Abstract
The neighbourhoods of cytochrome P450 (CYP) genes in deuterostome genomes, as well as those of the cnidarians Nematostella vectensis and Acropora digitifera and the placozoan Trichoplax adhaerens were examined to find clues concerning the evolution of CYP genes in animals. CYP genes created by the 2R whole genome duplications in chordates have been identified. Both microsynteny and macrosynteny were used to identify genes that coexisted near CYP genes in the animal ancestor. We show that all 11 CYP clans began in a common gene environment. The evidence implies the existence of a single locus, which we term the ‘cytochrome P450 genesis locus’, where one progenitor CYP gene duplicated to create a tandem set of genes that were precursors of the 11 animal CYP clans: CYP Clans 2, 3, 4, 7, 19, 20, 26, 46, 51, 74 and mitochondrial. These early CYP genes existed side by side before the origin of cnidarians, possibly with a few additional genes interspersed. The Hox gene cluster, WNT genes, an NK gene cluster and at least one ARF gene were close neighbours to this original CYP locus. According to this evolutionary scenario, the CYP74 clan originated from animals and not from land plants nor from a common ancestor of plants and animals. The CYP7 and CYP19 families that are chordate-specific belong to CYP clans that seem to have originated in the CYP genesis locus as well, even though this requires many gene losses to explain their current distribution. The approach to uncovering the CYP genesis locus overcomes confounding effects because of gene conversion, sequence divergence, gene birth and death, and opens the way to understanding the biodiversity of CYP genes, families and subfamilies, which in animals has been obscured by more than 600 Myr of evolution.
Keywords: cytochrome P450, ohnologues, evolution, animal P450s, synteny, CYP clans
1. Introduction
(a). CYP clans, problems in understanding animal CYP evolution, and a way forward
The cytochrome P450s (CYPs) constitute one of the most diverse eukaryotic gene families, with a dizzying complexity within and between species. CYP enzymes use molecular oxygen to modify substrate structure, critical in a huge number of physiological, ecological and toxicological processes. Generally, anerobes and some microaerophiles, such as the parasites Plasmodium or Giardia, lack CYP genes [1], but in aerobic organisms CYP numbers range from two genes in Schizosaccharomyces pombe to as many as 400 genes in some plants such as potato [2] or grapevine, cottonwood, soybean and rice [3]. Currently, in animals, the range is 35 CYP genes in the sponge Amphimedon queenslandica to approximately 235 in the cephalochordate Branchiostoma floridae (lancelet or amphioxus). CYP genes are classified into clans, families and subfamilies based on phylogenetics as well as sequence identity [4]. Orthologues, co-orthologues and paralogues of CYP genes are often difficult to properly classify as sequences can differ substantially between species, because of indels and gene conversion [5–7]. Likewise, the numbers of genes in a given subfamily often differ even between closely related species, for example, differences in the mouse, rat and human CYP2D and CYP2J clusters, because of differences in deletion or expansion (or ‘blooms’) in the number of genes ([8,9] see also [10]). This has confounded discerning relationships and especially the path of CYP gene evolution in animals. We address these issues in this study.
Animal CYPs display a molecular phylogeny with 11 distinct clades, each containing one or more CYP gene families. In the late 1990s, reviews of metazoan CYP gene evolution [11,12] introduced the concept of clans to describe these deep branching clusters, although at that time no animal genome sequence had been completed, and several animal CYP families (and clans) were not yet known. By 2003, there were 18 CYP families known in vertebrates, although CYP39 was not yet identified in fish [13]. CYP16 in the CYP26 Clan was the last vertebrate family recognized (D. Nelson 2010, unpublished data). The 11th clan, CYP Clan 74, originally known only from land plants, was first observed in animals as an expressed sequence tag (EST) from lancelet.1 Lee et al. [14] reported the first crystal structure of a plant CYP74, and identified animal CYP74 Clan members in the cephalochordate B. floridae, the sea anemone Nematostella vectensis, the coral Acropora millepora and the placazoan Trichoplax adhaerens [14]. This discovery was the crowning touch that led to the 11 animal CYP clans currently known. A list of the 196 animal CYP families and their clan membership can be found in the electronic supplementary material, table S1.
One of the key objectives concerning animal CYP evolution is to understand the origin of these 11 clans. Did the ancestor of all animals have one or more than one CYP gene? How did the 11 clans evolve from the earliest CYP(s)? Comparison of the amino acid sequences does not answer these questions because the branches between the clans are so deep. Only CYP Clans 3 and 4 appear to be sisters, while the relationships among the other nine clans have been elusive. The problem is similar to defining the relationships among the eukaryotic megagroups, unikonts, bikonts and excavates2 [17]. Tree-building methods are currently not able to resolve the deepest branches with high confidence [15]. Yoshida et al. [18] showed that eukaryotic CYP51 (sterol 14α demethylase) was probably orthologous to prokaryotic CYP51, and therefore a strong candidate for the original eukaryotic CYP. An alternative view is that bacterial CYP51 resulted from a lateral transfer from plants [19]. This would place the first origin of CYP51 in early eukaryotes, with a subsequent divergence into a cycloartenol branch and a lanosterol branch. This idea is in conflict with Cavalier-Smith [20], who views actinobacteria as the probable precursors of the neomuran ancestor of eukaryotes in part because CYP51 is found in actinobacteria. The idea that plant, animal and fungal lineages all evolved separate CYP collections de novo apparently from a CYP51 was explicitly stated by Nelson [11]. This idea is independent of an actinobacterial or early eukaryotic origin for CYP51.
Although the idea that CYP51 was the starting point has been around for some time, there has not been any consideration of the pathway(s) from CYP51 to the contemporary CYP clans, families and subfamilies. The accumulation of high-quality animal genome assemblies offers a new approach to CYP origins based on synteny. Gene neighbourhoods and gene structure can give additional evolutionary information not contained in coding sequence alone [21–23]. This is illustrated in several studies of genes in specific CYP families [24]. In this study, we examine CYP genes in their syntenic context, to discern the path by which the existing clans, including their families and subfamilies, can be linked to the original locus.
(b). The 2R hypothesis and its implications for CYP evolution
The concept of whole genome duplication (WGD) during animal evolution is an essential background for considering CYP gene evolution [25]. Several gene families provide evidence for two rounds (2R) of WGD in chordates, the most famous example being the clustered Hox genes. Each round of WGD results in the formation of duplicate genes, known as ohnologues [25,26]. Many ohnologues are lost as a tetraploid genome reduces the number of genes back down close to the original diploid number, while some are retained and acquire different functions (subfunctionalization of the original pre-duplicated gene, or neofunctionalization [25,27,28]).
Analysis of the Ciona genome confirmed the 2R hypothesis [29], as did the recent sequencing of the amphioxus (lancelet) genome [23]. Each region in the lancelet genome typically can be mapped in mammalian genomes to four paralogons, large paralogous regions originally derived from duplicated chromosomes ostensibly created in the two rounds of WGD. The ancestral karyotype has been reconstructed with 17 chordate linkage groups (CLGs). The human genome contains 135 segments that map 90 per cent of the genome onto these CLGs [23]. In addition to the 2R WGD, a third round (3R) has occurred in the vertebrate line leading to ray-finned (actinopterygian) fishes including zebrafish and medaka [30,31], but not lobe-finned (sarcopterygian) fishes such as the coelacanth [30].
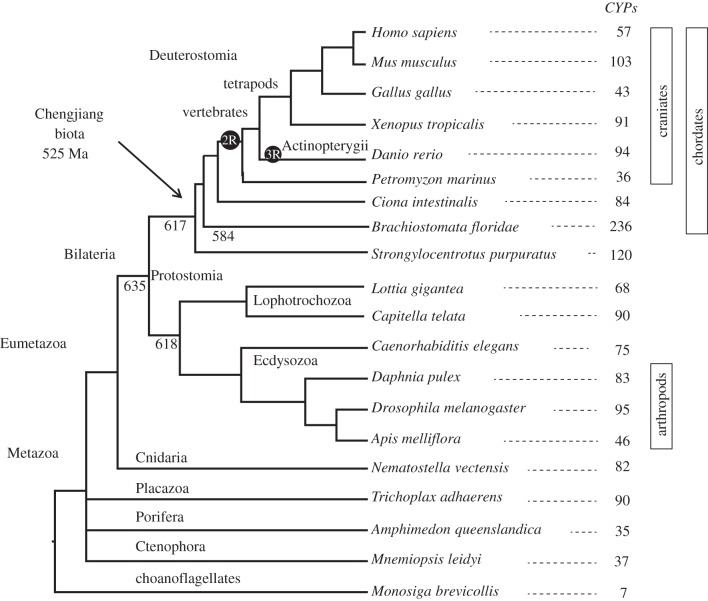
Evolutionarily, the 2R events have been bracketed in time between the divergence of tunicates from vertebrates and the origin of the gnathostomes (jawed vertebrates). Recent analyses argue that both 2R events preceded the cyclostome–gnathostome split [32,33] (figure 1). Absolute geological dates for the 2R events are not known owing to insufficient fossil evidence. Fossils of chordates (Haikouella lanceolata), hemichordates (Yunnanozoon lividum) and vertebrates (Myllokunmingia fengjiaoa and Haikouichthys ercaicunensis, now considered a single species) are found in the Chengjiang biota, dated to 525 Ma [37,38]. Therefore, the oldest divergence among deuterostomes (Xenambulacraria3 versus chordates) is older than 525 Ma. The vertebrate in this collection is thought to post-date the origin of the hagfish [37]. If the placement of the 2R events in figure 1 is correct, then the 2R duplications would have to be older than 525 Ma. A firm younger boundary for 2R is the minimum date for the split between the actinopterygians (ray-finned fish) and the sarcopterygians (lobe-finned fish and tetrapods), which was no later than 419 Ma [39]. This date probably should move back to the divergence of sharks from the bony fishes, but sharks do not leave good fossils. The oldest putative shark scales are from the Ordovician Harding Sandstone (approx. 450 Ma [40]). Therefore, the 2R events are at least 450 Ma old and are probably older than 525 Ma. Molecular sequence analysis places the origin of deuterostomes earlier than the fossil evidence, perhaps as much as 643–845 Ma. These estimates require some assumptions about rates of change and choices of calibration points so they are not as firm as fossil dates [41,42]. This time frame corresponds to the snowball Earth hypothesis that may have triggered key events in animal evolution [43,44].
Figure 1.
Metazoan phylogeny showing the 2R and 3R WGD events and total number of CYP coding genes in each genome. This phylogeny is based on the best current knowledge of metazoan evolution [34]. Gene counts were determined based on protein predictions from the respective genome projects (see §2). Phylogenetic divisions are based on currently accepted consensus; where no consensus exists a polytomy is displayed. 525 Ma is the minimum age for vertebrates based on the vertebrate fossils in the Chengjiang biota. Other estimated divergence times are from Edgecombe et al. [36].
It is now possible to determine which genes in vertebrate genomes that are mapped to the CLGs arose by WGD from a common ancestor in the original non-duplicated genome. After each tetraploidization, approximately half of new genes are randomly lost to restore a diploid condition [23], meaning that both copies have been retained for relatively few of the duplicated genes. However, a few CYP genes that were duplicated diverged to create new CYP families and subfamilies. Duplicated genes arising from WGD events are called ohnologues in recognition of Ohno [25–27], who articulated this concept. Large chromosomal segments that contain ohnologues are called paralogons [45]. Vertebrate genomes appear to be composed of four paralogons for each ancestral segment that is preserved from the last common ancestor before 2R.
Deuterostomes and protostomes together form Bilateria, one of the major extant lineages of animals (Bilateria, Cnidaria, Ctenophora, Placozoa and Porifera), although Porifera is possibly paraphyletic [46]. The relative order of animal radiation (phylogenetic topology and rooting) is still a matter of scientific dispute, hindered in part by incomplete sampling, but also complicated by a mixture of short and long branch lengths in the extant lineages [36,47].
There is currently no consensus as to which of the extant basal metazoan lineages (Porifera, Ctenophora or Placozoa) is the earliest diverging metazoan [34,36,48]. The recent results from the Mnemiopsis leidyi genome support an early divergence of Porifera and Ctenophora before the remaining groups (Placozoa, Cnidaria and Bilateria, together called Parahoxozoa; [46]). Continuing genome sequencing from the critical species will probably resolve the question in the near future. However, the current understanding is sufficient for the analysis here.
(c). Use of synteny to track the origins of CYP clans
Orthologous regions of vertebrate genomes often retain the same gene order over long segments. A block of a few genes in the same order is called microsynteny, and shared microsynteny in different species assures a common genomic ancestral gene order is responsible [22,49–51]. In WGD events and segmental duplications, the gene order in both of the duplicate regions initially is identical to the parent sequence. After duplication, many paralogue pairs may lose one copy. When both are retained, over time the syntenic relationships may change as genes move when chromosomes undergo various rearrangements. This makes detection of the duplication difficult. However, the signal is much easier to detect when a duplicated genome is compared with an unduplicated genome, such as that of the lancelet. Up to four paralogous segments from the 2R WGD events map back to one original locus, and the original gene neighbourhood is revealed. Invertebrate genomes such as Trichoplax, sea urchin, sponge and Nematostella are also unduplicated when compared with vertebrate genomes, so comparison of vertebrate genome regions with these unduplicated genomes can help one to reveal the original gene neighbourhoods for genes of interest. (Note, however, that few insect genes follow the 4 : 1 rule [52,53]). A recent study on microsynteny in metazoans did identify 795 groups of genes conserved across multiple animal taxa, so the occurrence of these groups is perhaps more common than previously thought [51]. Identification of syntenic relationships can be used to infer evolutionary history of the genes. It is this synteny approach that we apply to analysis of the evolutionary paths of the animal CYP genes, with the goal of defining the gene content of the original loci for each CYP clan. As we argue below, these separate loci can be traced to one original CYP locus.
2. Material and methods
The UCSC browser [54–56] was used to BLAT search [57] for CYP genes and their neighbours in animal genomes including most of the deuterostome genomes on the browser. Special emphasis was given to human, chicken (Gallus gallus), anole (Anolis carolinensis), frog (Xenopus tropicalis), medaka (Oryzias latipes), lancelet (B. floridae), tunicate (Ciona intestinalis) and sea urchin (Strongylocentrotus purpuratus). In addition, the JGI genome browsers were used for Trichoplax adhaerens [58] and N. vectensis [59], and the OIST browser for the coral Acropora digitifera [60]. Labelled screenshots of many of the results are provided as electronic supplemental material (figures S1–S37). The genomes used in the analysis of the Bilateria are from deuterostomes; the protostome genomes have very little conserved synteny remaining so they are not useful in this analysis. Data from the ctenophore comb jelly (or sea walnut, Mnemiopsis leidyi) genome were provided by A. Baxevanis and C. Schnitzler.
The analysis by Putnam et al. [23] was invaluable for identifying the human paralogons of CYP containing genes or their neighbours. Their supplemental figures S19 and S20 and table S14 were crucial for finding paralogous regions derived from the ancestral CLGs. Nucleotide coordinates from the UCSC Genome Browser were obtained for each human CYP. The segment ID was looked up in table S14 of Putnam, following which figures S19 or S20 were used to find the CLG that matched that segment ID and the other paralogons that matched that CLG [23].
We considered genes on the same chromosome/scaffold with less than five intervening genes to be syntenous, an observation in line with the findings of Irimia et al. [51] that three to 10 intervening genes produced a false discovery rate (less than 0.0002 per gene pair) similar to the less than or equal to four gene distance that they used [51].
Phylogenetic analyses were conducted on datasets consisting of previously published CYP gene sets [61–65], those genomes identified above, and additional complete CYP gene complements from numerous different genomes obtained from JGI (Helobdella robusta, Capitella teleta, Lottia gigantea) and other genome projects (Amphimedon queenslandica, M. ledyii). CYP sequences were predicted using a combination of hidden Markov model (HMM) searches using HMMer (v. 2.3.2 and v. 3.0b3 [66–68]). Sequences were aligned using Muscle (v. 3.8) [69] and masked using custom-written Perl scripts based on the alignment quality scores. Maximum-likelihood phylogenies were constructed using RAxML (v. 7.2.6) [70–73] and presented using Figtree (v. 1.3.1). Multiple inferences of the ML tree resulted in the same overall topology, although the short branches (tips) are not stable to multiple ML inferences. This type of dataset is not robust to the bootstrap assumptions; it has characters compatible with, but not informative for a node, large numbers of taxa and potential co-evolving sites [74].
3. Results
(a). Distribution and phylogeny of metazoan CYPs
The numbers of CYPs observed in metazoan genomes ranges widely, from 35 (A. queenslandica) to 2364 (B. floridae; figure 1). Insect genomes separately display a similar range of CYP numbers (approx. 40–170; R. Feyereisen 2012, personal communication). There is no obvious correlation of relative divergence time with the total number of CYPs, or of CYP families, in a particular genome. Indeed, the size of a gene family undergoing birth–death evolution, as is the case for the CYP superfamily ([75,76] see also [10]), does not appear to be related to divergence time. Instead, gene family size and distribution appear to be describable by a power-law function (see [10] and references therein). Further work will be necessary to characterize these distributions.
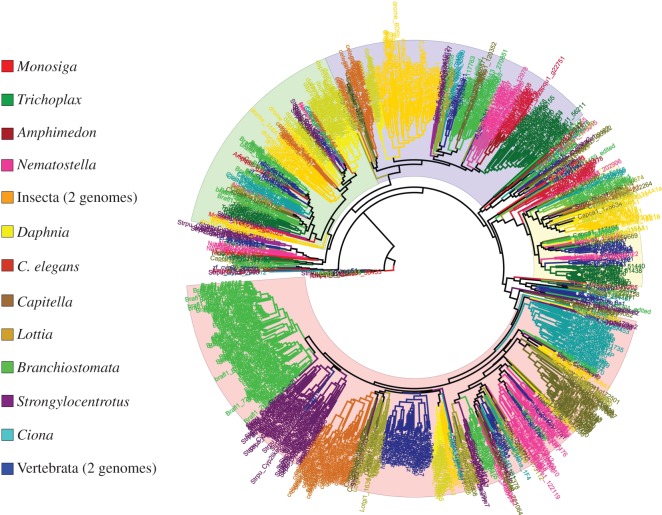
Phylogenetic analyses of large collections of metazoan CYPs support the previous designation of the clan structure (figure 2), and further demonstrate the existence of large ‘blooms’ of CYP families and subfamilies [75]. Certain genomes contain large numbers of CYPs in distinct families and subfamilies that are closely related phylogenetically, appearing as ‘blooms’ of related CYP genes (figure 2). These CYP genes are often the product of tandem duplication and are thus located in close proximity to one another in the genome. Detailed analyses of these regions for several genomes have been previously published (e.g. Mus musculus and Homo sapiens [9], S. purpuratus [61] and Danio rerio [63]).
Figure 2.
Maximum-likelihood phylogeny of CYP protein sequences. Branches represent individual sequences, and are coloured according to species or lineage (e.g. Insecta, Vertebrata). The tree is rooted with CYP51, and all CYP51 genes in various genomes cluster at or near the root. Large highlighted blocks indicate the major clans—Clan 2 (pink), Clan 3 (blue), Clan 4 (green) and Mito Clan (yellow). While the clan ordering is robust to different alignments, the relative position of specific subclans and individual families is sensitive to alignment and does not display high bootstrap values (not shown).
The clan structure in figure 2 is robust to different alignments and protein sequence sets. However, the relative position of specific smaller clans and individual families is sensitive to alignment and does not display high bootstrap values (not shown). Hence, determining the evolutionary origin of CYP clans requires additional information, supplied by the analysis of conserved micro- and macrosynteny.
(b). Ohnologue CYPs
Table 1 lists the CYP gene families arising from the two WGD in the vertebrate line (with human as example), and those appearing in fish following the teleost-specific 3R WGD event [30]. These genes were identified from the location of CYP genes on the human–cephalochordate paralogons that were defined by Putnam et al. [23]. Duplicated genes resulting from these WGD events are in the same clan, and usually within the same family. Therefore, the 2R and 3R events are too recent to account for the origin of animal CYP clans. As detailed below, the CYP clans predate the origin of Cnidaria, and indeed may predate the origin of Metazoa.
Table 1.
Whole genome duplicated CYPs.
| original gene | duplicated gene | event |
|---|---|---|
| CYP1A | CYP1D | 2R |
| CYP2D precursor | CYP2K/CYP2W | 2R |
| CYP7A | CYP8B | 2R |
| CYP11A | CYP11C | 2R |
| CYP26B | CYP26A/C | 2R |
| CYP27B | CYP27A/C | 2R |
| CYP4F precursor | CYP4T/CYP4ABXZ | 2R |
| CYP17A1 | CYP17A2 | 2R |
| CYP7A1 | CYP7D1 | 3R |
| CYP19A1 | CYP19A2 | 3R |
(c). Gene neighbours and CYP clans
CYP genes in the 19 vertebrate CYP families were mapped back to their paralogous regions in lancelet using the human–paralogon map of Putnam et al. [23]. Often, these paralogons do not contain any CYP sequences, but they can be used to identify genes that were in the neighbourhood of an ancestral CYP. Over several 100 Myr, a human (or other vertebrate) CYP may not have a recognizable orthologue in a phylogenetically distant genome such as Trichoplax or Nematostella, as the sequences may have diverged too far to be classified even to the same CYP family. This is not necessarily true for genes closely linked to a CYP gene, as they may include highly conserved sequences serving essential functions. Thus, some genes may have highly conserved and clearly recognizable 1 : 1 orthologues, from human to Trichoplax. If a gene linked to a CYP gene in human matches strongly to a single gene in a distant genome, and that gene also is in the vicinity of a CYP gene, then one may conclude that those two CYP genes shared the same ancestral neighbourhood. This is the argument from macrosynteny. If the two CYP genes also happen to be in the same CYP clan, one may conclude that those CYP genes probably share a common ancestor. The probability increases when more than one homologous gene maps to the same region as a CYP gene. The synteny mapping approach did reveal unexpected linkages between many CYP clans as described in detail in the following sections.
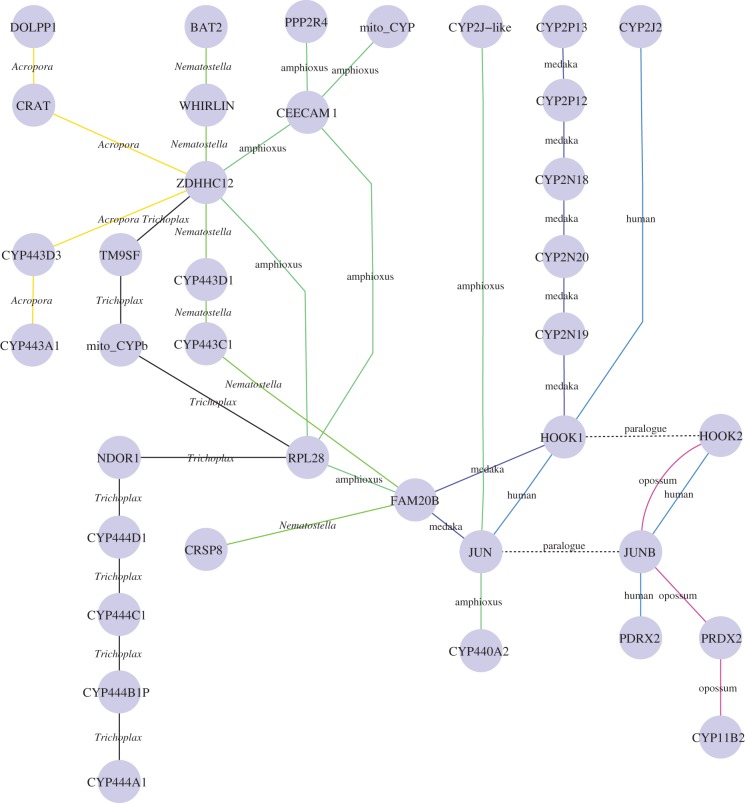
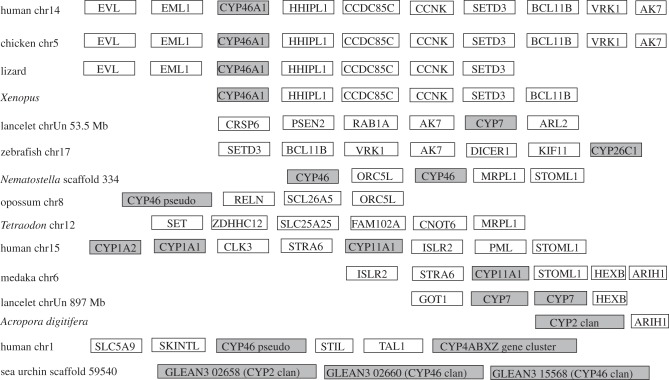
Figure 3 shows an example of synteny analysis. The gene ZDHHC12 is found in all 10 species included in that figure. In the cnidarians N. vectensis and A. digitifera, the ZDHHC12 gene is adjacent to Clan 74 CYPs. In the placazoan T. adhaerens ZDHHC12 is one gene from a mito Clan CYP and eight genes from four Clan 74 CYP genes (see the electronic supplementary material, figure S10). The gene pair CRAT DOLPP1 is adjacent to ZDHHC12 in Acropora and a similar gene association is found in lancelet, chicken and human. These associations suggest that this syntenic region has been conserved over more than 600 Myr of evolution.
Figure 3.
Synteny mapping with emphasis on ZDHHC12, CRAT and DOLPP1 and their neighbours. These three genes maintain syntenic relationships from Acropora to human. CYP Clan 74, mito Clan and Clan 2 P450s are linked to these genes and/or their neighbours. Connecting lines are colour-coded by species and they indicate a neighbour relationship, though not all genes are shown.
Other genes in the same neighbourhood provide indirect linkages to other genomic segments. Thus, in Trichoplax and in lancelet,5 RPL28 is adjacent to FAM20B. FAM20B is 1.2 Mb from a mito Clan CYP in lancelet, and is adjacent to the HOOK1/CYP2J/CYP2N/CYP2P locus in the fish medaka (figure 3). The genes JUN/JUNB and HOOK1/2 are tied to this same region in all vertebrates examined to date. These genes are close to another CYP Clan 74 member (CYP440A2) and a CYP2J-like gene in lancelet. JUN/JUNB and HOOK1/2 are also close to CYP2J2 in human and CYP11B2 (a mito Clan gene) in opossum. Nematostella has CRSP8 and BAT2 in this region. These genes also appear in human and chicken. We conclude that these syntenic genes in different species, from mammals to coral, define an ancestral genomic region. This single figure shows three CYP clans (Clans 2, 74 and mito) that we predict were present in this ancestral animal locus.
(d). Extreme age of animal CYP clans
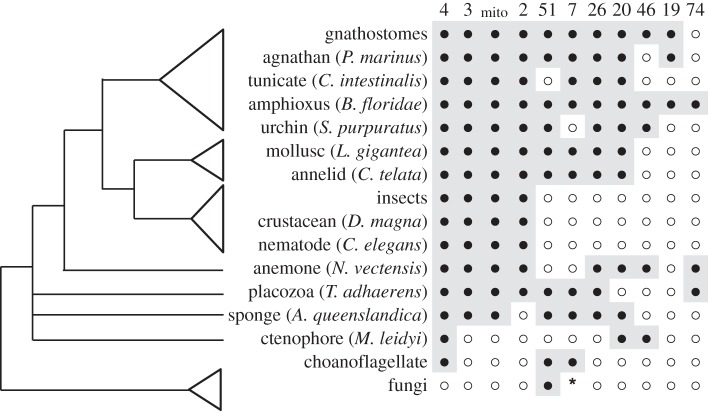
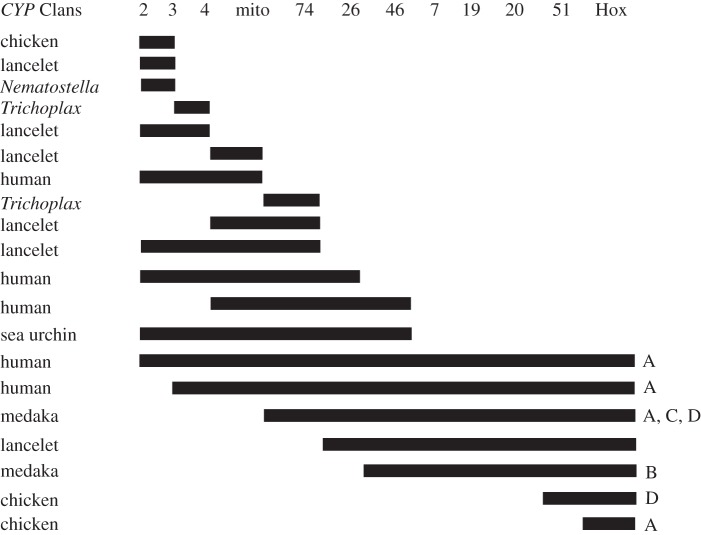
The 11 animal CYP clans (Clans 2, 3, 4, 7, 19, 20, 26, 46, 51, 74 and mitochondrial or mito) are shown in figure 4. Not all animals have all 11. Thus, the Ecdysozoa (insects, crustaceans, nematodes) only have Clans 2, 3, 4 and mito. However, other protostomes may have additional clans; thus, four more clans (7, 20, 26 and 51) appear in molluscs and annelids. Clan 74 was originally described as a land plant CYP clan, but now has been found in cnidarians, Trichoplax and lancelet. This raises the question of Clan 74 origin. CYP74 sequences exist in both land plants and some animals, but not in green algae or fungi6. In fact, as discussed below, CYP74 is likely to have originated in marine animals and only later transferred horizontally to land plants. The CYP7 and CYP19 (aromatase) families are chordate-specific, but they are extremely sequence divergent from other CYP clans, which suggests that they are either rapidly evolving or that they may be much older than the chordate line. Gene duplication within chordates would be unlikely to produce new clans, as suggested in the discussion of ohnologues above. CYP39 is considered part of Clan 7, and CYP39-like sequences are known from Trichoplax, sponges and Monosiga brevicollis (a choanoflagellate). (CYP Clan 7 genes are observed in some filamentous fungi (electronic supplementary material, figure S23), but the limited distribution may indicate a lateral transfer.) Trichoplax, which is thought to have diverged between cnidarians and bilaterians, has eight clans (it is missing Clans 19, 20 and 46). Clan 46 is found in Nematostella and Clan 20 is found in Nematostella and sponges, which are thought to predate Trichoplax. Therefore, 10 of 11 CYP clans were in existence at least by the time of cnidarian origins. The 2R events, which were discussed above, failed to create any new CYP clans, so the clans are older than the 2R duplications. Because two cnidarians are described in the Chengjiang biota including Xianguangia sinica, a primitive sea anemone [38], the CYP clans are presumably older than 525 Ma. Once again, molecular evidence places the origin of Cnidaria much earlier than the Chengjiang biota [41,42].
Figure 4.
Distribution of CYP clans in animals and fungi. Presence of a clan is indicated by a closed circle. Absence is indicated by an open circle. The asterisk denotes a probable lateral gene transfer from animals to the filamentous fungi. Some clan losses are evident, particularly in insects and Crustacea (Ecdysozoa), which are known to lack CYP51 and thus require dietary cholesterol. Note also that CYP Clan 19 does not appear until cephalochordates.
These facts argue for a relatively rapid origin of the animal CYP clans followed by a much slower divergence within the clans.
The following sections present evidence for the linkage of the CYP clans to each other in a common neighbourhood near the Hox genes before the advent of sponges (Clans 51, 3, 4, 7, 20, 26, mito) or cnidarians (Clans 2, 46, 74). CYP19 does not appear until chordates, but linkage evidence suggests that it was part of the original locus. These sections detail the paths connecting the CYP clans (via families and subfamilies).
(e). The CYP4V and CYP2C connection
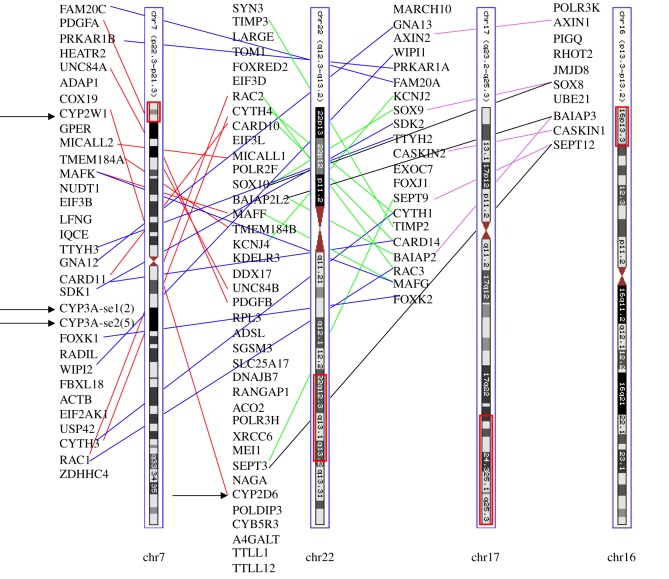
Attempts to trace the origins of CYP genes backward in time by mapping the locations of neighbouring genes turned up some unexpected connections between the CYP4s (Clan 4) and CYP2s (Clan 2). The CYP4V2 orthologue in medaka (O. latipes) lies next to TLR3, SORBS2 and PDLIM3. In human, a similar syntenic region contains CYP4V2, FAM149A, TLR3, SORBS2 and PDLIM3 (figure 5, lines H, I). There are three SORBS genes in human and each is adjacent to a PDLIM gene. SORBS1 and SORBS3 are on paralogons as defined in Putnam et al. [23]. SORBS2 is not mapped to a CLG in that paper but it is presumably on a third human paralogon. The human CYP2C gene cluster is next to ACSM6, PDLIM1 and SORBS1 (figure 5, line J). A similar gene arrangement is seen in chicken and lizard (figure 5, lines K, L), though in the lizard the contig ends at HELLS, just before where a CYP2C gene would be expected. Nematostella has a SORBS1- and PDLIM7-like gene pair on scaffold_597 illustrating an ancient association between SORBS and PDLIM genes, but the scaffold is too small to see whether there might be an adjacent CYP gene. Zebrafish have PDLIM1, PDLIM3A and PDLIM3B. PDLIM3A is next to TLR3 and CYP4V2, similar to the medaka. Zebrafish PDLIM1 is 11 genes from ATOH7 and 20 genes (400 kb) from CHUK. Line L in figure 5 shows that in the lizard CHUK is two genes from the SORBS1 PDLIM1 pair, and line M shows that CHUK is 380 kb from the HoxB gene cluster in medaka, and 3.5 Mb from CYP26A1. The close proximity of these CYP genes to PDLIMx and SORBSx indicates the ancestors of CYP4V and CYP2C were close neighbours. The possibility that CYP2C is an ohnologue of CYP4V can be ruled out because the CYP Clans 2 and 4 are older than the duplication of the SORBS1 PDLIM1 gene pair, which probably occurred during the 2R WGD events. This implies that CYP4V and a CYP2C precursor were neighbours before the SORBS1 PDLIM1 gene pair duplicated. Following duplication, one of the CYP genes was lost from each of the descendant regions, separating CYP2C from CYP4V. We will discuss the linkage of these genes with CYP26A1 later.
Figure 5.
All 11 CYP clans can be linked directly or indirectly to Hox gene clusters. P450 genes are boxed. Hox genes are aligned down the centre with chr and Mb locations given. Non-Hox regions are labelled as non-Hox. Orthologues and ohnologues are aligned as much as possible to highlight sytenic relationships as seen with EXOC6 in lines M and N. Not all genes in the region are shown because of space limitations.
(f). CYP2D and CYP2K/CYP2W are ohnologues whose ancestor was near the CYP Clan 3 ancestor in early animals
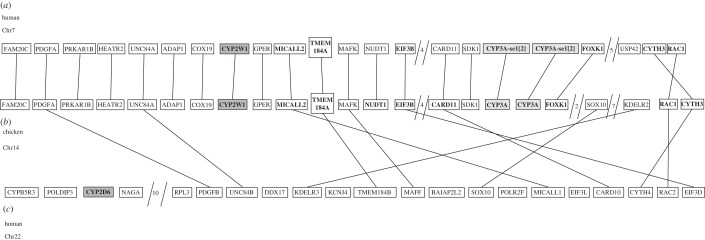
The gene neighbourhoods around CYP2D6 and CYP2W1 in human and some CYP2K genes in fish are descended from a common ancestral region duplicated in a 2R WGD event. Figure 6 shows a map of some of the genes in this region, with ohnologues in the two human regions a and c connected by lines. Figure 7 shows the paralogons for this region, with connections between ohnologues (electronic supplementary material, figures S1 and S2 show the expanded gene neighbourhoods depicted in abbreviated form in figures 6a,c and 7; electronic supplementary material, figures S3 and S4 show these regions in mouse and rat, respectively). These genes will be central to defining the original CYP2 gene neighbourhood in very early animals. Many of these genes appear also in figure 5. The chicken CYP2W1 region is abbreviated in figure 5 as line A and the human CYP2W1 region is part of line B.
Figure 6.
Genes in the vicinity of human CYP2W1, CYP2D6 and chicken CYP2W1. The human CYP2W1 region is telomeric. Connecting lines between (b) and (c) mark ohnologues. Not all genes from the regions are included.
Figure 7.
Human paralogons covering the CYP2D6 and CYP2W1 regions, including two CYP3 pseudogenes (arrows mark P450s). Probable ohnologues are connected by lines. The regions shown are boxed in red in the ideograms.
The CYP2W1 region is closely associated with the original CYP3A gene location. The evolution of CYP3 genes in 16 vertebrates has been studied by Qiu et al. [77]. Note that in chicken the CYP3A genes lie between SDK1 and FOXK1. This is the condition seen in opossum, platypus, chicken and lizard. This location is called CYP3HR1 in Qiu et al. [77]. The original CYP3A gene moved in human to a new location called CYP3HR2, but it left behind traces as two small pseudogenes between SDK1 and FOXK1. SDK1 is still next to intact CYP3 genes in horse, but not in human, rhesus, cow, mouse, rat, guinea pig or dog, although like human, the dog has CYP3 pseudogenes in this location.
Acropora digitifera (reef-building coral) has a CYP Clan 3 gene next to NPTXR, a gene in human only two genes from UNC84B. UNC84B is also in a CYP3 paralogon (figure 6c and 7, chr 22). This result argues for some preservation of synteny in this region going back to the time of the common ancestor of bilaterians and cnidarians.
The zebrafish CYP3C1 gene is next to WIPI2 FOXK1 and this gene is syntenic with the land animal CYP3A gene cluster at CYP3HR1 (see the electronic supplementary material, figure S5). The zebrafish CYP3A65 gene has moved to a new location, so even though CYP3C1 is syntenic with tetrapod CYP3As, it has diverged in sequence enough so that it has lost identity as a CYP3A. Lancelet retains CYP3A- and CYP2D-like genes as neighbours with only one gene separating them (see the electronic supplementary material, figure S6). This intervening gene is plant-like, resembling 4-coumarate coenzyme A ligase (4CL) and it is not found in vertebrates. However, COG3 is six genes from the Clan 3 gene in lancelet. COG3 is found between CHUK and SORBS1 PDLIM1 in the lizard (figure 5, line L).
The sea anemone has 19 Clan 3 members. One, in particular,7 is useful for this analysis because it is adjacent to a small cluster of at least three Clan 2 genes.8 Trichoplax has 39 Clan 3 members. One 450 kb region has 22 Clan 3 CYPs in several blocks and four adjacent 4CL genes (scaffold_5:880272–1300000), showing that an association between CYP Clan 3 and 4CL has been preserved from Trichoplax to lancelet. The four 4CL genes are bracketed by three CYPs before and nine CYPs after. The last CYP in this large cluster is a Clan 4 member, 15 genes away from the CYP Clan 3 genes. There is a 4CL gene in Nematostella9, but it is on the edge of the scaffold and adjacent genes downstream are not in view. The data presented here unite members of the Clans 2, 3 and 4 into a single locus in very early animals. Even today, in animals such as chicken, lancelet and Nematostella, Clan 2 and Clan 3 members are still close neighbours. Direct linkages between CYP genes from different clans near one another on the same chromosome are summarized in figure 8.
Figure 8.
Direct linkage between different CYP clans including Hox clusters (A–D). Each bar indicates a neighbour relationship between the clan members listed on the top of the figure, in the species listed on the left.
(g). Other CYP Clan 4 members
The oldest CYP4 genes in chordates seem to be in the subfamilies CYP4V and CYP4F. A detailed evolutionary study of Clan 4 genes from 28 species has been conducted, noting a major bifurcation of CYP4 subfamilies into CYP4V and related sequences (figure 1, node F, [35]), and all other vertebrate CYP4 subfamilies CYP4A, B, F, T, X and Z [35]. Synteny involving the CYP4V subfamily was discussed above. Here, we give attention to CYP4A, B, F, T, X and Z. The study of Kirischian and Wilson [35] splits this set into CYP4F (node D) and CYP4A, B, T, X and Z (node G). The CYP4F cluster on human chr 19 is on a paralogon with the CYP4ABXZ cluster. Therefore, the CYP4ABXZ and CYP4F gene clusters are possible descendents of ohnologues that were created in the 2R WGDs from a CYP4 gene in the chordate ancestor. This would explain the bifurcation into node G (CYP4A, B, T, X, Z) and node D (CYP4F) in fig. 1 of Kirischian and Wilson [35]. The Kirischian and Wilson study described difficulty in determining the relationships between the CYP4T and CYP4B subfamilies and suggested additional studies would be needed. Examination of the macrosynteny surrounding the CYP4T genes of fish and X. tropicalis supports a syntenic relationship with the mammalian CYP4ABXZ locus, although this is hard to demonstrate conclusively because of much expansion and chromosomal rearrangement in tetrapods. The genes near the fish CYP4T genes are very spread out in human. A comparison of the fugu CYP4T5 genomic region (300 kb) with the human genome shows that every gene in the fugu region matches to a human gene on chr 1, though the human segment is 21.4 Mb (see the electronic supplementary material, figure S36). The size difference of these regions is partly because of the genome size difference, as the fugu genome is 365 Mb, whereas the human genome is 10 times larger, at about 3 Gb [78]. The genes in lancelet surrounding a CYP4T homologue (KIF2A CYP4T PTCH1 MKNK1) also match the human region but in a much narrower 2.4 Mb window. This is consistent with CYP4T being in node G in fig. 1 of Kirischian and Wilson [35] and being an ohnologue derived from a CYP4F ancestor.
The only CYP4F-like gene in Nematostella is adjacent to CHD7. In lancelet, the CHD7 gene is 375 kb (four genes) from a small CYP2 gene cluster, whereas in chicken, lizard and human CHD7 is near CYP7A1 (as shown for human in the electronic supplementary material, figure S22). The lancelet genome has Clan 2 genes that are less than 100 kb (i.e. two genes SOX5, BCAT1) away from Clan 4 genes (see the electronic supplementary material, figure S7). These neighbour relationships with CHD7 link Clan 4 with Clan 2 and Clan 7 ancestors in early animals.
The ctenophore M. leidyi (comb jelly or sea walnut) has two Clan 4 genes adjacent to one another on AGCP01004322.1. Mnemiopsis is a very early branching animal, probably diverging before cnidarians, so it may provide valuable clues to the early neighbours of Clan 4. A Mnemiopsis gene most like CRIP2 is only 3.5 kb upstream of the CYP Clan 4 genes. In medaka, CRIP2 is seven genes from DICER1 and 11–12 genes from AK7 and VRK1. These genes have tight linkages to members of CYP Clans 26 and 46 (figure 9). These observations link Clan 4 to Clans 2, 7, 26 and 46. Section 3h links CYP Clan 4 to CYP Clan 74.
Figure 9.
Synteny mapping of CYP16 family members showing linkages to CYP26 family members, the CYP Clan 46 and the CYP Clan 2 via IDE and DICER1 and to the CYP7 and mito Clans via HEXB. P450 genes are in shaded boxes. CYP16B1 has a direct linkage to CYP20 in lancelet.
(h). FAM20, PDRX and CYP Clan 74
The synteny relationships depicted in figures 3, 5 and 6, and electronic supplementary material, figures S8–S15 support a neighbourhood in the past that included members of Clans 2, 3, 4, 7, mito, 51 and Clan 74. In figure 6, the CYP2W1 region begins with FAM20 C in human and chickens and it includes CYP3 genes/pseudogenes. FAM20 paralogues are significant markers for synteny analysis. A FAM20B-like gene is next to two Clan 74 members in Nematostella (CYP443C1 and CYP443D1; figure 3 and electronic supplementary material, figure S8). Note that whirlin (WHRN) is four genes from those Clan 74 members (figure 3) and in lancelet WHRN/DFNB31 appears 1 Mb (approx. nine genes) from a Hox cluster that is 300 kb (four genes) from CYP442A2, another Clan 74 member (figure 5, line Y). FAM20B in medaka is next to JUN and PRDX6 on the left and 220 kb from HOOK1 and five CYP2 family genes on the right (figure 3 and electronic supplementary material, figure S9). Trichoplax has four Clan 74 genes in tandem (CYP441A1, CYP441B1P, CYP441C1 and CYP441D1) and they are four genes away from a PRDX6-like gene and seven genes away from a mitochondrial clan CYP (figure 3 and electronic supplementary material, figure S10). The gene CRSP8/MED27 in Nematostella also is adjacent to FAM20B. CRSP8/MED27 and WHRN are only five genes apart on medaka chr 9, so these genes have been associated since cnidarians. In human, JUN is adjacent to FGGY HOOK1 and CYP2J2. In the lancelet, JUN is next to the Clan 74 gene CYP440A2 on the left and a CYP2J-like gene on the right (figure 3 and electronic supplementary material, figure S11). The gene ATOH7 is also seen in this region and in figure 5, line F near CYP51. A FAM20C-like gene in lancelet is about 14 genes from a PRDX1-like gene and 9–10 genes from AK3 and RAD23A (see the electronic supplementary material, figure S12). RAD23A is nine genes from HOOK2 JUNB PRDX2 in human (see the electronic supplementary material, figure S13), which is on a paralogon of the HOOK1 CYP2J2 region. Thus, the FAM20 and PDRX paralogues link many CYP clans through indirect linkages.
These observations also support a close association between Clan 74 CYPs and CYP2W/K/D and CYP2J-like genes (Clan 2) in early animals. A Clan 4 gene and the Clan 74 member CYP440A6 are adjacent in lancelet (see the electronic supplementary material, figure S14). CYP2, CYP4 and CYP440A8 (Clan 74) are found in a 500 kb window in lancelet (electronic supplementary material, figure S15). A single Clan 2 founder gene is inferred, which duplicated and diverged to give rise to the multiple CYP2 loci in vertebrates and other animals. The mitochondrial clan CYPs seen in Trichoplax and in lancelet (figure 3) draw the mito Clan into this unique ancestral region, which we name the ‘cytochrome P450 genesis locus’.
(i). Mitochondrial clan CYPs
The mitochondrial CYP clan in vertebrates includes the CYP11, CYP24 and CYP27 families. Additional mito CYP families are found in invertebrates, including CYP12 and CYP49 in insects. The mito Clan appears as a monophyletic group descended from one common ancestor in the animals, as no other eukaryotic organisms have mitochondrial CYPs. Mito Clan genes are nuclear-encoded, and contain mitochondrial-targeting sequences and not hydrophobic N-terminal anchor domains. The original founder mitochondrial CYP probably acquired a mitochondrial-targeting sequence early in the history of animals. Of note, however, is that in some species, a few CYPs of more recent origin have bimodal endoplasmic reticulum and mitochondrial targeting, including CYP1A [79,80] and CYP2E1 [79,81].
Vertebrate CYP11A and CYP11B/11C are ohonologues that duplicated during 2R (table 1). These genes are on parlogons defined by the CLK2 and CLK3 genes. CLK2 is adjacent to the CYP11C1 gene in fish (fugu, zebrafish, medaka, stickleback and coelacanth; see electronic supplementary material, table S2), whereas CLK3 is between CYP11A1 and the CYP1A1–CYP1A2 pair in tetrapods. CYP11B1 evolved from CYP11C1 of fish as shown by the indirect linkage to BAI1: stickleback has two CLK2 genes, CLK2b adjacent to CYP11C1 and CLK2a adjacent to BAI1, while BAI1 is eight genes from CYP11B1 in humans (see electronic supplementary material, table S2). Additional evidence for tetrapod CYP11B being orthologous to fish CYP11C1 comes from the genes DGAT1 and SCAMP3. DGAT1 is adjacent to CYP11B1 in turtle and frog, and two genes from SCAMP3 in fugu, linking SCAMP3 to CYP11B. In tetrapods, SCAMP3 is next to CLK2, and, as shown earlier, CLK2 is adjacent to CYP11C1 in fish.
The CYP11-related gene seen in modern lancelet (39% identical to catfish CYP11A1) is approximately eight genes from TNPO2. The opossum has a gene block TNPO2 ASNA1 HOOK2 JUNB PRDX2 RANSEH2A ATP6VOD1 CYP11B1 that exhibits shared synteny with a similar gene set in lancelet. Therefore, the lancelet ancestor had a CYP11-like gene that is orthologous to vertebrate ohnologues CYP11A and CYP11C/CYP11B. The new synteny evidence thus counters the argument of Markov & Laudet [82] that CYP11 is strictly vertebrate, though the enzymatic activity of this lancelet P450 is not yet known and it may not have side-chain cleavage activity.
The evolution of CYP11 is of critical importance because it is an essential enzyme in the steroidogenesis pathway. Baker [83] discussed the evolution of steroids and their nuclear receptors. CYP11 sequences are rare prior to the emergence of teleost fish, although a fragment of CYP11A is known from little skate (WGS AESE012655227.1 Leucoraja erinacea). Searches for CYP11 in lamprey or hagfish have so far failed to find a candidate. Therefore, the lancelet CYP11 sequence is a key sequence and its function needs to be determined.
Linking various other CYPs to each of these CYP11 paralogues ultimately links lancelet CYP Clans 11, 2, 4, 7 and 74 together. Thus, CYP11A1 is only seven genes from CYP1A1 and CYP1A2 (in Clan 2) on human chr 15 and on chicken chr 10. In medaka, CYP11A1 is only three genes from HEXB (see the electronic supplementary material, figure S16); HEXB is adjacent to two CYP7 genes in the lancelet, linking CYP7 to CYP11. In opossum, a CYP11B orthologue is three genes from the PDRX2 JUNB HOOK2 locus (figure 3 and electronic supplementary material, figure S17) that is a paralogon of the JUN HOOK1 CYP2J locus (based on data in Putnam et al. [23]).
A CYP11-like gene in the lancelet is four genes away from a Clan 4 member (electronic supplementary material, figure S18). CYP4T1 in zebrafish is adjacent to ARNT on chr 16. In medaka, ARNT is three genes from CYP11B (figure 5, line E, chr 16). The Nematostella mito Clan member XM_001640379 is two genes from a SLIT1-like gene. SLIT1 is adjacent to the Clan 74 member CYP440A1v1 in lancelet (see the electronic supplementary material, figure S19). These close neighbours link CYP11 to the CYP genesis locus as well as to genes in the lancelet Clans 4, 7 and 74. CYP27 shows an indirect linkage to CYP20 via the gene PDE1B. CYP20 is six genes from PDE1B in chicken and PDE1B is 3.2 Mb from CYP27B1 in human (figure 5, lines T and W). The CYP51 section below also links CYP27 to CYP51.
(j). CYP51
CYP51 (lanosterol 14 α-demethylase; Clan 51) is adjacent to lanosterol synthase (LSS) in the lancet genome. These enzymes function sequentially in the steroid biosynthetic pathway and thus CYP51 and LSS constitute a functional gene pair. This arrangement is also evident in Trichoplax, which has two CYP51 genes flanking LSS (scaffold 3: 300–323 kb), and also in sponge (contig 13514, 61–65 kb). By using this apparently ancestral link of CYP51 to LSS, we can link other CYP genes through adjacent orthologues and paralogues. Even though CYP51 and LSS are not linked in vertebrates, the association of a CYP gene with LSS ties that gene back to the P450 genesis locus that contained both CYP51 and LSS. For example, CYP27A is 85 kb (seven intervening genes) from LSS in medaka (see the electronic supplementary material, figure S20). CYP27A is the only CYP in a 10 Mb window centred on LSS. This region also contains a Hox cluster (see §3k).
Additional genes linked to LSS or CYP51 provide evidence for their membership in the P450 genesis locus. In sea urchin, LSS is four genes from ZDHHC12, a gene that is adjacent to two Clan 74 genes in Nematostella (figure 3 and electronic supplementary material, figure S8) and Trichoplax (figure 3 and electronic supplementary material, figure S10). A ZDHHC12-like gene is adjacent to RPL28, FAM20B in lancelet (see figure 4 and §3h). METTL6 is two genes from CYP51 in the lancelet. In Ciona, METTL6 is two genes from B4GALT4, and B4GALT5 is adjacent to PTGIS/CYP8A1 in human, opossum, lizard and platypus. The gene ATOH7 is found in lancelet two genes from CYP51. In medaka, the genes EIF3L, ATOH7, PKD2, PDLIM1 and PRDX3 are found in an approximately 30 gene 763 kb window. EIF3L is seen in figure 6c in the CYP2D6 region. Lines F and G in figure 5 have ATOH7 and PKD2 near CYP51 and the HoxB cluster. PDLIM1/3 is seen in lines H to L, figure 5, near CYP4V and CYP2C genes. PRDX genes were discussed earlier in relation to Clans 74 and 2. The chicken has a direct linkage of CYP51 with the HoxA cluster (figure 5, line D, chr 2). The presence of HDAC9 and TWIST1 in chicken also links the CYP51 gene to the HoxA cluster in medaka (figure 5, line E), which has a direct linkage to CYP11B. TWIST1 is one gene away from ATOH7 and three genes from CYP51 in the lancelet (figure 5, line F). These facts support a close neighbourhood of CYP51 with Clans 2, 3, 4, 7, mito and 74 in an early animal ancestor. The presence of PDE11A1 (figure 5, lines E and W) further links CYP20 to this neighbourhood.
(k). Clan 7 (CYP7, CYP8, CYP39) and Hox genes near the ancient CYP locus
CYP7 occurs twice in lancelet between a HEXB pair and VPS24 GOT1 (electronic supplementary material, figure S21). The two CYP7 genes are on opposite strands and both are incompletely known because of gaps in the genome sequence to date. As mentioned earlier, CYP11A1 in medaka is only three genes from a HEXB gene (see the electronic supplementary material, figure S16). The gene sequence SLIT1 PRDX3 … GOT1 VPS24 is found at another location in the lancelet. Note that SLIT1 and PDRX1 were associated with a CYP Clan 74 member (electronic supplementary material, figure S19), and many PDRX genes are linked to CYP Clan 2 genes.
Three CYP7 sequences are present in C. intestinalis. CYP7c is adjacent to PIP4K2C, as is CYP7B1 in zebrafish. In human, PIP4K2C is nine genes from CYP27B1 (figure 5, line T, chr 12). The PIP5K2C gene is only three genes from CYP27B1 in medaka. In opossum, a PIP4K2C homologue is next to ARMC3. ARMC1 is next to CYP7B1 in human, chicken, frog and fugu. Thus, CYP7 is linked to CYP27.
Further linkages bring in the Hox genes. In chicken, PIP4K2C is approximately 300 kb from a Hox gene cluster on chr 27. A Clan 74 gene in lancelet (CYP442A2) is only four genes (300 kb) from the single Hox gene cluster (figure 5, line Y). PNPO is seven genes from the HoxB gene cluster in human and the WNT3, WNT9B pair is another 18 genes from PNPO (figure 5, line G, chr 17). WNT3, PDK1 and PNPO are near the HoxB cluster in medaka with five genes between PNPO and HoxB5 (figure 5, line M). PNPO is adjacent to CYP51 in the lancelet (figure 5, line F). WNT1 and ARF1 are near the HoxC cluster in medaka (figure 4, line R). (An association between WNT genes and Hox clusters was noted previously by Putnam et al. [23].) Medaka has a HoxA gene cluster on chr 16, 260 kb (11 genes) from TWIST1 (figure 5, line E). LSS, CYP51, PNPO, ATOH7, METTL6 and TWIST1 are all adjacent in the lancelet (figure 5, line F). Another Hox cluster in medaka (chr 19 17.5 Mb) is flanked by WNT3, PKD2, PNPO on the one side and the genes CHUK, CASKIN1, METTL9, KDELR2 and FOXK1 on the other side (figure 5, line M). This last set of genes is found in the paralogons with the CYP2W1/CYP2D6 and CYP3A in some vertebrates (figure 6c). In human and chicken, CHD7 is six and seven genes from CYP7A1 (see the electronic supplementary material, figure S22). CHD7 is adjacent to Nematostella XM_001639310, a Clan 4 CYP sequence. In lancelet, CHD7 is 375 kb from a CYP2 gene cluster. Additional evidence for Clan 2 linkage to Clan 7 includes VPS24 being adjacent to CYP7 in lancelet. In chicken, VPS24 is adjacent to KDM3A/JMJD1A and about seven genes from EIF2AK3. EIF2AK3 is adjacent to the Clan 7 member CYP7.b in Ciona (electronic supplementary material, figure S24). EIF2AK1 is in the CYP2W1 paralogon (figure 6a), providing an indirect linkage of Clans 2 and 7. These observations link Clan 7 to CYP2W/CYP2D, CYP3, CYP4, CYP27, CYP51, CYP74 and the Hox gene clusters.
The region surrounding human CYP7B1 on chr 8 is on a paralogon with the PDE7B, MYB and EYA4 genes on chr 6 from 133.5 to 136.5 Mb. The human paralogon has the ohnologues EYA1 and PDE7A found near CYP7B1 (see the electronic supplementary material, figure S22). In Trichoplax. MYB is next to the trox-2 (Gsx) gene, the only Hox/ParaHox-type gene in Trichoplax and proposed as a possible ancestral ProtoHox gene [84]. The linkage provides another tie between Clan 7 and the Hox genes.
The reconstructed CLG include all the Hox gene clusters in CLG16 [23]. CYP genes near these clusters would have been duplicated with the clusters during the 2R WGD events. Human CYP27B1 is 4 Mb from the HoxC cluster and CYP27A1 and CYP27C1 are on the q arm of chr 2 with the HoxD cluster, although the CYP27A and C genes and HoxD are spread out over 92 Mb. Human CYP51A1 is on chr 7 with the HoxA cluster, but they are on opposite sides of the centromere. No human CYPs remain near HoxB, though a FOXK1 homologue is 1.1 Mb from the HoxB cluster on chr 19 in medaka and CYP26A1 is 4 Mb from that same HoxB cluster (figure 5, line M). CYP3 genes are adjacent to FOXK1 in chicken. These facts are consistent with CYP27B being an ohnologue with the CYP27A/C pair ancestor. The lancelet Hox gene cluster has these genes near it: KCNB1 KCNJ4 SOX10 UBE2S LFNG SLC26A11 HIBADH TTLL12 WHRN, with other genes intermingled (figure 5, line Y). Most of these genes are found in figure 5, in the CYP2W/CYP2D paralogons. KCNB1 is next to CYP8A1 in human and HIBADH is seen near the HoxA clusters in human medaka and chicken (figure 5, lines B–E).
The CYP8 family genes appear to have originated between lancelet and jawless fishes. No CYP8 genes were found in the lancelet genome, but at least three intron-containing CYP8B-like genes are present in lamprey (Petromyzon marinus). Interestingly, there are no CYP8A-like genes in lamprey, yet bony fish genomes have clearly recognizable CYP8A genes, with introns in the same locations as the introns in the CYP8B-like genes in lamprey.10 CYP8B genes in fish have no introns, suggesting retrotransposition of ancestral CYP8A mRNA into early fish genomes. The CYP8B genes of fish are more like the CYP8 genes in lamprey than they are like the CYP8A genes in fish. These data suggest that the original CYP8 gene was CYP8B-like. The retrotransposed gene with no introns retains this character, whereas the intron-containing gene diverged to form the CYP8A ancestor. Thus, CYP8A was derived from a CYP8B-like precursor. The CYP8A gene is near five genes that have clear paralogues near the CYP7A and CYP7B genes in human (see the electronic supplementary material, figure S22). These paralogues are not found near the CYP8B gene, consistent with a retrotransposition event.
Lampreys make petromyzonol and the pheromone petromyzonol 24-sulphate that guides lamprey migration back to their spawning grounds [85]. Petromyzonol 24-sulphate (7alpha, 12alpha, 24-trihydroxy-5alpha-cholan-3-one 24-sulphate) is hydroxylated on the 7, 12 and 24 positions. CYP8B1 hydroxylates the CYP12 position in bile acid synthesis. Therefore, the earliest lamprey CYP8 probably made this pheromone. CYP8A is prostagandin I2 synthase (PTGIS). PGI2 or prostacyclin appears to be a novel biochemical made only after lampreys split from Euteleostomes (bony vertebrates). It is probable that CYP8 arose from CYP7A by WGD duplication and divergence. Dehal & Boore [29] suggested that regions containing CYP7A and CYP8A were on paralogons or chromosome pieces quadruplicated in the 2R WGD process leading to vertebrates (see the electronic supplementary material, figure S22). This would be a logical way of explaining the origin of CYP8 genes between lancelet and lampreys.
CYP39 is found in the sponge A. queenslandica, indicating that the CYP39 family is very old. Monosiga brevicollis (a single-celled choanoflagellate) also has a CYP39-like gene. If it is correct that CYP39 belongs in Clan 7, then Clan 7 also is very old and probably was present in the CYP genesis locus very early in the history of animals. CYP39A1 is 3 Mb (18 genes) from CYP2AC1P in human. CYP39A1 is only seven genes from the CYP2AC gene cluster in lizard (flanked by MUT and RHAG). MUT is only eight genes from CYP46A1 in medaka.
CYP7 is found only in chordates and it may have diverged from a CYP39 precursor. There are, however, CYP7-like genes in fungi (see the electronic supplementary material, figure S23), which raises a question of the origin of the fungal CYP7-like genes. Is this a case of convergent evolution or lateral transfer, or is Clan 7 older than Opisthokonta (fungi + animals)? The evidence for CYP7 being linked to the CYP genesis locus argues for Clan 7 originating with animals and not before. We suggest, therefore, that the fungal Clan 7 genes are probably derived from a lateral transfer from animals to fungi, and specifically to some filamentous fungi.
If CYP7 diverged from CYP39 by segmental duplication only in the chordate lineage, then the gene neighbours of CYP7 should be more like the gene neighbours of CYP39. As mentioned earlier, CYP7 genes are adjacent to HEXB and PIP4K2C, which associate themselves with CYP11 and CYP27 sequences (both mito Clan members). There is no evidence linking the CYP7 and CYP39 neighbourhoods. This would argue against CYP7 deriving from CYP39 during chordate evolution. An older origin for CYP7 would require many gene losses to produce the current picture.
The CYP39A region in lancelet includes the genes OSCP1/c1orf102, CYP39A, NUDC. In human, the OSCP1 gene maps to a region on chr 1 that is a paralogon of the CYP7A1, CYP7B1, CYP11B1, CYP11B2 region of human chr 8. The paralogon relationship shows that CYP7, CYP11 and CYP39 genes were present in a single region in the common ancestor before the 2R WGD event. However, the CYP39 gene is in a different location in human. It should be borne in mind that lancelet has three CYP7 genes. The ancestor certainly had both CYP7 and CYP39 genes, indicating that CYP7 is not the product of the 2R WGD.
The OSCP1 homologue on medaka chr 16 is about 700 kb from a CYP4T12/CYP4T13P gene/pseudogene pair. The only OSCP1 homologue detected in Nematostella is three genes from a CYP3-like gene pair, which would be in Clan 3. This links CYP39 to CYP Clan 3 and CYP Clan 4 in the animal ancestor. Both CYP39 and CYP7 seem to be very old. Both map back to a similar CYP-rich neighbourhood. CYP7 does not appear to be significantly newer than CYP39. This suggests that there was a loss of CYP7 in sponges, cnidarians, protostomes and ambulacrarians (hemichordates + echinoderms). This may change as more genome data become available. The possibility exists that CYP7 and CYP39 are in separate clans, and if so, this would make 12 animal CYP clans.
(l). The CYP Clan 26 and the use of retinoids in development
The CYP26 enzymes hydroxylate retinoic acid and thereby inactivate retinoid signalling, creating sharp retinoid boundaries crucial in the developing hindbrain [86], spinal cord [87] and retina [88]. The CYP Clan 26, which includes CYP16s, is quite old in animals. CYP16s are found in Trichoplax and cnidarian genomes and even among sponge ESTs (EC374157, Oscarella carmela). The CYP16 gene has been lost in mammals and is absent from the zebrafish genome, but found in other fish and many invertebrates (D. Nelson 2010, unpublished data).
Figure 9 shows synteny around the CYP26 and CYP16 genes. The Trichoplax CYP16 gene is six genes from HEXB and is the only Clan 26 member in Trichoplax. As noted earlier, HEXB is adjacent to CYP7 genes in the lancelet and only three genes away from CYP11A1 in medaka (figure 9). Gene order around CYP16 in Trichoplax includes the SLC26A11 orthologue, in the order SLC26A11 CYP16 DCLRE1A ADD2 HEXB. In human, ADD2 is 1.5 Mb from CYP26B1. In lancelet, CYP16B is two genes from DCLRE1A. In lancelet, CYP16B also is 585 kb (10 genes) away from CYP20. The lancelet TIMP1/2 orthologue lies between CYP20 and DCLRE1A, and it is found in the paralogons in figure 7 between CYTH1 and CARD14 on chr 17. TIMP3 is on chr 22 in figure 7. The best hit to the SLC26A11 gene in lancelet is four genes from a CYP2U-like gene in Clan 2. This lancelet region is similar to the single Hox cluster in lancelet and it contains GNA12, ADSL, LFNG, UBE2S and SOX10 (figure 5, lines Y and Z), which is similar to the CYP2W1 and CYP2D6 regions in vertebrates (figure 6c). Furthermore, CYP26A1 is only about 13 genes from the CYP2C cluster in human (approx. 1.6 Mb). Examination of zebrafish chr 17 and zebra finch chr 5 reveals CYP26C1 and CYP46A1 are at opposite ends of a conserved block of genes. The linkages in figure 9 cover sequences in CYP Clans 2, 4, 7, 20, 26, 46 and mito. These associations place CYP26 with other CYP genes discussed earlier, in the CYP genesis locus.
(m). CYP Clan 46
Synteny in the neighbourhood around CYP46 is summarized in figure 10. CYP46 is four genes from SETD3 in human, chicken, anole lizard and frog. SETD3 is only nine genes from CYP26C1 in medaka, two to four genes from AK7 and VRK1 in chicken and six to eight genes distant from AK7 VRK1 in human. As mentioned earlier, a CYP7 gene is adjacent to AK7 in lancelet. VRK1 is only two genes from SETD3 in zebrafish.
Figure 10.
Synteny mapping of the CYP46 clan to CYP7 and CYP26 clans via AK7, to ZDHHC12 via MRPL1 (see figure 2 for ZDHHC12 linkages), to CYP2 and mito clans via STOML1. CYP46 is connected by direct linkages to CYP Clans 2 and 4.
In medaka, the CYP46A1 gene is eight genes away from MUT. MUT is adjacent to the very large CYP2AC subfamily gene cluster in X. tropicalis. MUT is also next to CYP2AC1P in human and in some other vertebrates. In Nematostella, there are two CYP46-related genes. They are separated by an ORC5L homologue. The best hit for ORC5L in M. leidyi (a ctenophore) is 800 bp from a CYP Clan 46 gene. ORCL5 is 4.4 Mb from the CYP3A cluster in human. MRPL1 is adjacent to one of the CYP46 genes in Nematostella. MRPL1 is four genes from ZDHHC12 in Tetraodon nigroviridis (freshwater puffer), and figure 3 shows that ZDHHC12 has many linkages to the 2, 74 and mito Clans. In medaka, CYP11A1 is only 16 kb away from HEXB and ARIH1. HEXB is adjacent to two CYP7 sequences in lancelet, and ARIH1 is next to a CYP Clan 2 member in the coral Acropora. A CYP46 pseudogene, CYP46A-se1[12:13:14], is only six genes from the CYP4ABXZ locus on human chr 1. In sea urchin, the sequences GLEAN3_02660 and 15568 are two adjacent CYP Clan 46 sequences. They are also next to a CYP Clan 2 gene, GLEAN3_02658, on scaffold 59540 (see the electronic supplementary material, figure S25). These associations link CYP Clan 46 with the 2, 4, 7, 26, 74 and mito Clans.
(n). ARF gene linkages to CYPs
ARF-like GTPases apparently shared a close association with CYPs in the animal ancestor. ARF and ARL genes are ancient eukaryotic 21 kDa GTPases [89,90]. ARL genes do not appear to have a syntenous link to CYPs. By contrast, there is a frequent association of CYP genes and their neighbours with ARF genes. Animals have three classes of ARF genes (I, II and III). Phylogenies of ARF proteins predict that the first animals had two ARFs, one class I + II and one class III (also known as ARF6) that gave rise to all animal ARFs [91]. The ARF class (I + II) genes have 12 associations with CYP genes or their close neighbours. These ARF–CYP linkages are described below.
A CYP Clan 46 gene (gw.166.6.1 [Nemve1:11772]) in Nematostella is found only 3.5 kb from an ARF1 homologue. Another ARF1-like sequence in the lancelet is found next to HHIPL1. Human HHIPL1 is adjacent to CYP46A1. This permutation suggests that all three genes were neighbours in a common ancestor. Another ARF1-like gene is only four genes from CYP51 in Trichoplax. An ARF4-related gene in lancelet is adjacent to a Clan 2 member (see the electronic supplementary material, figure S26). In sea urchin, an ARF2-related gene is only two genes away from HOOK1 (see the electronic supplementary material, figure S27); HOOK1 is adjacent to the CYP2J locus in all vertebrates (exemplified in zebrafish [63]). Furthermore, a Clan 3 member is only three genes distant from an ARF1 homologue in the lancelet (electronic supplementary material, figure S28).
ARF1 in medaka is 420 kb from CYP26A1 (figure 5, line M). An ARF4-like gene in medaka is six genes (176 kb) from CYP8B1. An ARF1-like gene in medaka is 333 kb from the HoxC cluster (figure 5, line R). Finally, an ARF homologue in lancelet is found adjacent to the cluster containing CYP7s described earlier (PDE1B HEXB CYP7 CYP7 VSP24 GOT1; electronic supplementary material, figure S21). PDE1B is six genes (270 kb) from CYP20 in chicken, whereas PDE11A is 2 Mb distant (figure 5, line W). These associations link ARF genes to Clans 2, 3, 7, mito, 20, 26 and 46, and to the Hox clusters.
The ARF6 connections with CYP genes are less abundant. Because class I + II ARFs and class III ARFs (ARF6) are both found in fungi, it seems that only one of these ARF classes could logically be associated with the P450 genesis locus. Ciona has an ARF6 homologue three genes from a CYP Clan 2 member (electronic supplementary material, figure S30). The ARF6 orthologue in lancelet (97% identical to human ARF6) is adjacent to PDE11A1, which as stated above is close to CYP20 in chicken. In medaka, PDE11A1 is 3 Mb from CYP11B (figure 5, lines W and E). Trichoplax has an ARF6-like gene that is 11 genes (320 kb) from a Clan 4 member. An indirect linkage in Rhesus macaque has ARF6 next to a PDLIM1 pseudogene (electronic supplementary material, figure S31). As described earlier, PDLIM1 is near the CYP2C genes, whereas PDLIM3 is near CYP4V2 orthologues (figure 5, lines H–L). A similar PDLIM pseudogene is found in the same place in human. Because this is a pseudogene, and it does not appear to be on a paralogon of PDLM1 or PDLIM2 regions, the linkage to ARF6 could be accidental. Another example of a spurious link between ARF6 and CYP genes is found near the CYP3 cluster in human. Thus, CYP3 is adjacent to a zinc finger protein ZNF498 in many eutherian mammals but not in marsupials, or in other vertebrates. Humans have an ARF6 pseudogene between CYP3A5 and ZNF498. Although this is a very tight association between ARF6 and CYP3 genes, the pseudogene is found only in great apes: human, chimp and orangutan, not in mouse, rat, dog, cow or horse. This had to be a recent insertion, so it is not relevant to early CYP neighbourhoods. Furthermore, the CYP3A cluster in human has moved from its original location close to the CYP2W1 gene as described earlier. Thus, in some rare cases, linkages of ARF6 genes with CYP genes appear to be chance occurrences, but this does not diminish the strength of most ARF–CYP linkages as evolutionarily conserved.
(o). CYP19
The origin of CYP19 (aromatase) remains a mystery, as CYP19 sequences are distinct from other CYP clans and offer no clues to the parent origin. Aromatase synthesizes oestrogen from testosterone via a three-step demethylation [92]. This process is at the end of a long steroid pathway initiated with the CYP51-mediated synthesis of cholesterol that also includes CYP11A1 for side-chain cleavage and CYP17 for 17/20 lyase and hydroxylase activities [93].
A gene duplication to create CYP19 would have had to precede lancelet, which has CYP19. This would be earlier than the 2R WGDs, although, some sequence relatedness would be expected between CYP19 and its parent clan if the duplication were early in chordate evolution. It seems unlikely that a duplication within deuterostomes or even bilaterians would create a new CYP clan, because the 2R WGDs (more than 525 Ma) failed to create even a new family (except apparently CYP8). Therefore, it is plausible that CYP19 was an original member of the CYP genesis locus and it was lost in many lineages just like Clan 7. A mechanism to explain multiple CYP clan losses may require that the lost genes were adjacent in a tight cluster. For example, insects, crustaceans and C. elegans, in the Ecdysozoa, all lack the same seven CYP clans. Possibly, these genes were adjacent and lost together in a block, a difficult hypothesis to test.
The lancelet has three different CYP19 sequences in two different subfamilies (CYP19B1, CYP19C1 and CYP19C2), the subfamilies are 41 per cent identical to each other and both are approximately 40 per cent identical to human CYP19A1. Mizuta & Kubokawa [94] cloned a CYP19C3 orthologue from ovaries of the related species Branchiostoma belcheri, but the gene was not heterologously expressed and assayed. Radioimmunoassay of ovarian steroids did show oestradiol was being synthesized, but which of the CYP19 genes was responsible is not known. Comparing the CYP19 location in vertebrates and the lancelet shows that they are different, suggesting that CYP19 has moved to a new location in vertebrates, which would not be relevant to connecting CYP19 to the CYP genesis locus. This possibility was a concern of Castro et al. [95] who examined the synteny around the CYP19 genes in vertebrates, but they did not have the lancelet synteny data. The lancelet has kept the original location of CYP19, and the genes surrounding it are informative.
The lancelet region in the electronic supplementary material, figure S32 contains CYP19B1v1 with an adjacent pseudogene, in a region that also has two TGFB-like genes. The orthologue of one gene (TGFB1) is found four genes from the CYP2ABFGST cluster in human. The lancelet region also has TTLL5 that is adjacent to TGFB3 in human. TGFB3 is not near a CYP, but it is on a paralogon to the CYP2ABFGST cluster. TGFB2 in human is also on a third paralogon to the CYP2ABFGST cluster. TGFB2 in lancelet is next to CYP4T1.
The lancelet CYP19B1v1 gene is adjacent to a NOP14 gene, and to EXOC6 on the other side. In medaka, there is only one gene similar to NOP14, located four genes from RAD23B. In Ciona, EXOC6 is about 300 kb from a RAD23-like gene on chr 8. A RAD23-like gene occurs in the CYP genesis locus often near PRDX genes (see §3h). In human, the NOP14 region on chr 4p is a paralogon of the CYP26B- containing segment on human chr 2 and the CYP26A and CYP26C segment on human chr 10. EXOC6 is also adjacent to the CYP26C1–CYP26A1 gene pair in human, chicken, lizard and frog. CYP26A1 is next to EXOC6 in medaka (figure 5, line M), but CYP26C1 has moved to a new location. The presence of genes in line M that are also found in the CYP2W1/CYP3A paralogon ties CYP26 and CYP19 to the CYP2 and CYP3 Clans. After the 2R WGD events, CYP26B was duplicated to form a new CYP26A/C precursor (table 1). The gene EXOC6 was also duplicated next to it and is now EXOC6B. In zebrafish, EXOC6B is adjacent to CYP26B (chr 7 approx. 16.23 Mb; though only the N-terminal of the gene is present in the UCSC browser danRer5 assembly). EXOC6B is adjacent to CYP26B1 in frog, chicken and human. These synteny relationships are indicators that CYP19 and CYP26 were close neighbours in the past.
Ciona intestinalis has EXOC6 adjacent to COG3 on chr 8. COG3 in lizard is next to the SORBS1 PDLIM1 HELLS trio (figure 5, line L). These genes are adjacent to CYP2C in human and chicken (figure 5, lines J, K), but the lizard contig ends here. COG3 matches a gene in lancelet that is six genes from a CYP cluster containing CYP3 and CYP2 sequences (see the electronic supplementary material, figure S6). Further, synteny evidence links CYP19 in lancelet to CYP2U1 in human (see the electronic supplementary material, figure S37). The CYP2U1 gene on human chr 1 is flanked by PAPSS1 on one side and HADHSC and LEF1 on the other side. These same three genes flank the CYP19C genes in lancelet (see the electronic supplementary material, figure S37). These observations link CYP19 to the CYP Clans 2 and 3.
Additional support is given by the gene LCT, which is adjacent to NOP14 in the lancelet. In medaka, this gene is situated between FAIM on one side and HDAC9 TWIST1 on the other side (see the electronic supplementary material, figure S33, chr 21). FAIM is found in figure 5, line Y next to the lancelet Hox cluster. HDAC9 and TWIST1 are seen in figure 5, lines B and D–F near CYP51, the HoxA cluster in chicken and medaka and the CYP11B gene in medaka. These genes are part of the CYP2W1 paralogon on human chr 7.
The gene EML1 is another neighbour of the CYP19B1v1 gene in lancelet (see the electronic supplementary material, figure S32). EML1 is adjacent to CYP46 in human. There is one other location for the CYP19B1v2 gene in lancelet. However, the CYP19B1v1 and v2 genes are 99 per cent identical and probably represent alleles in the genome assembly. Both have tail-to-tail NOP14 neighbours. Other neighbouring genes differ, but there are gaps in the assembly so some genes may be missing. CYP19B1v2 has NOP14 on one side and ABCC4/CFTR on the other side. ABCC4/CFTR is one gene from WNT2 in human. The related gene ABCC3 is also found in the CYP2W1 paralogon of chicken. These observations have placed the CYP19 ancestor into the same region as precursors of the 2, 3, 4, 26, 46, 51 and mito Clans.
(p). CYP20
The genes flanking CYP20 in lancelet are RPL27, AAAS and SLC11A2 (see the electronic supplementary material, figure S34). AAAS is 800 kb from the HoxC cluster in X. tropicalis and 700 kb from the chr 12 HoxC cluster in human. CYP27B1 is on the opposite side of this HoxC cluster in human (figure 5, line T). The SLC11A2 gene is 900 kb from CYP27B1 on medaka chr 5, but it is only five genes (250 kb) from CYP27B1 in X. tropicalis. Opossum has SLC11A1 on chr 7 380 kb from CYP27A1. In human, SLC11A1 is on chr 2q in the same chromosomal region as CYP27A (385 kb away) and CYP20 (15.1 Mb away). It appears that CYP27, CYP20 and SLC11A were duplicated during the 2R WGD, and CYP27B became linked to SLC11A2, as seen in medaka and X. tropicalis, while CYP27A is linked to SLC11A1. Apparently, CYP20 moved away from the original site and lost the duplicate copy near CYP27B. RPL27 is two genes from RPL3 in zebrafish and it is four genes from a RAC paralogue. Both RPL3 and RAC2 are found in the paralogon containing CYP2D6 (figure 7, chr 22). The human chr 7 paralogon includes RAC1, CYP2W1 and the original vertebrate CYP3 locus (figure 7). PDE1A is 300 kb from CYP20 in chicken. PDE1C is 870 kb from CYP8A1 in stickleback, and PDE1B is 3.3 Mb from CYP27B1 in human. The PDE1A,B,C genes in human are on paralogons containing the Hox D, C and A clusters, respectively [23].
More ancient neighbours are found in Nematostella. There are two adjacent CYP20s in Nematostella. The best match to the right-hand-side neighbour (fgenesh1_pm.scaffold_58000009 [Nemve1:229175]) is MCM9 in lancelet. Surprisingly, this is adjacent to a CYP2 gene cluster with nine members (see the electronic supplementary material, figure S35). The gene on the other side of CYP20 in Nematostella matches with RAB18. RAB18 is 370 kb from a four gene CYP20 cluster in lancelet (see the electronic supplementary material, figure S34). RAB18 is two genes from ABI1 in medaka, and ABI2 is adjacent to CYP20 in human, and in opossum, lizard, frog and medaka. The medaka RAB18 is approximately 700 kb from PDRX6 and FGGY. FGGY is two genes from CYP2J2 in human. ABI was duplicated in the 2R WGD events and CYP20 followed ABI2. This shows that RAB18 to ABI1/2 tracks the CYP20 trajectory from Nematostella to human. RAB18 is close to ARMC4 in medaka, chicken, opossum and human. In Ciona, ARMC4 is only 90 kb from a pair of CYP4 genes on chr 9. CYP20 is 1.6 Mb from DICER1 in medaka. DICER1 is three genes from CYP26C1 in zebrafish (figure 9). CYP20 is 685 kb from CHD7 in fugu. CHD7 has links to CYP7 in vertebrates (electronic supplementary material, figure S22) and to Clan 2 genes in lancelet. Through these interactions, the CYP20 gene is also associated with the 2, 3, 4, 7, 26 and mito Clan neighbourhoods.
4. Discussion
The current abundance of animal genomes and the concept of CYP clans, inferred from deep branching in molecular phylogenies [4,96], have made it possible to trace linkages among the CYP families and clans, through analysis of syntenic relationships. The nature of relationships among the CYP clans has been difficult to determine from phylogenetic trees, except for Clans 3 and 4, which consistently cluster together, suggesting that they shared a common ancestor. CYP Clan 3 and Clan 4 members are present in cnidarians and even in sponges (figure 4), thus the divergence of these two clans occurred very early in animal evolution. In fact, figure 4 shows all the clans except Clan 19 have sequence evidence indicating that they were present at least by the origin of cnidarians. In most cases, however, the percent identity of sequences in different clans is in the low 20 per cent range, and thus comparison of the CYP sequences alone does not permit further insights into evolutionary relationships.
The examination of synteny allows new relationships to be detected based on shared neighbours. Applying this approach to members of all 11 CYP clans uncovered heretofore unknown linkages among CYP clans. Some clans have more detectable linkages than others, but in the end all 11 clans can be tied to each other through shared linkages. As an illustration, CYP20 is not linked to CYP7, but it is linked to CYP27 in the mito Clan. CYP11 in the mito Clan is linked to CYP7, so CYP7 is linked indirectly to CYP20. Our analysis indicates that all the CYP clans shared one common neighbourhood, which we call the cytochrome P450 genesis locus. This has never been hinted at before and it is a novel result of the synteny analysis.
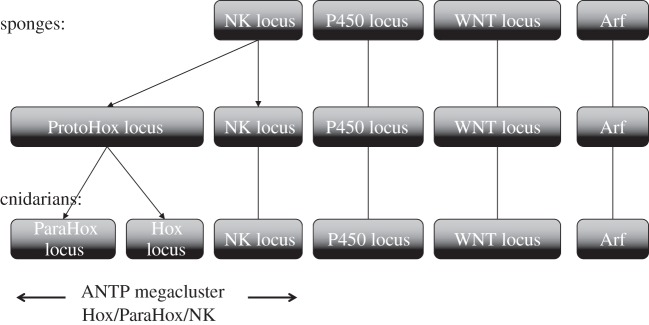
Some of the genes in this neighbourhood that we call the CYP genesis locus are quite well known, including the Hox gene cluster and several WNT genes. The proximity of the WNT genes to the Hox genes has been noted before [23]. Other genes also occur close to the genesis locus. Thus, an ANTP megacluster consisting of Hox, ParaHox and NK genes has been proposed [97]. The sponge genome still maintains an NK gene cluster with six genes [98], but there are no Hox or ParaHox genes in this genome. Although not described in §3, further linkage of the sponge NK genes to CYPs is provided by the KIF11 gene that is adjacent to the NK5/6/7b gene in sponges (ACUQ01000781) and is also adjacent to CYP26C1 in zebrafish (figure 5, line P) and very close to CYP46 in lancelet (figure 9). Because there are CYP genes in the A. queenslandica sponge genome but no Hox genes, the CYP gene cluster appears to predate the Hox gene cluster that seemingly arose from an NK gene. Other sponge genomes may hold additional clues to the genesis locus gene environment.
A CYP3 Clan member (XM_002107799.1, 39% identical to CYP3A4 human) is 10 genes from an NK2 gene cluster in Trichoplax (from 4.32 to 4.36 Mb on scaffold 1), suggesting the ancestral CYP gene cluster was linked to the NK gene cluster.
Together, the WNT, Hox, NK and CYP genes represent a developmental locus in early animals (figure 11). An NK-like homeobox domain is found approximately 14 kb from a CYP46 Clan member in the ctenophore M. leidyi. EXOC6 is 13 genes from another Trichoplax NK cluster containing NK5, NK6 and Hex. EXOC6 has been linked to CYP19 and CYP26. The forces acting to keep the NK genes, the Hox genes, and WNT genes together may also have kept this block of CYP genes from dispersing after their formation through cis duplication. Studies on fish ParaHox gene clusters support the existence of interdigitated control regions in the clusters that select against their loss [99]. Irimia et al. proposed the existence of Genomic Regulatory Blocks, with transcriptional enhancers for developmental genes being embedded in neighbouring genes, thus keeping them together [51]. Furthermore, the gene functions may interact. For example, retinoic acid regulates Hox gene expression, and CYP26 is a retinoic acid hydroxylation enzyme that controls retinoic acid concentration. Other CYP substrates/products may influence Hox, ParaHox, NK or Wnt gene expression.
Figure 11.
The P450 genesis locus in animals.
Consequences of gene clustering may also involve gene losses. The loss of seven CYP clans in insects, crustaceans and nematodes (all ecdysozoans) may have occurred by a block deletion of the clustered CYP genes. Such a major loss of CYP genes all at once could be expected to have profound effects on ecdysozoan development. It has been known since the time of Geoffroy de St Hilaire that the arthropod dorsoventral axis is inverted compared with vertebrates, indicating a major developmental shift in this group [100–102]. Could it be related to the simultaneous loss of so many CYP genes?
The fact that all CYP clans can be traced back to this neighbourhood has other implications as well. The CYP7, 19 and 74 Clans appear to originate in the genesis locus and not from outside it. Otherwise, it would not be possible to find linkages to the locus. This is illustrated by vertebrate CYP19, which apparently moved to a new location from that seen in the lancelet. The vertebrate location of CYP19 is unrelated to the genesis locus and cannot be traced back to it. If the CYP19 Clan existed in the genesis locus as predicted, its function would probably not be the same as modern CYP19 aromatase enzymes. The novel aromatase biochemistry of CYP19 emerged in chordates, as is seen in the lancelet. Evidence for CYP11 and CYP19 sequences in lancelet was presented earlier, and steroid nuclear receptors are not found before the chordates [93].
An animal origin for CYP74 is surprising, since the first CYP74 genes all were found in land plants, however, CYP74s are not found in algae or fungi. The dating of early metazoan divergences is a controversial subject; for this discussion, we use dates taken from Berney & Pawlowski11 [103] and from Blair [41] and Hedges et al. [42]. The earliest appearance of the CYP74 Clan is in Trichoplax, thought to have arisen between 733 and 592 Ma. This predates the appearance of land plant fossil spores at 475 Ma [104,105] by approximately 117–258 Myr. How did a CYP74 gene make the transition from marine animals to land plants? Presumably a vector would be required, such as a bacterium that could obtain the CYP74 gene from marine animals. Indeed, two CYP74-like genes have been identified in bacteria (see [14] electronic supplementary material, figure S18). These are Methylobacterium nodulans and Methylobacterium sp. 4–46, plant symbiotic bacteria involved in nitrogen fixation in legumes, ideal candidates for gene transfer to plants. Lateral gene transfer to early land plants is known to have occurred for T globins [106] and for glutamine synthase IIB [107]. Two additional examples that are involved in CYP pathways are PAL and 4CL. The first gene in the general phenylpropanoid pathway of plants, phenylalanine ammonia lyase (PAL) was acquired by a horizontal gene transfer from soil bacteria early during land colonization [108]. The gene 4-coumarate coenzyme A ligase (4CL), found next to CYP Clan 3 genes in lancelet and Trichoplax, is the third gene of the plant phenylpropanoid pathway [109], acting after CYP73A1, a CYP that encodes cinnamate 4-hydroxylase [110]. The 4CL genes were thought to be plant-specific rather like CYP74, but as mentioned above they exist in the lancelet, Trichoplax and Nematostella, as does CYP74 [14]. Analysis of the purple sea urchin detected two more of these 4CL genes [111]. A bacterial version of 4CL has been found in the soil bacterium Streptomyces coelicolor [112]. This raises the possibility that 4CL may have a similar history to PAL and CYP74.
CYP7 and CYP19 are chordate-specific. CYP7 and CYP39 are currently placed in the same clan, but CYP39 sequences are detected in the choanoflagellate M. brevicollis, sponges and Trichoplax, while CYP7 is not seen until lancelet. Possibly, CYP7 and CYP39 should be placed in different clans; the relationship between them is fairly weak. This would allow CYP7 to be newly evolved. However, CYP Clan 7 members can be linked to the genesis locus, suggesting they are in fact very old as well. A more recently derived chordate-specific CYP Clan 7 gene such as CYP8 should show a strong resemblance to another Clan 7 CYP, and it does. This argues against a de novo appearance of CYP 7 only in chordates. Instead, we suggest there have been gene losses from Clan 7 in pre-chordate animals. A similar argument applies to CYP19.
The analysis here suggests that early appearance of CYP clans could have resulted from tandem duplication of a progenitor CYP. The time window of animal origins appears to be fairly limited, perhaps 632–863 Myr [103–114]. Thus, establishing the order in which CYP clans arose via tandem duplication of an initial CYP gene may be difficult. A Clan 4 member appears in M. brevicollis, though this may be the precursor of the Clans 3 and 4. A CYP39-related gene (7 Clan) is seen in M. brevicollis, so the Clans 4 and 7 may have been very early members in the genesis locus. Monosiga brevicollis also has several CYPs most like plant CYP710/CYP61, CYP704, CYP711 and CYP745, gene families that we presume were lost on the way to animals. The ctenophore M. leidyi has the first known 20 and 46 Clan members. The placement of sponges and placozoans in evolution is not certain, but we assume in figure 4 that sponges were earlier (see [35]). Sponges have the first observed 26 and mito Clan members. Trichoplax has the first Clan 2 and Clan 74 members. All clans except 19 predate the origin of Cnidaria. The CYP Clan 19 has no sequence evidence until lancelet, but for reasons mentioned earlier, it must have a much older origin because of linkage with many other CYP clans in addition to its very divergent sequence. Gene loss has certainly occurred in some lineages and this may confuse the interpretation of first appearances. Additional genomes, especially from sponges, may help to more precisely clarify the origin of some CYP clans/families such as CYP2, CYP7, CYP19, CYP46 and CYP74.
Earlier attempts to discern the evolution of animal CYPs strongly suggested that all animal CYPs derived from a single CYP51 gene, based largely on the presence of a CYP51 in bacteria, plants, fungi and animals [11,18,115,116]. It has been argued that the bacterial CYP51 may have arisen from a lateral transfer from plants, but this does not alter our discussion here on animal P450s [19]. One could envision that, as a source of sterols important in cell membrane function, CYP51 could be an essential early enzyme. On the other hand, the catalytic function of extant CYP51s is sufficiently specialized that one could also argue that some other less specialized function may have characterized the progenitor CYP. Identifying a precursor to the progenitor CYP would help to determine its role and identity. Regardless of the identity of the CYP occupying the initial locus, the recognition of the CYP genesis locus as the starting point for animal CYP evolution opens the way for more detailed analysis of each clan to understand the origin of specific families. As more genome assemblies become available, reconstruction of ancestral genomes becomes possible as already achieved with the CLGs. The history of each CYP is really a vector through time that tracks gene duplications and losses, lateral transfers, WGDs and chromosomal rearrangements. Armed with more and more detailed genome histories, we will be better able to trace individual gene histories. The limiting factor becomes what time has erased, leaving no trace for us to read.
5. Definitions
- Microsynteny:
The preservation of gene order on a nearly 1 : 1 basis without large insertions, gaps or rearrangements.
- Macrosynteny:
The retention of orthologues in large chromosomal regions at higher than statistically expected levels. These genes are not necessarily close together and they may be spread over large regions, as in electronic supplementary material, figure S36 where 10 orthologue/co-orthologue pairs are spread over a 21 Mb region.
- Co-orthologues:
Tandemly duplicated genes that arose from a single orthologous ancestral gene. The current loci in two different species may not contain the same number of members, but each is a co-orthologue of the other species orthologous gene(s). Example: the CYP4T gene in fish and CYP4A, B, X and Z genes in human.
- Paralogons:
Multiple regions created by whole genome duplication (WGD). The two WGD events in chordates resulted in four paralogons for each original segment in the pre-duplicated protochordate genome. Originally, these were equivalent to whole duplicated chromosomes, but over time they become smaller segments owing to chromosomal rearrangements.
- Ohnologues:
Genes duplicated in a WGD event that survive to the present day. Most ohnologue-duplicated genes get lost as the tetraploid genome reduces the number of genes back down close to the original diploid number. Some are retained and acquire new functions.
- CYP clan:
A CYP clan is a clade of genes. Clans are relatively deep branchings on phylogenetic trees. There are eleven total clans of animal CYPs but only four clans in most arthropods and nematodes and five in ticks. Plants have a different set of CYP clans, except for CYP51 and CYP74 that are in common.
- Unikonts:
A proposed eukaryotic taxonomic group defined by the presence of only one flagellum; includes opsithokonts and amoebozoas.
- Bikonts:
A eukaryotic taxonomic group defined by cells with two emergent flagella; includes Archaeplastida (plants and relatives), Excavata, Rhizaria and Chromalveolata.
- Excavates (Excavata):
A deep branch of eukaryotes containing many parasitic, often ‘amitochondrial’ species; includes Giardia, Trypanosoma, Euglena, Trichomonas.
Acknowledgements
This work was supported in part by NSF grant no. IOS-1242275 to D.R.N. This study was supported in part by a NOAA Sea grant no. NA10OAR4170086 (to J.J.S. and J.V.G.), NIH grant nos. 5R01-ES015912 (J.J.S.) and F32-ES012794 (J.V.G.), a Superfund Basic Research Center grant no. 2P42ES07381 (J.J.S.), and by the Ocean Life and Deep Ocean Exploration Institutes of the Woods Hole Oceanographic Institution. The authors have no competing interests to declare.
Endnotes
(GenBank BI386673 dated 20 May 2003). This sequence was posted to the Cytochrome P450 Homepage in 2005, though it was not recognized as a CYP74 Clan member at that time.
Unikonts, bikonts and excavates are three monophyletic divisions of eukaryotes determined by multi-gene tree building methods, using large concatenated sequence alignments [15]. Morphologically unikonts (animals, fungi, Amoebozoa) and bikonts (plants, alveolates, stramenopiles, Rhizaria) have one or two flagella at a specific point in their life cycle. Excavates have a characteristic ventral feeding groove on their cell surface (examples are Jakobids, Euglena, Giardia, trypanosomes). Cavalier–Smith argues that excavates are bikonts [16].
Xenambulacraria = hemichordates+echinoderms+xenoturbellarids.
The 236 P450 gene count for Branchiostoma (lancelet) seems high compared to other chordates. The paper of Putnam et al. [23] argues for a fairly strict paralogon relationship to vertebrates, so the vertebrate P450s should have an ohnolog precursor in lancelet. A large expansion is not expected. Part of the increase may be due to alleles being counted as different genes. The P450s of lancelet have not been systematically named yet so this possibility cannot be confirmed now. Another possibility is dramatic gene blooms in some clans. Note in figure 2 the very large sector in the CYP2 clan between 08.00 and 09.00 o'clock that looks like a large gene bloom. Blooms of P450s have been discussed by Sezutsu et al. [10].
RPL28 occurs in two different genomic locations in lancelet shown in figure 3.
Fungi have about 15–20 clans that are not completely defined yet. CYP51 and CYP7 Clan members are seen in fungi. The distribution of 7 Clan members is only found in some filamentous fungi strongly suggesting a lateral transfer from animals to fungi. Therefore, it is shown as an asterisk in figure 4. The other fungal clans are distinct from the animal clans.
(fgenesh1_pg.scaffold_275000001).
Fugu Ensembl:ENSTRUG00000004549, zebrafish Ensembl:ENSDARG00000060094.
Dates for early animal divergence nodes: choanoflagellates versus all other animals 863 Ma (711–1052); sponges versus ctenophores, Trichoplax, cnidarians and bilaterians 812 Ma (671–985); ctenophores versus Trichoplax, cnidarians and bilaterians 733 Ma (603–893); Trichoplax versus cnidarians and bilaterians 592 Ma (551–696).
References
- 1.Wisedpanichkij R, Grams R, Chaijaroenkul W, Na-Bangchang K. 2011. Confutation of the existence of sequence-conserved cytochrome P450 enzymes in Plasmodium falciparum. Acta Trop. 119, 19–22 10.1016/j.actatropica.2011.03.006 (doi:10.1016/j.actatropica.2011.03.006). [DOI] [PubMed] [Google Scholar]
- 2.The tomato genome consortium 2012. The genomes of tomato and its closest wild relative provide insights on evolution of plant genomes. Nature 485, 635–641 10.1038/nature11119 (doi:10.1038/nature11119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young ND, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524 10.1038/nature10625 (doi:10.1038/nature10625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson DR, et al. 1996. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6, 1–42 10.1097/00008571-199602000-00002 (doi:10.1097/00008571-199602000-00002) [DOI] [PubMed] [Google Scholar]
- 5.Goldstone HM, Stegeman JJ. 2006. A revised evolutionary history of the CYP1A subfamily: gene duplication, gene conversion, and positive selection. J. Mol. Evol. 62, 708–717 10.1007/s00239-005-0134-z (doi:10.1007/s00239-005-0134-z) [DOI] [PubMed] [Google Scholar]
- 6.Cohen MB, Feyereisen R. 1995. A cluster of cytochrome P450 genes of the CYP6 family in the house fly. DNA Cell Biol. 14, 73–82 10.1089/dna.1995.14.73 (doi:10.1089/dna.1995.14.73) [DOI] [PubMed] [Google Scholar]
- 7.Heim MH, Meyer UA. 1992. Evolution of a highly polymorphic human cytochrome P450 gene cluster: CYP2D6. Genomics 14, 49–58 10.1016/S0888-7543(05)80282-4 (doi:10.1016/S0888-7543(05)80282-4) [DOI] [PubMed] [Google Scholar]
- 8.Kubota A, Stegeman JJ, Goldstone JV, Nelson DR, Kim EY, Tanabe S, Iwata H. 2011. Cytochrome P450 CYP2 genes in the common cormorant: evolutionary relationships with 130 diapsid CYP2 clan sequences and chemical effects on their expression. Comp. Biochem. Physiol. Toxicol. Pharmacol. 153, 280–289 10.1016/j.cbpc.2010.11.006 (doi:10.1016/j.cbpc.2010.11.006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. 2004. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14, 1–18 10.1097/00008571-200401000-00001 (doi:10.1097/00008571-200401000-00001) [DOI] [PubMed] [Google Scholar]
- 10.Sezutsu H, Le Goff G, Feyereisen R. 2013. Origins of P450 diversity. Phil. Trans. R. Soc. B 368, 20120428. 10.1098/rstb.2012.0428 (doi:10.1098/rstb.2012.0428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson DR. 1999. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 369, 1–10 10.1006/abbi.1999.1352 (doi:10.1006/abbi.1999.1352) [DOI] [PubMed] [Google Scholar]
- 12.Nelson DR. 1998. Metazoan cytochrome P450 evolution. Comp. Biochem. Physiol. 121, 15–22 [DOI] [PubMed] [Google Scholar]
- 13.Nelson DR. 2003. Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Arch. Biochem. Biophys. 409, 18–24 10.1016/S0003-9861(02)00553-2 (doi:10.1016/S0003-9861(02)00553-2) [DOI] [PubMed] [Google Scholar]
- 14.Lee DS, Nioche P, Hamberg M, Raman CS. 2008. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 455, 363–368 10.1038/nature07307 (doi:10.1038/nature07307) [DOI] [PubMed] [Google Scholar]
- 15.Burki F, Shalchian-Tabrizi K, Pawlowski J. 2008. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol. Lett. 4, 366–369 10.1098/rsbl.2008.0224 (doi:10.1098/rsbl.2008.0224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalier-Smith T. 2006. Cell evolution and Earth history: stasis and revolution. Phil. Trans. R. Soc. B 361, 969–1006 10.1098/rstb.2006.1842 (doi:10.1098/rstb.2006.1842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derelle R, Lang BF. 2012. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol. Biol. Evol. 29, 1277–1289 10.1093/molbev/msr295 (doi:10.1093/molbev/msr295) [DOI] [PubMed] [Google Scholar]
- 18.Yoshida Y, Aoyama Y, Noshiro M, Gotoh O. 2000. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem. Biophys. Res. Commun. 273, 799–804 10.1006/bbrc.2000.3030 (doi:10.1006/bbrc.2000.3030) [DOI] [PubMed] [Google Scholar]
- 19.Rezen T, Debeljak N, Kordis D, Rozman D. 2004. New aspects on lanosterol 14alpha-demethylase and cytochrome P450 evolution: lanosterol/cycloartenol diversification and lateral transfer. J. Mol. Evol. 59, 51–58 10.1007/s00239-004-2603-1 (doi:10.1007/s00239-004-2603-1) [DOI] [PubMed] [Google Scholar]
- 20.Cavalier-Smith T. 2002. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 52, 7–76 [DOI] [PubMed] [Google Scholar]
- 21.Catchen JM, Conery JS, Postlethwait JH. 2009. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 19, 1497–1505 10.1101/gr.090480.108 (doi:10.1101/gr.090480.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Housworth EA, Postlethwait J. 2002. Measures of synteny conservation between species pairs. Genetics 162, 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putnam NH, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 10.1038/nature06967 (doi:10.1038/nature06967) [DOI] [PubMed] [Google Scholar]
- 24.Verslycke T, Goldstone JV, Stegeman JJ. 2006. Isolation and phylogeny of novel cytochrome P450 genes from tunicates (Ciona spp.): a CYP3 line in early deuterostomes? Mol. Phylogenet. Evol. 40, 760–771 10.1016/j.ympev.2006.04.017 (doi:10.1016/j.ympev.2006.04.017) [DOI] [PubMed] [Google Scholar]
- 25.Ohno S. 1970. Evolution by gene duplication. Berlin, Germany: Springer [Google Scholar]
- 26.Wolfe K. 2000. Robustness: it's not where you think it is. Nat. Genet. 25, 3–4 10.1038/75560 (doi:10.1038/75560) [DOI] [PubMed] [Google Scholar]
- 27.Postlethwait JH. 2007. The zebrafish genome in context: ohnologs gone missing. J. Exp. Zool. B Mol. Dev. Evol. 308, 563–577 10.1002/jez.b.21137 (doi:10.1002/jez.b.21137) [DOI] [PubMed] [Google Scholar]
- 28.Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. 2004. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 20, 481–490 10.1016/j.tig.2004.08.001 (doi:10.1016/j.tig.2004.08.001) [DOI] [PubMed] [Google Scholar]
- 29.Dehal P, Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3, e314. 10.1371/journal.pbio.0030314 (doi:10.1371/journal.pbio.0030314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer A, Van de Peer Y. 2005. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27, 937–945 10.1002/bies.20293 (doi:10.1002/bies.20293) [DOI] [PubMed] [Google Scholar]
- 31.Siegel N, Hoegg S, Salzburger W, Braasch I, Meyer A. 2007. Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications. BMC Genomics 8, 312. 10.1186/1471-2164-8-312 (doi:10.1186/1471-2164-8-312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuraku S. 2008. Insights into cyclostome phylogenomics: pre-2R or post-2R. Zool. Sci. 25, 960–968 10.2108/zsj.25.960 (doi:10.2108/zsj.25.960) [DOI] [PubMed] [Google Scholar]
- 33.Kuraku S, Meyer A, Kuratani S. 2009. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol. Biol. Evol. 26, 47–59 10.1093/molbev/msn222 (doi:10.1093/molbev/msn222) [DOI] [PubMed] [Google Scholar]
- 34.Hejnol A, et al. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. B 276, 4261–4270 10.1098/rspb.2009.0896 (doi:10.1098/rspb.2009.0896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirischian NL, Wilson JY. 2012. Phylogenetic and functional analyses of the cytochrome P450 family 4. Mol. Phylogenet. Evol. 62, 458–471 10.1016/j.ympev.2011.10.016 (doi:10.1016/j.ympev.2011.10.016) [DOI] [PubMed] [Google Scholar]
- 36.Edgecombe GD, Giribet G, Dunn CW, Hejnol A, Kristensen RM, Neves RC, Rouse GW, Worsaae K, Sorensen MV. 2011. Higher-level metazoan relationships: recent progress and remaining questions. Org. Divers. Evol. 11, 151–172 10.1007/s13127-011-0044-4 (doi:10.1007/s13127-011-0044-4) [DOI] [Google Scholar]
- 37.Xian-Guang H, Aldridge RJ, Siveter DJ, Siveter DJ, Xiang-hong F. 2002. New evidence on the anatomy and phylogeny of the earliest vertebrates. Proc. R. Soc. Lond. B 269, 1865–1869 10.1098/rspb.2002.2104 (doi:10.1098/rspb.2002.2104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xian-Guang H, Aldridge RJ, Bergström J, Siveter DJ, Siveter DJ, Xiang-hong F. 2003. The Cambrian fossils of Chengjiang, China: the flowering of early animal life. London, UK: Blackwell Publishing Ltd [Google Scholar]
- 39.Zhu M, Zhao W, Jia L, Lu J, Qiao T, Qu Q. 2009. The oldest articulated osteichthyan reveals mosaic gnathostome characters. Nature 458, 469–474 10.1038/nature07855 (doi:10.1038/nature07855) [DOI] [PubMed] [Google Scholar]
- 40.Sansom IJ, Smith MM, Smith MP. 1996. Scales of thelodont and shark-like fishes from the Ordovician of Colorado. Nature 379, 628–630 10.1038/379628a0 (doi:10.1038/379628a0) [DOI] [Google Scholar]
- 41.Blair JE. 2009. Animals (Metazoa) In The timetree of life (eds Hedges SB, Kumar S.), pp. 223–230 Oxford, UK: Oxford University Press [Google Scholar]
- 42.Hedges SB, Blair JE, Venturi ML, Shoe JL. 2004. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 4, 2. 10.1186/1471-2148-4-2 (doi:10.1186/1471-2148-4-2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffman PF, Kaufman AJ, Halverson GP, Schrag DP. 1998. A neoproterozoic snowball earth. Science 281, 1342–1346 10.1126/science.281.5381.1342 (doi:10.1126/science.281.5381.1342) [DOI] [PubMed] [Google Scholar]
- 44.Hoffman PF. 2008. Snowball Earth: status and new developments. GEO (IGC Special Climate Issue) 11, 44–46 [Google Scholar]
- 45.Coulier F, Popovici C, Villet R, Birnbaum D. 2000. MetaHox gene clusters. J. Exp. Zool. 288, 345–351 (doi:10.1002/1097-010X(20001215)288:4<345::AID-JEZ7>3.0.CO;2-Y) [DOI] [PubMed] [Google Scholar]
- 46.Ryan JF, Pang K, Mullikin JC, Martindale MQ, Baxevanis AD. 2010. The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa. Evodevo 1, 9. 10.1186/2041-9139-1-9 (doi:10.1186/2041-9139-1-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pett W, Ryan JF, Pang K, Mullikin JC, Martindale MQ, Baxevanis AD, Lavrov DV. 2011. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: insight from mtDNA and the nuclear genome. Mitochondrial DNA 22, 130–142 10.3109/19401736.2011.624611 (doi:10.3109/19401736.2011.624611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn CW, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749 10.1038/nature06614 (doi:10.1038/nature06614) [DOI] [PubMed] [Google Scholar]
- 49.Woods IG, et al. 2005. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 15, 1307–1314 10.1101/gr.4134305 (doi:10.1101/gr.4134305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kevei Z, Seres A, Kereszt A, Kalo P, Kiss P, Toth G, Endre G, Kiss GB. 2005. Significant microsynteny with new evolutionary highlights is detected between Arabidopsis and legume model plants despite the lack of macrosynteny. Mol. Genet. Genomics 274, 644–657 10.1007/s00438-005-0057-9 (doi:10.1007/s00438-005-0057-9) [DOI] [PubMed] [Google Scholar]
- 51.Irimia M, et al. 2012. Extensive conservation of ancient microsynteny across metazoans due to cis-regulatory constraints. Genome Res. (doi:10.1101/gr.139725.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughes AL, Friedman R. 2003. 2R or not 2R: testing hypotheses of genome duplication in early vertebrates. J. Struct. Funct. Genom. 3, 85–93 10.1023/A:1022681600462 (doi:10.1023/A:1022681600462) [DOI] [PubMed] [Google Scholar]
- 53.Friedman R, Hughes AL. 2003. The temporal distribution of gene duplication events in a set of highly conserved human gene families. Mol. Biol. Evol. 20, 154–161 [DOI] [PubMed] [Google Scholar]
- 54.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12, 996–1006 10.1101/gr.229102 (doi:10.1101/gr.229102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujita PA, et al. 2011. The UCSC genome browser database: update 2011. Nucleic Acids Res. 39, D876–D882 10.1093/nar/gkq963 (doi:10.1093/nar/gkq963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhn RM, et al. 2009. The UCSC genome browser database: update 2009. Nucleic Acids Res. 37, D755–D761 10.1093/nar/gkn875 (doi:10.1093/nar/gkn875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kent WJ. 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12, 656–664 10.1101/gr.229202 (doi:10.1101/gr.229202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srivastava M, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454, 955–960 10.1038/nature07191 (doi:10.1038/nature07191) [DOI] [PubMed] [Google Scholar]
- 59.Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 10.1126/science.1139158 (doi:10.1126/science.1139158) [DOI] [PubMed] [Google Scholar]
- 60.Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 10.1038/nature10249 (doi:10.1038/nature10249) [DOI] [PubMed] [Google Scholar]
- 61.Goldstone JV, et al. 2006. The chemical defensome: environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev. Biol. 300, 366–384 10.1016/j.ydbio.2006.08.066 (doi:10.1016/j.ydbio.2006.08.066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldstone JV. 2008. Environmental sensing and response genes in Cnidaria: the chemical defensome in the sea anemone Nematostella vectensis. Cell Biol. Toxicol. 24, 483–502 10.1007/s10565-008-9107-5 (doi:10.1007/s10565-008-9107-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldstone JV, McArthur AG, Kubota A, Zanette J, Parente T, Jonsson ME, Nelson DR, Stegeman JJ. 2010. Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish. BMC Genom. 11, 643. 10.1186/1471-2164-11-643 (doi:10.1186/1471-2164-11-643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baldwin WS, Marko PB, Nelson DR. 2009. The cytochrome P450 (CYP) gene superfamily in Daphnia pulex. BMC Genom. 10, 169. 10.1186/1471-2164-10-169 (doi:10.1186/1471-2164-10-169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feyereisen R. 2006. Evolution of insect P450. Biochem. Soc. Trans. 34, 1252–1255 10.1042/BST0341252 (doi:10.1042/BST0341252) [DOI] [PubMed] [Google Scholar]
- 66.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195. 10.1371/journal.pcbi.1002195.g001 (doi:10.1371/journal.pcbi.1002195.g001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11, 431. 10.1186/1471-2105-11-431 (doi:10.1186/1471-2105-11-431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14, 755–763 10.1093/bioinformatics/14.9.755 (doi:10.1093/bioinformatics/14.9.755) [DOI] [PubMed] [Google Scholar]
- 69.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. 10.1186/1471-2105-5-113 (doi:10.1186/1471-2105-5-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 71.Ott M, Zola J, Aluru S, Stamatakis A. 2007. Large-scale maximum likelihood-based phylogenetic analysis on the IBM BlueGene/L. In ACM/IEEE Supercomputing conference 2007 (ed. Verastegui B.), New York, NY: Association for Computing Machinery [Google Scholar]
- 72.Stamatakis A. 2006. Phylogenetic models of rate heterogeneity: a high performance computing perspective. In Proceedings of 20th IEEE/ACM Intl Parallel and Distributed Processing Symposium, Rhodos, Greece. Piscataway, NJ: IEEE. [Google Scholar]
- 73.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 10.1080/10635150802429642 (doi:10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 74.Soltis PS, Soltis DE. 2003. Applying the bootstrap in phylogeny reconstruction. Stat. Sci. 18, 256–267 [Google Scholar]
- 75.Feyereisen R. 2011. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta 1814, 19–28 10.1016/j.bbapap.2010.06.012 (doi:10.1016/j.bbapap.2010.06.012) [DOI] [PubMed] [Google Scholar]
- 76.Thomas JH. 2007. Rapid birth-death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet. 3, e67. 10.1371/journal.pgen.0030067 (doi:10.1371/journal.pgen.0030067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qiu H, et al. 2008. CYP3 phylogenomics: evidence for positive selection of CYP3A4 and CYP3A7. Pharmacogenet. Genom. 18, 53–66 10.1097/FPC.0b013e3282f313f8 (doi:10.1097/FPC.0b013e3282f313f8) [DOI] [PubMed] [Google Scholar]
- 78.Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. 1993. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature 366, 265–268 10.1038/366265a0 (doi:10.1038/366265a0) [DOI] [PubMed] [Google Scholar]
- 79.Avadhani NG, Sangar MC, Bansal S, Bajpai P. 2011. Bimodal targeting of cytochrome P450s to endoplasmic reticulum and mitochondria: the concept of chimeric signals. FEBS J. 278, 4218–4229 10.1111/j.1742-4658.2011.08356.x (doi:10.1111/j.1742-4658.2011.08356.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhagwat SV, Biswas G, Anandatheerthavarada HK, Addya S, Pandak W, Avadhani NG. 1999. Dual targeting property of the N-terminal signal sequence of P4501A1. Targeting of heterologous proteins to endoplasmic reticulum and mitochondria. J. Biol. Chem. 274, 24 014–24 022 10.1074/jbc.274.34.24014 (doi:10.1074/jbc.274.34.24014) [DOI] [PubMed] [Google Scholar]
- 81.Neve EP, Ingelman-Sundberg M. 2001. Identification and characterization of a mitochondrial targeting signal in rat cytochrome P450 2E1 (CYP2E1). J. Biol. Chem. 276, 11 317–11 322 10.1074/jbc.M008640200 (doi:10.1074/jbc.M008640200) [DOI] [PubMed] [Google Scholar]
- 82.Markov GV, Laudet V. 2011. Origin and evolution of the ligand-binding ability of nuclear receptors. Mol. Cell. Endocrinol. 334, 21–30 10.1016/j.mce.2010.10.017 (doi:10.1016/j.mce.2010.10.017) [DOI] [PubMed] [Google Scholar]
- 83.Baker ME. 2005. Xenobiotics and the evolution of multicellular animals: emergence and diversification of ligand-activated transcription factors. Integr. Comp. Biol. 45, 172–178 10.1093/icb/45.1.172 (doi:10.1093/icb/45.1.172) [DOI] [PubMed] [Google Scholar]
- 84.Schierwater B, Kamm K, Srivastava M, Rokhsar D, Rosengarten RD, Dellaporta SL. 2008. The early ANTP gene repertoire: insights from the placozoan genome. PloS ONE 3, e2457. 10.1371/journal.pone.0002457 (doi:10.1371/journal.pone.0002457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnson NS, Yun SS, Thompson HT, Brant CO, Li W. 2009. A synthesized pheromone induces upstream movement in female sea lamprey and summons them into traps. Proc. Natl Acad. Sci. USA 106, 1021–1026 10.1073/pnas.0808530106 (doi:10.1073/pnas.0808530106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. 2007. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development 134, 177–187 10.1242/dev.02706 (doi:10.1242/dev.02706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emoto Y, Wada H, Okamoto H, Kudo A, Imai Y. 2005. Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev. Biol. 278, 415–427 10.1016/j.ydbio.2004.11.023 (doi:10.1016/j.ydbio.2004.11.023) [DOI] [PubMed] [Google Scholar]
- 88.McCaffery P, Wagner E, O'Neil J, Petkovich M, Drager UC. 1999. Dorsal and ventral retinal territories defined by retinoic acid synthesis, break-down and nuclear receptor expression. Mech. Dev. 82, 119–130 10.1016/S0925-4773(99)00022-2 (doi:10.1016/S0925-4773(99)00022-2) [DOI] [PubMed] [Google Scholar]
- 89.Kahn RA, Volpicelli-Daley L, Bowzard B, Shrivastava-Ranjan P, Li Y, Zhou C, Cunningham L. 2005. Arf family GTPases: roles in membrane traffic and microtubule dynamics. Biochem. Soc. Trans. 33, 1269–1272 10.1042/BST20051269 (doi:10.1042/BST20051269) [DOI] [PubMed] [Google Scholar]
- 90.Munro S. 2005. The Arf-like GTPase Arl1 and its role in membrane traffic. Biochem. Soc. Trans. 33, 601–605 10.1042/BST0330601 (doi:10.1042/BST0330601) [DOI] [PubMed] [Google Scholar]
- 91.Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. 2006. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172, 645–650 10.1083/jcb.200512057 (doi:10.1083/jcb.200512057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nebert D, Wikvall K, Miller WL. 2013. Human cytochromes P450 in health and disease. Phil. Trans. R. Soc. B 368, 20120431. 10.1098/rstb.2012.0431 (doi:10.1098/rstb.2012.0431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Markov GV, Tavares R, Dauphin-Villemant C, Demeneix BA, Baker ME, Laudet V. 2009. Independent elaboration of steroid hormone signaling pathways in metazoans. Proc. Natl Acad. Sci. USA 106, 11 913–11 918 10.1073/pnas.0812138106 (doi:10.1073/pnas.0812138106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mizuta T, Kubokawa K. 2007. Presence of sex steroids and cytochrome P450 genes in amphioxus. Endocrinology 148, 3554–3565 10.1210/en.2007-0109 (doi:10.1210/en.2007-0109) [DOI] [PubMed] [Google Scholar]
- 95.Castro LF, Santos MM, Reis-Henriques MA. 2005. The genomic environment around the Aromatase gene: evolutionary insights. BMC Evol. Biol. 5, 43. 10.1186/1471-2148-5-43 (doi:10.1186/1471-2148-5-43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nebert DW, Nelson DR, Feyereisen R. 1989. Evolution of the cytochrome P450 genes. Xenobiotica 19, 1149–1160 10.3109/00498258909043167 (doi:10.3109/00498258909043167) [DOI] [PubMed] [Google Scholar]
- 97.Garcia-Fernandez J. 2005. The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 6, 881–892 10.1038/nrg1723 (doi:10.1038/nrg1723) [DOI] [PubMed] [Google Scholar]
- 98.Larroux C, Fahey B, Degnan SM, Adamski M, Rokhsar DS, Degnan BM. 2007. The NK homeobox gene cluster predates the origin of Hox genes. Curr. Biol. 17, 706–710 10.1016/j.cub.2007.03.008 (doi:10.1016/j.cub.2007.03.008) [DOI] [PubMed] [Google Scholar]
- 99.Mulley JF, Chiu CH, Holland PW. 2006. Breakup of a homeobox cluster after genome duplication in teleosts. Proc. Natl Acad. Sci. USA 103, 10 369–10 372 10.1073/pnas.0600341103 (doi:10.1073/pnas.0600341103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lacalli TC. 1995. Dorsoventral axis inversion. Nature 373, 110–111 10.1038/373110c0 (doi:10.1038/373110c0) [DOI] [PubMed] [Google Scholar]
- 101.Lacalli TC. 1996. Dorsoventral axis inversion: a phylogenetic perspective. BioEssays 18, 251–254 10.1002/bies.950180313 (doi:10.1002/bies.950180313) [DOI] [Google Scholar]
- 102.Geoffroy de St. Hilaire E. 1822. Considérations générales sur la vertébré. Mém. Mus. Hist. Nat. 9, 89–119 [Google Scholar]
- 103.Berney C, Pawlowski J. 2006. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc. R. Soc. B 273, 1867–1872 10.1098/rspb.2006.3537 (doi:10.1098/rspb.2006.3537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wellman CH. 2010. The invasion of the land by plants: when and where? New Phytol. 188, 306–309 [DOI] [PubMed] [Google Scholar]
- 105.Wellman CH, Osterloff PL, Mohiuddin U. 2003. Fragments of the earliest land plants. Nature 425, 282–285 10.1038/nature01884 (doi:10.1038/nature01884) [DOI] [PubMed] [Google Scholar]
- 106.Vinogradov SN, Hoogewijs D, Arredondo-Peter R. 2011. What are the origins and phylogeny of plant hemoglobins? Commun. Integr. Biol. 4, 443–445 10.4161/cib.4.4.15429 (doi:10.4161/cib.4.4.15429) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghoshroy S, Binder M, Tartar A, Robertson DL. 2010. Molecular evolution of glutamine synthetase II: phylogenetic evidence of a non-endosymbiotic gene transfer event early in plant evolution. BMC Evol. Biol. 10, 198. 10.1186/1471-2148-10-198 (doi:10.1186/1471-2148-10-198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Emiliani G, Fondi M, Fani R, Gribaldo S. 2009. A horizontal gene transfer at the origin of phenylpropanoid metabolism: a key adaptation of plants to land. Biol. Direct. 4, 7. 10.1186/1745-6150-4-7 (doi:10.1186/1745-6150-4-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vogt T. 2010. Phenylpropanoid biosynthesis. Mol. Plant 3, 2–20 10.1093/mp/ssp106 (doi:10.1093/mp/ssp106) [DOI] [PubMed] [Google Scholar]
- 110.Batard Y, Schalk M, Pierrel MA, Zimmerlin A, Durst F, Werck-Reichhart D. 1997. Regulation of the cinnamate 4-hydroxylase (CYP73A1) in Jerusalem artichoke tubers in response to wounding and chemical treatments. Plant Physiol. 113, 951–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goel M, Mushegian A. 2006. Intermediary metabolism in sea urchin: the first inferences from the genome sequence. Dev. Biol. 300, 282–292 10.1016/j.ydbio.2006.08.030 (doi:10.1016/j.ydbio.2006.08.030) [DOI] [PubMed] [Google Scholar]
- 112.Kaneko M, Ohnishi Y, Horinouchi S. 2003. Cinnamate:coenzyme A ligase from the filamentous bacterium Streptomyces coelicolor A3(2). J. Bacteriol. 185, 20–27 10.1128/JB.185.1.20-27.2003 (doi:10.1128/JB.185.1.20-27.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yin L, Zhu M, Knoll AH, Yuan X, Zhang J, Hu J. 2007. Doushantuo embryos preserved inside diapause egg cysts. Nature 446, 661–663 10.1038/nature05682 (doi:10.1038/nature05682) [DOI] [PubMed] [Google Scholar]
- 114.Chernikova D, Motamedi S, Csuros M, Koonin EV, Rogozin IB. 2011. A late origin of the extant eukaryotic diversity: divergence time estimates using rare genomic changes. Biol. Direct. 6, 26. 10.1186/1745-6150-6-26 (doi:10.1186/1745-6150-6-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rozman D, Stromstedt M, Tsui LC, Scherer SW, Waterman MR. 1996. Structure and mapping of the human lanosterol 14alpha-demethylase gene (CYP51) encoding the cytochrome P450 involved in cholesterol biosynthesis; comparison of exon/intron organization with other mammalian and fungal CYP genes. Genomics 38, 371–381 10.1006/geno.1996.0640 (doi:10.1006/geno.1996.0640) [DOI] [PubMed] [Google Scholar]
- 116.Lamb DC, Kelly DE, Kelly SL. 1998. Molecular diversity of sterol 14alpha-demethylase substrates in plants, fungi and humans. FEBS Lett. 425, 263–265 10.1016/S0014-5793(98)00247-6 (doi:10.1016/S0014-5793(98)00247-6) [DOI] [PubMed] [Google Scholar]