Abstract
The plasma membrane is still one of the enigmatic cellular structures. Although the microscopic structure is getting clearer, not much is known about the organization at the nanometre level. Experimental difficulties have precluded unambiguous approaches, making the current picture rather fuzzy. In consequence, a variety of different membrane models has been proposed over the years, on the basis of different experimental strategies. Recent data obtained via high-resolution single-molecule microscopy shed new light on the existing hypotheses. We thus think it is a good time for reviewing the consistency of the existing models with the new data. In this paper, we summarize the available models in ten propositions, each of which is discussed critically with respect to the applied technologies and the strengths and weaknesses of the approaches. Our aim is to provide the reader with a sound basis for his own assessment. We close this chapter by exposing our picture of the membrane organization at the nanoscale.
Keywords: singe molecule microscopy, FRET, single particle tracking, membrane rafts, plasma membrane, lipids
1. Introduction
The cellular plasma membrane is a fascinating organelle. Representing the interface to the outside world defines the majority of its tasks: it is a barrier for polar substances, so that the cytosol is efficiently shielded from the extracellular milieu; it is a docking point for external stimuli, hosting receptors for growth, differentiation, uptake, etc.; it represents the contact zone to other cells in body tissue, and carries the required adhesion molecules, but also the connection to filaments for providing the mechanical support; the cell membrane is an electrical insulator with switchable resistance, enabling the controlled influx or efflux of specific ions for electrical signalling; finally, the two-dimensional fluid surface speeds up chemical reactions, as the time for two reaction partners to meet is substantially shortened compared with three-dimensional volumes.
What is the structure that would best fulfil such a variety of tasks? The cornerstone of our picture of the membrane structure has been the fluid mosaic model by Singer and Nicholson from 1972, which for the first time combined membrane fluidity with the proposition that membrane proteins were dispersed randomly in the lipid bilayer [1,2]. We now know, however, that the plasma membrane hosts thousands of lipid species and more than one-third of the proteome, making it an extremely complex matrix. It is the plethora of potential lipid–lipid, protein–protein and lipid–protein interactions that renders theoretical predictions and molecular modelling of membranes so difficult. But also experimentalists suffer from hurdles imposed by the nature of the plasma membrane itself. Depending on the experimental approach, researchers made surprising observations that led to a confusing repertoire of concepts and ideas, most of which had more relation to the way the experiment was actually performed than to the nature of the investigated object. Already in the middle 1970s, indications had arisen for a segregation of lipids into domains of different fluidity and composition [3]. Nowadays, abbreviations, acronyms and terms such as detergent-resistant membranes (DRMs), glycosphingolipid-enriched membrane domains (GEMs), membrane/lipid rafts, pickets, fences, transient confinement zones, nanodomains, hotspots, etc., are part of the scientific literature, addressing probably the same, but also probably different structures. In particular, it was the concept of lipid rafts—later apologetically termed membrane rafts—that has laid out the framework for interpreting interactions in membranes [4]. After years of (re)search, however, hard evidence is still missing, culminating in a rather vague consensus definition from 2006:
‘Membrane rafts are small (10–200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein–protein and protein–lipid interactions’. [5, p. 1597]
In this review, we survey previous studies with respect to their conclusions about plasma membrane structures. A special focus will be laid on novel high-resolution and ultrasensitive microscopy methods, which have generated a wealth of new quantitative insights in recent years. The review is organized as follows: we start by summarizing recent studies in relation to ten propositions, and discuss the strengths and weaknesses of these approaches. Next, we expose our view of the plasma membrane organization. We finish by providing a brief technology glossary.
2. Proposition 1: detergent-resistant membranes correspond to structures in the cell membrane
(a). In a nutshell
First indications for the existence of structural domains within the plasma membrane of cells came from biochemical extraction of the membrane, together with subsequent sucrose density gradient centrifugation, which made it possible to qualitatively distinguish a distinct part of the plasma membrane that is not soluble in mild detergent [6]. Investigation of such DRMs further yielded their protein content, which was found to be consistently different from the remaining fractions of the membrane [7,8]. Taken together, those studies were one experimental basis for the formulation of the ‘raft theory’, in which the existence of platforms within the plasma membrane, membranes of organelles and membranes of transport vesicles has been postulated [4]. Besides their initial importance for the proposition of plasma membrane domains, detergent extraction methods are still widely used to study the recruitment of membrane proteins to the insoluble or soluble part of the plasma membrane.
(b). Technology
Most researchers use the non-ionic detergent Triton X-100 for cell lysis on ice [6]; alternatively, other detergents have been used to further discriminate the DRMs [9] or to enable the analysis at 37°C [10]. The lysates are placed in a centrifuge tube with a step gradient of sucrose and centrifuged at 100 000g for approximately 3 h (see Brown [11] for a protocol). Low-density DRMs can be separated from high-density cytoskeletal proteins, cytoplasmic proteins and mixed micelles.
(c). Pros
There is a striking number of studies in which changes in a protein's DRM association were found to correlate with changes in the protein's functional state [12–14]. Even if the detergent massively alters the phase state of the membranes, the differential partitioning into soluble and non-soluble phases is remarkable. In this view, a DRM raft is not a physical entity; instead DRM association specifies a physico-chemical property of the investigated protein.
(d). Concerns
There are principal limitations to this in vitro method. First, artefacts due to re-localization of proteins during detergent extraction have been reported [15,16]. Second, to avoid degradation processes, solubilization of cells is commonly performed at low temperatures, which most likely has an effect on the phase state of the membrane and may alter its physical properties [17,18]. Third and most importantly, the presence of high amounts of detergent changes the thermodynamic properties of the lipid mixture and most likely affects or even generates the observed lipid domains [19].
In addition, there is a wealth of potential parameters that may influence a protein's partitioning to detergent-stabilized phases, which are neither predictable nor interpretable. Palmitoylation, dimerization or association with the membrane skeleton, for example, can be expected to alter the solubility per se, without the need for a hypothetical ‘raft structure’.
3. Proposition 2: lipids and proteins hop between periodic compartments generated by the membrane cytoskeleton
(a). In a nutshell
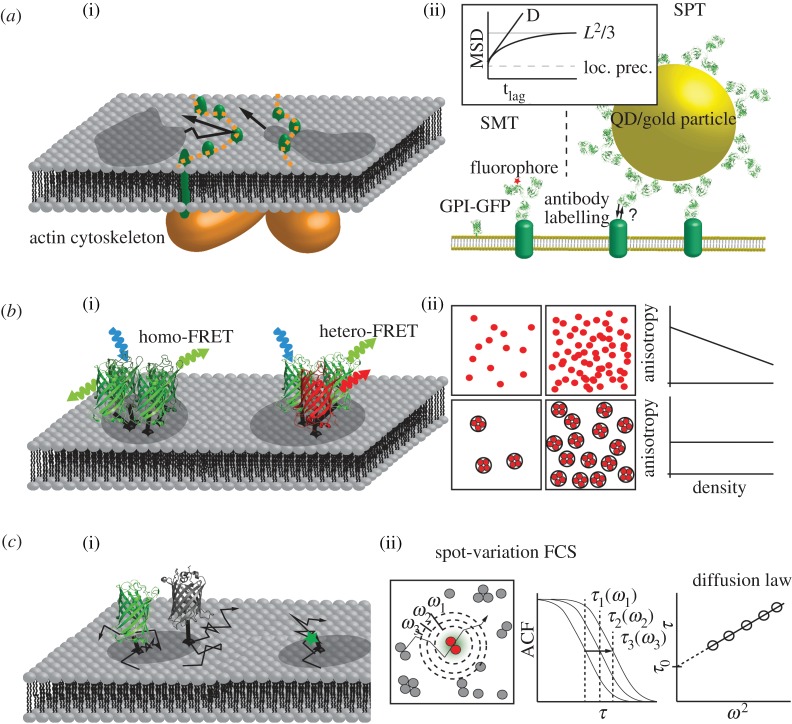
Results from single-particle (SPT) or single-molecule tracking (SMT) of plasma membrane constituents are frequently used to extract information on rafts. In particular, work from the laboratory of Aki Kusumi pushed the field forward. In a review article, he proposed that rafts are short-lived structures with a lifetime of 1 ms or less [20]. Kusumi's hypothesis was based on the picket-fence model of the plasma membrane developed by the group over the years [21–25]. By following the motion of gold-labelled proteins or lipids in the live cell plasma membrane, the researchers found hop diffusion between adjacent compartments induced by the underlying cortical actin meshwork. They postulated that actin-anchored proteins line up like pickets along the actin fences, thereby transmitting the structural information from the cytosolic to the exoplasmic leaflet of the lipid bilayer. In this model, the pickets would act as size-exclusion barriers to the motion of objects, thereby particularly limiting the transit of rafts. Because raft markers (e.g. glycosylphosphatidylinositol (GPI)-anchored proteins) showed the same hop frequency as non-raft markers (e.g. the lipid dioleyl-phosphatidylethanolamine), Kusumi et al. concluded that rafts have to be dissolved before the transit through the pickets, and therefore appear to be extremely short-lived (figure 1a).
Figure 1.
(a(i)) Proposition 2: Kusumi's picket-fence model is shown. Trans-membrane proteins (pickets in green) associated to the underlying actin network hinder the free diffusing membrane rafts (darker regions). The diffusion of the membrane rafts in this model is explained by their fast dissociation and association rates that allow for passing of the obstacles (fences). (a(ii)) The key methods supporting this model are SMT and SPT. For SMT, proteins are directly labelled with fluorophores (green fluorescent protein; GFP) or via an antibody; SPT uses 40 nm gold or quantum dots labelled via an antibody. SPT is limited by the fact that multiple interactions of one label cannot be excluded. The inset depicts the data analysis and interpretation. Tracks are analysed and mean square displacement (MSD) versus time lag (tlag) is plotted to differentiate between different modes of diffusion. From the y-intercept, the localization precision can be derived, while the slope of the curve is the diffusion coefficient D. In the case of hindered diffusion, the curve saturates to L2/3, where L is the size of the structure. (b(i)) Proposition 3: Förster resonance energy transfer (FRET) was used for estimating raft association by measuring the proximity of proteins. Homo-FRET can be measured via fluorescence anisotropy, hetero-FRET via spectral changes. (b(ii)) Fluorescence depolarization experiment for different arrangements of fluorescently labelled GPI-anchored proteins. The upper case depicts GPI-anchored proteins in a random distribution, where the anisotropy drops with increasing concentration. In the lower case, GPI-anchored proteins are associated with membrane rafts defining the number of proteins in close proximity. When the concentration is increased, fluorescence anisotropy stays constant. (c(i)) Proposition 4: trapping of fluorescently labelled GPI-anchored proteins or sphingomyelin to membrane rafts. A GPI-anchored protein (here indicated by a GFP anchored to a lipid) diffuses into a raft, where it gets trapped for a certain time. Photobleaching during the recording sequence yields an increasing number of invisible molecules (indicated by a dark GFP). (c(ii)) Analysis of the diffusion law recorded with sv-FCS. By increasing the excitation spot area ω2, the fluorophore's transition times τD increase accordingly, recognizable in the autocorrelation curves (ACF). Fitting each ACF returns τD(ω2), the diffusion time as function of the laser spot radius ω. From the y-intercept, the diffusion law offset, τ0, is obtained.
(b). Technology
Colloidal gold probes (diameter 40 nm) were used as labels, which can be detected owing to light scattering as dark spots on transmission light microscopy images [26]. Cross-correlation with a known kernel allows for calculating the bead positions [27] to an accuracy of less than 20 nm even at a frame rate of 50 000 frames per second (fps) [24]. Proteins were labelled via monoclonal antibodies, to which the gold particles were linked via secondary antibodies [24]. Fabs were used instead of full antibodies to minimize protein cross-linking. For lipid tracking, fluorescein-conjugated phosphoethanolamine was inserted into the plasma membrane and linked via anti-fluorescein antibody to the gold probes [21]. The trajectories of the gold particles on the membrane were analysed based on the time-dependence of the mean square displacement. For most trajectories (around 90%), clear deviations from free Brownian motion were apparent and could be fitted by a confined diffusion model, yielding the compartment size in x- and y-direction, Lx and Ly, the free mobility within the domain, Dmicro, and the long range mobility, Dmacro [28]. The mean residence time τ was calculated from τ = LxLy/4Dmacro. Typical values for domain sizes and residence times are around 40–100 nm and 2–20 ms, respectively [22].
Kusumi and colleagues identified the membrane skeleton as the confining structure. Proteins bound to the actin cytoskeleton line up along the filaments like the pickets of a fence. The pickets represent barriers to the diffusion of tracers owing to direct steric repulsion and hydrodynamic friction [29,30].
(c). Pros
The picket-fence model is very attractive, as it solves a long-lasting puzzle in membrane biophysics: the motion of proteins and lipids in the plasma membrane is reduced by more than a factor of 10 compared with synthetic membranes or membrane blebs [22]. The single-particle tracking data were obtained on living cells, in most cases on the top part of the cell distal from the glass slide, so that the labelled molecules and the membrane itself were hardly affected by the presence of the supporting coverslip. Indeed, the influence of actin on the mobility of transmembrane proteins has also been confirmed by other studies (see also §§4 and 11).
(d). Concerns
Our first concern relates to the size of the label, which represents the major caveat of this approach (see figure 1a(ii)). The hop diffusion model is based on high-speed tracking of gold-labelled proteins or lipids at a time resolution of approximately 20 µs. It is well known, however, that the label strongly influences the observed mobility, yielding a reduction by a factor of 5 compared with single-molecule tracking [24]; stronger cross-linking using antibody-labelled gold particles led to immobilization of GPI-anchored proteins by stimulating the interaction with the actin cytoskeleton [31]. Even more so, a different study showed that labelling with quantum dots, which are similar in size to the gold probes, reduced the mobility and altered the mode of motion of membrane proteins substantially [32].
Unfortunately, there is currently no alternative method for resolving the putative rapid diffusion of tracers within the corrals. By pushing the technology to its limits, our group has achieved single fluorophore tracking at illumination times of approximately 300 µs [33]; Semrau et al. [34] came up with a new idea to improve time resolution using partially overlapping two-colour excitation, further improving the time resolution of single-fluorophore tracking to approximately 100 µs. Yet, those methods are still an order of magnitude too slow for direct identification of jumps.
Despite their limitations, single-fluorophore tracking experiments by our group indicated that the hop diffusion model may not be generalizable. We tracked the Fab-labelled GPI-anchored protein CD59 at a time resolution of 1 ms (illumination time of 300 µs, delay time 700 µs). Under these conditions, hop diffusion should be visible as an increased offset of the mean square displacement. Careful statistical analysis of all datasets, however, yielded no indication for a significant confinement on short time and length scales [33].
As a second concern, the connection to membrane rafts is rather indirect: essentially, the argumentation is based on the observation that raft and non-raft proteins hop at the same speed. Yet, taking the lateral mobility of a membrane protein as a measure of the size of the diffusing structure is problematic in general [35]; particularly, it is unclear in this model how the transition of a raft-like structure through the putative pickets would proceed, rendering a priori assumptions problematic.
4. Proposition 3: GPI-anchored proteins form nanoclusters of a few molecules in the plasma membrane
(a). In a nutshell
By determining the fluorescence anisotropy as a measure of homo-FRET, the group of Mayor showed in a series of papers that GPI-anchored proteins are distributed non-randomly in the live cell plasma membrane [36–38]. Upon analysing the dependence of the FRET signal on photobleaching and by fitting with a multi-parameter model, the authors concluded that the investigated proteins associate to small ‘nanoclusters’ consisting of only up to four molecules [37] (figure 1b). These clusters were distributed non-randomly in the plasma membrane, and concentrated to optically resolvable domains [36]. From the photobleaching behaviour, the presence of immobile nanoclusters was concluded, which were continuously reformed in situ at 37°C; such formation was not observed at room temperature. Mayor's group favour a model in which cell surface proteins are moved by dynamic actin to transient nanodomains defined by stable phases rich in actin asters [39]. Similar data were obtained in Hans Gerritsen's laboratory by homo-FRET analysis via time-resolved imaging of the anisotropy decay; GPI-anchored green fluorescent protein (GFP) was found to assemble in small clusters of two to three molecules [40,41].
The results were in also agreement with data obtained by the group of Garcia-Parajo via scanning near-field optical microscopy (SNOM) on fixed monocytes. In their study, the monovalent bacterial toxin pro-aerolysin was used to label GPI-anchored proteins in general. The proteins were found to be either monomeric (approx. 70%) or associated to nanoclusters of two to four molecules [42].
(b). Technologies
Mayor's group measured homo-FRET by the reduction in fluorescence anisotropy. Most data were recorded on the GPI-anchor of the human folate receptor, either labelled with GFP or with a fluorescent folate analogue. In general, FRET may arise from accidental encounters of randomly dispersed proteins, or from proteins that are closely packed in small domains. In the first case, one would expect a dependence of the FRET signal on the surface density of probe molecules. Mayor and colleagues, however, found concentration-independent FRET values, which were taken as indication for the presence of nanoclusters; in this view, a higher average probe concentration on the cell surface leads to a higher number of clusters, but not to a higher probe density within the clusters (figure 1b(ii)).
Garcia-Parajo and co-workers developed SNOM to a level that it can be used in aqueous solution [43] and have applied it since to various cell biological systems [42,44,45]. The small aperture of approximately 80–100 nm allows for high-resolution imaging on flat membrane regions (approx. 80 nm); the high sensitivity allows for detection even of single molecules (figure 2b(ii)). Naturally, the SNOM approach does not allow for further determination of the underlying kinetics. Nanoclusters happened to be dispersed at a sufficiently low surface density so that individual clusters could be imaged as separated structures with SNOM, whereas they were too crowded for being resolved with standard diffraction-limited optics [42].
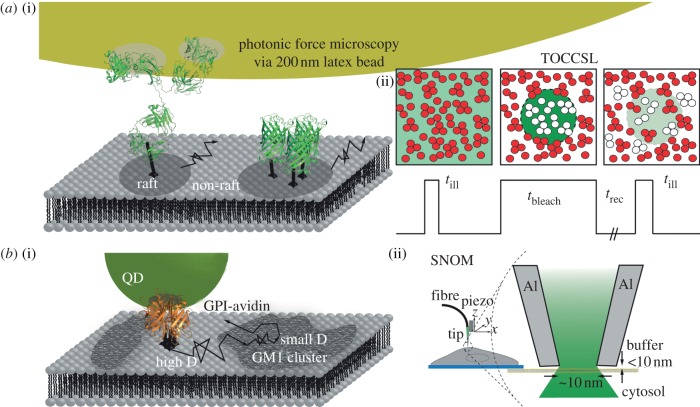
Figure 2.
(a(i)) Proposition 5 and 6: some proteins co-diffuse as parts of nanoplatforms. Drag forces experienced by membrane proteins were measured with photonic force microscopy by linking the protein of interest to 200 nm latex beads. The proteins were tagged via antibodies, keeping the antibody to latex sphere ratio low, and the latex bead was trapped in a focussed laser beam. Alternatively, the co-diffusion of fluorescently labelled proteins within rafts can be measured via TOCCSL. (a(ii)) Sketch of the TOCCSL method. A small area of the cell membrane is photobleached with a high power bleach pulse (tbleach). During the recovery time (trec) non-bleached proteins can diffuse into the bleached area. In the post-bleach image, single proteins are well resolvable as individual spots and can be analysed for brightness, mobility and co-localization. (b(i)) Proposition 7: there is lipid-mediated connectivity in the plasma membrane. For example, GPI-anchored proteins were found to interact with lipid clusters induced by cross-linking GM1; their diffusion was found to be slower within the clusters than outside. (b(ii)) SNOM makes use of nano-engineered glass fibre tips with apertures of 80–100 nm, coated with a metal layer for heat dissipation. This tip is placed close to the cell membrane so that only a small region is illuminated, thereby increasing the optical resolution.
(c). Pros
The homo-FRET data were recorded on the top membrane of live cells. The labels were small ligands or GFP; cross-validation with different labels confirmed the results. FRET per se measures the average degree of clustering, without the need for clusters to persist for longer times. FRET thus allows for capturing extremely transient phenomena. With the SNOM technique, although applied after fixation, clusters of GPI-anchored proteins could for the first time be directly imaged.
(d). Concerns
After the first homo-FRET papers, other groups repeated the experiments with hetero-FRET, but they could not confirm the data [46–48]. The inconsistency is not fully understood yet, but most likely originates from the difference in the way the labelling was achieved: the homo-FRET approach senses all nanodomains, whereas hetero-FRET senses only nanodomains in which two differently labelled proteins were co-localized; if nanodomains carry only a few labelled proteins on average, the likelihood for co-localization of the two colours becomes low. A second concern related to the FRET studies is the rather indirect nature of the experiment. Although concentration-independent FRET values provide a valid argument for clustering per se, quantification of the cluster size is strongly model-dependent.
For the SNOM experiments, one concern relates to cell fixation prior to the experiment, the effect of which is difficult to predict; for example, GPI-anchored proteins were found to exhibit residual mobility after paraformaldehyde fixation [49]. In addition, the interaction with the near-field tip may affect the obtained images: scanning forces of approximately 300 pN were typically used [42,43]; for comparison, only 50 pN is known to deform the cell surface in atomic force microscopy [50]. Finally, the total size of the near-field tip is actually larger than the aperture owing to the aluminium coating, limiting the applicability of SNOM to rather flat regions; invaginations may be flattened out or—if not sufficiently elastic—generate blurry images.
5. Proposition 4: some proteins and lipids are transiently immobilized to small domains
(a). In a nutshell
From the late 1990s on, researchers attempted to extract information on membrane constituents by analysing their mobility. In a comprehensive survey of membrane proteins, Kenworthy et al. [51] found no trend when comparing the mobility of raft versus non-raft proteins, and took that as evidence for ruling out the existence of raft-platforms in the membrane.
The diffusion constant per se, however, may be too indirect to unambiguously measure properties of the diffusing object. The group of Marguet thus developed a new method based on fluorescence correlation spectroscopy (FCS), which allows a closer investigation of the diffusional behaviour of membrane constituents. In this FCS variant termed spot-variation FCS (sv-FCS), the size of the laser-focus is systematically varied [52,53]. Analysis of the diffusion time as function of the spot size yields the so-called diffusion law, where positive offsets are regarded as indication for transient confinement to rafts. The method has been applied and further improved by others, yielding consistent results [54,55]. With this approach, cholesterol-dependent positive offsets in the diffusion law were indeed observed for GFP-GPI [52] and fluorescent sphingomyelin [54]. The offset originates from the trapping of the probe to small areas (less than 20 nm in the case of Eggeling et al. [54]), which are kept in place by the actin cytoskeleton [56] (figure 1c).
The data were further confirmed by a new, single fluorophore tracking variant introduced by Eggeling and co-workers [57]. Moreover, the data are consistent with older results from our laboratory, in which we found that the motion of fluorescently labelled lipids was indeed not free: a fully saturated lipid was confined to half micron-sized domains in smooth muscle cells [58]; an oxidized phospholipid was observed to halt for seconds at non-resolvably small domains [59].
(b). Technology
In sv-FCS, the width of the laser spot is systematically changed, FCS curves are recorded for each spot size and fitted with diffusion models (one-component, two-component or anomalous diffusion). When the autocorrelation decays τD are plotted versus the laser spot area ω2, a straight line through the origin indicates the free diffusion of the probe (τD = ω2/4D). A negative offset of the line is the consequence of confined diffusion [60], a positive offset originates from probe retardations or immobilizations [61].
In the original studies, one caveat has been the difficulty to approach the y-axis intercept, because the smallest feasible laser spot size is limited by diffraction to approximately 0.05 µm2. In consequence, the noise in the recorded decay times led to large uncertainties in the offsets of the curves. This caveat was solved by using nanometric apertures [62] or a stimulated emission depletion (STED) beam for excitation [54]; with the new techniques, the older studies could essentially be confirmed.
In addition, the Eggeling group developed a novel ‘confocalized’ single-fluorophore tracking method [57]. A focused laser beam is used for excitation, and the emitted signal is projected onto three densely packed fibres connected to avalanche photodiodes, which provides the spatial information. Streams of single photons are recorded and can be a posteriori grouped into packages, thereby specifying the effective illumination time (0.5–2 ms were reported, with a spatial accuracy of 10–20 nm).
(c). Pros
Data were recorded on the top membrane of live cells; the labels were small (GFP, dye-labelled lipids). The results were confirmed by various laboratories on different cell types, indicating a general mechanism.
(d). Concerns
While it appears attractive to interpret the positive offset of FCS diffusion laws by a domain partitioning model, numerous alternative effects may complicate the picture. We have recently pointed out that the diffusion law offset is a consequence of probe retardation [61], which may not only occur owing to the transient partitioning into a domain of different phase state; for instance, binding to immobile proteins or larger assemblies would also lead to a positive diffusion law offset, and even plasma membrane topology may be misinterpreted as transient immobilization [63].
This makes the interpretation of effects upon drug treatment difficult. Let us briefly discuss the two most common approaches for discriminating lipid-mediated interactions from effects induced by the membrane skeleton, i.e. the reduction of membrane cholesterol levels and the depolymerization of the actin cytoskeleton. Cholesterol is required for forming liquid-ordered phases in model membranes [64] and has therefore been believed to represent an essential component of raft-type interactions in cell membranes [4]. Various ways have been introduced to extract cholesterol from cell membranes, to convert it, or to interfere with its synthesis [52,65]. The absence of cholesterol is thought to disassemble raft domains and thereby release the fluorescent probe, which would result in a reduction of the diffusion law offset. Also the effects of actin on domain formation and dynamics are not fully understood, and include the stabilization of transient fluctuations [66], the generation of confinement zones (see §3) or the immobilization by (in)direct binding to the membrane skeleton. Again, the effect would be the release of the fluorescent probe and a reduced diffusion law offset. It is thus not surprising that experiments revealed essentially the same effects of the two drug treatments—cholesterol reduction and actin degradation—on the nanoscale mobility of fluorescence labelled lipids [56].
In the case of single-molecule tracking in general (and the confocalized method in particular), care should be taken when estimating the size of a confinement zone based on a molecule's frame-to-frame jump distance. The unknown and unresolvable diffusion of the probe within the domain during the illumination leads to a collapse of the measured trajectory at the domain centre; in consequence, domains appear smaller than they actually are [33,67,68]. Using equation 3 from Wieser et al. [33], we estimated the physical size of a square domain, given an experimental observation of 6 or 12 nm achieved by illuminating the sample for illumination times till as used in the study of Sahl et al. [57]. While for quasi-immobile probes with D <10–3 µm2 s−1 the effect is negligible, it may get significant when the probe diffuses rapidly (D > 1 µm2 s−1): even domains of 30 nm diameter would be in accordance with detected confinement sizes between 6 and 12 nm.
6. Proposition 5: some proteins diffuse as integral parts of nanoplatforms
(a). In a nutshell
A key paper on rafts originated from a collaboration from the Hörber and Simons group [69]. In this work, beads were coupled to individual membrane proteins and the local viscous drag was measured with a special optical tweezers arrangement (figure 2a). Because viscous drag depends on the size of the diffusing object, Hörber and coworkers were interested whether raft and non-raft proteins show different behaviour, both under control conditions and upon depletion of cholesterol. Consistently, the raft markers yielded a drag reduction upon cholesterol depletion, whereas no effect was observed for the non-raft proteins. The differential behaviour was taken as indication that indeed some of the tracked molecules were part of raft structures. In this study, local viscous drags were found to be constant over time-scales of minutes, indicating that the associated rafts were rather long-lived. A radius of 26 nm was estimated, using the Saffman–Delbrück relation for protein diffusion [70].
(b). Technology
Latex beads with a diameter of 200 nm were coated with antibody at low concentrations and bound to the cell surface molecules; ideally, each bead was coupled to a single protein molecule diffusing in the plasma membrane. The beads were kept in the focus of a laser-optical trap, and the excursions within the trap's energy profile were recorded with subnanometre and microsecond resolution (the method was termed ‘photonic force microscopy’). The autocorrelation function of the particle's position in a harmonic potential decays exponentially with a correlation time τ = γ/κ, where γ and κ denote the local viscous drag and the stiffness of the potential, respectively. From the measured τ and the known trap stiffness, γ could be calculated (or the diffusion constant D = kbT/γ, kb the Boltzmann constant, T the temperature).
The viscous drag can be related to the size of the diffusing object. The underlying idea is that the viscosity experienced by the transmembrane region of the protein is much higher than the viscosity of the bead in the aqueous extracellular environment; the damping is thus dominated by the membrane part. Saffman & Delbrück [70] calculated the viscous drag on a cylindrical particle with radius r in a lipid bilayer of known thickness and viscosity, so that the radius of the moving object can be estimated. Hörber et al. found a substantial cholesterol-dependence for the viscous drag of GPI-anchored proteins, while for an artificial putative non-raft protein no cholesterol-dependence was observed.
(c). Pros
Photonic force microscopy is a live-cell method; all data were recorded on the top membrane.
(d). Concerns
It is difficult to ensure that each bead was linked to only one protein in the plasma membrane. Moreover, the rather large bead may get stuck in the extracellular matrix, thereby indicating a too high value for the viscous drag and thus too large platform sizes. Finally, the Saffman–Delbrück equation was recently challenged by experimental data, which showed a much stronger size-dependence of the viscous drag [35], rendering the size estimate of 26 nm problematic.
7. Proposition 6: some proteins and lipids co-diffuse over seconds time-scale
(a). In a nutshell
We developed a method termed TOCCSL which senses the co-diffusion of membrane constituents on seconds time-scales [71,72]. When analysing the movement of GPI-anchored mGFP in the live cell plasma membrane, we observed significant homo-association (up to 60% dimers; figure 2a) [72]. Similar homo-association was found for the fluorescent lipid analogue Bodipy-GM1. The association was lost when cholesterol was depleted. Moreover, raising the temperature to 39°C substantially reduced clustering. The data indicate a model of stable lipid-mediated interactions in the plasma membrane.
Recently, the Groves laboratory published a study yielding similar results on shorter time-scales: they assessed the co-diffusion of green- and red-labelled proteins via cross-correlation FCS. Co-diffusion was observed for the GFP- and Cherry-labelled Lck membrane anchor, and for the GFP- and Cherry-labelled RhoA-anchor, but no co-diffusion for different anchor types [73]. The weak cross-correlation signal indicates low occupation of clusters. The results are also in very good agreement with a recent study published by the Kusumi group, in which the homo-association of the GPI-anchored protein CD59 was investigated [74]: the molecule formed dimers with a lifetime more than hundred milliseconds; cholesterol extraction released the interaction, which was interpreted as the consequence of lipid-mediated stabilization of the dimer. Interestingly, no heterodimers between different GPI-anchored proteins were observed, indicating a role of the protein ectodomain for stabilization.
(b). Technology
A region of the cell membrane is photobleached so that it does not contain any active fluorophore. Owing to diffusion, fluorescence recovers until ultimately the photobleached region will be filled again, like in a conventional fluorescence recover after photobleaching (FRAP) experiment. In TOCCSL, however, the initial phase of the recovery is analysed, in which the first fluorescently labelled molecules enter the photobleached area. On such images, the molecules are visible as well-separated diffraction-limited spots. The spots can be further analysed for their brightness as a measure of the molecular association [72]; also two-colour analysis can be performed to study co-diffusion of different species [75]. We summarized the technique and its variants in a recent review article [76].
(c). Pros
Stable molecular association was part of original raft concepts, but has been abandoned by most groups in favour of more transient interactions in the last years [5]; Owing to technical limitations, however, the question was not solved convincingly. TOCCSL measures directly that stable lipid-mediated association of membrane proteins occurs.
(d). Concerns
Inherently, the TOCCSL method can only sense mobile membrane constituents; thus, no information is available on immobile molecules. Moreover, there is a selection for molecules showing higher mobility, as they will recover first; slowly moving molecules or clusters cannot be analysed. Finally, the time window for analysis is rather small: diffusion is not sufficiently fast to record images immediately after the photobleaching pulse (up to approx. 100 ms); by contrast, if the recovery time is chosen too long (greater than 5 s), the signals will start overlapping, making single-molecule analysis impossible.
8. Proposition 7: GPI-anchored proteins interact with clustered lipids in the plasma membrane
(a). In a nutshell
Pinaud et al. [77] developed a tetrameric GPI-anchored avidin (Av-GPI), which, upon stable expression in HeLa cells, was labelled with biotinylated quantum dots for single-particle tracking (SPT). Cholera toxin subunit B (CTxB) was used to stain GM1 in the live cell plasma membrane (in the meantime, it became clear that CTxB actually induces GM1 clustering [45,78]). At the ensemble level, Pinaud et al. detected the co-localization of Av-GPI with CTxB-induced GM1 clusters. Via SPT, they observed single Av-GPI complexes entering and leaving GM1 clusters; association times in the order of tens of seconds were determined (figure 2b). In the clusters, the Av-GPI mobility was reduced by one to two orders of magnitude. Interestingly, GM1 domains were found to be located in close proximity to—but clearly distinct from—caveolae.
Van Zanten et al. [45] used confocal microscopy and SNOM on monocytes and dendritic cells to investigate a similar question concerning the relation between CTxB-induced GM1 clusters and the GPI-anchored protein CD55, yielding similar but not identical results. With confocal microscopy, they observed co-localization of CD55 and GM1, which disappeared in the SNOM images; instead, a close proximity of CD55 and GM1 domains within 100 nm was observed.
(b). Technology
Pinaud et al. [79] fused the GPI-anchoring signal sequence of CD14 with chicken avidin, which formed stable tetramers in the plasma membrane of HeLa cells. Small (diameter 13 nm) biotinylated quantum dots developed by the Shimon Weiss laboratory were used as labels for SPT. Quantum dots are preferred labels by many groups, as they do not photobleach and thus allow for tracking over extended periods of time. Data were recorded at 100 ms time intervals at the bottom membrane of the cells in total internal reflection fluorescence (TIRF) configuration, yielding a localization precision of 30 nm. The statistical step-size distributions of each trajectory were analysed to discriminate fast and slow modes of motion [80].
SNOM was performed as described earlier (§4). A primary and fluorescent secondary antibody was used to label CD55.
(c). Pros
SPT experiments were conducted with labels much smaller than commonly used (diameter of 13 nm versus e.g. the 40 nm particles used by Kusumi's group). The idea of measuring the interaction between a lipid-anchored protein and clustered lipids brought up a new level of complexity.
(d). Concerns
For the SPT experiments, the influence of the label on the diffusion properties is difficult to estimate. Pinaud et al. used Western blot analysis to demonstrate Av-GPI tetramerization. It is not clear, however, how the probe aggregates in the plasma membrane. In particular, the used quantum dots carry multiple biotins and thus may further cross-link the Av-GPI tetramers on the membrane. Experiments were performed at the bottom membrane of the cells, where spatial constraints may affect the motion of the particle.
In the SNOM approach, cell fixation may have altered the nanostructural assembly. Specifically, fixation with paraformaldehyde was shown to be not sufficient to fully immobilize GPI-anchored proteins [49], so that the labelling with primary and secondary antibodies might have facilitated protein clustering and thereby induced its clustering.
9. Proposition 8: patches of liquid-ordered and disordered phase coexist in the plasma membrane
(a). In a nutshell
For many years, the term ‘raft’ has been suspected to refer to relatively ordered liquid phases embedded in a contiguous relatively disordered liquid phase within lipid membranes. As a first indication for this, detergent-resistant liposomes were found to be in liquid ordered phase [4,81]. Synthetic membranes made of 1 : 1 : 1 DOPC/brain sphingomyelin/cholesterol mixtures showed lateral separation in ordered and disordered phase [82]; later, this mixture has become the ‘canonical raft mixture’, and has been studied extensively since (for a review, see [64]).
Whether such phases do exist also in the natural cell membrane has become a matter of debate. Swamy et al. [83] have observed phase coexistence in the live cell plasma membranes of various cell types using electron spin resonance; interestingly, throughout the cell types, they found a predominant ordered phase. The groups of Tobias Baumgart and Kai Simons introduced plasma membrane blebs (also termed giant plasma membrane vesicles) as models for studying the phase state of the plasma membrane [78,84–86]. They found that the membranes can segregate into micrometre-sized fluid domains with characteristic protein and lipid partitioning, and with characteristic order differences between the coexisting phases. Analysis of domain size fluctuations revealed that the plasma membrane composition is close to a miscibility critical point [87].
Further results demonstrating the coexistence of phases of different order come from the use of the lipophilic dye Laurdan to study order differences in biomembranes [88]. Laurdan spectra are sensitive to the polarity of the local environment; the characteristic red-shifted emission in polar environments can be used to quantify the membrane order around the probe. In special cases, macroscopic differences in cell membrane order were observed [89–91]. More recently, Sanchez et al. [92] addressed nanoscale phase segregation in live cell membranes by combining Laurdan spectroscopy with FCS. Fluctuations in the Laurdan spectra due to the movement of ordered domains in a continuous disordered matrix were recorded (figure 3a). Depending on the cell type, domain radii between 25 and 300 nm were calculated.
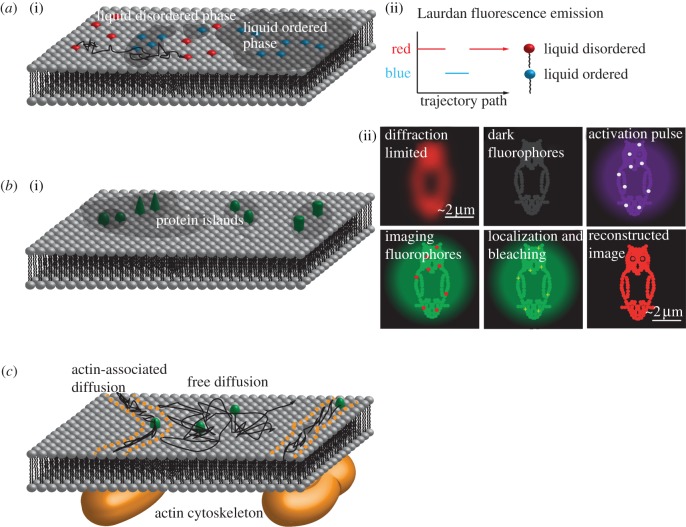
Figure 3.
(a(i)) Proposition 8: liquid ordered and liquid disordered phases coexist in the plasma membrane. Laurdan is a lipophilic dye that changes fluorescence emission upon contact with water molecules; it is frequently used to probe the lipid packing and thereby the membrane order. (a(ii)) Single diffusing Laurdan dyes change their emission from red to blue when they diffuse from liquid disordered to an ordered phase. (b(i)) Proposition 9: different membrane proteins are associated and form so-called protein islands; the proteins are trapped within islands for at least 4–10 s. (b(ii)) PALM relies on the repeated switching and localization of photo-activatable fluorophores. A minute fraction of inactive fluorophores is activated by illumination with a short wavelength. The active fluorophores are imaged at single-molecule level, and photobleached. Activation, imaging and photobleaching are repeated until most fluorophores have been recorded. On the individual frames, the single dye molecules can be localized with a precision of approximately 20 nm; all positions yield the final high-resolution image. (c) Proposition 10: SMT and SPT can be used to measure the influence of cortical actin on the diffusion of membrane proteins. For actin-associated diffusion, the tracked particles show a one-dimensional diffusion along the underlying actin fibres (e.g. CD36), for other proteins (e.g. the Fcε-receptor) free diffusion in the voids of the actin meshwork was observed.
(b). Technologies
Protocols for producing plasma membrane spheres were described and discussed in detail in Sezgin et al. [85]. Researchers currently work out appropriate conditions for the isolation of the plasma membrane spheres. For example, protein palmitoylation gets lost when using DTT for membrane blebbing, thereby altering protein partitioning [93]. Also, the transition temperature for phase separation depends on the chemicals used for isolation, and is very low (below 5°C) when neither cross-linking nor reducing chemicals were used [86].
Most data on phase states in lipid membranes were obtained via the analysis of Laurdan-stained samples. Laurdan incorporates into membranes owing to its aliphatic tail of 12 carbons; the fluorescent moiety is close to the head-group region, where it senses the accessibility and mobility of the surrounding water molecules: essentially, more ordered membrane environments correlate with a lower polarity and thus a blue-shifted emission. Commonly, researchers specify the generalized polarization (GP = (Fblue − Fred)/(Fblue + Fred)) from the emission recorded in the different colour channels. The obtained GP value may be affected by fragmentation into subdomains, yielding a weighted average. Note that GP values do not correspond to order parameters obtained, e.g. via NMR.
For the FCS experiment, Sanchez et al. [92] made the assumption that Laurdan moves rapidly in and out of domains, so that this movement is too fast to be resolvable; the signal fluctuations arise from the movement of whole domains within the confocal laser spot. Like in conventional FCS experiments, the decay and the amplitude of the autocorrelation function provide information on the diffusion constant and the number of diffusing objects. The obtained size estimates are nearest neighbour distances.
(c). Pros
Plasma membrane spheres contain the natural lipid compositions including membrane proteins, and are therefore preferential to mixtures of synthetic lipids used in earlier studies. Laurdan spectroscopy is currently the only method that provides access to the structure of the lipid bilayer in a live-cell context; as an additional advantage, the obtained results on Laurdan are not influenced by preferential probe partitioning.
(d). Concerns
Plasma membrane spheres are not native plasma membranes: the membrane leaflet asymmetry is lost, mixtures are equilibrated and proteins/lipids are disconnected from the underlying cortical actin network. In addition, there is a strong influence on protein oligomerization and palmitoylation by the chemicals used for isolation. Finally, there are reports indicating that phases can be induced by lipid photooxidation when observing lipid membranes under the microscope [94,95].
In case of Laurdan imaging, GP measures the water partitioning in the bilayer's head-group region. In addition to phase changes, however, there may be additional causes for differences in the water content of the polar headgroup region, for example membrane bending in cellular protrusion [96]. A further concern relates to the high probe concentration needed for the experiment. Although the surface density in the plasma membrane was not directly measured in the study by Sanchez et al. [92], we can give a rough estimate: the idea of the authors was to label each domain with sufficient probe concentration so that the loading fluctuations get negligible; let us assume 100 molecules per domain, yielding 10 per cent shot noise fluctuations. In a domain of 25 nm radius, we would thus expect a molar fraction of approximately 3 per cent Laurdan compared with bulk lipid, which appears sufficiently high to affect the bilayer properties.
A general concern related to the concept of small phases may be placed here. According to Gibb's phase rule, the number of coexisting phases, P, is given by p = C–F, with C the number of coexisting components, and F the degrees of freedom (here the concentrations; we assumed temperature and pressure to be fixed). Given the thousands of lipid and protein species, we thus may expect numerous different phases coexisting in the plasma membrane. Yet, the phase rule is valid only for large systems; on the small scale of a cell, however, interfacial energy at the domain boundaries also provides significant contributions to the overall energy. It is thus more appropriate to denote the situation as one-phase regime with microscopic heterogeneities [97]. At the present time, such mesoscale collective phenomena of multicomponent systems are far from being understood theoretically.
10. Proposition 9: membrane proteins are assembled within membrane domains (‘protein islands’)
(a). In a nutshell
Studies on plasma membrane sheets using electron microscopy (EM) and immunogold staining have indicated the presence of various protein domains [98,99]; in particular, using standard staining protocols, EM images typically showed brighter and darker regions. On the basis of EM images of MDCK, RBL, CHO and T cells, the Mark Davis group came up with the hypothesis that all membrane proteins were actually assembled within the dark patches of up to approximately 200 nm in size [100] (figure 3b); the domains contained essentially all proteins that could be biotinylated on sulfhydryl or carboxyl groups. Lillemeier et al. [100] compared putative raft versus non-raft marker proteins, using the first 10 N-terminal amino acids of the tyrosine-kinase Lck and a mutant with both palmitoylation sites mutated to alanine, respectively. Although raft and non-raft proteins were found in close proximity, they occupied different subregions within a given protein island. A stain for cholesterol revealed significantly enhanced cholesterol concentration in the protein islands compared to the remaining lipid matrix. No information could be obtained for GPI-anchored proteins, which were not labelled by the assay.
The protein island model was confirmed in live cells on the basis of the more special scenario of proteins involved in T cell signalling, including the T cell receptor and LAT [101]. For this, Lillemeier et al. [100] reverted to a single-molecule photoactivation approach (PALM). Using fast camera readout, they recorded at rates of 100–250 fps, and combined 1000 frames for a single PALM image; thereby, the recording of a single super-resolution image required only 4–10 s. Interestingly, FCS revealed high mobility of the investigated proteins, indicating that molecules move in and out of domains, where they get transiently trapped. The protein island model in T cells was recently challenged by a study of Williamson et al. [102], who claimed that the observed clusters of LAT were actually vesicles close to the plasma membrane.
(b). Technology
For EM, cells were allowed to adhere to polylysine-coated or MHC-coated EM grids. A polylysine-coated coverslip was placed on top of the cells and ripped by separation of the coverslip from the grid, leaving the bottom membranes attached to the grid. Cells were fixed with paraformaldehyde, and proteins were labelled with specific antibodies or streptavidin bound to gold particles [100].
PALM was performed on T cells adhered to polylysine- or MHC-coated surfaces, and on T cells bound to functionalized lipid bilayers [101]. Experiments were performed using the photoswitchable protein PSCFP2, a monomeric GFP homologue that fluoresces in the cyan part of the spectrum when excited at 402 nm; upon intense illumination at 405 nm it irreversibly photoconverts, and an excitation peak at 490 nm (emission at 511 nm) arises [103].
(c). Pros
The clusters are clearly visible on both EM and PALM images, making the methods very direct.
(d). Concerns
It is difficult to estimate whether the paraformaldehyde fixation required for EM had an influence on the protein distributions (see again [49]). In the case of PALM, cluster analysis is hampered by overcounting, i.e. some molecules may be reversibly photocycled so that they are counted several times, mimicking the presence of clusters [104]. The presence of vesicles close to the membrane may complicate the interpretation [102].
11. Proposition 10: actin compartmentalizes the plasma membrane
(a). In a nutshell
There are reports showing the influence of the actin membrane skeleton on the mobility and functionality of membrane proteins [105–107] (figure 3c). Andrews et al. [106] studied the mobility of the Fcε-receptor labelled with quantum dots on RBL cells. On overlays with GFP-actin images, they observed the receptor moving preferentially in the voids of the actin meshwork. The Batista group studied the mobility of Fab-labelled B cell receptor (BCR) at the single molecule level [105]; also in this case, clear correlations with the actin membrane skeleton were observed, with single BCR molecules showing reduced mobility in actin-rich regions of the cell. Moreover, when actin was degraded by latrunculin A or cytochalasin D, BCR mobility increased concomitantly with an increase in the cellular calcium signals. Finally, Jaqaman et al. [107] studied the diffusional motion of CD36—a class B scavenger receptor responsible, e.g., for the uptake of oxidized low-density lipoprotein (LDL)—in the plasma membrane of macrophages. They observed diffusion along linear trajectories induced by the cortical actomyosin meshwork. Confinement to the linearly shaped subareas of the membrane was speculated to enhance the clustering of CD36 and in consequence its binding affinity for oxidized LDL.
(b). Technologies
The papers used SPT with various labelling approaches (Quantum dots in recent studies [106,107], dye-labelled Fabs in Jaqaman et al. [107], primary/secondary dye-labelled Fabs in Treanor et al. [105]). The role of actin was directly visualized using GFP-actin [106], Lifeact-GFP [105], or addressed indirectly via depolymerizing drugs [107].
(c). Pros
The single-particle/molecule tracking approaches are very direct, particularly when overlaid with the actin images.
(d). Concerns
Similar to §10, it is difficult to rule out the presence of membrane-proximal vesicles, which may have contributed to the data. The usage of quantum dots may be problematic, as discussed in §3.
12. Conclusions
By now, an overwhelming number of studies provide evidence for heterogeneity in the plasma membrane. From the described propositions, however, there is no clear picture emerging. What is the difficulty? First, most experiments were conducted on different proteins or lipids using different cellular systems, which makes a direct comparison difficult. Second, the different approaches have different strengths and weaknesses (listed as pros and concerns above). We do not doubt the results per se, but some of the interpretations may be ambiguous. It is indeed hard to discriminate fact from artefact: many results could not have been obtained with an alternative technology, precluding independent confirmations. Third, we face a semantic problem: after performing a special experiment, the data are usually interpreted in the framework of a particular model. Terminologies are not only used to describe the data, but also to underscore and highlight the findings. A variety of putative raft characterizations was thereby introduced in the literature, including size, lifetime, mobility, phase state and composition.
Having discussed the work and interpretations mainly of others, it is time now to state our beliefs about the consequences of the above propositions (our view has been supported by a recent study published after preparation of this manuscript [74]):
— we believe that if lipids get immobilized for a few tens of milliseconds (§5), possibly by binding to immobile proteins (or protein clusters), the same will happen en route during the lipid's diffusional path by binding to mobile proteins (or protein clusters). In consequence, most lipids are to some extent associated with proteins. Indeed, this picture is supported by Molecular Dynamics simulations of membrane proteins (the KV1.2 potassium channel) in a fluid lipid bilayer [30]: in this case, hardly any lipid was found to be unaffected by the presence of the protein. The idea is along the lines of the lipid shell model proposed by Anderson & Jacobson [108], and supported by §§6 and 7;
— we further believe that there is an intrinsic tendency of natural membranes to phase separate (§9), or—at the nanoscopic level—that there is a connectivity over length scales exceeding the size of one molecule. Unfortunately, for systems with a compositional complexity of a cell membrane, a full thermodynamic description is not available yet. We still may expect nanoscale domains, but showing larger fluctuations than classical phases owing to the lower amount of particles contained within a domain. Similar to phases, such domains would form cohesive matrices, so that the likelihood of a molecule to stay within the domain is higher than to leave the domain;
— we believe that the membrane skeleton provides a template for protein and lipid organization. The evidence for an influence of the cortical actin on the motion and association of proteins and lipids is strong (§§3, 4 and 11); also the transient immobilizations observed via sv-FCS were linked to the actin cytoskeleton (§5). The observed effects of actin may be diverse in their origin: direct interactions with large cytoplasmic domains of proteins, binding of actin to lipids in the cytosolic membrane leaflet, and indirect mechanisms transmitted via actin-binding membrane proteins could play a role. In any case, the pinning of proteins including their associated lipid shells will stabilize membrane heterogeneities [66]; the newly formed structure will behave similar to a lipid or protein cluster induced by cross-linking (§8);
— taken together, it appears as if protein interactions were not only transmitted directly by molecular contact, but also indirectly via lipid connectivity. The total pair-wise interaction energy will thus depend both on the protein's amino acid chains and on the lipids that are complexed with the proteins, thereby adding up the effects. Transient clusters will be formed if the interaction energies happen to be low, and more stable clusters if the interaction energies are high. Specific assembly of proteins may be held in place via actin, thereby generating the observed spatial heterogeneities (§10).
13. Technology glossary (see table 1)
Table 1.
Key parameters characterizing the different propositions. n.a., not applicable.
| prop. | system | time resolution | spatial resolution |
|---|---|---|---|
| 1 | in vitro | n.a. | n.a. |
| 2 | live cells | 0.05 ms | <20 nm |
| 3 | live cells (FRET); fixed cells (SNOM) | n.a. | ∼ 5 nm (FRET); 80 nm (SNOM) |
| 4 | live cells | ∼10 ms (sv-FCS); 0.5 ms (SMT) | ∼20 nm (sv-FCS with STED); 10–20 nm (SMT)a |
| 5 | live cells | 300 ms | 100 nm |
| 6 | live cells | ∼500 ms | 10–20 nma |
| 7 | live cells | 100 ms | 30 nma |
| 8 | live cells (Laurdan) | n.a. | 300 nm |
| blebs | 300 nm | ||
| 9 | live cells | 4–10 s | 25 nma |
| 10 | live cells | <100 ms | ∼30 nma |
aSingle-molecule localization precision.
(a). Förster resonance energy transfer (figure 1b)
If two dyes approach each other closer than the Förster length of a few nanometres, their energy levels begin to couple, so that excitation energy can be transferred between the molecules [109]. FRET is thus a sensitive method to measure molecular proximity. The Förster length defines the distance at which half of the energy gets transferred. It scales with the overlap of the donor's emission spectrum with the acceptor's absorption spectrum: the measurement mode is termed hetero-FRET if two different dyes are used as donor–acceptor pair, and homo-FRET if the same dye is used. Typical Förster lengths are around 5–7 nm. Homo-FRET can be measured via the fluorescence anisotropy, which decreases owing to the scrambling of the polarization upon energy transfer.
(b). Scanning near-field optical microscopy (figure 2b)
SNOM is based on scanning a very small light source with dimensions smaller than the wavelength (typically 50–100 nm) very close (typically less than 10 nm) to the specimen (for review see [110,111]). When applied to cells, resolution of approximately 80 nm has been reported [111]. SNOM was one of the first techniques used for single-molecule detection [112], and allows for measurements in aqueous solution [112]. Owing to the low time resolution (typical scanning speeds are a few µm s−1), however, cells have to be fixed before imaging.
(c). Single-molecule photoswitching microscopy (figure 3b)
Super-resolution microscopy concepts based on the stochastic photoswitching of fluorophores from an inactive to an active state were introduced in 2006 and acronymed i.a. PALM, fPALM and STORM [113–115]. In these measurement modes, photo-activatable or photo-switchable fluorophores are stochastically turned on, imaged and switched off or photobleached; the whole process is repeated until most of the fluorophores of the sample have been addressed. The obtained positions are mapped, yielding an image with a resolution limited only by the localization precision [116].
(d). Single-molecule/particle tracking (figure 1a)
The molecule of interest is linked via a specific ligand to an object that provides contrast in light microscopy, e.g. a fluorescent or scattering particle, or a single dye molecule. Samples are imaged via fluorescence or transmission light microscopy (in case of scatterers), and movies are recorded at rates of approximately 10 up to 50 000 fps. Many researchers use stroboscopic illumination, so that the illumination time can be set independently of the readout speed: illumination should be long enough to collect sufficient photons for high signal-to-noise ratio imaging, but short enough to virtually freeze the motion of the molecules during illumination. The position of the particle or the fluorophore is encoded by the centroid of the photon distribution and can be determined to accuracy much below the diffraction limit by fitting with the point spread function [117–120]. Various algorithms were published for tracking the particles over consecutive images within a sequence [121–126]. Data are analysed either at the single-molecule level, or by pooling the information of many molecules, for example by analysing the mean square displacement as a function of the time-lag. For reviews, see [126–128].
(e). Spot-variation fluorescence correlation spectroscopy (figure 1c)
In FCS, the fluctuations of the fluorescence signal generated within a confocal laser spot are analysed, typically by calculating the autocorrelation function (for review, see [129]). Because the transit of fluorescently labelled molecules through the laser focus generates characteristic fluctuations, FCS is frequently used for analysing the mobility of membrane constituents. Marguet and colleagues introduced an innovative extension to FCS by varying the diameter of the laser focus, ω, termed sv-FCS [52,53]. In this case, the average residence time of the diffusing molecules within the laser spot, τ, shows a characteristic functional dependence on ω; for example, confined diffusion or diffusion with transient immobilizations generates a linear curve with a negative or positive offset τ0, respectively [60,61]. Eggeling et al. [54] combined sv-FCS with STED microscopy to obtain structural insights below the diffraction limit.
(f). Thinning out clusters while conserving the stoichiometry of labelling (figure 2a)
We recently devised a single-molecule methodology to detect long-term associations of molecules moving within a membrane [71,76]. Similar to FRAP assays, a small area of the membrane is fully photobleached. Owing to the fluidity of the membrane, the fluorescence signal recovers. At the onset of the recovery process (within the first few hundred milliseconds up to seconds), single well-separated spots can be observed and further analysed: for example, the brightness can be used to obtain information on the stoichiometric composition of the object [72,130], and two-colour microscopy can be performed to study co-localization [75].
(g). Total internal reflection fluorescence microscopy
When the excitation beam falls under a shallow angle onto the interface between a high and low refractive medium, total reflection occurs, so that only a narrow region of the low refractive medium proximal to the interface gets excited. When applied to cells, this imaging mode allows for selective imaging of the bottom plasma membrane and for efficient reduction of background signal from the rest of the cell. The excitation intensity decays exponentially with a typical decay length of approximately 0.2λ, where λ is the wavelength of the excitation beam [131]. TIR excitation can be implemented in any fluorescence microscope when using a high numerical aperture objective (NA > 1.4) for illumination and detection [131].
Acknowledgements
We thank Mario Brameshuber for comments on the manuscript. This work was supported by the GEN-AU project of the Austrian Federal Ministry for Science and Research, and the Austrian Science Fund (FWF projects Y250-B10 and I301-B12). E.K. was supported by a FEBS long-term Fellowship.
References
- 1.Singer SJ. 1992. The structure and function of membranes: a personal memoir. J. Membr. Biol. 129, 3–12 10.1007/BF00232051 (doi:10.1007/BF00232051) [DOI] [PubMed] [Google Scholar]
- 2.Singer SJ, Nicolson GL. 1972. The fluid mosaic model of the structure of cell membranes. Science 175, 720–731 10.1126/science.175.4023.720 (doi:10.1126/science.175.4023.720) [DOI] [PubMed] [Google Scholar]
- 3.Morrisett JD, Pownall HJ, Plumlee RT, Smith LC, Zehner ZE. 1975. Multiple thermotropic phase transitions in Escherichia coli membranes and membrane lipids. A comparison of results obtained by nitroxyl stearate paramagnetic resonance, pyrene excimer fluorescence, and enzyme activity measurements. J. Biol. Chem. 250, 6969–6976 [PubMed] [Google Scholar]
- 4.Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387, 569–572 10.1038/42408 (doi:10.1038/42408) [DOI] [PubMed] [Google Scholar]
- 5.Pike LJ. 2006. Rafts defined. J. Lipid Res. 47, 1597–1598 10.1194/jlr.E600002-JLR200 (doi:10.1194/jlr.E600002-JLR200) [DOI] [PubMed] [Google Scholar]
- 6.Brown DA, Rose JK. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68, 533–544 10.1016/0092-8674(92)90189-J (doi:10.1016/0092-8674(92)90189-J) [DOI] [PubMed] [Google Scholar]
- 7.Melkonian KA, Chu T, Tortorella LB, Brown DA. 1995. Characterization of proteins in detergent-resistant membrane complexes from Madin–Darby canine kidney epithelial cells. Biochemistry 34, 16 161–16 170 10.1021/bi00049a031 (doi:10.1021/bi00049a031) [DOI] [PubMed] [Google Scholar]
- 8.Lisanti MP, Scherer PE, Vidugiriene J, Tang Z, Hermanowski-Vosatka A, Tu YH, Cook RF, Sargiacomo M. 1994. Characterization of caveolin-rich membrane domains isolated from an endothelial-rich source: implications for human disease. J. Cell Biol. 126, 111–126 10.1083/jcb.126.1.111 (doi:10.1083/jcb.126.1.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roper K, Corbeil D, Huttner WB. 2000. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat. Cell Biol. 2, 582–592 10.1038/35023524 (doi:10.1038/35023524) [DOI] [PubMed] [Google Scholar]
- 10.Drevot P, Langlet C, Guo XJ, Bernard AM, Colard O, Chauvin JP, Lasserre R, He HT. 2002. TCR signal initiation machinery is pre-assembled and activated in a subset of membrane rafts. EMBO J. 21, 1899–1908 10.1093/emboj/21.8.1899 (doi:10.1093/emboj/21.8.1899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown DA. 2002. Isolation and use of rafts. Curr. Protoc. Immunol. 51, 11.10.11–11.10.23 [DOI] [PubMed] [Google Scholar]
- 12.Paladino S, Sarnataro D, Pillich R, Tivodar S, Nitsch L, Zurzolo C. 2004. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J. Cell Biol. 167, 699–709 10.1083/jcb.200407094 (doi:10.1083/jcb.200407094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hundt M, et al. 2006. Impaired activation and localization of LAT in anergic T cells as a consequence of a selective palmitoylation defect. Immunity 24, 513–522 10.1016/j.immuni.2006.03.011 (doi:10.1016/j.immuni.2006.03.011) [DOI] [PubMed] [Google Scholar]
- 14.Bate C, Williams A. 2011. Monoacylated cellular prion protein modifies cell membranes, inhibits cell signaling, and reduces prion formation. J. Biol. Chem. 286, 8752–8758 10.1074/jbc.M110.186833 (doi:10.1074/jbc.M110.186833) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peirce M, Metzger H. 2000. Detergent-resistant microdomains offer no refuge for proteins phosphorylated by the IgE receptor. J. Biol. Chem. 275, 34 976–34 982 10.1074/jbc.M005819200 (doi:10.1074/jbc.M005819200) [DOI] [PubMed] [Google Scholar]
- 16.Kurzchalia TV, Hartmann E, Dupree P. 1995. Guilt by insolubility: does a protein's detergent insolubility reflect a caveolar location? Trends Cell Biol. 5, 187–189 10.1016/S0962-8924(00)88990-4 (doi:10.1016/S0962-8924(00)88990-4) [DOI] [PubMed] [Google Scholar]
- 17.London E, Brown DA. 2000. Insolubility of lipids in Triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim. Biophys. Acta 1508, 182–195 10.1016/S0304-4157(00)00007-1 (doi:10.1016/S0304-4157(00)00007-1) [DOI] [PubMed] [Google Scholar]
- 18.Li XM, Smaby JM, Momsen MM, Brockman HL, Brown RE. 2000. Sphingomyelin interfacial behavior: the impact of changing acyl chain composition. Biophys. J. 78, 1921–1931 10.1016/S0006-3495(00)76740-3 (doi:10.1016/S0006-3495(00)76740-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heerklotz H. 2002. Triton promotes domain formation in lipid raft mixtures. Biophys. J. 83, 2693–2701 10.1016/S0006-3495(02)75278-8 (doi:10.1016/S0006-3495(02)75278-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusumi A, Koyama-Honda I, Suzuki K. 2004. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic 5, 213–230 10.1111/j.1600-0854.2004.0178.x (doi:10.1111/j.1600-0854.2004.0178.x) [DOI] [PubMed] [Google Scholar]
- 21.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157, 1071–1081 10.1083/jcb.200202050 (doi:10.1083/jcb.200202050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murase K, et al. 2004. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 86, 4075–4093 10.1529/biophysj.103.035717 (doi:10.1529/biophysj.103.035717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki K, Ritchie K, Kajikawa E, Fujiwara T, Kusumi A. 2005. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 88, 3659–3680 10.1529/biophysj.104.048538 (doi:10.1529/biophysj.104.048538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umemura YM, Vrljic M, Nishimura SY, Fujiwara TK, Suzuki KG, Kusumi A. 2008. Both MHC class II and its GPI-anchored form undergo hop diffusion as observed by single-molecule tracking. Biophys. J. 95, 435–450 10.1529/biophysj.107.123018 (doi:10.1529/biophysj.107.123018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34, 351–378 10.1146/annurev.biophys.34.040204.144637 (doi:10.1146/annurev.biophys.34.040204.144637) [DOI] [PubMed] [Google Scholar]
- 26.Geerts H, De Brabander M, Nuydens R, Geuens S, Moeremans M, De Mey J, Hollenbeck P. 1987. Nanovid tracking: a new automatic method for the study of mobility in living cells based on colloidal gold and video microscopy. Biophys. J. 52, 775–782 10.1016/S0006-3495(87)83271-X (doi:10.1016/S0006-3495(87)83271-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gelles J, Schnapp BJ, Sheetz MP. 1988. Tracking kinesin-driven movements with nanometre-scale precision. Nature 331, 450–453 10.1038/331450a0 (doi:10.1038/331450a0) [DOI] [PubMed] [Google Scholar]
- 28.Kusumi A, Sako Y, Yamamoto M. 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65, 2021–2040 10.1016/S0006-3495(93)81253-0 (doi:10.1016/S0006-3495(93)81253-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusumi A, Suzuki KG, Kasai RS, Ritchie K, Fujiwara TK. 2011. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 36, 604–615 10.1016/j.tibs.2011.08.001 (doi:10.1016/j.tibs.2011.08.001) [DOI] [PubMed] [Google Scholar]
- 30.Niemela PS, Miettinen MS, Monticelli L, Hammaren H, Bjelkmar P, Murtola T, Lindahl E, Vattulainen I. 2010. Membrane proteins diffuse as dynamic complexes with lipids. J. Am. Chem. Soc. 132, 7574–7575 10.1021/ja101481b (doi:10.1021/ja101481b) [DOI] [PubMed] [Google Scholar]
- 31.Suzuki KG, Fujiwara TK, Sanematsu F, Iino R, Edidin M, Kusumi A. 2007. GPI-anchored receptor clusters transiently recruit Lyn and Ga for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177, 717–730 10.1083/jcb.200609174 (doi:10.1083/jcb.200609174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nechyporuk-Zloy V, Dieterich P, Oberleithner H, Stock C, Schwab A. 2008. Dynamics of single potassium channel proteins in the plasma membrane of migrating cells. Am. J. Physiol. Cell Physiol. 294, C1096–C1102 10.1152/ajpcell.00252.2007 (doi:10.1152/ajpcell.00252.2007) [DOI] [PubMed] [Google Scholar]
- 33.Wieser S, Moertelmaier M, Fuertbauer E, Stockinger H, Schütz GJ. 2007. (Un)confined diffusion of CD59 in the plasma membrane determined by high-resolution single molecule microscopy. Biophys. J. 92, 3719–3728 10.1529/biophysj.106.095398 (doi:10.1529/biophysj.106.095398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semrau S, Pezzarossa A, Schmidt T. 2011. Microsecond single-molecule tracking (µsSMT). Biophys. J. 100, L19–L21 10.1016/j.bpj.2010.12.3721 (doi:10.1016/j.bpj.2010.12.3721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gambin Y, Lopez-Esparza R, Reffay M, Sierecki E, Gov NS, Genest M, Hodges RS, Urbach W. 2006. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl Acad. Sci. USA 103, 2098–2102 10.1073/pnas.0511026103 (doi:10.1073/pnas.0511026103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goswami D, Gowrishankar K, Bilgrami S, Ghosh S, Raghupathy R, Chadda R, Vishwakarma R, Rao M, Mayor S. 2008. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 135, 1085–1097 10.1016/j.cell.2008.11.032 (doi:10.1016/j.cell.2008.11.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma P, Varma R, Sarasij RC, Ira Gousset K, Krishnamoorthy G, Rao M, Mayor S. 2004. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116, 577–589 10.1016/S0092-8674(04)00167-9 (doi:10.1016/S0092-8674(04)00167-9) [DOI] [PubMed] [Google Scholar]
- 38.Varma R, Mayor S. 1998. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394, 798–801 10.1038/29563 (doi:10.1038/29563) [DOI] [PubMed] [Google Scholar]
- 39.Gowrishankar K, Ghosh S, Saha S, C R, Mayor S, Rao M. 2012. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 149, 1353–1367 10.1016/j.cell.2012.05.008 (doi:10.1016/j.cell.2012.05.008) [DOI] [PubMed] [Google Scholar]
- 40.Bader AN, Hofman EG, Voortman J, van Bergen En Henegouwen PM, Gerritsen HC. 2009. Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophys. J. 97, 2613–2622 10.1016/j.bpj.2009.07.059 (doi:10.1016/j.bpj.2009.07.059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bader AN, Hofman EG, van Bergen En Henegouwen PM, Gerritsen HC. 2007. Imaging of protein cluster sizes by means of confocal time-gated fluorescence anisotropy microscopy. Opt. Express 15, 6934–6945 10.1364/OE.15.006934 (doi:10.1364/OE.15.006934) [DOI] [PubMed] [Google Scholar]
- 42.van Zanten TS, Cambi A, Koopman M, Joosten B, Figdor CG, Garcia-Parajo MF. 2009. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc. Natl Acad. Sci. USA 106, 18 557–18 562 10.1073/pnas.0905217106 (doi:10.1073/pnas.0905217106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koopman M, de Bakker BI, Garcia-Parajo MF, van Hulst NF. 2003. Shear force imaging of soft samples in liquid using a diving bell concept. Appl. Phys. Lett. 83, 5083–5085 10.1063/1.1634385 (doi:10.1063/1.1634385) [DOI] [Google Scholar]
- 44.de Bakker BI, Bodnar A, van Dijk EM, Vamosi G, Damjanovich S, Waldmann TA, van Hulst NF, Jenei A, Garcia-Parajo MF. 2008. Nanometer-scale organization of the alpha subunits of the receptors for IL2 and IL15 in human T lymphoma cells. J. Cell Sci. 121, 627–633 10.1242/jcs.019513 (doi:10.1242/jcs.019513) [DOI] [PubMed] [Google Scholar]
- 45.van Zanten TS, Gomez J, Manzo C, Cambi A, Buceta J, Reigada R, Garcia-Parajo MF. 2010. Direct mapping of nanoscale compositional connectivity on intact cell membranes. Proc. Natl Acad. Sci. USA 107, 15 437–15 442 10.1073/pnas.1003876107 (doi:10.1073/pnas.1003876107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenworthy AK, Edidin M. 1998. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 Å using imaging fluorescence resonance energy transfer. J. Cell Biol. 142, 69–84 10.1083/jcb.142.1.69 (doi:10.1083/jcb.142.1.69) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kenworthy AK, Petranova N, Edidin M. 2000. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell 11, 1645–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glebov OO, Nichols BJ. 2004. Lipid raft proteins have a random distribution during localized activation of the T-cell receptor. Nat. Cell Biol. 6, 238–243 10.1038/ncb1103 (doi:10.1038/ncb1103) [DOI] [PubMed] [Google Scholar]
- 49.Tanaka KAK, et al. 2010. Membrane molecules mobile even after chemical fixation. Nat. Methods 7, 865–866 10.1038/nmeth.f.314 (doi:10.1038/nmeth.f.314) [DOI] [PubMed] [Google Scholar]
- 50.Muller DJ, Dufrene YF. 2011. Atomic force microscopy: a nanoscopic window on the cell surface. Trends Cell Biol. 21, 461–469 10.1016/j.tcb.2011.04.008 (doi:10.1016/j.tcb.2011.04.008) [DOI] [PubMed] [Google Scholar]
- 51.Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J. 2004. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 165, 735–746 10.1083/jcb.200312170 (doi:10.1083/jcb.200312170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenne PF, Wawrezinieck L, Conchonaud F, Wurtz O, Boned A, Guo XJ, Rigneault H, He HT, Marguet D. 2006. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 25, 3245–3256 10.1038/sj.emboj.7601214 (doi:10.1038/sj.emboj.7601214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wawrezinieck L, Rigneault H, Marguet D, Lenne PF. 2005. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 89, 4029–4042 10.1529/biophysj.105.067959 (doi:10.1529/biophysj.105.067959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eggeling C, et al. 2009. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162 10.1038/nature07596 (doi:10.1038/nature07596) [DOI] [PubMed] [Google Scholar]
- 55.Humpolikova J, Gielen E, Benda A, Fagulova V, Vercammen J, Vandeven M, Hof M, Ameloot M, Engelborghs Y. 2006. Probing diffusion laws within cellular membranes by Z-scan fluorescence correlation spectroscopy. Biophys. J. 91, L23–L25 10.1529/biophysj.106.089474 (doi:10.1529/biophysj.106.089474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mueller V, et al. 2011. STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophys. J. 101, 1651–1660 10.1016/j.bpj.2011.09.006 (doi:10.1016/j.bpj.2011.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sahl SJ, Leutenegger M, Hilbert M, Hell SW, Eggeling C. 2010. Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proc. Natl Acad. Sci. USA 107, 6829–6834 10.1073/pnas.0912894107 (doi:10.1073/pnas.0912894107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schütz GJ, Kada G, Pastushenko VP, Schindler H. 2000. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J. 19, 892–901 10.1093/emboj/19.5.892 (doi:10.1093/emboj/19.5.892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhode S, Grurl R, Brameshuber M, Hermetter A, Schutz GJ. 2009. Plasma membrane fluidity affects transient immobilization of oxidized phospholipids in endocytotic sites for subsequent uptake. J. Biol. Chem. 284, 2258–2265 10.1074/jbc.M807591200 (doi:10.1074/jbc.M807591200) [DOI] [PubMed] [Google Scholar]
- 60.Destainville N. 2008. Theory of fluorescence correlation spectroscopy at variable observation area for two-dimensional diffusion on a meshgrid. Soft Matter 4, 1288–1301 10.1039/b718583a (doi:10.1039/b718583a) [DOI] [PubMed] [Google Scholar]
- 61.Ruprecht V, Wieser S, Marguet D, Schutz GJ. 2011. Spot variation fluorescence correlation spectroscopy allows for superresolution chronoscopy of confinement times in membranes. Biophys. J. 100, 2839–2845 10.1016/j.bpj.2011.04.035 (doi:10.1016/j.bpj.2011.04.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wenger J, Conchonaud F, Dintinger J, Wawrezinieck L, Ebbesen TW, Rigneault H, Marguet D, Lenne PF. 2006. Diffusion analysis within single nanometric apertures reveals the ultrafine cell membrane organization. Biophys. J. 92, 913–919 10.1529/biophysj.106.096586 (doi:10.1529/biophysj.106.096586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adler J, Shevchuk AI, Novak P, Korchev YE, Parmryd I. 2010. Plasma membrane topography and interpretation of single-particle tracks. Nat. Methods 7, 170–171 10.1038/nmeth0310-170 (doi:10.1038/nmeth0310-170) [DOI] [PubMed] [Google Scholar]
- 64.Goni FM, Alonso A, Bagatolli LA, Brown RE, Marsh D, Prieto M, Thewalt JL. 2008. Phase diagrams of lipid mixtures relevant to the study of membrane rafts. Biochim. Biophys. Acta 1781, 665–684 10.1016/j.bbalip.2008.09.002 (doi:10.1016/j.bbalip.2008.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lasserre R, et al. 2008. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat. Chem. Biol. 4, 538–547 10.1038/nchembio.103 (doi:10.1038/nchembio.103) [DOI] [PubMed] [Google Scholar]
- 66.Machta BB, Papanikolaou S, Sethna JP, Veatch SL. 2011. Minimal model of plasma membrane heterogeneity requires coupling cortical actin to criticality. Biophys. J. 100, 1668–1677 10.1016/j.bpj.2011.02.029 (doi:10.1016/j.bpj.2011.02.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ritchie K, Shan XY, Kondo J, Iwasawa K, Fujiwara T, Kusumi A. 2005. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 88, 2266–2277 10.1529/biophysj.104.054106 (doi:10.1529/biophysj.104.054106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Destainville N, Salome L. 2006. Quantification and correction of systematic errors due to detector time-averaging in single-molecule tracking experiments. Biophys. J. 90, L17–L19 10.1529/biophysj.105.075176 (doi:10.1529/biophysj.105.075176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pralle A, Keller P, Florin EL, Simons EL, Horber JK. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148, 997–1008 10.1083/jcb.148.5.997 (doi:10.1083/jcb.148.5.997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saffman PG, Delbruck M. 1975. Brownian motion in biological membranes. Proc. Natl Acad. Sci. USA 72, 3111–3113 10.1073/pnas.72.8.3111 (doi:10.1073/pnas.72.8.3111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moertelmaier M, Brameshuber M, Linimeier M, Schütz GJ, Stockinger H. 2005. Thinning out clusters while conserving stoichiometry of labeling. Appl. Phys. Lett. 87, 263903. 10.1063/1.2158031 (doi:10.1063/1.2158031) [DOI] [Google Scholar]
- 72.Brameshuber M, et al. 2010. Imaging of mobile long-lived nanoplatforms in the live cell plasma membrane. J. Biol. Chem. 285, 41 765–41 771 10.1074/jbc.M110.182121 (doi:10.1074/jbc.M110.182121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Triffo SB, Huang HH, Smith AW, Chou ET, Groves JT. 2012. Monitoring lipid-anchor organization in cell membranes by PIE-FCCS. J. Am. Chem. Soc. 134, 10 833–10 842 10.1021/ja300374c (doi:10.1021/ja300374c) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki KG, Kasai RS, Hirosawa KM, Nemoto YL, Ishibashi M, Miwa Y, Fujiwara TK, Kusumi A. 2012. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat. Chem. Biol. 8, 774–783 10.1038/nchembio.1028 (doi:10.1038/nchembio.1028) [DOI] [PubMed] [Google Scholar]
- 75.Ruprecht V, Brameshuber M, Schütz GJ. 2010. Two-color single molecule tracking combined with photobleaching for the detection of rare molecular interactions in fluid biomembranes. Soft Matter 6, 568–581 10.1039/b916734j (doi:10.1039/b916734j) [DOI] [Google Scholar]
- 76.Brameshuber M, Schutz GJ. 2012. Detection and quantification of biomolecular association in living cells using single-molecule microscopy. Methods Enzymol. 505, 159–186 10.1016/B978-0-12-388448-0.00017-6 (doi:10.1016/B978-0-12-388448-0.00017-6) [DOI] [PubMed] [Google Scholar]
- 77.Pinaud F, Michalet X, Iyer G, Margeat E, Moore HP, Weiss S. 2009. Dynamic partitioning of a glycosyl-phosphatidylinositol-anchored protein in glycosphingolipid-rich microdomains imaged by single-quantum dot tracking. Traffic 10, 691–712 10.1111/j.1600-0854.2009.00902.x (doi:10.1111/j.1600-0854.2009.00902.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lingwood D, Ries J, Schwille P, Simons K. 2008. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl Acad. Sci. USA 105, 10 005–10 010 10.1073/pnas.0804374105 (doi:10.1073/pnas.0804374105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pinaud F, King D, Moore HP, Weiss S. 2004. Bioactivation and cell targeting of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides. J. Am. Chem. Soc. 126, 6115–6123 10.1021/ja031691c (doi:10.1021/ja031691c) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schütz GJ, Schindler H, Schmidt T. 1997. Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys. J. 73, 1073–1080 10.1016/S0006-3495(97)78139-6 (doi:10.1016/S0006-3495(97)78139-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown DA, London E. 1998. Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164, 103–114 10.1007/s002329900397 (doi:10.1007/s002329900397) [DOI] [PubMed] [Google Scholar]
- 82.Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, Gratton E. 2001. Lipid rafts reconstituted in model membranes. Biophys. J. 80, 1417–1428 10.1016/S0006-3495(01)76114-0 (doi:10.1016/S0006-3495(01)76114-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swamy MJ, Ciani L, Ge M, Smith AK, Holowka D, Baird B, Freed JH. 2006. Coexisting domains in the plasma membranes of live cells characterized by spin-label ESR spectroscopy. Biophys. J. 90, 4452–4465 10.1529/biophysj.105.070839 (doi:10.1529/biophysj.105.070839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. 2007. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl Acad. Sci. USA 104, 3165–3170 10.1073/pnas.0611357104 (doi:10.1073/pnas.0611357104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sezgin E, Kaiser HJ, Baumgart T, Schwille P, Simons K, Levental I. 2012. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 7, 1042–1051 10.1038/nprot.2012.059 (doi:10.1038/nprot.2012.059) [DOI] [PubMed] [Google Scholar]
- 86.Levental I, Grzybek M, Simons K. 2011. Raft domains of variable properties and compositions in plasma membrane vesicles. Proc. Natl Acad. Sci. USA 108, 11 411–11 416 10.1073/pnas.1105996108 (doi:10.1073/pnas.1105996108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Veatch SL, Cicuta P, Sengupta P, Honerkamp-Smith A, Holowka D, Baird B. 2008. Critical fluctuations in plasma membrane vesicles. ACS Chem. Biol. 3, 287–293 10.1021/cb800012x (doi:10.1021/cb800012x) [DOI] [PubMed] [Google Scholar]
- 88.Parasassi T, De Stasio G, d'Ubaldo A, Gratton E. 1990. Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence. Biophys. J. 57, 1179–1186 10.1016/S0006-3495(90)82637-0 (doi:10.1016/S0006-3495(90)82637-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kindzelskii AL, Sitrin RG, Petty HR. 2004. Cutting edge: optical microspectrophotometry supports the existence of gel phase lipid rafts at the lamellipodium of neutrophils: apparent role calcium signaling. J. Immunol. 172, 4681–4685 [DOI] [PubMed] [Google Scholar]
- 90.Gaus K, Gratton E, Kable EP, Jones AS, Gelissen I, Kritharides L, Jessup W. 2003. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl Acad. Sci. USA 100, 15 554–15 559 10.1073/pnas.2534386100 (doi:10.1073/pnas.2534386100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gaus K, Chklovskaia E, Fazekas de St Groth B, Jessup W, Harder T. 2005. Condensation of the plasma membrane at the site of T lymphocyte activation. J. Cell Biol. 171, 121–131 10.1083/jcb.200505047 (doi:10.1083/jcb.200505047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanchez SA, Tricerri MA, Gratton E. 2012. Laurdan generalized polarization fluctuations measures membrane packing micro-heterogeneity in vivo. Proc. Natl Acad. Sci. USA 109, 7314–7319 10.1073/pnas.1118288109 (doi:10.1073/pnas.1118288109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. 2010. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl Acad. Sci. USA 107, 22 050–22 054 10.1073/pnas.1016184107 (doi:10.1073/pnas.1016184107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao J, Wu J, Shao H, Kong F, Jain N, Hunt G, Feigenson G. 2007. Phase studies of model biomembranes: macroscopic coexistence of Lα + Lβ, with light-induced coexistence of Lα + Lo phases. Biochim. Biophys. Acta 1768, 2777–2786 10.1016/j.bbamem.2007.07.009 (doi:10.1016/j.bbamem.2007.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ayuyan AG, Cohen FS. 2006. Lipid peroxides promote large rafts: effects of excitation of probes in fluorescence microscopy and electrochemical reactions during vesicle formation. Biophys. J. 91, 2172–2183 10.1529/biophysj.106.087387 (doi:10.1529/biophysj.106.087387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee JCM, Law RJ, Discher DE. 2001. Bending contributions hydration of phospholipid and block copolymer membranes: unifying correlations between probe fluorescence and vesicle thermoelasticity. Langmuir 17, 3592–3597 10.1021/la001678v (doi:10.1021/la001678v) [DOI] [Google Scholar]
- 97.Heimburg T. 2007. Thermal biophysics of membranes. Weinheim, Germany: Wiley-VCH [Google Scholar]
- 98.Prior IA, Muncke C, Parton RG, Hancock JF. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160, 165–170 10.1083/jcb.200209091 (doi:10.1083/jcb.200209091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson BS, Pfeiffer JR, Oliver JM. 2000. Observing FcεRI signaling from the inside of the mast cell membrane. J. Cell Biol. 149, 1131–1142 10.1083/jcb.149.5.1131 (doi:10.1083/jcb.149.5.1131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. 2006. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl Acad. Sci. USA 103, 18 992–18 997 10.1073/pnas.0609009103 (doi:10.1073/pnas.0609009103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. 2010. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 10.1038/ni.1832 (doi:10.1038/ni.1832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Williamson DJ, Owen DM, Rossy J, Magenau A, Wehrmann M, Gooding JJ, Gaus K. 2011. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat. Immunol. 12, 655–662 10.1038/ni.2049 (doi:10.1038/ni.2049) [DOI] [PubMed] [Google Scholar]
- 103.Chudakov DM, Verkhusha VV, Staroverov DB, Souslova EA, Lukyanov S, Lukyanov KA. 2004. Photoswitchable cyan fluorescent protein for protein tracking. Nat. Biotechnol. 22, 1435–1439 10.1038/nbt1025 (doi:10.1038/nbt1025) [DOI] [PubMed] [Google Scholar]
- 104.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. 2011. Identification of clustering artifacts in photoactivated localization microscopy. Nat. Methods 8, 527–528 10.1038/nmeth.1627 (doi:10.1038/nmeth.1627) [DOI] [PubMed] [Google Scholar]
- 105.Treanor B, Depoil D, Gonzalez-Granja A, Barral P, Weber M, Dushek O, Bruckbauer A, Batista FD. 2010. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity 32, 187–199 10.1016/j.immuni.2009.12.005 (doi:10.1016/j.immuni.2009.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Andrews NL, Lidke KA, Pfeiffer JR, Burns AR, Wilson BS, Oliver JM, Lidke DS. 2008. Actin restricts FcεRI diffusion and facilitates antigen-induced receptor immobilization. Nat. Cell Biol. 10, 955–963 10.1038/ncb1755 (doi:10.1038/ncb1755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jaqaman K, Kuwata H, Touret N, Collins R, Trimble WS, Danuser G, Grinstein S. 2011. Cytoskeletal control of CD36 diffusion promotes its receptor and signaling function. Cell 146, 593–606 10.1016/j.cell.2011.06.049 (doi:10.1016/j.cell.2011.06.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anderson RG, Jacobson K. 2002. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296, 1821–1825 10.1126/science.1068886 (doi:10.1126/science.1068886) [DOI] [PubMed] [Google Scholar]
- 109.Vogel SS, Thaler C, Koushik SV. 2006. Fanciful FRET. Sci. STKE 2006, re2. 10.1126/stke.3312006re2 (doi:10.1126/stke.3312006re2) [DOI] [PubMed] [Google Scholar]
- 110.Dunn RC. 1999. Near-field scanning optical microscopy. Chem. Rev. 99, 2891–2928 10.1021/cr980130e (doi:10.1021/cr980130e) [DOI] [PubMed] [Google Scholar]
- 111.van Zanten TS, Cambi A, Garcia-Parajo MF. 2010. A nanometer scale optical view on the compartmentalization of cell membranes. Biochim. Biophys. Acta 1798, 777–787 10.1016/j.bbamem.2009.09.012 (doi:10.1016/j.bbamem.2009.09.012) [DOI] [PubMed] [Google Scholar]
- 112.Betzig E, Chichester RJ. 1993. Single molecules observed by near-field scanning optical microscopy. Science 262, 1422–1425 10.1126/science.262.5138.1422 (doi:10.1126/science.262.5138.1422) [DOI] [PubMed] [Google Scholar]
- 113.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 10.1126/science.1127344 (doi:10.1126/science.1127344) [DOI] [PubMed] [Google Scholar]
- 114.Rust M, Bates M, Zhuang X. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 10.1038/nmeth929 (doi:10.1038/nmeth929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hess ST, Girirajan TP, Mason MD. 2006. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 10.1529/biophysj.106.091116 (doi:10.1529/biophysj.106.091116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shroff H, Galbraith CG, Galbraith JA, Betzig E. 2008. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423 10.1038/nmeth.1202 (doi:10.1038/nmeth.1202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bobroff N. 1986. Position measurement with a resolution and noise-limited instrument. Rev. Sci. Instrum. 57, 1152–1157 10.1063/1.1138619 (doi:10.1063/1.1138619) [DOI] [Google Scholar]
- 118.Pertsinidis A, Zhang Y, Chu S. 2010. Subnanometre single-molecule localization, registration and distance measurements. Nature 466, 647–651 10.1038/nature09163 (doi:10.1038/nature09163) [DOI] [PubMed] [Google Scholar]
- 119.Thompson RE, Larson DR, Webb WW. 2002. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82, 2775–2783 10.1016/S0006-3495(02)75618-X (doi:10.1016/S0006-3495(02)75618-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. 2003. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 10.1126/science.1084398 (doi:10.1126/science.1084398) [DOI] [PubMed] [Google Scholar]
- 121.Serge A, Bertaux N, Rigneault H, Marguet D. 2008. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods 5, 687–694 10.1038/nmeth.1233 (doi:10.1038/nmeth.1233) [DOI] [PubMed] [Google Scholar]
- 122.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 10.1038/nmeth.1237 (doi:10.1038/nmeth.1237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mashanov GI, Molloy JE. 2007. Automatic detection of single fluorophores in live cells. Biophys. J. 92, 2199–2211 10.1529/biophysj.106.081117 (doi:10.1529/biophysj.106.081117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoon JW, Bruckbauer A, Fitzgerald WJ, Klenerman D. 2008. Bayesian inference for improved single molecule fluorescence tracking. Biophys. J. 94, 4932–4947 10.1529/biophysj.107.116285 (doi:10.1529/biophysj.107.116285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tvarusko W, Bentele M, Misteli T, Rudolf R, Kaether C, Spector DL, Gerdes HH, Eils R. 1999. Time-resolved analysis and visualization of dynamic processes in living cells. Proc. Natl Acad. Sci. USA 96, 7950–7955 10.1073/pnas.96.14.7950 (doi:10.1073/pnas.96.14.7950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wieser S, Schütz GJ. 2008. Tracking single molecules in the live cell plasma membrane: Do's and Don't's. Methods 46, 131–140 10.1016/j.ymeth.2008.06.010 (doi:10.1016/j.ymeth.2008.06.010) [DOI] [PubMed] [Google Scholar]
- 127.Saxton MJ, Jacobson K. 1997. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 10.1146/annurev.biophys.26.1.373 (doi:10.1146/annurev.biophys.26.1.373) [DOI] [PubMed] [Google Scholar]
- 128.Ruprecht V, Axmann M, Wieser S, Schütz GJ. 2011. What can we learn from single molecule trajectories? Curr. Prot. Pept. Sci. 12, 714–724 10.2174/138920311798841753 (doi:10.2174/138920311798841753) [DOI] [PubMed] [Google Scholar]
- 129.Elson EL. 2011. Fluorescence correlation spectroscopy: past, present, future. Biophys. J. 101, 2855–2870 10.1016/j.bpj.2011.11.012 (doi:10.1016/j.bpj.2011.11.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Madl J, Weghuber J, Fritsch R, Derler I, Fahrner M, Frischauf I, Lackner B, Romanin C, Schutz GJ. 2010. Resting state orai1 diffuses as homotetramer in the plasma membrane of live mammalian cells. J. Biol. Chem. 285, 41 135–41 142 10.1074/jbc.M110.177881 (doi:10.1074/jbc.M110.177881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Axelrod D. 1989. Total internal reflection fluorescence microscopy. Methods Cell Biol. 30, 245–270 10.1016/S0091-679X(08)60982-6 (doi:10.1016/S0091-679X(08)60982-6) [DOI] [PubMed] [Google Scholar]