Abstract
Single-molecule (SM) spectroscopy has been an exciting area of research offering significant promise and hope in the field of sensor development to detect targets at ultra-low levels down to SM resolution. To the experts and developers in the field of surface-enhanced Raman spectroscopy (SERS), this has often been a challenge and a significant opportunity for exploration. Needless to say, the opportunities and excitement of this multidisciplinary area impacts span the fields of physics, chemistry and engineering, along with a significant thrust in applications constituting areas in medicine, biology, environment and agriculture among others. In this review, we will attempt to provide a quick snapshot of the basics of SM-SERS, nanostructures and devices that can enable SM Raman measurement. We will conclude with a discussion on SERS implications in biomedical sciences.
Keywords: single molecule, surface-enhanced Raman spectroscopy, nanotechnology, hot spot, biological sciences
1. Introduction
Surface-enhanced Raman spectroscopy (SERS) is based on the phenomena in which molecules adsorbed onto noble metal surfaces generate a million-fold or even higher enhanced signal under favourable conditions [1–4]. Since its first observation in the 1970s [5–7], scientists and engineers have been fascinated by its complexity and possibility. In spite of the numerous advances, the technique has remained elusive for the most part. With the surge in micro- and nanofabrication techniques and the tremendous interest in nanoparticle synthesis, significant developments in both fundamental and applied research in SERS have been possible by scientists and entrepreneurs.
Theoretical studies have indicated that the basic enhancement mechanisms for SERS include chemical enhancement (CM) [8–10] and electromagnetic enhancement (EM), with the EM playing a predominant role in enhancement [11–17]. Based on this concept, SERS-active substrates have usually been found to support plasmonic fields and enhance the Raman signal with exquisite sensitivity. The unique and versatile advantages of SERS include the tremendous multiplexing capacity for simultaneous target detection due to the narrow width of the vibrational Raman bands, quantification based on a specific SERS fingerprint of the corresponding labels, conformation and structural studies of the targets, high photostability and optimal contrast possible with the red or near-infrared (NIR) excitation to minimize background owing to autofluorescence from biological samples, such as blood, cells or tissue. More importantly, only a single laser excitation is necessary to excite the Raman reporters for multiplex detection compared with the multiple wavelengths necessary for multiplex fluorescence signal. Owing to the high sensitivity, less interference from the environment and amplified fingerprint of the SERS signals, the application of SERS to obtain an in depth understanding of a variety of systems, particularly those that are biological in nature, has proved to be invaluable.
Leveraging the findings from SERS, single-molecule (SM)-SERS was first observed in 1997 by two independent groups using silver and gold nanoparticles as SERS substrates together with a dye for enhanced Raman signal recording [18,19]. The enhancement factor (EF) for SM-SERS was estimated to be as high as 1013–1014, sufficiently large in principle to induct Raman spectroscopy into the coveted club of SM spectroscopies. It should be noted that recent experimental and theoretical results have claimed the limit of the EM to be less than 1011 [20]. Such independent reports on SM-SERS triggered a renewed interest in this technique to expand the technology and applications to pathways traversed by its fluorescence counterpart. The statistics from the ISI database depicted in figure 1 shows continued interest in SM-SERS. A brief survey of the literature will indicate that development spans the entire realm of SM-SERS science, which is especially encouraging. Stimulated and coherent Raman techniques are also in place, predominantly to examine deeper sections and will not be a part of this discussion.
Figure 1.
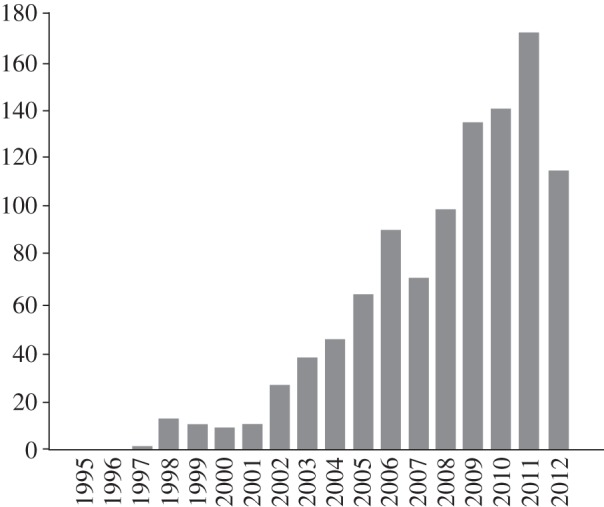
Statistical graph to show publications based on ‘SM-SERS’ by year.
Our intent in this review is to discuss the basics of SM-SERS and summarize its applications in biomedical sciences. Specifically, we will introduce the relevant physical and chemical concepts relevant to the approaches to achieve SM-SERS and the key role of hot spots. Topics such as the basics of SM-SERS (§2), the nanostructure for SM-SERS and their role in enhancing Raman scattering of surface molecules (§3) are discussed. The importance of devices and systems for SM Raman sensing will be discussed in §4. We conclude by highlighting key biological applications in §5.
2. Basics of single-molecule surface-enhanced Raman spectroscopy
Owing to the persistent efforts of various groups, SM-SERS is now becoming a well-established technique and a subfield of SERS as well as a technique in the broader scope of SM laser spectroscopy. Cues to the conditions under which SM-SERS could be observed are being increasingly well understood, and have been discussed and reviewed in several works [21–24]. Herein, only the basic properties and the most important aspects and features will be summarized and highlighted.
As discussed in the initial study in 1997, measurement of low concentration is simple and easily understood and has its origins in SM fluorescence measurements. Monitoring SMs is based on the simple idea of affording the measurement at ultra-low concentration of the analyte [18,19]. However, the problem in developing tools to measure low concentration is the non-uniform distribution of molecules on the metal surface, where molecules need to be adsorbed to create hot spots to produce a signal at the so-called SM resolution. Briefly, SM-SERS is not just a property of the molecule, but is a combination of the response of the molecular microcosm and the localized plasmon-supporting nanostructure, both of which contribute to the success of this field [25]. The bi-analyte approach was then developed to resolve problems that might be encountered in detecting low concentrations by using two distinguishable SERS analytes, reported first by Le Ru et al. [26], and normally aided by suitable statistical analysis of the fluctuations [27]. The simultaneous use of two analyte molecules enables a clear confirmation of signals from a single or few molecules and eliminates most of the uncertainties associated with the Raman signal relating to low molecule concentrations in previous experiments. This approach provides a relatively simple and general direction to assess the concentration at which the largest number of SERS can be sampled [26]. By combining the Langmuir–Blodgett approach with the bi-analyte SERS method, an individual fingerprint could be distinguished at SM resolution by SERS [22,28]. A more detailed explanation of the advantages of the bi-analyte approach is provided in the study of Etchegoin & Le Ru [21]. More recently, Dieringer and colleagues presented a convincing frequency domain proof for SM detection using isotopologues of rhodamin 6G, which provides a clear evidence for single-molecule SERS detection owing to frequency rather than intensity correlation [29,30]. This approach has been applied and substantially extended by Etchegoin and co-workers to analyse two or multi-analytes and extract SM events from a large dataset [31–35].
The second very important aspect for SM-SERS is the EF and the distribution of EM. It is well known that SM-SERS was achieved due to the presence of hot spots, which are regions with highly localized plasmon emitting to show a significant electromagnetic coupling effect between particles, typically the gap junctions between metallic nanostructures. For SM detection by SERS, an EF of approximately 108 is required or desirable. A more important consideration is the distribution of hot spots (or EF), which should be uniform enough to produce a reproducible SERS signal. Figure 2 shows an example of the spatial EF distribution in the gap region between two colloidal gold nanoparticles by finite-element modelling [21,25]. As indicated, the EF value can vary by an order of magnitude over distances comparable to a few molecular dimensions, which is a universal property of gap hot spots resulting in a long tail probability distribution that determines most of the basic characteristics of SM-SERS statistics. More recently, Le Ru et al. [36] proposed a simple scheme based on selective adsorption of the target analyte at the SERS hot spots for detection of every single target molecule in solution and verified by comparing the average and maximum (SM) SERS EF.
Figure 2.
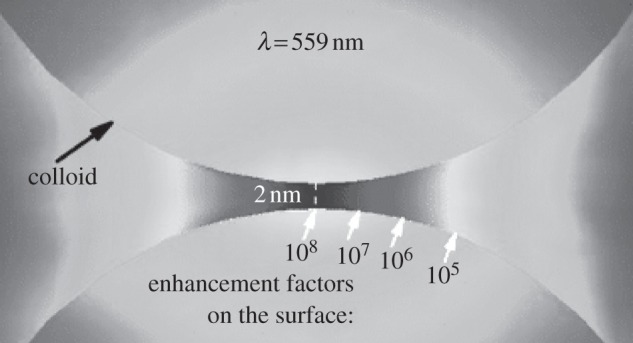
Enhancement factor distribution in the region of the gap (2 nm) between two gold colloids (radii = 30 nm) calculated in the electrostatic approximation with finite-element modelling (adapted from [21]).
Another very intriguing characteristic of SM-SERS is the temporal fluctuation, observed in the past work [18,19]. While SM-SERS has been found to exhibit drastic temporal fluctuations in both line intensity and frequency [37,38], the vibrational mode emission from SM-SERS has been shown to undergo a characteristic intermittent behaviour that might encode both the dynamics of the molecule and the details of its interaction with the environment [39]. This phenomenon is also called as the SERS blinking effect [40–42]. However, it is still not clear whether it is a defined aspect of SM-SERS since it is an instantaneous process and cannot be ‘turned off’. Therefore, the nature and the origin of SM-SERS blinking effect is still a hot topic in this field. One typical explanation for this phenomenon is that the blinking contains both a thermo-activated component and a light-induced component, which means that the blinking is caused from thermally activated diffusion of individual molecules on the particle surface coupled with photo-induced electron transfer and structural relaxation of surface active sites [42]. Therefore, an investigation of blinking effect (spectra fluctuations) will help in addressing some of the fundamental issues, such as the statistical ageing and entanglement of vibrational modes, etc. [43–45]. Detailed knowledge of the fundamental dynamic processes in molecular systems, the charge or electron-transfer mechanism regulating the molecule–metal interaction under light excitation might be helpful in addressing blinking effects [46–49].
3. Nanostructures for single-molecule surface-enhanced Raman spectroscopy
It is well known that SERS actually is a nanostructure-enhanced Raman scattering phenomenon [50]. Therefore, SERS signals (EFs) produced on the nanostructure are strongly related to the size, shape, aggregation state and the type of nanostructures. Typically, silver nanostructures usually generate higher enhanced signal than gold with the same size and shape at the visible laser excitation, while less in the red laser excitation, which depends on the localized surface plasmon resonance [51]. The comparison of EFs based on various size, shape and hot spots has been discussed in a recent review [52–54] and will not be discussed here.
As discussed in §2, an EF of at least approximately 108 is required in most situations or desirable for SM-SERS detection. To achieve this EF, nanostructures with hot spots are the prerequisite as demonstrated since the first observation of SM-SERS in 1997 [18,19]. A class of silver and gold nanostructures that has provided large EFs is colloidal aggregates [55], as well as metal evaporated films [56] and, most recently, modified scanning tunnelling microscopy (STM) tips [57]. Although the easiest route to achieve SM-SERS is to use the aggregated metal colloid, which could be obtained by simply changing the solvent and the ionic strength, the aggregation state is hard to control because signals from randomly aggregated nanoparticles often suffer from uncertainty in the size of aggregates and geometry. In contrast, composite nanostructures with well-defined geometry are a good alternative to achieve controlled and reproducible signal. As reported by our group, periodic and dynamic three-dimensional gold nanoparticle–DNA network structures have been fabricated for SERS-based quantification, which could also be grown at the cell surface marker sites and applied for the detection of cancer stem cells [58,59]. Electron-beam lithography technique is an excellent tool for the fabrication of composite nanostructures with desired geometry, and has been used for single pathogen detection [60]. Meanwhile, significant efforts have been spent on the chemical synthesis of composite nanostructures with well-defined geometry, such as dimers, trimers and complex nanoassemblies. Metal nanoparticle dimer is the simplest discrete assembly, which exhibits one hot spot between two particles upon laser illumination. Synthesis of dimers could be achieved either by biomolecular-directed assembly or by using chemical linkers. Loweth et al. [61] and Park et al. [62] have demonstrated the assembly of gold nanoparticles into dimers, trimers and larger structures based on a DNA-programmed procedure. Chemical linkers, such as the rigid, multivalent thiol-linkers [63], phenylacetylene [64,65], thiol-terminated hydrophobic ligand [66] and so on [67] have been demonstrated for the fabrication of dimers with a relatively high yield. For efficient SERS applications, Xia and co-workers proposed a simple and one-step method to generate dimers without any additional assembly steps, which were obtained by introducing a small amount of sodium chloride into the reaction solution. The formation of dimers was achieved due to the change in the colloidal stability [68]. However, the grand challenge is in the synthesis of dimer metal nanoparticles with high yield because a population of the mixed nanoclusters (monomer, dimer and trimer) is often generated. Most recently, Lim et al. [69] developed a high-yield synthetic method for the synthesis of SERS-active gold–silver core–shell nanodumbbells (GSND, or heterodimers) for SM detection. As indicated in figure 3a, GSND were synthesized by the hybridization of DNA onto the gold nanoparticle surface first, where the Raman dye resides between the two-particle gap junctions. And then a silver shell layer was allowed to grow around the heterodimer structure to form the GSND particle as shown by high-resolution transmission electron microscopy (HR-TEM; figure 3b). The interparticle distance between the two particles and the shell thickness of the silver layer is clearly evident in the image. Figure 3c illustrates the SM behaviour using GSND particle surface with the shell thickness of 5 nm. It should be noted that an on–off cycle for Raman signals, indicative of the presence of an SM, was clearly observed (figure 3c). Meanwhile, recent studies have emerged on the use of metallic junctions in the so-called nanoparticles on metal film or nanoparticles on mirror system for SM-SERS. For example, the generation of hot spot with silver nanocubes for SM-SERS on gold film was reported by Xia and co-workers [70]. Park and co-workers [71,72] report the SM-SERS study, particularly the charge transfer enhancement in SM on the junction between individual gold nanoparticle–aminobenzenethiol–gold film. Notably, tip-enhanced Raman scattering (TERS) is another typical nanoparticle–metal film system for SM-SERS study since most of the TERS studies were achieved on the junction between the nanoparticles on the tips and the metal substrate. This will be discussed in detail in §4.
Figure 3.
(a) Schematic of the fabrication of GSND particles. (b) Typical HR-TEM images of the synthesized GSND particles with 5 nm Ag shell thicknesses. Here ds–s and dc–c indicates the distances between two particle surfaces and cores, respectively. (c) Single-molecule behaviours from the GSND with a 5 nm Ag shell. Blinking SERS spectra taken from the GSND (accumulation time = 1 s, 100 times) (adapted from [69]).
Nanoassemblies with multiple hot spots have also been fabricated according to colloidal chemistry reported by Schlücker's group [73]. The electromagnetic field and the EF around the nanoassemblies were simulated and calculated by the finite-element method, which indicated that the EF value on the nanoassemblies can be as high as 1010, demonstrating its potential to be used as a SERS substrate for SM sensing. With the advancement of nanotechnology, it was believed that more and more SM-SERS substrate could be fabricated with well-defined geometry for reproducible and uniform SERS signal generation. A review of SERS based on molecularly-mediated assembly of plasmonic particles can be found in a recent report [74].
4. Devices/systems for single-molecule surface-enhanced Raman spectroscopy
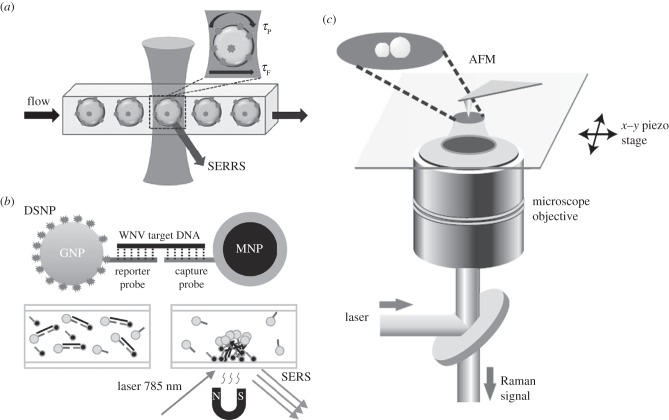
To achieve SM-SERS, several systems were devised and employed. Examples include microfluidic (figure 4a) or opto-electrofluidic approaches with the advantages of fast and dynamic on-demand generation of SERS-active sites for sensitive signal, and a magnetic pull-down system (figure 4b) with the advantages of concentrated samples for sensitive detection. As indicated in figure 4a, by implementing a flow-based system with higher throughput, the detected event can be accomplished in a very short analysis time [75]. Due to the advantages of the microfluidic system in SERS, immunoassays for protein detection have been developed based on microfluidics by SERS [76,77]. By using a magnetic pull-down system, proteins and DNA from viruses were detected effectively and such a system was very beneficial for SM or single-particle SERS study [78,79].
Figure 4.
(a) Systems for the SM-SERS detection such as microfluidic system, (b) magnetic pull-down and (c) AFM-correlated nano-Raman spectroscopy (adapted from [69,75,79]). GNP, gold nanoparticle; MNP, model paramagnetic nanoparticle.
Most recently, with the development of optical tools and electron microscopy, super optical resolution imaging and STEM imaging have also contributed to SM-SERS development. As reported by Stranahan et al. by using super-resolution imaging as a powerful new tool the centroid position of the SERS field could be mapped to within 10 nm resolution, revealing a spatial relationship between the SM-SERS centroid position and the highest SERS intensity [80]. Meanwhile, by employing electron-energy-loss spectroscopy (EELS) in a scanning transmission electron microscope (STEM), Mirsaleh-Kohan et al. [81] obtained maps of the localized surface plasmon models of SM-SERS-active nanostructures, in which the SM characteristics were confirmed by the bianalyte approach. Another more promising and most widely used system for SM and single-particle SERS is the combination of Raman microscopy with atomic force microscopy (AFM) as indicated in figure 4c, where, an AFM-coupled nano-Raman spectroscopy setup is described for nanoscale detection [69].
By combing the STM/AFM with Raman microcopy, a new subfield of SERS called TERS was developed, which was first reported by the Zenobi group in 2000 for the observation of the Raman spectrum from brilliant cresyl blue upon moving a silverized AFM tip into the focal region and in contact with a thin dye layer [82]. The attractive properties of TERS lie in its ultrasensitivity (down to SM) and the visualized image of the substrate at nanometre resolution. More details on TERS can be found in the earlier studies [83–85]. Furthermore, by using silver tips and the monolayer of the same dye made by span-coating on Au substrate, Zenobi's group observed the SM-TERS signal from brilliant cresyl blue in 2007 [86]. They claimed that they can attain SM sensitivity because the Raman band from brilliant cresyl blue shows fluctuations in intensity and to a minor extent also in frequency [83,86], which is a possible indicator for SM behaviour as discussed in §2 (the characteristics of SM-SERS). Another popular dye used for SM-TERS study is malachite green isothiocyanate [58,87,88], which has a strong adsorption band centred around the excitation line at 632.8 nm and exhibits strong fluorescence as reported by Domke et al. [89]. Therefore, dyes are a specific class of molecules for SM-SERS or SM-TERS investigation because of the resonance contribution. However, TERS has also demonstrated SM sensitivity in detecting small molecules such as biological samples (DNA strands, proteins and membranes) [83,90]. It should be pointed out that the key indicators to achieve SM-TERS are the modification/fabrication of the tips and the controlled distance between the tips and the substrate with molecule adsorption [83]. For example, Zhang et al. [86] proved the near-field property of TERS by recording the distance dependence of TERS, which exhibits a strong signal increase when the tip–sample distance is lowered below 30 nm. Owing to the advantages of TERS in SM detection, significant applications in biology could be forthcoming.
5. Surface-enhanced Raman spectroscopy in biology
Owing to the significant advantages of SERS, including the extreme sensitivity, inherent molecular specificity and narrow bandwidth, the technique has made significant inroads in many fields, including catalysis, environmental science, basic sciences and medicine. The sensitivity of SM-SERS has been very beneficial in exploring a range of topics, for instance, electrochemistry, resonant Raman spectra, conduction, heating property and homogeneous Raman broadening. In particular, it is reasonable to state that SERS has enhanced biology. For example, the detection of proteins and DNA, in vivo or in vitro, to observe targeting has been achieved because of the unique advantage of SERS over fluorescence in biology, including multiplex detection, photostability, and particularly the optimal contrast enabled by the use of red to NIR excitation to minimize autofluorescence from biological species, such as blood, tissue and cells. In this section, we will summarize the application of SERS for multiplex and quantitative in vitro and in vivo studies in biological sciences, and monitoring of environmental species and ions that are implicated in pollution and contamination.
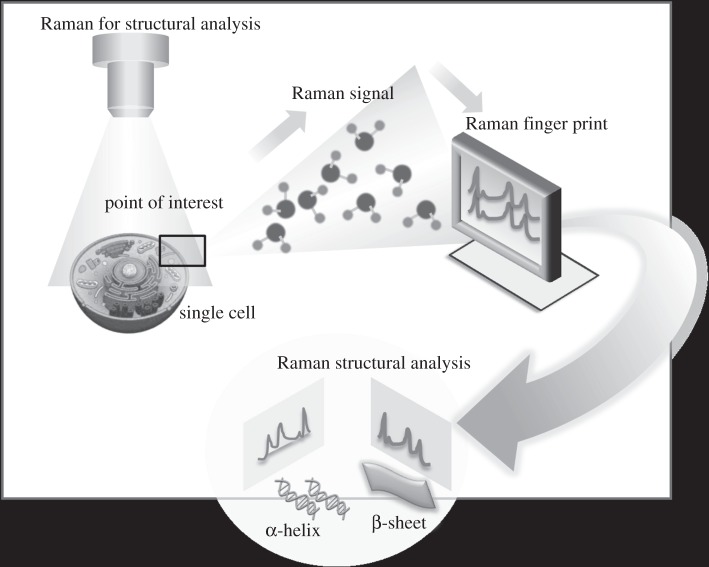
It should be noted that SERS has been used extensively for label-free fingerprinting and in conjunction with appropriate Raman active labels for detection and quantification. Label-free detection is attractive and has the advantages of simplicity and direct observation, as demonstrated by several applications in biology. Of these, one of the most promising applications of SM-SERS is in the rapid detection and identification of individual DNA bases with Raman structural specificity and characterization of individual base pairs in DNA fragments without the use of fluorescent or radioactive labels, as reported by Kneipp et al. in 1998 [91], where adenine was detected by using silver colloid (label-free). Meanwhile, single HIV-1 DNA and single base extension reaction has been detected for DNA methylation analysis [92,93]. Ultrasensitive detection of proteins such as haemoglobin was also reported by using the aggregated silver colloid [94]. Organisms such as pathogens were detected successfully by a label-free approach with high sensitivity using the highly resonant plasmonic nanostructures that can give rise to the formation of multiple hot spots. For instance, pathogens were detected with silver nanospheres as SERS particles, which exhibit multiple hot spots within the structure [95]. Figure 5 demonstrates the possibility of using SM-SERS to examine the intracellular conformation and structure of biological components, a field that has not been significantly explored up to now. Intracellular growth of gold nanoislands can serve as an excellent SERS agent for label-free detection and monitoring of intracellular molecular components, with potential benefit in direct and rapid detection [96]. Such systems may eventually have significant impact in cancer diagnostics through rapid genetic screening or label-free detection of dilute low molecular weight biomarkers.
Figure 5.
SM-SERS for structural and conformational studies.
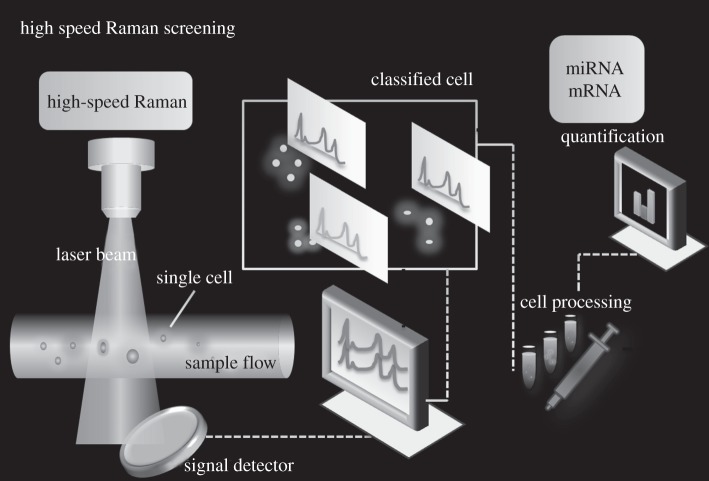
SERS labels have been more beneficial in ultrasensitive detection because of the high Raman cross section of Raman reporters (dyes) and the selectivity achieved by target elements, such as antibodies, aptamers and DNA. Several groups have reported on the detection of protein, DNA and other components in vitro based on SERS labels [97–101]. For example, our group has reported a SERS aptasensor approach from the nanorod–nanopaticle junction for protein detection by labelling the junction with a Raman dye [102]. Quantitative SERS for gene expression estimation has also been accomplished [103] using Raman multiplexers developed for alternative gene splicing [104]. Screening based on SERS labels shows excellent promise in rapid multiplex detection and targeting as shown in figure 6. In this concept schematic, we envision a highly sensitive and high-speed Raman system for simultaneous sorting of cells based on the exquisite cellular signatures paralleling the speed of fluorescence sorting (by flow cytometry) for simultaneous analysis of proteins, DNA or other cellular targets at SM sensitivity in single cells for quantification. Several reports on the application of SERS for multiplex and quantitative monitoring or detection of whole cells and organisms using this highly sensitive and high-speed Raman system have been reported. In 2008, Nie's group illustrated an in vivo multiplex targeting approach using SERS probes with NIR laser excitation [105], which is free from autofluorescence. Meanwhile, Zavaleta et al. [106] have demonstrated the multiplexed imaging in living mice with encoded SERS nanoparticles with multiplexing ability for molecular imaging. The ability of Raman spectroscopy to separate the spectral fingerprints of up to 10 different types of SERS particles in a living mouse has been demonstrated [106]. Dynamic monitoring of the biodistribution of SERS particles in zebrafish embryos for in vivo and multiplex imaging [107] has also been attempted. In the future, these might move to the SM realm with the incorporation of powerful software tools as in fluorescence super-resolution microscopies.
Figure 6.
SERS-based high-speed Raman for simultaneous sorting of cells based on cellular fingerprint and SM quantification of targets.
SERS has also been demonstrated as a powerful tool for environmental monitoring, to detect contaminants in the ppb–ppm range to monitor the interaction of chemistry with the biology or the ecology of various systems. For example, the intracellular bioreduction of chromate-decorated gold nanoparticles and intracellular grown gold nanoislands by Shewanella oneidensis MR-1 have been monitored by Raman and SERS imaging at very high sensitivity [108,109]. Lead ion detection has been achieved by fabricating the electromagnetic coupling between the gold nanoparticles and the surface plasmon polarization substrate (gold surface) based on DNAzyme. The detection limit for lead ion was demonstrated at 20 pM sensitivity [110]. SERS could play a very significant role in environment monitoring because very few systems could afford the sensitivity offered by chemical fingerprinting. Onsite SM-SERS devices, with reusable enhancer templates, could be constructed for monitoring toxic substances that might be present in trace levels.
6. Conclusions and perspectives
In summary, the basic properties of SM-SERS and the nanostructures, devices and systems used for detection have been discussed and summarized. SM-SERS is now accepted as a well-established subfield of SERS and could emerge as a separate field because of the significant understanding of the plasmon coupling effect and hot spot formation. The implications of SERS in biology, including advances in in vivo and in vitro, have been highlighted based on the properties of Raman signal generation and enhancement. Although significant improvements have been possible in the synthesis of complex and highly efficient plasmonic SERS nanoparticles with controllable sizes and shapes, challenges in the preparation of stable, well-defined nanoassemblies with consistent hot spots exist, implying an opportunity for further research. Despite the explosion in the fabrication methods of nanostructures, both chemically synthesized and patterned substrates that are tailored to specific applications might be necessary for routine use, as SM sensors. A tremendous opportunity exists in quantitative SERS and for the detection of analytes at ultra-low concentrations to advance healthcare. Attempts in hyperspectral and high-speed Raman imaging to provide chemical data rivalling the speed and sensitivity possible by fluorescence methods could be envisioned. The entrepreneurial efforts of scientists and engineers involved in Raman research will be critical for translation. Funding as expected will drive the research and a more significant contribution from around the globe can be foreseen.
References
- 1.Aroca R. 2006. Surface-enhanced vibrational spectroscopy. New York, NY: John Wiley & Sons, Inc [Google Scholar]
- 2.Kneipp K, Moskovits M, Kneipp H. 2006. Surface enhanced Raman scattering: physics and applications, topics in applied physics, vol. 103 Berlin, Germany: Springer [Google Scholar]
- 3.Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. 2008. Surface-enhanced Raman spectroscopy. Annu. Rev. Anal. Chem. 1, 601–626 10.1146/annurev.anchem.1.031207.112814 (doi:10.1146/annurev.anchem.1.031207.112814) [DOI] [PubMed] [Google Scholar]
- 4.Schlücher S. 2010. Surface enhanced Raman spectroscopy: analytical, biophysical and life science applications. Weinheim, Germany: Wiley-VCH [Google Scholar]
- 5.Fleischmann M, Hendra PJ, Mcquillan AJ. 1974. Raman-spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 26, 163–166 10.1016/0009-2614(74)85388-1 (doi:10.1016/0009-2614(74)85388-1) [DOI] [Google Scholar]
- 6.Jeanmaire DL, Van Duyne RP. 1977. Surface Raman spectroelectrochemistry. I. Heterocyclic, aromatic, and aliphatic-amines adsorbed on anodized silver electrode. J. Electroanal. Chem. 84, 1–20 10.1016/S0022-0728(77)80224-6 (doi:10.1016/S0022-0728(77)80224-6) [DOI] [Google Scholar]
- 7.Albrecht MG, Creighton JA. 1977. Anomalously intense Raman-spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 99, 5215–5217 10.1021/ja00457a071 (doi:10.1021/ja00457a071) [DOI] [Google Scholar]
- 8.Otto A, Billmann J, Eickmans J, Erturk U, Pettenkofer C. 1984. The ‘adatom model’ of SERS (surface enhanced Raman-scattering): the present status. Surf. Sci. 138, 319–338 10.1016/0039-6028(84)90251-6 (doi:10.1016/0039-6028(84)90251-6) [DOI] [Google Scholar]
- 9.Bruckbauer A, Otto A. 1998. Raman spectroscopy of pyridine adsorbed on single crystal copper electrodes. J. Raman Spectrosc. 29, 665–672 (doi:10.1002/(SICI)1097-4555(199808)29:8<665::AID-JRS288>3.3.CO;2-Y) [DOI] [Google Scholar]
- 10.Arenas JF, Woolley MS, Otero JC, Marcos JI. 1996. Charge-transfer processes in surface-enhanced Raman scattering. Franck-Condon active vibrations of pyrazine. J. Phys. Chem. 100, 3199–3206 10.1021/jp952240k (doi:10.1021/jp952240k) [DOI] [Google Scholar]
- 11.Etchegoin P, Le Ru E. 2009. Principles of surface-enhanced Raman spectroscopy and related plasmonic effects. Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 12.Kahl M, Voges E. 2000. Analysis of plasmon resonance and surface-enhanced Raman scattering on periodic silver structures. Phys. Rev. B 61, 14 078–14 088 10.1103/PhysRevB.61.14078 (doi:10.1103/PhysRevB.61.14078) [DOI] [Google Scholar]
- 13.Moskovits M, Dilella DP, Maynard KL. 1988. Surface Raman-spectroscopy of a number of cyclic aromatic molecules adsorbed on silver: selection-rules and molecular-reorientation. Langmuir 4, 67–76 10.1021/la00079a012 (doi:10.1021/la00079a012) [DOI] [Google Scholar]
- 14.Moskovits M. 1985. Surface-enhanced spectroscopy. Rev. Mod. Phys. 57, 783–826 10.1103/RevModPhys.57.783 (doi:10.1103/RevModPhys.57.783) [DOI] [Google Scholar]
- 15.Moskovits M. 2005. Surface-enhanced Raman spectroscopy: a brief retrospective. J. Raman. Spectrosc. 36, 485–496 10.1002/jrs.1362 (doi:10.1002/jrs.1362). [DOI] [Google Scholar]
- 16.Schatz GC. 1984. Theoretical studies of surface enhanced Raman scattering. Acc. Chem. Res. 17, 370–376 10.1021/ar00106a005 (doi:10.1021/ar00106a005) [DOI] [Google Scholar]
- 17.Young MV, Van Duyne RP. 2006. Electromagnetic mechanism of SERS. In Surface-enhanced Raman scattering , vol. 103 (eds Kneipp K, Moskovits M, Kneipp H.), p. 19 Berlin, Germany: Springer [Google Scholar]
- 18.Nie S, Emory SR. 1997. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science 275, 1102. 10.1126/science.275.5303.1102 (doi:10.1126/science.275.5303.1102) [DOI] [PubMed] [Google Scholar]
- 19.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. 1997. Single molecule detection using surface-enhanced Raman scattering (SERS). Phys. Rev. Lett. 78, 1667–1670 10.1103/PhysRevLett.78.1667 (doi:10.1103/PhysRevLett.78.1667) [DOI] [Google Scholar]
- 20.Pettinger B. 2010. Single-molecule surface- and tip-enhanced Raman spectroscopy. Mol. Phys. 108, 2039–2059 10.1080/00268976.2010.506891 (doi:10.1080/00268976.2010.506891) [DOI] [Google Scholar]
- 21.Etchegoin PG, Le Ru EC. 2008. A perspective on single molecule SERS: current status and future challenges. Phys. Chem. Chem. Phys. 10, 6079–6089 10.1039/b809196j (doi:10.1039/b809196j) [DOI] [PubMed] [Google Scholar]
- 22.Pieczonka NPW, Aroca RF. 2008. Single molecule analysis by surfaced-enhanced Raman scattering. Chem. Soc. Rev. 37, 946–957 10.1039/b709739p (doi:10.1039/b709739p) [DOI] [PubMed] [Google Scholar]
- 23.Pieczonka NPW, Moula G, Skarbek AR, Aroca RF. 2010. Single-molecule and trace detection by SERS, ch. 4 In Surface enhance Raman spectroscopy, analytical, biophysical and life science applications (ed. Schluker S.), pp. 87–101 Weinheim, Germany: Wiley-VCH; 10.1002/9783527632756.ch4 (doi:10.1002/9783527632756.ch4) [DOI] [Google Scholar]
- 24.Le Ru EC, Etchegoin PG. 2012. Single-molecule surface-enhanced Raman spectroscopy. Annu. Rev. Phys. Chem. 63, 65–87 10.1146/annurev-physchem-032511-143757 (doi:10.1146/annurev-physchem-032511-143757) [DOI] [PubMed] [Google Scholar]
- 25.Le Ru EC, Etchegoin PG, Meyer M. 2006. Enhancement factor distribution around a single surface-enhanced Raman scattering hot spot and its relation to single molecule detection. J. Chem. Phys. 125, 204701. 10.1063/1.2390694 (doi:10.1063/1.2390694) [DOI] [PubMed] [Google Scholar]
- 26.Le Ru EC, Meyer M, Etchegoin PG. 2006. Proof of single-molecule sensitivity in surface enhanced Raman scattering (SERS) by means of a two-analyte technique. J. Phys. Chem. B. 110, 1944–1948 10.1021/jp054732v (doi:10.1021/jp054732v) [DOI] [PubMed] [Google Scholar]
- 27.Etchegoin PG, Meyer M, Blackie E, Le Ru EC. 2007. Statistics of single-molecule surface-enhanced Raman scattering signals: fluctuation analysis with multiple analyte techniques. Anal. Chem. 79, 8411–8515 10.1021/ac071231s (doi:10.1021/ac071231s) [DOI] [PubMed] [Google Scholar]
- 28.Goulet PJG, Aroca RF. 2007. Distinguishing individual vibrational fingerprints: single-molecule surface enhanced resonance Raman scattering from one-to-one binary mixtures in Langmuir–Blodgett monolayers. Anal. Chem. 79, 2728–2734 10.1021/ac062059f (doi:10.1021/ac062059f) [DOI] [PubMed] [Google Scholar]
- 29.Dieringer JA, Lettan RB, Scheidt KA, Van Duyne RP. 2007. A frequency domain existence proof of single-molecule surface-enhanced Raman spectroscopy. J. Am. Chem. Soc. 129, 16 249–16 256 10.1021/ja077243c (doi:10.1021/ja077243c) [DOI] [PubMed] [Google Scholar]
- 30.Kleinman SL, Ringe E, Valley N, Wustholz KL, Phillips E, Scheidt KA, Schatz GC, Van Duyne RP. 2011. Single-molecule surface-enhanced Raman spectroscopy of crystal violet isotopologues: theory and experiment. J. Am. Chem. Soc. 133, 4115–4122 10.1021/ja110964d (doi:10.1021/ja110964d) [DOI] [PubMed] [Google Scholar]
- 31.Blackie E, Le Ru EC, Meyer M, Timmer M, Burkett B, Northcote P, Etchegoin PG. 2008. Bi-analyte SERS with isotopically edited dyes. Phys. Chem. Chem. Phys. 10, 4147–4153 10.1039/b803738h (doi:10.1039/b803738h) [DOI] [PubMed] [Google Scholar]
- 32.Etchegoin PG, Lacharmoise PD, Le Ru EC. 2009. Influence of photostability on single-molecule surface enhanced Raman scattering enhancement factors. Anal. Chem. 81, 682–688 10.1021/ac802083z (doi:10.1021/ac802083z) [DOI] [PubMed] [Google Scholar]
- 33.Blackie EJ, Le Ru EC, Etchegoin PG. 2009. Single-molecule surface-enhanced Raman spectroscopy of nonresonant molecules. J. Am. Chem. Soc. 131, 14 466–14 472 10.1021/ja905319w (doi:10.1021/ja905319w) [DOI] [PubMed] [Google Scholar]
- 34.Etchegoin PG, Le Ru EC. 2010. Resolving single molecules in surface-enhanced Raman scattering within the inhomogeneous broadening of Raman peaks. Anal. Chem. 82, 2888–2892 10.1021/ac9028888 (doi:10.1021/ac9028888) [DOI] [PubMed] [Google Scholar]
- 35.Etchegoin PG, Le Ru EC, Meyer M. 2009. Evidence of natural isotopic distribution from single-molecule SERS. J. Am. Chem. Soc. 131, 2713–2716 10.1021/ja808934d (doi:10.1021/ja808934d) [DOI] [PubMed] [Google Scholar]
- 36.Le Ru EC, et al. 2011. A scheme for detecting every single target molecule with surface-enhanced Raman spectroscopy. Nano Lett. 11, 5013–5019 10.1021/nl2030344 (doi:10.1021/nl2030344) [DOI] [PubMed] [Google Scholar]
- 37.Weiss A, Haran G. 2001. Time-dependent single-molecule Raman scattering as a probe of surface dynamics. J. Phys. Chem. B. 105, 12 348–12 354 10.1021/jp0126863 (doi:10.1021/jp0126863) [DOI] [Google Scholar]
- 38.Bizzarri AR, Cannistraro S. 2003. Temporal fluctuations in the SERRS spectra of single iron-protoporphyrin IX molecule. Chem. Phys. 290, 297–306 10.1016/S0301-0104(03)00158-7 (doi:10.1016/S0301-0104(03)00158-7) [DOI] [Google Scholar]
- 39.Bizzarri AR, Cannistraro S. 2005. Levy statistics of vibrational mode fluctuations of single molecules from surface-enhanced Raman scattering. Phys. Rev. Lett. 94, 068303. 10.1103/PhysRevLett.94.068303 (doi:10.1103/PhysRevLett.94.068303) [DOI] [PubMed] [Google Scholar]
- 40.Maruyama Y, Ishikawa M, Futamata M. 2004. Thermal activation of blinking in SERS signals. J. Phys. Chem. B. 108, 673–678 10.1021/jp035838y (doi:10.1021/jp035838y). [DOI] [Google Scholar]
- 41.Otto A. 2001. Theory of first layer and single molecule surface-enhanced Raman scattering (SERS). Phys. Status Solidif. 188, 1455–1470 (doi:10.1002/1521-396X(200112)188:4<1455::AID-PSSA1455>3.0.CO;2-4) [DOI] [Google Scholar]
- 42.Emory SR, Jensen RA, Wenda T, Han MY, Nie SM. 2006. Re-examining the origins of spectral blinking in single-molecule and single-nanoparticle SERS. Faraday Discuss. 132, 249–259 10.1039/b509223j (doi:10.1039/b509223j) [DOI] [PubMed] [Google Scholar]
- 43.Barkai E, Jung YJ, Silbey R. 2004. Theory of single-molecule spectroscopy: beyond the ensemble average. Ann. Rev. Phys. Chem. 55, 457–507 10.1146/annurev.physchem.55.111803.143246 (doi:10.1146/annurev.physchem.55.111803.143246) [DOI] [PubMed] [Google Scholar]
- 44.Allegrini P, Grigolini P, West BJ. 1996. Dynamical approach to Levy processes. Phys. Rev. E. 54, 4760–4767 10.1103/PhysRevE.54.4760 (doi:10.1103/PhysRevE.54.4760) [DOI] [PubMed] [Google Scholar]
- 45.Brokmann X, Hermier JP, Messin G, Desbiolles P, Bouchard JP, Dahan M. 2003. Statistical aging and nonergodicity in the fluorescence of single nanocrystals. Phys. Rev. Lett. 90, 120601/1. 10.1103/PhysRevLett.90.120601 (doi:10.1103/PhysRevLett.90.120601) [DOI] [PubMed] [Google Scholar]
- 46.Adams DM, et al. 2003. Charge transfer on the nanoscale: current status. J. Phys. Chem. B. 107, 6668–6697 10.1021/jp0268462 (doi:10.1021/jp0268462) [DOI] [Google Scholar]
- 47.Bizzarri AR, Cannistraro S. 2005. SERS and tunneling spectroscopy investigation of iron-protoporphyrin IX adsorbed on a silver tip. J. Phys. Chem. B. 109, 16 571–16 574 10.1021/jp053979p (doi:10.1021/jp053979p) [DOI] [PubMed] [Google Scholar]
- 48.Holman MW, Liu R, Adams DM. 2003. Single-molecule spectroscopy of interfacial electron transfer. J. Am. Chem. Soc. 125, 12 649–12 654 10.1021/ja0343104 (doi:10.1021/ja0343104) [DOI] [PubMed] [Google Scholar]
- 49.Joachim C, Gimzewski JK, Aviram A. 2000. Electronics using hybrid-molecular and mono-molecular devices. Nature 408, 541–548 10.1038/35046000 (doi:10.1038/35046000) [DOI] [PubMed] [Google Scholar]
- 50.Aroca RF, Alvarez-Puebla RA, Pieczonka N, Sanchez-Cortez S, Garcia-Ramos JV. 2005. Surface-enhanced Raman scattering on colloidal nanostructures. Adv. Colloid. Inter. Sci. 116, 45–61 10.1016/j.cis.2005.04.007 (doi:10.1016/j.cis.2005.04.007) [DOI] [PubMed] [Google Scholar]
- 51.Le Ru RC, Etchegoin PG. 2008. Principles of surface-enhanced Raman spectroscopy: and related plasmonic effects. Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 52.Banholzer MJ, Millstone JE, Qin LD, Mirkin CA. 2008. Rationally designed nanostructures for surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 37, 885–897 10.1039/b710915f (doi:10.1039/b710915f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko H, Singamaneni S, Tsukruk W. 2008. Nanostructured surfaces and assemblies as SERS media. Small 4, 1576–1599 10.1002/smll.200800337 (doi:10.1002/smll.200800337) [DOI] [PubMed] [Google Scholar]
- 54.Camden JP, Dieringer JA, Zhao J, Van Duyne RP. 2008. Controlled plasmonic nanostructures for surface-enhanced spectroscopy and sensing. Acc. Chem. Res. 41, 1653–1661 10.1021/ar800041s (doi:10.1021/ar800041s) [DOI] [PubMed] [Google Scholar]
- 55.Markel VA, Shalaev VM, Zhang P, Huynh W, Tay L, Haslett TL, Moskovits M. 1999. Near-field optical spectroscopy of individual surface-plasmon modes in colloid clusters. Phys. Rev. B 59, 10 903–10 909 10.1103/PhysRevB.59.10903 (doi:10.1103/PhysRevB.59.10903) [DOI] [Google Scholar]
- 56.Shubin VA, Sarychev AK, Clerc JP, Shalaev VM. 2000. Local electric and magnetic fields in semicontinuous metal films: beyond the quasistatic approximation. Phys. Rev. B: Condens. Matter Mater. Phys. 62, 11 230–11 244 10.1103/PhysRevB.62.11230 (doi:10.1103/PhysRevB.62.11230) [DOI] [Google Scholar]
- 57.Domke KF, Zhang D, Pettinger B. 2006. Toward Raman fingerprints of single dye molecules at atomically smooth Au(111). J. Am. Chem. Soc. 128, 14 721–14 727 10.1021/ja065820b (doi:10.1021/ja065820b) [DOI] [PubMed] [Google Scholar]
- 58.Lee K, Irudayaraj J. 2009. Periodic and dynamic 3-D gold nanoparticle-DNA network structures for surface-enhanced Raman spectroscopy-based quantification. J. Phys. Chem. C. 113, 5980–5983 10.1021/jp809949v (doi:10.1021/jp809949v) [DOI] [Google Scholar]
- 59.Lee K, Drachev V, Irudayaraj J. 2011. DNA-gold nanoparticle reversible networks grown on cell surface marker sites: application in diagnostics. ACS Nano. 5, 2109–2117 10.1021/nn1030862 (doi:10.1021/nn1030862) [DOI] [PubMed] [Google Scholar]
- 60.Yan B, Thubagere A, Premasiri WR, Ziegler LD, Dal Negro L, Reinhard BM. 2009. Engineered SERS substrates with multiscale signal enhancement: nanoparticle cluster arrays. ACS Nano. 3, 1190–1202 10.1021/nn800836f (doi:10.1021/nn800836f) [DOI] [PubMed] [Google Scholar]
- 61.Loweth CJ, Caldwell WB, Peng X, Alivisatos AP, Schultz PG. 1999. DNA-based assembly of gold nanocrystals. Angew. Chem. Int. Ed. 38, 1808–1812 (doi:10.1002/(SICI)1521-3773(19990614)38:12<1808::AID-ANIE1808>3.3.CO;2-3) [DOI] [PubMed] [Google Scholar]
- 62.Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. 2008. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 10.1038/nature06508 (doi:10.1038/nature06508) [DOI] [PubMed] [Google Scholar]
- 63.Sardar R, Heap TB, Shumaker-Parry JS. 2007. Versatile solid phase synthesis of gold nanoparticle dimers using an asymmetric functionalization approach. J. Am. Chem. Soc. 129, 5356–5357 10.1021/ja070933w (doi:10.1021/ja070933w) [DOI] [PubMed] [Google Scholar]
- 64.Novak JP, Feldheim DL. 2000. Assembly of phenylacetylene-bridged silver and gold nanoparticle arrays. J. Am. Chem. Soc. 122, 3979–3980 10.1021/ja000477a (doi:10.1021/ja000477a) [DOI] [Google Scholar]
- 65.Brousseau LC, Novak JP, Marinakos SM, Feldheim DL. 1999. Assembly of phenylacetylene-bridged gold nanocluster dimers and trimers. Adv. Mater. 11, 447–451 (doi:10.1002/(SICI)1521-4095(199904)11:6<447::AID-ADMA447>3.0.CO;2-I) [DOI] [Google Scholar]
- 66.Wang X, Li G, Chen T, Yang M, Zhang Z, Wu T, Chen H. 2008. Polymer-encapsulated gold-nanoparticle dimers: facile preparation and catalytical application in guided growth of dimeric ZnO-nanowires. Nano Lett. 8, 2643–2647 10.1021/nl080820q (doi:10.1021/nl080820q) [DOI] [PubMed] [Google Scholar]
- 67.Guerrini L, Izquierdo-Lorenzo I, Rodriguez-Oliveros R, Sanchez-Gil JA, Sanchez-Cortes S, Garcia-Ramos JV, Domingo C. 2010. α,ω-aliphatic diamines as molecular linkers for engineering Ag nanoparticle clusters: tuning of the interparticle distance and sensing application. Plasmonics 5, 273–286 10.1007/s11468-010-9143-x (doi:10.1007/s11468-010-9143-x) [DOI] [Google Scholar]
- 68.Li W, Camargo PHC, Lu X, Xia Y. 2009. Dimers of silver nanospheres: facile synthesis and their use as hot spots for surface-enhanced Raman scattering. Nano Lett. 9, 485–490 10.1021/nl803621x (doi:10.1021/nl803621x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lim DK, Jeon KS, Kim HM, Nam JM, Suh YD. 2010. Nanogap-engineerable Raman-active nanodumbbells for single-molecule detection. Nat. Mat. 9, 60–67 10.1038/NMAT2596 (doi:10.1038/NMAT2596) [DOI] [PubMed] [Google Scholar]
- 70.Rycenga M, Xia XH, Moran CH, Zhou F, Qin D, Li ZY, Xia YN. 2011. Generation of hot spots with silver nanocubes for single-molecule detection by surface-enhanced Raman scattering. Angew. Chem. 50, 5473–5477 10.1002/anie.201101632 (doi:10.1002/anie.201101632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park WH, Kim ZH. 2010. Charge transfer enhancement in the SERS of a single molecule. Nano Lett. 10, 4040–4048 10.1021/nl102026p (doi:10.1021/nl102026p) [DOI] [PubMed] [Google Scholar]
- 72.Park WH, Ahn SH, Kim ZH. 2008. Surface-enhanced Raman scattering from a single nanoparticle–plane junction. Chem. Phys. Chem. 9, 2491–2494 10.1002/cphc.200800563 (doi:10.1002/cphc.200800563) [DOI] [PubMed] [Google Scholar]
- 73.Gellner M, Steinigeweg D, Ichilmann S, Salehi M, Schutz M, Kompe K, Haase M, Schlucker S. 2011. 3D self-assembled plasmonic superstructures of gold nanospheres: synthesis and characterization at the single-particle level. Small 7, 3445–3451 10.1002/smll.201102009 (doi:10.1002/smll.201102009) [DOI] [PubMed] [Google Scholar]
- 74.Guerrini L, Graham D. 2012. Molecularly-mediated assemblies of plasmonic nanoparticles for Surface-Enhanced Raman Spectroscopy applications. Chem. Soc. Rev. 41, 7085–7107 10.1039/C2CS35118H (doi:10.1039/C2CS35118H) [DOI] [PubMed] [Google Scholar]
- 75.Cecchini MP, Stapountzi MA, McComb DW, Albrecht T, Edel JB. 2011. Flow-based autocorrelation studies for the detection and investigation of single-particle surface-enhanced resonance Raman spectroscopic events. Anal. Chem. 83, 1418–1424 10.1021/ac102925h (doi:10.1021/ac102925h) [DOI] [PubMed] [Google Scholar]
- 76.Hwang H, Kim SH, Yang SM. 2011. Microfluidic fabrication of SERS-active microspheres for molecular detection. Lab Chip 11, 87–92 10.1039/c0lc00125b (doi:10.1039/c0lc00125b) [DOI] [PubMed] [Google Scholar]
- 77.Huh YS, Chung AJ, Erickson D. 2009. Surface enhanced Raman spectroscopy and its application to molecular and cellular analysis. Microfluid. Nanofluid. 6, 285–297 10.1007/s10404-008-0392-3 (doi:10.1007/s10404-008-0392-3) [DOI] [Google Scholar]
- 78.Zhang H, Harpster MH, Wilson WC, Johnson PA. 2012. Surface-enhanced Raman scattering detection of DNAs derived from virus genomes using Au-coated paramagnetic nanoparticles. Langmuir 28, 4030–4037 10.1021/la204890t (doi:10.1021/la204890t) [DOI] [PubMed] [Google Scholar]
- 79.Zhang H, Harpster MH, Park HJ, Johnson PA, Wilson WC. 2011. Surface-enhanced Raman scattering detection of DNA derived from the West Nile virus genome using magnetic capture of Raman-active gold nanoparticles. Anal. Chem. 83, 254–260 10.1021/ac1023843 (doi:10.1021/ac1023843) [DOI] [PubMed] [Google Scholar]
- 80.Stranahan SM, Willets KA. 2010. Super-resolution optical imaging of single-molecule SERS hot spots. Nano Lett. 10, 3777–3784 10.1021/nl102559d (doi:10.1021/nl102559d) [DOI] [PubMed] [Google Scholar]
- 81.Mirsaleh-Kohan N, et al. 2012. Single-molecule surface-enhanced Raman scattering: can STEM/EELS image electromagnetic hot spots? J. Phys. Chem. Lett. 3, 2303–2309 10.1021/jz300967q (doi:10.1021/jz300967q) [DOI] [PubMed] [Google Scholar]
- 82.Stockle RM, Suh YD, Deckert V, Zenobi R. 2000. Nanoscale chemical analysis by tip-enhanced Raman spectroscopy. Chem. Phys. Lett. 318, 131–136 10.1016/S0009-2614(99)01451-7 (doi:10.1016/S0009-2614(99)01451-7) [DOI] [Google Scholar]
- 83.Yeo BS, Stadler J, Schmid T, Zenobi R, Zhang WH. 2009. Tip-enhanced Raman spectroscopy: its status, challenges and future directions. Chem. Phys. Lett. 472, 1–13 10.1016/j.cplett.2009.02.023 (doi:10.1016/j.cplett.2009.02.023) [DOI] [Google Scholar]
- 84.Domke KF, Petinger B. 2010. Studying surface chemistry beyond the diffraction limit: 10 years of TERS. Chem. Phys. Chem. 11, 1365–1373 10.1002/cphc.200900975 (doi:10.1002/cphc.200900975) [DOI] [PubMed] [Google Scholar]
- 85.Pettinger B, Schambach P, Villag omez CJ, Scott N. 2012. Tip-enhanced Raman spectroscopy: near-fields acting on a few molecules. Annu. Rev. Phys. Chem. 63, 379–399 10.1146/annurev-physchem-032511-143807 (doi:10.1146/annurev-physchem-032511-143807) [DOI] [PubMed] [Google Scholar]
- 86.Zhang W, Yeo BS, Schmid T, Zenobi R. 2007. Single molecule tip-enhanced Raman spectroscopy with silver tips. J. Phys. Chem. C 111, 1733–1738 10.1021/jp064740r (doi:10.1021/jp064740r) [DOI] [Google Scholar]
- 87.Ren B, Picardi G, Pettinger B, Schuster R, Ertl G. 2005. Tip-enhanced Raman spectroscopy of benzenethiol adsorbed on Au and Pt single-crystal surfaces. Angew. Chem. Int. Edit. 44, 139–142 10.1002/anie.200460656 (doi:10.1002/anie.200460656) [DOI] [PubMed] [Google Scholar]
- 88.Pettinger B, Ren B, Picardi G, Schuster R, Ertl G. 2004. Nanoscale probing of adsorbed species by tip-enhanced Raman spectroscopy. Phys. Rev. Lett. 92, 096101. 10.1103/PhysRevLett.92.096101 (doi:10.1103/PhysRevLett.92.096101) [DOI] [PubMed] [Google Scholar]
- 89.Domke KF, Zhang D, Pettinger B. 2007. Tip-enhanced Raman spectra of picomole quantities of DNA nucleobases at Au (111). J. Am. Chem. Soc. 129, 6708–6709 10.1021/ja071107q (doi:10.1021/ja071107q) [DOI] [PubMed] [Google Scholar]
- 90.Yeo BS, Maedler S, Schmid T, Zhang W, Zenobi R. 2008. Tip-enhanced Raman spectroscopy can see more: the case of cytochrome C. J. Phys. Chem. C. 112, 4867–4873 10.1021/jp709799m (doi:10.1021/jp709799m) [DOI] [Google Scholar]
- 91.Kneipp K, Kneipp H, Kartha VB, Manoharan R, Deinum G, Itzkan I, Dasari RR, Feld MS. 1998. Detection and identification of a single DNA base molecule using surface-enhanced Raman scattering (SERS). Phys. Rev. E. 57, R6281–6284 10.1103/PhysRevE.57.R6281 (doi:10.1103/PhysRevE.57.R6281) [DOI] [Google Scholar]
- 92.Hu J, Zheng PC, Jiang J-H, Shen G-L, Yu R-Q, Liu G-K. 2010. Sub-attomolar HIV-1 DNA detection using surface-enhanced Raman spectroscopy. Analyst 135, 1084–1089 10.1039/b920358c (doi:10.1039/b920358c) [DOI] [PubMed] [Google Scholar]
- 93.Hu J, Zhang C. 2012. Single base extension reaction-based surface enhanced Raman spectroscopy for DNA methylation assay. Biosensors. Bioelectron. 31, 451–457 10.1016/j.bios.2011.11.014 (doi:10.1016/j.bios.2011.11.014) [DOI] [PubMed] [Google Scholar]
- 94.Xu HX, Bjerneld EJ, Käll M, Börjesson L. 1999. Spectroscopy of single hemoglobin molecules by surface enhanced Raman scattering. Phys. Rev. Lett. 83, 4357–4360 10.1103/PhysRevLett.83.4357 (doi:10.1103/PhysRevLett.83.4357) [DOI] [Google Scholar]
- 95.Wang Y, Lee K, Irudayaraj J. 2010. Silver nanosphere SERS probes for sensitive identification of pathogens. J. Phys. Chem. C 114, 16 122–16 128 10.1021/jp1015406 (doi:10.1021/jp1015406) [DOI] [Google Scholar]
- 96.Shamsaie A, Jonczyk M, Sturgis J, Irudayaraj J. 2007. Intracellularly grown gold nanoparticles as potential surface-enhanced Raman scattering probes. J. Biomed. Opt. 12, 020502. 10.1117/1.2717549 (doi:10.1117/1.2717549) [DOI] [PubMed] [Google Scholar]
- 97.Doering WE, Piotti ME, Natan MJ, Freeman RG. 2007. SERS as a foundation for nanoscale, optically detected biological labels. Adv. Mater. 19, 3100–3108 10.1002/adma.200701984 (doi:10.1002/adma.200701984) [DOI] [Google Scholar]
- 98.Schlücker S, Kustner B, Punge A, Bonfig R, Marx A, Strobel P. 2006. Immuno-Raman microspectroscopy: in situ detection of antigens in tissue specimens by surface-enhanced Raman scattering. J. Raman. Spectrosc. 37, 719–721 10.1002/jrs.1534 (doi:10.1002/jrs.1534) [DOI] [Google Scholar]
- 99.Porter MD, Lipert RJ, Sioperko LM, Wang G, Narayanan R. 2008. SERS as a bioassay platform: fundamentals, design, and applications. Chem. Soc. Rev. 37, 1001–1011 10.1039/b708461g (doi:10.1039/b708461g) [DOI] [PubMed] [Google Scholar]
- 100.Schlücker S, Kiefer W. 2009. Selective detection of proteins and nucleic acids with biofunctionalized SERS labels. In Frontiers in molecular spectroscopy (ed. Laane J.), pp. 26–35 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 101.Schlücker S. 2009. SERS microscopy: nanoparticle probes and biomedical applications. Chem. Phys. Chem 10, 1344–1354 10.1002/cphc.200900119 (doi:10.1002/cphc.200900119) [DOI] [PubMed] [Google Scholar]
- 102.Wang Y, Lee K, Irudayaraj J. 2010. SERS aptasensor from nanorod–nanoparticle junction for protein detection. Chem. Commun. 46, 613–615 10.1039/b919607b (doi:10.1039/b919607b) [DOI] [PubMed] [Google Scholar]
- 103.Sun L, Irudayaraj J. 2009. Quantitative surface-enhanced Raman for gene expression estimation. Biophys. J. 96, 4709–4716 10.1016/j.bpj.2009.03.021 (doi:10.1016/j.bpj.2009.03.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sun L, Yu C, Irudayaraj J. 2008. Raman multiplexers for alternative gene splicing. Anal. Chem. 80, 3342–3349 10.1021/ac702542n (doi:10.1021/ac702542n) [DOI] [PubMed] [Google Scholar]
- 105.Qian XM, Peng XH, Ansari DO, Nie SM. 2008. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotech. 26, 83–90 10.1038/nbt1377 (doi:10.1038/nbt1377) [DOI] [PubMed] [Google Scholar]
- 106.Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, Natan MJ, Gambhir SS. 2009. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proc. Natl Acad. Sci. USA 106, 13 511–13 516 10.1073/pnas.0813327106 (doi:10.1073/pnas.0813327106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Y, Seebald J, Szeto D, Irudayaraj J. 2010. Biocompatibility and biodistribution of surface-enhanced Raman scattering nanoprobes in zebrafish embryos: in vivo and multiplex imaging. ACS Nano. 4, 4039–4053 10.1021/nn100351h (doi:10.1021/nn100351h) [DOI] [PubMed] [Google Scholar]
- 108.Ravindranath S, Henne T, Thompson D, Irudayaraj J. 2011. Raman chemical imaging of chromate reduction sites in a single bacterium using intracellularly grown gold nanoislands. ACS Nano. 5, 4729–4736 10.1021/nn201105r (doi:10.1021/nn201105r) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ravindranath S, Henne K, Thompson D, Irudayaraj J. 2011. Single molecule in vivo analysis of toll-like receptor 9 and CpG DNA interaction. PLoS ONE 6, e16634. 10.1371/journal.pone.0017991 (doi:10.1371/journal.pone.0017991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y, Irudayaraj J. 2011. SERS aptasensor from nanorod–nanoparticle junction for protein detection. Chem. Commun. 47, 4394–4396 10.1039/c0cc04140h (doi:10.1039/c0cc04140h) [DOI] [PubMed] [Google Scholar]