Abstract
The current rapid rate of human-driven environmental change presents wild populations with novel conditions and stresses. Theory and experimental evidence for evolutionary rescue present a promising case for species facing environmental change persisting via adaptation. Here, we assess the potential for evolutionary rescue in wild vertebrates. Available information on evolutionary rescue was rare and restricted to abundant and highly fecund species that faced severe intentional anthropogenic selective pressures. However, examples from adaptive tracking in common species and genetic rescues in species of conservation concern provide convincing evidence in favour of the mechanisms of evolutionary rescue. We conclude that low population size, long generation times and limited genetic variability will result in evolutionary rescue occurring rarely for endangered species without intervention. Owing to the risks presented by current environmental change and the possibility of evolutionary rescue in nature, we suggest means to study evolutionary rescue by mapping genotype → phenotype → demography → fitness relationships, and priorities for applying evolutionary rescue to wild populations.
Keywords: wildlife, vertebrates, conservation, genetic rescue, global changes
1. Introduction
Facing increasing human-driven changes, several populations and species now experience a mismatch between locally adapted traits and novel conditions, leading to an increase in mortality, and a decrease in abundance [1,2]. In response, many researchers seek to identify mechanisms that may allow species and populations to persist under changed conditions [3,4]. As the primary mechanism for species persistence during environmental changes has been adaptation by natural selection, biologists have encouraged the integration of evolutionary ecology concepts into conservation biology [3,5]. Despite being steeped in evolutionary theory, Conservation biology [6] has historically focused primarily on preserving neutral genetic variation [7,8], rather than evaluating adaptations which may prevent extinction. Preserving neutral genetic variation, however, ignores the possible link between phenotypes and demography which is potentially a major concern when protecting populations [9]. A growing body of evidence suggests that evolutionary rescue (henceforth ER), which does link evolutionary potential (genetic variation) to demographic change can occur in theory [10,11] and in the laboratory [12] (but see [13]). Nonetheless, it remains unclear whether ER is prevalent in nature, and under what circumstances evolution by natural selection may prevent extirpation or extinction despite environmental change.
The gravity of current environmental change and the risk it presents to species of conservation concern justifies the review of the evidence for ER in nature and a discussion of possible approaches for studying applied ER. Here, we first present the general requirements for ER. We then review the limited empirical research on this topic to assess its importance for persistence of wild vertebrates, with emphasis on species of conservation concern. We focus on studies of adaptive tracking and genetic rescue in vertebrate populations that provide convincing evidence that the mechanisms of ER can occur in nature. Finally, we discuss the application of ER in conservation and describe approaches available to study ER under field conditions.
2. Requirements for evolutionary rescue
Classically, ER has been considered a special case of adaptive tracking that is too slow to prevent an initial decline in population size. Documentation of ER requires three conditions. Initially, (i) there must be a decline in population growth (λ < 1) owing to changes in the environment [10,11]. Subsequently, (ii) the population will require a rapid change in the phenotype, either via selection-induced changes in the standing genetic variation or through novel advantageous mutations. Without a change in the phenotype the population will go extinct. With ER, (iii) a recovery in population growth should be observed. A population rescue could also result from within-generation changes in the phenotype owing to plasticity or other ecological factors [12], and we refer to these as ecological rescues. Although the mechanisms underlying the change in phenotypic distribution differ between ER and ecological rescue, both result in a U-shaped temporal trend of population size [10]. A U-shaped trend might, however, not be evident in all the cases of rescue. Environmental changes may be gradual, persistent and directional, resulting in either adaptation which is sufficiently rapid [13–15] such that population fitness remains less than unity, or failure to adapt resulting in extinction. The U-shaped trend might also be masked by density dependence or other demographic processes that buffer population fitness in ER when environmental change is gradual or constant [16]. Here, we focus primarily on ER; however, in many empirical examples ecological and evolutionary mechanisms of response might occur together and disentangling their effects is probably tricky.
3. Evidence for evolutionary rescue in vertebrates
Genetic adaptations to drastic environmental changes have been reported in a number of taxa in natural conditions (bacteria [17], plants [18], mammals [19], birds [20] and invertebrates [21]; see also [22] for a review). It has been shown that evolution can occur on an ecological time-scale (see [5,23,24] for examples). Thus, ER could, in theory, be an important mechanism by which wild species can adapt to human-induced changes [25,26]. Although ER is theoretically possible and has been shown to prevent extinction in laboratory studies [25], we are unaware of similar examples in wild vertebrate populations. Arguably, the main reason why ER remains largely undocumented in nature is that obtaining data on both population dynamics and evolutionary changes is logistically difficult. Few studies have obtained the required data on genotype, phenotype and population parameters [27], and none of these study populations faced extinction.
In this article, we argue that the best support for the potential for ER to aid population recovery in nature come from pesticide resistance in rats and biological control in European rabbits, adaptive tracking and genetic rescue. The next section (§3a) discusses each of these cases and underlines their relevance for our understanding of ER.
(a). Anti-vitamin K resistance in rodents and biological control in rabbits
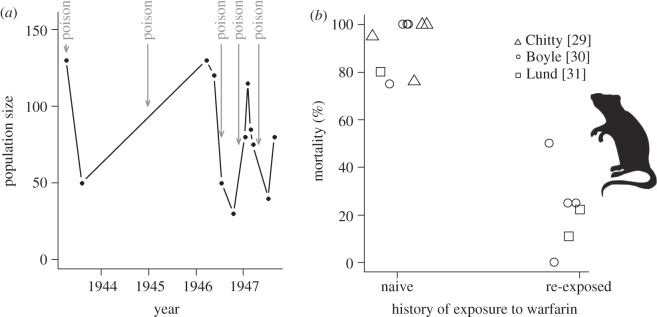
For rodent populations (e.g. Rattus norvegicus, R. rattus), the drastic environmental change consisted of exposure to anti-vitamin K pesticides (warfarin and bromadiolone) which caused mortality from lethal bleeding. Early warfarin use was presumed to have resulted in sharp declines in rat abundance (some evidence presented in figure 1a, although rat population data was unavailable prior to 1942). It was not long until individuals from rat populations thought to have been previously exposed to warfarin were observed with purportedly lower mortality rates upon re-exposure than naive populations (figure 1b) [29–31]. Adaptation was thought to have occurred partly because of selection on standing genetic variation and to the occurrence of up to six new mutations [32,33], which are preventing anti-vitamin K blocking [34,35]. Resistance to warfarin provides evidence that new mutation can rescue a population from new environmental constraint.
Figure 1.
(a) Effect of warfarin treatment on abundance of rats (Rattus norvegicus) in Baltimore, ML, USA (adapted from Jackson [28]). (b) Mortality rates of rats from populations of apparently non-resistant versus resistant individuals from Baltimore, Scotland and Denmark (triangle, circle and square, respectively). Populations purported to have been previously exposed to warfarin appeared to have exhibited lower mortality rates upon re-exposure.
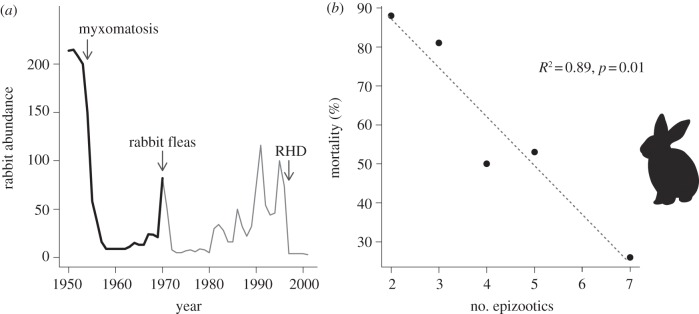
Adaptation also allowed the European rabbit to ‘escape’ biological control in Australia. Here, the drastic environmental change consisted of exposure to the myxoma virus (genus Leporipoxvirus) [19]. Initially, the virus killed 99 per cent of animals infected [36]. The virus spread quickly across the continent leading to a marked decline in rabbit abundance (figure 2). As predicted from theory [39], there was rapid selection for a less virulent myxoma strain; one which caused a lower mortality rate (≤90%) and took longer to cause mortality [38]. In parallel, rabbits evolved resistance to myxoma. Laboratory experiments showed that individuals from previously exposed populations were able to recover from myxoma infection that was always lethal in naive rabbits [38]. Because it involved coevolution, the case of rabbits and myxoma virus is not as clear an instance of ER as the anti-vitamin K resistance in rats. The virus now constitutes a sustained environmental stress, mostly by decreasing juvenile survival [40], but it is no longer an extinction threat for rabbits. Another example of environmental change for Australian rabbits is haemorrhagic disease virus from which the population has yet to recover to pre-exposure population sizes (figure 2) [41,42].
Figure 2.
(a) Fluctuations in rabbit abundance in Australia between 1950 and 2000 during three critical attempts at biological control: 1950 myxomatosis; 1970 rabbit fleas which increased the efficacy of myxomatosis transmission; and 1995 rabbit haemorrhagic disease (RHD). Adapted from Saunders et al. [37]. (b) Changing mortality rate (%) for rabbit populations repeatedly exposed to myxomatosis. The significant decrease in mortality is often used as evidence of coevolution between host and virus. Adapted from Best & Kerr [38].
These examples satisfy the three conditions of ER and support the contention that evolution can rescue wild populations from drastic environmental changes. However, while the ER concept has been proposed in a context where species are rescued from extinction [25], these two examples involve very abundant and highly fecund mammals: not traits associated with most vertebrates facing extinction or extirpation.
(b). Adaptive tracking
Adaptive tracking is the response to gradual, rather than abrupt, environmental change in selection on standing genetic variation that offsets population decline (e.g. the characteristic U-shaped longitudinal trajectory is lacking). However, in contrast to the examples above, it only provides partial evidence for ER in nature. Adaptive tracking has been quantified in several long-term studies, including the Galapagos finches (Geospiza spp.) [43,44], great tits (Parus major) [45,46] and red squirrels (Tamisciurus hudsonius) [47,48]. These population have all tracked some form of environmental change, such as gradual and sustained climate change [46,47] or abrupt, if not sustained, stochastic weather events [49]. Adaptive tracking, however, is only relevant to ER if being unable to track the environmental change leads to extinction. In the previous examples, there is no direct evidence that failing to track the environmental changes would have resulted in extinction. The demographic impacts of maladaptation are indeed rarely evaluated (but see [50]). A case study comes from a pied flycatcher (Ficedula hypoleuca) population which appears maladapted to its changing environment [51,52]. This population seems unable to fully track the changing phenology of their primary food source as a result of warmer spring temperatures. Flycatchers are unable to time their arrival date in the breeding grounds with the emergence of caterpillars. Consequently, the population has declined [51] and shows no sign of rescue.
(c). Genetic rescue
Perhaps the best evidence that ER can be an important mechanism for vertebrate population persistence come from genetic rescue (reviewed in [53–55]) that improves the fitness of local and often small populations by introducing novel genetic material, often through ‘artificial’ gene flow [54]. Unlike ER, however, genetic rescue does not always result from directional changes in mean phenotype. Instead, the novel genetic material may increase mean fitness by increasing heterozygosity and reducing inbreeding depression, with no change to ecologically relevant phenotypes. For genetic rescue, new alleles are typically drawn from standing genetic variation over a larger geographical scale than the target population. Artificial immigration risks diluting local adaptations and might lead to outbreeding depression (see [56] for discussion), several examples of genetic rescue have been deemed successful.
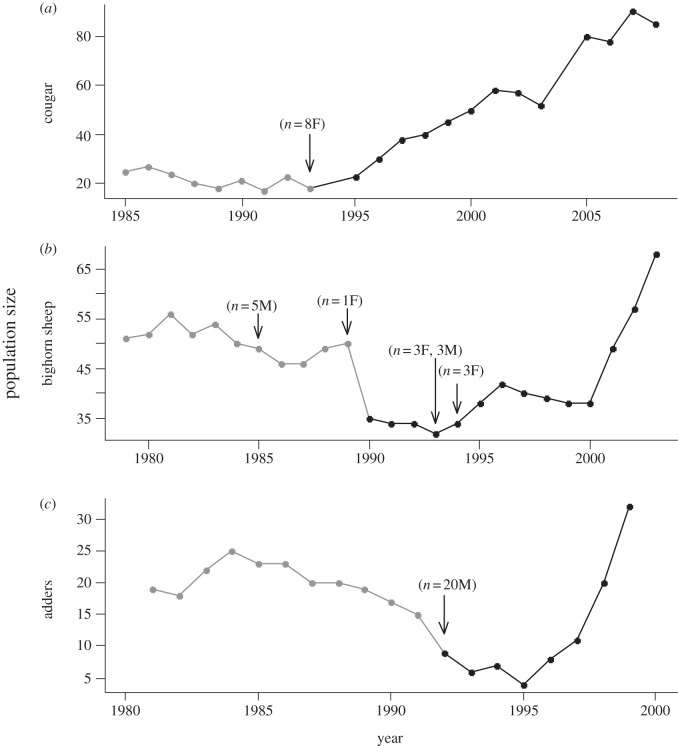
Most notably, genetic rescue has been credited with the restoration of populations of cougars in Florida (Puma concolor [55]), bighorn sheep (Ovis canadensis [57,58]) and adders (Viper berus [59]; figure 3). In all cases, populations were isolated and inbred. Deleterious alleles had reduced the reproductive success of individuals, leading to a decline in the population size. Because natural immigration was nearly impossible, researchers introduced novel genetic material by translocating individuals from other populations. Following admixture of local and introduced individuals, offspring production and survival increased, and populations recovered (see references above and figure 3). These examples illustrate the short time-scales (less than 10 years, which for bighorn sheep is approximately two generations) required for ‘new genes’ to reduce genetic load and allow a population to increase. More importantly, they underline the importance for conservation of the link between genetically based traits and fitness (e.g. male dominance rank, lamb birth weight, breeding date, see [57] for details). As population growth is a function of the number of individuals that survive and reproduce, quantifying the effect of ‘new genes’ on the probability of persistence is critical to evaluate the population-scale consequences of changes in the distribution of adaptive traits. It should be noted, however, that the effectiveness of genetic rescue is reduced if deteriorating ecological conditions persist (see [61] for example).
Figure 3.
Population size through time, including accumulation of deleterious alleles (grey) and increased reproductive success attributed to the introduction of new individuals and novel alleles (black arrows) for three examples of genetic rescue: (a) Cougars in Florida (Puma concolor) with the introduction of female immigrants from Texas (adapted from Hedrick & Fredrickson [55]); (b) novel genetic material introduced from male and female bighorn sheep into an isolated population (Ovis canadensis; adapted from Hogg et al. [57]); and (c) for an adder (Vipera berus) population (adapted from Madsen et al. [60]).
4. Applications: the importance of evolutionary rescue in conservation
Evolutionary rescue is of fundamental importance in evolutionary biology, but is it relevant to conservation? If it is relevant, would its inclusion change the practice of conservation? Many accepted practices in conservation are partly or mostly aimed at avoiding the erosion of genetic diversity, including the protection of corridors to allow gene flow, sustain large population sizes and avoid artificial selection [62]. The question we address here, however, is whether ER may assist the conservation of species facing human-induced environmental changes, such as global warming, the introduction of exotic competitors or diseases, or habitat alterations.
Although this question is of fundamental importance, so far only laboratory and theoretical work can be used to address it. For example, experimental research has suggested that in large populations, with minimal stochastic effects, ER will take approximately 25 generations [25]. As Bell & Gonzalez [25] underscored, exclusion of environmental stochasticity probably resulted in underestimation of the population size required for ER in variable environments. This was also emphasized by Willi & Hoffman [63] who documented population persistence using data from laboratory populations of Drosophila birchii facing environmental change. Although ER was not observed as all populations eventually went extinct, larger populations nonetheless tended to persist longer because of lower stochasticity and greater genetic variation [63].
Clearly, many factors will affect the time required for ER to occur, e.g. the extent of stress, standing variation, mating system. Here, we take the Bell & Gonzalez [25] estimate of 25 generation as a basis to reflect on the possible importance of rescue for wild species facing abrupt anthropogenic threats. Similar times, approximately 26 and 30 generations, have been observed for time to pesticide resistance in laboratory populations of bulb mites, Rhizogluphus robini [64] and Oriental fruit flies, Bactrocera dorsalis [65], respectively. For many long-lived species on the IUCN Red List, 25 generations represent a very long time. For example, estimates of time to ER would be approximately 141 years for the critically endangered African wild ass (Equus asinus) and approximately 597 years for vulnerable African elephants (Loxodonta africana; generation times from Gaillard et al. [66]). The contrast in the life histories of these species of conservation concern and Australian rabbits, where ER was documented, is striking. The rabbit, which can theoretically produce 11 litters per year [67] has one of the fastest life-histories among mammals [68]. Predicted time to rescue may be as little as approximately 4 years [66], although evidence from the attempted eradication of rabbits in Australia suggests that it may have taken 10–12 years (figure 2). This very crude assessment suggests that for most vertebrates of conservation concern confronted with strong environmental stress, predicted time to ER could be long in the absence of large amounts of within-population genetic variation. Very few declining species, with typically small population size, reduced genetic variation and strong stochasticity are likely to persist for that long under stressful conditions, especially given that endangered species typically face multiple and interacting threats [56].
Evolutionary rescue may be more probable when changes in the environment are gradual [16], albeit what constitutes ‘gradual’ will be scale-dependent and relate to generation time of species. Gradual changes, such as climate change, require the mean population phenotype of a trait under selection to constantly track a moving optimal phenotype, creating an inevitable lag because traits are never perfectly heritable [15]. Theoretical studies suggest that a population's trait cannot adapt at rates more than 10 per cent of the phenotypic standard deviation per generation [13,14]; furthermore, a population with a large difference between its current mean phenotype and a new optimal phenotype will be less likely to adapt to stochastic events [69].
The speed of adaption will greatly depend not only on the occurrence of new mutations but also on the standing additive genetic variation for adaptive traits and on constraints originating from genetic correlations among traits [70]. One could argue that the yeast used in the experiment [25] were evolving at a slow rate owing to lack of genetic variation resulting from use of bottlenecked populations with very low initial diversity and that 25 generations might not be a conservative figure. Some studies have even shown that evolutionary changes can occur within a generation in the field [49]; whether similar tempo of evolution is common is still unknown. The number of genes controlling the expression of key traits will also impact the speed of adaptation. Adaptation should be faster if strong selection is acting on oligogenic variation (see [71,72]) than if a number of genes affect the trait of interest. It is also becoming clear that phenotypic plasticity can lead to faster adaptive responses to environmental change [73,74]. Plastic responses for a trait might reduce the fitness costs of environmental change and increase population persistence despite slow genetic change [73,74]. For example, a review of human-induced rate of phenotypic changes concluded that changes are greater in human-disturbed contexts than in natural contexts, mainly through phenotypic plasticity [75].
The practice of conservation biology requires a blend of natural and social science, because few conservation initiatives are likely to succeed unless they have public support. Suggesting a widespread role for ER in conservation may lead to a false sense of security about the ability of endangered species to adapt to human-induced environmental change. Many groups in society would welcome an adaptationist argument as a reason to ignore human-induced environmental change. For example, it has been suggested that polar bears (Ursus maritimus) in western Hudson Bay may ‘adapt’ to climate change by increasing their consumption of terrestrial prey and possibly hibernate more like brown bears (Ursus arctos [76]). With a generation time of approximately 12 years [77] and accepting the estimate of 25 generations for ER (and also assuming limited additive genetic variation and plasticity), polar bears would require approximately 300 years to adapt. During those 300 years, the effects of climate change would have to be weak enough to allow the persistence of the population, and other anthropogenic threats would have to be eliminated. It is thus probable that, unless plasticity is especially effective in allowing adaptation, given the current population trend and unsustainable hunting quotas, polar bears will probably disappear from western Hudson Bay over a much shorter time-scale [77].
5. Future direction
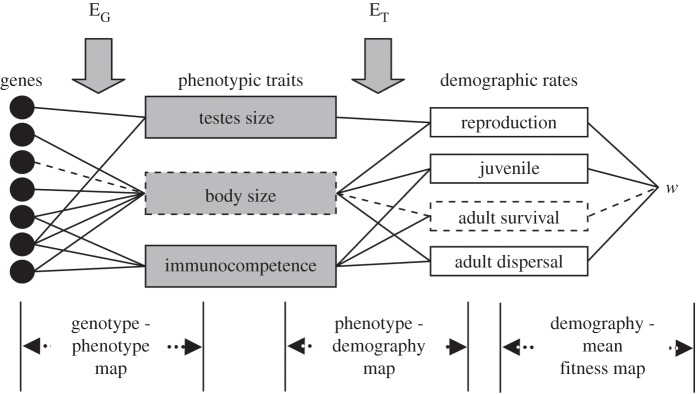
It is unlikely that ‘failed’ ER studies would be published as anything other than examples of extinction or extirpation. As such, any species extinction can be considered an example of failed ER. Given the number of extinctions observed in recent years [78], ER may be the exception rather than the norm. In this section, we discuss how to understand, test for and eventually predict evolutionary changes in the wild. To better understand the importance of ER for species persistence in a changing world, we require long-term studies with detailed information on genotype, phenotype, demography and population growth. As with other evolutionary changes, to understand the population level consequence of changing allele frequency we must know the links between the different levels of biological organisation illustrated in figure 4. Below, we describe the advances in methods and approaches which can elucidate these links.
Figure 4.
A schematic of the knowledge required to improve our understanding of evolutionary changes and of the potential for evolutionary rescue to occur in the wild. This demographic framework shows how multiple alleles can interact to produce phenotypic traits. These phenotypic traits influence demographic rates and fitness (w). A factor that affects the links between genes and phenotypic traits includes the influence of the environment on gene expression (EG). A factor that affects the link between phenotypic traits and demographic rates includes environmental effects on trait expression (ET). Although many pathways exist, the dotted lines denotes one route from allele→trait→demographic rate→w. Reproduced from [79] with permission.
(a). The genotype–phenotype map
A promising avenue to improve our understanding of adaptation to environmental changes (the basis for ER) is to better quantify the relevant genotype–phenotype relationships. To do so, one could explore cases of genetic rescue to detail the fate of newly introduced alleles in recipient populations and identify which genes affect key quantitative traits subject to natural selection (as on the left-end side of figure 4). Application of quantitative genetics methods and of high throughput genomics highlights promising ways for understanding the genotype–phenotype relationship. For instance, the animal model [80] is now commonly used in the wild to estimate additive genetic variation, a critical component of the evolutionary potential of a trait. To our knowledge, no examples of the application of the animal model to ER yet exist; however, several authors have highlighted its potential [81–83]. High throughput genomic techniques with large sets of microsatellites loci and/or high-density SNP data provide helpful tools to achieve this objective [80]. For example, a recent study of bighorn sheep used 195 loci genotyped for 219 individuals and genomic positioning of loci on a linkage map to assess the effects of genetic rescue in this species [58]. The authors identified several genomic regions that probably affected fitness-related traits. Interestingly, a relatively small number of markers (30 loci) allowed predicting correctly the major effects of the rescue through their contribution to life-history trait variation [58]. Applying these techniques to understand the genetic basis of evolutionary responses to human-driven change will certainly help us understand by which mechanisms (e.g. plasticity, selection on standing genetic variation, mutation) rescue is more likely to occur.
(b). The phenotype–demography map
The second step in building knowledge on adaptation is to understand how genotype and phenotype affect the performance of organisms. The main approaches to explore these links are selection analyses [84]—as in the middle section of figure 4. Since the seminal book by Endler [85], several studies have assessed the strength and variability of viability and fertility selection in the wild [86]. Clearly, it is unrealistic to monitor all possible phenotypes in anticipation of a catastrophic environmental change. On the basis of the available evidence from the many studies of natural selection, however, one could potentially target an a priori subset of phenotypes as possible candidates to evolve under the pressure of novel stress. For example, to persist through climate change, evolution or plasticity in phenological traits [21] are more likely to be important, while response to exploitation might lead to changes in morphology and life-history traits (bighorn sheep [87], Atlantic cod, Gadus morhua [88]). By understanding which traits are more likely to affect individual performance, one should be able to predict the population consequences of changes in genetically based traits.
(c). The demography–mean fitness map
To predict whether populations will recover after an evolutionary response to an environmental change, one needs to quantify the magnitude of these effects on population dynamics. Despite the formative role demography has played in the development of evolutionary theory [89,90], the development of approaches linking genotype and phenotype to vital rates and population growth is very recent [91–94]. This has led to the emergence of the new field of evolutionary demography [95], which builds upon the phenotype–demography map constructed for different stages in a population to estimate the proportion of variance in population growth that can be explained by changes in phenotype or genotype in the population of interest (as in the right-end of figure 4; [96,97]). Evolutionary demography studies will therefore be very helpful to parse the relative importance of ecological versus evolutionary changes for population growth.
It is impractical to map genotype–fitness for all species of conservation concern. What then are those who manage these populations to do? First, managers need to prioritize which links in the map are most relevant to their position in the conservation chain. For example, captive breeding programmes will probably try initially to map genotype–phenotype then phenotype–demography [98]. Alternatively, for exploited populations [99,100] managers should first understand the phenotype–demography then the demography–fitness links. These links will inform which harvest practices will be least detrimental to population viability. For populations which are candidates for genetic rescue, the critical link to assess will probably be genotype–phenotype, focusing on life-history traits [54]. It is important to note that when the objective is to promote ER and species conservation, the cost of prioritizing some links in the genotype–fitness map will be failure to detect that ER has occurred, not failure for ER to occur. Ultimately, ER brings to conservation biology a focus on the link between evolutionary potential and demographic change, and reinforces that for populations the best defence to buffer against environmental stresses is to focus on maintaining sustainable population sizes, ensuring demographic stability, preserving genetic variation affecting life-history traits, and protecting and enhancing gene flow among populations.
6. Concluding remarks
Empirical knowledge on the occurrence of ER in nature is rare. It is thus difficult to generalize about its importance for conservation. Given that laboratory and theoretical evidence suggests that ER occurs mostly in relatively large populations in controlled environments, we argue that in most cases ER is improbable in species and populations targeted by conservation programmes. These populations are often small (<50 individuals) and have low genetic variation. Additionally, if the time required for beneficial mutations to establish themselves is more than 25 generations, then many vertebrates would require time-scales of over 100 years, which is less probable than their expected persistence time given the rate of anthropogenic change. As pointed out by Smith [101], ‘if a species died out because its members could not survive the immediate consequences of a meteorite impact, or because the island to which it was confined sank beneath the sea, no capacity for rapid evolution would have saved it’. Thus, ER is not a panacea for management and conservation. However, ER is a possibility for cases where additive genetic variation for adaptation is present, and thus may be a more prevalent mechanism to prevent common species from becoming uncommon. Additionally, the alleles which facilitate ER are more likely to result from standing genetic variation or through translocation of new individuals/genes than from novel mutations. As such, ER should enter the conservation biology toolbox; albeit not become a tool which permits adaptationist complacency. Ultimately, ER is a race between environmental and evolutionary processes. It seems clear that sometimes it can occur in nature, but the crucial question we must address is which one of these two forces is more likely to win in an increasingly changing world.
Acknowledgements
D.G., M.F.B. and F.P. are funded by NSERC Discovery Grants. F.P. holds the Canada Research Chair in Evolutionary Demography and Conservation. Initial ideas for this article were born in a working group funded by the Québec Center for Biodiversity Science. We would also like to thank O. Ronce, T. E. Reed, and one anonymous referee for thoughtful reviews of previous versions of our manuscript.
References
- 1.Butchart SHM, et al. 2010. Global biodiversity: indicators of recent declines. Science 328, 1164–1168 10.1126/science.1187512 (doi:10.1126/science.1187512) [DOI] [PubMed] [Google Scholar]
- 2.Loehle C, Eschenbach W. 2012. Historical bird and terrestrial mammal extinction rates and causes. Diversity and Distributions 18, 84–91 10.1111/j.1472-4642.2011.00856.x (doi:10.1111/j.1472-4642.2011.00856.x) [DOI] [Google Scholar]
- 3.Stockwell CA, Hendry AP, Kinnison MT. 2003. Contemporary evolution meets conservation biology. Trends Ecol. Evol. 18, 94–101 10.1016/S0169-5347(02)00044-7 (doi:10.1016/S0169-5347(02)00044-7) [DOI] [Google Scholar]
- 4.Kinnison MT, Hendry AP, Stockwell CA. 2007. Contemporary evolution meets conservation biology II: impediments to integration and application. Ecol. Res. 22, 947–954 10.1007/s11284-007-0416-6 (doi:10.1007/s11284-007-0416-6) [DOI] [Google Scholar]
- 5.Hendry AP, et al. 2011. Evolutionary principles and their practical application. Evol. Appl. 4, 159–183 10.1111/j.1752-4571.2010.00165.x (doi:10.1111/j.1752-4571.2010.00165.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meine C, Soule M, Noss RF. 2006. ‘A mission-driven discipline’: the growth of conservation biology. Conserv. Biol. 20, 631–651 10.1111/j.1523-1739.2006.00449.x (doi:10.1111/j.1523-1739.2006.00449.x) [DOI] [PubMed] [Google Scholar]
- 7.Crandall KA. 2009. A multifaceted approach to species conservation. Anim. Conserv. 12, 105–106 10.1111/j.1469-1795.2009.00254.x (doi:10.1111/j.1469-1795.2009.00254.x) [DOI] [Google Scholar]
- 8.Mace GM, Purvis A. 2008. Evolutionary biology and practical conservation: bridging a widening gap. Mol. Ecol. 17, 9–19 10.1111/j.1365-294X.2007.03455.x (doi:10.1111/j.1365-294X.2007.03455.x) [DOI] [PubMed] [Google Scholar]
- 9.Lande R. 1988. Genetics and demography in biological conservation. Science 241, 1455–1460 10.1126/science.3420403 (doi:10.1126/science.3420403) [DOI] [PubMed] [Google Scholar]
- 10.Gomulkiewicz R, Holt RD. 1995. When does evolution by natural selection prevent extinction? Evolution 49, 201–207 10.2307/2410305 (doi:10.2307/2410305) [DOI] [PubMed] [Google Scholar]
- 11.Boulding EG, Hay T. 2001. Genetic and demographic parameters determining population persistence after a discrete change in the environment. Heredity 86, 313–324 10.1046/j.1365-2540.2001.00829.x (doi:10.1046/j.1365-2540.2001.00829.x) [DOI] [PubMed] [Google Scholar]
- 12.Lavergne S, Evans MEK, Burfield IJ, Jiguet F, Thuiller W. 2012. Are species' responses to global change predicted by past niche evolution? Phil. Trans. R. Soc. B 368, 20120091. 10.1098/rstb.2012.0091 (doi:10.1098/rstb.2012.0091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch M, Lande R. 1993. Evolution and extinction in response to environmental change. In Biotic interactions and global change (eds Karieva PM, Kingsolver JG, Huey RB.), pp. 234–250 Sunderland, MA: Sinauer Associates [Google Scholar]
- 14.Burger R, Krall C. 2004. Quantitative genetic models and changing environments. In Evolutionary conservation biology (eds Ferrière R, Dieckmann U, Couvet D.), pp. 171–186 Cambridge, UK: Cambridge University Press [Google Scholar]
- 15.Reed TE, Schindler DE, Waples RS. 2011. Interacting effects of phenotypic plasticity and evolution on population persistence in a changing climate. Conserv. Biol. 25, 56–63 10.1111/j.1523-1739.2010.01552.x (doi:10.1111/j.1523-1739.2010.01552.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell G, Gonzalez A. 2011. Adaptation and evolutionary rescue in metapopulations experiencing environmental deterioration. Science 332, 1327–1330 10.1126/science.1203105 (doi:10.1126/science.1203105) [DOI] [PubMed] [Google Scholar]
- 17.Levin BR, Lipsitch M, Perrot V, Schrag S, Antia R, Simonsen L, Moore Walker N, Stewart FM. 1997. The population genetics of antibiotic resistance. Clin. Infect. Dis. 24, S9–S16 10.1093/clinids/24.Supplement_1.S9 (doi:10.1093/clinids/24.Supplement_1.S9) [DOI] [PubMed] [Google Scholar]
- 18.Jasieniuk M, Brûlé-Babel AL, Morrison IN. 1996. The evolution and genetics of herbicide resistance in weeds. Weed Sci. 44, 176–193 [Google Scholar]
- 19.Fenner F. 2010. Deliberate introduction of the European rabbit, Oryctolagus cuniculus, into Australia. Rev. Scient. Techn.-Office Int. Epizoot. 29, 103–111 [DOI] [PubMed] [Google Scholar]
- 20.Bearhop S, Fiedler W, Furness RW, Votier SC, Waldron S, Newton J, Bowen GJ, Berthold P, Farnsworth K. 2005. Assortative mating as a mechanism for rapid evolution of a migratory divide. Science 310, 502–504 10.1126/science.1115661 (doi:10.1126/science.1115661) [DOI] [PubMed] [Google Scholar]
- 21.Bradshaw WE, Holzapfel CM. 2006. Evolutionary response to rapid climate change. Science (Washington) 312, 1477–1478 10.1126/science.1127000 (doi:10.1126/science.1127000) [DOI] [PubMed] [Google Scholar]
- 22.Gomulkiewicz R, Shaw R. 2012. Evolutionary rescue beyond the models. Phil. Trans. R. Soc. B 368, 20120093. 10.1098/rstb.2012.0093 (doi:10.1098/rstb.2012.0093). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll SP, Hendry AP, Reznick DN, Fox CW. 2007. Evolution on ecological time-scales. Funct. Ecol. 21, 387–393 10.1111/j.1365-2435.2007.01289.x (doi:10.1111/j.1365-2435.2007.01289.x) [DOI] [Google Scholar]
- 24.Barrett RDH, Schluter D. 2008. Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 10.1016/j.tree.2007.09.008 (doi:10.1016/j.tree.2007.09.008) [DOI] [PubMed] [Google Scholar]
- 25.Bell G, Gonzalez A. 2009. Evolutionary rescue can prevent extinction following environmental change. Ecol. Lett. 12, 942–948 10.1111/j.1461-0248.2009.01350.x (doi:10.1111/j.1461-0248.2009.01350.x) [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez A, Ronce O, Ferriere R, Hochberg ME. 2012. Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Phil. Trans. R. Soc. B 368, 20120404. 10.1098/rstb.2012.0404 (doi:10.1098/rstb.2012.0404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clutton-Brock T, Sheldon BC. 2010. Individuals and populations: the role of long-term, individual-based studies of animals in ecology and evolutionary biology. Trends Ecol. Evol. 25, 562–573 10.1016/j.tree.2010.08.002 (doi:10.1016/j.tree.2010.08.002) [DOI] [PubMed] [Google Scholar]
- 28.Jackson WB. 1998. Ecology of pest rodents in the urban environment. In Modern trends in ecology and environment (ed. Ambasht RS.). Leiden, The Netherlands: Backhuys Publishers [Google Scholar]
- 29.Chitty D. 1954. The study of the brown rat and its control by poison. In Control of rats and mice (eds Chitty D, Southern HN.). Oxford, UK: Oxford University Press [Google Scholar]
- 30.Boyle CM. 1960. Case of apparent resistance of Rattus norvegicus berkenhout to anticoagulant poisons. Nature 188, 517. 10.1038/188517a0 (doi:10.1038/188517a0) [DOI] [Google Scholar]
- 31.Lund M. 1964. Resistance to warfarin in the common rat. Nature 203, 778. 10.1038/203778a0 (doi:10.1038/203778a0) [DOI] [PubMed] [Google Scholar]
- 32.Pelz H-J, et al. 2005. The genetic basis of resistance to anticoagulants in rodents. Genetics 170, 1839–1847 10.1534/genetics.104.040360 (doi:10.1534/genetics.104.040360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishizuka M, Tanikawa T, Tanaka KD, Heewon M, Okajima F, Sakamoto KQ, Fujita S. 2008. Pesticide resistance in wild mammals—mechanisms of anticoagulant resistance in wild rodents. J. Toxicol. Sci. 33, 283–291 10.2131/jts.33.283 (doi:10.2131/jts.33.283) [DOI] [PubMed] [Google Scholar]
- 34.Lasseur R, Longin-Sauvageon C, Videmann B, Billeret M, Berny P, Benoit E. 2006. Warfarin resistance in a French strain of rats. J. Biochem. Mol. Toxicol. 19, 379–385 10.1002/jbt.20104 (doi:10.1002/jbt.20104) [DOI] [PubMed] [Google Scholar]
- 35.Rost S, Pelz H-J, Menzel S, MacNicoll A, Leon V, Song K-J, Jakel T, Oldenburg J, Muller C. 2009. Novel mutations in the VKORC1 gene of wild rats and mice—a response to 50 years of selection pressure by warfarin? BMC Genet. 10, 4. 10.1186/1471-2156-10-4 (doi:10.1186/1471-2156-10-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenner F, Fantini B. 1999. Biological control of vertebrate pests: the history of myxomatosis—an experiment in evolution. Wallingford, UK: CABI [Google Scholar]
- 37.Saunders G, Cooke B, McColl K, Shine R, Peacock T. 2010. Modern approaches for the biological control of vertebrate pests: an Australian perspective. Biol. Control 52, 288–295 10.1016/j.biocontrol.2009.06.014 (doi:10.1016/j.biocontrol.2009.06.014) [DOI] [Google Scholar]
- 38.Best SM, Kerr PJ. 2000. Coevolution of host and virus: the pathogenesis of virulent and attenuated strains of Myxoma virus in resistant and susceptible European rabbits. Virology 267, 36–48 10.1006/viro.1999.0104 (doi:10.1006/viro.1999.0104) [DOI] [PubMed] [Google Scholar]
- 39.May RM, Anderson RM. 1983. Epidemiology and genetics in the coevolution of parasites and hosts. Proc. R. Soc. Lond. B 219, 281–313 10.1098/rspb.1983.0075 (doi:10.1098/rspb.1983.0075) [DOI] [PubMed] [Google Scholar]
- 40.Cooke BD. 1983. Changes in the age structure and size of populations of wild rabbits in South Australia following the introduction of European rabbit fleas, Spilopsullus cuniculi (Dale), as vectors for myxomatosis. Austral. Wildl. Res. 10, 105–120 10.1071/WR9830105 (doi:10.1071/WR9830105) [DOI] [Google Scholar]
- 41.Mutze G, Cooke B, Alexander P. 1998. The initial impact of rabbit hemorrhagic disease on European rabbit populations in South Australia. J. Wildl. Dis. 34, 221–227 [DOI] [PubMed] [Google Scholar]
- 42.Cooke BD. 2002. Rabbit haemorrhagic disease: field epidemiology and the management of wild rabbit populations. Rev. Scient. Techn. -Office Int. Epizoot. 21, 347–358 [DOI] [PubMed] [Google Scholar]
- 43.Grant PR, Grant BR. 1995. Predicting microevolutionary responses to directional selection on heritable variation. Evolution 49, 241–251 10.2307/2410334 (doi:10.2307/2410334) [DOI] [PubMed] [Google Scholar]
- 44.Grant PR, Grant BR. 2002. Unpredictable evolution in a 30-year study of Darwin's finches. Science 296, 707–711 10.1126/science.1070315 (doi:10.1126/science.1070315) [DOI] [PubMed] [Google Scholar]
- 45.Garant D, Kruuk LEB, Wilkin TA, McCleery RH, Sheldon BC. 2005. Evolution driven by differential dispersal within a wild bird population. Nature 433, 60–65 10.1038/nature03051 (doi:10.1038/nature03051) [DOI] [PubMed] [Google Scholar]
- 46.Charmantier A, McCleery RH, Cole LR, Perrins C, Kruuk LEB, Sheldon BC. 2008. Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science 320, 800–803 10.1126/science.1157174 (doi:10.1126/science.1157174) [DOI] [PubMed] [Google Scholar]
- 47.Reale D, McAdam AG, Boutin S, Berteaux D. 2003. Genetic and plastic responses of a northern mammal to climate change. Proc. R. Soc. Lond. B 270, 591–596 10.1098/rspb.2002.2224 (doi:10.1098/rspb.2002.2224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berteaux D, Réale D, McAdam AG, Boutin S. 2004. Keeping pace with fast climate change: can arctic life count on evolution? Integr. Comp. Biol. 44, 140–151 10.1093/icb/44.2.140 (doi:10.1093/icb/44.2.140) [DOI] [PubMed] [Google Scholar]
- 49.Grant BR, Grant PR. 1993. Evolution of Darwin's finches caused by a rare climatic event. Proc. R. Soc. B 251, 111–117 10.1098/rspb.1993.0016 (doi:10.1098/rspb.1993.0016) [DOI] [Google Scholar]
- 50.Gianapp PL, Reed TE, Verhulst S, Visser ME. 2012. Predicting demographically sustainable rates of adaptation: can great tit breeding time keep pace with climate change? Phil. Trans. R. Soc. B 368, 20120289. 10.1098/rstb.2012.0289 (doi:10.1098/rstb.2012.0289). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Both C, Bouwhuis S, Lessells CM, Visser ME. 2006. Climate change and population declines in a long-distance migratory bird. Nature 441, 81–83 10.1038/nature04539 (doi:10.1038/nature04539) [DOI] [PubMed] [Google Scholar]
- 52.Both C, Visser ME. 2001. Adjustment to climate change is constrained by arrival date in a long-distance migrant bird. Nature 411, 296–298 10.1038/35077063 (doi:10.1038/35077063) [DOI] [PubMed] [Google Scholar]
- 53.Ingvarsson PK. 2001. Restoration of genetic variation lost—the genetic rescue hypothesis. Trends Ecol. Evol. 16, 62–63 10.1016/S0169-5347(00)02065-6 (doi:10.1016/S0169-5347(00)02065-6) [DOI] [PubMed] [Google Scholar]
- 54.Tallmon D, Luikart G, Waples R. 2004. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 19, 489–496 10.1016/j.tree.2004.07.003 (doi:10.1016/j.tree.2004.07.003) [DOI] [PubMed] [Google Scholar]
- 55.Hedrick P, Fredrickson R. 2010. Genetic rescue guidelines with examples from Mexican wolves and Florida panthers. Conserv. Genet. 11, 615–626 10.1007/s10592-009-9999-5 (doi:10.1007/s10592-009-9999-5) [DOI] [Google Scholar]
- 56.Edmands S. 2007. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 16, 463–475 10.1111/j.1365-294X.2006.03148.x (doi:10.1111/j.1365-294X.2006.03148.x) [DOI] [PubMed] [Google Scholar]
- 57.Hogg JT, Forbes SH, Steele BM, Luikart G. 2006. Genetic rescue of an insular population of large mammals. Proc. R. Soc. B 273, 1491–1499 10.1098/rspb.2006.3477 (doi:10.1098/rspb.2006.3477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller J, Poissant J, Hogg J, Coltman D. 2012. Genomic consequences of genetic rescue in an insular population of bighorn sheep (Ovis canadensis). Mol. Ecol. 21, 1583–1596 10.1111/j.1365-294X.2011.05427.x (doi:10.1111/j.1365-294X.2011.05427.x) [DOI] [PubMed] [Google Scholar]
- 59.Madsen T, Ujvari B, Olsson M. 2004. Novel genes continue to enhance population growth in adders (Vipera berus). Biol. Conserv. 120, 145–147 10.1016/j.biocon.2004.01.022 (doi:10.1016/j.biocon.2004.01.022) [DOI] [Google Scholar]
- 60.Madsen T, Shine R, Olsson M, Wittzell H. 1999. Conservation biology: restoration of an inbred adder population. Nature 402, 34–35 10.1038/46941 (doi:10.1038/46941) [DOI] [Google Scholar]
- 61.Adams JR, Vucetich LM, Hedrick PW, Peterson RO, Vucetich JA. 2011. Genomic sweep and potential genetic rescue during limiting environmental conditions in an isolated wolf population. Proc. R. Soc. B 278, 3336–3344 10.1098/rspb.2011.0261 (doi:10.1098/rspb.2011.0261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferriére R, Diekmann O, Couvet D. 2004. Evolutionary conservation biology. Cambridge, UK: Cambridge University Press [Google Scholar]
- 63.Willi Y, Hoffmann AA. 2009. Demographic factors and genetic variation influence population persistence under environmental change. J. Evol. Biol. 22, 124–133 10.1111/j.1420-9101.2008.01631.x (doi:10.1111/j.1420-9101.2008.01631.x) [DOI] [PubMed] [Google Scholar]
- 64.Chen JS, Lo KC. 1990. Toxicity of organophosphorus insecticides to diazinon-reversely-selected, pesticide-pressure-relaxed and field-resistant strains of bulb mite Rhizoglyphus robini (Acari: Acaridae). Exp. Appl. Acarol. 8, 243–252 10.1007/BF01202135 (doi:10.1007/BF01202135) [DOI] [Google Scholar]
- 65.Hsu J-C, Feng H-T, Wu W-J. 2004. Resistance and synergistic effects of insecticides in Bactrocera dorsalis (Diptera: Tephritidae) in Taiwan. J. Econ. Entomol. 97, 1682–1688 10.1603/0022-0493-97.5.1682 (doi:10.1603/0022-0493-97.5.1682) [DOI] [PubMed] [Google Scholar]
- 66.Gaillard J-M, Yoccoz NG, Lebreton J-D, Bonenfant C, Devillard S, Loison A, Pontier D, Allaine D. 2005. Generation time: a reliable metric to measure life-history variation among mammalian populations. Am. Nat. 166, 119–123 10.1086/430330 (doi:10.1086/430330) [DOI] [PubMed] [Google Scholar]
- 67.Cheeke PR. 1986. Potentials of rabbit production in tropical and subtropical agricultural systems. J. Anim. Sci. 63, 1581–1586 [Google Scholar]
- 68.Dobson FS, Oli MK. 2007. Fast and slow life histories of mammals. Ecoscience 14, 292. 10.2980/1195-6860(2007)14[292:FASLHO]2.0.CO;2 (doi:10.2980/1195-6860(2007)14[292:FASLHO]2.0.CO;2) [DOI] [Google Scholar]
- 69.Bürger R, Lynch M. 1997. Adaptation and extinction in changing environments. In Environmental stress, adaptation and evolution (eds Bijlsma R, Loeschcke V.), pp. 209–239 Basel, Switzerland: Birkhauser-Verlag; [DOI] [PubMed] [Google Scholar]
- 70.Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates [Google Scholar]
- 71.Bell G. 2008. Selection: the mechanisms of evolution. Oxford, UK: Oxford University Press [Google Scholar]
- 72.Bell G. 2010. Fluctuating selection: the perpetual renewal of adaptation in variable environments. Phil. Trans. R. Soc. B 365, 87–97 10.1098/rstb.2009.0150 (doi:10.1098/rstb.2009.0150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price TD, Qvarnström A, Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. Lond. B 270, 1433–1440 10.1098/rspb.2003.2372 (doi:10.1098/rspb.2003.2372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chevin L-M, Lande R, Mace GM. 2010. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 8, e1000357. 10.1371/journal.pbio.1000357 (doi:10.1371/journal.pbio.1000357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hendry AP, Farrugia TJ, Kinnison MT. 2008. Human influences on rates of phenotypic change in wild animal populations. Mol. Ecol. 17, 20–29 10.1111/j.1365-294X.2007.03428.x (doi:10.1111/j.1365-294X.2007.03428.x) [DOI] [PubMed] [Google Scholar]
- 76.Dyck MG, Soon W, Baydack RK, Legates DR, Baliunas S, Ball TF, Hancock LO. 2007. Polar bears of western Hudson Bay and climate change: are warming spring air temperatures the ‘ultimate’ survival control factor? Ecol. Complexity 4, 73–84 10.1016/j.ecocom.2007.03.002 (doi:10.1016/j.ecocom.2007.03.002) [DOI] [Google Scholar]
- 77.COSEWIC 2008. COSEWIC assessment and update status report on the polar bear (Ursus maritimus). Ottawa, Canada: Committee on the Status of Endangered Wildlife in Canada [Google Scholar]
- 78.Brooks TM, Mittermeier RA, da Fonseca GAB, Gerlach J, Hoffmann M, Lamoreux JF, Mittermeier CG, Pilgrim JD, Rodrigues ASL. 2006. Global biodiversity conservation priorities. Science 313, 58–61 10.1126/science.1127609 (doi:10.1126/science.1127609) [DOI] [PubMed] [Google Scholar]
- 79.Coulson T, Benton T, Lundberg P, Dall SRX, Kendall BE. 2006. Putting evolutionary biology back in the ecological theatre: a demographic framework mapping genes to communities. Evol. Ecol. Res. 8, 1155–1171 [Google Scholar]
- 80.Hoffmann AA, Willi Y. 2008. Detecting genetic responses to environmental change. Nat. Rev. Genet. 9, 421–432 10.1038/nrg2339 (doi:10.1038/nrg2339) [DOI] [PubMed] [Google Scholar]
- 81.Coltman DW. 2008. Molecular ecological approaches to studying the evolutionary impact of selective harvesting in wildlife. Mol. Ecol. 17, 221–235 10.1111/j.1365-294X.2007.03414.x (doi:10.1111/j.1365-294X.2007.03414.x) [DOI] [PubMed] [Google Scholar]
- 82.Hansen MM, Oliveri I, Waller DM, Nielson EE, The GeM Working Group 2012. Monitoring adaptive genetic responses to environmental change. Mol. Ecol. 21, 1311–1329(doi:10.1111/j.1365-294X.2011.05463.x) [DOI] [PubMed] [Google Scholar]
- 83.Markert J, Champlin DM, Gutjahr-Gobell R, Grear JS, Kuhn A, McGreevy TJ, Roth A, Bagley MJ, Nacci DE. 2010. Population genetic diversity and fitness in multiple environments. BMC Evol. Biol. 10, 205. 10.1186/1471-2148-10-205 (doi:10.1186/1471-2148-10-205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lande R, Arnold SJ. 1983. The measurement of selection on correlated characters. Evolution 37, 1210–1226 10.2307/2408842 (doi:10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- 85.Endler JA. 1986. Natural selection in the wild. Princeton, NJ: Princeton University Press [Google Scholar]
- 86.Kingsolver JG, Hoekstra HE, Hoekstra JM, Berrigan D, Vignieri SN, Hill CE, Hoang A, Gibert P, Beerli P. 2001. The strength of phenotypic selection in natural populations. Am. Nat. 157, 245–261 10.1086/319193 (doi:10.1086/319193) [DOI] [PubMed] [Google Scholar]
- 87.Coltman DW, O'Donoghue P, Jorgenson JT, Hogg JT, Strobeck C, Festa-Bianchet M. 2003. Undesirable evolutionary consequences of trophy hunting. Nature 426, 655–658 10.1038/nature02177 (doi:10.1038/nature02177) [DOI] [PubMed] [Google Scholar]
- 88.Olsen EM, Heino M, Lilly GR, Morgan MJ, Brattey J, Ernande B, Dieckmann U. 2004. Maturation trends indicative of rapid evolution preceded the collapse of northern cod. Nature 428, 932–935 10.1038/nature02430 (doi:10.1038/nature02430) [DOI] [PubMed] [Google Scholar]
- 89.Malthus TR. 1798. An essay on the principle of population. London, UK: J. Johnson [Google Scholar]
- 90.Darwin C. 1859. On the origin of species by means of natural selection. London, UK: John Murray [Google Scholar]
- 91.van Tienderen PH. 2000. Elasticities and the link between demographic and evolutionary dynamics. Ecology 81, 666–679 10.1890/0012-9658(2000)081[0666:EATLBD]2.0.CO;2 (doi:10.1890/0012-9658(2000)081[0666:EATLBD]2.0.CO;2) [DOI] [Google Scholar]
- 92.Hairston NG, Ellner SP, Geber MA, Yoshida T, Fox JA. 2005. Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 8, 1114–1127 10.1111/j.1461-0248.2005.00812.x (doi:10.1111/j.1461-0248.2005.00812.x) [DOI] [Google Scholar]
- 93.Ellner SP, Rees M. 2006. Integral projection models for species with complex demography. Am. Nat. 167, 410–428 10.1086/499438 (doi:10.1086/499438) [DOI] [PubMed] [Google Scholar]
- 94.Coulson T, Tuljapurkar S, Childs DZ. 2010. Using evolutionary demography to link life history theory, quantitative genetics and population ecology. J. Anim. Ecol. 79, 1226–1240 10.1111/j.1365-2656.2010.01734.x (doi:10.1111/j.1365-2656.2010.01734.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Metcalf CJE, Pavard S. 2007. Why evolutionary biologists should be demographers. Trends Ecol. Evol. 22, 205–212 10.1016/j.tree.2006.12.001 (doi:10.1016/j.tree.2006.12.001) [DOI] [PubMed] [Google Scholar]
- 96.Pelletier F, Clutton-Brock T, Pemberton J, Tuljapurkar S, Coulson T. 2007. The evolutionary demography of ecological change: linking trait variation and population growth. Science 315, 1571–1574 10.1126/science.1139024 (doi:10.1126/science.1139024) [DOI] [PubMed] [Google Scholar]
- 97.Ozgul A, Childs DZ, Oli MK, Armitage KB, Blumstein DT, Olson LE, Tuljapurkar S, Coulson T. 2010. Coupled dynamics of body mass and population growth in response to environmental change. Nature 466, 482–485 10.1038/nature09210 (doi:10.1038/nature09210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pelletier F, Réale D, Watters J, Boakes EH, Garant D. 2009. Value of captive populations for quantitative genetics research. Trends Ecol. Evol. 24, 263–270 10.1016/j.tree.2008.11.013 (doi:10.1016/j.tree.2008.11.013) [DOI] [PubMed] [Google Scholar]
- 99.Njiforti HL. 1996. Preferences and present demand for bushmeat in north Cameroon: some implications for wildlife conservation. Environ. Conserv. 23, 149–155 10.1017/S0376892900038534 (doi:10.1017/S0376892900038534) [DOI] [Google Scholar]
- 100.Milner-Gulland EJ, Shea K, Possingham H, Coulson T, Wilcox C. 2001. Competing harvesting strategies in a simulated population under uncertainty. Anim. Conserv. 4, 157–167 10.1017/S1367943001001184 (doi:10.1017/S1367943001001184) [DOI] [Google Scholar]
- 101.Maynard Smith J. 1989. The causes of extinction. Phil. Trans. R. Soc. Lond. B 325, 241–252 10.1098/rstb.1989.0086 (doi:10.1098/rstb.1989.0086) [DOI] [PubMed] [Google Scholar]