulating specific sequences [42,43] have enabled the ability to create customized DFR and/or F3′H mutants from any suitable carnation variety. RNAi-induced silencing of carnation DFR along with expression of a F3′5′H coding sequence from pansy and a petunia DFR gene in the pelargonidin variety Cerise Westpearl [63] successfully resulted in two products, the purple-coloured FLORIGENE Moonberry and the light-purple-coloured FLORIGENE Moonpearl, both recently launched onto the commercial market (table 2 and figure 3; see www.florigene.com). About 25 million stems of the ‘Moon’ series carnations are sold per year mainly in the USA, EU and Japan.
Over 46 construct combinations, including F3′5′H sequences from various sources such as pansy, blue salvia, petunia, Sollya sp. (bluebell creeper) and a variety of promoter and terminator sequences, were assessed. Of 200 white carnation cultivars analysed, 51 were found to be DFR mutants, 14 of these were transformed with six varieties resulting in candidate products and a final three of these leading to products. Of 118 pelargonidin-accumulating lines, 11 were transformed with six leading to candidate products and one leading to products (to date). Of 60 cyanidin lines that were identified, four were transformed with three resulting in candidate products and one leading to products (F. Brugliera 2010, unpublished data).
The anthocyanin pathway in carnations has now become a model pathway in visualizing competition between introduced F3′5′H and endogenous enzymes.
(d). Rose
Classically bred roses (R. hybrida) were produced by extensive interspecies hybridization of about eight wild species. The hybridization over hundreds of years has resulted in cultivated roses with red colour derived from cyanidin-based anthocyanins, orange colour derived from pelargonidin-based anthocyanins and occasionally carotenoids, and yellow colour from carotenoids. Wild-type roses do not accumulate anthocyanins derived from pelargonidin suggesting that classically bred roses that accumulate pelargonidin are mutant in the F3′H gene [64]. Roses are able to synthesize flavonols that also confer variations to the flower colour. However, roses hardly have intrinsic factors to generate blue flower colours because they do not produce delphinidin or flavones, rarely contain aromatically acylated anthocyanins [65] and tend to have lower vacuolar pH.
In petunia, carnation and even yeast, F3′5′H genes from various plant species were shown to be functional [59]. However, in roses, the petunia F3′5′H gene under the control of constitutive or floral-specific promoters isolated from flavonoid biosynthetic genes that were shown to be functional in petunia and carnation did not result in delphinidin in rose petals, presumably owing to instability of the F3′5′H transcripts as no or degraded transcripts were detected [59]. Butterfly pea- and gentian-derived F3′5′H-coding regions did not result in delphinidin either. Expression of salvia-, Lavendula sp. (lavender)- and bluebell creeper-derived F3′5′H in rose resulted in 1–10% delphinidin of total anthocyanidins, with no evident colour changes. However, upon the introduction of pansy-derived F3′5′H coding regions (two F3′5′H genes were isolated from pansy) under the control of a constitutive promoter (CaMV 35S), a significant amount (up to 60%) of the anthocyanins were derived from delphinidin and subsequent colour change was observed (figure 4a) [59].
Figure 4.
Transgenic rose accumulating delphinidin-based anthocyanins, mainly delphinidin 3,5-diglucoside, by expressing a pansy F3′5′H. (a) Transgenic rose flower expressing the pansy F3′5′H gene (right) derived from a red rose (left). The flower also contained comparable amounts of cyanidin 3,5-diglucoside and had a low vacuolar pH and thus did not show significant blueing. (b) A transgenic rose flower expressing the pansy F3′5′H gene with delphinidin 3,5-diglucosides derived from a pink rose with a higher vacuolar pH than the rose in (a), large amount of flavonols and smaller amount of cyanididin 3,5-diglucoside. The flower has a blue hue that has not been achieved by hybridization breeding [66]. (c) A wedding bouquet containing the novel coloured transgenic rose flowers.
In contrast to carnation, rose cultivars that were specifically deficient in DFR activity were not found. Rose cultivars having higher petal vacuolar pH, lower F3′H activity and higher amounts of flavonols in petals were selected therefore from hundreds analysed. About 20 out of 40 selected cultivars were then successfully transformed. The resultant flower colour and delphinidin contents were variable. Transgenic roses accumulating greater than 90 per cent delphinidin were obtained for some cultivars [66], with delphinidin 3,5-diglucoside as the dominant anthocyanin. The two transgenic lines showing most significant and similar colour change were selected for commercialization as cut flowers. After showing that their release was most unlikely to affect biodiversity in Japan [67,68], general release permission (commercial production and sales permissions) has been granted for them in Japan. It was also shown that the flower colour changes did not affect pollinators’ (mostly bees) visits to the flowers [67]. One of them has been commercialized since 2009 in Japan as Suntory blue rose Applause (figure 4b,c). Its colour and alluring fragrance (the fragrance is the same as the host rose) is appreciated by consumers in Japan where one stem costs about ¥3000 (40 US dollar when 1 dollar is 75 yen) at retail shops.
The flower colour is the bluest among available roses but ‘still lack the true Royal Blue touch’ (news in Nature Biotechnology vol. 27, December 2009). Because delphinidin 3,5-diglucosides alone in the rose vacuole does not confer a blue colour, further modification, such as production of strong co-pigments and elevation of vacuolar pH, should shift the flower colour towards blue [2]. Royal blue or sky blue roses would be very attractive from an aesthetic or commercial point of view although many efforts such as optimization of expression of many genes need to be made to achieve such colour.
Simple overexpression of the pansy-derived F3′5′H gene in rose does not always lead to exclusive accumulation of delphinidin-based anthocyanins due to competing endogenous enzymes (F3′H and DFR). A genetic construct designed to overexpress the pansy F3′5′H and Iris hollandica (iris) DFR genes and downregulate the endogenous rose DFR gene via RNAi-mediated silencing was introduced into rose cultivars. The resultant transgenic events produced flowers that exclusively accumulated anthocyanins derived from delphinidin (over 99%) with significant colour changes [66]. However, they exhibited growth retardation for unknown reasons and their commercialization has not been achieved.
4. Efforts to red
Contrary to roses and carnations, some plant species such as iris and gentian mainly accumulate delphinidin-based anthocyanins and exhibit blue/violet colour, but lack intense red or orange flower colours. Accumulation of anthocyanins derived from pelargonidin should result in such colours. There are primarily two reasons for absence of pelargonidin: DFR substrate specificity and dominant F3′H activity. DFRs of petunia [12], Nicotiana tabacum (tobacco) [12], Cymbidium hybrida (cymbidium) [69], African daisy [70] and probably many other species do not catalyse DHK to leucopelargonidin, the direct precursor of pelargonidin (figure 1). In the case of chrysanthemum, presence of F3′H activity hampers pelargonidin accumulation because feeding its inhibitor into the petals resulted in pelargonidin [71], and downregulation of chrysanthemum DFR in transgenic chrysanthemum resulted in pelargonidin-based pigments accumulating in the petals (F. Brugliera 2009, unpublished data).
Expression of DFR coding sequences isolated from Zea mays (maize) [72], gerbera [73] and rose [74] in petunia hybrid lines which normally accumulated DHK owing to the deficiency of F3′H and F3′5′H successfully led to pelargonidin biosynthesis and orange coloration. However, overexpression of the rose DFR gene in a petunia with F3′H and F3′5′H activities did not result in pelargonidin accumulation because the endogenous F3′H and F3′5′H were able to outcompete the introduced rose DFR for DHK. Similarly, pelargonidin-derived anthocyanins did not accumulate in lilac/violet African daisy (dominant in F3′5′H gene) upon expression of DFR coding sequences isolated from gerbera or strawberry. However, RNAi-mediated suppression of the F3′5′H gene in African daisy already expressing a gerbera DFR gene resulted in pelargonidin production and subsequent colour changes especially in young petals [70].
In petunia, cyanidin biosynthesis was redirected to pelargonidin biosynthesis by downregulating the petunia F3′H gene and expressing a rose DFR gene. The resultant petunia produced orange flowers [54]. However, an increase in flavonols was also detected, suggesting that downregulation of FLS may also be required for efficient accumulation of pelargonidin-based anthocyanins. This was done in tobacco where expression of DFR coding sequences isolated from gerbera and downregulation of the endogenous F3′H and FLS genes resulted in red flowers that accumulated anthocyanins derived predominantly from pelargonidin [75]. Nierembergia sp. (cup flower) exclusively accumulates delphinidin-derived anthocyanins in petals and produces only violet flower colours. Downregulation of the F3′5′H gene and overexpression of rose DFR resulted in only white flower colour owing to endogenous FLS activity [76]. However, downregulation of both F3′5′H and FLS genes and overexpression of rose DFR resulted in pink flower colour from pelargonidin-based anthocyanins [45].
The predominant anthocyanin in blue- or violet-coloured torenia is derived from delphinidin with a minor amount derived from cyanidin. Suppression of the F3′5′H gene led to an increase of cyanidin-based anthocyanins and further expression of the torenia F3′H gene elevated the amount of cyanidin and resulted in pink flower colours [15]. Pelargonidin production was achieved by suppression of the endogenous F3′5′H and F3′H genes (figure 5a). In order to elevate the pelargonidin amount, additional expression of a rose or Pelargonium sp. (geranium) DFR gene was necessary. Expression of the DFR coding region from geranium resulted in higher levels of pelargonidin-based anthocyanins (figure 5b) [77]. Transcription of double-stranded RNA of the target gene was used to downregulate the gene. Because small RNAs and its synthesis may have correlations with various biological phenomena, it was an open question as to whether double-stranded transcription may have a detrimental effect on the growth of the transgenic plants. Selected transgenic pink torenia lines were vegetatively propagated and subjected to a field trial. They grew as well as their host and the modified colour was consistent (figure 5c) [45].
Figure 5.
Transgenic pink torenia flowers accumulating pelargonidin that were generated from torenia accumulating delphinidin-based anthocyanins by (a) suppression of F3′5′H and F3′H genes: (i) blue torenia host, (ii) transgenic flower; and (b) additional expression of pelargonium DFR gene: (i) violet torenia host, (ii) transgenic flower. (c) Field trial of the pink torenia plants. Their growth was comparable with their hosts.
Cyclamen persicum Mill. (cyclamen) is a commercially important pot plant that lacks the orange–red colours derived from pelargonidin-based anthocyanins. Changes to the levels of cyanidin-derived anthocyanins with accompanying colour changes were achieved in two cultivars of cyclamen by suppressing the endogenous F3′5′H gene [78].
Blue gentian flowers accumulate gentiodelphin, a stable blue delphinidin-based anthocyanin modified with two aromatic acyl groups. Suppression of the endogenous F3′5′H gene resulted in a decrease in delphinidin and an increase of cyanidin levels and with a colour change to magenta [79]. Additional suppression of the aromatic acyltransferase that is involved in biosynthesis of gentiodelphin resulted in lilac or pale-blue flower colour [80].
Both in cyclamen and gentian, downregulation of the endogenous F3′H and F3′5′H genes and expression of a DFR gene resulting in a DFR with appropriate substrate specificity is expected to yield pelargonidin-derived anthocyanins as in the case of torenia mentioned earlier. However, accumulation of large amounts of pelargonidin-based anthocyanins and/or lowering vacuolar pH may be necessary to achieve significant and commercially viable colour change.
5. Engineering flavones
Flavones can be co-pigments that contribute to blueing and darkening of anthocyanins. However, at the same time, because flavones and anthocyanins share common precursors, their accumulation would be negatively correlated. When the torenia FNSII gene was expressed in a petunia with violet flowers owing to accumulation of anthocyanins derived from delphinidin and higher vacuolar pH, flavones were detected; however, the amount of anthocyanins decreased and the flower colour was paler [54]. Similarly, roses expressing the torenia FNSII gene did accumulate flavones but the flower colour became very pale. No co-pigment effect was observed (Y. Tanaka 2008, unpublished results). Similarly, the expression of a gentian FNSII gene in tobacco flowers resulted in flavone accumulation, dramatic decrease in anthocyanins and pale flower colour [81].
Suppression of the FNSII gene in torenia resulted in a decrease in flavones and accumulation of flavanones and, strangely, the level of detected anthocyanins also decreased and flower colour became paler. This suggested that in torenia, flavones may be necessary to stabilize anthocyanins [15].
6. Future challenges
Although it is possible to metabolically engineer the flavonoid biosynthetic pathway to alter the flavonoid and anthocyanin composition in petals by expressing or suppressing P450 genes in the pathway, it is apparent that the road to accumulate large amounts of specific flavonoids is not as straightforward. Protein–protein interaction among flavonoid biosynthetic enzymes and possibly specificity of anthocyanin transport machinery that has not yet been elucidated may well also need to be considered for specific flavonoid accumulation. It is still difficult to modify anthocyanidins to anthocyanins having the appropriate decorations and to regulate vacuolar pH and coexisting compounds and metal ions. Typical and practical difficulties for non-model species are low transformation frequencies, longer period for flowering (more than 1 year is common) and variable transgene expression. Conquering such technological difficulties and easing the heavy burden of regulation on genetically modified flowers will improve the chances of novel coloured flowers produced with engineered P450 genes becoming available at a commercial level.
Acknowledgements
The authors are grateful to all colleagues in Suntory Holdings Ltd. (Japan) and Florigene Pty Ltd (Australia), Florigene Europe (the Netherlands) and Paso de Luna (Colombia). Dr David Nelson is acknowledged for his contribution to P450 nomenclature for many years. The photo of figure 4c was kindly provided by Dr Okuhara from his wedding.
References
- 1.Tanaka Y, Sasaki N, Ohmiya A. 2008. Plant pigments for coloration: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749 10.1111/j.1365-313X.2008.03447.x (doi:10.1111/j.1365-313X.2008.03447.x) [DOI] [PubMed] [Google Scholar]
- 2.Yoshida K, Mori M, Kondo T. 2009. Blue flower color development by anthocyanins: from chemical structure to cell physiology. Nat. Prod. Rep. 26, 884–915 10.1039/B800165K (doi:10.1039/B800165K) [DOI] [PubMed] [Google Scholar]
- 3.Honda T, Saito N. 2002. Recent progress in the chemistry of polyacylated anthocyanins as flower color pigment. Heterocycles 56, 633–692 10.3987/REV-01-SR(K)2 (doi:10.3987/REV-01-SR(K)2) [DOI] [Google Scholar]
- 4.Tanaka Y. 2006. Flower colour and cytochromes P450. Phytochem. Rev. 5, 283–291 10.1007/s11101-006-9003-7 (doi:10.1007/s11101-006-9003-7) [DOI] [Google Scholar]
- 5.Shiono M, Matsugaki N, Takeda K. 2005. Phytochemistry: structure of the blue cornflower pigment. Nature 436, 791. 10.1038/436791a (doi:10.1038/436791a) [DOI] [PubMed] [Google Scholar]
- 6.Yoshida K, Kondo T, Okazaki Y, Katou K. 1995. Cause of blue petal colour. Nature 373, 291. 10.1038/373291a0 (doi:10.1038/373291a0) [DOI] [Google Scholar]
- 7.Rausher MD. 2006. The evolution of flavonoids and their genes. In The science of flavonoids (ed. Grotewold E.), pp. 175–211 Berlin, Germany: Springer [Google Scholar]
- 8.Stotz G, Forkmann G. 1981. Hydroxylation of the B-ring of flavonoids in the 3 - and 5-position with enzyme extracts from flowers of Verbena hybrida. Z. Naturforsch. 37c, 19–23 [Google Scholar]
- 9.Menting JGT, Scopes RK, Stevenson TW. 1994. Characterization of flavonoid 3′, 5′-hydroxylases in microsomal membrane fraction of Petunia hybrida flowers. Plant Physiol. 106, 633–642 10.1104/pp.106.2.633 (doi:10.1104/pp.106.2.633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holton TA, et al. 1993. Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366, 276–279 10.1038/366276a0 (doi:10.1038/366276a0) [DOI] [PubMed] [Google Scholar]
- 11.de Vetten N, ter Horst J, van Schaik HP, de Boer A, Mol J, Koes R. 1999. A cytochrome b5 is required for full activity of flavonoid 3′, 5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl Acad. Sci. USA 96, 778–783 10.1073/pnas.96.2.778 (doi:10.1073/pnas.96.2.778) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forkman G, Ruhnau B. 1987. Distinct substrate specificity of dihydroflavonol 4-reductase from flowers of Petunia hybrida. Z. Naturforsch. 42C, 1146–1148 [Google Scholar]
- 13.Winkel BSJ. 2004. Metabolic channeling in plants. Annu. Rev. Plant Biol. 55, 85–107 10.1146/annurev.arplant.55.031903.141714 (doi:10.1146/annurev.arplant.55.031903.141714) [DOI] [PubMed] [Google Scholar]
- 14.Brugliera F, Barri-Rewell G, Holton TA, Mason JG. 1999. Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J. 19, 441–451 10.1046/j.1365-313X.1999.00539.x (doi:10.1046/j.1365-313X.1999.00539.x) [DOI] [PubMed] [Google Scholar]
- 15.Ueyama U, Suzuki K, Fukuchi-Mizutani M, Fukui Y, Miyazaki K, Ohkawa H, Kusumi T, Tanaka Y. 2002. Molecular and biochemical characterization of torenia flavonoid 3′-hydroxylase and flavone synthase II and modification of flower color by modulating the expression of these genes. Plant Sci. 163, 253–263 10.1016/S0168-9452(02)00098-5 (doi:10.1016/S0168-9452(02)00098-5) [DOI] [Google Scholar]
- 16.Nelson D, Werck-Reichhart D. 2011. A P450-centric view of plant evolution. Plant J. 66, 194–211 10.1111/j.1365-313X.2011.04529.x (doi:10.1111/j.1365-313X.2011.04529.x) [DOI] [PubMed] [Google Scholar]
- 17.Seitz C, Eder C, Deiml B, Kellner S, Martens S, Forkmann G. 2006. Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol. Biol. 61, 365–381 10.1007/s11103-006-0012-0 (doi:10.1007/s11103-006-0012-0) [DOI] [PubMed] [Google Scholar]
- 18.Seitz C, Ameres S, Forkmann G. 2007. Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase. FEBS Lett. 581, 3429–3434 10.1016/j.febslet.2007.06.045 (doi:10.1016/j.febslet.2007.06.045) [DOI] [PubMed] [Google Scholar]
- 19.Ishiguro K, Masumi T, Tanaka Y. 2012. Functional analysis of Antirrhinum kelloggii flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes; critical role in flower color and evolution in the genus Antirrhinum. J. Plant Res. 125, 451–456 10.1007/s10265-011-0455-5 (doi:10.1007/s10265-011-0455-5) [DOI] [PubMed] [Google Scholar]
- 20.Falginella L, Castellarin SD, Testolin R, Gambetta GA, Morgante M, Di Gaspero G. 2010. Expansion and subfunctionalisation of flavonoid 3′,5′-hydroxylases in the grapevine lineage. BMC Genomics 11, 562. 10.1186/1471-2164-11-562 (doi:10.1186/1471-2164-11-562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rausher MD. 2008. Evolutionary transitions in floral color. Int. J. Plant Sci. 169, 7–21 10.1086/523358 (doi:10.1086/523358) [DOI] [Google Scholar]
- 22.Hoshino A, Morita Y, Choi JD, Saito N, Toki K, Tanaka Y, Iida S. 2003. Spontaneous mutations of the flavonoid 3′-hydroxylase gene conferring reddish flowers in the three morning glory species. Plant Cell Physiol. 44, 990–1001 10.1093/pcp/pcg143 (doi:10.1093/pcp/pcg143) [DOI] [PubMed] [Google Scholar]
- 23.Zufall RA, Rausher MD. 2003. The genetic basis of a flower color polymorphism in the common morning glory (Ipomoea purpurea). J. Hered. 94, 442–448 10.1093/jhered/esg098 (doi:10.1093/jhered/esg098) [DOI] [PubMed] [Google Scholar]
- 24.Nakatsuka T, Nishihara M, Mishiba K, Hirano H, Yamamura S. 2006. Two different transposable elements inserted in flavonoid 3′,5′-hydroxylase gene contribute to pink flower coloration in Gentiana scabra. Mol. Genet. Genomics 275, 231–241 10.1007/s00438-005-0083-7 (doi:10.1007/s00438-005-0083-7) [DOI] [PubMed] [Google Scholar]
- 25.Sato M, Kawabe T, Hosokawa M, Tatsuzawa F, Doi M. 2011. Tissue culture-induced flower-color changes in Saintpaulia caused by excision of the transposon inserted in the flavonoid 3′, 5′ hydroxylase (F3′5′H) promoter. Plant Cell Rep. 30, 929–939 10.1007/s00299-011-1016-z (doi:10.1007/s00299-011-1016-z) [DOI] [PubMed] [Google Scholar]
- 26.Takahashi R, Dubouzet JG, Matsumura H, Yasuda K, Iwashina T. 2010. A new allele of flower color gene W1 encoding flavonoid 3′5′-hydroxylase is responsible for light purple flowers in wild soybean Glycine soja. BMC Plant Biol. 10, 155. 10.1186/1471-2229-10-155 (doi:10.1186/1471-2229-10-155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toda K, Yang D, Yamanaka N, Watanabe S, Harada K, Takahashi R. 2002. A single-base deletion in soybean flavonoid 3′-hydroxylase gene is associated with gray pubescence color. Plant Mol. Biol. 50, 187–196 10.1023/A:1016087221334 (doi:10.1023/A:1016087221334) [DOI] [PubMed] [Google Scholar]
- 28.Hopkins R, Rausher MD. 2011. Identification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature 469, 411–414 10.1038/nature09641 (doi:10.1038/nature09641) [DOI] [PubMed] [Google Scholar]
- 29.Martens S, Forkmann G, Britsch L, Wellmann F, Matern U, Lukacin R. 2003. Divergent evolution of flavonoid 2-oxoglutarate-dependent dioxygenases in parsley. FEBS Lett. 544, 93–98 10.1016/S0014-5793(03)00479-4 (doi:10.1016/S0014-5793(03)00479-4) [DOI] [PubMed] [Google Scholar]
- 30.Stotz G, Forkmann G. 1981. Oxidation of flavanones to flavones with flower extracts of Antirrhinum majus (snapdragon). Z. Naturforsch. 36c, 737–741 [Google Scholar]
- 31.Akashi T, Aoki T, Ayabe S. 1998. Identification of a cytochrome P450 cDNA encoding (2S)-flavanone2-hydroxylase of licorice (Glycyrrhiza echinata L.; Fabaceae) which represents licodione synthase and flavone synthase II. FEBS Lett. 431, 287–290 10.1016/S0014-5793(98)00781-9 (doi:10.1016/S0014-5793(98)00781-9) [DOI] [PubMed] [Google Scholar]
- 32.Akashi T, Fukuchi-Mizutani M, Aoki T, Ueyama Y, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Ayabe S. 1999. Molecular cloning and biochemical characterization of a novel cytochrome P450, flavone synthase II, that catalyzes direct conversion of flavanones to flavones. Plant Cell Physiol. 40, 1182–1186) (doi:10.1093/oxfordjournals.pcp.a029505) [DOI] [PubMed] [Google Scholar]
- 33.Martens S, Forkmann G. 1999. Cloning and expression of flavone synthase II from Gerbera hybrids. Plant J. 20, 611–618 10.1046/j.1365-313X.1999.00636.x (doi:10.1046/j.1365-313X.1999.00636.x) [DOI] [PubMed] [Google Scholar]
- 34.Du Y, Chu H, Chu IK, Lo C. 2010. CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol. 154, 324–333 10.1104/pp.110.161042 (doi:10.1104/pp.110.161042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R. 2009. The C-glycosylation of flavonoids in cereals. J. Biol. Chem. 284, 17926–17934 10.1074/jbc.M109.009258 (doi:10.1074/jbc.M109.009258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yabuya T, Nakamura M, Iwashina T, Yamaguchi M, Takehara T. 1997. Anthocyanin-flavone copigmentaion in bluish purple flowers of Japanese garden iris (Irsi ensata Thunb.). Euphytica 98, 163–167 10.1023/A:1003152813333 (doi:10.1023/A:1003152813333) [DOI] [Google Scholar]
- 37.Fukui Y, Tanaka Y, Kusumi T, Iwashita T, Nomoto K. 2003. A rationale for the shift in colour towards blue in transgenic carnation flowers expressing the flavonoid 3′,5′-hydroxylase gene. Phytochemistry 63, 15–23 10.1016/S0031-9422(02)00684-2 (doi:10.1016/S0031-9422(02)00684-2) [DOI] [PubMed] [Google Scholar]
- 38.Akashi T, Aoki T, Ayabe S. 1999. Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol. 121, 821–828 10.1104/pp.121.3.821 (doi:10.1104/pp.121.3.821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steele CL, Gijzen M, Qutob D, Dixon RA. 1999. Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch. Biochem. Biophys. 367, 146–150 10.1006/abbi.1999.1238 (doi:10.1006/abbi.1999.1238) [DOI] [PubMed] [Google Scholar]
- 40.Jung W, Yu O, Lau SM, O'Keefe DP, Odell J, Fader G, McGonigle B. 2000. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 18, 208–212 10.1038/72671 (doi:10.1038/72671) [DOI] [PubMed] [Google Scholar]
- 41.Tanaka Y, Brugliera F, Chandler S. 2009. Recent progress of flower colour modification by biotechnology. Int. J. Mol. Sci. 10, 5350–5369 10.3390/ijms10125350 (doi:10.3390/ijms10125350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wesley SV, Helliwell CA, Smith NA. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590 10.1046/j.1365-313X.2001.01105.x (doi:10.1046/j.1365-313X.2001.01105.x) [DOI] [PubMed] [Google Scholar]
- 43.Wang M-B, Waterhouse PM. 2001. Application of gene silencing in plants. Curr. Opin. Plant Biol. 5, 146–150 10.1016/S1369-5266(02)00236-4 (doi:10.1016/S1369-5266(02)00236-4) [DOI] [PubMed] [Google Scholar]
- 44.Nakamura N, Fukuchi-Mizutani M, Suzuki K, Miyazaki K, Tanaka Y. 2006. RNAi suppression of the anthocyanidin synthase gene in Torenia hybrida yields white flowers with higher frequency and better stability than antisense and sense suppression. Plant Biotechnol. 23, 13–18 10.5511/plantbiotechnology.23.13 (doi:10.5511/plantbiotechnology.23.13) [DOI] [Google Scholar]
- 45.Tanaka Y, Brugliera F, Kalc G, Senior M, Dyson B, Nakamura N, Katsumoto Y, Chandler S. 2010. Flower color modification by engineering of the flavonoid biosynthetic pathway: practical perspectives. Biosci. Biotechnol. Biochem. 74, 1760–1769 10.1271/bbb.100358 (doi:10.1271/bbb.100358) [DOI] [PubMed] [Google Scholar]
- 46.Tanaka Y, Ohmiya A. 2008. Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 19, 190–197 10.1016/j.copbio.2008.02.015 (doi:10.1016/j.copbio.2008.02.015) [DOI] [PubMed] [Google Scholar]
- 47.Nishihara M, Nakatsuka T. 2011. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 33, 433–441 10.1007/s10529-010-0461-z (doi:10.1007/s10529-010-0461-z) [DOI] [PubMed] [Google Scholar]
- 48.Comai L, Moran P, Maslyar D. 1990. Novel and useful properties of a chimeric plant promoter combining CaMV35S and MAS elements. Plant Mol. Biol. 15, 373–381 10.1007/BF00019155 (doi:10.1007/BF00019155) [DOI] [PubMed] [Google Scholar]
- 49.Holton TA, Tanaka Y.1994. Transgenic flowering plants. International Patent Publication number WO94/28140.
- 50.Shimada Y, Nakano-Shimada R, Ohbayashi M, Okinaka Y, Kiyokawa S, Kikuchi Y. 1999. Expression of chimeric P450 genes encoding flavonoid-3′, 5′-hydroxylase in transgenic tobacco and petunia plants. FEBS Lett. 461, 241–245 10.1016/S0014-5793(99)01425-8 (doi:10.1016/S0014-5793(99)01425-8) [DOI] [PubMed] [Google Scholar]
- 51.Mori S, Kobayashi H, Hoshi Y, Kondo M, Nakano M. 2004. Heterologous expression of the flavonoid 3′,5′-hydroxylase gene of Vinca major alters flower color in transgenic Petunia hybrida. Plant Cell Rep. 22, 415–421 10.1007/s00299-003-0709-3 (doi:10.1007/s00299-003-0709-3) [DOI] [PubMed] [Google Scholar]
- 52.Okinaka Y, Shimada Y, Nakano-Shimada R, Ohbayashi M, Kiyokawa S, Kikuchi Y. 2003. Selective accumulation of delphinidin derivatives in tobacco using a putative flavonoid 3′,5′-hydroxylase cDNA from Campanula medium. Biosci. Biotechnol. Biochem. 67, 161–165 10.1271/bbb.67.161 (doi:10.1271/bbb.67.161) [DOI] [PubMed] [Google Scholar]
- 53.Togami J, et al. 2006. Molecular characterization of the flavonoid biosynthesis of Verbena hybrida and the functional analysis of verbena and Clitoria ternatea F3′5′H genes in transgenic verbena. Plant Biotechnol. 23, 5–11 10.5511/plantbiotechnology.23.5 (doi:10.5511/plantbiotechnology.23.5) [DOI] [Google Scholar]
- 54.Tsuda S, et al. 2004. Flower color modification of Petunia hybrida commercial varieties by metabolic engineering. Plant Biotechnol. 21, 377–386 10.5511/plantbiotechnology.21.377 (doi:10.5511/plantbiotechnology.21.377) [DOI] [Google Scholar]
- 55.Bloor SJ. 1998. A macrocyclic anthocyanin from red/mauve carnation flowers. Phytochemistry 49, 225–228 10.1016/S0031-9422(97)01051-0 (doi:10.1016/S0031-9422(97)01051-0) [DOI] [Google Scholar]
- 56.Gonnet J-F, Fenet B. 2000. ‘Cyclamen red’ color based on a macrocyclic anthocyanin in carnation flowers. J. Agric. Food Chem. 48, 22–25 10.1021/jf9907642 (doi:10.1021/jf9907642) [DOI] [PubMed] [Google Scholar]
- 57.Holton TA.1996. Transgenic plants exhibiting altered flower colour and methods for producing same. International Patent Publication number WO96/36716.
- 58.Mol J, Cornish E, Mason J, Koes R. 1999. Novel coloured flowers. Curr. Opin. Biotechnol. 10, 198–201 10.1016/S0958-1669(99)80035-4 (doi:10.1016/S0958-1669(99)80035-4) [DOI] [PubMed] [Google Scholar]
- 59.Brugliera F, Tanaka Y, Mason J.2004. Flavonoid 3′,5′-hydroxylase gene sequences and uses therefor. International Patent Publication number WO/2004/020637.
- 60.Brugliera F, Tull D, Holton TA, Karan M, Treloar N, Simpson K, Skurczynska J, Mason JG. 2000. Introduction of a cytochrome b5 enhances the activity of flavonoid 3′,5′-hydroxylase (a cytochrome P450) in transgenic carnation. Int. Plant Mol. Biol. Rep. (Suppl.) 18, S6–S8 [Google Scholar]
- 61.Tanaka Y, Katsumoto Y, Brugliera F, Mason J. 2005. Genetic engineering in floriculture. Plant Cell Tissue Org. 80, 1–24 10.1007/s11240-004-0739-8 (doi:10.1007/s11240-004-0739-8) [DOI] [Google Scholar]
- 62.Brugliera F.2009. Genetically modified plants with altered inflorescence. International Patent Publication number WO/2009/062259.
- 63.Brugliera F.2010. A plant with altered inflorescence. International Patent Publication number WO/2010/069004.
- 64.Mikanagi Y, Yokoi M, Ueda Y, Saito N. 1995. Flower flavonol and anthocyanin distribution in subgenus Rosa. Biochem. Syst. Ecol. 23, 183–200 10.1016/0305-1978(95)93849-X (doi:10.1016/0305-1978(95)93849-X) [DOI] [Google Scholar]
- 65.Mikanagi Y, Saito N, Yokoi M, Tatsuzawa F. 2000. Anthocyanins in flowers of genus Rosa, sections Cinnamomeae (=Rosa), Chinenses, Gallicanae and some modern garden roses. Biochem. Syst. Ecol. 28, 887–902 10.1016/S0305-1978(99)00127-1 (doi:10.1016/S0305-1978(99)00127-1) [DOI] [PubMed] [Google Scholar]
- 66.Katsumoto Y, et al. 2007. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 48, 1589–1600 10.1093/pcp/pcm131 (doi:10.1093/pcp/pcm131) [DOI] [PubMed] [Google Scholar]
- 67.Nakamura N, et al. 2011. Environmental risk assessment and field performance of rose (Rosa x hybrida) genetically modified for delphinidin production. Plant Biotechnol. 28, 251–261 10.5511/plantbiotechnology.11.0113a (doi:10.5511/plantbiotechnology.11.0113a) [DOI] [Google Scholar]
- 68.Nakamura N, Tems U, Fukuchi-Mizutani M, Chandler S, Matsuda Y, Takeuchi S, Matsumoto S, Tanaka Y. 2011. Molecular based evidence for a lack of gene-flow between Rosa x hybrida and wild Rosa species in Japan. Plant Biotechnol. 28, 245–250 10.5511/plantbiotechnology.10.1217a (doi:10.5511/plantbiotechnology.10.1217a) [DOI] [Google Scholar]
- 69.Johnson ET, Yi H, Shin B, Oh B-J, Cheong H, Choi G. 1999. Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J. 19, 81–85 10.1046/j.1365-313X.1999.00502.x (doi:10.1046/j.1365-313X.1999.00502.x) [DOI] [PubMed] [Google Scholar]
- 70.Seitz C, Vitten M, Steinbach P, Hartl S, Hirsche J, Rhthje W, Treutter D, Forkmann G. 2007. Redirection of anthocyanin synthesis in Osteospermum hybrida by a two-enzyme manipulation strategy. Phytochemistry 68, 824–833 10.1016/j.phytochem.2006.12.012 (doi:10.1016/j.phytochem.2006.12.012) [DOI] [PubMed] [Google Scholar]
- 71.Schwinn KE, Markham KR, Giveno N. 1993. Floral flavonoids and the potential for pelargonidin biosynthesis in commercial chrysanthemum cultivars. Phytochemistry 35, 145–150 10.1016/S0031-9422(00)90523-5 (doi:10.1016/S0031-9422(00)90523-5) [DOI] [Google Scholar]
- 72.Meyer P, Heidemann I, Forkmann G, Saedler H. 1987. A new petunia flower colour generated by transformation of a mutant with a maze gene. Nature 330, 677–678 10.1038/330677a0 (doi:10.1038/330677a0) [DOI] [PubMed] [Google Scholar]
- 73.Helariutta Y, Elomaa P, Kotilainen H, Seppanen P, Teeri TH. 1993. Cloning of cDNA for dihydroflavonol 4-reductase (DFR) and charaterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol. 22, 183–193 10.1007/BF00014927 (doi:10.1007/BF00014927) [DOI] [PubMed] [Google Scholar]
- 74.Tanaka Y, Fukui Y, Fukuchi-Mizutani M, Holton TA, Higgins E, Kusumi T. 1995. Molecular cloning and characterization of Rosa hybrida dihydroflavonol 4-reductase. Plant Cell Physiol. 36, 1023–1031 [DOI] [PubMed] [Google Scholar]
- 75.Nakatsuka T, Abe Y, Kakizaki Y, Yamamura S, Nishihara M. 2007. Production of red-flowered plants by genetic engineering of multiple flavonoid biosynthetic genes. Plant Cell Rep. 26, 1951–1959 10.1007/s00299-007-0401-0 (doi:10.1007/s00299-007-0401-0) [DOI] [PubMed] [Google Scholar]
- 76.Ueyama Y, Katsumoto Y, Fukui Y, Fukuchi-Mizutani M, Ohkawa H, Kusumi T, Iwashita T, Tanaka Y. 2006. Molecular characterization of the flavonoid biosynthetic pathway and flower color modification of Nierembergia sp. Plant Biotechnol. 23, 19–24 10.5511/plantbiotechnology.23.19 (doi:10.5511/plantbiotechnology.23.19) [DOI] [Google Scholar]
- 77.Nakamura N, Fukuchi-Mizutani M, Fukui Y, Ishiguro K, Suzuki K, Tanaka Y. 2010. Generation of red flower varieties from blue Torenia hybrida by redirection of the flavonoid pathway from delphinidin to pelargonidin. Plant Biotechnol. 27, 375–383 10.5511/plantbiotechnology.10.0610a (doi:10.5511/plantbiotechnology.10.0610a) [DOI] [Google Scholar]
- 78.Boase MR, Lewis DH, Davies KM, Marshall GB, Patel D, Schwinn KE, Deroles SC. 2010. Isolation and antisense suppression of flavonoid 3′, 5′-hydroxylase modifies flower pigments and colour in cyclamen. BMC Plant Biol. 10, 107. 10.1186/1471-2229-10-107 (doi:10.1186/1471-2229-10-107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakatsuka T, Mishiba K, Abe Y, Kubota A, Kakizaki Y, Yamamura S, Nishihara M. 2008. Flower color modification of gentian plants by RNAi-mediated gene silencing. Plant Biotechnol. 25, 61–68 10.5511/plantbiotechnology.25.61 (doi:10.5511/plantbiotechnology.25.61) [DOI] [Google Scholar]
- 80.Nakatsuka T, Mishiba K, Kubota A, Abe Y, Yamamura S, Nakamura N, Tanaka Y, Nishihara M. 2010. Genetic engineering of novel flower colour by suppression of anthocyanin modification genes in gentian. J. Plant Physiol. 167, 231–237 10.1016/j.jplph.2009.08.007 (doi:10.1016/j.jplph.2009.08.007) [DOI] [PubMed] [Google Scholar]
- 81.Nakatsuka T, Nishihara M, Mishiba K, Yamamura S. 2006. Heterologous expression of two gentian cytochrome P450 genes can modulate the intensity of flower pigmentation in transgenic tobacco plants. Mol. Breed. 17, 91–99 10.1007/s11032-005-2520-z (doi:10.1007/s11032-005-2520-z) [DOI] [Google Scholar]