Abstract
The first eukaryote genome revealed three yeast cytochromes P450 (CYPs), hence the subsequent realization that some microbial fungal genomes encode these proteins in 1 per cent or more of all genes (greater than 100) has been surprising. They are unique biocatalysts undertaking a wide array of stereo- and regio-specific reactions and so hold promise in many applications. Based on ancestral activities that included 14α-demethylation during sterol biosynthesis, it is now seen that CYPs are part of the genes and metabolism of most eukaryotes. In contrast, Archaea and Eubacteria often do not contain CYPs, while those that do are frequently interesting as producers of natural products undertaking their oxidative tailoring. Apart from roles in primary and secondary metabolism, microbial CYPs are actual/potential targets of drugs/agrochemicals and CYP51 in sterol biosynthesis is exhibiting evolution to resistance in the clinic and the field. Other CYP applications include the first industrial biotransformation for corticosteroid production in the 1950s, the diversion into penicillin synthesis in early mutations in fungal strain improvement and bioremediation using bacteria and fungi. The vast untapped resource of orphan CYPs in numerous genomes is being probed and new methods for discovering function and for discovering desired activities are being investigated.
Keywords: microbes, biotechnology, cytochrome P450, bioactive, resistance
1. Microbial cytochromes P450, sterols and the early metabolism of molecular oxygen
Cytochromes P450 (CYPs) were proposed in the 1970s to have been one of the ways early microbes might have detoxified the first atmospheric oxygen [1], a hypothesis that can lead to much speculation and interest. The observed reactions that CYPs can undertake and that do not require oxygen, obviously points to the possible existence and function of ancestral CYPs before atmospheric molecular oxygen appeared on the Earth about 2.4 Ga. In geological analysis, the presence of sterols that reflect eukaryotic life goes back into the Precambrian and they produce geochemical fossils, steranes, that are detected in rocks as far back as 2.7 Ga [2]. The reliability of these molecules to be as old as the rocks they are contained in has been refuted and the concomitant proof of eukaryotes existing at this point in geological time. The earliest evidence for eukaryotes was revised to 1.78–1.68 Ga [2]. The appearance of oxygen is often called the ‘great oxidation event’, but recently Waldbauer et al. [3] provided evidence that micro-aerobic conditions could have supported steroid biosynthesis before the earliest detectable atmospheric oxygen. An ancestral activity for CYPs is accorded to CYP51 which undertakes an early 14α-demethylation reaction of the first sterol in the pathway, lanosterol, for some micro-organisms and higher eukaryotes; or as in the ‘plant route’ where obtusifoliol (synthesized via cycloartenol) is the CYP51 substrate. Linking this data on the earliest sterols detected and the prerequisite for CYP51 early in sterol biosynthesis, it would seem reasonable to suggest placing the first CYP51 appearance at 1.78–1.68 Ga according to Rasmussen et al. [2], or earlier. Even if eukaryotes are not as old as this and appeared 850 Ma [4], CYP51 appearance would have been soon after the evolution of the first sterols, as one of the first modifying enzymes of sterol biosynthesis. This would have been in an early eukaryote or possibly a bacterial ancestor. Subsequent to the other steranes detected, further geochemical fossils, 24-isopropylcholestanes, have been used to point to the origin of the Metazoa (marine desmosponges) in rocks to greater than 635 Ma [5].
Sterols were first reported in a bacterium Methyloccocus capsulatus by Bird et al. [6] and the first bacterial gene (and protein) involved in sterol biosynthesis was identified in this organism, a CYP51 [7]. A route to sterol biosynthesis via lanosterol in this bacterium adds to the discussion on the evolution of sterol biosynthesis that is normally associated with eukaryotes. The M. capsulatus pathway may have resulted from horizontal gene transfer, although it is conceivable that a prokaryote ancestor of eukaryotes might have developed sterol biosynthesis [8]. The M. capsulatus pathway of sterol biosynthesis contains proteins encoded by overlapping squalene monoxygenase and oxidosqualene cyclase genes that are required before the CYP51 step [8]. Another published pathway of bacterial sterol biosynthesis, leading only to lanosterol and parkeol, was found in Gemmata obscuriglobus [9], while in the myxobacterium Stigmatella aurantiaca, the production of cycloartenol and the gene responsible was observed [10]. Neither of these diverse bacteria contain a CYP51 activity, but bioinformatic analysis supports the presence of a CYP51 biosynthetic gene and a sterol pathway in another myxobacterium Plesiocystis pacifica [11]. Other homologues of sterol biosynthetic genes are detected in bacteria that seem to have been recruited to other functions after gene transfer and for some it is possible to concentrate sterols from the growth media [12].
The sterol pathway requires oxygen at an early point (at squalene epoxidation); this would have been the first step of sterol biosynthesis ‘locking up’ oxygen, but not into sterol. The sterol C4-demethylations and C5-desaturation also add to the extension of the pathway that has been suggested to be driven by oxygen increases [13]. The alternative and not exclusive view is that each addition to the pathway produced a more complex sterol that conferred a selective advantage to micro-organisms regarding their membrane function. The latter is presumably reflected by the addition of a series of reductases to the pathway tailoring the sterol molecule. Subsequent sterol evolution in micro-organisms and then multi-cellular organisms presumably led to the varied end products found in some organisms, while for others it led to the loss of sterol biosynthesis entirely since it could be obtained externally, e.g. from the diet as for insects and nematodes.
2. Microbial cytochromes P450
The study of microbial cytochromes P450 is of natural academic interest, offering the potential to develop model systems that could assist the understanding of human cytochromes P450 and their role in health. Moreover, the early divergence of microbial CYPs and their extreme biodiversity, which are being revealed through modern genome sequencing techniques, have excited as much interest in their roles in the natural world, as in the consideration of critical applications that may be exploited for future biotechnological needs. As it is now recognized that only a small fraction of microbial life can be cultured, the expansion of metagenomics also contributes in expanding the known diversity of CYPs, such as demonstrated in Craig Ventner's investigations [14]. We hope to reveal here the wide relevance of these genes and proteins for key aspects of what are often called ‘Grand Challenges’, i.e. food security, biorefinery and industrial biotechnology (to create a sustainable future) and for improving and sustaining health.
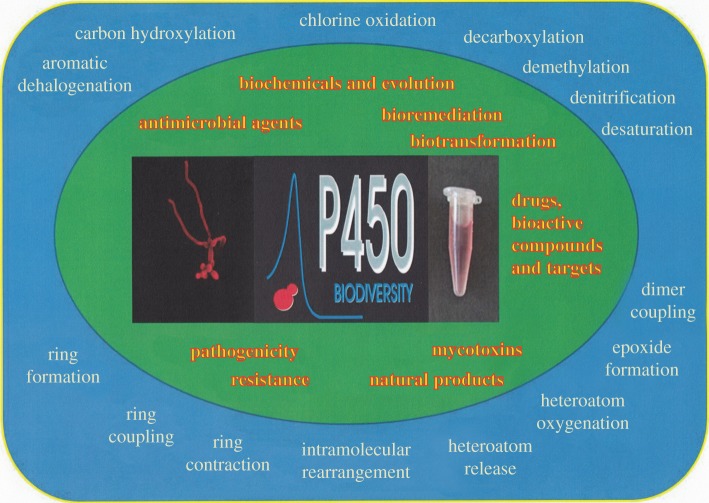
As in macrobiological systems, CYPs of micro-organisms undertake a wide variety of reaction types (figure 1), the signature of these haemoproteins is to act as mono-oxygenases, but many reactions are reported [15]. These CYPs are key for microbial natural product production, as biocatalysts, for bioremediation and as drug and agrochemical targets.
Figure 1.
Areas of fundamental interest and application for microbial CYPs and reactions associated with them.
That cytochrome P450 investigations were dominated in the years after their first identification [16,17] by mammalian cytochrome P450 was to be expected given the important biosynthetic roles of human forms in, for example, endogenous functions such as steroid biosynthesis and bile acid production, as well as drug/xenobiotic metabolism and its role in pharmacology and toxicology (xenobiotic metabolism). However, cytochrome P450 studies in yeast were initiated during the 1960s [18] and the early expectation was that small numbers of CYPs were present. Not surprisingly therefore, the initial family numbers for lower eukaryotes that were allocated were a modest 51–69, compared with 1–49 for animals. The system for nomenclature of CYP families and subfamilies is dealt with elsewhere in this issue, but is based on amino acid sequence identity (greater than 40% identity placed in same CYP family; greater than 55% in same subfamily). The system was rapidly overwhelmed in the genomic era, with allocated numbers 501–699 soon taken and as the CYP family numbers for lower eukaryotes grew, numbers onwards beyond 5001 have had to be assigned. A single fungal species can often contain more than a hundred CYP genes compared with the 57 found in man and to date more than 2500 DNA sequences have been identified as fungal CYPs.
Many bacteria contain no cytochromes P450; this is true of Eubacteria, for example Escherichia coli, and the Archaea, and early researchers may be forgiven for not anticipating any or few to be discovered. However, bacterial CYPs have also increased substantially in family numbers, with more than 1000 named (CYP101 upwards). The actinomycetes often contain tens of CYP genes in their CYPome and are implicated in the production of many natural products used in medicine (e.g. in streptomycetes [19,20]) and various lipid metabolism in mycobacteria, where CYPs represent potential drug targets [21–23]. Myxobacteria that are sources of natural products such as epothilone anti-cancer drugs are also rich in CYPs [24], but even among actinomycetes some organisms can have low CYP numbers, such as the probiotic organism Bifidobacterium longum that has one CYP of unknown function [25]. Possibly this low number reflects the niche in the intestine as with E. coli that has no CYPs.
The manuscript signalling the beginning of bacterial CYP studies is generally attributed to Appleby [26], while a CYP from Pseudomonas putida called P450CAM , or CYP101, permitted growth on camphor and became a paradigm for protein studies and on genetics of a catabolic plasmid from the Gunsalus laboratory [27–30]. Subsequently, this soluble CYP was the first for which a crystal structure was obtained [31], with the number of new structures increasing dramatically enabling templates for many to be modelled, particularly those hampered by the membrane anchoring properties of the majority of CYPs.
As with considerations of animal and plant P450, where evolution is related in part to producing deterrents/attractants and detoxification of food materials, so too is the biodiversity of microbial CYPs frequently bound up with using different carbon sources for growth. For example, CYPs can act to undertake initial oxidation of carbon sources such as naturally occurring alkanes by Candida spp. In contrast, the bacterial CYP177A1 permits growth on the explosive RDX (hexahydro-1,3.5-trinitro-1,3.5-triazine) and subsequent phytoremediation [32]. During the 1970s, single-cell protein production from oil was considered and cytochrome P450 was implicated in the growth of Candida tropicalis for this purpose, where it undertook the initial metabolism of these carbon sources [33]. Biotechnological applications were also discovered subsequently to involve CYPs, such as for some of the first industrial biotransformations in the 1950s undertaken by filamentous fungi in the production of corticosteroids by using their abilities to undertake steroid 11-hydroxylation, reactions difficult to achieve chemically [34]. More recently, it was realized that mutation in CYP504 was responsible for channelling metabolites into penicillin production, instead of 2-hydroxylation of phenyl acetate, during some of the earliest titre improvements in the Wisconsin strains by industrial strain improvement [35]. With the identification of CYP sequences comes the challenge of identifying protein function, and it is of interest that an increasing number of microbial CYPs are being associated with secondary metabolic pathways undertaking oxidative decoration of natural products that may be exploited in fine chemical production.
3. Microbial cytochromes P450 and their electron donors
Microbial CYPs have revealed diverse classes [36] that will be useful in biotechnology. Ten classes have currently been assigned. Most bacterial CYPs are soluble and are so-called Class I and driven by ferredoxin and ferredoxin reductases. There are often many genes for both these latter proteins encoded in bacterial genomes and few complete studies on functional redundancy have been performed such as for Streptomyces coelicolor A3(2) CYP105D5 [37]. This class also includes the archetypal CYP101 or P450CAM from P. putida described above.
Class II includes most eukaryote CYPs localized to the endoplasmic reticulum and some other membranes via an N-terminal anchor and associated with a separate NADPH-cytochrome P450 reductase (CPR). This is not the only electron donor in microbial Class II systems as in yeast a knockout of the gene is still viable and continues to show CYP51 activity in sterol biosynthesis that is required for viability [38,39]. The cytochrome b5 system has been attributed with this sustained activity since a double knockout is lethal. Other roles of yeast CPR have been revealed in cell division giving rise to polarity defects indicating a role for CPR in bud site selection, bud growth, cell wall assembly, mating and invasive growth [40,41]. The structure of yeast CPR has been determined in a form that retains the ability to drive CYP activity, the only such example to date [42]. CPR is usually encoded by only one gene in most eukaryotes except plants [43] and some filamentous fungi [44]. The roles of these multiple CPRs is of clear interest and in aspergilli and some other fungi is also accompanied by duplicated CYPs, e.g. CYP51 that impinges on antifungal therapy with CYP51 azole drug inhibitors [44–46]. In Cochliobolus lunatus, the cytochrome P450 system is of biotechnological interest as the fungus is used in preparation of corticosteroids and CYPs have been associated with detoxification of plant defence compounds [47]. Lah et al. [48] reported two CPRs in this fungus where one was associated with primary metabolism and the other with xenobiotic detoxification. Interestingly, when reconstituted with a CYP53 the two CPRs produced different products suggesting another level of control of metabolism and presumably an effect of CPR binding on the CYP substrate bound structure.
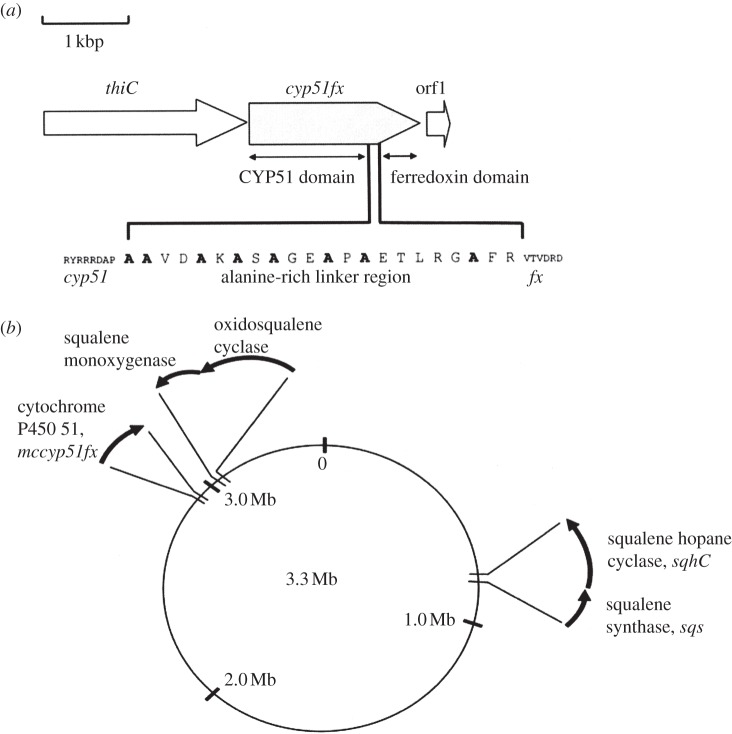
Class III CYPs are to date bacterial in origin and use flavin mononucleotide (FMN) containing flavodoxin as the reduction partner, first found for CYP176A1 (P450cin) metabolizing cineol [49]. Class IV is also of microbial origin found in an acidothermophilic archaeon. CYP119 in Sulfolbus solfataricus can obtain electrons from a non-NAD(P)H-dependent reductase [50] and is active at high temperature and pressure. Class V CYPs are represented currently by CYP51 from M. capsulatus and contains a C-terminal fusion via a short linker to a (3Fe-4S) ferredoxin domain (figure 2). This CYP was the first bacterial sterol biosynthetic gene/protein reported [7] and was discussed above.
Figure 2.
(a) The structure of the linker region for the first enzyme of bacterial sterol biosynthesis to be studied, CYP51 from M. capsulatus, together with the chromosomal loci of genes (b) for sterol biosynthesis and hopanoid biosynthesis [8].
CYP177A1 is another new class of cytochrome P450 (Class VI) encoding a natural fusion between an N-terminal flavodoxin domain and a C-terminal haem domain identified by Seth-Smith et al. [51]. It was identified in Rhodococcus rhodochrous (strain 11Y) and it allows this bacterium to obtain nitrogen from a high-explosive pollutant called RDX (see earlier). Class VII CYPs have been reported as natural fusions of CYP116B2, also called P450 RhF, from Rhodococcus sp. NCIMB 9784. This protein consists of a C-terminal reductase domain containing FMN and NADPH binding motifs and a ferredoxin-like domain with a N-terminal haem linked to the C-terminus via a linker [52,53].
Class VIII CYP systems consist of an N-terminal haem domain fused to a eukaryotic-like CPR-like domain at the C-terminus. The first example emerged from the Fulco laboratory for CYP102A1 (P450BM-3), a protein from Bacillus megaterium that metabolizes fatty acids [54]. A similar membrane-bound fungal system was also discovered in Fusarium oxysporum CYP505 [55] and subsequently in a number of filamentous fungal genomes [45]. Class IX CYP was assigned to CYP55 (P450nor) also from F. oxysporum and identified in the Shoun laboratory. This CYP is a nitric oxide reductase and a soluble eukaryotic CYP that derives electrons from NADH [56]. The last CYP class X assigned to date includes CYPs that catalyse reactions without a redox partner, first associated with plant allene oxide synthase [57]. The reactions of this class that includes animal and plant CYPs involve an intramolecular transfer system and examples include divinyl ether synthase and thromboxane synthase [36,57].
Without doubt the number of classes will increase from the current knowledge of the ‘tip of the iceberg’ of metagenomic and culturable microbial genomes. Bacterial CYPs exhibiting peroxygenase activities have been discovered that bind H2O2 with high affinity and turn over fatty acid at a high rate; the first identified was CYP152A1 from Bacillus subtilis [58]. These could comprise an additional class and they seem not to require a reduction system for oxygen activation. In the Aspergillus nidulans CYPome, the identification of PpoA (CYP6001A1) as a fusion protein of a fatty acid dioxygenase/peroxidize and a cytochrome P450 was made after characterization of the heterologous protein produced in E. coli. It displayed the characteristic 450 nm carbon monoxide binding spectrum and is involved in regulating the balance of sexual and asexual life cycles by metabolizing fatty acids to produce precocious sexual inducer factors [59]. Another cytochrome P450 has also been found recently with more than one activity, but not necessarily reflecting a different CYP class. CYP170 was identified as possessing a hydroxylase activity participating in albaflavenone biosynthesis in S. coelicolor A3(2), as well as having a separate farnesene biosynthetic terpene synthase active site [60].
4. Microbial cytochromes P450 and the environment
Some of the largest numbers of CYPs in individual microbes are found in white- and brown-rot fungi. In Phanerochaete chrysosporium, about 150 CYPs were identified thus revealing the scale of unravelling orphan CYP function that faces the community, as only a few have defined functions or (useful) activities associated with them (e.g. CYP51, [61]). The detection of cytochrome P450 in this fungus and demonstration of catalytic activity in the metabolism of polycyclic aromatic hydrocarbons [62] has been followed by examination of CYP gene expression patterns upon exposure to an inducer of this class of compound, to reveal those CYPs with this functional profile [63]. This fungus is of further interest with respect to commercial bioremediation and the fungus' ability to degrade lignin with breakdown also associated with CYP activities [64,65]. These CYP activities are less well investigated than the extracellular peroxidizes and laccases involved in this process. The importance of CYPs in the breakdown of plant material in the environment in white-rot fungi is true also of brown-rot fungi [66] where an even larger number of CYP genes have been discovered [67]. Two hundred and fifty CYP genes were identified in the Postia placenta genome of which 184 were isolated as full-length cDNA and expressed in S. cerevisiae to probe activity that included stilbene as a CYP substrate.
The large biodiversity of CYPs is not only found in these wood-rot fungi, but in other saprophytic organisms, e.g. Aspergillus oryzae [68], and in soil actinomycetes such as the 18 CYPs in S. coelicolor A3(2) [19] and approximately 40 in Mycobacterium smegmatis [12,69]. The ability of these organisms to biotransform xenobiotics is well known, including for instance, the ability of aspergilli in industrial steroid hydroxylation. As for some human CYPs that have evolved to assist detoxification of natural products in the diet and which now are known to contribute centrally to drug metabolism, some of these microbial CYPs have the capability to metabolize drugs and have been used to produce mammalian drug metabolites. These metabolites have been needed for pharmacological and toxicological evaluation in the drug discovery process particularly in the days before heterologous human cytochromes P450 were available commercially.
Some microbial CYPs may have been engaged in detoxification as well as synthesis of natural products and in microbial systems of fungi and bacteria CYPs can contribute to bioremediation. For example, one study examined Rhodococcus and Gordonia strains for the ability to degrade petrol additives methyl tert-butyl ether, ethyl tert-butyl ether and tert-amyl butyl ether and found an involvement of CYP249 [70].
Another very important role of fungal CYPs in the cycling of nutrients includes CYP55 of fungi, a fungal de-nitrification enzyme contributing to the nitrogen cycle [71]. De-nitrification is a process where nitrate or nitrite is reduced to a gaseous form of nitrogen (N2 or N2O) and fungi lacking nitrous oxide reductase produce nitrous oxide, a greenhouse gas, as a final product [72]. Studies have revealed fungi are a major contributor to de-nitrification in grasslands, semi-arid and forest soils with implications for soil fertility [73].
Dependent upon the ecological niche new and interesting roles for CYPs have been discovered, for example, insects that ‘farm’ fungi to provide a source of sterol [74]. CYPs also play a part in fungal pathogenicity against insects. The insect cuticle comprises a mixture of lipids, including long-chain alkanes, alkenes, wax esters and fatty acids, and recently the involvement of CYPs in breaking down this barrier was shown for Beauveria bassiana [75]. Eight CYPs from seven different CYP families were associated with metabolism of cuticle lipid. These included CYP52s, originally identified as alkane and fatty acid metabolizing CYPs in Candida spp. [33], but also a member of a new family of CYPs, CYP5337, as well as new members of subfamilies from the CYP53 family. Candida spp. contain multiple CYP52s with overlapping substrate specificity shown by biochemical and genetic methods [76,77], and it was suggested that the CYPs identified as being induced by cuticle lipids would also show such overlap [75]. Manipulating these activities could increase virulence and has the potential to assist in bio-control of insects using fungal pathogens.
5. Food security and microbial cytochromes P450
Microbial CYPs have been revealed to be important for food security issues and one of the main reasons is that many fungal pathogens of crops are controlled by inhibitors of CYP51 that block ergosterol biosynthesis [78,79]. These were first introduced in the early 1970s. About 40 per cent of the agrochemical market is for fungicides and to maintain and increase yields will require their continued use for the foreseeable future. There have been concerted screens to develop new fungicides but, as in the pharmaceutical area, this has led to limited success. The strobolurin fungicides were successfully introduced and target the respiration pathway by inhibiting cytochrome bc1, but several plant pathogens have developed resistance reaching high levels [80]. The continued success of CYP51 inhibitors (also known as azole fungicides or demethylase inhibitors) has led to renewed interest in discovery of new agents also set against a backdrop of European regulation to improve the safety of pesticides.
The azole antifungals represented an older era of drug and agrochemical discovery. Although they were introduced without knowledge of their mode of action in the 1970s, it was later that they were recognized to inhibit firstly sterol biosynthesis among other cellular effects [78] and then more specifically, fungal cytochrome P450 undertaking sterol 14α-demethylation [81]. They are central to antifungal therapy clinically, where the immuno-compromised are at risk, and despite attempts to discover better agents are central to control of many plant pathogens. In both the clinic and the field, the emergence of resistance is a problem.
The gene encoding the particular target emerged from studies on Saccharomyces cerevisiae and was named CYP51 in the nomenclature [82]. It was found to be an essential gene for viability, a result confirming it as the target after the various biochemical correlations suggesting this. We now appreciate this CYP activity is ancient and other CYP51s are found in sterol-producing animals, plants, protists, but rarely in bacteria, producing 14α-demethylated sterols [69,83]. The 14α-demethylation involves three sequential monooxygenase events with release of formic acid [84–86].
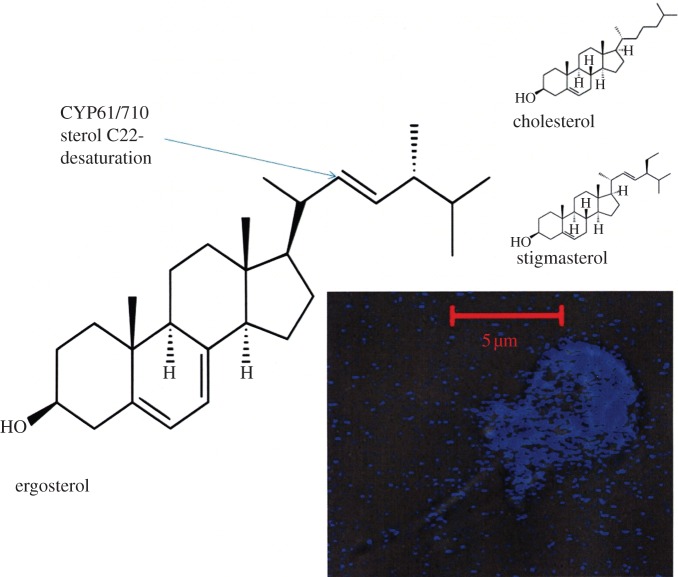
As proposed in 1995 [87], another cytochrome P450 CYP61 (sterol 22-desaturase) represents an ancient activity for the superfamily being present as CYP710 in plants [88]. Homologues in lower eukaryotes have also been assigned CYP710 status, although they often make the ‘fungal sterol’ ergosterol. These two CYP families should be unified, as is the case for CYP51s with a conserved function. One of the first uses of amino acid sequence information to assign function to a gene in a newly sequenced genome was made for CYP61 and the suggestion that CYP61 arose from CYP51 during the successive addition to the sterol biosynthetic pathway during its evolution in unicellular organisms. Subsequently, in 1997, the activity and sensitivity to azole drugs was characterized [87–90]. We wait to see whether CYP61/CYP710 emerges in sponge genomes, as it is present in the choanoflagellates (figure 3), ancestors of fungi and animals [91], and sponges have been reported to contain 22-desaturated sterols [92] that require CYP61/CYP710. To date, it has not been found in the first sponge genome to be sequenced from Amphimedon queenslandia (D. Nelson 2011, personal communication).
Figure 3.
Ergosterol chemical structure is shown illustrating the C22-desaturation undertaken by CYP61/CYP710. For comparison, the molecular structures of cholesterol and a phytosterol, stigmasterol, are also given. Inset is a micrograph of the filipin-stained unicellular choanoflagellate Monosiga ovate (filipin binds to ergosterol). The presence of ergosterol in the closest living relatives of metazoans and as the most common end product in protists, fungi and some algae, suggests the evolution of ergosterol was most likely an early event in eukaryote evolution, or was repeatedly gained over time.
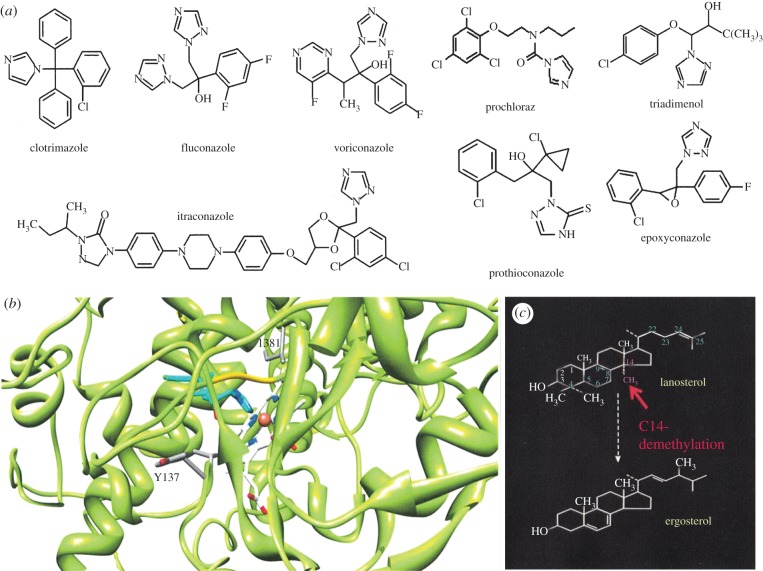
The genes encoding the enzymes that undertake the ancient CYP51 activity are undergoing evolution in the field as a response to use of azole inhibitors that bind to this target. Even with resistance, crop management can be maintained by altering fungicides and using mixtures. Azole inhibitors of CYP51 act by binding via the N-3 of imidazole or N-4 of triazole to the haem as a sixth ligand, while the N-1 substituent group binds to the amino acid tertiary structure of the active site and substrate channel (figure 4) [20]. To date, there are no fungal CYP51 structures, although the structure of human CYP51 was recently elucidated [93].
Figure 4.
(a) A variety of azole structures, including clotrimazole, fluconazole, voriconazole and itraconazole (medical antifungal drugs) and the agrochemical demethylase inhibitors prochoraz, triadimenol, epoxyconazole and prothioconazole. (b) The molecular model of triadimenol bound to M. graminicola CYP51 with locations of resistance residues Y137 and I381. (c) Ergosterol biosynthesis from lanosterol highlighting the 14α-methyl group removed by CYP51.
The decreased sensitivity of the wheat pathogen Mycosphaerella graminicola to azole fungicides appears to be mainly due to mutation in CYP51 and this is true for other important plant pathogens. A complex and increasing number of mutations have been revealed over time within the same gene, indeed it is now impossible to find wild-type CYP51s in the field in the UK. Yeast complementation tests revealed these mutations resulted in functional, but resistant, protein activity [94]. The substitutions L50S, Y459D and Y461H and a two-amino-acid deletion mutant Delta Y459/G460 all complemented yeast with their activity and conferred resistance. Another change, I381V, inactivated protein activity, but this was restored when found in combination with other mutations at Y459/Y461 [94]. Mutation at S524T was also recently shown to confer resistance and to rescue activity in combination with Y137F and V136A mutations. Combinations of recent alterations detected in the field of D134G, V136A, Y461S and S524T also reduced sensitivity to the two main commercial fungicides currently used, epoxiconazole and prothioconazole [95].
A structural rationale has recently emerged for these CYP51 resistance mutations using a multiple homology method, individually identified for each variant, rather than a single structural scaffold. Using this approach, the development of resistance could be explained when considering multiple mutant CYP51s, through modified structure and increases in the binding pocket volume, while also explaining the regained sensitivity to some earlier fungicides in some recent mutants studied using yeast complementation [94,95]. This approach provides a potential tool to manage fungicide use and explore potential for resistance for new fungicides [96].
The participation of CYPs in biosynthetic pathways of natural product synthesis can, in the context of food security, be a problem because of the synthesis of mycotoxins. Mycotoxins are synthesized by many phytopathogenic and food spoilage fungi, including Aspergillus, Fusarium and Penicillium spp. These can have significant health effects: the carcinogenic effect of aflatoxin, and nuts and grains need to be monitored for mycotoxins. For example, two CYP619s have been found to participate in the biosynthesis of patulin in Aspergillus clavatus among a cluster of 15 genes [97]. CYP619C3 catalysed the hydroxylation on m-cresol while CYP619C2 undertook the hydroxylation of m-hydroxybenzylalcohol and m-cresol to gentisyl alcohol and 2,5-dihydroxytoluene, although the latter is not a known precursor of patulin. The earliest discovery of mycotoxin biosynthetic CYPs resulted in the discovery of the CYP58 family. This was identified in studies on the TRI4 gene of Fusarium sporotrichioides catalysing the first oxidation during trichothecene biosynthesis, where it is involved in apotrichodiol biosynthesis [98]. The CYP65 family was also identified as being encoded by TRI11 in this pathway in F. sporotrichioides [99]. The CYP59 family, one of the earliest fungal CYP families identified, was also found to be involved in sterigmatocystin biosynthesis in A. nidulans [100].
Apart from mycotoxin biosynthetic pathways, ergot fungi are also well known for their effects on contaminating food causing ergotism after, for example, the consumption of contaminated rye bread. The pathway includes a CYP required for the conversion of clavines to d-lysergic acid among a cluster of ergot alkaloid genes [101]. Mindful of Paracelsus' famous quote, these ‘poisons’ have also been turned to pharmaceutical use in migraine treatment and in inducing contractions in the uterus to stop bleeding after childbirth.
Bacterial CYPs may also be important in aspects of food security, but that is less clear than for the fungal examples described. Streptomyces scabies causes potato scab disease; the completed genome has multiple CYPs and their potential roles in pathogenicity and as targets of agrochemicals deserves attention. The main phytotoxin produced by S. scabies is thaxtomin and biosynthesis of thaxtomin cyclic dipeptide phytotoxins requires CYP for postcyclization hydroxylation of the cyclic dipeptide [102].
Beyond bacteria and fungi, there are other important microbial pathogens, including the fungal-like oomycetes that have filamentous growth. The potato blight and sudden oak death pathogens are examples of Phytophthoras that are organisms that do not synthesize sterols, but alternative CYPs are potential targets [103]. The soybean pathogen P. sojae has a CYPome of 30 CYPs, while the sudden oak death pathogen P. ramorum contains 24 CYPs. Other examples of oomycete pathogens include the legume pathogen Aphanomyces euteiches, which does have a sterol biosynthetic pathway, including a CYP51, the proven fungicide target [104]. The suggestion in this paper that similar products to the azole fungicides could be introduced for the Saprolegniales would also be useful in fish farming where these pathogens are problematic and dealt with, until recently, using toxic malachite green.
6. Industrial biotechnology and a sustainable future
Many of the topics and themes running through this paper and volume overlap and reference to important developments in industrial biotechnology already touched on. Hence the initial importance of CYPs in biotransformations was achieved using fungal CYP activities in whole-cell biotransformations to allow corticosteroids to be produced. The impetus for this may have been in the belief that the Luftwaffe experimented with these compounds and their derivatives to enhance pilot performance in the Second World War [105] stimulating organic chemistry research in a manner similar to the push for penicillin production, via fermentation and strain improvement, in the development of biotechnology. The regio- and stereo-specificity in steroid biotransformation by individual fungal species varied, presumably reflecting the CYPs concerned [34]. Despite this historical importance, only recently has a fungal steroid hydroxylase been identified from Rhizopus oryzae [106]. As in aspergilli, the 11α-hydroxylation of the steroid skeleton can simplify steroid drug production. CYP509C12 was identified among 48 putative CYPs as being inducible by addition of progesterone, and its activity confirmed by heterologous expression in fission yeast. This could lead the way for altered methods in the preparation of steroids using alternative microbes or enzymes.
One major example of such an alternative approach was the metabolic pathway engineering of the ergosterol pathway in S. cerevisiae to divert it towards corticosteroid production [107]. This required manipulation of activities other than CYPs and the yeast pathway was diverted by expression of a plant and eight mammalian proteins, including CYP11A1, CYP11B1, CYP17A1 and CYP21A1. A recent paper also discusses the alteration of yeast to a form producing cholesterol efficiently instead of ergosterol [108]. Other ambitious examples of pathway engineering that required CYP as an important player are the production of the anti-malarial drug precursor artemisinic acid using yeast [109] and taxol precursor in E. coli [110].
The promise of cytochromes P450 in industrial biotechnology extends to bioremediation purposes described earlier and in chemical production using whole cells. Statin biotransformation has also been used to modify a fungal natural product to produce a major drug. Mevastatin is produced by Penicillium citrinum and hydroxylation of mevastatin by Streptomyces carbophilus is used to produce the anti-cholesterol drug pravastatin [111].
Another current application of whole-cell biotransformations is for the production of dicarboxylic acids from fatty acids and alkanes. These chemicals are versatile intermediates that could be used to produce polymers, to replace oil-based products in a bio-refinery model and lead to further high-value end products. The ability for CYP52A3 to undertake the reaction cascade to dicarboxylic acids was first reported in the 1990s [112]. A variety of Candida spp. are known to have CYP52 family gene sequences, but not all CYP52s produce diacid. The ability of C. tropicalis to produce α,ω-dicarboxylic acid was enhanced by blocking β-oxidation so that these products were not used for growth, and this has formed the basis of an industrial process [113]. CYP52 in Candida spp. can also have involvement and potential application in the production of sphorolipids that have diverse applications, including as surfactants [114].
The advantage of whole-cell biotransformation is the presence of a cellular system to generate electrons for these reactions and the presence of the CPR system needed to deliver these to CYPs. An alternative to this is to use purified proteins in a reconstituted system, but the main issue currently is the value of the product set against the cost of the process and, most importantly, knowledge of the specific biotransformation for each CYP. Indeed, whole-cell CYPome analysis using heterologous gene expression in yeast has been used to screen for activities across hundreds of fungal CYPs [82]. These studies used yeast expression with co-expression of CPR. In a similar study on a smaller CYPome from the actinomycete Nocardia farcinica, a novel high-throughput ligation independent system was developed for E. coli in which the CYPs were fused to a reductase domain of the fusion protein of CYP116B (P450RhF) and for which approximately half the number of CYPs could be screened subsequently and yielded activities [115]. It is rare that all CYPs examined can be expressed well in high-throughput on the first pass, and subsequent investigations may be needed to achieve high-level expression for the recalcitrant CYPs. High-throughput methods are needed to reveal orphan CYP activities in genomes and metagenomes for biotechnology that will augment the green biotechnology approaches that are emerging. Such an approach will also be important for revealing the endogenous functions of orphan CYPs. The main alternative method currently to identifying endogenous activity and function of CYPs is the metabolomic comparison of wild-type and mutant microbes where radioisotopic comparisons may assist. For example, for CYP154A1 from S. coelicolor, the substrate was identified in this way as a dipentaenone and then shown to be metabolized by CYP154A1 via an intra molecular cyclization without oxidation/reduction in a novel reaction for CYPs [116].
The main area where purified CYPs are used commercially and produced as biotechnology reagents is in toxicology related to chemicals, agrochemicals and pharmaceuticals. Here the fate of xenobiotics with respect to human (mammalian) CYP metabolism is investigated, in part, as a registration requirement for the use of the drugs/compounds and also as a determination of those metabolites activities in relation to parent substrates. In other settings besides drug metabolism, e.g. for flavours and fragrances, CYPs could enhance properties and add value [117]. The metabolic fate of xenobiotics is revealed for the main CYPs undertaking metabolism, and the metabolites can be prepared for further studies. These CYPs and CPR are produced in microbial systems usually, and it is common in E. coli systems to modify the N-terminus to achieve heterologous protein expression [118], with and without the membrane anchor [119].
For other industrial applications, the supply of pure enantiomers as substrates and the use of expensive cofactors, or cofactor recycling, may not justify the use of pure enzymes currently, but this is an active area of investigation. Feasibility studies on immobilizing CYP systems have been published using purified CYP and colloidal liquid aphrons [120] and mammalian membrane-bound CYPs have been incorporated into nanodiscs together with CPR to investigate activity/application [121]. Cofactor recycling has also been investigated in experiments using CYP105D1 [122], and activity in biotransformations has also been studied using CYP activities driven by electrodes. Cobalt (III) sepulchrate was used as a mediator and platinum electrodes allowed activity of CYPs to be achieved albeit with reduced activity [123,124].
Of course, genetic manipulation and process engineering can be used to enhance whole-cell biocatalysts as well as enzymes. Cell walls and membranes and endogenous transporters can provide barriers, while product purification from a whole cell can present process issues when compared with cleaner single enzyme systems. Whether using whole cells or pure enzymes, the improvement of activity using directed evolution has also been applied to CYPs, and CYP102A1 or P450BM3 has been a favourite for this as it has a turnover much higher than typical eukaryotic CYPs. Instead of values around 1, often reported for mammalian CYPs, it can turnover arachidonic acid at 17 100 min−1 [125]. This is presumably because it is very efficiently coupled to the reductase as a fusion protein. Early directed evolution experiments altered P450 for more efficient use of peroxide to support activity, including for P450BM3 [126,127]. Subsequent experiments altered P450BM3 [128] to include metabolism of the gaseous alkanes propane and butane. CYP156A6 from Mycobacterium sp. strain HXN-1500 has also been improved for terminal butane oxidation [129]. The abundance of methane could be used to replace petroleum as a feedstock for industry and can also be produced sustainably by anaerobic digestion from biomass. It is interesting to speculate that CYPs might be developed to help achieve that goal. If not, these methods will still be used as routes to drug preparations, fine chemicals, biorefinery and bioremediation in a manner that will contribute towards a sustainable future.
The identification of lipids and fatty acids as CYP substrates has been a feature of this and many publications on the superfamily. Recently, a CYP152 was identified as a novel fatty acid decarboxylase from Jeotgalicoccus sp. ATCC 8456 [130]. The ability of this CYP to produce 1-alkenes from fatty acids has enormous potential for fuel and chemical feedstock production and is being exploited by LS9 Inc. in California, who discovered this property. Other CYP152s from other bacteria are highly active with hydrogen peroxide and this was true of the fatty acid decarboxylase discovered.
7. Microbes, cytochromes P450 and human health
Microbial CYPs are intimately associated with many of the natural products used as medicines. By metabolic engineering and in future synthetic biology they will be incorporated into strategies to produce, or enhance, known and new drugs sustainably using microbes [109,110]. Among bacterial CYPs probably the earliest antibiotic biosynthesis CYP gene and protein studied in detail was P450 eryF (CYP107A1) involved in C6-hydroxylation of the macrolide 6-deoxyerythronolide B during erythromycin biosynthesis in the bacterium Saccharopolyspora ertherea [131,132]. As mentioned above, many pathways of bacterial and fungal secondary metabolism contain CYPs involved in multi-step oxidation, rearrangements, epoxidations and heteroatom oxidation. Microbial CYPs can contribute other activities in secondary metabolism, such as carbon–carbon bond formation in flaviolin polymer biosynthesis involving CYP158 [133]. These diverse reactions are important for the activity of the final compounds and their diversity (table 1).
Table 1.
Examples of bacterial and fungal CYPs with identified activity/function. For some pathways, e.g. sterigmatocystin biosynthesis, more than one CYP is involved in the pathway.
| microbe | CYP | activity/function |
|---|---|---|
| microbial eukaryotes | ||
| Saccharomyces cerevisiae | CYP51 | sterol biosynthesis (14-demethylation) |
| Candida tropicalis | CYP52A1 | alkane and fatty acid catabolism |
| Fusarium oxysporum | CYP55A1 | denitrification |
| Saccharomyces cerevisiae | CYP56A1 | dityrosine formation for spore wall |
| Nectria haematococca | CYP57A1 | phytoalexin detoxification |
| Aspergillus nidulans | CYP59A1, CYP60A2, CYP60B1,CYP62A1 | sterigmatocystin biosynthesis/mycotoxin |
| Rhizopus oryzae | CYP509C1 | steroid 11α-hydroxylase |
| Chlamydomonas rheinhard | CYP710 (CYP61) | sterol biosynthesis (22-desaturation) |
| bacteria | ||
| Pseudomas putida | CYP101A1 | catabolism of camphor |
| Bacillus megaterium | CYP102A1 | fatty acid catabolism |
| Streptomyces avermitils | CYP105P1, CYP105D6 | filipin biosynthesis/antifungal |
| Saccharopolyspora erythrea | CYP107A1 | erythromycin biosynthesis/antibacterial |
| Streptomyces lavendulae | CYP107N1, CYP160A1, CYP105F1 | mitomycin c biosynthesis/antitumour |
| Streptomyces hygroscopius | CYP122A2 | rapamycin biosynthesis/immunosuppressant/anti-ageing? |
| Mycobacterium tuberculosis | CYP125A1 | catabolism of cholesterol |
| Jeotgalicoccus sp. ATCC 8456 | CYP152A3 | decarboxylation of fatty acid |
| Streptomyces fradiae | CYP105L, CYP113B1, CYP154B1 | tylosin biosynthesis/veterinary antibacterial |
| Streptomyces noursei | CYP105H1, CYP161A1 | nystatin biosynthesis/antifungal |
| Streptomyces spheroides | CYP163A1 | novobiocin biosynthesis/antibacterial |
| Sorangium cellulosum | CYP167A1 | epothilone biosynthesis/antitumour |
| Streptomyces avermitils | CYP171A1 | avermectin biosynthesis/anthelmintic and insecticide |
Streptomycetes produce about 70 per cent of the antibiotics by type, although in market value the fungal penicillin and cephalosporin antibiotics are dominant. The CYPome of S. coelicolor A3(2) revealed 18 CYPs and, as in other genomes subsequently, unsuspected secondary metabolism pathways involving orphan CYPs often organized as operons [33]. In the case of CYP158A2, a three-gene operon was identified encoding a Type III polyketide synthase. In other examples, CYPs of S. coelicolor A3(2) appeared in operons with a non-ribosomal peptide synthase (CYP105N1, 33) and another (CYP170A1) adjacent to a terpene synthase, subsequently determined to be involved in albaflavanone biosynthesis [134]. The operons can often provide clues to enzyme activities and by combining experimental approaches, e.g. gene knockouts, expressing whole pathways, or purifying individual enzymes to produce substrates for testing, may thereby reveal function of individual proteins. More problematic are those CYPs not associated with operons and at loci that give no clue to function, and a number of S. coelicolor CYP functions remain to be determined.
Antibacterials other than erythromycin also require CYPs for biosynthesis, including vancomycin, important in combating antimicrobial resistance. CYP146 is the fourth gene in the vancomycin gene cluster from Amycolatopsis mediterranei and is a carrier-protein binding CYP involved in β-hydroxytyrosine formation [135]. In addition to antibacterials are metabolites requiring CYP for their synthesis and which target other pathogens, such as for the anthelmintic avermectin synthesis in S. avermitilis [136]. The polyene antifungals nystatin from Streptomyces noursei and amphotericin B from Streptomyces nodosus also require CYP for their biosynthesis. Gene knockouts of the CYP in S. nodosus altered the polyene produced and deleting/adding CYPs could produce new antibiotic types [137]. Similar studies on two cytochromes P450s, NysL and NysN, from S. noursei produced new analogues [138]. CYPs are also involved in anti-neoplastic natural product synthesis, such as in the actinomycin D gene cluster in Streptomyces chrysomallus [139]. Other bacteria, such as the myxobacterium, produce natural products and EpoK, CYP167A1 is involved in biosynthesis of the promising anti-cancer epithilones A and B [140].
Fungal secondary metabolism frequently has CYPs involved and these may occur in gene clusters associated with polyketide synthases [44] and, for example, lovastatin biosynthesis in Aspergillus terreus [141]. Lovastatin is a simvastatin precursor that is a semi-synthetic cholesterol-lowering agent. The lovA gene encodes a cytochrome P450 clustered with the lovB polyketide synthase. LovA performed two previously elusive oxidative reactions, introduction of a 4α,5-double bond and C-8 hydroxylation. An interesting experimental development in an investigation of programming new polyketide synthases, coupled to co-expression of two CYPs, led to the resurrection of an extinct metabolite, basianin, in Aspergillus oryzae [142]. Future manipulations of microbes may allow new secondary products to be produced in a manner analogous to the streptomycete and fungal examples above.
Apart from the health benefits CYPs provide in terms of naturally occurring drugs and platform chemicals for future pharmaceuticals, the microbial CYPs have proved to be drug targets themselves, via the CYP51 antifungal inhibitors [69,143,144]. As mentioned earlier, the azole antifungals are a main plank of therapy in the clinic with many products also available over the counter in pharmacies. Orally active azole antifungals began to be introduced with ketoconazole in the 1980s after previous topical agents had been commercialized, such as econazole, miconazole and clotrimazole. Owing to toxicity issues and because of drug–drug interactions with drug metabolizing CYPs that can disturb the clearance of drugs, better agents were required. Fluconazole offered better safety and higher solubility by the late 1980s.
Concurrent with this antifungal drug discovery the HIV pandemic was emerging. Candidosis in HIV patients was often the first presentation of their condition and, together with prophylactic azole treatment over years, saw the first serious emergence of clinical antifungal drug resistance (figure 5) [145,146]. Fluconazole was not very effective in managing another fungal disease, aspergillosis, but itraconazole and voriconazole were shown to be useful later. Itraconazole resistance in aspergillosis emerged during the 1990s [147] and with it the possibility of the interplay between exposure to agrochemical azoles acting as a factor in resistance arising in the clinic [148]. There is a difference between concerns about the spread of resistance in fungi from that in bacteria, as the mechanisms in fungi involve chromosomal genes and do not involve highly transmissible plasmids. Newer azoles have come through to the clinic, such as posaconazole, and in the absence of alternative targets to CYP51 being found, further azole compounds are coming into clinical trial. Prior to azoles, the therapy used was amphotericin B-based which can lead to renal problems if prolonged. For topical use, nystatin was available. Alternative β-glucan synthase inhibitors have also emerged as drugs, e.g. caspfungin and micafungin [149].
Figure 5.
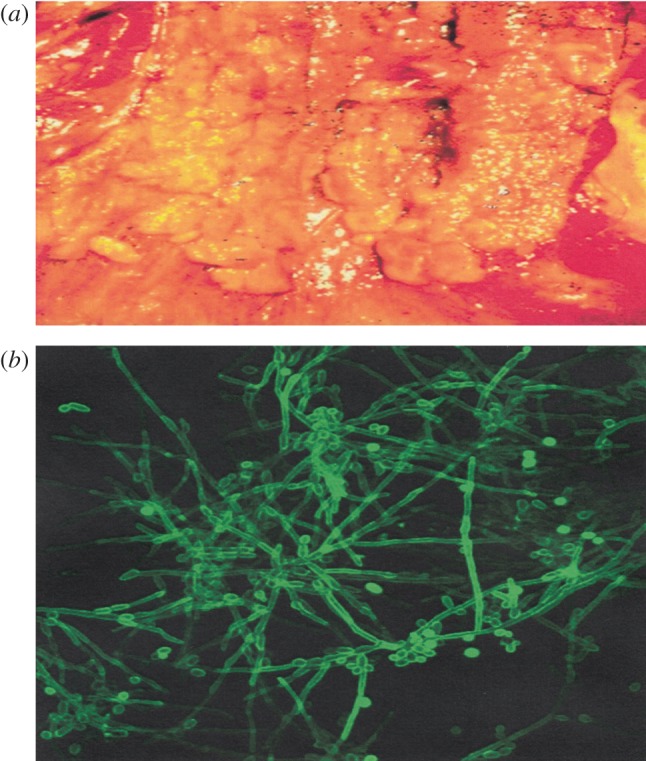
(a) Infection of oesophagus with C. albicans and (b) a photomicrograph of dimorphic C. albicans in hyphal growth that is associated with pathogenicity.
Fungal infection has been increasing due to the rise of immuno-compromised conditions such as chemotherapy, organ transplantation and HIV infection [143]. Currently, the market is predicted to grow to $8.4 billion by 2016. The current top three diseases requiring antifungal therapy by value are aspergillosis, skin and nail infections (by dermatophytes) and candidosis. Other very serious pathogens exist, including species such as Cryptococcus neoformans, Histoplasma capsulatum and increasingly in immuno-compromised patients species not seen until quite recently, such F. oxysporum and even S. cerevisiae. Sometimes the infections are generally found in specific geographical locations, such as Paracoccidiodes brasilensis in Central and South America and Penicillium marneffei in southeast Asia. The Centre for Disease Control has recently also highlighted cryptococcosis as a major cause of mortality in sub-Saharan Africa associated with AIDS, possibly more than from mycobacterial infection.
The cost of antifungal therapy in the third world is also an issue where the incidence of HIV is high. Currently, in the antifungal market voriconazole sells $820 M per annum, caspofungin $600 M per annum, liposomal amphotericin B (ambisome) $500 M per annum, with posaconazole and micafungin at $500 M per annum when combined (http://www.aspergillus.org.uk/updates/NewsletterSeptember2011.html). Another important consumer niche for azole antifungals is the anti-dandruff shampoo nizoral containing ketoconazole that controls the Malassezia globosa responsible for it [150]. Azoles may also gain use in the growing interest in a link between severe asthma and fungal exposure [151].
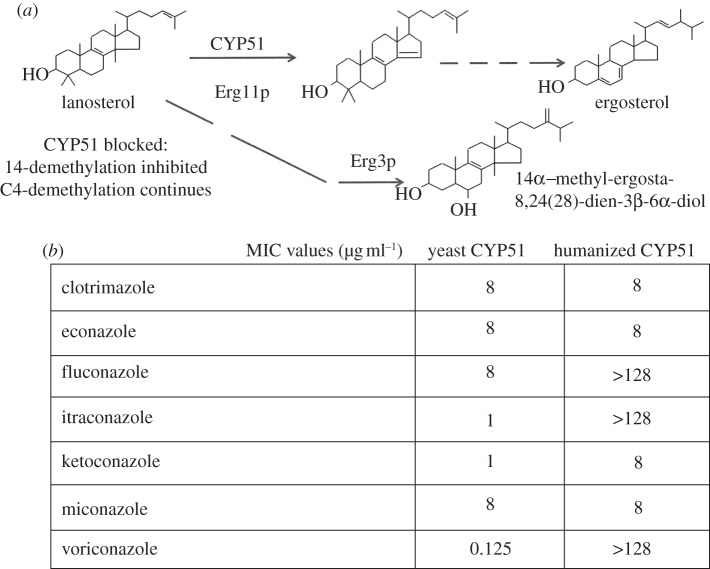
The mechanism of the azole antifungal activity is through the N-3 of imidazole or N-4 of triazole compounds coordinating with the haem of the CYP51 of human pathogens, as for plant pathogens. This interaction with human CYP51 has been given a firmer basis from the structure published, and the therapeutic basis of treatments is the selectivity of the drugs in inhibiting fungal and not human CYP51. This can be demonstrated using purified enzymes, but also in useful screening tools at the level of whole yeast where the CYP51 is replaced at the chromosomal locus with the human CYP51 [152]. Simple growth tests reveal the discrimination between the forms with humanized yeast remaining relatively resistant to an increasing extent during the development of orally active azoles. Voriconazole showed more than 1000-fold increase in the concentration needed to inhibit growth in humanized yeast, whereas clotrimazole showed no change in minimum inhibitory concentration between the strains (figure 6). Selectivity is vital for drug safety and interactions with other human CYPs involved in drug metabolism and endogenous biosynthetic functions are also very important considerations.
Figure 6.
(a) The sterol pathway of azole treatment in C. albicans illustrating the accumulation of 14α-methyl-ergosta-8,24(28)-dien-3β-6α-diol as the end product that is replaced by 14α-methylfecosterol in erg3 mutants and allows growth to continue. (b) The minimum inhibitory concentrations (MICs) are shown for two yeast strains containing S. cerevisiae CYP51 and one replaced by an ORF encoding human CYP51. This microbial tool reveals the selectivity of different azole compounds by making growth comparisons with the greatest selectivity and difference in MIC, shown for voriconazole.
Resistance mechanisms involving CYP51 amino acid changes have been discussed for plant pathogens and are also found in human pathogens. Of interest are the number of residues associated with fluconazole resistance in CYP51 in C. albicans, the different amino acids associated with resistance in A. fumigatus and those seen in plant pathogens [69]. Obviously, the different CYP51 proteins and the different azoles they are exposed to, together with different environments, will select different molecular events. For C. albicans, new mutations from the clinic were observed in the late 1990s and by genetic screens in yeast transformants these were associated with resistance [153–155] as well as observations of the emergence of resistance in matched sets of sequential isolates from patients [153].
After demonstration that R467K was a substitution in C. albicans causing fluconazole resistance [153], a number of other mutations were observed in resistant strains and placed in an early molecular model for CYP51 [154]. The mutations included another apart from R467K, namely G464S, that is also part of the haem binding domain and which might cause resistance by altering the position of the bound azole above the haem. These mutations were observed to cause resistance in yeast transformants together with another common mutation Y132H [155] and S405F. As in azole-resistant plant pathogens some isolates also contained combinations of mutations. These were often in a homozygous state in this diploid pathogen suggesting gene conversion had occurred in the second allele conferring higher resistance. Biochemical analysis has provided support for the altered affinity of the mutant proteins for azole drugs such as for G464S [156] and more recently I471T, an amino acid adjacent to the cysteine at 470 that acts as a haem ligand. In the case of I471T, the resistance was attributed to a combination of increased turnover, increased affinity for substrate and a reduced affinity for fluconazole in the presence of substrate allowing the enzyme to remain active at higher fluconazole concentrations [157].
The molecular basis of resistance and in turn the best therapeutic response to it are still under investigation. Shortly after the outset of fluconazole resistance in C. albicans being studied, it was realized that not only diverse point mutations in CYP51 were involved in resistance, but also in multiple mechanisms [158,159]. This is not yet clear for resistance in filamentous fungi. The other mechanisms clearly identified in C. albicans are increased expression of drug transporters, particularly CDR1, through mutation in a transcription factor [160] and altering the pathway of 14α-demethylated sterols accumulating under treatment away from a toxic 14α-methyl-3,6-diol end product to 14α-methylfecosterol that C. albicans can grow on [161,162]. This latter mechanism involves suppression of the effect of azoles through mutation in the sterol C5-desaturase (Erg3). It is presumed the enzyme undertakes a faulty desaturation producing the 6-OH group of the 14-methylated sterol. The sterol 6-OH of the 14α-methyl-3,6-diol is proposed to interfere with phospholipid interactions in membrane function. Azole resistance of this type has been referred to as infinite resistance. Although the literature refers frequently to the accumulation of 14-methylated sterols being the cause of arrest under azole treatment this is contrary to the concept of the value of sterols, and oxidosqualene cyclase, to organisms when sterols evolved to allow a primitive sterol pathway [162]. Not all 14α-methyl sterols are inconsistent with growth and genetic studies indicate lanosterol supports growth in yeast [163]; we have found recently a Candida glabrata that grows with lanosterol as a main sterol (SL Kelly and DE Kelly 2011, unpublished observation).
Other mechanisms of resistance have been observed in C. albicans, including increased transcription of CYP51, but this has been examined by heterologous hyper-overexpression and only changes resistance by a small amount compared with the several orders of magnitude increase in CYP51 [144]. Aneuploidy and isochromosome formation in the locus of CYP51 have been associated with resistance [164], but small changes in CYP51 expression itself may not confer much resistance as between haploid and diploid S. cerevisiae. Studies in other fungi have mainly revealed resistance mechanisms associated with altered CYP51 and in A. fumigatus these are found in one of two CYP51s in the genome. Both CYP51A and CYP51B were shown by yeast complementation to function in 14α-demethylation [46], but only CYP51A and not CYP51B exhibits alterations associated with azole resistance [148]. Together with protein affinity studies it appears the CYP51A is most resistant inherently and therefore selection pressure is exerted on this form [45]. However, it would appear unlikely that CYP51B was unable to become resistant in any scenario and this requires further investigation. The mutations observed in CYP51 associated with resistance are at positions not found in C. albicans and include G54, L98 and M220 [148]. Other mutations have been observed, for instance G138C, Y431C and G434C and also increased CYP51A expression in pan-azole-resistant A. fumigatus [165].
In other microbial pathogens, CYPs may in future be targeted to treat patients. Lessons from resistance to antifungals and more CYP51 and other protein structures to inform drug development will be useful. CYP51 is the target in fluconazole therapy of Acanthamoeba polyphaga causing eye infections associated with contact lens use [166]. Amoebae can also cause serious disease in the immuno-compromised and harmful bacteria, such as methicillin-resistant Staphlococcus aureus, can be carried by them offering further opportunity for application. CYP51 has also emerged as a potential target for agents to treat Leishmania and trypanosome parasites by inhibiting ergosterol biosynthesis. Structures of these CYP51s have emerged to assist with new drug development for Chagas disease and sleeping sickness [167,168]. However, CYP5122A1 in Leishmania donovani has recently emerged as a novel potential drug target as it was found to be essential for survival [169]. Another major area where CYPs may be targets for new drugs is with mycobacteria where the target is not CYP51 [12,20], but potentially other essential features for growth or pathogenicity such as cholesterol catabolism by Mycobacterium tuberculosis CYP125 and CYP142 [170,171]. In Mycobacterium leprae, a massive decay of CYP and other genes has occurred through evolution with partial gene sequences still evident. The retention of CYP164A1 suggests this could be important and another drug target [172].
8. Concluding comments
The snapshot of microbial CYPs here reveals a fascinating diversity of functions and activities that have and will be of great use to humanity and the natural world. They have evolved specialized roles in helping organisms to compete in their niche. CYPs also promise to be a key to biotechnological adaptation to a post-oil-based economy producing chemical platforms for fuels, plastics and other products and combating global warming. In developing control agents for pathogens CYP-inhibitors help with human disease and increased food production. Better, more selective agents that degrade quickly after use will be important to reduce environmental effects and toxicities. New catalysts will emerge and new genomes and metagenomes, from Antarctic ice lakes to the soils of the Tundra, will contain thousands of CYPs, but the challenge will be to identify key biochemistry, how to develop screens for them and how to translate this knowledge. There remains much natural science to investigate and to fascinate.
Acknowledgements
We are grateful for financial support from the ERDF and Welsh Government-funded project BEACON, Biotechnology and Biological Sciences Research Council (BBSRC) and from NIH (USA) (RO1 GM069970). We are indebted to Dr David Nelson for his long-standing efforts in the nomenclature of CYPs and discussions. Also to Nicola Rolley for microscopy of M. ovata, to Dr Steve Smith for microscopy of fungi and to Dr Jonathan Mullins for the molecular model of M. graminicola CYP51.
References
- 1.Wickramashighe RH, Villee CA. 1975. Early role during chemical evolution for cytochrome P450 in oxygen detoxification. Nature 256, 509–510 10.1038/256509a0 (doi:10.1038/256509a0) [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR. 2008. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 455, 1101–1104 10.1038/nature07381 (doi:10.1038/nature07381) [DOI] [PubMed] [Google Scholar]
- 3.Waldbauer JR, Newman DK, Summons RF. 2011. Microaerobic steroid biosynthesis and the molecular fossil record of Archean Life. Proc. Natl Acad. Sci. USA 108, 13 409–13 414 10.1073/pnas.1104160108 (doi:10.1073/pnas.1104160108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalier-Smith T. 2010. Deep phylogeny, ancestral groups and the four ages of life. Phil. Trans. R. Soc. B 365, 111–132 10.1098/rstb.2009.0161 (doi:10.1098/rstb.2009.0161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Love GD, et al. 2009. Fossil steroids record the appearance of Desmospongiae during the Cryogenian period. Nature 457, 718–721 10.1038/nature07673 (doi:10.1038/nature07673) [DOI] [PubMed] [Google Scholar]
- 6.Bird CW, Lynch JM, Pirt FJ, Reid WW, Brooks CJW, Middleditch BS. 1971. Steroids and squalene in Methylococcus capsulatus grown on methane. Nature 230, 473–474 10.1038/230473a0 (doi:10.1038/230473a0) [DOI] [PubMed] [Google Scholar]
- 7.Jackson CJ, Lamb DC, Marczylo T, Warrilow AGS, Manning NJ, Lowe DJ, Kelly DE, Kelly SL. 2002. A novel sterol 14α-demethylase/ferredoxin fusion protein (MCCYP51FX) from Methylococcus capsulatus represents a new class of the cytochrome P450 superfamily. J. Biol. Chem. 277, 46 959–46 965 10.1074/jbc.M203523200 (doi:10.1074/jbc.M203523200) [DOI] [PubMed] [Google Scholar]
- 8.Jackson CJ, Lamb DC, Warrilow AGS, Parker JE, de Melo NR, Kelly DE, Kelly SL. 2008. P450s in microbial sterol biosynthesis and drug targets. Acta Chim. Slov. 55, 58–62 [Google Scholar]
- 9.Pearson A, Budin M, Brooks JJ. 2003. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl Acad. Sci. USA 100, 15 352–15 357 10.1073/pnas.2536559100 (doi:10.1073/pnas.2536559100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bode HB, Zeggel B, Silakowski B, Wenzel SC, Reichenbach H, Muller R. 2003. Steroid biosynthesis in prokaryotes: identification of myxobacterial steroids and cloning the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca. Mol. Microbiol. 47, 471–481 10.1046/j.1365-2958.2003.03309.x (doi:10.1046/j.1365-2958.2003.03309.x) [DOI] [PubMed] [Google Scholar]
- 11.Desmond E, Gribaldo S. 2009. Phylogenomics of sterol biosynthesis: insights into the origin, evolution and diversity of a key eukaryotic feature. Genome Biol. Evol. 1, 364–381 10.1093/gbe/evp036 (doi:10.1093/gbe/evp036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson CJ, Lamb DC, Marczylo TH, Parker JE, Manning NJ, Kelly DE, Kelly SL. 2003. Conservation and cloning of CYP51: a sterol 14-demethylase from Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 301, 558–563 10.1016/S0006-291X(02)03078-4 (doi:10.1016/S0006-291X(02)03078-4) [DOI] [PubMed] [Google Scholar]
- 13.Galea AM, Brown AJ. 2009. Special relationship between sterols and oxygen: were sterols an adaptation to aerobic life? Free Radic. Biol. Med. 47, 880–889 10.1016/j.freeradbiomed.2009.06.027 (doi:10.1016/j.freeradbiomed.2009.06.027) [DOI] [PubMed] [Google Scholar]
- 14.Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 10.1126/science.1093857 (doi:10.1126/science.1093857) [DOI] [PubMed] [Google Scholar]
- 15.Lamb DC, Waterman MR, Kelly SL, Guenguerich FP. 2007. Cytochromes P450 and drug discovery. Curr. Opin. Biotechnol. 18, 504–512 10.1016/j.copbio.2007.09.010 (doi:10.1016/j.copbio.2007.09.010) [DOI] [PubMed] [Google Scholar]
- 16.Omura T, Sato R. 1962. A new cytochrome in liver microsomes. J. Biol. Chem. 237, 1375–1376 [PubMed] [Google Scholar]
- 17.Estabrook RW, Cooper DY, Rosenthal O. 1963. The light reversible carbon monoxide inhibition of the steroid C21-hydroxylase system of the adrenal cortex. Biochem. Z. 338, 741. [PubMed] [Google Scholar]
- 18.Lindenmeyer A, Smith L. 1964. Cytochromes and other pigments of baker's yeast grown aerobically and anaerobically. Biochem. Biophys. Acta 93, 445–461 10.1016/0304-4165(64)90329-0 (doi:10.1016/0304-4165(64)90329-0) [DOI] [PubMed] [Google Scholar]
- 19.Lamb DC, Skaug T, Song H, Jackson CJ, Podust L, Waterman MR, Kell DB, Kelly DE, Kelly SL. 2002. The cytochrome P450 complement (CYPome) of Streptomyces coelicolor A3(2). J. Biol. Chem. 277, 24 000–24 005 10.1074/jbc.M111109200 (doi:10.1074/jbc.M111109200) [DOI] [PubMed] [Google Scholar]
- 20.Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson CJ, Omura S, Waterman MR, Kelly SL. 2003. Cytochrome P450 complement of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem. Biophys. Res. Commun. 307, 610–619 10.1016/S0006-291X(03)01231-2 (doi:10.1016/S0006-291X(03)01231-2) [DOI] [PubMed] [Google Scholar]
- 21.Jackson CJ, Lamb DC, Kelly DE, Kelly SL. 2000. Bactericidal and inhibitory effects of azole antifungal compounds on Mycobacterium smegmatis. FEMS Microbiol. Lett. 192, 159–162 10.1111/j.1574-6968.2000.tb09375.x (doi:10.1111/j.1574-6968.2000.tb09375.x) [DOI] [PubMed] [Google Scholar]
- 22.Guardiola-Diaz HM, Foster L-A, Mushrush D, Vaz ADN. 2001. Azole-antifungal binding to a novel cytochrome P450 from Mycobacterium tuberculosis: implications for the treatment of tuberculosis. Biochem. Pharmacol. 61, 1463–1470 10.1016/S0006-2952(01)00571-8 (doi:10.1016/S0006-2952(01)00571-8) [DOI] [PubMed] [Google Scholar]
- 23.Ahmad Z, Sharma S, Khuller GK. 2006. Azole antifungals as novel chemotherapeutic agents against murine tuberculosis. FEMS Microbiol. Lett. 261, 181–186 10.1111/j.1574-6968.2006.00350.x (doi:10.1111/j.1574-6968.2006.00350.x) [DOI] [PubMed] [Google Scholar]
- 24.Khatri Y, Hannemann F, Ewen KM, Pistorius D, Perlova O, Kagawa N, Brachmann AO, Müller R, Bernhardt R. 2010. The CYPome of Sorangium cellulosum So ce56 and identification of CYP109D1 as a new fatty acid hydroxylase. Chem. Biol. 17, 1295–1305 10.1016/j.chembiol.2010.10.010 (doi:10.1016/j.chembiol.2010.10.010) [DOI] [PubMed] [Google Scholar]
- 25.Sela DA, et al. 2008. The genome sequence of Bifidiobacterium longum subsp infantis . Proc. Natl Acad. Sci. USA 105, 18 964–18 969 10.1073/pnas.0809584105 (doi:10.1073/pnas.0809584105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Appleby CA. 1967. A soluble haemoprotein P450 from nitrogen-fixing rhizobium bacteroids. Biochem. Biophys. Acta 147, 399–402 10.1016/0005-2795(67)90423-0 (doi:10.1016/0005-2795(67)90423-0) [DOI] [PubMed] [Google Scholar]
- 27.Katagiri M, Ganguli BN, Gunsalus IC. 1968. A soluble cytochrome P450 functional in methylene hydroxylation. J. Biol. Chem. 243, 3543–3546 [PubMed] [Google Scholar]
- 28.Tyson CA, Lipscomb JD, Gunsalus IC. 1972. The role of putidaredoxin and P450 cam in methylene hydroxylation. J. Biol. Chem. 247, 5777–5784 [PubMed] [Google Scholar]
- 29.Rheinwald JG, Chakrabarty AM, Gunsalus IC. 1973. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc. Natl Acad. Sci. USA 70, 885–889 10.1073/pnas.70.3.885 (doi:10.1073/pnas.70.3.885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sligar SG, Debrunner PG, Lipscomb JD, Namtvedt MJ, Gunsalus IC. 1974. A role of the putidaredoxin COOH-terminus in P-450cam (cytochrome m) hydroxylations. Proc. Natl Acad. Sci. USA 71, 3906–3910 10.1073/pnas.71.10.3906 (doi:10.1073/pnas.71.10.3906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. 1985. The 2.6A crystal structure of Pseudomonas putida cytochrome P450 . J. Biol. Chem. 260, 16 122–16 130 [PubMed] [Google Scholar]
- 32.Rylott EL, Jackson RG, Edwards J, Womack GL, Seth-Smith SMB, Rathbone DA, Strand SE, Bruce NC. 2006. An explosive-degrading cytochrome P450 activity and its targeted application for the phytoremediation of RDX. Nat. Biotechnol. 24, 216–219 10.1038/nbt1184 (doi:10.1038/nbt1184) [DOI] [PubMed] [Google Scholar]
- 33.Lebeault JM, Lode ET, Coon MJ. 1971. Fatty acid and hydrocarbon hydroxylation in yeast: role of cytochrome P-450 in Candida tropicalis. Biochem. Biophys. Res. Commun. 42, 413–419 10.1016/0006-291X(71)90386-X (doi:10.1016/0006-291X(71)90386-X) [DOI] [PubMed] [Google Scholar]
- 34.Smith KE, Ahmed F, Antoniou T. 1993. Microbial transformation of steroids. Biochem. Soc. Trans. 21, 1077–1080 10.1042/bst0211077 (doi:10.1042/bst0211077) [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Saiz M, Barredo JL, Moreno MA, Fernandez-Canon JM, Penalva MA, Diez B. 2001. Reduced function of a phenylacetate-oxidising cytochrome P450 caused by strong genetic improvement in early phylogeny of penicillin-producing strains. J. Bacteriol. 183, 5465–5471 10.1128/JB.183.19 (doi:10.1128/JB.183.19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hannemann F, Bichet A, Ewen KM, Bernhardt R. 2007. Cytochrome P450 systems–biological variations of electron transport chains. Biochim. Biophys. Acta 1770, 330–344 10.1016/j.bbagen.2006.07.017 (doi:10.1016/j.bbagen.2006.07.017) [DOI] [PubMed] [Google Scholar]
- 37.Chun YJ, Shimada T, Sanchez-Ponce R, Martin MV, Lei L, Zhao B, Kelly SL, Waterman MR, Lamb DC, Guengerich FP. 2007. Electron transport pathway for a Streptomyces cytochrome P450: cytochrome P450 105D5-catalyzed fatty acid hydroxylation in Streptomyces coelicolor A3(2). J. Biol. Chem. 282, 17 486–17 500 10.1074/jbc.M700863200 (doi:10.1074/jbc.M700863200) [DOI] [PubMed] [Google Scholar]
- 38.Sutter TC, Loper JC. 1989. Disruption of the Saccharomyces cerevisiae gene for NADPH-cytochrome P450 reductase causes increased sensitivity to ketoconazole. Biochem. Biophys. Res. Commun. 160, 1257–1266 10.1016/S0006-291X(89)80139-1 (doi:10.1016/S0006-291X(89)80139-1) [DOI] [PubMed] [Google Scholar]
- 39.Venkateswarlu K, Lamb DC, Kelly DE, Manning NJ, Kelly SL. 1998. The N-terminal membrane domain of yeast NADPH-cytochrome P450 (CYP) oxidoreductase is not required for catalytic activity in sterol biosynthesis or in reconstitution of CYP activity. J. Biol. Chem. 273, 4492–4496 10.1074/jbc.273.8.4492 (doi:10.1074/jbc.273.8.4492) [DOI] [PubMed] [Google Scholar]
- 40.Lamb DC, Kelly DE, Manning NJ, Kaderbhai MA, Kelly SL. 1999. Biodiversity of the P450 catalytic cycle: yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction. FEBS Lett. 462, 283–288 10.1016/S0014-5793(99)01548-3 (doi:10.1016/S0014-5793(99)01548-3) [DOI] [PubMed] [Google Scholar]
- 41.Tiedje C, Holland DG, Just U, Hofken T. 2007. Proteins involved in sterol synthesis interact with Ste20 and regulate cell polarity. J. Cell Sci. 120, 3613–3624 10.1242/jcs.009860 (doi:10.1242/jcs.009860) [DOI] [PubMed] [Google Scholar]
- 42.Lamb DC, Kim Y, Yermalitskaya LV, Yermalitsky VN, Lepesheva GI, Kelly SL, Waterman MR, Podust LM. 2005. A second FMN binding site in yeast NADPH-cytochrome P450 reductase suggests a mechanism of electron transfer by diflavin reductases. Structure 14, 51–61 10.1016/j.str.2005.09.015 (doi:10.1016/j.str.2005.09.015) [DOI] [PubMed] [Google Scholar]
- 43.Mizutani M, Ohta D. 1998. Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 116, 357–367 10.1104/pp.116.1.357 (doi:10.1104/pp.116.1.357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelly D, Krasevec N, Mullins J, Nelson DR. 2009. The CYPome (cytochrome P450 complement) of Aspergillus nidulans. Fungal Genet. Biol. 46, S53–S61 10.1016/j.fgb.2008.08.010 (doi:10.1016/j.fgb.2008.08.010) [DOI] [PubMed] [Google Scholar]
- 45.Warrilow AG, Melo N, Martel CM, Parker JE, Nes WD, Kelly SL, Kelly DE. 2010. Expression, purification, and characterization of Aspergillus fumigatus sterol 14-alpha demethylase (CYP51) isoenzymes A and B. Antimicrob. Agents Chemother. 54, 4225–4234 10.1128/AAC.00316-10 (doi:10.1128/AAC.00316-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martel CM, Parker JE, Warrilow AG, Rolley NJ, Kelly SL, Kelly DE. 2010. Complementation of a Saccharomyces cerevisiae ERG11/CYP51 (sterol 14α-demethylase) doxycycline-regulated mutant and screening of the azole sensitivity of Aspergillus fumigatus isoenzymes CYP51A and CYP51B. Antimicrob. Agents Chemother. 54, 4920–4923 10.1128/AAC.00349-10 (doi:10.1128/AAC.00349-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vitas M, Smith KE, Plavec J, Kesselmeier J, Pajic T, Ferlan A, Zigon RD, Kelly SL, Komel R. 1999. Induction of steroidal hydroxylase activity by plant defence compounds in the filamentous fungus Cochliobolus lunatus. Chemosphere 38, 853–863 10.1016/S0045-6535(98)00229-X (doi:10.1016/S0045-6535(98)00229-X) [DOI] [PubMed] [Google Scholar]
- 48.Lah L, et al. 2011. The versatility of the fungal cytochrome P450 monooxygenase system is instrumental in xenobiotic detoxification. Mol. Microbiol. 81, 1374–1389 10.1111/j.1365-2958.2011.07772.x (doi:10.1111/j.1365-2958.2011.07772.x) [DOI] [PubMed] [Google Scholar]
- 49.Hawkes DB, Adams GW, Burlingame AL, Ortiz de Montellano PR, De Voss JJ. 2002. Cytochrome P450(cin) (CYP176A), isolation, expression, and characterization. J. Biol. Chem. 277, 27 725–27 732 10.1074/jbc.M203382200 (doi:10.1074/jbc.M203382200) [DOI] [PubMed] [Google Scholar]
- 50.Puchkaev AV, Ortiz de Montellano PR. 2005. The Sulfolobus solfataricus electron donor partners of thermophilic CYP119: an unusual non-NAD(P)H-dependent cytochrome P450 system. Arch. Biochem. Biophys. 434, 169–177 10.1016/j.abb.2004.10.022 (doi:10.1016/j.abb.2004.10.022) [DOI] [PubMed] [Google Scholar]
- 51.Seth-Smith SM, Rosser SJ, Basran A, Travis ER, Dabbs ER, Nicklin S, Bruce NC. 2002. Cloning, sequencing, and characterization of the hexahydro-1,3,5-trinitro-1,3,5-triazine degradation gene cluster from Rhodococcus rhodochrous. Appl. Environ. Microbiol. 68, 4764–4771 10.1128/AEM.68.10.4764-4771.2002 (doi:10.1128/AEM.68.10.4764-4771.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts GA, Grogan G, Greter A, Flitsch SL, Turner NJ. 2002. Identification of a new class of cytochrome P450 from Rhodococcus sp. J. Bacteriol. 184, 3898–3908 10.1128/JB.184.14 (doi:10.1128/JB.184.14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts GA, Celik A, Hunter DJ, Ost TW, White JH, Chapman SK, Turner NJ, Flitsch SL. 2003. A self-sufficient cytochrome P450 with a primary structural organisation that includes flavin domain and a (2Fe-2S) redox center. J. Biol. Chem. 278, 48 914–48 984 10.1074/jbc.M309630200 (doi:10.1074/jbc.M309630200) [DOI] [PubMed] [Google Scholar]
- 54.Wen LP, Fulco AJ. 1987. Cloning of the gene encoding a catalytically self-sufficient cytochrome P450 fatty-acid monooxygenase induced by barbiturates in Bacillus megaterium and its functional expression and regulation in heterologous (Escherichia coli) and homologous (Bacillus megaterium) hosts. J. Biol. Chem. 262, 6676–6682 [PubMed] [Google Scholar]
- 55.Kitazume T, Takaya N, Nakayama N, Shoun H. 2000. Fusarium oxysporum fatty-acid subterminal hydroxylase is a membrane-bound eukaryotic counterpart of Bacillus megaterium cytochrome P450BM3. J. Biol. Chem. 275, 39 734–39 740 10.1074/jbc.M005617200 (doi:10.1074/jbc.M005617200) [DOI] [PubMed] [Google Scholar]
- 56.Tomura D, Obika K, Fukamizu A, Shoun H. 1994. Nitric oxide reductase cytochrome P-450 gene, CYP55, of the fungus Fusarium oxysporum containing a potential binding-site for FNR, the transcription factor involved in the regulation of anaerobic growth of Escherichia coli. J. Biochem. 116, 88–94 [DOI] [PubMed] [Google Scholar]
- 57.Song WC, Funk CD, Brash AR. 1993. Molecular cloning of an allene oxide synthase: a cytochrome P450 specialised for the metabolism of fatty acid hydroperoxides. Proc. Natl Acad. Sci. USA 90, 8519–8523 10.1073/pnas.90.18.8519 (doi:10.1073/pnas.90.18.8519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsunaga I, Ueda A, Sumimoto T, Ichihara K, Ayata M, Ogura H. 2001. Site-directed mutagenesis of the putative distal helix of peroxygenase cytochrome P450. Arch. Biochem. Biophys. 394, 45–53 10.1006/abbi.2001.2512 (doi:10.1006/abbi.2001.2512) [DOI] [PubMed] [Google Scholar]
- 59.Brodhun F, Gobel C, Hornung E, Feussner I. 2009. Identification of PpoA from Aspergillus nidulans as a fusion protein of a fatty acid haem dioxygenase/peroxidise and a cytochrome P450. J. Biol. Chem. 284, 11 792–11 805 10.1074/jbc.M809152200 (doi:10.1074/jbc.M809152200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao B, Lei L, Vassylyev DG, Lin X, Cane DE, Kelly SL, Yuan H, Lamb DC, Waterman MR. 2009. Crystal structure of albaflavanone monooxygenase containing a moonlighting terpene synthase. J. Biol. Chem. 284, 36 711–36 719 10.1074/jbc.M109.064683 (doi:10.1074/jbc.M109.064683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warrilow A, Ugochukwu C, Lamb D, Kelly D, Kelly S. 2008. Expression and characterization of CYP51, the ancient sterol 14-demethylase activity for cytochromes P450 (CYP), in the white-rot fungus Phanerochaete chrysosporium. Lipids 43, 1143–1153 10.1007/s11745-008-3239-5 (doi:10.1007/s11745-008-3239-5) [DOI] [PubMed] [Google Scholar]
- 62.Masaphy S, Levanon D, Henis Y, Venkateswarlu K, Kelly SL. 1996. Evidence for cytochrome P-450 and P-450 mediated benzo(a)pyrene hydroxylation in the white-rot fungus Phanerochaete chrysosporium. FEMS Lett. 135, 51–55 10.1016/0378-1097(95)00428-9 (doi:10.1016/0378-1097(95)00428-9) [DOI] [PubMed] [Google Scholar]
- 63.Syed K, Doddapaneni H, Subramanian V, Lam YW, Yadav JS. 2010. Genome to function characterisation of novel fungal P450 monooxygenases oxidising polycyclic aromatic hydrocarbons (PAHs). Biochem. Biophys. Res. Commun. 399, 492–497 10.1016/j.bbrc.2010.07.094 (doi:10.1016/j.bbrc.2010.07.094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doddapaneni H, Subramanian V, Yadav JS. 2005. Physiological regulation, xenobiotic induction and heterologous expression of P450 monoxygenase gene pc-3 (CYP63A3), a new member of the CYP63 gene cluster in the white-rot fungus Phanerochaete chrysosporium . Curr. Microbiol. 50, 292–298 10.1007/s00284-005-4480-2 (doi:10.1007/s00284-005-4480-2) [DOI] [PubMed] [Google Scholar]
- 65.Matsuzaki F, Wariishi H. 2005. Molecular characterisation of cytochrome P450 catalyzing hydroxylation of benzoates from the white-rot fungus Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 334, 1184–1189 10.1016/j.bbrc.2005.07.013 (doi:10.1016/j.bbrc.2005.07.013) [DOI] [PubMed] [Google Scholar]
- 66.Eastwood DC, et al. 2011. The plant cell-wall decomposing machinery underlies the functional diversity of forest fungi. Science 333, 762–765 10.1126/science.1205411 (doi:10.1126/science.1205411) [DOI] [PubMed] [Google Scholar]
- 67.Ide M, Ichinose H, Wariishi H. 2011. Molecular identification and functional characterisation of cytochrome P450 monooxygenases from the brown-rot basidiomycete Postia placenta. Arch. Microbiol. (doi:10.1007/s00203-011-0753-2) [DOI] [PubMed] [Google Scholar]
- 68.Nazmul H, Nazir KH, Ichinose H, Wariishe H. 2010. Molecular characterization and isolation of cytochrome P450 genes from the filamentous fungus Aspergillus oryzae . Arch. Microbiol. 192, 395–408 10.1007/S00203-010-0562-z (doi:10.1007/S00203-010-0562-z) [DOI] [PubMed] [Google Scholar]
- 69.Kelly SL, Kelly DE, Jackson CJ, Warrilow AG, Lamb DC. 2005. The diversity and importance of cytochrome P450 in microbes. In Cytochrome P450 (ed. Ortiz de Montellano P.), pp. 585–617 New York, NY: Plenum Press [Google Scholar]
- 70.Malandain C, Fayolle-Gulchard F, Vogel TM. 2010. Cytochromes P450-mediated degradation of fuel oxygenates by environmental isolates. FEMS Microbiol. Ecol. 72, 289–296 10.1111/j.1574-6941.2010.00847.x (doi:10.1111/j.1574-6941.2010.00847.x) [DOI] [PubMed] [Google Scholar]
- 71.Kobayashi M, Matsuo Y, Takimoto A, Suzuki S, Maruo F, Shoun H. 1996. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 271, 16 263–16 267 10.1074/jbc.271.27.16263 (doi:10.1074/jbc.271.27.16263) [DOI] [PubMed] [Google Scholar]
- 72.Ma WK, Farrell RE, Siciliano SD. 2008. Soil formate regulates the fungal nitrous oxide emission pathway. Appl. Environ. Microbiol. 74, 6690–6696 10.1128/AEM.00797-08 (doi:10.1128/AEM.00797-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hayatsu M, Tago K, Saito M. 2008. Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci. Plant Nutr. 54, 33–45 10.1111/j.1747-0765.2007.00195.x (doi:10.1111/j.1747-0765.2007.00195.x) [DOI] [Google Scholar]
- 74.Mueller UG, Gerado N. 2002. Fungus-farming insects: multiple origins and diverse evolutionary histories. Proc. Natl Acad. Sci. USA 99, 15 247–15 249 10.1073/pnas.242594799 (doi:10.1073/pnas.242594799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedrini N, Zhang S, Juarez MP, Keyhani NO. 2010. Molecular characterization and expression analysis of a suite of cytochrome P450 enzymes implicated in insect hydrocarbon degradation in the entomopathogenic fungus Beauveria bassiana. Microbiology 156, 2549–2557 10.1099/mic.0.039735-0 (doi:10.1099/mic.0.039735-0) [DOI] [PubMed] [Google Scholar]
- 76.Zimmer T, Ohkuma M, Ohta A, Takagi M, Schunck WJ. 1996. The CYP52 multigene family of Candida maltosa encodes functionally diverse n-alkane-inducible cytochromes P450. Biochem. Biophys. Res. Commun. 224, 784–789 10.1006/bbrc.1996.1100 (doi:10.1006/bbrc.1996.1100) [DOI] [PubMed] [Google Scholar]
- 77.Ohkuma M, Zimmer T, Iida T, Schunck WH, Ohta A, Takagi M. 1998. Isozyme function of n-alkane-inducible cytochromes P450 in Candida maltose revealed by sequential gene disruption. J. Biol. Chem. 273, 3948–3953 10.1074/jbc.273.7.3948 (doi:10.1074/jbc.273.7.3948) [DOI] [PubMed] [Google Scholar]
- 78.Ragsdale NN, Sisler HD. 1972. Inhibition of ergosterol synthesis in Ustilago maydis by the fungicide triarimol. Biochem. Biophys. Res. Commun. 46, 2048–2053 10.1016/0006-291X(72)90757-7 (doi:10.1016/0006-291X(72)90757-7) [DOI] [PubMed] [Google Scholar]
- 79.Parker JE, Warrilow AG, Cools HJ, Martel CM, Nes WR, Fraaije BA, Lucas JA, Kelly DE, Kelly SL. 2011. Mechanism of binding of prothioconazole to Mycosphaerella graminicola CYP51 differs from that of other azole antifungals. Appl. Environ. Microbiol. 77, 1460–1465 10.1128/AEM.01332-10 (doi:10.1128/AEM.01332-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Torriani SF, Brunner PC, McDonald BA, Sierotzki H. 2009. QoI resistance emerged independently at least 4 times in European populations of Mycosphaerella graminicola. Pest. Manag. Sci. 65, 155–162 10.1002/ps.1662 (doi:10.1002/ps.1662) [DOI] [PubMed] [Google Scholar]
- 81.Aoyama Y, Yoshida Y, Hata S, Nishino T, Katsuki H. 1983. Buthiobate: a potent inhibitor for yeast cytochrome P450 catalysing 14α-demethylation of lanosterol. Biochem. Biophys. Res. Commun. 115, 642–647 10.1016/S0006-291X(83)80192-2 (doi:10.1016/S0006-291X(83)80192-2) [DOI] [PubMed] [Google Scholar]
- 82.Kalb VF, Woods CW, Turi TG, Dey CR, Sutter TR, Loper JC. 1987. Primary structure of P450 lanosterol demethylase gene from Saccharomyces cerevisiae. DNA 6, 529–537 [DOI] [PubMed] [Google Scholar]
- 83.Lamb DC, Kelly DE, Kelly SL. 1998. Molecular diversity of sterol 14α-demethylase substrates in plants, fungi and humans. FEBS Lett. 425, 263–265 10.1016/S0014-5793(98)00247-6 (doi:10.1016/S0014-5793(98)00247-6) [DOI] [PubMed] [Google Scholar]
- 84.Akhtar M, Alexander K, Boar RB, McGhie JF, Barton DHR. 1978. Chemical and enzymic studies on the characterisation of intermediates during the removal of the 14α methyl group in cholesterol biosynthesis. Biochem. J. 169, 449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aoyama Y, Yoshida Y, Sato R. 1984. Yeast cytochrome P450 catalysing lanosterol 14α-demethylation. II. Lanosterol metabolism by purified P45014DM and by intact microsomes. J. Biol. Chem. 259, 1661–1666 [PubMed] [Google Scholar]
- 86.Shyadehi AZ, Lamb DC, Kelly SL, Kelly DE, Schunck WH, Wright JN, Corina D, Akhtar M. 1996. The mechanism of the acyl-carbon bond cleavage reaction catalyzed by recombinant sterol 14 α-demethylase of Candida albicans (other names are: lanosterol 14 alpha-demethylase, P-45014DM, and CYP51). J. Biol. Chem. 271, 12 445–12 450 10.1074/jbc.271.21.12445 (doi:10.1074/jbc.271.21.12445) [DOI] [PubMed] [Google Scholar]
- 87.Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Parks LW, Kelly DE. 1995. Purification and reconstitution of activity of Saccharomyces cerevisiae P450 61, a sterol 22-desaturase. FEBS Lett. 377, 217–220 10.1016/0014-5793(95)01342-3 (doi:10.1016/0014-5793(95)01342-3) [DOI] [PubMed] [Google Scholar]
- 88.Morikawa T, et al. 2006. Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. Plant Cell 18, 1008–1022 10.1105/tpc.105.036012 (doi:10.1105/tpc.105.036012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelly SL, Lamb DC, Baldwin BC, Corran AJ, Kelly DE. 1997. Characterisation of Saccharomyces cerevisiae CYP61, a sterol delta 22-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 272, 9986–9988 10.1074/jbc.272.15.9986 (doi:10.1074/jbc.272.15.9986) [DOI] [PubMed] [Google Scholar]
- 90.Kelly SL, Lamb DC, Kelly DE. 1997. Sterol 22-desaturase, cytochrome P450 61, possesses activity in xenobiotic metabolism. FEBS Lett. 412, 233–235 10.1016/S0014-5793(97)00785-0 (doi:10.1016/S0014-5793(97)00785-0) [DOI] [PubMed] [Google Scholar]
- 91.Kodner RB, Summons RE, Pearson A, King N, Knoll AH. 2008. Sterols in a unicellular relative of the metazoans. Proc. Natl Acad. Sci. USA 105, 9897–9902 10.1073/pnas.0803975105 (doi:10.1073/pnas.0803975105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hagemann A, Voigt O, Worheide G, Thiel V. 2008. The sterols of calcareous sponges (Calcarea, Porifera). Chem. Phys. Lipids 156, 26–32 10.1016/j.chemphyslip.2008.07.001 (doi:10.1016/j.chemphyslip.2008.07.001) [DOI] [PubMed] [Google Scholar]
- 93.Strushkevich N, Usanov SA, Park HW. 2010. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 397, 1067–1078 10.1016/j.jmb.2010.01.075 (doi:10.1016/j.jmb.2010.01.075) [DOI] [PubMed] [Google Scholar]
- 94.Cools HJ, Parker JE, Kelly DE, Lucas JA, Fraaije BA, Kelly SL. 2010. Heterologous expression of mutated eburicol 14alpha-demethylase (CYP51) proteins of Mycosphaerella graminicola to assess effects on azole fungicide sensitivity and intrinsic protein function. Appl. Environ. Microbiol. 76, 2866–2872 10.1128/AEM.02158-09 (doi:10.1128/AEM.02158-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cools HJ, Mullins JG, Fraaije BA, Parker JE, Kelly DE, Lucas JA, Kelly SL. 2011. Impact of recently emerged sterol 14{alpha}-demethylase (CYP51) variants of Mycosphaerella graminicola on azole fungicide sensitivity. Appl. Environ. Microbiol. 77, 3830–3837 10.1128/AEM.00027-11 (doi:10.1128/AEM.00027-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mullins JG, Parker JE, Cools HJ, Togawa RC, Lucas JA, Fraaije BA, Kelly DE, Kelly SL. 2011. Molecular modelling of the emergence of azole resistance in Mycosphaerella graminicola. PLoS ONE 6, e20973. 10.1371/journal.pone.0020973 (doi:10.1371/journal.pone.0020973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Artigot MP, Loiseau N, Laffitte J, Mas-Reguieg L, Tadrist S, Oswald IP, Puel O. 2009. Molecular cloning and functional characterization of two CYP619 cytochrome P450s involved in biosynthesis of patulin in Aspergillus clavatus. Microbiology 155, 1738–1747 10.1099/mic.0.024836-0 (doi:10.1099/mic.0.024836-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hohn TM, Desjardins AE, McCormick SP. 1995. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monoxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet. 248, 95–102 10.1128/AEM.68.6.2959-2964.2002 (doi:10.1128/AEM.68.6.2959-2964.2002) [DOI] [PubMed] [Google Scholar]
- 99.Alexander NJ, Hohn TM, McCormick SP. 1998. The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase reuired for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 64, 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Keller NP, Segner S, Bhatnagar D, Adams TH. 1995. stcS, a putative P-450 monooxygenase, is required for the conversion of versicolorin A to sterigmatocystin in Aspergillus nidulans . Appl. Environ. Microbiol. 61, 3628–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haarmann T, Ortel I, Tudzynski P, Keller U. 2006. Identification of the cytochrome P450 monoxygenase that bridges the clavinet and ergoline alkaloid pathways. ChemBioChem. 7, 645–652 10.1002/cbic.200500487 (doi:10.1002/cbic.200500487) [DOI] [PubMed] [Google Scholar]
- 102.Healy FG, Krasnoff SB, Wach M, Gibson DM, Loria R. 2002. Involvement of a cytochrome P450 monooxygenase in thaxtomin A biosynthesis by Streptomyces acidiscabies . J. Bacteriol. 184, 2019–2029 10.1128/JB.184.7.2019-2029 (doi:10.1128/JB.184.7.2019-2029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tyler BM, et al. 2009. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266 10.1126/science.1128796 (doi:10.1126/science.1128796) [DOI] [PubMed] [Google Scholar]
- 104.Gaulin E, Bottin A, Dumas B. 2010. Sterol biosynthesis in oomycete pathogens. Plant Signal. Behav. 5, 258–260 10.4161/psb.5.3.10551 (doi:10.4161/psb.5.3.10551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Birch AJ. 1992. Steroid hormones and the Luftwaffe. A venture into fundamental strategic research and some of its consequences: the Birch reduction becomes a birth reduction. Steroids 57, 363–377 [DOI] [PubMed] [Google Scholar]
- 106.Petric S, Hakki R, Bernhardt R, Zigon D, Cresnar B. 2010. Discovery of a steroid 11α-hydroxylase from Rhizopus oryzae and its biotechnological application. J. Biotechnol. 150, 428–437 10.1016/j.jbiotec.2010.09.928 (doi:10.1016/j.jbiotec.2010.09.928) [DOI] [PubMed] [Google Scholar]
- 107.Szczebara FM, et al. 2003. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 21, 143–149 10.1038/nbt775 (doi:10.1038/nbt775) [DOI] [PubMed] [Google Scholar]
- 108.Souza CM, Schwabe TME, Pichler H, Ploier B, Leitner E, Guan XL, Wenk MR, Riezman I, Riezman H. 2011. A stable yeast strain efficiently producing cholesterol instead of ergosterol is functional for tryptophan uptake, but not weak organic acid resistance. Metab. Eng. 13, 555–569 10.1016/j.ymben.2011.06.006 (doi:10.1016/j.ymben.2011.06.006) [DOI] [PubMed] [Google Scholar]
- 109.Ro DK, et al. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943 10.1038/nature04640 (doi:10.1038/nature04640) [DOI] [PubMed] [Google Scholar]
- 110.Ajikumar PK, et al. 2010. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330, 70–74 10.1126/science.1191652 (doi:10.1126/science.1191652) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matsuoka T, Miyakoshi S, Tanzawa K, Nakahara K, Hosobuchi M, Serizawa N. 1989. Purification and characterisation of cytochrome P450sca from Streptomyces carbophilus. Eur. J. Biochem. 184, 707–713 10.1111/j.1432-1033.1989.tb15070.x (doi:10.1111/j.1432-1033.1989.tb15070.x) [DOI] [PubMed] [Google Scholar]
- 112.Scheller U, Zimmer T, Becher D, Schauer F, Schunck WH. 1998. Oxygenation cascade in conversion of n-alkanes to alpha,omega-dioic acids catalyzed by cytochrome P450 52A3. J. Biol. Chem. 273, 32 528–32 534 10.1074/jbc.273.49.32528 (doi:10.1074/jbc.273.49.32528) [DOI] [PubMed] [Google Scholar]
- 113.Craft DL, Madduri KM, Eshoo M, Wilson CR. 2003. Identification and characterization of the CYP52 family of Candida tropicalis ATCC 20336, important for the conversion of fatty acids and alkanes to α,ω-dicarboxylic acids. Appl. Environ. Microbiol. 69, 5983–5991 10.1128/AEM.69.10.5983-5991.2003 (doi:10.1128/AEM.69.10.5983-5991.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Van Bogaert IN, De Mey M, Develter D, Soetaert W, Vandamme EJ. 2009. Importance of the cytochrome P450 monooxygenase CYP52 family for the sphorolipid-producing yeast Candida bombicola. FEMS Yeast Res. 9, 87–94 10.1111/j.1567-1364.2008.00454.x (doi:10.1111/j.1567-1364.2008.00454.x) [DOI] [PubMed] [Google Scholar]
- 115.Sabbadin F, Hyde R, Robin A, Hilgarth EM, Delenne M, Flitsch S, Turner N, Grogan G, Bruce NC. 2010. LICRED: a versatile drop-in vector for rapid generation of redox-self sufficient cytochrome P450s. ChemBioChem 11, 987–994 10.1002/cbic.201000104 (doi:10.1002/cbic.201000104) [DOI] [PubMed] [Google Scholar]
- 116.Cheng Q, Lamb DC, Kelly SL, Lei L, Guenguerich FP. 2010. Cyclization of a cellular dipentaeone by Streptomyces coelicolor cytochrome P450 154A1 without oxidation/reduction. J. Am. Chem. Soc. 143, 15 173–15 175 10.1021/ja107801v (doi:10.1021/ja107801v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Urlacher VB, Makhsumkhanov A, Schmid RD. 2005. Biotransformation of β-ionone by engineered cytochrome P450 BM-3. Appl. Microbiol. Biotechnol. 70, 53–59 10.1007/s00253-005-0028-4 (doi:10.1007/s00253-005-0028-4) [DOI] [PubMed] [Google Scholar]
- 118.Barnes HJ, Arlotto MP, Waterman MR. 1991. Expression and enzymatic activity of recombinant cytochrome P450 17α-hydroxylase in Escherichia coli. Proc. Natl Acad. Sci. USA 88, 5597–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cosme J, Johnson EF. 2000. Engineering a microsomal cytochrome P450 2C5 to be a soluble, monomeric enzyme. Mutations that alter aggregation, phospholipid dependence of catalysis, and membrane binding. J. Biol. Chem. 275, 2545–2553 10.1074/jbc.275.4.2545 (doi:10.1074/jbc.275.4.2545) [DOI] [PubMed] [Google Scholar]
- 120.Lamb SB, Lamb DC, Kelly SL, Stuckey DC. 1998. Cytochrome P450 immobilisation as a route to bioremediation/biocatalysis. FEBS Lett. 431, 343–346 10.1016/S0014-5793(98)00771-6 (doi:10.1016/S0014-5793(98)00771-6) [DOI] [PubMed] [Google Scholar]
- 121.Nath A, Atkins WM, Sligar SG. 2007. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 10.1021/bi602371n (doi:10.1021/bi602371n) [DOI] [PubMed] [Google Scholar]
- 122.Taylor M, Lamb DC, Cannell RJ, Dawson M, Kelly SL. 2000. Cofactor recycling with immobilised heterologous cytochrome P450 105D1 (CYP105D1). Biochem. Biophys. Res. Commun. 279, 708–711 10.1006/bbrc.2000.4002 (doi:10.1006/bbrc.2000.4002) [DOI] [PubMed] [Google Scholar]
- 123.Estabrook RW, Faulkener KM, Shet MS, Fisher CW. 1996. Applications of electrochemistry for P450-catalyzed reactions. Methods Enzymol. 272, 44–51 10.1016/S0076-6879(96)72007-4 (doi:10.1016/S0076-6879(96)72007-4) [DOI] [PubMed] [Google Scholar]
- 124.Sadeghi SJ, Fantuzzi A, Gilardi G. 2011. Breakthrough in P450 bioelectrochemistry and future perspectives. Biochem. Biophys. Acta 1814, 237–248 10.1016/bbapap.2010.07.010 (doi:10.1016/bbapap.2010.07.010) [DOI] [PubMed] [Google Scholar]
- 125.Noble MA, Miles CS, Chapman SK, Lysek DA, MacKay AC, Reid GA, Hanzlik RP, Munro AW. 1999. Roles of key active-site residues in flavocytochrome P450 BM-3 (CYP102). Biochem. J. 37, 15 799–15 807 10.1021/bj980462d (doi:10.1021/bj980462d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Joo H, Lin Z, Arnold FH. 1999. Laboratory evolution of peroxide-mediated cytochrome P450 hydroxylation. Nature 399, 670–673 10.1038/21395 (doi:10.1038/21395) [DOI] [PubMed] [Google Scholar]
- 127.Cirino PC, Arnold FH. 2002. Regioselectivity and activity of cytochrome P450 BM-3 and mutant F87A in reactions driven by hydrogen peroxide. Adv. Synth. Catal. 344, 932–937 10.1002/1615-4169(200210 (doi:10.1002/1615-4169(200210)) [DOI] [Google Scholar]
- 128.Glieder A, Farinas ET, Arnold FH. 2002. Laboratory evolution of a soluble, self-sufficient, highly active alkane hydroxylase. Nat. Biotechnol. 20, 1135–1139 10.1038/nbt744 (doi:10.1038/nbt744) [DOI] [PubMed] [Google Scholar]
- 129.Koch DJ, Chen MJ, Van Bellen JB, Arnold FJ. 2008. In vivo evolution of butane oxidation by terminal alkane hydroxylases AlkB and CYP153A6. Appl. Environ. Microbiol. 75, 337–344 10.1128/AEM.01758-08 (doi:10.1128/AEM.01758-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A. 2011. Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 77, 1718–1727 10.1128/AEM.02580-10 (doi:10.1128/AEM.02580-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Andersen AF, Tatsuka K, Gunji H, Ishiyama T, Hutchinson CR. 1993. Substrate specificity of a 6-deoxyerythromycin B hydroxylase, a bacterial cytochrome P450 of erythromycin A biosynthesis. Biochemistry 32, 1905–1913 10.1021/bi00059a004 (doi:10.1021/bi00059a004) [DOI] [PubMed] [Google Scholar]
- 132.Nagano S, Cupp-Vickery JR, Poulos TL. 2005. Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T: investigation of the proton transfer system in P450eryF. J. Biol. Chem. 280, 22 102–22 107 10.1074/jbc.M501732200 (doi:10.1074/jbc.M501732200) [DOI] [PubMed] [Google Scholar]
- 133.Zhao B, et al. 2005. Binding of two flaviolin substrate molecules, oxidative coupling and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 CYP158A2. J. Biol. Chem. 280, 11 599–11 607 10.1074/jbc.M410933200 (doi:10.1074/jbc.M410933200) [DOI] [PubMed] [Google Scholar]
- 134.Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE. 2008. Biosynthesis of the sesquiterpene antibiotic albaflavone in Streptomyces coelicolor A3(2). J. Biol. Chem. 283, 8183–8189 10.1074/jbc.M710421200 (doi:10.1074/jbc.M710421200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cryle MJ, Meinhart A, Schlichting I. 2010. Structural characterization of OxyD, a cytochrome P450 involved in beta-hydroxytyrosine formation in vancomycin biosynthesis. J. Biol. Chem. 285, 24 562–24 574 10.1074/jbc.M110.131904 (doi:10.1074/jbc.M110.131904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmtic macrolide avermectin in Streptomyces avermitilis. Proc. Natl Acad. Sci. USA 96, 9509–9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Byrne B, Carmody M, Gibson E, Rawlings B, Caffrey P. 2003. Biosynthesis of doxyamphotericins and deoxyamphoteronolides by engineered strains of Streptomyces nodosus. Chem. Biol. 10, 1215–1224 10.1016/j.chembiol.2003.12.001 (doi:10.1016/j.chembiol.2003.12.001) [DOI] [PubMed] [Google Scholar]
- 138.Brautaset T, Sletta H, Degnes KF, Sekurova ON, Bakke I, Volokhan O, Andreassen T, Ellingsen TE, Zotchev SB. 2011. New nystatin-related antifungal polyene macrolides with altered polyol region generated via biosynthetic engineering of Streptomyces noursei. Appl. Environ. Microbiol. 77, 6636–6643 10.1128/AEM.05780-11 (doi:10.1128/AEM.05780-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Keller U, Lang M, Crnovcic I, Pfennig F, Schauwecker F. 2010. The actinomycete biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J. Bacteriol. 192, 2583–2595 10.1128/JB.01526-09 (doi:10.1128/JB.01526-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ogura H, Nishida CR, Hoch UR, Perera R, Dawson JH, Ortiz de Montellano PR. 2004. EpoK, a cytochrome P450 involved in biosythesis of the anticancer agents epothilones A and B. Substrate mediated rescue of a P450 enzyme. Biochemistry 43, 14 712–14 721 10.1021/bi048980d (doi:10.1021/bi048980d) [DOI] [PubMed] [Google Scholar]
- 141.Burriuso J, Nguyen DT, Li JW, Roberts JN, MacNevin G, Chaytor JL, Marcus SL, Vederas JC, Ro DK. 2011. Double oxidation of the cyclic nonaketide dihydromonacolin L to monacolin J by a single cytochrome P450 monooxygenase, LovA. J. Am. Chem. Soc. 133, 8078–8081 10.1021/ja201138v (doi:10.1021/ja201138v) [DOI] [PubMed] [Google Scholar]
- 142.Fisch KM, et al. 2011. Rational domain swaps decipher programming in fungal highly reducing polyketide synthases and resurrect an extinct metabolite. J. Am. Chem. Soc. 133, 16 635–16 641 10.1021/ja206914q (doi:10.1021/ja206914q) [DOI] [PubMed] [Google Scholar]
- 143.Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8, 76–81 10.1016/S1471-4914(02)02280-3 (doi:10.1016/S1471-4914(02)02280-3) [DOI] [PubMed] [Google Scholar]
- 144.Kelly SL, Arnoldi A, Kelly DE. 1993. Molecular genetic analysis of azole antifungal mode of action. Biochem. Soc. Trans. 21, 1034–1038 10.1054/drup.1999.0112 (doi:10.1054/drup.1999.0112) [DOI] [PubMed] [Google Scholar]
- 145.Venkateswarlu K, Denning DW, Manning NJ, Kelly SL. 1995. Resistance to fluconazole in Candida albicans from AIDS patients correlated with reduced intracellular accumulation of drug. FEMS Lett. 131, 337–341 10.1111/j.1574-6968.1995.tbc07797.x (doi:10.1111/j.1574-6968.1995.tbc07797.x) [DOI] [PubMed] [Google Scholar]
- 146.Sanglard D, Kuchler K, Ischer F, Pagani JL, Monod M, Bile J. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39, 2378–2386 10.1128/AAC.39.11.2378 (doi:10.1128/AAC.39.11.2378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Denning DW, Venkateswarlu K, Oakley KL, Anderson NJ, Manning NJ, Stevens DA, Warnock DW, Kelly SL. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41, 1364–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Verweij PE, Snelders E, Kema GHJ, Mellado E, Melchers WJG. 2009. Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect. Dis. 9, 789–795 10.1016/S1473-3099(09)70265-8 (doi:10.1016/S1473-3099(09)70265-8) [DOI] [PubMed] [Google Scholar]
- 149.Sucher AJ, Chahine EB, Balcer HE. 2009. Echinocandins: the newest class of antifungals. Ann. Pharmacother. 43, 1647–1657 10.1345/aph.1M237 (doi:10.1345/aph.1M237) [DOI] [PubMed] [Google Scholar]
- 150.Xu J, et al. 2007. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human pathogens. Proc. Natl Acad. Sci. USA 104, 18 730–18 735 10.1073/pnas.0606756104 (doi:10.1073/pnas.0606756104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Denning DW, O'Driscoll BR, Hogaboam CM, Bowyer P, Niven RM. 2006. The link between fungi and severe asthma: a summary of the evidence. Eur. Respir. J. 27, 615–626 10.1183/09031936.06.0074705 (doi:10.1183/09031936.06.0074705) [DOI] [PubMed] [Google Scholar]
- 152.Parker JE, Merkamm M, Manning NJ, Pompon D, Kelly SL, Kelly DE. 2008. Differential azole antifungal efficacies contrasted using a Saccharomyces cerevisiae strain humanized for sterol 14α-demethylase at the homologous locus. Antimicrob. Agents Chemother. 52, 3597–3603 10.1128/AAC.00517-08 (doi:10.1128/AAC.00517-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.White TC. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α-demethylase in Candida albicans. Antimicrob. Agents Chemother. 41, 1488–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Loffler J, Kelly SL, Hebart H, Schumacher U, Lass-Florl C, Einsele H. 1997. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Lett. 151, 263–268 10.1111/j.1574-6968.1997.tb12580.x (doi:10.1111/j.1574-6968.1997.tb12580.x) [DOI] [PubMed] [Google Scholar]
- 155.Sanglard D, Ischer F, Koymans L, Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42, 241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kelly SL, Lamb DC, Loeffler J, Einsele H, Kelly DE. 1999. The G464S amino acid substitution in Candida albicans sterol 14α-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem. Biophys. Res. Commun. 262, 174–179 10.1006/bbrc.1999.1136 (doi:10.1006/bbrc.1999.1136) [DOI] [PubMed] [Google Scholar]
- 157.Warrilow AGS, Martel CM, Parker JE, Melo N, Lamb DC, Nes WD, Kelly DE, Kelly SL. 2010. Azole binding properties of Candida albicans sterol 14-α demethylase (CaCYP51). Antimicrob. Agents Chemother. 54, 4235–4245 10.1128/AAC.00587-10 (doi:10.1128/AAC.00587-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.White TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41, 1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Franz R, Kelly SL, Lamb DC, Kelly DE, Ruknke M, Morschhauser J. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42, 3065–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Coste A, Turner V, Ischer F, Morschhauser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans . Genetics 172, 2139–2156 10.1534/genetics.105.054767 (doi:10.1534/genetics.105.054767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Watson PF, Rose ME, Ellis SW, England H, Kelly SL. 1989. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem. Biophys. Res. Commun. 164, 1170–1175 10.1016/0006-291X(89)91792-0 (doi:10.1016/0006-291X(89)91792-0) [DOI] [PubMed] [Google Scholar]
- 162.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AGS, Rolley N, Kelly DE, Kelly SL. 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54, 4527–4533 10.1128/AAC.00348-10 (doi:10.1128/AAC.00348-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gachotte D, Pierson CA, Lees ND, Barbuch R, Koegel C, Bard M. 1997. A yeast sterol auxotroph (erg25) is rescued by addition of azole antifungals and reduced levels of heme. Proc. Natl Acad. Sci. USA 94, 11 173–11 178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans . Science 313, 367–370 10.1126/science.1128242 (doi:10.1126/science.1128242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Albarag AM, Anderson M, Howard SJ, Robson GD, Warn PA, Sanglard D, Denning DW. 2011. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob. Agents Chemother. 55, 5113–5121 10.1128/AAC.00517-11 (doi:10.1128/AAC.00517-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Amolis SP, Heney C. 1999. Acanthamoebae keratitis with live isolates treated with cryosurgery and fluconazole. Am. J. Ophthamol. 127, 718–720 10.1016/S0002-9394(98)00426-7 (doi:10.1016/S0002-9394(98)00426-7) [DOI] [PubMed] [Google Scholar]
- 167.Lepesheva GL, et al. 2010. Crystal structures of Trypanosoma brucei sterol 14α-demethylase and implications for selective treatment of human infections. J. Biol. Chem. 285, 1773–1780 10.1074/jbc.M109.067470 (doi:10.1074/jbc.M109.067470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen CK, Leung SSF, Guilbert C, Jacobson MP, McKerrow JH, Podust L. 2010. Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and fluconazole. PLoS Negl. Trop. Dis. 4, e651. 10.1371/journal.pntd.000651 (doi:10.1371/journal.pntd.000651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Verma S, Mehta A, Shaha C. 2011. CYP5122A1, a novel cytochrome P450 is essential for survival of Leishmania donovani. PLoS ONE 6, e25273. 10.1371/journal.pone.0025273 (doi:10.1371/journal.pone.0025273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.McLean KJ, Lafite P, Levy C, Cheesman MR, Mast N, Pikuleva IA, Leys D, Munro AW. 2009. The structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection. J. Biol. Chem. 284, 35 524–35 533 10.1074/jbc.M109.032706 (doi:10.1074/jbc.M109.032706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Quellet H, Johnston JB, Ortiz de Montellano PR. 2011. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis . Trends Microbiol. 19, 530–539 10.1016/j.tim.2011.07.009 (doi:10.1016/j.tim.2011.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Warrilow AGS, Jackson CJ, Parker JE, Marczylo TH, Kelly DE, Lamb DC, Kelly SL. 2009. Identification, characterisation and azole binding properties of CYP164A2, a homolog of ML2088, the sole cytochrome P450 of Mycobacterium leprae. Antimicrob. Agents Chemother. 53, 1157–1164 10.1128/AAC.01237-08 (doi:10.1128/AAC.01237-08) [DOI] [PMC free article] [PubMed] [Google Scholar]