Abstract
Bemisia tabaci (Hemiptera: Aleyrodidae) is a globally distributed pest composed of at least 34 morphologically indistinguishable cryptic species. At least seven species of endosymbiont have been found infecting some or all members of the complex. The origin(s) of the associations between specific endosymbionts and their whitefly hosts is unknown. Infection is normally vertical, but horizontal transmission does occur and is one way for new infections to be introduced into individuals. The relationships between the different members of the cryptic species complex and the endosymbionts have not been well explored. In this study, the phylogenies of different cryptic species of the host with those of their endosymbionts were compared. Of particular interest was whether there was evidence for both coevolution and horizontal transmission. Congruence was observed for the primary endosymbiont, Portiera aleyrodidarum, and partial incongruence in the case of two secondary endosymbionts, Arsenophonus and Cardinium and incongruence for a third, Wolbachia. The patterns observed for the primary endosymbiont supported cospeciation with the host while the patterns for the secondary endosymbionts, and especially Wolbachia showed evidence of host shifts and extinctions through horizontal transmission rather than cospeciation. Of particular note is the observation of several very recent host shift events in China between exotic invader and indigenous members of the complex. These shifts were from indigenous members of the complex to the invader as well as from the invader to indigenous relatives.
Introduction
A large number of herbivorous insects including the phloem feeding insects of the Hemiptera suborder Sternorrhyncha (aphids, whiteflies, psyllids, scales and mealybugs) harbour endosymbiotic bacteria [1]. The phloem is nutrient deficient, and the metabolites produced by some of these bacteria fortify their diet [2]. These bacteria, referred to as the primary endosymbionts (P-endosymbionts), are confined to specialized host cells called bacteriocytes (or mycetocytes) which form the bacteriome [3] and are transmitted vertically from mother to offspring [4]. In addition, insects have more recently acquired a number of endosymbiotic bacteria which in many cases live outside the bacteriocytes [3]–[5]. These bacteria are designated as secondary endosymbionts (S-endosymbionts). Their relationship with the host may be either facultative or obligatory and as well as being transmitted vertically to offspring, may also be transmitted horizontally through either direct and indirect contact with other infected individuals [4], [6], [7].
Cospeciation studies of the genetic relationships between P-endosymbionts and their hosts usually reveal high levels of congruence. This suggests an ancient infection of an insect ancestor by a bacterium followed by its vertical transmission and subsequent cospeciation with the host [8]. In contrast, there is usually a lack of evolutionary congruence between the S-endosymbionts and their hosts, suggesting both multiple infections over time as well as horizontal transfer between unrelated hosts [4], [7], [9]–[13]. Despite the knowledge that S-endosymbionts can be transmitted horizontally, there is little data as to whether the level of horizontal transmission across the different S-endosymbionts is equivalent. One approach used to infer the transmission and evolutionary history of endosymbiont infections is to contrast the evolutionary relationships between the endosymbionts and their hosts.
Endosymbiont infections in arthropods are common [14] and amongst these, the whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae), contains the greatest known diversity with seven so far being described [8], [15], [16,]. Studies of the Aleyrodidae show that a distinct lineage within the gammaproteobacteria of P-endosymbiont is always present [17], [18] and has been given the provisional designation Candidatus ‘Portiera aleyrodidarum’ [19]. In addition, B. tabaci is also infected with several species of S-endosymbionts, Candidatus ‘Hamiltonella defensa’ (Enterobacteriaceae), Arsenophonus, Cardinium (Bacteroidetes), Fritschea bemisiae (Simkaniaceae), Rickettsia and Wolbachia [13], [16]–[18], [20]–[24]. Our knowledge of the function of these S-endosymbionts in B. tabaci is limited, but evidence suggests that within populations, the composition of S-endosymbionts varies both temporally and spatially [5], [8], [13], [16], [18], [20], [25]–[28]. Further, S-endosymbiont infections have been shown to carry a fitness benefit [24] as well as cost [29], [30].
Our most recent understanding of B. tabaci is that it is a cryptic species complex of at least 34 morphologically indistinguishable species [31]–[34] that exhibit complete or partial mating isolation [35]–[37]. In the case of complete mating isolation copulation does not usually occur whereas for partial mating isolation, the resulting fitness of the progeny is substantially reduced relative to the parents. The complex has a global distribution with a distinct geographic structure in terms of genetic relatedness, Sub-Saharan Africa, New World, Africa/Mediterranean/Middle East/Asia Minor, Indian Ocean/East Africa and Asia/Australia [31], [32], [38]. Two members of the complex, Middle East - Asia Minor 1 (MEAM1, commonly referred to as biotype B in the literature) and Mediterranean (MED, commonly referred to as biotype Q in the literature) have spread well beyond their respective home ranges through the trade in ornamental plants [38], [39] and both have invaded parts of Asia over the last 20 years [32]. As well as the invaders, Asia has at least 15 indigenous members of the complex; AsiaI, AsiaII_1, AsiaII_2, AsiaII_3, AsiaII_4, AsiaII_5, AsiaII_6, AsiaII_7, AsiaII_8, AsiaII_9, AsiaII_10, AsiaIII, China1, China2, China3 [31], [33]. The question therefore arises as to what is the cause of this level of species diversification. S-endosymbiont diversity is known to vary both within and between different members of the complex [5], [8], [13], [16], [18], [20], [25]–[28] and it is possible that they have contributed to the development of this diversity.
The presence of several cryptic species in the B. tabaci complex might be the result of reproductive isolation induced by S-endosymbionts (e.g. Wolbachia, Arsenophonus, Cardinium) [27], [40]–[42]. All S-endosymbionts are susceptible to horizontal transmission [11], [12], however the high prevalence of Wolbachia [14] across so many species that are unable to copulate suggests it may be particularly prone to horizontal transfer.
One approach to explore whether horizontal transfer is occurring is to compare the evolutionary relationships of the host with those of their endosymbionts. This is usually done using tree-based methods which compare the branching structure to determine whether tree topologies are more similar and if more codivergence events are present than would be expected by chance. Commonly used tree based methods include reconciliation analysis (TreeMap) [43] and generalized parsimony (TreeFitter) [44]. TreeMap is used to find optimal reconstructions of the history of association by maximizing cospeciation events and minimizing host shifts. It uses the Jungles analysis [45] so as to consider all potentially optimal solutions and so enables host shifts to be considered. TreeFitter assigns costs against the four types of co-phylogenetic events (cospeciation, duplication, sorting and host shift) [43], [46] and the optimal solution(s) is the one with the lowest global cost of reconstruction. Cospeciation is the joint speciation of two organisms with a close ecological association (usually a mutualistic or symbiotic relationship). In other words, cospeciation events occur when host and parasitoid species co-diverge and so display parallel cladogenesis. When incongruence occurs this signals the absence of cospeciation and instead suggests that host switching, sorting or duplication may be involved. Host switching events occur when a species successfully colonises a host species other than its current host. Duplication, or intrahost speciation, occurs when a species lineage diverges without the stimulus of host speciation and results in several closely related species on the descendant host lineage. Sorting events occur when species are entirely, or apparently, removed from host species. These reflect events where a species is predicted to occur, but does not [43], [47]. TreeMap and TreeFitter enable the null hypothesis that the two phylogenies are related randomly to be tested by comparing the scores of optimal reconstructions (number of cospeciation events for TreeMap and global cost for TreeFitter) with those of phylogenies that have been obtained randomly through the use of the permutation procedure. Because these programs require fully resolved trees, all combinations of the obtained tree topologies between the host and its endosymbionts need to be tested so as to account for phylogenetic uncertainty. TreeFitter also allows assignment of different costs to the four types of events and by varying these costs, the effect on the test results can then be compared. In addition to tree-based methods, host and parasitoid phylogenies can be assessed for similarity using distance-based and data based methods [48]–[53], but these latter methods are only valid when the genetic data being compared is homologous and as the data in this study are derived from different gene regions, only tree-based methods are appropriate [48], [54], [55]. This approach was therefore adopted to explore the evolutionary relationships between B. tabaci, its P-endosymbiont and three of its S-endosymbionts, Arsenophonus, Cardinium and Wolbachia and to so determine whether the patterns observed were more indicative of cospeciation and horizontal transfer events.
Results
Prevalence of endosymbionts among Asian B. tabaci cryptic species
All 570 individuals (30 individuals per location) collected from 19 different locations were infected with the species of primary endosymbiont, P. aleyrodidarum (Table S1). In case of the S-endosymbionts, 76.3% individuals of different B. tabaci cryptic species were positive for Wolbachia, 34.2% for Arsenophonus and 10.5% for Cardinium (Table S1). Out of a total of 19 collections, 16 positive collections were sequenced for primary endosymbionts and Wolbachia, ten for Arsenophonus and five for Cardinium based on B. tabaci cryptic species, geographical locations and plant hosts of B. tabaci cryptic species. Of the 16 different Wolbachia sequenced for the wsp gene, 11 were from members of the complex that are indigenous to Asia, two AsiaII_1, two AsiaII_7, five AsiaI and two China1 and five to invasive members of the complex, three MED and two MEAM1. For the Wolbachia ftsZ gene, 6 were from species indigenous to Asia, one China1, four Asia1, one AsiaII_7 and five that are invaders, three MED and two MEAM1 (Table S1). Of the 10 different Arsenophonus sequenced, three were from AsiaI, three from AsiaII_1, two from AsiaII_6, one from AsiaII_7 and one from MED while for the five Cardinium, three were from AsiaII_1, one from AsiaII_6 and one from MED (Table S1). All three individuals from each infected sample location were infected with the same haplotype of endosymbiont i.e. there was no variation within location.
Phylogenetic analysis of host B. tabaci cryptic species and their endosymbionts
The phylogenetic reconstruction for B. tabaci complex from Asia and the clusters assigned to each of the different species is shown in Fig. 1A and is consistent with previously published trees [31], [32]. The relationship between the different P. aleyrodidarum is shown in Fig. 1B. The relationships for the most part mirror those of B. tabaci (Fig. 1B).
Figure 1. Phylogenies of the host B. tabaci cryptic species and their P-endosymbionts.
A. Phylogenetic tree reconstruction based on mtCOI sequences (length = 830 bp) of host B. tabaci cryptic species using ML analysis under the HKY+G substitution model. The bootstrap values are indicated. Bemisia afer (GU 220055) is used as outgroup. Accession numbers for Btab1-19 submitted to GenBank are JX428681–JX428696. All mtCOI sequences of host B. tabaci cryptic species used in this study were clustered with other blast references sequences from GenBank and their ML phylogenetic reconstruction is shown Fig. S1A. B. Phylogenetic tree reconstruction based on 16S rRNA gene sequences (length = 1100 bp) of P-endosymbionts of B. tabaci cryptic species using ML analysis under the HKY+G substitution model. The bootstrap values are indicated. Trialeurodes vaporariorum (AF400483) is used as the outgroup. Accession numbers for Ptab1-Ptab16 submitted to GenBank are JX428713–JX428731. All 16S sequences of P-endosymbionts used in this study were clustered with other blast references sequences from GenBank and their ML phylogenetic reconstruction is shown Fig.. S1B.
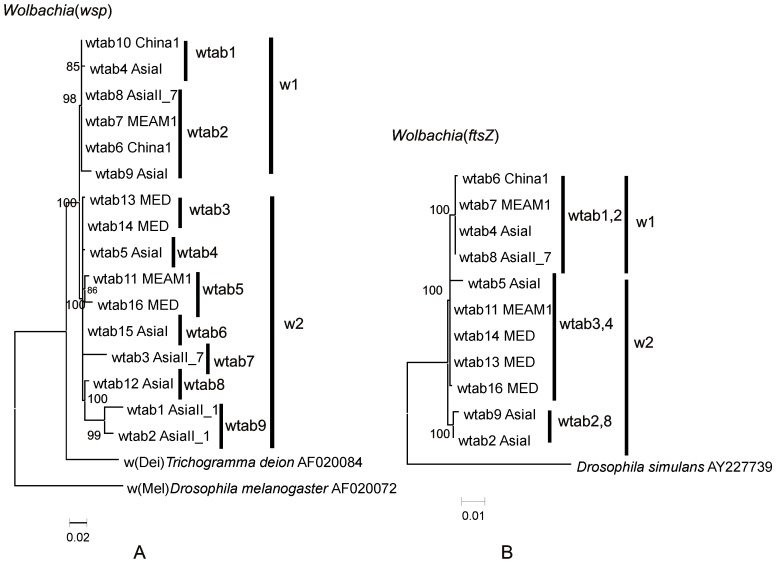
The 16 Wolbachia were grouped into two main clusters, W1 and W2 (Fig. 2 A, B). All Wolbachia sequences found in this study belonged to supergroup B (Fig. 2 A, B). The relationships between different Wolbachia based on their wsp and ftsZ sequences are shown in Fig. 2 A and B, respectively. The wsp tree has nine well supported clades (bootstraps >70%), wtab1-wtab9. Clades wtab1, 4, 6–9 consist of Wolbachia from indigenous Asian members of the complex only and wtab3 and wtab5 consist of invader members only whereas wtab2 has Wolbachia from both indigenous Asian and invader members of the B. tabaci complex (Fig. 2 A).
Figure 2. Phylogenies of the S-endosymbiont Wolbachia.
A. Phylogenetic tree reconstruction based on wsp gene sequences (length = 480 bp) of Wolbachia of B. tabaci cryptic species using maximum-likelihood analysis under the T92 substitution model. The bootstrap values are indicated. Trichogramma deion (AF020084) and Drosophila melanogaster (AF020072) are used as outgroups. Accession numbers for wtab1-16 submitted to GenBank are JX428697–JX428712. All wsp sequences of Wolbachia used in this study were clustered with other blast references sequences from GenBank and their ML phylogenetic reconstruction is shown Fig. C. B. Phylogenetic tree reconstruction based on ftsZ gene sequences (length = 850) of Wolbachia of B. tabaci cryptic species using ML analysis under the TN93 substitution model. The bootstrap values are indicated. Drosophila simulans in case of ftsZ gene (AY227739) is used as the outgroup. Accession numbers for sequences used in the tree are JX428732–JX428742. All ftsZ sequences of Wolbachia used in this study were clustered with other blast references sequences from GenBank and their ML phylogenetic reconstruction is shown Fig. S1D.
The ftsZ tree has three well supported clades (bootstraps >70%). The Wolbachia from different clades of wsp tree were clustered together in same clade here in ftsZ tree (Fig. 2 A). Out of three clades of ftsZ tree, the top clade consists of Wolbachia from indigenous Asian members as well as one from invader member of the complex whereas the middle clade consists of Wolbachia from invader members along with one from indigenous member and the bottom clade consists of Wolbachia from only indigenous Asian members of the complex (Fig. 2 A).
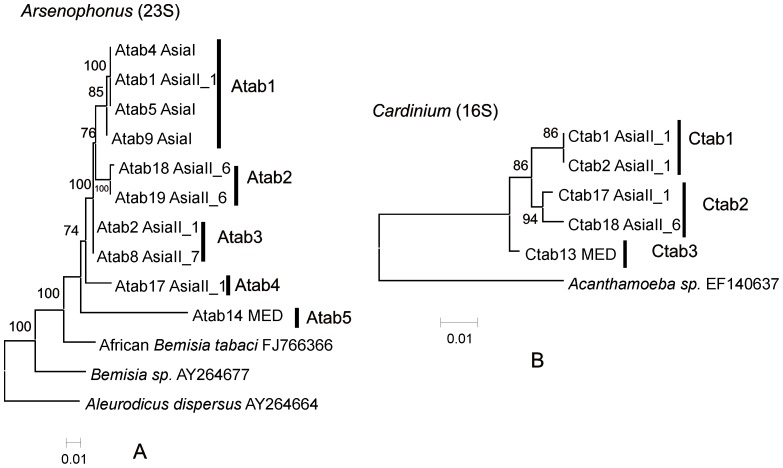
All the Arsenophonus grouped into two well supported clusters Atab1-Atab5 (Fig. 3A). Atab5 had a single representative from MED whereas Atab1-Atab4 represented individuals found in indigenous Asian B. tabaci cryptic species.
Figure 3. Phylogenies of the S-endosymbionts Arsenophonus and Cardinium.
A. Phylogenetic tree reconstruction based on 23S rRNA gene sequences (length = 550 bp) of Arsenophonus of B. tabaci cryptic species using ML analysis under the HKY+G substitution model. The bootstrap values are indicated. African B. tabaci (FJ66366), Bemisia sp. (AY264677) and Aleurodicus disperses (AY264664) are used as the outgroups. Accession numbers for sequences used in the tree are JX428666–JX428675. All 23S sequences of Arsenophonus used in this study were clustered with other blast references sequences from GenBank and their ML phylogenetic reconstruction is shown Fig. S1E. B. Phylogenetic tree reconstruction based on 16S rRNA gene sequences (length = 400) of Cardinium of B. tabaci cryptic species using MLd analysis under the K2 substitution model. The bootstrap values are indicated. Acanthamoeba sp. (EF140637) is used as the out group. Accession numbers for sequences used in the tree are JX428676–JX428680. All 16S sequences of Cardinium used in this study were clustered with other blast references sequences from GenBank and their ML phylogenetic reconstruction is shown Fig. S1F.
Each of the five different Cardinium was belonging to one of two clusters, one containing two groups Ctab1 and Ctab2 and the other Ctab3. Ctab3 was from the invasive MED whereas the others represented individuals from indigenous Asian B. tabaci cryptic species.
Co-phylogenies of B. tabaci host and its endosymbionts
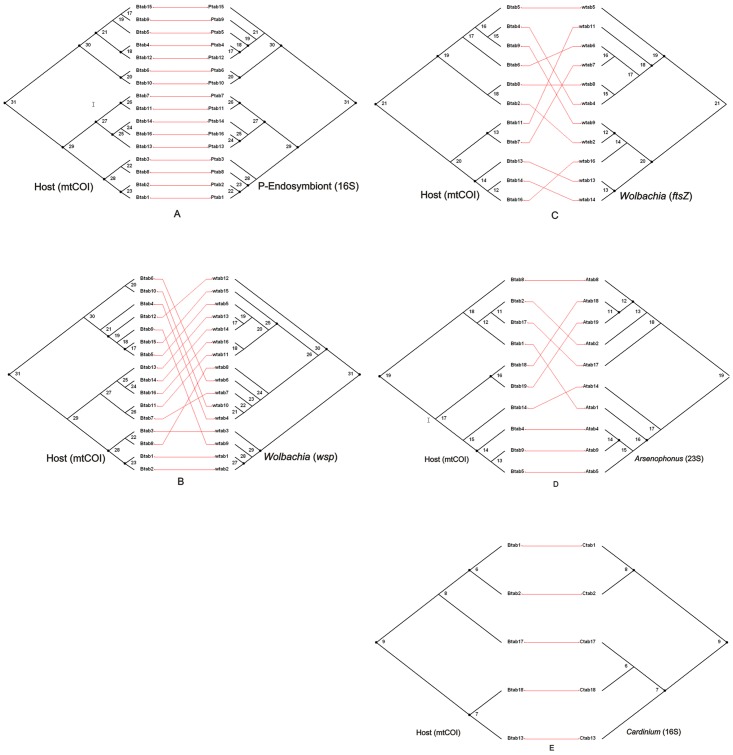
There was high congruence between the B. tabaci mtCOI and P. aleyrodidarum 16S rRNA gene phylogenies (Fig. 4A). In contrast, there is less agreement between the B. tabaci mtCOI and the S-endosymbiont phylogenies. The Wolbachia and B. tabaci phylogenies were incongruent (Fig. 4B, C) whereas the phylogenies for Cardinium and Arsenophonus showed partially congruence with B. tabaci (Fig. 4D and E).
Figure 4. Comparisons of B. tabaci cryptic species and endosymbionts ML phylogenies.
Black dots show cospeciation points. A. Host B. tabaci mtCOI versus P-endosymbiont, P. aleyrodidarum 16S rRNA gene. B. Host B. tabaci mtCOI versus S-endosymbiont, Wolbachia wsp. C. Host B. tabaci mtCOI versus S-endosymbiont, Wolbachia ftsZ genes. D. Host B. tabaci mtCOI versus S-endosymbionts, Arsenophonus 23S rRNA gene. E. Host B. tabaci mtCOI versus S-endosymbionts, Cardinium 16S rRNA gene.
Analysis of the single optimal topology for mtCOI with the ML and MP topologies for endosymbionts using TreeFitter at a p-value significance cut-off of 0.05 showed 12 cospeciation events and only two host switching and sorting events between B. tabaci and their P-endosymbionts (Fig. S2A). The two observed events of host switching in P-endosymbionts were actually between populations of same species (Fig. S2A). In the case of Wolbachia while there was evidence to support six to eight cospeciation events (six in the case of ftsZ and eight in the case of wsp), there was evidence to support a considerable number of random host switches (3–7) and sorting events (13–14) (Fig. S2 B, C). This was also the case for Arsenophonus, five cospeciation events, four host switches and four sorting events (Fig. S2D) and for Cardinium, three cospeciation events, one host switch and one sorting events (Fig. S2E).
Geographical correlations of B. tabaci cryptic species and its endosymbionts
There were no significant correlations between host genetic variation and geographical distances in the case of both invasive and indigenous cryptic species and their P-endosymbionts (r = −0.23, P = 0.097 for indigenous; r = −0.112, P = 0.80 for invasive). Likewise, there are no significant correlations between S-endosymbionts genetic variation (measured separately from invasive and indigenous host cryptic species) and geographical distances between the collection site in case of Wolbachia using ftsZ gene (r = −0.072 P = 0.081 for indigenous; r = 0.232 P = 0.60 for invasive) or using wsp gene (r = −0.27, P = 0.197 for indigenous; r = 0.203, P = 0.66 for invasive) as well as in case of Arsenophonus (r = 0.083,P = 0.75 for indigenous) or Cardinium (r = −0.01, P = 1.00 for indigenous) suggesting that their genotypes are not geographically restricted.
Host mtCOI correlation with P- and S-endosymbionts
The P-endosymbiont, Wolbachia and Arsenophonus were significantly correlated with host mtCOI (Table 1). The correlation was highest for the P-endosymbiont followed by Arsenophonus then Wolbachia (Table 1). The small dataset for Cardinium lead to no significant correlation (Table 2). Wolbachia (wsp gene) was least, but with significant correlation with host genetic diversities while Wolbachia (fstZ gene) was not correlated with host genetic variations (Table 1).
Table 1. The mantel correlations between the genetic differences between B. tabaci sequences and that of their endosymbionts.
| Endosymbionts Genes | n | Lower 95% CI | correlation | Upper 95% CI | P |
| P-Endosymbionts (16S rRNA) | 16 | 0.75 | 0.78 | 0.83 | 0.0001 |
| Arsenophonus (23S rRNA) | 10 | 0.36 | 0.50 | 0.60 | 0.0001 |
| Wolbachia (wsp) | 16 | 0.10 | 0.17 | 0.26 | 0.0156 |
| Wolbachia (fstZ) | 11 | −0.30 | −0.025 | 0.26 | 0.8531 |
| Cardinium (16S rRNA) | 5 | −0.30 | 0.33 | 0.82 | 0.3610 |
Not all samples had all the endosymbionts resulting in different sample sizes (n). The lower and upper 95% confidence intervals (CI) of estimates are also given.
Table 2. Summary of co-phylogenetic comparisons of host species and their endosymbionts ML phylogenies.
| Endosymbionts Genes | Cospeciation | Duplication | Host switch | Sorting |
| P-Endosymbionts (16S rRNA) | 12 | 0 | 2 | 2 |
| Arsenophonus (23S rRNA) | 5 | 0 | 4 | 4 |
| Wolbachia (wsp) | 8 | 0 | 7 | 13 |
| Wolbachia (fstZ) | 6 | 1 | 3 | 14 |
| Cardinium (16S rRNA) | 3 | 0 | 1 | 1 |
The reconstruction shows the co-phylogenetic analysis between the mtCOI gene of B. tabaci cryptic species with the genes of their primary endosymbionts and secondary endosymbionts.
Overall, both distance and tree based analyses produced a very similar result. In the tree based analysis, there was high cospeciation in the case of P-endosymbionts as compared to S-endosymbionts and there are more host switches for the S-endosymbionts, especially in Wolbachia (Table 2, Fig. 4). Similarly in the distance based analysis, P-endosymbionts are highly correlated with their host as compared to S-endosymbionts with Wolbachia showing the lowest level of correlation (Table 1).
Discussion
The results confirmed a very high degree of co-cladogenesis between B. tabaci and P. aleyrodidarum; this was expected and has been shown in numerous studies [56]–[58] and suggests a single infection of a whitefly ancestor with a bacterium followed by vertical transmission to the progeny. In contrast, the phylogenies of the S-endosymbionts were largely incongruent with that of the host. One explanation is that P-endosymbionts are always found within the bacteriocytes [4] which are highly specific cells with a limited distribution within the host. This suggests that being limited to the bacteriocytes makes it more difficult for horizontal transmission and this is supported by the very low number of host switches and sorting events. In contrast, S-endosymbionts are not limited to the bacteriocytes [4], [6] and the much smaller number of cospeciation events and much higher numbers of host switches and sorting events suggest that being able to reside in multiple cell types makes it easier for horizontal transmission to occur. Of the S-endosymbionts, Wolbachia showed a lack of congruence whereas Arsenophonus and Cardinium showed partial congruence. This suggests that of these S-endosymbionts, Wolbachia may be particularly adapted to undergoing horizontal transmission [11], [12]. An alternative explanation is that P-endosymbionts are essential and so there may be extremely strong selection on both host and bacterium to maintain the association [2]. In contrast, S-endosymbionts are either parasites or mutualists. In the case of the former, the association is likely to be unstable due to selection for host resistance while in the later, the association is subject to changes in ecological circumstance and so lost when environmental change selects against the association [7], [59].
It is generally assumed that vertical transmission of Wolbachia predominates within species and that horizontal transfer is a rare event [60], [61]. As a consequence, one would expect the association between the Wolbachia genome and the mitochondrial genome to be non-random i.e. in linkage disequilibrium as both are vertically transmitted from mother to offspring [62]. However, if horizontal transmission is occurring then one would expect the relationship between the two genomes to become decoupled; the extent of which would be indicated by the level of incongruence between the two phylogenies. Our results show a considerable degree of congruence suggesting that horizontal transmission was occurring at a rate sufficient to mask linkage disequilibrium.
The lack of congruence between mitochondrial and Wolbachia gene genealogies cannot be explained in terms of a single infection event followed by co-divergence within the species. Instead, it is strongly compatible with extensive shuffling within the species. Overall, based on the Wolbachia/mtCOI data, the pattern that emerges is a complex history of infection in B. tabaci that is shaped by both vertical and horizontal transmission plus frequent turnover (loss and replacement) within a single species. The higher infection frequency in B. tabaci and the common occurrence of multiple infections in B. tabaci are good arguments that horizontal transfers occurs between different cryptic species of B. tabaci. High rates of horizontal transmission are one explanation for the frequency of Wolbachia infections that have been observed both across the B. tabaci complex as well as in arthropods in general [13], [14], [47]. Moreover, B. tabaci has been shown to harbor different Wolbachia [13]. Theoretically, it has been shown that two different Wolbachia strains can coexist stably in parapatric host populations [63] and that bidirectional cytoplasmic incompatibility reduces the gene flow of locally adapted alleles and selects for pre-mating isolation [63], [64]. Research indicates that different Wolbachia may contribute to reproductive isolation through cytoplasmic incompatibility [65]. It may therefore be that cytoplasmic incompatibility in combination with relatively high rates of horizontal transmission has been promoting the high level of diversity observed across the B. tabaci cryptic species complex. In addition to Wolbachia, Cardinium and Arsenophonus are also known to manipulate host reproduction in a wide range of insect species including B. tabaci [27], [59].
Extensive collections of MEAM1 across its home range (Middle East and Asia Minor) and invaded range have not provided any good evidence that the invading MEAM1 was infected with Wolbachia prior to its invasion of China [8], [15], [16], [31], [32], [38], [66]. There is one study [67] that identifies MEAM1 infected with Wolbachia in Israel, but this observation is in doubt as extensive sampling across Israel has since been unable to find any evidence for Wolbachia infections in MEAM1 (Zchori Fein personnel communication). MEAM1 most likely invaded China in the mid-1990s and studies investigating the presence of S-endosymbiont infections in MEAM1 in mainland China reported no presence of Wolbachia at least up to 2005 whereas studies from 2007 onwards have reported varying levels of infections with Wolbachia [13], [18], [28], [66]–[68, Ahmed MZ et al. unpublished data]. The presence of similar Wolbachia in both Asian indigenous (AsiaI and China1) and exotic members of the B. tabaci complex provides good support for the argument that the Wolbachia in MEAM1 in China was acquired from the indigenous Asian members of the complex and that this has occurred within the last 6 years. On the other hand, the other exotic invader found in China, MED, was first detected in China in 2003 [69]. MED was known to have been infected with Wolbachia before its invasion of China [13], [18] and our study shows a close association between the Wolbachia from Chinese MED and that found in the Mediterranean home range. The observation that some of the Wolbachia infecting AsiaI in China are less related to those from other indigenous Asian Wolbachia and more related to Wolbachia of Mediterranean origin suggests that there has been a recent horizontal transfer of Wolbachia from the invading MED to the indigenous AsiaI and that this has occurred since 2003.
Our study showed that the indigenous Asia1 also harbored another Wolbachia infection that was the same as the one found in MEAM1. However, this also occurred in the other B. tabaci species to have invaded China, MED. This also occurs in MED from Egypt and suggests a second horizontal transfer event in China from MED to Asia1 as well as MEAM1 and again this transfer has occurred within the last 10 years.
Intra- and interspecific horizontal transfers of Wolbachia occur between organisms that interact in close confinement [70]. In general, intraspecific horizontal transfer is more successful than interspecific transfer [70] and is most likely due to incompatibilities between Wolbachia and hosts' nuclear/cytoplasmic background [12], [71]. The apparently frequent interspecific horizontal transmission between different members of the B. tabaci complex suggests that there is sufficient similarity between the different members of the complex to support the Wolbachia from related members of the complex. This is supported by the numerous observations that have been made in regards to courtship interactions between different members of the complex [72]. While copulation seldom, if ever, occurs, there is still a complex courtship process which suggests that the different species are still sufficiently close that interspecies mate recognition persists.
The observation of a number of apparent horizontal transfer events between different members of the B. tabaci complex raises the question as to the mechanism that may be enabling this to occur. The horizontal transfer of Wolbachia from an infected whitefly to other insects as a result of feeding on the same leaf substrate has been suggested as one mechanism [73]. For this to be possible, the bacterium needs to be small enough to pass through the salivary duct. Recent research involving mosquitoes suggests that the bacterium may be too large to pass through the salivary duct making the prospect of transmission through the plant unlikely [74].
Another possible route for horizontal transmission is parasitoids, of which numerous species parasitise the different members of the B. tabaci complex [75]. One study has shown that Rickettsia from B. tabaci were readily able to infect the larvae of two species of parasitoid within the whitefly host, but were unable to be vertically transmitted as the bacteria were unable to infect the parasitoid's oocytes [76]. Gueguen et al. [15] also found evidence for horizontal transfer between different members of the B. tabaci complex and suggested parasitoids may be involved. In contrast, it has been argued that transfer from parasitoids to hosts was quite unlikely [12], but as discussed above, the systems being considering did not involve closely related hosts and so the cytoplasmic backgrounds were likely to be quite different.
It has also been suggested that endosymbiont communities can be used to resolve taxonomic distinction within the B. tabaci complex [15], [20], [77]. As has been shown in aphids [78], our results indicate that the P-endosymbionts may be a useful means of considering the taxonomic relationships within the B. tabaci complex. However, the low level genetic linkage between the host and their S-endosymbionts, especially Wolbachia, due to frequent host shifts through horizontal transfer suggest that they have little role to play in resolving the taxonomy of the B. tabaci complex.
Conclusion
Endosymbionts of B. tabaci cryptic species show evidence for variable degrees of horizontal transmission. Primary endosymbionts are almost entirely vertically transmitted whereas the secondary endosymbionts and especially Wolbachia, show evidence of horizontal transfers between different species within the B. tabaci species complex. This also suggests that secondary endosymbiont genetic variation may not reflect host genetic variation and should not be used to infer taxonomic relationships within the host species complex. This is not surprising as secondary endosymbionts are facultative in their association with the host and so not always present within all individuals within a species. Wolbachia shows evidence a number of horizontal host shifts across the complex with three occurring within the last 20 years. This suggests that it may move readily between different members of the complex and most likely via parasitoid activity. If this is the case, then it is possible that Wolbachia may be playing a role in driving the process of speciation with the B. tabaci species complex and may help explain the high level of species level diversity that is being increasingly observed.
Materials and Methods
Whiteflies sampling
Bemisia tabaci were collected from 19 locations in China between 2007 and 2010. All collections were undertaken by the Department of Entomology, South China Agricultural University, China and no any specific permission was required to collect any of these samples. The identity of the species was determined by comparing their mitochondrial cytochrome oxidase one (mtCOI) against published consensus sequences [31] and using the assignment rules from that study [31], [79].
DNA extraction and PCR amplification
DNA was extracted from individual adult females using a previously published method [13], [18]. The mtCOI was amplified using the primer pair C1-J-2195/TL2-N-3014 [80]. The Portiera 16S ribosomal RNA gene was amplified using the 28F and 1098R primer pair [8] while Wolbachia wsp was amplified using the wsp81F and wsp691R primers [10] and ftsZ using Wolbachia specific primers [9], the Cardinium 16S rRNA gene was amplified by Ch-F and Ch-R primers [81] and the Arsenophonus 23S rRNA gene was amplified using Ars23SF and Ars23R [23]. Amplification for the respective primer sets followed previously published methods [10], [23], [80], [81].
Sequence alignments, measurement of genetic distances and phylogenetic analysis
Several recent studies have shown that exchange of genetic information can occur among Wolbachia supergroups [82], [83] and/or among strains within the same individual [84]. It has been demonstrated that recombination occurs across the whole Wolbachia genome [83] and in most of genes including wsp, gltA, dnaA, ftsZ and groEL [83], [85], [86]. As a result the two of the most frequently used genes (wsp and ftsZ) were used for genotyping Wolbachia, but because of the concern that recombination might influence the patterns observed, recombination analyses were undertaken to assess the level of recombination and identify portions of amplified fragment where recombination had taken place. All amplified gene products were cloned and three clones then sequenced. An additional three clones were sequenced if any sequence variation was detected in the first three clones. The sequences were first analyzed and aligned with DNAStar (Lasergene V5.0) and ClustalX 1.83 [87]. Genetic distances were calculated using the Kimura 2-parameter model in MEGA [88]. The phylogenetic relationships among the different cryptic species and their endosymbionts were analyzed by Maximum Likelihood (ML) using MEGA5 and PAUP*4.0b10 [88], [89]. The tree is drawn to scale, and branch lengths are measured in number of substitutions per site using MEGA5. Bootstrap values >70% from 1,000 iterations are shown.
Recombination analysis
The consequences of incorrect phylogenetic signals caused by recombination can be avoided by first detecting recombination events prior to phylogenetic analysis. Two methods were used to consider recombination. The first was the bootscan method, implemented in the program Simplot [90] to construct replicate trees in a sliding window approach. The threshold level of 70% of the permutated trees in a given window for the detection of a recombination was used. The second used a non-phylogenetic method to apply dynamic programming to minimize the mutation and recombination cost between the various sequences used. This method was implemented in the software RECCO [91] which tests the hypothesis of no recombination and in line with Maydt and Lengauer [91] a significance level of 0.05 as the cutoff was used. The number of permutations was set at 1000, gap extension cost to 0.2, max α for the permutation test to 1 and the significance level to P≤0.05. The parameter α, representing the ratio of mutation cost to recombination cost was set to 0.2, the methods used to calculate mutation and recombination costs were respectively, Hamming and Delta Dirac. Those sequences where significant recombination was identified were not used in the subsequent phylogenetic analyses. Moreover wsp sequences have a number of hypervariable regions which on the basis of Braig et al. [92] and Nirgianaki et al. [20] were excluded from the analysis; this reduced the number of bases available for analysis from 625 to 480.
Co-phylogenetic analysis
The Mantel test on the genetic distance matrices of cospeciating hosts and parasitoids was used to test for significant association between the matrices [48]. TreeFitter was then used to compare the single optimal topology derived from the combined mtCOI sequences of each B. tabaci species and each of ML and MP topologies derived from the combined gene sequences of P- and S-endosymbionts. All tests were performed based on 999 permutations. The support for cospeciation, host-switching, lineage sorting and duplication was explored using the exact search and best reconstructions option in TreeMap 2.0b. Only branches in the ML tree that were supported by at least 85% bootstrap support were considered [93].
Correlation analysis
To test whether the genetic differences between B. tabaci increased with geographic distance, Mantel correlations between B. tabaci genetic differences and mapped distances between collections were undertaken using the mantel function in the ecodist package [94] in R version 2.13.0 [R Development Core Team 2011]. The same approach was then used to analyze how well endosymbiont genetic variability correlated with differences in whitefly genetic variability. Geographical distances between the locations from where the whiteflies were collected were calculated using their GPS coordinates.
Supporting Information
Phylogenetic tree reconstruction of B. tabaci and its endosmbionts based on various gene sequences. A. Phylogenetic tree reconstruction based on mtCOI gene sequences (length = 830 bp) of host B. tabaci cryptic species using maximum-likelihood analysis under the HKY+G substitution model. The bootstrap values are indicated. B. Phylogenetic tree reconstruction based on 16S rRNA gene sequences (length = 1100 bp) of P-endosymbionts of B. tabaci cryptic species using maximum-likelihood analysis under the JC+G substitution model. The bootstrap values are indicated. C. Phylogenetic tree reconstruction based on wsp gene sequences (length = 480 bp) of Wolbachia of B. tabaci cryptic species using maximum-likelihood analysis under the T92+G substitution model. The bootstrap values are indicated. D. Phylogenetic tree reconstruction based on ftsZ gene sequences (length = 850) of Wolbachia of B. tabaci cryptic species using maximum-likelihood analysis under the TN93+G+I substitution model. E. Phylogenetic tree reconstruction based on 23S rRNA gene sequences (length = 550 bp) of Arsenophonus of B. tabaci cryptic species using maximum-likelihood analysis under the HKY+G substitution model. The bootstrap values are indicated. F. Phylogenetic tree reconstruction based on 16S rRNA gene sequences (length = 400) of Cardinium of B. tabaci cryptic species using maximum-likelihood analysis under the K2+G substitution model. The bootstrap values are indicated.
(DOC)
The exact search at best reconstructions using TreeMap 2.0b show the ML phylogenetic comparison of endosymbionts over their hosts. Endosymbionts displayed in black and the hosts in light grey; in reconstruction; cospeciation events are shown by (•), duplications by(▪) and host switches by (→). A. Host B. tabaci and its P-endosymbionts. B. Host B. tabaci and its Wolbachia (wsp). C. Host B. tabaci and its Wolbachia (ftsZ). D. Host B. tabaci and its Arsenophonus (23S). E. Host B. tabaci and its Cardinium (16S).
(DOC)
Asian B. tabaci cryptic species population sampled for S-endosymbionts.
(DOC)
Funding Statement
This research was funded by the Program for New Century Excellent Talents in University 2011, National Basic Research Program of China (973 Project, 2009CB119203) and the China National Natural Science Foundation (31071732). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Moran NA, Baumann P (2000) Bacterial endosymbionts in animals. Curr Opin Microbiol 3: 270–275. [DOI] [PubMed] [Google Scholar]
- 2. Baumann P (2005) Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Ann Rev Microbiol 59: 155–189. [DOI] [PubMed] [Google Scholar]
- 3. Moran NA, Telang A (1998) Bacteriocyte-associated symbionts of insects: a variety of insect groups harbor ancient prokaryotic endosymbionts. BioScience 48: 295–304. [Google Scholar]
- 4.Buchner P (1965) Endosymbiosis of animals with plant microorganisms, p. 332–338. John Wiley and Sons Interscience, New York, N.Y.
- 5. Costa HS, Westcot DM, Ullman DE, Rosell R, Brown JK, et al. (1995) Morphological variation in Bemisia endosymbionts. Protoplasma 189: 194–202. [Google Scholar]
- 6.Baumann PN, Moran A, Baumann L (2000) Bacteriocyte-associated endosymbionts of insects. In M. Dworkin (ed.), The prokaryotes. [Online.] Springer, New York, N.Y. Available: http://link.springer.de/link/service/books/10125.
- 7. Feldhaar H (2011) Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36: 533–543. [Google Scholar]
- 8. Zchori-Fein E, Brown JK (2002) Diversity of prokaryotes associated with Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Ann Entomol Soc Am 95: 711–718. [Google Scholar]
- 9. Werren JH, Zhang W, Guo LR (1995) Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc R Soc Lond B Biol Sci 261: 55–63. [DOI] [PubMed] [Google Scholar]
- 10. Zhou W, Rousset F, O'Neill SL (1998) Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc R Soc Lond B Biol Sci 265: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russell JA, Latorre A, Sabater-Muñoz B, Moya A, Moran NA (2003) Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol 12: 1061–1075. [DOI] [PubMed] [Google Scholar]
- 12. Vavre F, Fleury F, Lepetit D, Fouillet P, Bouletreau M (1999) Phylogenetic evidence for horizontal transmission of Wolbachia in host- parasitoid associations. Mol Biol Evol 1: 61711–1723. [DOI] [PubMed] [Google Scholar]
- 13. Ahmed MZ, Ren SX, Mandour NS, Qiu BL (2010) Prevalence of Wolbachia supergroups A and B in the sweetpotato whitefly Bemisia tabaci and its natural enemies. J Eco Entomol 103: 1848–1059. [DOI] [PubMed] [Google Scholar]
- 14. Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH (2008) How many species are infected with Wolbachia? — a statistical analysis of current data. FEMS Microbiol Lett 281: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gueguen G, Vavre F, Gnankine O, Peterschmitt M, Charif D, et al. (2010) Endosymbiont metacommunities, mtDNA diversity and the evolution of the Bemisia tabaci (Hemiptera: Aleyrodidae) species complex. Mol Ecol 19: 4365–4378. [DOI] [PubMed] [Google Scholar]
- 16. Chiel E, Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Katzir N, et al. (2007) Biotype-dependent secondary symbionts communities in sympatric populations of Bemisia tabaci. . Bull Entomol Res 97: 407–413. [DOI] [PubMed] [Google Scholar]
- 17. Clark MA, Baumann L, Munson MA, Baumann P, Campbell BC, et al. (1992) The eubacterial endosymbionts of whiteflies (Homoptera: Aleyrodidae) constitute a lineage distinct from the endosymbionts of aphids and mealybugs. Curr Microbiol 25: 119–123. [Google Scholar]
- 18. Ahmed MZ, Xue X, Li XX, Ren SX, Jin GH, et al. (2010) Prevalence of endosymbionts in Bemisia tabaci populations and their in vivo sensitivity to antibiotics. Curr Microbiol 61: 322–328. [DOI] [PubMed] [Google Scholar]
- 19. Thao ML, Baumann P (2004) Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl Environ Microbiol 70: 3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nirgianaki A, Banks GK, Frohlich DR, Veneti Z, Braig HR, et al. (2003) Wolbachia infections of the whitefly Bemisia tabaci . Curr Microbiol 47: 93–101. [DOI] [PubMed] [Google Scholar]
- 21. Everett KDE, Thao M, Horn M, Dyszynski GE, Baumann P (2005) Novel chlamydiae in whiteflies and scale insects:endosymbionts ‘Candidatus Fritschea bemisiae’ strain Falk and ‘Candidatus Fritschea eriococci’ strain Elm. Int J Syst Evol Microbiol 55: 1581–1587. [DOI] [PubMed] [Google Scholar]
- 22. Weeks AR, Velten R, Stouthamer R (2003) Incidence of a new sex-ratio-distorting endosymbiotic bacterium among arthropods. Proc R Soc Lond B Biol Sci 270: 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thao ML, Baumann P (2004) Evidence for multiple acquisition of Arsenophonus by whitefly species (Sternorrhyncha: Aleyrodidae). Curr Microbiol 48: 140–144. [DOI] [PubMed] [Google Scholar]
- 24. Himler AG, Adachi-Hagimori TA, Bergen JE, Kozuch A, Kelly SE, et al. (2011) Rapid spread of a bacterial symbionts in an invasive whitefly is driven by fitness benefits and female bias. Science 332: 254–256. [DOI] [PubMed] [Google Scholar]
- 25. Ruan YM, Liu SS (2005) Detection and phylogenetic analysis of prokaryotic endosymbionts in Bemisia tabaci . Acta Entomol Sin 48: 859–865. [Google Scholar]
- 26. Ruan YM, Xu J, Liu SS (2006) Effects of antibiotics on fitness of the B biotype and a non-B biotype of the whitefly Bemisia tabaci . Entomol Experiment Appl 121: 159–166. [Google Scholar]
- 27. Thierry M, Becker N, Hajri A, Reynaud B, Lett JM, et al. (2011) Symbiont diversity and non-random hybridization among indigenous (Ms) and invasive (B) biotypes of Bemisia tabaci . Mol Ecol 20: 2172–2187. [DOI] [PubMed] [Google Scholar]
- 28. Chu D, Gao CS, De Barro P, Zhang YJ, Wan FH, et al. (2011) Further insights into the strange role of bacterial endosymbionts in whitefly, Bemisia tabaci: Comparison of secondary symbionts from biotypes B and Q in China. Bull Entomol Res 101: 477–486. [DOI] [PubMed] [Google Scholar]
- 29. Kontsedalov S, Zchori-Fein E, Chiel E, Gottlieb Y, Inbar M, et al. (2008) The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manag Sci 64: 789–792. [DOI] [PubMed] [Google Scholar]
- 30. Ghanim M, Kontsedalov S (2009) Susceptibility to insecticides in the Q biotype of Bemisia tabaci is correlated with bacterial symbiont densities. Pest Manag Sci 65: 939–942. [DOI] [PubMed] [Google Scholar]
- 31. Dinsdale A, Cook L, Riginos C, Buckley YM, Barro PD (2010) Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann Entomol Soc Am 103: 196–208. [Google Scholar]
- 32. De Barro PJ, Liu SS, Boykin LM, Dinsdale AB (2011) Bemisia tabaci: A statement of species status. Ann Rev Entomol 56: 1–19. [DOI] [PubMed] [Google Scholar]
- 33. Hu J, De Barro P, Zhao H, Wang J, Nardi F, et al. (2011) An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS One 6 (1) e16061 doi:10.1371/journal.pone.0016061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boykin LM, Armstrong KF, Kubatko L, De Barro P (2012) Species delimitation and global biosecurity. Evol Bioinform 8: 1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu J, De Barro PJ, Liu SS (2010) Reproductive incompatibility among genetic groups of Bemisia tabaci supports the proposition that the whitefly is a cryptic species complex. Bull Entomol Res 100: 359–366. [DOI] [PubMed] [Google Scholar]
- 36. Wang P, Sun DB, Qiu BL, Liu SS (2011) The presence of six putative species of the whitefly Bemisia tabaci complex in China as revealed by crossing experiments. Insect Sci 18: 67–77. [Google Scholar]
- 37. Liu SS, Colvin J, De Barro PJ (2012) Species concepts as applied to the whitefly Bemisia tabaci systematics: how many species are there? J Integ Agri 11: 176–186. [Google Scholar]
- 38. De Barro P, Ahmed MZ (2011) Genetic networking of the Bemisia tabaci cryptic Species complex reveals pattern of biological invasions. PLoS One 6 (10) e25579 doi:10.1371/journal.pone.0025579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dalton R (2006) The Christmas invasion. Nature 443: 898–900. [DOI] [PubMed] [Google Scholar]
- 40. Shoemaker DD, Katju V, Jaenike J (1999) Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria . Evolution 53: 1157–1164. [DOI] [PubMed] [Google Scholar]
- 41.Werren JH (1998) Wolbachia and speciation. In: Howard D, Berlocher S (eds) Endless forms: species and speciation, Oxford: Oxford University Press, 245–260.
- 42. Bordenstein SR, O'Hara FP, Werren JH (2001) Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia . Nature 409: 707–710. [DOI] [PubMed] [Google Scholar]
- 43. Page RDM (1994) Parallel phylogenies: reconstructing the history of host-parasite assemblages. Cladistics 10: 155–173. [Google Scholar]
- 44. Ronquist F (1995) Reconstructing the history of host-parasite associations using generalized parsimony. Cladistics 11: 73–89. [DOI] [PubMed] [Google Scholar]
- 45. Charleston MA (1998) Jungles: A new solution to the host/parasite phylogeny reconciliation problem. Math Biosci 149: 191–223. [DOI] [PubMed] [Google Scholar]
- 46. Page RDM, Charleston MA (1998) Trees within trees: phylogeny and historical associations. Trends Ecol Evol 13: 356–359. [DOI] [PubMed] [Google Scholar]
- 47. Paterson AM, Banks J (2001) Analytical approaches to measuring cospeciation of host and parasites: through a glass, darkly. Intr J Parasitol 31: 1012–1022. [DOI] [PubMed] [Google Scholar]
- 48. Light JE, Hafner M (2008) Codivergence in Heteromyid rodents (Rodentia: Heteromyidae) and their sucking lice of the genus Fahrenholzia (Phthiraptera: Anoplura). Syst Biol 57: 449–465. [DOI] [PubMed] [Google Scholar]
- 49.Legendre P (2001) Test of host-parasite coevolution: Program ParaFit user's guide. D'epartement de Sciences Biologiques, Universit'e de Montr'eal, Montr'eal.
- 50. Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order of Hominoidea. J Mol Evol 29: 170–179. [DOI] [PubMed] [Google Scholar]
- 51. Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16: 1114–1116. [Google Scholar]
- 52. Huelsenbeck JP, Rannala B, Larget B (2000) A Bayesian framework for the analysis of cospeciation. Evolution 54: 352–364. [DOI] [PubMed] [Google Scholar]
- 53. Johnson KP, Drown MD, Clayton DH (2001) A data based parsimony method of cophylogenetic analysis. Zooligica Scr 30: 79–87. [Google Scholar]
- 54.Light JE (2005) Host-parasite cophylogeny and rates of evolution in two rodent-louse assemblages. PhD dissertation University of Michigan.
- 55. Light JE, Hafner MS (2007) Disparate rates of evolution in sympatric lineages of chewing lice on pocket gophers. Mol Phy Evol 45: 997–1013. [DOI] [PubMed] [Google Scholar]
- 56. Peek AS, Feldman RA, Lutz RA, Vrijenhoek RC (1998) Cospeciation of chemoautotrophic bacteria and deep sea clams. PNAS 95: 9962–9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, et al. (2000) Cospeciation of psyllids and their prokaryotic endosymbionts. Appl Environ Microbiol 66: 2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baumann L, Thao ML, Funk CJ, Falk BW, James CK, et al. (2004) Sequence analysis of DNA fragments from the genome of the primary endosymbiont of Bemisia tabaci. . Curr Microbiol 48: 77–81. [DOI] [PubMed] [Google Scholar]
- 59. Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6: 741–751. [DOI] [PubMed] [Google Scholar]
- 60. Keller GP, Windsor DM, Saucedo JM, Werren JH (2004) Reproductive effects and geographical distributions of two Wolbachia strains infecting the Neotropical beetle, Chelymorpha alternans Boh. (Chrysomelidae, Cassidinae). Mol Ecol 13: 2405–2420. [DOI] [PubMed] [Google Scholar]
- 61. Shoemaker DD, Dyer KA, Ahren M, Mcabee K, Jaenike J (2004) Decreased diversity but increased substitution rate in host mtDNA as a consequence of Wolbachia endosymbiont. Genetics 168: 2049–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hurst GD, Jiggins FM (2005) Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc R Soc Lond B Biol Sci 272: 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Telschow A, Yamamura N, Werren JH (2005) Bidirectional cytoplasmic incompatibility and the stable coexistence of two Wolbachia strains in parapatric host populations. J Theor Biol 235: 265–274. [DOI] [PubMed] [Google Scholar]
- 64. Telschow A, Hammerstein P, Werren JH (2002) Effects of Wolbachia on genetic divergence between populations: mainland-island model. Integr Comp Biol 42: 340–351. [DOI] [PubMed] [Google Scholar]
- 65. Breeuwer JAJ, Werren JH (1990) Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature 346: 558–560. [DOI] [PubMed] [Google Scholar]
- 66. Chu D, Cong B, Zhang YJ, Xu BY, Wu QJ, et al. (2005) Detection and phylogenetic analysis of Wolbachia in different Bemisia tabaci biotypes. Acta Entomol Sin 48: 518–525. [Google Scholar]
- 67. Li ZX, Lin HZ, Guo XP (2007) Prevalence of Wolbachia infection in Bemisia tabaci . Curr Microbiol 54: 467–471. [DOI] [PubMed] [Google Scholar]
- 68. Guo XP, Li ZX (2008) Wolbachia extensively harbored by Bemisia tabaci in China. Acta Microbiol Sin 48: 63–67. [PubMed] [Google Scholar]
- 69. Chu D, Zhang YJ, Brown JK, Cong B, Xu BY, et al. (2006) The introduction of the exotic Q biotype of Bemisia tabaci from the Mediterranean region into China on ornamental crops. Fla Entomol 89: 168–174. [Google Scholar]
- 70. Huigens ME, de Almeida RP, Boons PAH, Luck RF, Stouthamer R (2004) Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc R Soc Lond B Biol Sci 271: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Heath BD, Butcher RD, Whitfield WG, Hubbard SF (1999) Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol 9: 313–316. [DOI] [PubMed] [Google Scholar]
- 72. Liu SS, De Barro PJ, Xu J, Luan JB, Zang LS, et al. (2007) Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318: 1769–1772. [DOI] [PubMed] [Google Scholar]
- 73. Sintupachee S, Milne JR, Poonchaisri S, Baimai V, Kittayapong P (2006) Closely Related Wolbachia Strains within the Pumpkin Arthropod Community and the Potential for Horizontal Transmission via the Plant. Microb Ecol 51: 294–301. [DOI] [PubMed] [Google Scholar]
- 74. Moreira LA, Saig E, Turley AP, Ribeiro JM, O'Neill SL, et al. (2009) Human probing behavior of Aedes aegypti when infected with a life-shortening strain of Wolbachia . PLoS Negl Trop Dis 3: e568 doi:10.1371/journal.pntd.0000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li SJ, Xue X, Ahmed MZ, Ren SX, Du YZ, et al. (2011) Host plants and natural enemies of Bemisia tabaci (Hemiptera: Aleyrodidae) in China. Insect Sci 18: 101–120. [Google Scholar]
- 76. Chiel E, Zchori-Fein E, Inbar M, Gottlieb Y, Adachi-Hagimori T, et al. (2009) Almost there: Transmission routes of bacterial symbionts between trophic levels. PLoS One 4 (3) e4767 doi:10.1371/journal.pone.0004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stahlhut J (2010) The endosymbiont community as taxonomic character: a novel approach to resolving the Bemisia tabaci complex. Mol Ecol 19: 4102–4104. [DOI] [PubMed] [Google Scholar]
- 78. Munson MA, Baumann P, Clark MA, Baumann L, Moran NA, et al. (1991) Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Biotechnol 173: 6321–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ahmed MZ, De Barro PJ, Olleka A, Ren SX, Mandour NS, et al. (2012) Use of consensus sequences to identify members of the Bemisia tabaci (Hemiptera: Aleyrodidae) cryptic species complex in Egypt and Syria. J Appl Entomol 136: 510–519. [Google Scholar]
- 80. Simon C, Frati F, Beckenbach A, Crespi B, Liu H, et al. (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene-sequences and a compilation of conserved polymerase chain-reaction primers. Ann Entomol Soc Am 87: 651–701. [Google Scholar]
- 81. Zchori-Fein E, Perlman SJ (2004) Distribution of the bacterial symbiont Cardinium in arthropods. Mol Ecol 13: 2009–2016. [DOI] [PubMed] [Google Scholar]
- 82. Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, et al. (2006a) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis . Appl Environ Microbiol 72: 7098–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Baldo L, Bordenstein S, Wernegreen JJ, Werren JH (2006b) Widespread recombination throughout Wolbachia genomes. Mol Biol Evol 23: 437–449. [DOI] [PubMed] [Google Scholar]
- 84. Von der Schulenburg JHG, Hurst GDD, Huigens TME, Van Meer MMM, Jiggins FM, et al. (2000) Molecular evolution and phylogentic utility of Wolbachia ftsZ and wsp gene sequences with special reference to the origin of male-killing. Mol Biol Evol 17: 584–600. [DOI] [PubMed] [Google Scholar]
- 85. Jiggins FM, von der Schulenburg JH, Hurst GD, Majerus ME (2001) Recombination confounds interpretations of Wolbachia evolution. Proc R Soc Lond B Biol Sci 268: 1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Werren JH, Bartos JD (2001) Recombination in Wolbachia . Curr Biol 11: 431–435. [DOI] [PubMed] [Google Scholar]
- 87. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucl Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swofford DL (2002) PAUP.* Phylogenetic Analysis Using Parsimony (*And Other Methods), version 4, Sinauer Associates, Sunderland, MA.
- 90. Salminen MO, Carr JK, Burke DS, McCutchan FE (1995) Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retrovir 11: 1423–1425. [DOI] [PubMed] [Google Scholar]
- 91. Maydt J, Lengauer T (2006) Recco: recombination analysis using cost optimization. Bioinformatics 22: 1064–1071. [DOI] [PubMed] [Google Scholar]
- 92. Braig HR, Zhou W, Dobson S, O'Neill SL (1998) Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis . J Bacteriol 180: 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Leikoski N, Fewer DP, Sivonen K (2009) Widespread occurrence and lateral transfer of the cyanobactin biosynthesis gene cluster in Cyanobacteria. Appl Eviron Microbiol 75: 853–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Goslee SC, Urban DL (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Statist Soft 22: 1–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree reconstruction of B. tabaci and its endosmbionts based on various gene sequences. A. Phylogenetic tree reconstruction based on mtCOI gene sequences (length = 830 bp) of host B. tabaci cryptic species using maximum-likelihood analysis under the HKY+G substitution model. The bootstrap values are indicated. B. Phylogenetic tree reconstruction based on 16S rRNA gene sequences (length = 1100 bp) of P-endosymbionts of B. tabaci cryptic species using maximum-likelihood analysis under the JC+G substitution model. The bootstrap values are indicated. C. Phylogenetic tree reconstruction based on wsp gene sequences (length = 480 bp) of Wolbachia of B. tabaci cryptic species using maximum-likelihood analysis under the T92+G substitution model. The bootstrap values are indicated. D. Phylogenetic tree reconstruction based on ftsZ gene sequences (length = 850) of Wolbachia of B. tabaci cryptic species using maximum-likelihood analysis under the TN93+G+I substitution model. E. Phylogenetic tree reconstruction based on 23S rRNA gene sequences (length = 550 bp) of Arsenophonus of B. tabaci cryptic species using maximum-likelihood analysis under the HKY+G substitution model. The bootstrap values are indicated. F. Phylogenetic tree reconstruction based on 16S rRNA gene sequences (length = 400) of Cardinium of B. tabaci cryptic species using maximum-likelihood analysis under the K2+G substitution model. The bootstrap values are indicated.
(DOC)
The exact search at best reconstructions using TreeMap 2.0b show the ML phylogenetic comparison of endosymbionts over their hosts. Endosymbionts displayed in black and the hosts in light grey; in reconstruction; cospeciation events are shown by (•), duplications by(▪) and host switches by (→). A. Host B. tabaci and its P-endosymbionts. B. Host B. tabaci and its Wolbachia (wsp). C. Host B. tabaci and its Wolbachia (ftsZ). D. Host B. tabaci and its Arsenophonus (23S). E. Host B. tabaci and its Cardinium (16S).
(DOC)
Asian B. tabaci cryptic species population sampled for S-endosymbionts.
(DOC)