Abstract
Evolutionary rescue occurs when a population genetically adapts to a new stressful environment that would otherwise cause its extinction. Forecasting the probability of persistence under stress, including emergence of drug resistance as a special case of interest, requires experimentally validated quantitative predictions. Here, we propose general analytical predictions, based on diffusion approximations, for the probability of evolutionary rescue. We assume a narrow genetic basis for adaptation to stress, as is often the case for drug resistance. First, we extend the rescue model of Orr & Unckless (Am. Nat. 2008 172, 160–169) to a broader demographic and genetic context, allowing the model to apply to empirical systems with variation among mutation effects on demography, overlapping generations and bottlenecks, all common features of microbial populations. Second, we confront our predictions of rescue probability with two datasets from experiments with Saccharomyces cerevisiae (yeast) and Pseudomonas fluorescens (bacterium). The tests show the qualitative agreement between the model and observed patterns, and illustrate how biologically relevant quantities, such as the per capita rate of rescue, can be estimated from fits of empirical data. Finally, we use the results of the model to suggest further, more quantitative, tests of evolutionary rescue theory.
Keywords: adaptive mutation, stochastic demography, diffusion approximation, experimental evolution, resistance emergence
1. Introduction
Forecasts of future rates of species extinction are three to four orders of magnitude higher than known background rates of extinction in the fossil record [1]. Such forecasts of biodiversity loss have been criticized for not taking into account the capacity of organisms to adapt to their changing environment [2]. Evolutionary rescue describes the process by which a population, initially confronted with an environment causing its decline, is saved from extinction through genetic changes that recover growth. Emergence of resistance to chemotherapy (antibiotics, antivirals, pesticides, etc.) is also an important example of evolutionary rescue, well studied both empirically (reviewed in MacLean et al. [3]) and theoretically [4]. Several theoretical models have addressed the joint evolutionary and demographic processes leading to evolutionary rescue when the environment deteriorates gradually [5,6] or abruptly [6–8]. The very same process has also been modelled in more epidemiologically oriented models [4]. Rescue or demise depends on a race between population decline and adaptation: genotypes that adapt the population to the new environment must reach a substantial frequency before the population becomes extinct. These models predict that the probability of evolutionary rescue decreases with stress intensity and increases with initial population size or with the abundance of genetic variation available to fuel adaptation to the new conditions (reviewed in Bell [7]).
Forecasting extinction requires the development of a validated quantitative theory of evolutionary rescue. This includes, as a case of special interest, forecasting the emergence of resistance in diseases and pests affecting human health and economy. Current models of evolutionary rescue make different quantitative predictions about the probability of evolutionary rescue because of differences in assumptions regarding the genetic basis of adaptation to stress, and the stochastic process governing population dynamics. For instance, the pioneering model of Gomulkiewicz & Holt [8] used the infinitesimal model from quantitative genetics to describe the genetic basis for fitness, which is more appropriate when adaptation to stress is caused by a large pool of alleles at many loci, already present in the population at the onset of stress. Orr & Unckless [9] studied evolutionary rescue in the opposite case where adaptation is conveyed by a single genetic variant. A narrow genetic basis for adaptation has indeed been found in many cases of drug resistance [3,10]. They further compared (see also Ribeiro & Bonhoeffer [4]) the contribution from genetic variants, either present before the onset of stress (pre-existing mutation), or afterwards, during the period of population decline (de novo mutation). While the heuristic model by Gomulkiewicz & Holt [8] describes the deterministic growth of the rescued population above a threshold critical population size, the model by Orr & Unckless [9] explicitly describes the stochasticity inherent to the establishment of beneficial variants (see also [11]).
Empirical validation of evolutionary rescue theory is still in its infancy. Experimental evolution offers a potentially powerful method for this validation [12]. In particular, rapid evolution in microcosms allows the study of replicated trajectories of adaptation to evaluate the probability of rescue versus extinction. This is rarely possible in natural populations where only a single realization of any of these stochastic outcomes is observable. Usually, however, extinction has been considered as a nuisance in experimental evolution. Only recently, several studies have tackled the challenge of describing the probability of evolutionary rescue, using fast-reproducing organisms in microcosms with various stresses causing decline, such as yeast adapting to saline conditions [13,14], virus adapting to high temperature [15], flour beetles adapting to a new host [16,17] or bacteria adapting to antibiotic stress [18]. The study of the emergence of resistance to chemotherapy in microbes also has a very long and fruitful history (reviewed in recent studies [3,19]), which relates directly to evolutionary rescue. Bell & Gonzalez [13] showed a clear threshold in population size below which rescue was very unlikely in Saccharomyces cerevisiae undergoing salt stress, as predicted by theory [9]. Ramsayer et al. [20] studied adaptation of Pseudomonas fluorescens to antibiotic stress; they found that rescue probability increased with the genetic diversity at the onset of stress [17], giving qualitative support to other facets of evolutionary rescue theory.
In spite of this progress, tightening the link between theory and experiments requires evolutionary rescue models to use empirically measurable parameters. Models also need to account for certain peculiarities of microbial populations. In microbes undergoing limited recombination, clonal interference reduces polymorphism. Thus, the assumption of an asexual population with a narrow genetic basis for rescue (as in Orr & Unckless [9]) is a good way to describe the genetics of rescue in these cases. Furthermore, microbes often display continuous growth with overlapping generations, whereas most existing theories on evolutionary rescue (including Orr & Unckless [9]) assume discrete-time geometric growth or decay (but see Knight et al. [21]). Generalizing rescue predictions to a larger set of life cycles is now needed. For example, microbial populations experience periods of progressive growth or decline, but also frequent bottlenecks both in nature and in the laboratory, where serial transfers are routinely used. How bottlenecks affect the probability and dynamics of rescue is not known. Finally, even if a single variant generates the rescue in any given population, different variants may become fixed in different replicate populations, because they are sampled from a spectrum of potential mutation effects. This variability of outcomes has so far been ignored in analytical models where a single resistance fitness class is considered [4,9].
The aim of this paper is to propose methods to foster a better quantitative link between experimental evolution and predictions of evolutionary rescue theory. First, we generalize the predictions by Orr & Unckless [9] to a broader demographic and genetic context, with a full description of the stochasticity of the rescue process. This allows us to apply the model to empirical systems with variable mutation effects on growth rates, overlapping generations and the inclusion of bottlenecks. We show that the results obtained by Orr & Unckless [9] generalize, at least approximately, to more general demographic assumptions, in the absence of density-dependence. Second, we use the results of the model to discuss possible empirical tests of rescue theory, either by (mostly qualitative) tests of some predictions on existing datasets, or by discussing empirical methods to test other aspects of evolutionary rescue. We also discuss how models and data on the stochastic dynamics of rescued populations could enhance our understanding of the underlying evolutionary processes.
2. Methods
(a). Datasets
We compare the predictions from our model with data from experiments on evolutionary rescue, using two species: S. cerevisiae [13] and P. fluorescens [20]. These experiments illustrate aspects of population dynamics and experimental protocols that we aim to address in our theoretical developments. They also allow us to evaluate the general qualitative relevance of our analytical predictions.
Bell & Gonzalez [13] studied the effect of population size on the probability of rescue in bottlenecked populations of yeast under salt stress. From a large mass culture, derived from a single clone in permissive conditions, replicate populations were diluted to a wide range of starting population sizes (10–107 cells) and then exposed to a saline (NaCl) environment or a salt-free control. This allowed a test of the effect of the inoculum size on the probability of rescue. High concentrations of NaCl were used such that populations initially declined in the new environment as a result of the combined effects of reduced growth in the presence of salt and dilution during transfers. In the control experiment, the wild-type was grown and serially diluted in the absence of NaCl. Final population density was measured by optical density; populations were considered extinct when the optical density was equal to or below the mean value of control wells containing no cells.
In a recent study, Ramsayer et al. [20] investigated the dynamics of evolutionary rescue in naturally declining populations of P. fluorescens facing three bactericidal doses of streptomycin (50, 100 and 200 μg ml−1 plus a control). Unlike in Bell & Gonzalez [13], replicate populations started from high density (≈ 108 cells) and without any further bottlenecking during the experiment. To investigate the impact of initial standing variation on evolutionary rescue, two types of starting populations were compared. For the ‘clonal’ population type, 10 single clones were grown to high stationary density, from each of which replicate populations were established and exposed to the antibiotic. For the ‘admixture’ populations, the starting population was a mixture from eight large populations that had been maintained in replicate vials over 14 serial transfers in a permissive environment. Replicate admixture populations were established from this mixture. Ten replicates per population origin and antibiotic dose were tested, as well as two control antibiotic-free populations. Population sizes were measured by plating constant volume samples and counting colony-forming units (CFUs) at times t = 0, 4, 9, 22, 30, 53 h after introduction of the antibiotic. Populations were considered extinct when no CFUs were detected in any of the samples at t = 53 h.
(b). Analytical predictions
As in the experiments described earlier, the scenario envisioned by our model is that of a single isolated asexual population confronted with a new environment causing its decline. An initial number of individuals No is introduced in this new environment (inoculum size). We model the probability that such a population initially decaying at rate ro < 0 is saved from extinction by the occurrence and the establishment of genetic variants showing positive growth (r > 0). We derive analytical predictions from stochastic models of population dynamics, using branching processes and diffusion approximations. A key assumption of our model is that the fates of individual genetic variants are independent of each other (as in previous studies [8,9]), which precludes the analytical treatment of density-dependent growth (but see Uecker & Hermisson [22]). This may be a reasonable approximation if interactions between individuals are negligible in declining populations. We will discuss how comparison with experimental data could allow us to validate this approximation.
We consider the case of continuously growing/decaying populations with overlapping generations such as microbial populations, but our analytical results apply to a broader range of demographic dynamics. We use a Feller diffusion process [23], also called ‘continuous state branching process’ (see the electronic supplementary material, appendix S1), to approximate the demographic process. This approximation reduces all the complexity of the life cycle into two key parameters: the mean reproductive output r (growth rate or Malthusian fitness), and the variance of this output σ (for use of similar diffusion approximation in age-structured populations, see Shpak [24]). This diffusion approximation and its range of validity are more detailed in the electronic supplementary material, appendix S1. In short, our approach, based on diffusion, is general in that it can approximate many life histories, but is still limited (i) to density independent and (ii) ‘smooth’ demographic processes (without instantaneous jumps in population size), and (iii) to moderately growing or decaying genotypes (|r|/σ ≪ 1). Here, we will mostly illustrate our predictions in the case of a birth–death process, where cells divide by binary fission at rate b and die at rate d, at exponentially distributed time intervals. In that case, the growth rate is r = b − d, and the reproductive variance is σ = b + d = r + 2d. We also show consistency with the results of Orr & Unckless [9] who assumed a discrete-time model.
Mutations occur as a Poisson process with given rate per time units: the process of growth, mutation, etc., are measured in consistent units (hours, days, etc.). Note that, as mutations occur every division event, the per unit time mutation rate is proportional to the birth rate (see details in Martin & Gandon [25]). In all scenarios, rescue mutations all arise in the same constant genotypic background (the dominant ‘wild-type’), so that a constant mutation rate can be used, except when multiple mutational steps are considered (see details in the corresponding section and electronic supplementary material, appendix S1). Mutations create a set of alleles with an arbitrary distribution of effects on growth rates r and reproductive variance σ. We call ‘resistant’ a genetic variant that shows positive growth (r > 0), and ‘rescue’ a variant that is resistant and has established, meaning that it has escaped initial stochastic loss and increased to substantial population size. Each genetic variant is either already present in the population at the onset of stress (pre-existing mutations) or appears afterwards by mutation (de novo mutations). We derive predictions for the probability of rescue in both scenarios. We further consider different demographic scenarios likely to occur in evolutionary rescue experiments: either the population is exponentially decaying in the new environment [20], or it declines owing to the combined effects of bottlenecks and decreased growth [13]. We also examine two scenarios concerning pre-existing mutations: either variants were generated from a single clone in a previous phase of expansion in the permissive environment, as in the ‘clonal’ treatment of [20] and in Bell & Gonzalez [13], or they were segregating in a population at mutation–selection balance before exposure to stress, as could be assumed in the ‘admixture’ treatment of Ramsayer et al. [20].
In the main text, we present the most simple and testable approximations, whereas the appendices in the electronic supplementary material provide a thorough description of how these approximations are derived. All analytical derivations were obtained with Mathematica v. 8.0 [26].
(c). Simulation check for a birth–death process
The analytical predictions were tested in exact simulations of a birth–death process, i.e. with cell division and death occurring at exponentially distributed times. In our simulations, mutation produced a spectrum of birth rates b, with a fixed death rate d. The simulation algorithm is described in Martin & Gandon [25], with demographic dynamics simulated by the exact Gillespie stochastic simulation algorithm [27]. Mutation is birth-dependent (it occurs only in cells that divide), and mutants show continuous variation in birth rates, generated by a Gaussian phenotype-to-fitness landscape. The landscape model here was merely used to produce mutations with a continuous spectrum of effects. This algorithm was optimized and encoded in C; matrix operations and random number generation were performed using the GNU Scientific Library [28]. Analysis of the simulations was performed with R [29].
(d). Data analysis
Analytical predictions provide a framework to analyse experiments that study how various factors affect rescue probability. We evaluated the fit of our predictions on the effect of inoculum size No on rescue probability in the yeast salt-stress experiment [13]. We then compared the observed versus the expected outcome of different evolutionary histories on rescue probability, in naturally decaying P. fluorescens populations under antibiotic stress [20]. Generalized linear models (GLMs) were performed with Mathematica v. 8.0 [26].
3. Results
(a). Theory: probability of evolutionary rescue in various scenarios
As only resistance mutations (r > 0) can achieve substantial frequency, we consider only the rate of mutation to resistance. Resistant mutants may appear in the ‘permissive’ environment (before the onset of stress), or in the stressful environment. We therefore denote the mutation rate to resistance in each condition by uP and uS, respectively. We denote by an overbar ( ) the mean of any quantity x among resistant genotypes produced by mutation.
) the mean of any quantity x among resistant genotypes produced by mutation.
(i). General result
As we will see below, a striking similarity emerges from the various scenarios considered, where the number of mutations destined to rescue the population is, in general, Poisson distributed. This is valid, in general, only when the per capita rate of mutation to rescue is small. Furthermore, the rate of the Poisson distribution can always be written  i.e. it is proportional to the number of individuals present at the onset of stress, No. The factor
i.e. it is proportional to the number of individuals present at the onset of stress, No. The factor 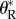 can thus be denoted a rate of rescue per inoculated individual. It will depend on the particular scenario. The derivations that follow will consist of showing this Poisson limit and providing closed form expressions for
can thus be denoted a rate of rescue per inoculated individual. It will depend on the particular scenario. The derivations that follow will consist of showing this Poisson limit and providing closed form expressions for  for each subcase, as a function of various biological parameters. The probability of an evolutionary rescue
for each subcase, as a function of various biological parameters. The probability of an evolutionary rescue  is the probability that at least one rescue mutation appears before extinction, and is given by one minus the probability of no rescue, i.e. the zero class of the Poisson (
is the probability that at least one rescue mutation appears before extinction, and is given by one minus the probability of no rescue, i.e. the zero class of the Poisson ( ). Thus, in what follows, all expressions for rescue probability take the form
). Thus, in what follows, all expressions for rescue probability take the form
| 3.1 |
Intuitively, the simple relationship to No reflects the fact that we assume independence between the fate of different lineages present at the onset of stress (no density – dependence).
Table 1 summarizes the explicit expressions for  in each scenario, with the corresponding figures showing the simulation checks.
in each scenario, with the corresponding figures showing the simulation checks.
Table 1.
Equations and simulation figures corresponding to each scenario treated in the study. The rate of rescue mutations  per individual inoculated (see equation (3.1)), for the various scenarios treated in this article. u is the rate of mutations with any effect on growth rate and uS (respectively uP) are the rate of mutation to resistance (r > 0 or mr > 0 with natural or bottleneck induced decay, respectively) in the stress (respectively in the permissive environment). |ro| or
per individual inoculated (see equation (3.1)), for the various scenarios treated in this article. u is the rate of mutations with any effect on growth rate and uS (respectively uP) are the rate of mutation to resistance (r > 0 or mr > 0 with natural or bottleneck induced decay, respectively) in the stress (respectively in the permissive environment). |ro| or  is the decay rate of the wild-type per unit time (or per cycle) with natural decay or bottleneck induced decay, respectively.
is the decay rate of the wild-type per unit time (or per cycle) with natural decay or bottleneck induced decay, respectively.  (or
(or 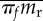 ) is the mean establishment probability (equation (3.2)) among random resistance mutations in these two cases. Here, we have assumed that resistance and its cost in the permissive environment are uncorrelated (see equation (3.6) versus (3.7)). The notations below are defined in more detail in the electronic supplementary materials, table and appendices.
) is the mean establishment probability (equation (3.2)) among random resistance mutations in these two cases. Here, we have assumed that resistance and its cost in the permissive environment are uncorrelated (see equation (3.6) versus (3.7)). The notations below are defined in more detail in the electronic supplementary materials, table and appendices.
| biological scenario |  |
simulation checks |
|---|---|---|
| de novo single step mutation, exponential decay (equation (3.3)) |  |
figure 2b and electronic supplementary materials figure S2 |
| de novo two-steps rescue, exponential decay (equation (S1.4)) |  |
None |
| pre-existent single step mutation, population initially at mutation–selection balance (equation (3.7)) |  |
figure 2a and electronic supplementary material, figure S3 |
| pre-existent single step mutation, population initiated by growth of a single clone from no → N1 (equation (3.9)) |  |
figure 2c |
| de novo single step mutation, decay from periodic bottlenecks (equation (3.4)) |  |
figure 2d and electronic supplementary material, figure S4 |
(ii). Rescue from de novo mutations
General probability of establishment. A ‘rescue’ mutant is a resistant mutant lineage that has risen to substantial copy number such that it is almost certain to avoid ultimate extinction (establishment). The probability of establishment, for a resistant mutant lineage with given demographic parameters (r,σ), initially present in a single copy is approximately [23]
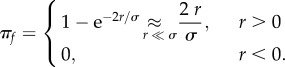 |
3.2 |
Quite intuitively, equation (3.2) predicts that the probability of establishment increases for mutants with large growth rate and small reproductive variance [24,30]. Throughout this paper, most simplifications are obtained when establishment is unlikely (πf = 0(r/σ) ≪ 1), an assumption already inherent in our use of diffusion approximations. Figure 1 shows πf as a function of r: equation (3.2) indeed approximates the exact πf in many cases, as long as r/σ is small (figure 1a,b). However, the diffusion approximation breaks down in the burst–death model with even moderately large burst sizes and small r/σ, because burst events introduce instantaneous jumps in the population size dynamics, inconsistent with the ‘smoothness’ assumption required for diffusion to be valid (see also electronic supplementary material, appendix S1).
Figure 1.

Accuracy of the Feller diffusion approximation for establishment probability. The probability of establishment (avoiding ultimate extinction) when started from a single individual is shown as a function of the ratio of Feller parameters (r/σ): (a) linear birth–death process with birth and death events occurring at rates b and d, for which r = b − d and σ = b + d, (b) discrete-time model with Poisson offspring distribution (Δnt ∼ Poisson(w nt)) for which r = w − 1 and σ = w ≈ 1, (c) linear burst–death process with bursts of size B occurring at rate b and death at rate d. This model assumes that an individual can either die or burst (producing B offspring) at exponentially distributed times, i.e. that both death and burst are Poisson processes with rate d and b, respectively (setting B = 1 retrieves case a.). For this model r = b B − d and σ = b B2 + d. In each case, the exact probability of establishment is compared to the Feller diffusion approximation: these computations (along with that of r and σ) are detailed for each case in electronic supplementary material, appendix S1, § 1. The approximation accurately captures the discrete-time model and the birth–death model, but it breaks down quickly for the burst–death model when burst sizes get large (although it always valid at low πf).
Continuously decaying populations. Let us first consider a clonal population consisting of a wild-type with negative growth rate ro < 0 that decays continuously to extinction in the absence of any genetic change. In this case, we can compute the exact probability that, before extinction of the wild-type, it produces a resistance mutation that establishes. We must take into account variation of (r,σ) among resistant mutants spawned by the wild-type genotype. Rescue mutations occur as an inhomogeneous Poisson process with instantaneous rate  where uS is the rate of mutation towards resistance in the stressful environment, Nt is the wild-type population at time t. The average probability of establishment among resistant mutants is
where uS is the rate of mutation towards resistance in the stressful environment, Nt is the wild-type population at time t. The average probability of establishment among resistant mutants is  which is πf in equation (3.2) averaged over the joint distribution of r and σ among these mutants. Note that the total number of rescue mutations spawned from the wild-type during its course to extinction is
which is πf in equation (3.2) averaged over the joint distribution of r and σ among these mutants. Note that the total number of rescue mutations spawned from the wild-type during its course to extinction is  where τE is the extinction time for a particular trajectory. The cumulated population size
where τE is the extinction time for a particular trajectory. The cumulated population size 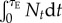 is a random variable that is distributed among replicate extinction trajectories. The resulting distribution of the number of rescue mutations, after averaging over this random variable, is a compound Poisson distribution, not simply a Poisson (see equation (S1.5) of electronic supplementary material, appendix S1). However, when the rate of rescue is not too large (
is a random variable that is distributed among replicate extinction trajectories. The resulting distribution of the number of rescue mutations, after averaging over this random variable, is a compound Poisson distribution, not simply a Poisson (see equation (S1.5) of electronic supplementary material, appendix S1). However, when the rate of rescue is not too large ( equation (3.1)), the Poisson distribution obtains as a limit of this exact distribution (see appendix S1 and electronic supplementary material, figures S1 and S2). A heuristic argument provides a simple derivation of the rate
equation (3.1)), the Poisson distribution obtains as a limit of this exact distribution (see appendix S1 and electronic supplementary material, figures S1 and S2). A heuristic argument provides a simple derivation of the rate 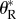 in this limit (the exact derivation is given in the electronic supplementary material, appendix S1). If we approximate that all replicate populations decay deterministically (exponentially) until extinction, the cumulated population size is then a constant, equal to its expected value No/|ro|, and the number of potential rescue mutations is approximately Poisson with rate
in this limit (the exact derivation is given in the electronic supplementary material, appendix S1). If we approximate that all replicate populations decay deterministically (exponentially) until extinction, the cumulated population size is then a constant, equal to its expected value No/|ro|, and the number of potential rescue mutations is approximately Poisson with rate  Then, the rate of rescue per inoculated individual is
Then, the rate of rescue per inoculated individual is
| 3.3 |
Here,  is the rate of rescue mutations per wild-type individual under stress, namely the corresponding rate of resistance mutations weighted by the probability that they establish. Equation (3.3) is accurate (figure 2b) unless the wild-type shows almost no decay (see the electronic supplementary material, figure S2), a case of little interest as rescue is effectively certain then. In agreement with previous theory, equation (3.3) predicts that the probability of a rescue increases when the initial population size is large (figure 2), the mutation rate towards resistance is high, the wild-type population does not decline too fast (small |ro|) and the mean establishment probability
is the rate of rescue mutations per wild-type individual under stress, namely the corresponding rate of resistance mutations weighted by the probability that they establish. Equation (3.3) is accurate (figure 2b) unless the wild-type shows almost no decay (see the electronic supplementary material, figure S2), a case of little interest as rescue is effectively certain then. In agreement with previous theory, equation (3.3) predicts that the probability of a rescue increases when the initial population size is large (figure 2), the mutation rate towards resistance is high, the wild-type population does not decline too fast (small |ro|) and the mean establishment probability  of resistant mutants is high.
of resistant mutants is high.
Figure 2.
Rescue probability from de novo mutations and standing variance. The probability of evolutionary rescue as a function of inoculum population size (No) is given for various values of the mutations rate to resistance uR. The dots are the results of exact simulations and the lines show the corresponding theory. We consider either rescue in the face of bottlenecks (a,c) or in naturally decaying populations (b,d), and we consider three possible scenario: rescue from standing variance with a population at mutation–selection balance (a), de novo rescue (b), rescue from standing variance with a population having undergone unlimited growth from a single clone (c), and rescue from de novo mutations in the presence of bottlenecks (d). In every case U = 0.001 or 0.0001 (black versus grey curves, respectively) per cell division, the death rate is d = 1 and the decay rate ( in d) is ro = −0.1 (squares) or −0.2 (triangles). In (a), the mutation selection balance was reached after 4000 unit times. In (c), the population grew from 10 individuals to 107 individuals and then was diluted to obtain the No reported on the graph. In (d), the dilution factor for bottlenecks was D = 1/100 and the wild-type growth rate during growth phases was ro = −0.1, with τ set to obtain the desired mo values.
in d) is ro = −0.1 (squares) or −0.2 (triangles). In (a), the mutation selection balance was reached after 4000 unit times. In (c), the population grew from 10 individuals to 107 individuals and then was diluted to obtain the No reported on the graph. In (d), the dilution factor for bottlenecks was D = 1/100 and the wild-type growth rate during growth phases was ro = −0.1, with τ set to obtain the desired mo values.
The mean establishment probability depends on the distribution of the ratio r/σ among resistant mutants (see equation (3.2) and electronic supplementary material, appendix S1). This suggests that information about the distribution of growth rates among resistant mutants is not sufficient to predict the probability of rescue. For precise quantitative tests, experiments should also seek to evaluate the extent of reproductive variance σ and its joint variation with r among resistant mutants. If most resistance mutations are only weakly growing (0 < r ≪ σ), and mutations affect only r but not σ (σ is then a numerical constant, still to be estimated), then  where
where  is the average growth rate among resistant mutants. This simplification can be justified, for example, for a birth–death process with a high turnover rate (high birth and death rates that compensate each other). In this case, demographic stochasticity is important as r = b − d ≪ σ = b + d. Mutations affecting both b and d contribute the same variance to r and σ, but the impact of this variance is smaller, relative to the mean value, on r than on σ . Finally, note that this simplified version of our equation (3.3) corresponds to eqn (3) of Orr & Unckless [9], who assumed a single allele (
is the average growth rate among resistant mutants. This simplification can be justified, for example, for a birth–death process with a high turnover rate (high birth and death rates that compensate each other). In this case, demographic stochasticity is important as r = b − d ≪ σ = b + d. Mutations affecting both b and d contribute the same variance to r and σ, but the impact of this variance is smaller, relative to the mean value, on r than on σ . Finally, note that this simplified version of our equation (3.3) corresponds to eqn (3) of Orr & Unckless [9], who assumed a single allele ( ) and a discrete generation model (Poisson offspring distribution, σ ≈ 1 for all genotypes, see electronic supplementary material, appendix S1).
) and a discrete generation model (Poisson offspring distribution, σ ≈ 1 for all genotypes, see electronic supplementary material, appendix S1).
Rescue in multiple steps. The earlier-mentioned treatment deals only with rescue produced by a single mutation event. Rescue can also occur in multiple steps, e.g. with a first intermediate step destined to be lost (‘transient’ mutation), but that produces the final rescuer genotype before its extinction. This process (in two steps) is modelled in appendix S1 (§2). The resulting rate of two-step rescue scales with the total rate of genomic mutations (with arbitrary effect on r) up until extinction:  (equation (S1.7)). The first ‘transient’ mutation is most likely to be one with r ≈ 0 (critical process; electronic supplementary material, appendix S1). Indeed, these mutants are the ones that wander the longest time before extinction, thus allowing them to produce secondary rescues. Non-resistant mutants (r < 0) all get quickly extinct, whereas resistant mutants (r > 0) either produce a single step rescue or get extinct early when still at low numbers. It is difficult to be more explicit without introducing a particular model describing the combined effect of multiple mutations on growth rates [31].
(equation (S1.7)). The first ‘transient’ mutation is most likely to be one with r ≈ 0 (critical process; electronic supplementary material, appendix S1). Indeed, these mutants are the ones that wander the longest time before extinction, thus allowing them to produce secondary rescues. Non-resistant mutants (r < 0) all get quickly extinct, whereas resistant mutants (r > 0) either produce a single step rescue or get extinct early when still at low numbers. It is difficult to be more explicit without introducing a particular model describing the combined effect of multiple mutations on growth rates [31].
Rescue with bottlenecks. The process that we have described so far is one where the wild-type population decays at a roughly constant rate until extinction. However, extinction may also take place in populations with non-steady decay. We consider here the case where the wild-type alternates periods of positive growth with catastrophic drops in population size (bottlenecks). We model bottlenecks at regular time intervals (every τ time units) with a constant dilution factor 0 < D < 1 (the portion of the population that is kept). This is particularly relevant to microbe studies, where serial transfers are often used to refresh the medium, and it indeed corresponds to the protocol used by Bell & Gonzales [13]. It should also be a good approximation to the case of rescue in natural populations undergoing randomly distributed catastrophic drops in population size [32]. As explained in electronic supplementary material, appendix S2, our treatment is based on the approach proposed by Wahl & Gerrish [33], and complementary to theirs, by allowing for long-term decay or growth, and assuming high reproductive stochasticity during growth phases (πf ≪ 1) (see also [34]).
In this context, the demographic processes are intrinsically discrete at the time-scale of cycles, although the underlying demography during the growth phase is continuous at smaller time-scale (measured in our basic time units). We denote mr the growth rate of a given mutant, over a cycle, i.e. its Malthusian fitness at the time-scale of cycles. It must satisfy mr = log(D) + rτ, where r is the growth rate during the growth phase (in our basic time units). The wild-type has negative Malthusian fitness  over the long run, so that it will get extinct in the absence of rescue mutations. Only those mutants with mr > 0 (called ‘resistant’) can survive ultimate extinction and thus generate a rescue (equation (S2.6)). In order to do so, they must (i) escape stochastic loss during growth phases; and (ii) escape loss during bottlenecks. From the electronic supplementary material, appendix S2 (equation (S2.6)), the probability of such ultimate establishment for a mutant present in single copy at the onset of a growth phase is shown to be
over the long run, so that it will get extinct in the absence of rescue mutations. Only those mutants with mr > 0 (called ‘resistant’) can survive ultimate extinction and thus generate a rescue (equation (S2.6)). In order to do so, they must (i) escape stochastic loss during growth phases; and (ii) escape loss during bottlenecks. From the electronic supplementary material, appendix S2 (equation (S2.6)), the probability of such ultimate establishment for a mutant present in single copy at the onset of a growth phase is shown to be  for a mutant with positive long-term growth (mr > 0), where πf is again given in equation (3.2), and is null for a non-resistant mutant (mr < 0). Let us denote uSτ, the rate of mutation to resistance (mr > 0), over a cycle (over τ basic time units), per individual present at the start of the cycle, and in the stressful environment. The population is started at the onset of a growth phase with No individuals. We show in the electronic supplementary material, appendix S2 that the total number of rescue mutations produced during the course to extinction is again Poisson distributed with a rate of rescue per inoculated individual given by equation (S2.12)
for a mutant with positive long-term growth (mr > 0), where πf is again given in equation (3.2), and is null for a non-resistant mutant (mr < 0). Let us denote uSτ, the rate of mutation to resistance (mr > 0), over a cycle (over τ basic time units), per individual present at the start of the cycle, and in the stressful environment. The population is started at the onset of a growth phase with No individuals. We show in the electronic supplementary material, appendix S2 that the total number of rescue mutations produced during the course to extinction is again Poisson distributed with a rate of rescue per inoculated individual given by equation (S2.12)
| 3.4 |
This time,  is the rate of rescue mutations per individual over a cycle, where
is the rate of rescue mutations per individual over a cycle, where  is the mean probability, among random resistant mutants, of avoiding stochastic loss during the growth phase and over the following serial dilutions. Equation (3.4) is valid only if (i) the rescue mutants grow very rapidly between bottlenecks, which is necessarily the case when bottlenecks are severe (D ≪ πf), and if (ii) the reproductive stochasticity during growth phases is substantial so that πf ≪ 1. Figure 2d shows that this approximation is accurate, and electronic supplementary material, figure S4 suggests that this holds even with mild stochasticity and dilution factors (πf ≈ 0.33 and D = 1/10).
is the mean probability, among random resistant mutants, of avoiding stochastic loss during the growth phase and over the following serial dilutions. Equation (3.4) is valid only if (i) the rescue mutants grow very rapidly between bottlenecks, which is necessarily the case when bottlenecks are severe (D ≪ πf), and if (ii) the reproductive stochasticity during growth phases is substantial so that πf ≪ 1. Figure 2d shows that this approximation is accurate, and electronic supplementary material, figure S4 suggests that this holds even with mild stochasticity and dilution factors (πf ≈ 0.33 and D = 1/10).
The exact similarity of equations (3.3) and (3.4) is striking, given the key differences in the process under study. The only difference, apart from the change in time-scale, lies in the expression for the probability that a given resistance translates into a rescue: in equation (3.4), instead of
in equation (3.4), instead of  in equation (3.3). This simply reflects the fact that mutants must avoid extinction both during the growth phase in which they appear, and over the longer time-scale of growth-dilution cycles (mr). If mutations only affect r and not σ, we have
in equation (3.3). This simply reflects the fact that mutants must avoid extinction both during the growth phase in which they appear, and over the longer time-scale of growth-dilution cycles (mr). If mutations only affect r and not σ, we have  , where
, where  is the mean Malthusian fitness, and CVr is the coefficient of variation of r among random resistance mutations. We thus see that contrary to equation (3.3), rescue is not only determined by the mean growth rate
is the mean Malthusian fitness, and CVr is the coefficient of variation of r among random resistance mutations. We thus see that contrary to equation (3.3), rescue is not only determined by the mean growth rate  of resistance mutations but also by their variance (CVr).
of resistance mutations but also by their variance (CVr).
(iii). Rescue from standing variance: pre-existing mutations
General result. The inoculum population may also contain a certain proportion of resistant genotypes before the onset of stress. The proportion will depend on the state of the population before stress, and the corresponding probability of rescue from pre-existing mutations is dependent on the scenario assumed for this state. In electronic supplementary material, appendix S1 (equation (S1.9)), we show that, provided the probability of establishment of resistant genotypes in the stressful environment is small (πf ≪ 1), equation (3.1) applies, with a rate of rescue per inoculated individual
| 3.5 |
where E(nR) is the expected number of pre-existing resistance mutations in the inoculum, at the onset of stress, and  is the average establishment probability πf (equation (3.2)) among them. Note that, in general,
is the average establishment probability πf (equation (3.2)) among them. Note that, in general,  is distinct from the average
is distinct from the average 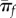 among de novo resistance mutations. Equation (3.5) is rather general, valid for an arbitrary state of the population before stress, an arbitrary distribution of mutation effects on growth rates before and after the stress. Only the expected number of pre-existing resistant genotypes before stress, and their mean establishment probability are required to predict the rescue probability from standing variance. To go further, we apply this result to two particular scenarios (detailed in electronic supplementary material, appendix S1): mutation–selection equilibrium prior to stress, and a population undergoing unlimited growth from a single clone, before facing the stress. Both scenarios correspond to frequently used settings in experimental evolution (e.g. contrast the ‘clonal’ and ‘admixture’ treatments in Ramsayer et al. [20]).
among de novo resistance mutations. Equation (3.5) is rather general, valid for an arbitrary state of the population before stress, an arbitrary distribution of mutation effects on growth rates before and after the stress. Only the expected number of pre-existing resistant genotypes before stress, and their mean establishment probability are required to predict the rescue probability from standing variance. To go further, we apply this result to two particular scenarios (detailed in electronic supplementary material, appendix S1): mutation–selection equilibrium prior to stress, and a population undergoing unlimited growth from a single clone, before facing the stress. Both scenarios correspond to frequently used settings in experimental evolution (e.g. contrast the ‘clonal’ and ‘admixture’ treatments in Ramsayer et al. [20]).
Mutation–selection equilibrium. We assume that a population has reached mutation–selection equilibrium before the stress occurs. Then, the population is culled to No individuals at the onset of stress. Alternatively, the population can be assumed to be at mutation–selection–drift equilibrium at constant size No all along (as in Orr & Unckless [9]). The expected number of pre-existing resistance mutations in both cases is E (nR) = NouP/cH, where cH is the harmonic mean of the cost of resistance mutations in the environment before stress. The cost of a given mutant is its selective disadvantage relative to the wild-type genotype, which is the genotype perfectly adapted to the previous environment. It is also this genotype that will dominate the population at the onset of stress. Here,  among pre-existing resistant variants is different from
among pre-existing resistant variants is different from 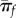 among de novo resistance mutations, whenever resistance and cost (r and c) are not independent among mutants (see §4 and electronic supplementary material, appendix S1). The resulting mean is
among de novo resistance mutations, whenever resistance and cost (r and c) are not independent among mutants (see §4 and electronic supplementary material, appendix S1). The resulting mean is  where
where  is the mean of
is the mean of  among random resistance mutations. The rate of rescue per inoculated individuals is
among random resistance mutations. The rate of rescue per inoculated individuals is
| 3.6 |
As for rescue from de novo mutations, the probability of rescue from standing variance increases with the size of the initial population. It increases with the rate of mutation towards resistance in the permissive environment and depends on the joint distribution of demographic rates of resistant mutants (r,σ) in the stressful environment, and of their cost c in the permissive environment. Equation (3.6) simplifies when the cost of a resistance, before stress, is independent of r or σ after the onset of stress. In this case, we can write  and the rate of rescue becomes
and the rate of rescue becomes  Denote the rate of rescue from de novo mutations
Denote the rate of rescue from de novo mutations 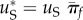 (from equation (3.3)). If we further assume that the mutation rate towards resistance is the same in permissive and stressful environments (uP = uS), we can then express the rate of a rescue from standing variance, as a function of the rate of de novo rescue mutations
(from equation (3.3)). If we further assume that the mutation rate towards resistance is the same in permissive and stressful environments (uP = uS), we can then express the rate of a rescue from standing variance, as a function of the rate of de novo rescue mutations  :
:
| 3.7 |
(Recall that cH is the harmonic mean of the cost, before stress, of those random de novo mutations that are resistant in the stress.) Mutations with a small cost before stress (and therefore at high frequency at the onset of stress) will dominate the harmonic mean cH and therefore the rescue process. Figure 2a shows the accuracy of equation (3.7) versus exact simulations. As for de novo rescue, we retrieve the corresponding result (eqn (5) of Orr & Unckless [9]) for a single allele (cH = c) and discrete demography, assuming a small πf (see electronic supplementary material, appendix S1).
Because the total number of rescue events from de novo mutations or from standing variants are both approximately Poisson distributed, the probability that a given rescue event is from either type is simply given by the relative weight of each Poisson parameter. In the case of a population at equilibrium before stress, and assuming independence between cost and resistance (equation (3.7)), the probability that the rescue mutation appeared after the onset of stress (de novo) as opposed to being present before is
| 3.8 |
where the right-hand-side limit is when mutation rates are equal across environments. We can see here that the relative contribution of each type depends on the rate of decay |ro| of the wild-type (i.e. the strength of the stress imposed). Provided the cost distribution is less affected by the stress than the decay rate (cH roughly constant as |ro| increases), we expect that rescue from standing variance will be more likely in more stressful conditions (larger |ro|). However, (i) the cost could be affected by the type of stress exerted; and (ii) this ignores the possible contribution of de novo rescue in multiple steps, which can be important under strong stresses [31].
Growth from a single clone. Our second scenario is intended to describe some microbe experiments where a single clone is grown to high density then put into a new stressful environment [13]. This may also be relevant to natural populations that have undergone a severe bottleneck and then grown back to a large population size before facing an abrupt environmental change. Under these conditions, the distribution of the number of resistance mutations at the end of the growth phase is given by models of fluctuation test experiments [35]. Although the distribution of the number of resistance mutations in this situation can be complex, its mean takes a simple form [35]. Assume that the population is grown from no clonal individuals to some constant level N1, then possibly diluted to achieve a given size No, prior to stress. The resulting expected number of resistance mutations in the inoculum is then E(nR) = No uP log(N1/no). As this number is directly proportional to the mutation rate to resistance uP independently of (r,σ), the mean fixation probability among pre-existing mutations is equal to that among de novo mutants  (see also electronic supplementary material, appendix S2). Applying our general equation (3.5) in this case, the rate of rescue (per inoculated individual per extinction) is then
(see also electronic supplementary material, appendix S2). Applying our general equation (3.5) in this case, the rate of rescue (per inoculated individual per extinction) is then
| 3.9 |
Figure 2c confirms the accuracy of the above expression versus exact simulations. Again, things simplify if we assume that the environmental change does not impact mutation rates (uP = uS) in which case equation (3.10) can be expressed in terms of the rate of de novo rescue, by setting  Contrary to the case of mutation–selection balance, the probability of rescue here does not depend on the fitness effects of resistant mutations in the permissive environment. As in equation (3.8), we find another simple rule for the probability of rescue from de novo versus pre-existing mutations:
Contrary to the case of mutation–selection balance, the probability of rescue here does not depend on the fitness effects of resistant mutations in the permissive environment. As in equation (3.8), we find another simple rule for the probability of rescue from de novo versus pre-existing mutations:
 |
3.10 |
As in equation (3.8), we expect that rescue from standing variance will be more likely in more stressful conditions (larger |ro|), all else being equal.
With bottlenecks. If we consider rescue from standing variance in the context of bottlenecked populations, all the results provided above equations (3.5)–(3.10) still hold, simply replacing  by its equivalent in bottlenecked populations
by its equivalent in bottlenecked populations  and ro by
and ro by  (equations (3.4) and (S2.6)).
(equations (3.4) and (S2.6)).
(b). Empirical tests: rescue probability versus inoculum size or evolutionary past
Equation (3.1) provides a theoretically based means to compare different rescue experiments by estimating  in each case: below, we show an empirical test of this equation and explain how such comparisons can be used.
in each case: below, we show an empirical test of this equation and explain how such comparisons can be used.
(i). Effect of inoculum size
Bell & Gonzalez [13] studied the effect of inoculum size on rescue probability, in bottlenecked populations of yeast in a saline environment. To do so, they grew a culture from a single clone to a constant large size N1 then diluted it to appropriate levels to study the effect of No on the probability of a rescue. In that case, de novo rescue should follow equation (3.4), while rescue from pre-existing mutations should follow equation (3.9) (table 1). Overall, we expect a relationship of the form  for some
for some  (equation (3.1)). We fitted this relationship to the yeast data by maximum likelihood (GLM with binomial error and link function L(p) = −log(1 − p)). Bell & Gonzalez also performed a control experiment, where the wild-type was inoculated in a non-stressful environment. Let PC (for ‘control’) be the probability that the population shows visible growth, when started with No individuals, in this control experiment. Let πcmc > 0 be the probability of persistence, when starting in single copy at the onset of a cycle, for the wild-type genotype in the control experiment (no stress apart from bottlenecks), characterized by index ‘c’. In the control experiment, the wild-type shows positive growth at rate rc and mc over time or cycles, respectively. Every single lineage is independent and ultimately goes extinct with some probability approximately 1 − πcmc (equation (S2.6) with πc = 1 − exp(−2rc/σ)), so the total probability of observing long-term growth, starting from No lineages, is simply:
(equation (3.1)). We fitted this relationship to the yeast data by maximum likelihood (GLM with binomial error and link function L(p) = −log(1 − p)). Bell & Gonzalez also performed a control experiment, where the wild-type was inoculated in a non-stressful environment. Let PC (for ‘control’) be the probability that the population shows visible growth, when started with No individuals, in this control experiment. Let πcmc > 0 be the probability of persistence, when starting in single copy at the onset of a cycle, for the wild-type genotype in the control experiment (no stress apart from bottlenecks), characterized by index ‘c’. In the control experiment, the wild-type shows positive growth at rate rc and mc over time or cycles, respectively. Every single lineage is independent and ultimately goes extinct with some probability approximately 1 − πcmc (equation (S2.6) with πc = 1 − exp(−2rc/σ)), so the total probability of observing long-term growth, starting from No lineages, is simply:  We again retrieve the same relationship to No (still because of the density-independence assumption), but with a higher expected rate, as it does not scale with mutation rates. We thus fitted the same relationship (
We again retrieve the same relationship to No (still because of the density-independence assumption), but with a higher expected rate, as it does not scale with mutation rates. We thus fitted the same relationship ( ) to the control experiments to estimate
) to the control experiments to estimate 
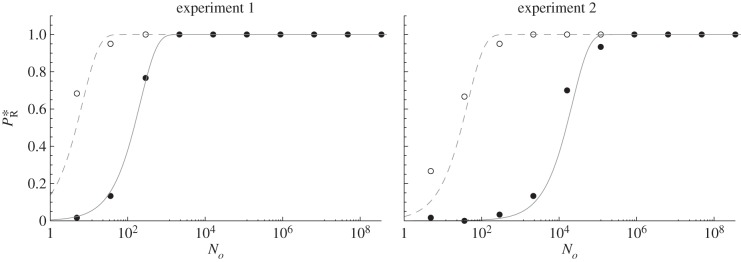
Figure 3 shows the result of the fit for both controls and stress treatments, with two replicate experiments in each case. The estimated parameters are given in table 2. The model fits the data well (goodness-of-fit tests based on deviance: p ≈ 1, table 2, the model is not significantly less fitted than the saturated model). Overall, while equation (3.1) is well supported by the available data, this prediction is somewhat too general to be strongly informative, being expected in all scenarios, and in controls. It does, however, suggest that each lineage derived from each inoculated individual has roughly independent mutational and demographic fates, which was a key assumption of the model. Interdependence between lineages may be at play in these experiments, but if so, here it has undetectable effect on the relationship between No and rescue probability. The parameters of this relationship have a clear biological interpretation:  estimates the overall rate of rescue per individual inoculated (in the stress, table 1), whereas θc estimates 2mcrc/σ in the control treatment, i.e. a measure of the probability of persistence from a single copy in this environment and for the wild-type genotype. The control experiment is therefore useful too, by giving an order of magnitude for the establishment probability of single mutations 2mcrc/σ. The estimates (0.14 and 0.02; table 2) suggest that our approximation r/σ ≪ 1 (at several points in the above theory) is reasonable, for this experiment at least. It is striking that the two replicate experiments gave widely differing estimates of θ, with a consistently lower value in experiment 1 than in experiment 2 (for both control and treatment). Day or batch effects could have induced a reduction in the growth rates of all genotypes (including control) or an increase in the reproductive variance.
estimates the overall rate of rescue per individual inoculated (in the stress, table 1), whereas θc estimates 2mcrc/σ in the control treatment, i.e. a measure of the probability of persistence from a single copy in this environment and for the wild-type genotype. The control experiment is therefore useful too, by giving an order of magnitude for the establishment probability of single mutations 2mcrc/σ. The estimates (0.14 and 0.02; table 2) suggest that our approximation r/σ ≪ 1 (at several points in the above theory) is reasonable, for this experiment at least. It is striking that the two replicate experiments gave widely differing estimates of θ, with a consistently lower value in experiment 1 than in experiment 2 (for both control and treatment). Day or batch effects could have induced a reduction in the growth rates of all genotypes (including control) or an increase in the reproductive variance.
Figure 3.
Relationship between inoculum size and rescue probability in yeast. The probability of a rescue 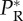 versus size of the inoculum No is shown for yeast populations faced with a salt stress plus dilutions or in a control experiment in standard yeast proteose dextrose medium [13]. The open circles are the results of the control experiment and the filled circles correspond to the salt treatment. The lines show the fit of equation (3.10), fitted using the corresponding MLE for θ as given in table 2: dashed lines: control with
versus size of the inoculum No is shown for yeast populations faced with a salt stress plus dilutions or in a control experiment in standard yeast proteose dextrose medium [13]. The open circles are the results of the control experiment and the filled circles correspond to the salt treatment. The lines show the fit of equation (3.10), fitted using the corresponding MLE for θ as given in table 2: dashed lines: control with 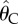 , plain lines: salt treatment (rescue) with
, plain lines: salt treatment (rescue) with  . The two plots correspond to two replicate experiments.
. The two plots correspond to two replicate experiments.
Table 2.
Estimates from the fit of  in figure 2. The deviance D of the model (2(log-likelihood(saturated model)–log-likelihood(model)) is used to test for the goodness of fit (saturated model: with binomial error and arbitrary P for each data point). If the model with
in figure 2. The deviance D of the model (2(log-likelihood(saturated model)–log-likelihood(model)) is used to test for the goodness of fit (saturated model: with binomial error and arbitrary P for each data point). If the model with  is as accurate as the saturated model then
is as accurate as the saturated model then  where p − 1 = 9, here, is the number of data points fitted (p=10) minus the number of parameters in the model evaluated (here only one parameter:
where p − 1 = 9, here, is the number of data points fitted (p=10) minus the number of parameters in the model evaluated (here only one parameter:  ). The p-value
). The p-value  provides a goodness of fit test of whether the model is significantly less accurate than the best fitting (saturated) model possible. In all cases here it is not (deviance is not significantly different from zero).
provides a goodness of fit test of whether the model is significantly less accurate than the best fitting (saturated) model possible. In all cases here it is not (deviance is not significantly different from zero).
| treatment | experiment | deviance D |  |
s.d. ( ) ) |
 |
|---|---|---|---|---|---|
| control | 1 | 0.52 | 0.023 | 0.027 | 0.99 |
| salt | 1 | 0.53 | 0.000045 | 0.000053 | 0.99 |
| control | 2 | 0.26 | 0.14 | 0.18 | 0.99 |
| salt | 2 | 0.008 | 0.0047 | 0.005 | 1 |
We hope that future studies will estimate the parameter 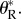 This will provide the basis for comparisons across experiments.
This will provide the basis for comparisons across experiments.
(ii). Effect of the evolutionary history before stress
In a recent experiment, Ramsayer et al. [20] studied the dynamics of evolutionary rescue in naturally declining populations of P. fluorescens facing three doses of streptomycin (50, 100 and 200 μg ml−1 plus a control). In addition, they studied the effect of the origin of the inoculum population, by comparing single ‘clonal’ populations and genetically diversified ‘admixture’ populations created from a mix of eight large cultures, putatively at evolutionary equilibrium (see §2). On this basis of protocol, de novo rescue should follow equation (3.3) in all treatments but the control, while the rescue from pre-existing mutations should follow equation (3.9) for the ‘clonal’ treatment and equation (3.6) or (3.7) for the ‘admixture’ treatment. In the latter case, we assume that stationary mutation–selection balance has been reached over 14 transfers prior to mixing, while the population had only undergone roughly 100 bacterial generations. This seems a reasonable approximation, and is surely the closest of the scenarios described above. There was no significant dose effect in this experiment, but a significantly higher rescue probability among the admixture treatment populations, relative to the clonal treatment [20]. These treatments differ only by the origin of the inoculum (their decay rates |ro| were indeed similar, data not shown). The observed effect must therefore come from differences in the rate of rescue from pre-existing mutations.
The quantitative impact of each treatment on rescue probabilities can be evaluated by looking at the inferred rate of rescue per individual inoculated  (from equation (3.1)), for each treatment. The initial population sizes were roughly equal in both treatments (No ≈ 3.18 × 109) and the proportions of rescued populations were
(from equation (3.1)), for each treatment. The initial population sizes were roughly equal in both treatments (No ≈ 3.18 × 109) and the proportions of rescued populations were  and
and  for the clonal and admixture treatments, respectively. The corresponding rates of rescue per inoculated individual give:
for the clonal and admixture treatments, respectively. The corresponding rates of rescue per inoculated individual give:  for the clonal treatment and
for the clonal treatment and  for the admixture treatment. According to our previous results, the rates of rescue from standing variance plus de novo mutations are
for the admixture treatment. According to our previous results, the rates of rescue from standing variance plus de novo mutations are  in the clonal treatment (equations (3.3)–(3.9)), and
in the clonal treatment (equations (3.3)–(3.9)), and  in the admixture treatment (equations (3.3)–(3.7)). Several parameters in these expressions can be estimated from the experiment. In the clonal treatment, populations were produced by inoculating 20 μl of stationary density cultures into 2 ml King's B medium (KB) grown from a single clone isolated on a Petri dish. The culture was grown back to stationary density, then 20 μl of this culture was inoculated into the stressful conditions. As a first (rough) approximation, we ignore possible variation within the first clonal 20 μl inoculum. If N1 is the stationary density (cells per ml) in KB, the initial number of cells grown was no = 0.02 N1 from the 20 μl clonal inoculum, the rescue population was initiated by diluting 2 ml of stationary culture to 20 μl (N1/no = 1/100) yielding an initial population size No = 0.02N1 = no. Overall, the factor in equation (3.9) is therefore log(N1/no) = log(100) = 4.6. For de novo rescue predictions, the distribution of |ro| among extinction trajectories can be evaluated by fitting exponential decay curves on replicate extinct population trajectories (data not shown). The resulting estimates give roughly 1/|ro|≈2.2, with potential variation within [1.2,9.5] (based the observed 95% CI for |ro| estimates). Putting these numbers together, we are left with three unknowns (
in the admixture treatment (equations (3.3)–(3.7)). Several parameters in these expressions can be estimated from the experiment. In the clonal treatment, populations were produced by inoculating 20 μl of stationary density cultures into 2 ml King's B medium (KB) grown from a single clone isolated on a Petri dish. The culture was grown back to stationary density, then 20 μl of this culture was inoculated into the stressful conditions. As a first (rough) approximation, we ignore possible variation within the first clonal 20 μl inoculum. If N1 is the stationary density (cells per ml) in KB, the initial number of cells grown was no = 0.02 N1 from the 20 μl clonal inoculum, the rescue population was initiated by diluting 2 ml of stationary culture to 20 μl (N1/no = 1/100) yielding an initial population size No = 0.02N1 = no. Overall, the factor in equation (3.9) is therefore log(N1/no) = log(100) = 4.6. For de novo rescue predictions, the distribution of |ro| among extinction trajectories can be evaluated by fitting exponential decay curves on replicate extinct population trajectories (data not shown). The resulting estimates give roughly 1/|ro|≈2.2, with potential variation within [1.2,9.5] (based the observed 95% CI for |ro| estimates). Putting these numbers together, we are left with three unknowns ( ) and two estimates (
) and two estimates ( ). However, if we neglect the variation in the mutation rate to resistance, per unit time, due to environmental change, then
). However, if we neglect the variation in the mutation rate to resistance, per unit time, due to environmental change, then 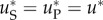 and we have two unknowns and two estimates:
and we have two unknowns and two estimates:  and
and  Solving with the mean or minimum/maximum values of 1/|ro| gives
Solving with the mean or minimum/maximum values of 1/|ro| gives  with range [2.9 × 10−11, 3.4 × 10−11] and
with range [2.9 × 10−11, 3.4 × 10−11] and  with range [0.0077, 0.0089].
with range [0.0077, 0.0089].
This is by no means a conclusive test of the theory, more an example of how it can be used to estimate several key quantities. It has the key drawback (i) of ignoring the effect of the environment on birth rates and consequently on mutation rates (uS = uP) and (ii) of neglecting rescue in two steps, so that the probability of de novo rescue is underestimated a priori. Yet, these rough estimates do seem at least of a reasonable order of magnitude. The estimate for 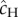 seems reasonable, based on the mean selection coefficients among random mutations, for Escherichia coli in optimal conditions [36] (
seems reasonable, based on the mean selection coefficients among random mutations, for Escherichia coli in optimal conditions [36] ( ). For
). For  a mutation rate to rescue of 10−11 is very small, but it is in fact not unrealistic. The rate of resistance to antibiotic stress in bacteria depends on the dose, the antibiotic and the species/genotype considered, but it often lies within [10−5 − 10−9]. Further, recalling that
a mutation rate to rescue of 10−11 is very small, but it is in fact not unrealistic. The rate of resistance to antibiotic stress in bacteria depends on the dose, the antibiotic and the species/genotype considered, but it often lies within [10−5 − 10−9]. Further, recalling that  is reduced by a factor
is reduced by a factor 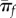 relative to the rate of resistance mutation, this rate may be of reasonable order. Overall, the observed discrepancy between the treatments is thus consistent with resistance costs of a few per cent and a very low resistance mutation rate.
relative to the rate of resistance mutation, this rate may be of reasonable order. Overall, the observed discrepancy between the treatments is thus consistent with resistance costs of a few per cent and a very low resistance mutation rate.
4. Discussion
In this paper, we have derived the probability of evolutionary rescue in various biological situations (summarized in table 1). The results are general for a density-independent demographic model with smooth variation (figure 1c); they allow for arbitrary variation in the demographic parameters (r,σ) among lineages produced by random mutation. We generalize several results already obtained by Orr & Unckless [9] in a discrete-time model of geometric growth and decay. All results were consistent with exact simulations where individuals either reproduced by binary fission or died (birth–death process). In general, this probability takes the form  (equation (3.1) and figure 2), where
(equation (3.1) and figure 2), where  is a rate of rescue mutation per inoculated individual. This is confirmed by empirical relationships between
is a rate of rescue mutation per inoculated individual. This is confirmed by empirical relationships between  and No in the yeast (table 2 and figure 3). To go further in the comparison of these predictions with experimental data, we now discuss several methodological issues concerning both evolutionary rescue experiments and modelling. We hope that these comments stimulate further experiments and strengthen the quantitative validation of models in this field of research.
and No in the yeast (table 2 and figure 3). To go further in the comparison of these predictions with experimental data, we now discuss several methodological issues concerning both evolutionary rescue experiments and modelling. We hope that these comments stimulate further experiments and strengthen the quantitative validation of models in this field of research.
(a). Assessing rescue probability
Measuring the probability of rescue is not that straightforward. First, one must choose the time period over which populations can be said to be either doomed or rescued. Such a decision can be better informed by measures of population size over time (rescue trajectories). One must also distinguish genetic rescue from plastic responses, which may be important [37] and will display different dynamics (detailed in Chevin et al. [38]).
(b). Measuring the key parameters
We have shown how rescue probabilities in different conditions could be used to infer key parameters of the process (rate of rescue mutations u* and harmonic mean of resistance cost cH, demographic stochasticity rc/σ). In order to perform such inferences, key parameters must be measured in rescue experiments: the inoculum size (No), the rate of decay of the wild-type under stress ( ), the dilution factor and dilution times imposed for bottlenecked populations (D and τ), and, in the case of growth from a single clone, the initial number of individuals and dilution applied, if any (no, N1, No).
), the dilution factor and dilution times imposed for bottlenecked populations (D and τ), and, in the case of growth from a single clone, the initial number of individuals and dilution applied, if any (no, N1, No).
More powerful tests of our theory would be possible by measuring these parameters by independent methods and comparing the two inferences. Measures of mutation rates to resistance prior to stress (uP) are possible using fluctuation tests (e.g. following the methods in recent studies [35,39]). However, quantitatively precise estimates may prove difficult to obtain: some estimates are biased by resistance cost c [35] and persistence probability πf (‘plating efficiency’ [39]), which must then be estimated too. The distribution of the cost of resistance (hence its harmonic mean cH) can be estimated by screening for random resistance mutations and then estimating their selection coefficient relative to the sensitive parent, in the permissive environment.
(c). High-throughput measurement of mutant parameters
What would also be required to fully test our predictions is a measure of the joint distribution of r and σ among a set of resistance mutations, in order to estimate  or
or  (i.e. the mean establishment probability among resistant mutants). This would ideally be carried out in high-throughput, with many replicate populations (and thus possibly many different mutants) jointly estimated. Measuring demographic stochasticity in microbial cells requires measuring the growth and death rate of cells (σ = r + d). Measuring death rates may be tricky, but the growth rate r of any given mutant can be obtained from time-series of plate counts [20] or of macroscopic measurements such as optical density [13]. However, plate counts are time consuming and thus limit the feasibility of high-resolution time-series, whereas optical density may not accurately estimate the cell density in environments causing high mortality (typical of rescue experiments), because dead cells can contribute to optical density for some time after death. Alternative high-throughput methods exist that measure both the rate of division and of death in bacterial populations, using fluorescence and luminescence signals from genetically marked strains [40]. This would allow estimation of joint distribution of (r,σ) among a set of screened resistant mutants, and thus parametrization of the predictions proposed in table 1.
(i.e. the mean establishment probability among resistant mutants). This would ideally be carried out in high-throughput, with many replicate populations (and thus possibly many different mutants) jointly estimated. Measuring demographic stochasticity in microbial cells requires measuring the growth and death rate of cells (σ = r + d). Measuring death rates may be tricky, but the growth rate r of any given mutant can be obtained from time-series of plate counts [20] or of macroscopic measurements such as optical density [13]. However, plate counts are time consuming and thus limit the feasibility of high-resolution time-series, whereas optical density may not accurately estimate the cell density in environments causing high mortality (typical of rescue experiments), because dead cells can contribute to optical density for some time after death. Alternative high-throughput methods exist that measure both the rate of division and of death in bacterial populations, using fluorescence and luminescence signals from genetically marked strains [40]. This would allow estimation of joint distribution of (r,σ) among a set of screened resistant mutants, and thus parametrization of the predictions proposed in table 1.
(d). Stochastic dynamics of rescue trajectories
Observed empirical dynamics of rescue could yield valuable tests of several of our model's assumptions, the first being the assumption of exponential decay, easily testable from the decay dynamics of doomed populations. These dynamics, which can be obtained from plate counts [20], or fluoro-luminometric measurements, could then potentially shed light on the relative contribution of different types of rescues (de novo single versus two steps versus pre-existing mutations). A full stochastic description of these dynamics, in the present analytical framework, will be the scope of future work.
(e). Studying other factors
In our data analysis, we focused on the effect of population size or evolutionary history of the population before stress. Other factors could be studied as well, and compared with the present predictions. The effect of the rate of decay could be studied by varying the intensity of stress, although one would have to keep in mind that not only |ro| varies in this case, but also  and
and  . Note also that according to whether stress decreases the birth rate or increases the death rate, it may affect (or not, respectively) the mutation rates to resistance uP and uS, e.g. with binary fission the mutation rate scales with the birth rate. The decay rate could also be studied in bottlenecked populations, where it could be varied using different dilutions D: with high dilution, fewer genotypes are potentially resistant (fewer have mr > 0). In general, qualitatively different stresses will be easier to compare by setting each stress level to obtain the same decay rate |ro|, so that the purely demographic impact of each stress is the same. This could be done, for example, with comparisons of multiple versus single antibiotics [41], or biotic versus abiotic stresses [15].
. Note also that according to whether stress decreases the birth rate or increases the death rate, it may affect (or not, respectively) the mutation rates to resistance uP and uS, e.g. with binary fission the mutation rate scales with the birth rate. The decay rate could also be studied in bottlenecked populations, where it could be varied using different dilutions D: with high dilution, fewer genotypes are potentially resistant (fewer have mr > 0). In general, qualitatively different stresses will be easier to compare by setting each stress level to obtain the same decay rate |ro|, so that the purely demographic impact of each stress is the same. This could be done, for example, with comparisons of multiple versus single antibiotics [41], or biotic versus abiotic stresses [15].
(f). More realistic demographic models for natural populations
Our model makes simplifying assumptions in order to analytically model a fully stochastic rescue process. A first assumption is that lineages are independent of each other, both genetically (no genetic exchange or sex) or demographically (no density-dependence). In natural populations, these assumptions may not hold due to the complexities of the environment, in particular when population dynamics depend on some key resource dynamics (e.g. host cells in the case of viruses [4]). Including density-dependence in this stochastic framework may be possible using the logistic stochastic process proposed in Lambert [42], or the diffusion analysis in Uecker & Hermisson [22]. The effect of sex should also be studied in these stochastic models. However, these assumptions may also not be so limiting, at least in some natural contexts: (i) when the genetic basis for the rescue is a single gene the system may behave almost as an asexual, (ii) when densities in the stress are low, density-independent growth may be driving the demographic dynamics. All these issues are a matter of experimental testing of course (for an example of rescue experiments with a sexual species, see Agashe and co-workers [16,17]).
A second key assumption of our model is that the environment changes abruptly from one where the population thrives to one where it starts to decline, the rate of decline remaining constant thereafter. A treatment of a more continuous environmental change is also necessary and would introduce time inhomogeneous processes, which are more challenging mathematically. Predictions from such extensions can be tested in controlled experiments where the stress increases/decreases at a desired rate.
Overall, we have presented a framework to address quantitative aspects of the process of evolutionary rescue, by comparing data from experimental evolution with simple yet relatively general predictions from stochastic process theory. We believe that an ongoing exchange between experimenters and theoreticians can help to better understand and predict the process and outcome of evolutionary rescue.
Acknowledgements
We thank Luis Miguel Chevin and David Waxman and three anonymous referees for their help and suggestions to improve the manuscript. Amaury Lambert pointed out two key results on Feller diffusions: the exponential distribution of the asymptotic process and the link between the cumulated size to extinction and the first passage time of a Brownian motion, via Lamperti transforms. Andrew Gonzalez kindly provided the data on the yeast rescue experiments. This work was funded by ANR EVORANGE (ANR-09-PEXT-011), G.M. and O.R. were funded by RTRA BIOFIS (INRA 065609) and G.M. was funded by PEPII (INSB-INEE-INSMI) from CNRS. This is publication ISEM 2012-130.
References
- 1.Millennium Ecosystem Assessment 2005. Ecosystems and human well-being: synthesis. Washington, DC: Island Press [Google Scholar]
- 2.Skelly DK, Joseph LN, Possingham HP, Freidenburg LK, Farrugia TJ, Kinnison MT, Hendry AP. 2007. Evolutionary responses to climate change. Conserv. Biol. 21, 1353–1355 10.1111/j.1523-1739.2007.00764.x (doi:10.1111/j.1523-1739.2007.00764.x) [DOI] [PubMed] [Google Scholar]
- 3.MacLean RC, Hall AR, Perron GG, Buckling A. 2010. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat. Rev. Genet. 11, 405–414 10.1038/nrg2778 (doi:10.1038/nrg2778) [DOI] [PubMed] [Google Scholar]
- 4.Ribeiro RM, Bonhoeffer S. 2000. Production of resistant HIV mutants during antiretroviral therapy. Proc. Natl Acad. Sci. USA 97, 7681–7686 10.1073/pnas.97.14.7681 (doi:10.1073/pnas.97.14.7681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch M, Lande R. 1993. Evolution and extinction in response to environmental change. In Biotic interactions and global climate change (eds Kareiva PM, Kingsolever JG, Huey RB.), pp. 234–250 Sunderland, MA: Sinauer Associates [Google Scholar]
- 6.Gomulkiewicz R, Houle D. 2009. Demographic and genetic constraints on evolution. Am. Nat. 174, E218–E229 10.1086/645086 (doi:10.1086/645086) [DOI] [PubMed] [Google Scholar]
- 7.Bell G. 2012. Evolutionary rescue and the limits of adaptation. Phil. Trans. R. Soc. B 368, 20120080. 10.1098/rstb.2012.0080 (doi:10.1098/rstb.2012.0080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomulkiewicz R, Holt RD. 1995. When does evolution by natural-selection prevent extinction. Evolution 49, 201–207 10.2307/2410305 (doi:10.2307/2410305) [DOI] [PubMed] [Google Scholar]
- 9.Orr HA, Unckless RL. 2008. Population extinction and the genetics of adaptation. Am. Nat. 172, 160–169 10.1086/589460 (doi:10.1086/589460) [DOI] [PubMed] [Google Scholar]
- 10.Babiker HA, Hastings IM, Swedberg G. 2009. Impaired fitness of drug-resistant malaria parasites: evidence and implication on drug-deployment policies. Expert Rev. Anti-Infective Ther. 7, 581–593 10.1586/eri.09.29 (doi:10.1586/eri.09.29) [DOI] [PubMed] [Google Scholar]
- 11.Holt RD, Gomulkiewicz R. 1997. The evolution of species niches: a population dynamic perspective. In Case studies in mathematical modelling: ecology, physiology, and cell biology (eds Othmer RG, Adler FR, Lewis MA, Dallon JC.), pp. 25–50 Englewood Cliffs, NJ: Prentice Hall [Google Scholar]
- 12.Bell G. 2008. Experimental evolution. Heredity 100, 441–442 10.1038/hdy.2008.19 (doi:10.1038/hdy.2008.19) [DOI] [PubMed] [Google Scholar]
- 13.Bell G, Gonzalez A. 2009. Evolutionary rescue can prevent extinction following environmental change. Ecol. Lett. 12, 942–948 10.1111/j.1461-0248.2009.01350.x (doi:10.1111/j.1461-0248.2009.01350.x) [DOI] [PubMed] [Google Scholar]
- 14.Bell G, Gonzalez A. 2011. Adaptation and evolutionary rescue in metapopulations experiencing environmental deterioration. Science 332, 1327–1330 10.1126/science.1203105 (doi:10.1126/science.1203105) [DOI] [PubMed] [Google Scholar]
- 15.Zhang QG, Buckling A. 2011. Antagonistic coevolution limits population persistence of a virus in a thermally deteriorating environment. Ecol. Lett. 14, 282–288 10.1111/j.1461-0248.2010.01586.x (doi:10.1111/j.1461-0248.2010.01586.x) [DOI] [PubMed] [Google Scholar]
- 16.Agashe D, Falk JJ, Bolnick DI. 2011. Effects of founding genetic variation on adaptation to a novel resource. Evolution 65, 2481–2491 10.1111/j.1558-5646.2011.01307.x (doi:10.1111/j.1558-5646.2011.01307.x) [DOI] [PubMed] [Google Scholar]
- 17.Agashe D. 2009. The stabilizing effect of intraspecific genetic variation on population dynamics in novel and ancestral habitats. Am. Nat. 174, 255–267 10.1086/600085 (doi:10.1086/600085) [DOI] [PubMed] [Google Scholar]
- 18.Perron GG, Gonzalez A, Buckling A. 2007. Source-sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc. R. Soc. B 274, 2351–2356 10.1098/rspb.2007.0640 (doi:10.1098/rspb.2007.0640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin BR, Lipsitch M, Perrot V, Schrag S, Antia R, Simonsen L, Moore Walker N, Stewart FM. 1997. The population genetics of antibiotic resistance. Clin. Infect. Dis. 24, S9–S16 10.1093/clinids/24.supplement_1.S9 (doi:10.1093/clinids/24.supplement_1.S9) [DOI] [PubMed] [Google Scholar]
- 20.Ramsayer J, Kaltz O, Hochberg ME. In press Evolutionary rescue in populations of Pseudomonas fluorescens across an antibiotic gradient. Evol. Appl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight TM, Barfield M, Holt RD. 2008. Evolutionary dynamics as a component of stage-structured matrix models: an example using Trillium grandiflorum. Am. Nat. 172, 375–392 10.1086/589898 (doi:10.1086/589898) [DOI] [PubMed] [Google Scholar]
- 22.Uecker H, Hermisson J. 2011. On the fixation process of a beneficial mutation in a variable environment. Genetics 188, 915–930 10.1534/genetics.110.124297 (doi:10.1534/genetics.110.124297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feller W. 1951. Diffusion processes in genetics. In Proceedings of the Second Berkeley Symposium on Mathematical Statikstics and Probability Berkeley, CA: University of California Press [Google Scholar]
- 24.Shpak M. 2007. Selection against demographic stochasticity in age-structured populations. Genetics 177, 2181–2194 10.1534/genetics.107.080747 (doi:10.1534/genetics.107.080747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin G, Gandon S. 2010. Lethal mutagenesis and evolutionary epidemiology. Phil. Trans. R. Soc. B 365, 1953–1963 10.1098/rstb.2010.0058 (doi:10.1098/rstb.2010.0058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfram Research I 2010. Mathematica edition: version 8.0. Champaign, IL: Wolfram Research [Google Scholar]
- 27.Gillespie DT. 1977. Exact stochastic simulation of coupled chemical-reactions. J. Phys. Chem. 81, 2340–2361 10.1021/j100540a008 (doi:10.1021/j100540a008) [DOI] [Google Scholar]
- 28.Galassi M., Davies J, Theiler J, Gough B, Jungman G, Alken P, Booth M, Rossi F. 2009. GNU scientific library reference manual, 3rd edn Bristol, UK: Network Theory [Google Scholar]
- 29.R Development Core Team. 2007. R: a language and environment for statistical computing. Vienna, Austria: R Development Core Team. [Google Scholar]
- 30.Lambert A. 2006. Probability of fixation under weak selection: a branching process unifying approach. Theor. Popul. Biol. 69, 419–441 10.1016/j.tpb.2006.01.002 (doi:10.1016/j.tpb.2006.01.002) [DOI] [PubMed] [Google Scholar]
- 31.Martin G, Aguilée R, Chevin LM, Ronce O. Evolutionary rescue over a fitness landscape. doi: 10.1534/genetics.118.300908. In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguilee R, Claessen D, Lambert A. 2009. Allele fixation in a dynamic metapopulation: founder effects versus refuge effects. Theor. Popul. Biol. 76, 105–117 10.1016/j.tpb.2009.05.003 (doi:10.1016/j.tpb.2009.05.003) [DOI] [PubMed] [Google Scholar]
- 33.Wahl LM, Gerrish PJ. 2001. The probability that beneficial mutations are lost in populations with periodic bottlenecks. Evolution 55, 2606–2610 [DOI] [PubMed] [Google Scholar]
- 34.Heffernan JM, Wahl LM. 2002. The effects of genetic drift in experimental evolution. Theor. Popul. Biol. 62, 349–356 10.1016/S0040-5809(02)00002-3 (doi:10.1016/S0040-5809(02)00002-3) [DOI] [PubMed] [Google Scholar]
- 35.Jaeger G, Sarkar S. 1995. On the distribution of bacterial mutants: the effects of differential fitness of mutants and non-mutants. Genetica 96, 217–223 10.1007/BF01439575 (doi:10.1007/BF01439575) [DOI] [Google Scholar]
- 36.Elena SF, Ekunwe L, Hajela N, Oden SA, Lenski RE. 1998. Distribution of fitness effects caused by random insertion mutations in Escherichia coli. Genetica 103, 349–358 10.1023/A:1017031008316 (doi:10.1023/A:1017031008316) [DOI] [PubMed] [Google Scholar]
- 37.Levin BR, Rozen DE. 2006. Opinion: non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4, 556–562 10.1038/nrmicro1445 (doi:10.1038/nrmicro1445) [DOI] [PubMed] [Google Scholar]
- 38.Chevin LM, Romain G, Richard G, Robert H, Simon F. 2012. Phenotypic plasticity in evolutionary rescue experiments. Phil. Trans. R. Soc. B 368, 20120089. 10.1098/rstb.2012.0089 (doi:10.1098/rstb.2012.0089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerrish P. 2008. A simple formula for obtaining markedly improved mutation rate estimates. Genetics 180, 1773–1778 10.1534/genetics.108.091777 (doi:10.1534/genetics.108.091777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehtinen J, Virta M, Lilius EM. 2003. Fluoro-luminometric real-time measurement of bacterial viability and killing. J. Microbiol. Methods 55, 173–186 10.1016/S0167-7012(03)00134-9 (doi:10.1016/S0167-7012(03)00134-9) [DOI] [PubMed] [Google Scholar]
- 41.Ward H, Perron GG, Maclean RC. 2009. The cost of multiple drug resistance in Pseudomonas aeruginosa. J. Evol. Biol. 22, 997–1003 10.1111/j.1420-9101.2009.01712.x (doi:10.1111/j.1420-9101.2009.01712.x) [DOI] [PubMed] [Google Scholar]
- 42.Lambert A. 2005. The branching process with logistic growth. Ann. Appl. Probability 15, 1506–1535 10.1214/105051605000000098 (doi:10.1214/105051605000000098) [DOI] [Google Scholar]