Abstract
Determining the organization of key molecules on the surface of live cells in two dimensions and how this changes during biological processes, such as signalling, is a major challenge in cell biology and requires methods with nanoscale spatial resolution and high temporal resolution. Here, we review biophysical tools, based on scanning ion conductance microscopy and single-molecule fluorescence and the combination of both of these methods, which have recently been developed to address these issues. We then give examples of how these methods have been be applied to provide new insights into cell membrane organization and function, and discuss some of the issues that will need to be addressed to further exploit these methods in the future.
Keywords: single-molecule fluorescence, scanning ion conductance microscopy, T-cell triggering, clathrin-mediated endocytosis
1. Introduction
The cell membrane of mammalian cells is only 7 nm thick separating the intracellular contents of the cell from its environment and protecting the cell from infection. The response of the cell to specific changes in its environment requires mechanisms to convert environmental changes to internal signals, and this function is generally performed by proteins embedded in the cell membrane responding to the binding of ligands or applied force. This includes many fundamental and biomedically relevant processes such as, for example, the triggering of the innate and adaptive immune responses, and the triggering of G protein-coupled receptors, which are major drug targets. Major questions in cell biology are to understand how the key proteins and molecules are organized in or just under the cell membrane, how they interact and reorganize in space and time to produce all the essential cellular functions, and how these functions can go awry in disease or infection. To address these questions, new biophysical methods are needed that are capable of determining the association between molecules, can deal with the fundamental heterogeneity that is present in complex biological systems and allow the organization of the cell membrane to be imaged on the nanoscale, below the optical diffraction limit. These methods need to be applied to the cell in its resting state and then have sufficient time resolution to follow the changes that take place during key biological processes on live cells.
There are several technical issues that arise in attempting to address these challenges. The first is having sufficient time resolution to follow the biological process of interest as well as sufficient spatial resolution to follow the changes that occur. The second issue is that some processes of interest occur only once and are irreversible, so one needs a method for rapid triggering of the events of interest. This is of particular importance if one wants to catch the first initial events that lead to a biochemical cascade or if it takes the cell a long time to return to its rest state, or if this never occurs. The third issue is one of sensitivity because some processes of interest can be initiated by single proteins in the cell membrane. The number of currently available techniques with sufficient sensitivity is limited to single-channel recording, single-molecule fluorescence, surface-enhanced Raman or scanning probe imaging. The fourth issue is that any changes followed need to be mapped back onto the overall cellular structure so that the results of experiments can be combined, so this implies measurement of at least two parameters simultaneously.
It is also worth considering the length scales involved and the time resolutions that might be needed. Individual proteins are of the order of 10 nm in diameter, and depending on function can be static and attached to the cell cytoskeleton or freely diffusing. The length scale for the organization of the cell membrane is more controversial with raft-like structures perhaps existing with a size of about 100 nm and then structures determined by the underlying cytoskeleton on the order of about 1 μm. This means that all this organization is undetectable using conventional optical imaging, which is limited by the optical diffraction limit. The timescales cover a wide range from the millisecond timescale for molecular motions or conformational changes to the minute timescale for structural and cytoskeletal reorganizations. Only methods that acquire data in a parallel fashion can avoid having to trade-off spatial and time resolution when imaging.
Over the past 10 years, we have developed two complementary methods for live-cell imaging that address some of these issues. The first method is a form of scanning probe microscopy, scanning ion conductance microscopy (SICM), which is capable of imaging the topography and function of living cells down to the level of individual molecular complexes [1,2]. The structures to be imaged need to be static or slowly changing for this approach, because the fastest rate for imaging a small region, about 1 × 1 μm, is a frame every 10 s. The second complementary method that we have developed is based on two-colour single-molecule fluorescence to detect and analyse associated molecules which are diffusing in the cell membrane [3]. We present an overview of these methods focusing on explaining how these methods work and what experiments they enable on live cells, by presenting some recent examples. We then discuss how these and other new biophysical methods may be applied to cell biology in the future.
2. Scanning ion conductance microscopy
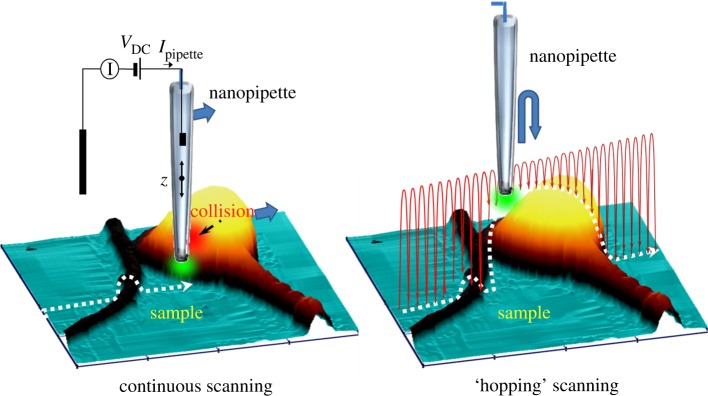
SICM was originally developed by Hansma et al. [4] and is a form of scanning probe microscopy based on a nanopipette in a conducting solution, normally physiological buffer with 150 mM NaCl (figure 1). The pipette is made by melting capillary glass normally under computer control using a commercial pipette puller. The pipette inner radius is typically between 10 and 50 nm and depends on the melting temperature of the glass used, so it is possible to produce finer pipettes from quartz. It is the pipette inner radius that determines the resolution of topographic images. The application of a voltage, typically a few hundred millivolts, between a silver chloride electrode in the pipette and in the bath leads to a flow of Na+ and Cl− ions to the electrodes. The ion flow is firstly limited by the small aperture at the tip of the pipette and is further reduced as the pipette approaches a surface, as ion flow is shut-down. This reduction in ion flow as the pipette approaches an ion impermeable surface is used for distance feedback. Typically, the pipette approaches the surface so that the ion current has been reduced by 0.1–1% from the limiting current, when far above the surface. At this point, the pipette is still about an inner radius away from the surface. In the original form of SICM, the pipette is then raster-scanned over the surface and the distance feedback control adjusts the pipette sample separation to keep the ion current reduction constant [5]. The amount the pipette needs to be moved, by a piezo actuator, is recorded and this allows the sample topography to be measured. The imaging time depends on the sample and scan size but is typically 10–20 min. The major advantages of this method are that the nanopipette can sense the presence of the surface when still an inner radius away, typically 10–50 nm, so there is no direct contact with the soft cell surface and the forces exerted on the cell when scanning are negligible. A common misconception, owing to the name, is that the method maps surface conductivity, but in the high-salt conditions used for imaging the Debye length is less than 1 nm, so the method is insensitive to differences in surface charge or the opening of ion channels in the cell membrane and the distance feedback responds only to changes in surface topography.
Figure 1.
Principle of SICM. In continuous scanning, the pipette is raster-scanned over the surface with continuous distance feedback control so the pipette stays a fixed distance above the surface. In contrast, in ‘hopping’ scanning, the pipette hops over the surface so it always approaches from above, eliminating surface collisions.
Like all scanning probe microscopies, because the SICM probe senses locally at the tip, it is not possible to scan highly convoluted surfaces where the side of the probe may touch before the tip has sensed the presence of a surface. To address this issue, we have recently modified SICM so that the distance feedback control is not continuous and so the pipette always approaches the surface from above; this is called hopping mode SICM [6]. The method has enabled us to scan complex neuronal networks, and in general allows highly complex cellular and tissue topography to be scanned. Because the pipette is no longer raster-scanned over the surface, it also becomes possible to use adaptive resolution where the pipette makes more or less measurements of the surface topography depending on the surface roughness. This has two advantages: first, it becomes possible to perform a lower resolution scan to identify the region of interest; second, the imaging time is reduced significantly. Hopping mode is compatible with the use of the nanopipette for local measurements of various cellular properties on the nanoscale as described below.
3. Local measurements combined with scanning ion conductance microscopy imaging
We have developed a family of local measurements based on the use of the nanopipette to probe the cell surface below it and the robust distance feedback of SICM on live cells. We have combined SICM with fluorescence detection so as to be able to identify specific labelled molecules on or near the cell surface. This was done initially in a near-field mode, by delivering light from the tip of a coated nanopipette [7]. However, it is more easily achieved in a far-field mode with the imaged confocal volume positioned just below the pipette tip [8]. Surface confocal mode allows one to image the fluorescence from labelled species on the cell surface in a single scan, because the surface is guided under the pipette tip using SICM distance feedback control. This removes problems in photobleaching fluorophores before they are imaged, an issue with conventional confocal microscopy that uses z slices to build up an image of the surface. We have used surface confocal microscopy to identify viral particles [8,9] and clathrin-coated pits (CCPs) on the cell surface [10,11].
The pipette can also be used to perform a series of nanoscale local assays on the sample, including single-channel recording, mechanical measurements by local pressure application, local voltage delivery of reagents and local chemical mapping. For single-channel recording, the pipette can be lowered onto the cell surface to form a high-resistance seal under computer control [12,13]. Electrophysiological recording can then be performed to determine if an ion channel is present in the cell membrane under the pipette. The advantage of this approach is that it can be entirely automated, which gives a success rate of forming a seal of better than 80 per cent. Because SICM allows one to image the cell surface prior to patching the surface, it then becomes possible to target a region of interest such as a microvillus, the synapse of a neuron or the body of a sperm and determine the nature and density of ion channels in this selected region.
The application of hydrostatic pressure to the pipette leads to this pressure developing at the pipette tip, when the pipette is held close to the cell surface [14,15]. On application of pressure, the soft cell deforms and the distance feedback control readjusts the pipette position to keep the separation from the surface constant. These experiments enable one to measure the mechanical properties of the cell without any direct contact between the probe and the surface.
The conical shape of the pipette means that almost the entire voltage drop occurs at the tip and that for the application of modest voltages to the pipette, it is possible to generate large electric fields up to 106 Vm−1 [16]. This high electric field at the pipette tip can be used to deliver charged molecules to the cell surface, owing to a combination of electrophoretic and dielectrophoretic forces. This enables controlled delivery of molecules onto a surface at defined positions, and by controlling the concentration of solution in the pipette and the applied voltage and dose time, delivery of individual molecules is possible [17–20]. This has been used to study the diffusion of molecules over the surface of a boar sperm and probe the nature of the barrier between different macrodomains on the surface, when combined with single-molecule tracking, and is discussed in more detail below [21]. We have also trapped fluorophores in the tip of the pipette due to the force generated by dielectrophoresis (induced dipole forces) [22]. This trapped dye is continually renewed from the bulk of the pipette, because the barrier of the trap is not perfect. If the fluorophore probes fluorescence changes with local pH or sodium, for example, the fluorescence at the pipette tip then reports back about the local pH or sodium ion concentration at the tip. The advantage of this approach is millisecond time response, and the capability to map chemical species with a resolution determined by the pipette inner radius.
A key advantage of the use of a nanopipette as the scanning probe is that it is possible to simply fabricate probes with many barrels using capillary glass with a septum down the centre, theta glass, or using glass with several capillaries fused together. Each barrel of the pipette has its own electrode and can be filled with a different reagent. One can then locally deliver one or another of these reagents by applying a voltage to the appropriate barrel. We have used this to write complex patterns in DNA and antibodies, controlling the amount delivered by the applied voltage or dosing time [17]. We have also added an electrode to the pipette tip, in a second barrel, so as to be able to perform simultaneous electrochemical measurements to map chemical species [23,24]. Both a silver electrode and carbon electrode have been used, allowing scanning electrochemical microscopy to be combined with SICM in a single probe.
4. Applications of scanning ion conductance microscopy to cell biology
SICM can be used to image unfixed, unstained, living cells with high resolution. This is most powerful when combined with a simultaneous functional measurement, as illustrated by our recent work. In the first example, the local delivery of reagents has been used to map β2 adrenergic receptors (β2ARs) on live heart cells using fluorescence to detect where local application of ligand from the pipette led to production of the secondary messenger cAMP, inside the cell [25]. In the second example, fluorescence was used to identify CCPs on the cell surface and then SICM was used to follow the topographic changes that take place during clathrin-mediated endocytosis [10]. In both cases, the functional molecular unit was not a single molecule but either a small cluster of receptors or a CCP containing 100–1000 clathrin molecules. This highlights that ultrasensitive but not necessarily single-molecule methods might be needed for some aspects of cell biology, reflecting the hierarchical organization of molecules into larger functional units.
The distribution of ion channels and receptors on the cell membrane is important for cell function, and the loss of particular distribution patterns is often associated with cell damage or disease. For example, the β1 and β2ARs are heterotrimeric G protein-coupled receptors found on the surface of cardiac muscle cells (cardiomyocytes). They mediate the heart's response to catecholamine hormones through the production of cAMP. Like with most G protein-coupled receptors, immunocytochemical or electron microscopy detection of native β2ARs receptors with antibodies has been limited by their low expression level and by insufficient antibody specificity. SICM topographical imaging and localized delivery through the nanopipette have been combined with fluorescence resonance energy transfer (FRET) microscopy to map the distribution of endogenous β1 and β2 adrenergic receptors and measure their function to compare healthy and heart failure-derived cardiomyocytes [25]. In these experiments, the nanopipette was positioned either in the T-tubule or on the cell crest, and receptor ligands applied in a highly localized fashion. It was possible to achieve very local stimulation by applying pressure to the pipette while constantly superfusing the cells with buffer solution. Use of antagonists allowed selective local stimulation of β1 or β2ARs, when combined with appropriate blockers. FRET microscopy was used to detect and measure the cAMP on its accumulation in the cell cytosol. By using this combined SICM FRET imaging method, it was possible to functionally localize the cAMP signalling of β1ARs and β2ARs to the surface structures of adult ventricular cardiomyocytes. β2ARs are present only in deep transverse tubules, whereas functional β1ARs are distributed across the entire cell surface. In diseased heart cells, β2ARs were redistributed from the transverse tubules to the cell crest, which significantly altered the cAMP signalling.
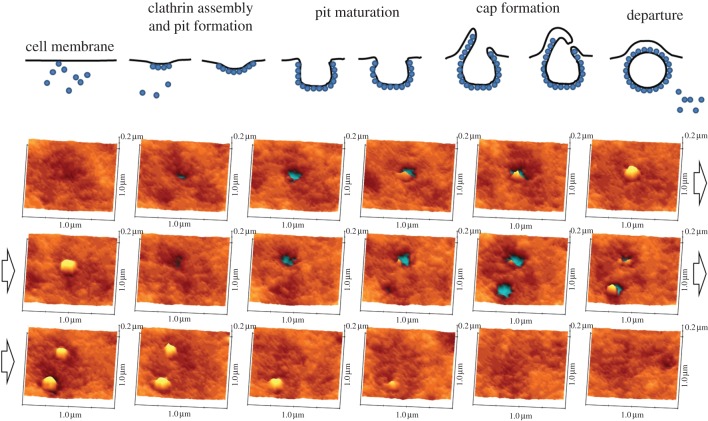
Clathrin-mediated endocytosis is a common mechanism for transport of molecules and small particles across the cell membrane where small CCPs, about 100 nm in diameter, are formed on the cell surface before the pits are closed to produce vesicles inside the cell. This process takes place on the timescale of a minute. Although optical microscopy techniques such as total internal reflection fluorescence (TIRF) microscopy have been used to measure the ensemble movement of clathrin–green fluorescent protein relative to the plasma membrane, they did not directly measure the topography of the cell surface [26,27]. By contrast, electron microscopy can provide high-resolution images of the pit structure but cannot follow the structural changes with time. We have used SICM to observe the topographical changes during the entire life cycle of individual CCPs in combination with simultaneous fluorescence measurements, taking images every of an area of 1 × 1 μm2 every 10 s, and hence, obtained complementary information about the process [10]. In this study, we directly observed the formation, steady state, disappearance, reappearance, splitting and fusion of pits. Splitting and fusion cannot be observed using TIRF imaging, because the pits are below the diffraction limit. By measuring a large number of pits, it was possible build up histograms of key parameters such as the changes in the pit depth and width with time and relate this to the changes in fluorescence signal owing to accumulation of clathrin. Interestingly, in addition to canonical symmetrical pit closure, we found that pits can close asymmetrically, with the formation of the protrusion at one side of the pit as shown in the time course in figure 2. Despite being nanoscale in size, the mechanism of CCP closure looks topographically similar to larger scale phago- and macropinocytosis. While similar structures have been observed in electron microscopy, it is not possible to establish whether they have a role in endocytosis without directly following the topographic changes of individual pits, as was done in this work. The approach of combined SICM topographic imaging and fluorescence confocal microscopy opens up possibilities for imaging the nanoscale topographical changes taking place on the apical cell surface, owing to specific labelled molecules, allowing key biological processes such as endocytosis, exocytosis and viral entry to be followed.
Figure 2.
SICM images of the life history of clathrin-coated pits (CCPs) showing cap formation prior to closure. An image was recorded every 10 s.
5. Single-molecule experiments using scanning ion conductance microscopy
We can use the pipette to perform single-molecule experiments on live cells. The nanopipette can be used to record single ion-channel activity at defined positions on the cell surface, including normally inaccessible regions such as the synapse of dendritic neurons, specific regions of a sperm and the T-tubule of cardiac myocytes [12,13]. We have used this to functionally map the distribution of ion channels on cardiac myocytes and shown that key ion channels are clustered in defined positions in the T-tubules, which is probably to help maintain reliable propagation of the action potential [13].
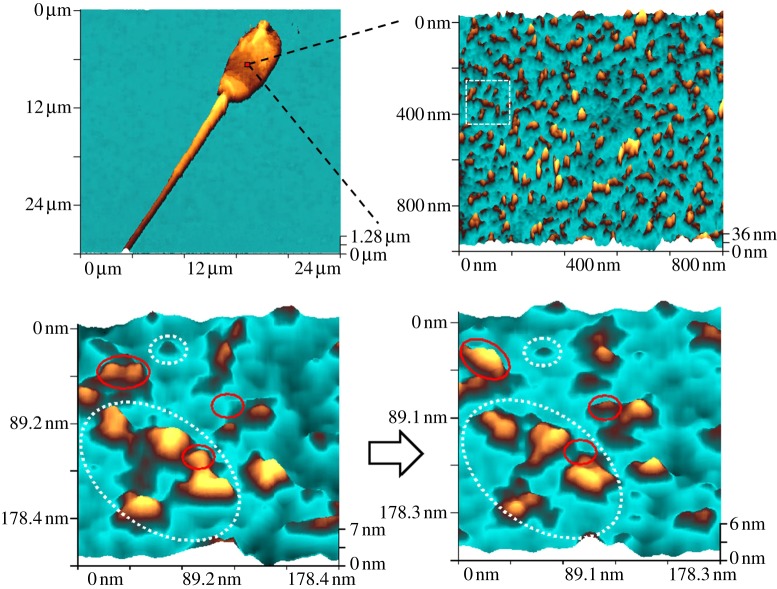
We have also demonstrated the capability of SICM to image individual protein complexes on the surface of live cells [28]. Spermatozoa were chosen as an example of a highly polarized cell which is relatively flat and whose membrane is compartmentalized into functionally and topographically different domains known as the anterior acrosome, equatorial segment, equatorial subsegment and postacrosome. Figure 3 shows images of increasing resolution of the equatorial segment and subsegment of a live boar spermatozoon that has initiated a spontaneous acrosome reaction in physiological buffer. The acrosome reaction leads to the formation of fusogenic protein complexes at the cell surface in preparation for binding to the egg. The irregular protrusions can be seen on the membrane surface in figure 3 and at higher resolution appear as projecting proteins or protein complexes. The diameter of the small and large protein complexes measured were approximately 14 nm and 30 nm, respectively.
Figure 3.
SICM images of a live boar sperm showing protein complexes on the cell surface. The dotted circles show static protein complexes, whereas the red circles show complexes undergoing dynamics in the 10 min between scans.
To investigate dynamics of single proteins further, we present zoomed images of a selected area. The images are taken 10 min apart. The majority of proteins are stable. However, the solid red lines show examples of conformational and rotational changes along with evidence for disassembly. There is some evidence for smaller proteins in our images that may be subunits, but this is close to the limit of current resolution. These experiments demonstrate that SICM can follow changes in the structure of the surface of the live cell at the resolution of individual protein complexes, provided that the surface is sufficiently flat, and that these images are very information rich and can reveal new protein dynamics on the nanoscale.
6. Combination with single-molecule fluorescence TIRF: tracking experiments
Single-molecule fluorescence to track molecules motion can be combined with SICM. This allows molecules to be rapidly and locally delivered to a defined region of the cell surface and then their diffusion over the cell membrane followed. Because the imaging time is limited by photobleaching of the fluorophore, this is an efficient method to probe diffusion across barriers in the cell membrane and such experiments can be used to understand how the cell membrane is organized. Experiments have been performed on polarized cells where diffusion barriers in the plasma membrane help segregate lipids and proteins into domains, where they are commensurate with specialized function. It is still not clear how these barriers are created and maintained. Boar spermatozoa can be used as model cells, and molecules can be delivered to a defined position on the cell surface using voltage driven delivery from the nanopipette and then tracked by performing TIRF on the apical surface of the flat boar sperm [21]. This allows the diffusion of single protein molecules and multi-molecular complexes to be followed between the three domains on the sperm head. The use of the nanopipette allowed repeated experiments to be performed on the same cell, to probe different macrodomains and subregions within the plasma membrane, and to investigate the existence of putative diffusion barriers. Because the topography of the cell surface is also known, it is then possible to correlate the measured trajectory to the presence of a particular cell structure or the underlying cytoskeleton. We also investigated exchange of molecules between different membrane domains. Using Alexa Fluor 555-cholera toxin B-subunit to label pre-existing GM1 ganglioside clusters in the cell membrane, we found that these relatively large structures diffused across the anterior acrosome–equatorial segment interface but could not get access to the postacrosome [29,30]. By contrast, smaller single lipid and protein molecules readily exchanged between all three domains, although they diffused more slowly on nearing and crossing to the postacrosome. The protein–cytoskeleton network thus appears to be able to form two types of diffusion interfaces on sperm heads, an ‘open’ interface and a ‘mass filter’ interface.
7. Fluorescence-based methods
(a). Principles of two-colour coincidence detection single-molecule fluorescence
Complementary to SICM, single-molecule fluorescence can be used to track and characterize individual labelled molecules diffusing over the cell membrane. Fluorescence analysis of single molecules, one by one, has been developed and applied widely to biomolecules over the past decade both in solution and on surfaces to probe biomolecular structure, dynamics and heterogeneity. Solution experiments are normally performed using a tightly focused Gaussian laser beam and confocal detection in order to achieve the smallest possible probe volume. Molecules can take different paths through the laser focus giving rise to variation in the excitation rate of the fluorophore and, hence, the fluorescence intensity detected. In order to address this issue, ratiometric methods have been developed where two fluorophores are attached to the same biomolecule, and the ratio of their fluorescence intensities is measured as they diffuse across the laser-excited volume. If the two fluorophores are sufficiently close as well as have overlapped emission and absorption spectra, then it is possible to excite the donor fluorophore and get FRET to the acceptor fluorophore so that only one laser is required. Such experiments are performed to measure the conformation of biomolecules, because the FRET efficiency depends on the donor–acceptor separation [31]. In the more general case, the two fluorophores are independently excited by two different spatially overlapped lasers and coincident fluorescent photons detected as the molecule diffuses across the laser-excited volume. Two-colour coincidence detection (TCCD) is based on continuous excitation of the confocal volume by both lasers [32]. The advantage of excitation of the two fluorophores with two independent lasers is that the fluorophores can be placed at any convenient position on the biomolecule and there is no requirement to place them close for FRET, which may not be possible if there is no information on the structure of the complex. The presence of coincident fluorescence in both channels allows us to sensitively detect associated molecules, and analysis of the intensity of these coincident fluorescent bursts allows us to determine the stoichiometries of the associated molecules [33]. TCCD is a novel method that removes the constraints on the labelling of biological molecules, allowing single-molecule methods to be applied to systems whose structure and stoichiometry are unknown, as well as allowing ultrasensitive detection of biomolecules [34] and the measurement of intramolecular dynamics [35]. TCCD has also been shown to be less sensitive to background fluorescence than single-colour excitation and capable of detecting femtomolar levels of molecular complex.
There were two major problems that became apparent as we started to apply TCCD to biological samples. First, it was not straightforward to perform reproducible control experiments where labelled components were non-associated, owing to the sample-to-sample variation in biological samples. In TCCD, there are chance coincident events that arise when two non-associated molecules happen to enter the probe at the same time. These events need to be removed from the data, so we can just determine and analyse the true coincident events that arise from associated molecules. This is critical to obtaining high sensitivity detection because for low fractions of associated molecules, below 1 per cent, the chance coincident events can be larger than the real coincident events. Because we could not determine the number of chance coincident events from control experiments, we devised two ways to do this directly from the experimental data alone [33]. One method is theoretical and based on the event rates of blue and red-labelled molecules. The second is based on red and blue time traces taken synchronously and desynchroinizing them so that the red trace is paired with a blue trace taken at a different time. Any coincident events that are determined from these traces are purely due to chance, allowing us to have the same red and blue event rates as the real experimental data and to determine the number of chance coincident events from the experimental data alone.
The second problem is to again use the experimental data alone to determine the correct threshold level above which we should count events. If set too high, then we will miss events and need to collect data for longer; and if set too low, then we will have problems with noise giving rise to events that are counted. In particular, the correct setting of the threshold is a particular problem with complex biological preparations or experiments on cells where the background level may change. To address this problem, we found that the fraction of associated molecules is a function of the red and blue threshold used and is maximum at the optimal threshold values for the blue and red channels [36]. This again allows us to use the experimental data alone to determine the correct threshold values for data analysis.
(b). Dynamic single-molecule colocalization
We extended the TCCD concept to two dimensions using a method we call dynamic single-molecule colocalization, or DySCo, which relies on quantitative video analysis of the fluorescence from two distinct fluorophores diffusing at the cell surface [37]. The advantages of tracking molecules are that there is no need to correct the data for chance coincidence events, because the probability of two non-associated molecules tracking within a short distance of one another for a number of frames is very low. In addition, we can detect immobile or slowly diffusing molecules that are unobservable using TCCD. Finally, very weakly expressed proteins can be analysed, whereas TCCD requires the event rate to be reasonably high to make data collection feasible. The strength of the DySCo method is its ability to quantify protein association, including both constitutive dimers and the much weaker interactions of transient dimers or of proteins whose diffusion is constrained by cytoskeleton-mediated corralling effects. These weaker interactions are likely to underpin signal transduction at the cell surface and are thus of great importance. An additional advantage of the method is that experiments at very low surface densities are possible, which will allow the investigation of weakly expressed membrane proteins, such as G protein-coupled receptors.
8. Application to neurodegenerative disease
One important biomedical problem is protein aggregation, which leads to neurodegenerative disease, including Alzheimer's and Parkinson's disease. In the initial stages of aggregation, small amounts of protein oligomers are formed that have been difficult to detect and analyse using ensemble bulk methods because of the transient and heterogeneous character of the species formed [38]. We have used the TCCD method to study the initial process of oligomerization of proteins, for instance, the early stages of neuroserpin polymerization [39], related to familial encephalopathy, and to examine the assembly of oligomeric species on the pathway to formation of amyloid fibrils of the SH3 domain of phosphatidylinositol-3-kinase [38] in vitro. The single-molecule experiments show that the species formed at the stage of the reaction where aggregates have previously been found to be maximally cytotoxic are a heterogeneous ensemble of oligomers with a median size of 38 ± 10 molecules. Our experiments provided direct evidence for a mechanism of amyloid aggregation in which the stable cross-β structure emerges via internal reorganization of disordered oligomers formed during the lag phase of the self-assembly reaction.
These experiments have recently been extended to examine the interactions between the extracellular chaperone clusterin and amyloid-β1–40 (Aβ1–40), a protein associated with Alzheimer's disease [40], and aggregation of α-synuclein, the protein whose misfolding and deposition is associated with Parkinson's disease [41]. We found that clusterin suppresses the aggregation of Aβ1–40 by interacting with the oligomers of the latter to form long-lived clusterin : Aβoligomer complexes to which monomers can no longer add, and that similar complexes are formed with oligomers that are released from fibrils. Under physiological conditions, these long-lived clusterin complexes will sequester and stabilize Aβ oligomers until they can be cleared by in vivo mechanisms, preventing the onset of Alzheimer's disease. In the case of α-synuclein, we used intermolecular FRET to identify a slow structural change of the initial oligomers to stable, proteinase-K resistant oligomers as the key step that leads ultimately to fibril formation. The structural conversion had a half-life of 1.5 days, indicating a high kinetic barrier for the conversion. These converted oligomers were found to be significantly more damaging to neurons and resulted in the production of intracellular reactive oxygen species. For both Aβ1–40 and α-synuclein, we observed an oligomer size distribution that peaked at dimers and then decayed away with size as predicted by recent modelling. In both cases in the absence of added soluble protein, the assembly process is reversed and fibrils disaggregate to form stable oligomers, directly confirming that fibrils could disaggregate at random positions to produce oligomers. These experiments now allow the possibility to add well characterized oligomers prepared in the test-tube to neurons and then follow the initial physiological changes as a function of time and oligomer dose. Such experiments should provide new insights into the molecular mechanism of how these oligomers damage neurons.
9. Application to T-cell biology
T lymphocytes play crucial roles in adaptive immune responses, where their surface interactions with antigen-presenting cells (APCs) are critical for the detection and subsequent elimination of pathogens. Many of these receptors have no intrinsic enzymatic activity, including the T-cell antigen receptor (TCR), which not only has to rely on extrinsic tyrosine kinases to initiate signalling, but also must do so while discriminating between ligands of differing quality. Furthermore, it has been shown that a single TCR complex can lead to T-cell triggering in the presence of co-receptor molecules [42], making it highly appropriate to study this phenomenon at the single-molecule level. The ligands for the TCR are antigenic peptide/major histocompatibility complexes (pMHC) expressed by the APC. Discrimination by the TCR is intimately linked to the longevity of the TCR/pMHC interaction, where the decision to trigger can be influenced by small changes in the binding kinetics. The TCR must initiate differential signalling pathways based on these differences, from titrating positive and negative selection during thymocyte development to causing a potent immune response in the periphery. These diverse mechanisms must all be intrinsic to the structure and organization of the antigen receptor triggering apparatus, and the enzymatic processes initiated by receptor engagement [43]. However, there is still no agreement about the structure and organization of the receptor triggering apparatus.
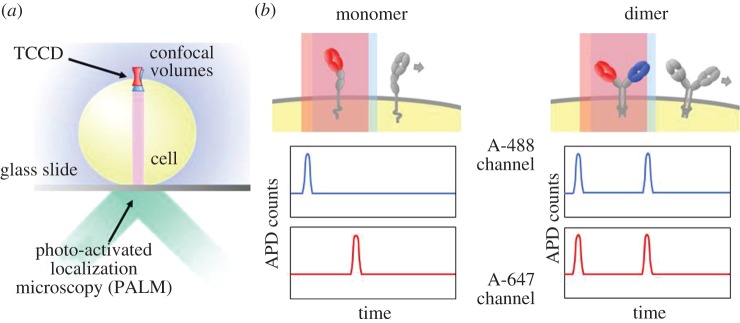
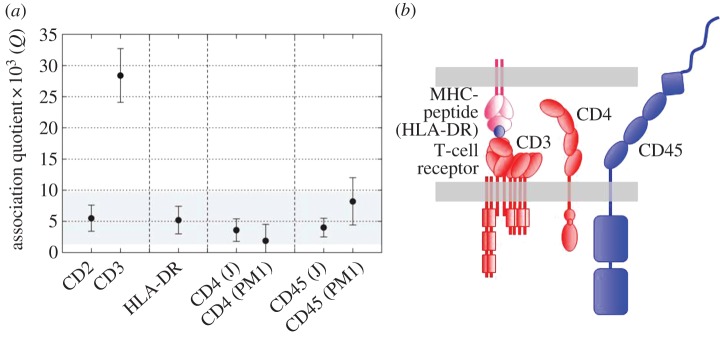
The TCR itself has been the most contentious and inferences about its stoichiometry have profoundly influenced thinking about triggering mechanisms [44], emphasizing why it is important to determine these stoichiometries and to do this for live cells. Initial attempts to resolve this issue for the TCR involved TCCD, wherein TCRs diffusing at the apical surface of T cells resting on a glass surface were examined [45]; the principle of the TCCD method applied to the cell surface is shown in figure 4a. In other experiments, DySCo was used [37]. Collectively, these studies strongly suggested that the TCR is monovalent [45], and the TCCD analysis has now been extended to other membrane components of the TCR triggering apparatus, i.e. CD4/Lck, CD45 and MHC class II. These proteins have all previously been claimed to form dimers, but gave strong signals for monomers in the TCCD analysis (figure 5a) [46]. The simple organization of the triggering apparatus of the T cell suggested by these studies is shown in figure 5b.
Figure 4.
Principle of TCCD on live cells. (a) Measurements are made on the apical cell surface in contrast to PALM experiments on the basal surface. (b) Associated molecules give rise to coincident bursts of red and blue fluorescence as they diffuse through the confocal volume excited by two overlapped lasers.
Figure 5.
TCCD analysis of a resting T cell. (a) All the components of the triggering apparatus of a T cell, i.e. MHC-peptide (HLA-DR), CD4 and CD45, give the same low value of association quotient (Q), a measure of the fraction of associated molecules, as the positive control monomeric protein, CD2. The dimer control, CD3, gives much higher levels of coincidence (each T-cell receptor contains two CD3 (epsilon) signalling subunits). The data are from James et al. [46]. (b) Cartoon of some of the likely stoichiometries of the triggering apparatus of the T cell, based on the TCCD experiments.
High-resolution measurements by others, based on photo-activation localization microscopy (PALM [47]), have, however, suggested that contrary to the findings of our TCCD and DySCo experiments, the receptor is instead pre-clustered in groups of 7–25 molecules in resting cells. A crucial difference between the TCCD and PALM experiments is that these were done at different locations, that is, at the apical surface of the T cell and the basal cell/glass interface, respectively (figure 4b). Using fluorescence recovery after photo-bleaching (FRAP) measurements, it has since been possible to show that contact with a functionalized glass surface in fact alters the behaviour of the TCR, indicating that it is important to make measurements at the apical surface of resting cells [46]. FRAP measurements at the apical surface showed that the TCR is largely if not completely monovalent or monomeric, whereas approximately 40 per cent of the receptors at the basal membrane contacting the glass surface become immobile in a signalling-dependent manner, explaining the discordant findings [46].
The ‘ground state’ measurements of the TCR triggering apparatus using TCCD and DySCo have been complemented with bioluminescence resonance energy transfer (BRET)-based experiments [46,48]. Although BRET requires heterologous protein expression, the luminescence detection mode allows molecular interactions to be studied at near-native expression levels so that the observations are relevant to the behaviour of the native proteins, which can also be studied directly using the TCCD method. The TCCD measurements discussed above showed with 95% confidence that all analysed proteins are monomeric [46]. When combined with the orthogonal BRET method for which, in the case of the analysis of the TCR, the upper limit on the fraction of oligomers was estimated to be 10 per cent, these two techniques allow us to be confident that these assignments represent the state of the proteins at the cell surface. An additional, unexpected outcome of the BRET experiments is the extent to which these membrane-bound molecules form non-specific associations within the membrane, owing to an increase in their effective concentrations versus cytosolic proteins, whose encounters are likely to be much less frequent [46]. This is likely to have important implications for the architecture of receptor signalling pathways.
Critically, this work establishes the ground state of the plasma membrane ex vivo and suggests that this state changes on surface contact, even on a non-activating surface, leading to signalling and receptor clustering. Therefore, measurements taken at the basal surface using TIRF can alter the biology being observed, which is an important message for all TIRF-based studies. Having established the organization of proteins for the resting T cell, the stage is set to start to follow the initial molecular reorganizations that lead to T-cell triggering. Determining the molecular mechanism is of great interest and may lead to new therapeutic treatments for autoimmune diseases.
10. Future outlooks
The work presented above shows that single-molecule experiments are now possible on live cells using SICM and single-molecule fluorescence and combinations of both of these methods. This allows the organization of the cell membrane to be probed, down to the level of individual molecules, and the changes in this organization to be followed, in time, during important biological processes. Furthermore, these methods can now be applied to important biomedical problems in immunology, neurodegenerative disease and heart disease, providing new important insights into the initial molecular events involved in these diseases. However, it is also important to note some of the limitations of these approaches. For example, all the studies presented have been on cells and extension of these methods to tissue is currently extremely challenging. Even when studying cells, fluorescence imaging methods that can be used on the apical cell surface are needed, to avoid any structural reorganization owing to the glass surface. Approaches based on the use of light sheets to control the plane of illumination, combined with imaging using a sensitive camera, have the potential to allow single-molecule imaging on this surface [49]. This approach also has the potential to extend single-molecule imaging from the cell surface to inside the cell, opening up the possibilities to study key nuclear processes. The photoactivatable fluorophores developed for super-resolution imaging, which can be switched into a fluorescent state [50], allow single-molecule tracking experiments to be performed at physiological concentrations of molecules or to control the time point at which the molecules are imaged, which is also an important advance for studies inside cells. Methods to image cellular function as well as the position of molecules or of the cell membrane are also needed. This can be done using fluorescent reporters inside the cell, as was done to detect triggering of β2ARs, or in a label-free mode by detecting, for example, ion-channel activity or chemical fluxes. Here perhaps lies one key challenge for the future, because it is likely that with nanoscale imaging a wide variety of structures or molecular associations may be detected and one needs a method to determine which are functional and which are chance associations, and have no functional importance. In addition, when studying low levels of proteins in the cell membrane, problems caused by small amounts of non-specific binding become more severe and need to be reduced by improved surface chemistry to ensure only specific binding is studied.
An important development in the past few years has been super-resolution fluorescence imaging, based on the excitation of single fluorophores, and this has the potential to provide ‘snap-shots’ of the organization of labelled proteins on the cell surface with better than 30 nm resolution. However, following how this organization changes with time is more challenging, because the method is based on the excitation and then photobleaching of individual fluorophores, so it is not possible to image the fluorophores again [51]. If this became possible, then it would allow series of ‘snap-shots’ to be taken and to directly follow the reorganization, which would be a significant advance. Another complementary super-resolution imaging method is stimulated emission microscopy (STED), which also offers much promise [52–54]. STED essentially offers a super-resolution form of confocal microscopy and so allows high speed and repetitive imaging of cellular organization below 100 nm resolution. This has been used to explore lipid organization on the cell surface [55]. The optimization of the fluorophores for STED has led to the reduction in the laser powers needed, reducing concerns about photodamage caused by the lasers [56].
Currently, the fastest rate of imaging using SICM is around 1 ×1 μm2 every 10 s. The relatively large distances, of the order of 100 nm, that the pipette needs to move to image the cell surface, to avoid direct contact, means that the speed the pipette can be moved and stopped limits the imaging rate. A total redesign of the instrument with a much smaller nanofabricated probe combined with higher resonance frequency piezos would be needed to increase the imaging rate above this limit. Alternatively, we have recently shown that it is possible to image relatively flat surfaces faster by only approximately following the overall surface topography and then reconstructing the true surface topography from the measured ion current and pipette position [57]. This method may allow faster scanning of small relatively flat regions of the cell surface.
Whatever methods are used for imaging, another key challenge is to obtain a coherent view of the cell membrane structure and will likely require the combined use of a number of complementary imaging methods. Differentiated cells with clear and reproducible cell structures such as sperm cells, cardiomyocytes, neurons or epithelial cells allow the merging of data taken on different cells of the same type. In addition, the underlying cell cytoskeleton provides another possible structure to which cell organization can be related. Studies of endogenous proteins are also required to ensure minimal perturbation of the membrane organization, and if studies are performed on the basal cell surface it is very important to ensure that there has been no artefactual reorganization on contacting the surface as discussed above. Next, nanoscale images need to be recorded on cells with reproducible structures under minimally perturbed conditions so that information from different studies can be combined. Only in this way can our understanding of the organization and function of the cell surface take full account of its complexity.
In conclusion, methods are now available that allow imaging of the cell surface with resolution comparable to scanning electron microscopy, can be performed on live cells under physiological conditions and can follow individual molecules on the cell membrane. This is an exciting advance and the challenge is now to apply these methods to gain new biological insights and select the biological problems where these new methods have most to offer.
Acknowledgements
The work described was funded by the EPSRC, BBSRC, MRC and Wellcome Trust. D.K. also received funding from the Frances and Augustus Newman foundation.
References
- 1.Klenerman D, Korchev Y. 2006. Potential biomedical applications of the scanned nanopipette. Nanomedicine (Lond.) 1, 107–114 10.2217/17435889.1.1.107 (doi:10.2217/17435889.1.1.107) [DOI] [PubMed] [Google Scholar]
- 2.Shevchuk AI, Novak P, Takahashi Y, Clarke R, Miragoli M, Babakinejad B, Gorelik J, Korchev YE, Klenerman D. 2011. Realizing the biological and biomedical potential of nanoscale imaging using a pipette probe. Nanomedicine (Lond.) 6, 565–575 10.2217/nnm.10.154 (doi:10.2217/nnm.10.154) [DOI] [PubMed] [Google Scholar]
- 3.Orte A, Clarke R, Klenerman D. 2010. Single-molecule two-colour coincidence detection to probe biomolecular associations. Biochem. Soc. Trans. 38, 914–918 10.1042/BST0380914 (doi:10.1042/BST0380914) [DOI] [PubMed] [Google Scholar]
- 4.Hansma PK, Drake B, Marti O, Gould SAC, Prater CB. 1989. The scanning ion-conductance microscope. Science 243, 641–643 10.1126/science.2464851 (doi:10.1126/science.2464851) [DOI] [PubMed] [Google Scholar]
- 5.Korchev YE, Bashford CL, Milovanovic M, Vodyanoy I, Lab MJ. 1997. Scanning ion conductance microscopy of living cells. Biophys. J. 73, 653–658 10.1016/S0006-3495(97)78100-1 (doi:10.1016/S0006-3495(97)78100-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novak P, et al. 2009. Nanoscale live-cell imaging using hopping probe ion conductance microscopy. Nat. Methods 6, 279–281 10.1038/nmeth.1306 (doi:10.1038/nmeth.1306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korchev YE, Raval M, Lab MJ, Gorelik J, Edwards CR, Rayment T, Klenerman D. 2000. Hybrid scanning ion conductance and scanning near-field optical microscopy for the study of living cells. Biophys. J. 78, 2675–2679 10.1016/S0006-3495(00)76811-1 (doi:10.1016/S0006-3495(00)76811-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorelik J, et al. 2002. Scanning surface confocal microscopy for simultaneous topographical and fluorescence imaging: application to single virus-like particle entry into a cell. Proc. Natl Acad. Sci. USA 99, 16 018–16 023 10.1073/pnas.252458399 (doi:10.1073/pnas.252458399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shevchuk AI, Hobson P, Lab MJ, Klenerman D, Krauzewicz N, Korchev YE. 2008. Imaging single virus particles on the surface of cell membranes by high-resolution scanning surface confocal microscopy. Biophys. J. 94, 4089–4094 10.1529/biophysj.107.112524 (doi:10.1529/biophysj.107.112524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shevchuk AI, et al. 2012. An alternative mechanism of clathrin-coated pit closure revealed by ion conductance microscopy. J. Cell Biol. 197, 499–508 10.1083/jcb.201109130 (doi:10.1083/jcb.201109130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shevchuk AI, Hobson P, Lab MJ, Klenerman D, Krauzewicz N, Korchev YE. 2008. Endocytic pathways: combined scanning ion conductance and surface confocal microscopy study. Pflugers Arch. 456, 227–235 10.1007/s00424-007-0410-4 (doi:10.1007/s00424-007-0410-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorelik J, et al. 2002. Ion channels in small cells and subcellular structures can be studied with a smart patch-clamp system. Biophys. J. 83, 3296–3303 10.1016/S0006-3495(02)75330-7 (doi:10.1016/S0006-3495(02)75330-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu Y, et al. 2002. High-resolution scanning patch-clamp: new insights into cell function. FASEB J. 16, 748–750 10.1096/fj.01-1024fje (doi:10.1096/fj.01-1024fje) [DOI] [PubMed] [Google Scholar]
- 14.Sanchez D, et al. 2008. Noncontact measurement of the local mechanical properties of living cells using pressure applied via a pipette. Biophys. J. 95, 3017–3027 10.1529/biophysj.108.129551 (doi:10.1529/biophysj.108.129551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sánchez D, et al. 2007. Localized and non-contact mechanical stimulation of dorsal root ganglion sensory neurons using scanning ion conductance microscopy. J. Neurosci. Methods 159, 26–34 10.1016/j.jneumeth.2006.06.018 (doi:10.1016/j.jneumeth.2006.06.018) [DOI] [PubMed] [Google Scholar]
- 16.Ying L, Bruckbauer A, Rothery AM, Korchev YE, Klenerman D. 2002. Programmable delivery of DNA through a nanopipet. Anal. Chem. 74, 1380–1385 10.1021/ac015674m (doi:10.1021/ac015674m) [DOI] [PubMed] [Google Scholar]
- 17.Rodolfa KT, Bruckbauer A, Zhou D, Korchev YE, Klenerman D. 2005. Two-component graded deposition of biomolecules with a double-barreled nanopipette. Angew. Chem. Int. Ed. Engl. 44, 6854–6859 10.1002/anie.200502338 (doi:10.1002/anie.200502338) [DOI] [PubMed] [Google Scholar]
- 18.Bruckbauer A, Zhou D, Kang DJ, Korchev YE, Abell C, Klenerman D. 2004. An addressable antibody nanoarray produced on a nanostructured surface. J. Am. Chem. Soc. 126, 6508–6509 10.1021/ja0317426 (doi:10.1021/ja0317426) [DOI] [PubMed] [Google Scholar]
- 19.Bruckbauer A, Zhou D, Ying L, Korchev YE, Abell C, Klenerman D. 2003. Multicomponent submicron features of biomolecules created by voltage controlled deposition from a nanopipet. J. Am. Chem. Soc. 125, 9834–9839 10.1021/ja035755v (doi:10.1021/ja035755v) [DOI] [PubMed] [Google Scholar]
- 20.Bruckbauer A, Ying L, Rothery AM, Zhou D, Shevchuk AI, Abell C, Korchev YE, Klenerman D. 2002. Writing with DNA and protein using a nanopipet for controlled delivery. J. Am. Chem. Soc. 124, 8810–8811 10.1021/ja026816c (doi:10.1021/ja026816c) [DOI] [PubMed] [Google Scholar]
- 21.Bruckbauer A, James P, Zhou DJ, Yoon JW, Excell D, Korchev Y, Jones R, Klenerman D. 2007. Nanopipette delivery of individual molecules to cellular compartments for single-molecule fluorescence tracking. Biophys. J. 93, 3120–3131 10.1529/biophysj.107.104737 (doi:10.1529/biophysj.107.104737) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piper JD, Clarke RW, Korchev YE, Ying L, Klenerman D. 2006. A renewable nanosensor based on a glass nanopipette. J. Am. Chem. Soc. 128, 16 462–16 463 10.1021/ja0650899 (doi:10.1021/ja0650899) [DOI] [PubMed] [Google Scholar]
- 23.Takahashi Y, Shevchuk AI, Novak P, Murakami Y, Shiku H, Korchev YE, Matsue T. 2012. Simultaneous noncontact topography and electrochemical imaging by SECM/SICM featuring ion current feedback regulation. J. Am. Chem. Soc. 132, 10 118–10 126 10.1021/ja1029478 (doi:10.1021/ja1029478) [DOI] [PubMed] [Google Scholar]
- 24.Takahashi Y, et al. 2011. multifunctional nanoprobes for nanoscale chemical imaging and localized chemical delivery at surfaces and interfaces. Angew. Chem. Int. Ed. 50, 9638–9642 10.1002/anie.201102796 (doi:10.1002/anie.201102796) [DOI] [PubMed] [Google Scholar]
- 25.Nikolaev VO, et al. 2010. Beta(2)-adrenergic receptor redistribution in heart failure changes cAMP compartmentation. Science 327, 1653–1657 10.1126/science.1185988 (doi:10.1126/science.1185988) [DOI] [PubMed] [Google Scholar]
- 26.Merrifield CJ, Feldman ME, Wan L, Almers W. 2002. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4, 691–698 10.1038/ncb837 (doi:10.1038/ncb837) [DOI] [PubMed] [Google Scholar]
- 27.Saffarian S, Kirchhausen T. 2008. Differential evanescence nanometry: live-cell fluorescence measurements with 10-nm axial resolution on the plasma membrane. Biophys. J. 94, 2333–2342 10.1529/biophysj.107.117234 (doi:10.1529/biophysj.107.117234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shevchuk AI, Frolenkov GI, Sánchez D, James PS, Freedman N, Lab MJ, Jones R, Klenerman D, Korchev YE. 2006. Imaging proteins in membranes of living cells by high-resolution scanning ion conductance microscopy. Angew. Chem. Int. Ed. Engl. 45, 2212–2216 10.1002/anie.200503915 (doi:10.1002/anie.200503915) [DOI] [PubMed] [Google Scholar]
- 29.Bruckbauer A, Dunne PD, James P, Howes E, Zhou D, Jones R, Klenerman D. 2010. Selective diffusion barriers separate membrane compartments. Biophys. J. 99, L1–L3 10.1016/j.bpj.2010.03.067 (doi:10.1016/j.bpj.2010.03.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones R, Howes E, Dunne PD, James P, Bruckbauer A, Klenerman D. 2010. Tracking diffusion of GM1 gangliosides and zona pellucida binding molecules in sperm plasma membranes following cholesterol efflux. Dev. Biol. 339, 398–406 10.1016/j.ydbio.2009.12.044 (doi:10.1016/j.ydbio.2009.12.044) [DOI] [PubMed] [Google Scholar]
- 31.Deniz AA, Dahan M, Grunwell JR, Ha TJ, Faulhaber AE, Chemla DS, Weiss S, Schultz PG. 1999. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Förster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA 96, 3670–3675 10.1073/pnas.96.7.3670 (doi:10.1073/pnas.96.7.3670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li HT, Ying LM, Green JJ, Balasubramanian S, Klenerman D. 2003. Ultrasensitive coincidence fluorescence detection of single DNA molecules. Anal. Chem. 75, 1664–1670 10.1021/ac026367z (doi:10.1021/ac026367z) [DOI] [PubMed] [Google Scholar]
- 33.Orte A, Clarke R, Balasubramanian S, Klenerman D. 2006. Determination of the fraction and stoichiometry of femtomolar levels of biomolecular complexes in an excess of monomer using single-molecule, two-color coincidence detection. Anal. Chem. 78, 7707–7715 10.1021/ac061122y (doi:10.1021/ac061122y) [DOI] [PubMed] [Google Scholar]
- 34.Li HT, Zhou DJ, Browne H, Balasubramanian S, Klenerman D. 2004. Molecule by molecule direct and quantitative counting of antibody–protein complexes in solution. Anal. Chem. 76, 4446–4451 10.1021/ac049512c (doi:10.1021/ac049512c) [DOI] [PubMed] [Google Scholar]
- 35.Li HT, Ren XJ, Ying LM, Balasubramanian S, Klenerman D. 2004. Measuring single-molecule nucleic acid dynamics in solution by two-color filtered ratiometric fluorescence correlation spectroscopy. Proc. Natl Acad. Sci. USA 101, 14 425–14 430 10.1073/pnas.0404295101 (doi:10.1073/pnas.0404295101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke RW, Orte A, Klenerman D. 2007. Optimized threshold selection for single-molecule two-color fluorescence coincidence spectroscopy. Anal. Chem. 79, 2771–2777 10.1021/ac062188w (doi:10.1021/ac062188w) [DOI] [PubMed] [Google Scholar]
- 37.Dunne PD, Fernandes RA, McColl J, Ji WY, James JR, Davis SJ, Klenerman D. 2009. DySCo: quantitating associations of membrane proteins using two-color single-molecule tracking. Biophys. J. 97, L5–L7 10.1016/j.bpj.2009.05.046 (doi:10.1016/j.bpj.2009.05.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orte A, Birkett NR, Clarke RW, Devlin GL, Dobson CM, Klenerman D. 2008. Direct characterization of amyloidogenic oligomers by single-molecule fluoresence. Proc. Natl Acad. Sci. USA 105, 14 424–14 429 10.1073/pnas.0803086105 (doi:10.1073/pnas.0803086105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiou A, Hägglöf P, Orte A, Chen AY, Dunne PD, Belorgey D, Karlsson-Li S, Lomas DA, Klenerman D. 2009. Probing neuroserpin polymerization and interaction with amyloid-beta peptides using single molecule fluorescence. Biophys. J. 97, 2306–2315 10.1016/j.bpj.2009.07.057 (doi:10.1016/j.bpj.2009.07.057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayan P, et al. 2012. The extracellular chaperone clusterin sequesters oligomeric forms of the amyloid-beta(1–40) peptide. Nat. Struct. Mol. Biol. 19, 79–83 10.1038/nsmb.2191 (doi:10.1038/nsmb.2191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cremades N, et al. 2012. Direct observation of the interconversion of normal and pathogenic forms of α-synuclein. Cell 149, 1048–1059 10.1016/j.cell.2012.03.037 (doi:10.1016/j.cell.2012.03.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. 2002. Direct observation of ligand recognition by T cells. Nature 419, 845–849 10.1038/nature01076 (doi:10.1038/nature01076) [DOI] [PubMed] [Google Scholar]
- 43.Smith-Garvin JE, Koretzky GA, Jordan MS. 2009. T Cell Activation. Annu. Rev. Immunol. 27, 591–619 10.1146/annurev.immunol.021908.132706 (doi:10.1146/annurev.immunol.021908.132706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis SJ, van der Merwe PA. 2006. The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 7, 803–809 10.1038/ni1369 (doi:10.1038/ni1369) [DOI] [PubMed] [Google Scholar]
- 45.James JR, White SS, Clarke RW, Johansen AM, Dunne PD, Sleep DL, Fitzgerald WJ, Davis SJ, Klenerman D. 2007. Single-molecule level analysis of the subunit composition of the T cell receptor on live T cells. Proc. Natl Acad. Sci. USA 104, 17 662–17 667 10.1073/pnas.0700411104 (doi:10.1073/pnas.0700411104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.James JR, et al. 2011. The T cell receptor triggering apparatus is composed of monovalent or monomeric proteins. J. Biol. Chem. 286, 31 993–32 001 10.1074/jbc.M111.219212 (doi:10.1074/jbc.M111.219212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. 2010. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 543. 10.1038/ni0610-543b (doi:10.1038/ni0610-543b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.James JR, Oliveira MI, Carmo AM, Iaboni A, Davis SJ. 2006. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat. Methods 3, 1001–1006 10.1038/nmeth978 (doi:10.1038/nmeth978) [DOI] [PubMed] [Google Scholar]
- 49.Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. 2011. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8, 417–423 10.1038/nmeth.1586 (doi:10.1038/nmeth.1586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Suarez M, Ting AY. 2008. Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 9, 929–943 10.1038/nrm2531 (doi:10.1038/nrm2531) [DOI] [PubMed] [Google Scholar]
- 51.Bo HA, Babcock H, Xiaowei ZA. 2010. Breaking the diffraction barrier: super-resolution imaging of cells. Cell 143, 1047–1058 10.1016/j.cell.2010.12.002 (doi:10.1016/j.cell.2010.12.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hell SW, Wichmann J. 1994. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 10.1364/OL.19.000780 (doi:10.1364/OL.19.000780) [DOI] [PubMed] [Google Scholar]
- 53.Hell SW. 2009. Microscopy and its focal switch. Nat. Methods 6, 24–32 10.1038/nmeth.1291 (doi:10.1038/nmeth.1291) [DOI] [PubMed] [Google Scholar]
- 54.Hell SW. 2007. Far-field optical nanoscopy. Science 316, 1153–1158 10.1126/science.1137395 (doi:10.1126/science.1137395) [DOI] [PubMed] [Google Scholar]
- 55.Eggeling C, et al. 2009. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162 10.1038/nature07596 (doi:10.1038/nature07596) [DOI] [PubMed] [Google Scholar]
- 56.Brakemann T, et al. 2011. A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching. Nat. Biotechnol. 29, 942–947 10.1038/nbt.1952 (doi:10.1038/nbt.1952) [DOI] [PubMed] [Google Scholar]
- 57.Zhukov A, Richards O, Ostanin VP, Korchev Y, Klenerman D. 2012. A hybrid scanning mode for fast scanning ion conductance microscopy (SICM) imaging. Ultramicroscopy 121, 1–7 10.1016/j.bbr.2011.03.031 (doi:10.1016/j.bbr.2011.03.031) [DOI] [PMC free article] [PubMed] [Google Scholar]