Abstract
Objective
To test whether a noninvasive urine-based multianalyte diagnostic readout assay that uses protein and DNA biomarkers can risk stratify patients with hematuria into those who are or are not likely to have bladder cancer and those who should receive standard care.
Patients and Methods
This prospective, observational, multicenter, single-assessment study was conducted between June 12, 2009, and April 15, 2011. Eligible patients presented with hematuria and as part of their evaluation underwent cystoscopy. Urine samples were analyzed for the presence of mutant FGFR3 and quantified matrix metalloproteinase 2 and the hypermethylation of TWIST1 and NID2. A patient's chance of having (positive predictive value [PPV]) or not having (negative predictive value [NPV]) cancer was determined by FGFR3 alone or by all 4 biomarkers, respectively.
Results
Cystoscopy/biopsy diagnosed 690 of 748 patients as negative and 58 as positive for bladder cancer. Of 21 patients identified by FGFR3 as highly likely to have cancer, 20 were also positive by cystoscopy/biopsy, resulting in a PPV of 95.2% (20 of 21), with specificity of 99.9% (689 of 690). The 4-marker combination identified 395 patients as having a low likelihood of cancer. Of these, 56.2% (388 of 690) also had negative biopsy/cystoscopy findings, resulting in an NPV of 98.2% (388 of 395). In total, 416 of the 748 patients with hematuria (55.6%) were identified with extremely high NPV and PPV to have or not have bladder cancer.
Conclusion
This multianalyte assay accurately stratified patients with high confidence into those who likely do or do not have bladder cancer. This test was developed to enhance and not to eliminate referrals for urologic evaluation.
Abbreviations and Acronyms: AUA, American Urological Association; CIDD, Clinical Intervention Determining Diagnostic; MADR, multianalyte diagnostic readout; MMP-2, matrix metalloproteinase 2; NPV, negative predictive value; PCR, polymerase chain reaction; PPV, positive predictive value
Urothelial cancer of the bladder is typically associated with asymptomatic hematuria (either gross or microscopic) and voiding symptoms.1 Up to 5% of patients with microscopic hematuria and approximately 10% with gross hematuria have urinary tract cancer.2-7 Despite the risk of urinary tract malignancy, the observed rate of referral to a urologist for men is approximately 13% to 47% and for women is approximately 27%.1-4 The poor referral rates for patients may, in part, be due to the fact that hematuria is often seen in other non–life-threatening urologic conditions, including urinary tract infections, benign prostatic hyperplasia, and urolithiasis.1,5 The lack of evaluation for cancer in patients with hematuria can result in delayed diagnosis and worse prognosis.1,3,6-8
One strategy to identify early disease is to ensure evaluation of patients at risk for bladder cancer.9 The American Urological Association (AUA) recommends a full urologic evaluation for patients with 2 of 3 properly collected urine samples that meet the criteria for microscopic hematuria and for high-risk patients (>40 years of age).10 Currently, primary care work-up of patients with hematuria involves urine dipstick testing, microscopic urinalysis, urine Gram stain/culture, and urine cytologic analysis.5 However, most of these methods have low sensitivity or specificity and an inability to detect early (low-load) cancers.11,12 A false-negative diagnosis can cause a wrong sense of security, and a false-positive diagnosis can result in patient anxiety.13
Although gross hematuria should always be evaluated by a urologist, a tool that might better stratify a patient's risk of bladder cancer and that eliminates the confusion created by false-positive and false-negative results would be helpful to primary care physicians in deciding how to better assess the risk faced by a patient. In this multicenter study, we evaluated a diagnostic assay that uses the Clinical Intervention Determining Diagnostic (CIDD) approach14 to stratify patients with hematuria into 3 groups: those with a high likelihood of having urothelial cancer, those who are unlikely to have cancer, and those who may be at risk and should continue to be routinely monitored for bladder cancer. Currently available biomarkers for detecting bladder cancer rely on a single cutoff value to define “cancer” vs “cancer free,” which results in either low specificity or low sensitivity (high false-positive or high false-negative results),15 and, therefore, fall short of providing clinically relevant results.16 The CIDD approach uses 2 biomarker cutoff values in 1 noninvasive assay (one that has high positive predictive value [PPV] and one that has high negative predictive value [NPV]) to triage the population, reducing ambiguity.14
We developed a noninvasive urine-based test that combines the robust performance of 3 DNA biomarkers (TWIST1, NID2, and FGFR3) with a protein biomarker (matrix metalloproteinase 2 [MMP-2]) into one multianalyte diagnostic readout (MADR) with high PPV and high NPV to identify patients with bladder cancer. The underlying idea is that an assay with high PPV could be used to identify patients who are likely to have cancer and who could benefit from accelerated intervention. Various studies have shown that sequence variations in FGFR3 are associated with bladder cancer development and that the mutant DNA can be detected in the urine of patients with cancer.17-21 Based on these studies and the one presented herein, a positive FGFR3 finding has high PPV for bladder cancer. Likewise, an assay with high NPV that is associated with high sensitivity could give the physician the confidence to report that a patient with assay-negative results has an extremely low likelihood of having cancer. In the case of the 4-marker combination, patients are considered negative for cancer when the results of all 4 biomarkers are negative. A third cohort of patients negative for FGFR3 sequence variations but not for any of the other 3 markers would not be considered to have elevated or reduced risk of bladder cancer and would be referred for standard care.
Patients and Methods
This was a prospective, observational, multicenter, single-assessment, diagnostic test study performed in 27 academic and community-based urology clinics between June 12, 2009, and April 15, 2011, that was designed to assess the ability of the MADR assay to stratify patients presenting with hematuria into those likely to have or not have bladder cancer. The study was conducted according to the principles established by the Declaration of Helsinki. Appropriate institutional review boards approved the protocol, and written informed consent was obtained from the patients.
Study Patients
Eligible patients (≥50 years of age) were undergoing evaluation for bladder cancer owing to a finding of gross or microscopic hematuria. Patients under 50 had a family history of bladder cancer and/or had more than 20-year history of smoking. All had an intact bladder. Patients were required to provide 25 mL of urine and were excluded from the study if they had any current cancer or history of cancer, an autoimmune disease, or a diagnosis of human immunodeficiency virus, hepatitis C, hepatitis B, or a sexually transmitted disease. Patients were also excluded if they had active tuberculosis or any other systemic infection.
The patient population was divided: one group was used to establish the cutoff values for the different biomarkers to maximize clinical performance (derivation group) and another group was used to validate the assay (validation group). For the derivation group, 252 eligible patients from this study (Predictive Biosciences study 2 [PBS-002]) were randomly chosen to establish a cutoff value for the 4 biomarkers: TWIST1, NID2, FGFR3, and MMP-2 protein. Six patients had inconclusive results; consequently, 246 patients were used for the analysis (Figure). It was estimated that approximately 10% of the randomly selected population would have bladder cancer. However, only 6 patients (2.4%) of the original derivation group had cancer, and this was not sufficient for determining cutoff values. Therefore, urine samples from 42 patients with known bladder cancer from a separate independent study (PBS-001) with similar demographic characteristics as the present trial were included in the derivation group, which brought the population of patients with bladder cancer to 16.7%.
FIGURE.
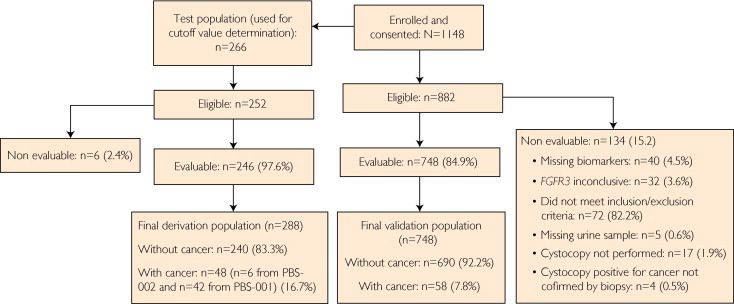
Flowchart diagram showing selection of patients for study of the performance of the noninvasive MADR diagnostic assay for urothelial cancer of the bladder in the evaluation of hematuria. MADR = mulitanalyte diagnostic readout; PBS 001 = Predictive Biosciences study 1; PBS 002 = Predictive Biosciences study 2.
Study Design
At the study visit, urine samples were collected for MADR assay, cystoscopy was performed, and demographic data were collected. The results of the MADR assay were blinded to the patients and physicians. Based on the results of cystoscopy and, when available, a biopsy report, patients were categorized as having or not having urothelial carcinoma of the bladder. A patient was considered negative for bladder cancer if the biopsy results indicated benign tissue or, in the absence of a biopsy, the cystoscopy showed no evidence of disease. All cancers were confirmed by biopsy. Patients with indeterminate cystoscopic findings and no biopsy results were omitted from the analysis.
MADR Assay
The MADR assay was performed by trained staff without knowledge of the cystoscopic or biopsy results. All the urine samples were collected before cystoscopy, bladder biopsy, or transurethral resection. Urine samples were stored at −80°C, and for DNA analysis, samples were stabilized with EDTA (25 mmol/L). For DNA analysis, genomic DNA was isolated using the QIAamp MinElute virus vacuum kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Purified DNA was stored in either buffer (AVE buffer; Qiagen) or water at −20°C.
MMP-2 Protein Quantitation
The urine level of MMP-2 was quantified by enzyme-linked immunosorbent assay using a modification of the manufacturer's instructions (R&D Systems, Minneapolis, MN). Samples were assayed in duplicate using an input of 50 μL of neat urine.
FGFR3 Mutation Assay
Genomic DNA isolated from 4 mL of urine was amplified in a multiplex, real-time polymerase chain reaction (PCR) with primers and dual-labeled fluorescent probes specific for human FGFR3 exons 7, 10, and 15 (Supplemental Table [available online at http://www.mayoclinicproceedings.org]) using the Roche LightCycler 480 system (Roche Applied Science, Indianapolis, IN) under standard conditions.
After purification of these PCR products, a second multiplexed PCR was performed to identify FGFR3 sequence variations using primers and probes listed in the Supplemental Table (available online at http://www.mayoclinicproceedings.org). The PCR contained primers encoding wild-type sequences with locked nucleic acid bases surrounding and including a known sequence variation site in exons 7, 10, or 15 and real-time PCR primers specific for each exon with dual-labeled fluorescent probes. The locked nucleic acid primers have a higher annealing affinity to wild-type DNA than to mutant DNA, resulting in inhibition of amplification of wild-type DNA and preferential amplification of mutant DNA. As a control for the amount of DNA included in each reaction, PCR amplification was performed in the absence of locked nucleic acid–containing primers. The ratio of the amplification cycles of positive controls (plasmids containing known FGFR3 sequence variations) to those of negative controls (wild-type DNA) was included in each assay to determine whether a sample did or did not contain a sequence variation. All the PCRs were performed in duplicate. Samples that did not have sufficient DNA after the initial real-time PCR were excluded from further analysis.
TWIST1 and NID2 Methylation Analysis
Conventional methylation-specific PCR was used to detect methylation of TWIST1 and NID2. A maximum of 2 μg of genomic DNA isolated from 8 mL of urine was bisulfite converted using an EpiTect bisulfite kit (Qiagen) following the manufacturer's directions. The resulting converted DNA was eluted into 30 μL of molecular-grade water and was stored at −20°C. Conventional methylation-specific PCR was performed using methylation-specific primers to the promoter regions of TWIST1 and NID2 (Supplemental Table, available online at http://www.mayoclinicproceedings.org) in a thermal cycler (C1000; Bio-Rad Laboratories, Hercules, CA) under standard conditions. Methylation thresholds for TWIST1 and NID2 were established by densitometry (units shown in thousands, K). To ensure that a negative experimental result was accurate, the minimum input of DNA (10 ng), as determined by unmethylated ACTIN-B quantitation, was required. Real-time PCR was performed using the Roche LightCycler 480 system under standard conditions. Samples negative for hypermethylation that did not meet the minimum DNA inputs were excluded from further analysis.
Statistical Analyses
The primary end point was the ability of the MADR assay to triage a patient population undergoing screening for bladder cancer due to the presence of hematuria using the 2–cutoff value CIDD approach (one cutoff value includes all the markers and the other includes only FGFR3). Performance of the combined 4-biomarker assay was focused on sensitivity and NPV for the purpose of identifying patients who are unlikely to have bladder cancer, and performance of FGFR3 alone was focused on specificity and PPV for the purpose of identifying patients with bladder cancer. The results of the MADR assay were compared with those of cystoscopy and biopsy. Secondary end points included subgroup analyses such as smokers vs nonsmokers and age associations. Retrospective analysis evaluated the sensitivity, specificity, and PPV of combined cytology and FGRF3 for determining whether a patient had bladder cancer.
Negative Predictive Value
The assessment of NPV used a composite of cutoff values for the 4 biomarkers. Cutoff values were selected to achieve approximately 90% sensitivity. A patient was classified as negative for cancer if the findings were below the cutoff value for all 4 biomarkers (TWIST1, <139 K; NID2, <680 K; MMP-2, <1.100 ng/mL; and FGFR3, negative). A patient was classified as indeterminate/positive for cancer if 1 of the 4 biomarkers did not meet the cutoff criteria. The NPV was calculated as follows: NPV = true-negative/all MADR assay negatives.
Positive Predictive Value
The evaluation of PPV used FGFR3 sequence variation status. A patient was considered positive for cancer if the FGFR3 result was positive. A patient was considered indeterminate/negative for cancer if the FGFR3 result was negative. The PPV was calculated as follows: PPV = true-positive/all FGFR3 assay positives. For the evaluation of PPV using a combination of FGFR3 sequence variation status and cytologic testing, a patient was considered positive for cancer if either FGFR3 or cytologic findings were positive. The PPV was calculated as follows: PPV = true-positive/all FGFR3 or cytology positive.
Results
Of the 1148 patients enrolled in the study, 266 were used for the derivation population to determine the optimum cutoff values for the MADR assay and 748 were evaluable and composed the validation population (Figure). The major reasons that patients were not evaluated included not meeting the inclusion or exclusion criteria or having 1 or more marker results that were inconclusive or missing (Figure).
Derivation Patient Group: Establishing Assay Cutoff Values
The derivation group of patients was used to establish marker combinations, cutoff values, and expected performance. The derivation group consisted of 240 patients with hematuria but not bladder cancer and 48 patients with bladder cancer.
The assay evaluated the performance of all 4 biomarkers (TWIST1, NID2, FGFR3, and MMP-2) individually and in combination. Individual marker cutoff values were established to maximize sensitivity (≥90%) and NPV when combined. Given FGFR3's high specificity and the large body of evidence linking FGFR3 sequence variations to bladder cancer,17-21 FGFR3 was used to identify patients who likely have cancer. For the purpose of identifying patients who are unlikely to have cancer, the maximum NPV of 99.5% was obtained using a combination of all 4 biomarkers with the following cutoff values: TWIST1, <139 K; NID2, <680 K; MMP-2, <1.100 ng/mL; and FGFR3, negative. This resulted in sensitivity of 94% and specificity of 65% (Table 1). Although the sensitivity of FGFR3 alone in this sample set was low, FGFR3 resulted in high PPV (71%) and specificity (99%) (Table 1). In total, 165 of the 294 patients with hematuria (56.1%) were identified as having extremely high NPV and PPV to have or not have bladder cancer.
TABLE 1.
MADR Assay Performance for the Derivation Population
| Biomarker | Negative assay criteria | NPV (%) | Sensitivity (% [range]) | POE (% [range]) |
|---|---|---|---|---|
| TWIST1 | <139 K | ND | 84 (71-94) | 82 (76-86) |
| NID2 | <680 K | ND | 33 (20-48) | 100 (99-100) |
| MMP-2 | <1.100 ng/mL | ND | 35 (22-51) | 74 (68-79) |
| FGFR3 | Negative | ND | 10 (3-23) | 99 (97-100) |
| All 4 biomarkers | 99.5 | 94 (83-99) | 65 (58-71) |
| Biomarker | PPV (%) | Specificity (% [range]) | POE (% [range]) | |
|---|---|---|---|---|
| FGFR3 | Positive | 71 | 99 (97-100) | 10 (3-23) |
MADR = multianalyte diagnostic readout; MMP-2 = matrix metalloproteinase 2; ND = not determined; NPV = negative predictive value; PPV = positive predictive value; POE = power of exclusion; POI = power of inclusion.
Validation Patient Group
Of the 748 evaluable patients, 690 (92.2%) did not have cancer and 58 (7.8%) were diagnosed as having bladder cancer by biopsy. Baseline demographic and disease characteristics in patients with and without bladder cancer are given in Table 2. Most patients with cancer had stage Ta tumors (noninvasive papillary carcinoma) (36 of 58 [62.1%]) and either grade 1 (28 of 58 [48.3%]) or grade 3 (25 of 58 [43.1%]) (Table 2).
TABLE 2.
| Biopsy/cystoscopy result |
||
|---|---|---|
| Characteristic | Positivec (n=58) | Negatived (n=690) |
| Age (y), mean (SD) | 71 (10.0) | 64 (9.6) |
| Age | ||
| <70 y | 29 (50.0) | 494 (71.6) |
| ≥70 y | 29 (50.0) | 196 (28.4) |
| Male sex | 48 (82.8) | 402 (58.3) |
| Smoking status | ||
| Yes | 47 (81.0) | 397 (57.5) |
| No | 11 (18.9) | 289 (41.9) |
| Unknown | 0 | 4 (0.6) |
| Hematuria | ||
| Gross | 43 (74.1) | 217 (31.4) |
| Microscopic | 15 (25.9) | 473 (68.5) |
| Determination of disease diagnosise | ||
| Biopsy | 58 (100) | 20 (2.9) |
| Cystoscopy | 0 | 670 (97.1) |
| TNM staging | ||
| Ta | 36 (62.1) | NA |
| Tis | 2 (3.4) | NA |
| T1 | 15 (25.9) | NA |
| T2 | 2 (3.4) | NA |
| T3 | 3 (5.2) | NA |
| Grade | ||
| 1 | 28 (48.3) | NA |
| 2 | 4 (6.9) | NA |
| 3 | 25 (43.1) | NA |
| Unknown | 1 (1.7) | NA |
NA = not applicable.
Data are given as No. (percentage) unless indicated otherwise.
Results by cystoscopy/biopsy = positive or evidence of disease.
Results by cystoscopy/biopsy = negative or no evidence of disease.
Determination of disease diagnosis refers to the method by which the determination of disease absence (cystoscopy negative) was made. The hierarchy of test results is biopsy, rigid cystoscopy, and flexible cystoscopy. The determination of disease presence (cystoscopy positive) can be made only by biopsy.
The combination of all 4 biomarkers identified 395 of the patients as unlikely to have cancer, of which 388 were confirmed negative by biopsy/cystoscopy (Table 3), resulting in a 98.2% NPV (Table 4). If the cancer prevalence was identical to that of the entire study population (6.4% prevalence) before samples were split into derivation and validation sets, then the prevalence-adjusted NPV would be 98.5%. FGFR3 mutant DNA was found in the urine of 21 patients, of which 20 were confirmed by biopsy (Table 5), indicating that FGFR3 had PPV of 95.2% and specificity of 99.9% (Table 6) and suggesting that these patients likely have cancer and should receive accelerated intervention. Of the 58 patients diagnosed by biopsy as having bladder cancer, 51 (87.5%) were above the 4-marker combination cutoff values. Twenty of these 58 patients (34.5%) had mutant FGFR3 DNA in their urine, indicating that they likely had bladder cancer (Table 5). Subgroup analysis did not find an association of MADR assay findings with smoking status or age (data not shown).
TABLE 3.
| Biopsy positive | Biopsy/cystoscopy negative | Total | |
|---|---|---|---|
| Combined biomarkers intermediate/positive | 51 (14.4) | 302 (85.6) | 353 |
| Combined biomarkers negative | 7 (1.8) | 388 (98.2) | 395 |
| Total | 58 (7.8) | 690 (92.2) | 748 |
MADR = multianalyte diagnostic readout.
Data are presented as No. (percentage).
TABLE 4.
MADR Assay Performance Characteristics (NPV) for the Validation Population
| Sensitivity | Specificity | NPV | PPV | |
|---|---|---|---|---|
| Combined biomarkers | ||||
| No. (%) | 51/58 (87.9) | 388/690 (56.2) | 388/395 (98.2) | ND |
| 95% CI | 76.0-94.5 | 52.4-60.0 | 96.1-99.3 | ND |
CI = confidence interval; MADR = multianalyte diagnostic readout; ND = not determined; NPV = negative predictive value; PPV = positive predictive value.
TABLE 5.
FGFR3 Assay Performance Compared With Cystoscopy/Biopsy for the Validation Populationa
| Biopsy positive | Biopsy/cystoscopy negative | Total | |
|---|---|---|---|
| FGFR3 positive | 20 (95.2) | 1 (4.8) | 21 |
| FGFR3 negative | 38 (5.2) | 689 (94.8) | 727 |
| Total | 58 (7.8) | 690 (92.2) | 748 |
Data are presented as No. (percentage).
TABLE 6.
FGFR3 Assay Performance Characteristics (PPV) for the Validation Population
| Sensitivity | Specificity | NPV | PPV | |
|---|---|---|---|---|
| FGFR3 | ||||
| No. (%) | 20/58 (34.5) | 689/690 (99.9) | ND | 20/21 (95.2) |
| 95% CI | 22.5-48.1 | 99.2-100.0 | ND | 76.2-99.9 |
CI = confidence interval; ND = not determined; NPV = negative predictive value; PPV = positive predictive value.
Since FGFR3 sequence variations are associated with noninvasive tumors of low stage and grade18 and since cytology has poor sensitivity for low-stage tumors but high sensitivity for high-grade and high-stage cancer,9 we asked whether urine cytologic findings might complement the FGFR3 results. This would result in more patients with bladder cancer receiving accelerated intervention. Cytologic data were available for 423 patients (24 with cancer and 399 without). In this subset of patients, FGFR3 had a sensitivity of 33.3% (8 of 24) and a specificity of 100% (Table 7). If cytologic results were limited to those having a negative or positive finding, cytologic testing alone had sensitivity of 29.2% (7 of 24) and specificity of 99.5% (397 of 399). Equivocal (atypical or suspicious) results (n = 41) were not included in these analyses. In combination, FGFR3 and cytologic analysis identified 58.3% (14 of 24) of patients with cancer for accelerated intervention, with specificity of 99.5% (397 of 399) and PPV of 87.5% (14 of 16) (Table 7).
TABLE 7.
FGFR3 and Cytology for PPV
| Sensitivity | Specificity | PPV | |
|---|---|---|---|
| FGFR3 | |||
| No. (%) | 8/24 (33.3) | 399/399 (100.0) | 8/8 (100.0) |
| 95% CI | 15.6-55.3 | 99.1-100.0 | 63.1-100.0 |
| Cytology | |||
| No. (%) | 7/24 (29.2) | 397/399 (99.5) | 7/9 (77.8) |
| 95% CI | 12.6-51.1 | 98.2-99.9 | 40.0-97.2 |
| FGFR3 + cytology | |||
| No. (%) | 14/24 (58.3) | 397/399 (99.5) | 14/16 (87.5) |
| 95% CI | 36.6-77.9 | 98.2-99.9 | 61.7-98.4 |
CI = confidence interval; PPV = positive predictive value.
Discussion
In this study, we used a multianalyte approach to stratify a population of patients with hematuria into groups that represent different patients' risk of having bladder cancer. The 4-marker combination had NPV of 98.2% for identifying patients who were not likely to have bladder cancer. FGFR3 alone had high PPV (95.2%) and high specificity (99.9%) for indicating patients who likely had bladder cancer and should receive accelerated intervention. Previous studies have also found that urine-based analysis of FGFR3 sequence variations is sensitive in detecting cancer of urothelial origin.17-21 This assay triaged 55.6% of the entire population presenting with hematuria as either positive or negative for bladder cancer. Although the remaining patients had intermediate results, these patients would be considered at standard risk for bladder cancer and could continue to be evaluated by standard methods. This approach of triaging patients into 3 groups may allow the physician to more selectively apply invasive procedures.
Although the AUA guidelines recommend that all patients who meet the hematuria criteria be evaluated by a urologist, and despite the risk of urinary tract malignancy, referral rates do not reflect compliance with the guidelines.1-4 The poor referral rates for patients may, in part, reflect the fact that there are multiple noncancerous underlying reasons for hematuria, some of which are not life threatening, making diagnosis and treatment decisions difficult.
The MADR assay is a noninvasive urine-based assay that stratifies patients with very low likelihood of bladder cancer from those of higher risk who should receive the standard of care or who are likely to have the disease and should receive accelerated intervention. The high NPV of the MADR assay (98.2%) provides physicians with certainty that a patient with a negative result has an extremely low likelihood of having cancer. This could translate into cystoscopy avoidance in properly selected patients, such as those with microhematuria and little to no risk factors for bladder cancer. This is supported by previous work that found that after a thorough evaluation, patients with hematuria who do not have bladder cancer have little risk of developing the disease.13,22,23 Although this may not obviate the need for evaluation of other sources of varying degrees of hematuria with traditional methods, it provides the physician with an additional tool that is associated with high certainty. Also, because of FGFR3's high PPV (95.2%), a positive FGFR3 finding may increase the rate at which a patient is referred to urology services and may potentially improve clinical outcomes. The MADR assay is unique because it stratifies the patient population and eliminates the confusion created by having high numbers of false-positive and false-negative results that are associated with many noninvasive assays.
Unlike other noninvasive assays, the MADR assay is not affected by the degree of hematuria or the presence of other urinary tract diseases.24 Moreover, it can reliably detect low-stage and low-grade disease and does not rely only on qualitative results or require large volumes of urine or intact cells.
Since the FGFR3 assay can identify patients with low-grade tumors, it might be used in conjunction with cytologic testing for diagnosing bladder cancer. This concept is supported by the findings that the combination of FGFR3 and cytology had specificity of 99.5% and PPV of 87.5%. Although the PPV for the combination of FGFR3 and cytology was lower than that for FGFR3 alone (95.2%), these results demonstrate the usefulness of combining assays that target tumors that differ in stage and grade. Incorporating a second molecular marker that has higher sensitivity and specificity than cytologic testing and that is not dependent on intact cells would complement FGFR3.
The AUA recommends that all patients at high risk for bladder cancer (eg, age >50 years, a ≥10-year smoking history, and ≥15 years of environmental exposure) and 1 positive urinalysis result for hematuria (3 red blood cells per high-power field) be referred to a urologist for evaluation.13 However, patients with hematuria represent a heterogeneous population with only approximately 5% being positive for bladder cancer, and the evidence supporting this recommendation is scanty. Referring all at-risk patients for a complete urologic assessment could be costly and could unnecessarily use limited resources.25,26 The MADR assay could help with stratification and better determine individual patient risk.
Conclusion
An assay to triage patients with hematuria for bladder cancer should be accurate and sufficiently simple so that samples can be collected and analyzed easily and rapidly. This unique approach uses biomarker combinations and cutoff values to achieve high NPV and PPV to stratify patient populations into 3 groups: those without bladder cancer, those who might have bladder cancer, and those who should receive the standard care. This study showed that the noninvasive urine-based MADR assay that uses the best performances of 4 biomarkers can accurately stratify patients into the different categories. The MADR test was developed with performance features to serve as an adjunct to the initial evaluation of patients with hematuria and not to supplant appropriate urologic referrals.
Acknowledgments
We thank Elizabeth Goodwin, PhD, and John Millholland, PhD, for editorial support; Ann Murphy Legg, RN for study design and oversight; and all the clinical collaborators on this study: James Bailen, MD, Yitzhak Berger, MD, Daniel Burzon, MD, James Cochran, MD, Jeffrey Dann, MD, Bernard Hertzmann, MD, Evan Goldfischer, MD, Daniel Saltzstein, MD, Neil Shore, MD, Mark White, MD, David Jablonski, MD, Caroline Ryan, MD, Joel Bass, MD, Eugene Kramolowsky, MD, Nicolas Franco, MD, Gary Goldberg, MD, Lewis Kriteman, MD, Lawrence Karsh, MD, Kenneth Kernen, MD, Victor Abraham, MD, Adam Stage, MD, Guy Bernstein, MD, Michael Perrotti, MD, Simon Mirelman, MD, Mark Schoenberg, MD, and Jed Kaminetsky, MD.
Footnotes
Potential Competing Interests: Dr Fernandez and Mr Shuber are employees of Predictive Biosciences Inc. Dr Karnes and Mayo Clinic received funding from Predictive Biosciences.
Supplemental Online Material
References
- 1.Nieder A.M., Lotan Y., Nuss G.R. Are patients with hematuria appropriately referred to urology?: a multi-institutional questionnaire based survey. Urol Oncol. 2010;28(5):500–503. doi: 10.1016/j.urolonc.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Khadra M.H., Pickard R.S., Charlton M., Powell P.H., Neal D.E. A prospective analysis of 1,930 patients with hematuria to evaluate current diagnostic practice. J Urol. 2000;163(2):524–527. [PubMed] [Google Scholar]
- 3.Johnson E.K., Daignault S., Zhang Y., Lee C.T. Patterns of hematuria referral to urologists: does a gender disparity exist? Urology. 2008;72(3):498–502. doi: 10.1016/j.urology.2008.01.086. [DOI] [PubMed] [Google Scholar]
- 4.Elias K., Svatek R.S., Gupta S., Ho R., Lotan Y. High-risk patients with hematuria are not evaluated according to guideline recommendations. Cancer. 2010;116(12):2954–2959. doi: 10.1002/cncr.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen R.A., Brown R.S. Clinical practice: microscopic hematuria. N Engl J Med. 2003;348(23):2330–2338. doi: 10.1056/NEJMcp012694. [DOI] [PubMed] [Google Scholar]
- 6.Singh R., Saleemi A., Walsh K., Popert R., O'Brien T. Near misses in bladder cancer: an airline safety approach to urology. Ann R Coll Surg Engl. 2003;85(6):378–381. doi: 10.1308/003588403322520717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messing E.M., Young T.B., Hunt V.B. Comparison of bladder cancer outcome in men undergoing hematuria home screening versus those with standard clinical presentations. Urology. 1995;45(3):387–396. doi: 10.1016/s0090-4295(99)80006-5. [DOI] [PubMed] [Google Scholar]
- 8.Chou R., Dana T. Screening adults for bladder cancer: a review of the evidence for the U.S. Preventive Services Task Force. Ann Intern Med. 2010;153(7):461–468. doi: 10.7326/0003-4819-153-7-201010050-00009. [DOI] [PubMed] [Google Scholar]
- 9.Lokeshwar V.B., Habuchi T., Grossman H.B. Bladder tumor markers beyond cytology: International Consensus Panel on bladder tumor markers. Urology. 2005;66(6, suppl 1):35–63. doi: 10.1016/j.urology.2005.08.064. [DOI] [PubMed] [Google Scholar]
- 10.Brausi M., Witjes J.A., Lamm D. A review of current guidelines and best practice recommendations for the management of nonmuscle invasive bladder cancer by the International Bladder Cancer Group. J Urol. 2011;186(6):2158–2167. doi: 10.1016/j.juro.2011.07.076. [DOI] [PubMed] [Google Scholar]
- 11.Wild P.J., Fuchs T., Stoehr R. Detection of urothelial bladder cancer cells in voided urine can be improved by a combination of cytology and standardized microsatellite analysis. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1798–1806. doi: 10.1158/1055-9965.EPI-09-0099. [DOI] [PubMed] [Google Scholar]
- 12.McDonald M.M., Swagerty D., Wetzel L. Assessment of microscopic hematuria in adults. Am Fam Physician. 2006;73(10):1748–1754. [PubMed] [Google Scholar]
- 13.Moyer V.A. Screening for bladder cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2011;155(4):246–251. doi: 10.7326/0003-4819-155-4-201108160-00008. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez C.A., Wszolek M.F., Loughlin K.R., Libertino J.A., Summerhayes I.C., Shuber A.P. A novel approach to using matrix metalloproteinases for bladder cancer. J Urol. 2009;182(5):2188–2194. doi: 10.1016/j.juro.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 15.Hong Y.M., Loughlin K.R. Economic impact of tumor markers in bladder cancer surveillance. Urology. 2008;71(1):131–135. doi: 10.1016/j.urology.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Duggan B., Williamson K. Molecular markers for predicting recurrence, progression and outcomes of bladder cancer (do the poster boys need new posters?) Curr Opin Urol. 2004;14(5):277–286. doi: 10.1097/00042307-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 17.van Oers J.M., Lurkin I., van Exsel A.J. A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clin Cancer Res. 2005;11(21):7743–7748. doi: 10.1158/1078-0432.CCR-05-1045. [DOI] [PubMed] [Google Scholar]
- 18.Zuiverloon T.C., van der Aa M.N., van der Kwast T.H. Fibroblast growth factor receptor 3 mutation analysis on voided urine for surveillance of patients with low-grade non-muscle-invasive bladder cancer. Clin Cancer Res. 2010;16(11):3011–3018. doi: 10.1158/1078-0432.CCR-09-3013. [DOI] [PubMed] [Google Scholar]
- 19.van Rhijn B.W., Lurkin I., Radvanyi F., Kirkels W.J., van der Kwast T.H., Zwarthoff E.C. The fibroblast growth factor receptor 3 (FGFR3) mutation is a strong indicator of superficial bladder cancer with low recurrence rate. Cancer Res. 2001;61(4):1265–1268. [PubMed] [Google Scholar]
- 20.van Oers J.M., Zwarthoff E.C., Rehman I. FGFR3 mutations indicate better survival in invasive upper urinary tract and bladder tumours. Eur Urol. 2009;55(3):650–657. doi: 10.1016/j.eururo.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Kompier L.C., van der Aa M.N., Lurkin I. The development of multiple bladder tumour recurrences in relation to the FGFR3 mutation status of the primary tumour. J Pathol. 2009;218(1):104–112. doi: 10.1002/path.2507. [DOI] [PubMed] [Google Scholar]
- 22.Messing E.M., Madeb R., Young T. Long-term outcome of hematuria home screening for bladder cancer in men. Cancer. 2006;107(9):2173–2179. doi: 10.1002/cncr.22224. [DOI] [PubMed] [Google Scholar]
- 23.Madeb R., Golijanin D., Knopf J. Long-term outcome of patients with a negative work-up for asymptomatic microhematuria. Urology. 2010;75(1):20–25. doi: 10.1016/j.urology.2009.06.107. [DOI] [PubMed] [Google Scholar]
- 24.Tilki D., Burger M., Dalbagni G. Urine markers for detection and surveillance of non-muscle-invasive bladder cancer. Eur Urol. 2011;60(3):484–492. doi: 10.1016/j.eururo.2011.05.053. [DOI] [PubMed] [Google Scholar]
- 25.Mohr D.N., Offord K.P., Owen R.A., Melton L.J., III Asymptomatic microhematuria and urologic disease: a population-based study. JAMA. 1986;256(2):224–229. [PubMed] [Google Scholar]
- 26.Feifer A.H., Steinberg J., Tanguay S., Aprikian A.G., Brimo F., Kassouf W. Utility of urine cytology in the workup of asymptomatic microscopic hematuria in low-risk patients. Urology. 2010;75(6):1278–1282. doi: 10.1016/j.urology.2009.09.091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


