Abstract
New specimens of the kleptoplastidal dinoflagellate Gymnodinium eucyaneum Hu were collected in China. We investigated the systematics of the dinoflagellate and the origin of its endosymbiont based on light morphology and phylogenetic analyses using multiple DNA sequences. Cells were dorsoventrally flattened with a sharply acute hypocone and a hemispherical epicone. The confusion between G. eucyaneum and G. acidotum Nygaard still needs to be resolved. We found that the hypocone was conspicuously larger than the epicone in most G. eucyaneum cells, which differed from G. acidotum, but there were a few cells whose hypocone and epicone were of nearly the same size. In addition, there was only one site difference in the partial nuclear LSU rDNA sequences of a sample from Japan given the name G. acidotum and G. eucyaneum in the present study, which suggest that G. eucyaneum may be a synonym of G. acidotum. Spectroscopic analyses and phylogenetic analyses based on nucleomorph SSU rDNA sequences and chloroplast 23 s rDNA sequences suggested that the endosymbiont of G. eucyaneum was derived from Chroomonas (Cryptophyta), and that it was most closely related to C. coerulea Skuja. Moreover, the newly reported kleptoplastidal dinoflagellates G. myriopyrenoides and G. eucyaneum in our study were very similar, and the taxonomy of kleptoplastidal dinoflagellates was discussed.
Introduction
Dinoflagellates are a diverse group of single-celled eukaryotic algae that occur in marine and freshwater all over the world [1]. Some have acquired chloroplasts via endosymbiosis [2]. This phenomenon provides insights into the Serial Endosymbiosis Theory that some algal groups arose via the ingestion and retention of photosynthetic, eukaryotic organisms and the subsequent reduction of their nonphotosynthetic organelles [3], [4], [5]. The origins and structures of endosymbionts are highly diverse. Karenia Hansen & Moestrup, Karlodinium Larsen, and Takayama de Salas have chloroplasts that originated from haptophyte algae, which are surrounded by three membranes but no other organelles remain from the endosymbiont [6], [7]. Durinskia Carty & Cox, Kryptoperidinium Lindemann, and Peridinium Ehrenberg contain chloroplasts derived from diatoms [8], [9]. The nucleus and mitochondria of the diatom remain Dinophyceae in the host cell where they are surrounded by a single membrane [10], [11]. The chloroplasts of Dinophysis Ehrenberg originated from a cryptophyte, probably Teleaulax Hill [12], [13], [14], [15], and they are surrounded by two membranes [16].
The retention time of plastids in dinoflagellates also varies greatly depending on the species involved and the conditions under which they are grown [17]. Some endosymbionts are permanent, whereas others are engulfed and temporarily retained in a functional state for a few weeks. The temporary retention of engulfed chloroplasts is known as “kleptoplastidy” and the endosymbionts (chloroplasts) are referred to as “kleptochloroplasts” [18]. The relationship between the endosymbiont and the host remains obscure. In a recent study of Dinophysis acuminata Claparède & Lachmann, it was observed that the kleptoplastids were serviced by nucleus-encoded proteins and horizontal gene transfer from the endosymbiont to the host nucleus was detected [19]. Therefore, studies of kleptoplastidy are very interesting and important for increasing our understanding of endosymbiosis and the evolution of algae.
A relatively small group of dinoflagellates have been described as having a blue-green coloration and researchers are keen to understand the source of their coloration [20]. Studies of Gymnodinum acidotum Nygaard, G. aeruginosum Stein, and G. myriopyrenoides Yamaguchi, Nakayama, Kai et Inouye had indicated that a cryptophycean endosymbiont was housed temporarily within the dinophycean cell, which was the source of the blue-green chloroplasts [20], [21], [22], [23], [24]. The discovery of nonphotosynthetic organelles in the endosymbionts in dinophycean cells suggested that these are examples of an early stage in the evolutionary process [23]. Thus, systematic studies of this group may be of great evolutionary interest. However, most previous studies are based on pigmentation and morphological observation, whereas the phylogenetic relationships among the blue-green group of dinoflagellates and their endosymbionts remain uncertain.
The blue-green freshwater dinoflagellate Gymnodinium eucyaneum Hu (Hu et al. 1980, as G. cyaneum; Hu 1983) was originally described from China as processing phycobilin like cryptomonads, suggesting that it probably contained a cryptophycean endosymbiont [25], [26]. At present, it has only been reported in China. In this study, we collected new specimens of G. eucyaneum from China and their cell morphology was observed by light microscopy, while the systematics of the dinoflagellates were investigated via phylogenetic analyses based on partial nuclear LSU rDNA sequences. To identify the origin of the endosymbiont, the nucleomorph SSU rDNA and chloroplast 23S rDNA sequences were determined, and the absorption spectrum of the phycocyanin was measured. The sequences of some cryptomonads were also determined for reference.
Results
Description
Gymnodinium eucyaneum (Hu, Yu et Zhang) Hu 1983, Hu, p.198–199; Gymnodinium cyaneum Hu, Yu et Zhang 1980, Hu et al., p. 651–653. Non Gymnodinium cyaneum Schiller 1955.
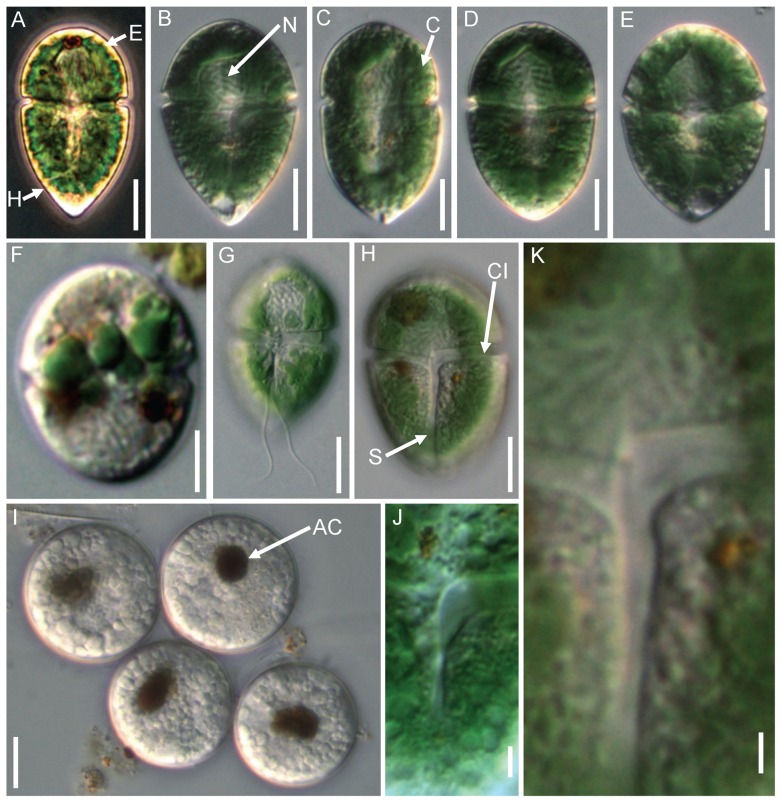
Unicellular, freshwater unarmored dinoflagellate. Cells were obviously dorsoventrally flattened, measuring 29∼48 μm in length, 16∼22 μm in width, and 12∼17 μm in thickness. In most cases, the hypocone was conspicuously larger than the epicone (Figs. 1A–D). The epicone was hemispherical and its length was approximately one-third of the total cell length (Figs. 1A–D). The hypocone was sharply acute (Figs. 1A–E). In a few cells, the hypocone and epicone were more or less the same size (Fig. 1E). The cingulum was wide, deeply excavated, and encircled the middle-upper part of the cell (Figs. 1G, H, J, K). There was no displacement of the cingulum and its ventral ends were at the same level, where both curved posteriorly at the junction with the sulcus (Figs. 1G, H, J, K). The sulcus was wide, expanding into the posterior part (Figs. 1G, H, J, K). Two flagella were inserted on the ventral side of the cingulum (Fig. 1G).
Figure 1. Micrographs of Gymnodinium eucyaneum.
Figs. A–E. Different cell shapes of the field samples. F Cells kept for 2∼4 weeks in the laboratory, showing that the chloroplast became smaller. G Ventral view showing the insertion of the flagella. H, J, K Ventral view showing the detail of the cingulum and sulcus. I Cysts each with a brownish accumulation of corpuscles. E: epicone; H: hypocone; N: nucleus; C: chloroplasts; CI: cingulum; S: sulcus; AC: accumulation of corpuscle. Scale bars: A–I = 10 μm; J–K = 2 μm.
A large spherical nucleus was situated in the anterior part of cell (Figs. 1B–E, G, H). Hundreds of granules of variable size were observed beneath the plasmalemma (Fig. 1). Numerous blue-green chloroplasts were located peripherally in the cell (Figs. 1A–H). Determining the actual number was difficult because they were very dense. The chloroplasts gradually became smaller when cells were retained in the lab (Fig. 1F). Colorless cysts with a brownish accumulation of corpuscle formed after 2∼4 weeks culture in filtered local water (Fig. 1I).
The voucher specimens examined were: HBI 3586× from Lake Donghu in Wuhan City, Hubei Province, collected by SX on April 18, 2012; HBI 3597× from a fishpond in Wuhan City, Hubei Province, collected by SX on May 3, 2012. The specimens are deposited in the Freshwater Algal Herbarium (HBI), Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, Hubei, China.
Spectroscopy
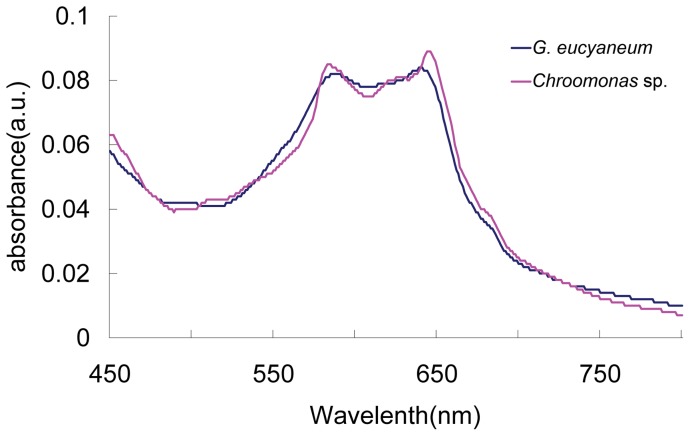
The absorption spectrum of the phycocyanin extracted from G. eucyaneum in this study is shown in Fig. 2. Two absorption peaks were observed at 641 nm and 585 nm. The peak at 641 nm was slightly higher than the peak at 585 nm.
Figure 2. Absorption spectrum of phycocyanin extracted from Gymnodinium eucyaneum samples and a Chroomonas sp. strain.
Phylogenetic Analyses
Host phylogeny based on the nuclear LSU rDNA
The LSU rDNA sequences aligned in this study contained 572 nucleotides for 38 taxa of dinoflagellates and their putative relatives. Of these nucleotides, 389 sites (68.0%) were variable and 309 sites (54.0%) were parsimoniously informative. The base frequencies were found to be homogeneous across taxa. The overall average pairwise distance was 0.253. The phylogenetic trees constructed by the ML and Bayesian analyses produced similar topologies to their composition, although only the Bayesian trees are presented. In the phylogenetic tree, the members of the genus Gymnodinium sensu stricto formed a well supported clade (0.96/76 for BA/ML) (Fig. 3). G. eucyaneum was present in this clade and it formed a robust subclade (1.00/100 for BA/ML) with G. acidotum. The interspecific pairwise divergence between them was 0.002 and there was only one site difference. Both clustered with G. palustre, G. myripyrenoidosum, and Amphidinium poecilochroum with high support (1.00/100 for BA/ML). The interspecific pairwise divergence between G. eucyaneum and G. myripyrenoidosum was 0.108. This group did not show strong affinity to any others.
Figure 3. Bayesian phylogenetic tree constructed from the nuclear LSU rDNA sequences.
The numbers on the nodes represent the posterior probabilities (PP)/bootstrap support values (BP) produced by the Bayesian inference and maximum-likelihood analyses. Values >0.50 for PP and >50 for BP are shown. The sequences obtained in our study are shaded in gray.
Endosymbiont phylogeny based on the nucleomorph SSU rDNA and chloroplast 23S rDNA
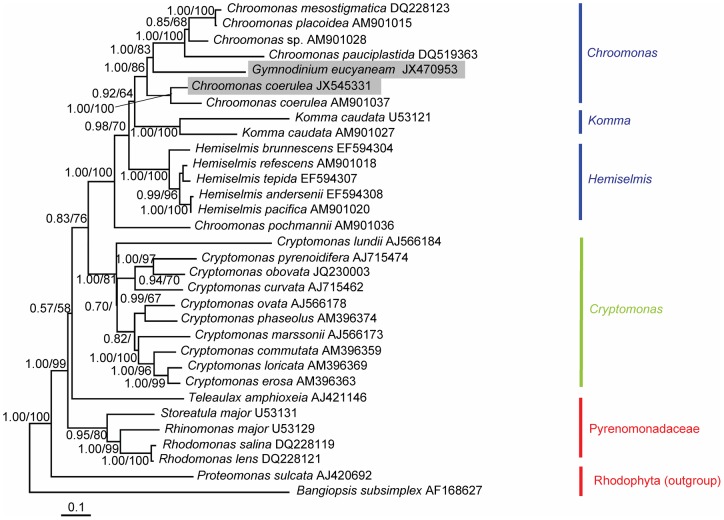
The nucleomorph SSU rDNA sequences aligned in this study contained 1872 nucleotides for 32 taxa of dinoflagellates, cryptomonads, and their putative relatives. Of these nucleotides, 741 sites (39.6%) were variable and 477 sites (25.5%) were parsimoniously informative. The base frequencies were found to be homogeneous across taxa. The overall average pairwise distance was 0.077. Three main clades were distinguished in the phylogenetic tree, which represented the blue-green, brownish-green, and red cryptomonad species (Fig. 4). The sequence of G. eucyaneum was positioned in the clade containing Chroomonas sp., C. mesostigmatica, C. placoidea, C. pauciplastida, and C. coerulea with high support (1.00/86 for BA/ML). The sequence of G. eucyaneum was most closely related to two C. coerulea sequences. One was from C. coerulea strain UTEX 2780 and the other was from C. coerulea collected from the lake where G. eucyaneum was collected in the present study. Another member of Chroomonas, C. pochmannii, was distantly related to this group. The Komma and Hemiselmis species, which are also blue-green in color, formed two robust groups (1.00/100 for BA/ML and 1.00/100 for BA/ML) with relatively distant relationships to G. eucyaneum.
Figure 4. Bayesian phylogenetic tree constructed from the nucleomorph SSU rDNA sequences.
The numbers on the nodes represent the posterior probabilities (PP)/bootstrap support values (BP) produced by the Bayesian inference and maximum-likelihood analyses. Values >0.50 for PP and >50 for BP are shown. The sequences obtained in our study are shaded in gray.
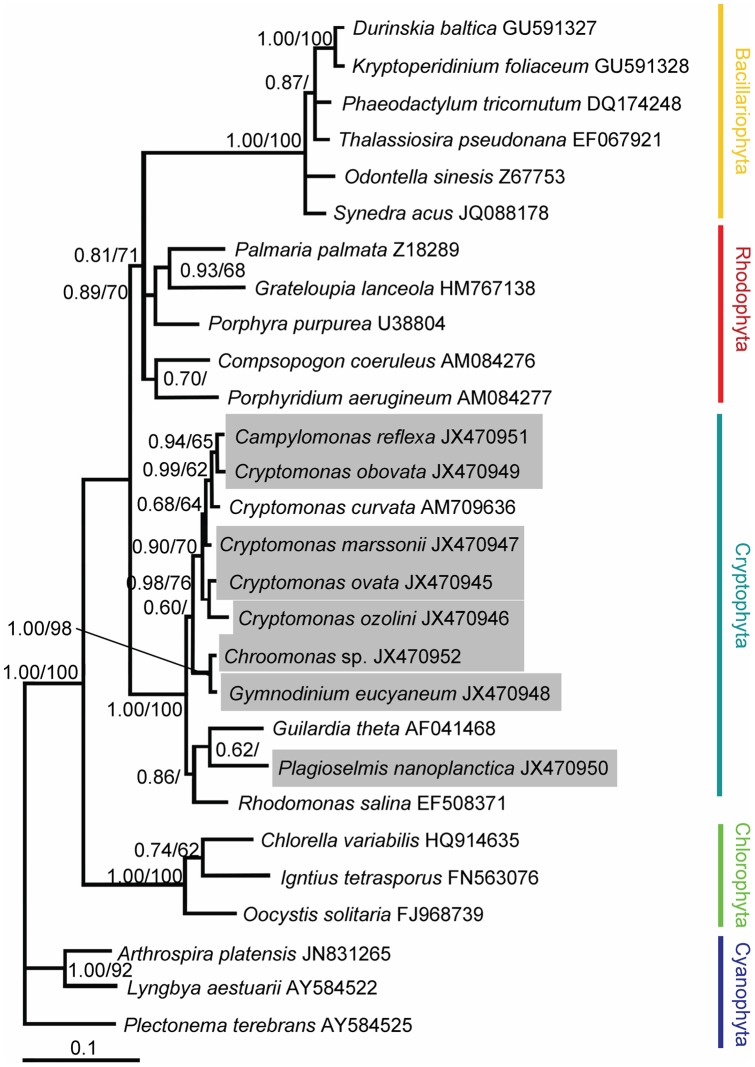
The chloroplast 23S rDNA sequences aligned in this study contained 976 nucleotides for 28 taxa of cryptomonads, dinoflagellates, diatoms, and other algae. Of these nucleotides, 292 sites (29.9%) were variable and 523 sites (22.2%) were parsimoniously informative. The base frequencies were found to be homogeneous across taxa. The overall average pairwise distance was 0.090. The algae from different phyla were well separated in the reconstructed phylogenetic tree (Fig. 5). The sequence of G. eucyaneum was positioned in the cryptomonad clade with high support (1.00/100 for BA/ML). The sequences of G. eucyaneum and C. coerulea from the same lake formed a robust lineage (1.00/98 for BA/ML) and the pairwise distance between them was 0.001. Several diatoms and dinoflagellates that contained endosymbiont derived from diatoms formed a well-supported clade (1.00/100 for BA/ML).
Figure 5. Bayesian phylogenetic tree constructed from the chloroplast 23S rDNA sequences.
The numbers on the nodes represent the posterior probabilities (PP)/bootstrap support values (BP) produce by the Bayesian inference and maximum-likelihood analyses. Values >0.50 for PP and >50 for BP are shown. The sequences obtained in our study are shaded in gray.
Discussion
Previous studies of Gymnodinium eucyaneum in China
Several studies have investigated G. eucyaneum in China since 1980 [25]–[29]. However, they are not widely known because they were written in Chinese. Evidence for the presence of phycobilin [25] and the ultrastructure of the chloroplasts [29] suggested that the chloroplasts of G. eucyaneum were derived from cryptophytes. Observations of the nucleus and nuclear substance [27] showed that one dinokaryon was present in all cells and the numbers of second eukaryotic nuclei ranged from 0–4 (rarely 7–10). This data may suggest that the second eukaryotic nuclei were temporary and that the chloroplasts of G. eucyaneum were “stolen” and could be lost. Similar report on the number of nuclei was made by Field and Rhodes [22].
Gymnodinium eucyaneum and G. acidotum
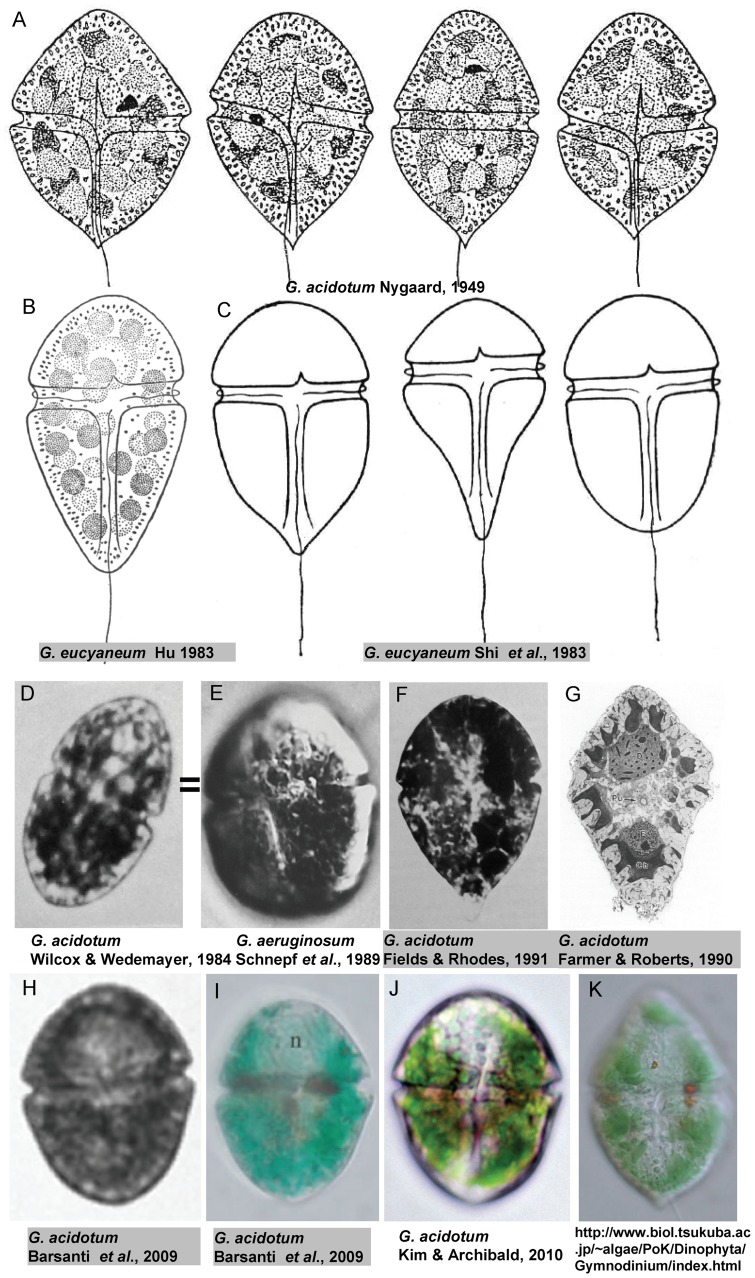
Since it was first described in China, G. eucyaneum has often been confused with another blue-green unarmored dinoflagellate, G. acidotum. Traditionally, unarmored dinoflagellates have been classified based mainly on the relative sizes of the epicone and hypocone [30]. According to their original descriptions, the epicone and the hypocone were nearly equal in G. acidotum [31] (Fig. 6A), whereas the hypocone was conspicuously larger than the epicone in G. eucyaneum [26], [27] (Figs. 6B, 6C). The hypocone was 1.3∼1.8 times as long as the epicone in G. eucyaneum according to our observations. Schnepf et al. (1989) suspected that the organism studied by Wedemayer (1984) under the name “Gymnodinium acidotum” was identical to G. aeruginosum [24] (Figs. 6D–E). After examining the relative sizes of the epicone and hypocone in the images provided in previous studies, we considered that the organisms studied by Farmer and Roberts (1990) [21], Fields and Rhodes (1991) [22], and Barsanti et al. (2009) [32] (Figs. 6F, G, H, I) under the name “G. acidotum” were different from the lectotype and G. acidotum in other studies (Figs. 2A, J, K), and they may be identical with the G. eucyaneum analyzed in our study. However, recent studies indicate that the classification based on the relative sizes of the epicone and hypocone does not reflect their phylogenetic relationships [7], [33], [34], [35]. In a recent study by Yamaguchi et al. (2011) [23], a partial nuclear LSU rDNA sequence of G. acidotum were included. There was only one site difference in the nuclear LSU rDNA sequence of G. acidotum in that study and G. eucyaneum in the present study. An image of G. acidotum was not provided in that paper, but the authors gave us the usage of a photo of G. acidotum collected from the same sample on their web site. (Fig. 6K) In the photo, the hypocone and the epicone were more or less the same size. While in our samples of G. eucyaneum, the epicone and hypocone were more or less the same size in some cells, although cells of this type were rare. We thought the relative size of epicone and hypocone may varies in different phases of lifecycle or in different habitats. Therefore, G. eucyaneum may be a synonym of G. acidotum. However, we prefer to postpone the synonymization until more morphological and molecular information on these organisms becomes available.
Figure 6. Drawings and photographs of Gymnodinium eucyaneum and G. acidotum.
The organisms shown in F–I were considered to be the same as G. eucyaneum in the present study.
Gymnodinium eucyaneum and other kleptoplastidal dinoflagellates
Recently, a new unarmored blue-green kleptoplastidal dinoflagellate, Gymnodinium myriopyrenoides Yamaguchi, Nakayama, Kai et Inouye was reported from Isonoura Beach in Japan [23]. Although the habitat of G. myriopyrenoides was marine and sand-dwelling, while G. eucyaneum was a freshwater species. we thought G. myriopyrenoides and G. eucyaneum were quite similar: in our phylogenetic analyses, the two were very close to each other; as to morphological characters, both of the two were dorsoventrally flattened and elongate-elliptical in ventral view; their epicone was conspicuously smaller than the hypocone; and they both had a wide, deeply incised cingulum with no displacement; both their endosymbionts came from blue-green cryptomonads. The symbiont of G. myriopyrenoides was found to be derived from Chroomonas or Hemiselmis via phylogenetic analyses based on plastid-encoded SSU rDNA, but it could not be identified at species level because plastid-encoded SSU rDNA sequeces of Chroomonas and Hemiselmis were insufficient. In the present research, the symbiont of G. eucyaneum was confirmed to be derived from Chroomonas via phylogenetic analyses based on nucleomorph SSU rDNA and spectrophotometric pigment analyses.
G. myriopyrenoides and G. eucyaneum, together with some other species of Gymnodinium and Amphidinium who also harbored blue-green kleptochloroplasts, such as G. aeruginosum, A. poecilochroum, A. latum and A. wigrense, formed a relatively distinct group in Dinophyceae in view of their kleptoplastidal behavior, morphological characters and close relationships in phylogenetic analyses. As mentioned above, recent ultrastructural and molecular phylogenetic studies revealed that the traditional taxonomy of unarmored dinoflagellates based mainly on the relative sizes of the epicone and hypocone was problematic. As revealed in ultrastructural and molecular phylogenetic studies, the genus Gymnodinium sensu Hansen er Moestrup and Amphidinium were polyphyletic [33], [34], [35], [36]. Yamaguchi et proposed to establish a new genus for these kleptoplastidal dinoflagellates based on morphological and molecular characters, and we considered this proposal was more reasonable than the traditional taxonomy.
Identification of the endosymbiont
Previous studies based on spectrophotometric pigment analyses suggested that the phycocyanins in G. eucyaneum resembled PC 645 and that G. eucyaneum may contain a blue-green cryptomonad endosymbiont [25], although the origin of the endosymbiont remained uncertain. Three cryptomonad genera contain blue-green chloroplasts [37], i.e., Hemiselmis, Komma, and Chroomonas, and Komma and Chroomonas both contain PC 645 [38]. The absorption spectrum of the phycocyanin extracted from G. eucyaneum in this study was almost the same as that extracted from G. eucyaneum in a previous study [25], which matched the PC 645 extracted from Chroomonas sp. strain CCMP 1221 [38]. In the phylogenetic analyses based on the nucleomorph SSU rDNA and chloroplast 23S rDNA in this investigation, the sequences of the endosymbiont were firmly included in the Chroomonas clade and they had relatively long distances from Komma and Hemiselmis. Thus, we suggest that the endosymbiont originated from Chroomonas. Furthermore, the sequence of the endosymbiont indicated that it was most closely related to two C. coerulea sequences. It is quite remarkable that C. coerulea and G. eucyaneum were collected from the same lake in the same month, in the present study. Thus, we suspect that the endosymbiont of G. eucyaneum detected in this study originated from C. coerulea. However, the species level identification could be problematic because the taxon sampling and taxonomic studies of Chroomonas were inadequate. In addition, DNA changes may have occurred after the endosymbiont was engulfed by the host, which may make the species level identification more complex.
In our study, G. eucyaneum survived for 2∼4 weeks without feeding, so the chloroplast was likely to be the nutritional source for the dinoflagellate host, which agreed with previous observations of starch grain accumulation in some kleptoplastidal dinoflagellates [39], [40]. The chloroplasts gradually became smaller and fewer, and host cell division was rarely seen, while attempts to establish clonal cultures failed. In a previous Chinese study, it was reported that the location and number of the nuclear substances (probably cryptomonad nuclei and nucleomorphs) varied greatly among the G. eucyaneum cells in the same samples, because the nuclear substances were randomly distributed to two daughter cells when host cell division occurred [27]. These studies suggest that the endosymbiont of G. eucyaneum is neither a food nor a permanent endosymbiont and that G. eucyaneum is a kleptoplastidal dinoflagellate.
Why cryptomonads?
In some dinoflagellates, the symbionts are derived from Chroomonas and other cryptomonads, such as Teleaulax. spp and Rhodomonas spp. [39], [41], [42], [43]. Thus, these cryptomonads must have unique characteristics that allow them to become symbionts of dinoflagellates. We propose four main reasons for this phenomenon, as follows.
First, cryptomonads are an appropriate size for engulfment by dinoflagellates.
Furthermore, the cell surface of cryptomonads is a delicate proteinaceous periplast rather than a cellulose wall, which is found only in cryptomonads, although the same term is applied to euglenoids but they have a different arrangement [44]. The periplast is vulnerable to rupturing or distortion [45], so it is probably easily disrupted and digested by the hosts after engulfment.
Moreover, the periplastidial compartment (periplastidial complex or chloroplast endoplasmic reticulum) may maintain a relatively appropriate intracellular environment for the endosymbiont in the host cells.
Finally, the chloroplast of cryptomonads may have the ability to adapt to new intracellular environments. It is widely believed that cryptomonads obtain their chloroplasts by ingesting a red algal-like photosynthetic endosymbiont [46], [47]. Thus, from an evolutionary viewpoint, the chloroplasts are fairly unique because they possess the remnants of a eukaryotic nucleus, the nucleomorph [48], [49], [50], [51]. Some genes may be encoded in the chloroplast or nucleomorph, which help the endosymbiont to adapt to the intracellular environment of the host. In addition, some alterations may have occurred in the genes or the ultrastructure of the chloroplasts, which help to integrate the newly engulfed organelles into the host cell. The nucleomorph appears to be important for the chloroplasts because it is retained in the host cell in most cases [20], [21], [22], [23], [24]. During the secondary endosymbiosis of cryptomonads, the genes encoded in the nucleomorph are highly compacted and most of the genes with metabolic functions are eliminated, leaving 30 genes for chloroplast-located proteins [52]. Gene transfer and reduction may occur once more in the nucleomorph after engulfment by the dinoflagellates. In a recent study of Dinophysis acuminata, horizontal gene transfer was detected from the kleptoplastidal chloroplast obtained from a cryptomonad to the host nucleus [21], which supports our hypothesis. In addition, endosymbiont genes were relocated to the nucleus via massive gene transfer in Karenia brevis, although the endosymbiont was a haptophyte instead of a cryptomonad, but the plastid still originated via a red algal secondary endosymbiosis [53]. The cryptomonad nucleus is probably not as important as the nucleomorph for the survival of the symbiont, because it is frequently lost in the host cell [20], [21], [24]. The lack of a cryptomonad nucleus in some dinoflagellates did not appear to affect the cell's ability to photosynthesize or move in response to varying levels of illumination [20]. In the present study, the genes encoded by the cryptomonad nucleus could not be amplified, although we tried many times, which suggested that the nucleus was probably lost.
All these unique characteristics might help the chloroplasts to adapt to new intracellular environments, although no clear evidence was available until now. Thus, it is necessary to study kleptoplastidal dinoflagellates and cryptomonads using comparative genomics and biochemistry methods to achieve a better understanding of the evolution of chloroplasts and algae.
Materials and Methods
Sample collection
Samples of G. eucyaneum were collected in April 2012 from Lake Donghu (30°32′55′′ N, 114°21′15′′ E) and from a fishpond in Wuhan, Hubei Province, China (30°28′34′′ N, 114°21′41′′ E) in May 2012, where it bloomed and accounted for more than 99% of the phytoplankton cell density. Samples of Cryptomonas obovata, Cr. marssonii, Cr. ozolini, Cr. ovata, C. coerulea, Campylomonas reflexa, and Plagioselmis nanoplanctica were collected from Lake Donghu in April 2012. Living cells were delivered immediately to the laboratory. Cells were kept cold during transportation. No specific permits were required for the described field studies. The locations where the samples collected from were not privately-owned or protected in any way, and the field studies did not involve endangered or protected species. Cells were isolated with the serial dilution pipetting technique [54] under an inverted microscope (CKX41; Olympus, Tokyo, Japan) and cultivated in sterilized lake water added with Bold's Basal Medium at a final concentration of 4%. Cells were grown at 17±2°C under a light: dark cycle of 14∶10 h (light intensity between 15 and 30 µmol photons m−2 s−1).
Morphological observation
Living cells and cells fixed with Lugol's solution at a final concentration of 1% were observed using differential interference contrast (DIC), phase contrast (PH), and epifluorescence microscopy (EFM) with a light microscope (Leica DM5000B, Wetzlar, Germany). Micrographs were captured using a digital camera (Leica DFC320, Wetzlar, Germany).
Spectrophotometry
Fishpond samples of G. eucyaneum and cultivated strain of C. coerulea were used. A 10 ml aliquot of the samples was pelleted for 3 min at 10,000×g and resuspended in phosphate buffer at pH 7.2 on three occasions. The cells were broken up by three cycles of freezing at −80°C and thawing. The cell debris was pelleted at 15,000×g. The absorption spectra of the supernatant was measured and recorded using a UV-1700 PharmaSpec spectrofluorometer (Shimadzu, Kyoto, Japan).
DNA extraction, PCR amplification, and DNA sequencing
Approximately 50 G. eucyaneum cells were isolated using a serial dilution pipetting technique [54] under an inverted microscope (CKX41; Olympus, Tokyo, Japan). The cryptomonads were isolated in the same way and cultivated in sterilized lake water with 4% Bold's Basal (BB) Medium added [55]. The total DNA was extracted using the CTAB method [56].
The partial nuclear LSU rDNA sequence was amplified using the primers described by Scholin et al. [57]. The conditions used for PCR amplification of the partial LSU rDNA sequence and thermal cycling were those described in Hansen et al. [6]. The specific primers for the nucleomorph SSU rDNA (CMsF, 5′-TGGCT CATTA CAACA GTTAT AG-3′; CMsR, 5′-AGGCA TTCCT CGTTC AAG-3′) and chloroplast 23S rDNA (Cr23S1F, 5′-CAATA GATGC CTGTA CCTTA AACC-3′; Cr23S1R, 5′-TGGAC CGAAC TGTCT CACG-3′) of the endosymbionts were designed based on the conserved areas of known sequences of cryptomonads. The amplification profile of the nucleomorph SSU rDNA consisted of an initial denaturation step at 94°C for 4 min, followed by 35 cycles of 1 min at 94°C, 1 min at 60°C, and 90 s at 72°C, with 10 min at 72°C for the final extension. The chloroplast 23S rDNA PCR amplification started with 4 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, 75 s at 72°C, ending with a final hold of 10 min at 72°C. All PCR amplicons were cleaned using an E.Z.N.A. Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA), before some PCR products were cloned into the pMD18-T vector (Takara, Dalian, China). All clones were sequenced using the universal sequencing primer M13 with an ABI 3700 sequencer (Applied Biosystems, Foster City, CA, USA). The sequences were deposited in GenBank under the Accession Nos JX470945∼JX470953, JX545331, and JQ639750.
Phylogenetic analyses
The sequences of putative relatives were downloaded from GenBank. Initially, the sequences were aligned using ClustalX 1.83 [58] and refined manually in SEAVIEW [59]. The final alignment of LSU rDNA sequences comprised a matrix of 38 sequences, including Perkinsus marinus (Perkinsozoa) as an outgroup taxon. A total of 32 cryptomonads, a dinoflagellate, and putative relative taxa were selected for the nucleomorph SSU rDNA sequence analyses. Species of Bangiophyceae were used as outgroup phylogenies because a previous study showed that members of the Bangiophyceae had close relationships with the nucleomorphs in cryptomonad cells [60]. The alignment of chloroplast 23S rDNA sequences comprised a matrix of 28 sequences including three blue-green algae as an outgroup.
Mutational saturation was evaluated in the variable positions of the alignments by plotting the pairwise distances against model-corrected distances for Tamura and Nei (1993) and Kimura (1980) models, which were estimated using MEGA (v.4.0) [61].
The phylogenies were estimated using the maximum-likelihood (ML) method in PAUP 4.0*(v. 4.0 beta) [62] and Bayesian inference (BI) in MrBayes (v. 3.1.2) [63]. For the ML analysis, ModelTest 3.06 [64] was used to select the evolutionarily best fit model given Akaike's Information Criterion (AIC). The best fit model for the LSU rDNA datasets was TrN+I+G. The best fit model for the nucleomorph SSU rDNA and chloroplast 23S rDNA datasets were GTR+I+G. In the ML analysis, a heuristic search option with random added sequences (100 replicates) and the tree bisection and reconnection branch-swapping algorithm were used for tree searching. A bootstrap analysis with 1000 replicates of the ML dataset was performed to estimate the statistical reliability. A Bayesian Markov Chain Monte Carlo analysis was conducted with seven Markov chains (six heated chains and one cold) for 5,000,000 generations, with the trees sampled every 1000 generations. The first 25% of the trees (burn-in samples) were discarded and the remaining samples were used to construct a Bayesian consensus tree and to infer the posterior probability.
Acknowledgments
We thank Prof. Lirong Song (Center for Algal Biology and Applied Research, Institute of Hydrobiology, Chinese Academy of Sciences, China) and Dr. Xiaoming Zhang (Graduate School of Life and Environmental Sciences, University of Tsukuba, Japan) for their cordial advices and helpful suggestions. We appreciate Dr. Haruyo Yamaguchi (National Institute for Environmental Studies, Tsukuba, JAPAN), Dr. Takeshi Nakawama and Prof. Isao Inouye (Graduate School of Life and Environmental Sciences, University of Tsukuba, Japan) helping us and giving us the permission to the usage of the photo of Gymnodinium acidotum.
Funding Statement
This work was financially supported by the National Natural Science Foundation of China (Grant Nos 30970501 and 31270252). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pochon X, Putnam HM, Burki F, Gates RD (2012) Identifying and Characterizing Alternative Molecular Markers for the Symbiotic and Free-Living Dinoflagellate Genus Symbiodinium. Plos One 7: e29816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnson MD (2011) The acquisition of phototrophy: adaptive strategies of hosting endosymbionts and organelles. Photosynth Res 107: 117–132. [DOI] [PubMed] [Google Scholar]
- 3. Taylor FJR (1974) Implications and extensions of the serial endosymbiosis theory of the origin of eukayotes. Taxon 23: 229–258. [Google Scholar]
- 4. Whatley JM, Whatley FR (1981) Chloroplast evolution. New Phytol 87: 233–247. [Google Scholar]
- 5. Gibbs SP (1981) The chloroplasts of some algal groups may have evolved from endosymbiotic eukaryotic algae. Ann N Y Acad Sci 361: 193–208. [DOI] [PubMed] [Google Scholar]
- 6. Hansen G, Daugbjerg N, Henriksen P (2000) Comparative study of Gymnodinium mikimotoi and Gymnodinium aureolum, comb. nov.( = Gyrodinium aureolum) based on morphology, pigment composition, and molecular data. J Phycol 36: 394–410. [Google Scholar]
- 7. de Salas CJS, Botes L, Nash G, Wright SW, Hallegraeff G (2003) Takayama gen. nov. (Gymnodiniales, Dinophyceae), a new genus of unarmed dinoflagellates with sigmoid apical grooves, including the description of two new species. J Phycol 39: 1233–1246. [Google Scholar]
- 8. Chesnick JM, Kooistra WHCF, Wellbrock U, Medlin LK (1997) Ribosomal RNA analysis indicates a benthic pinnate diatom ancestry for the endosymbionts of the dinoflagellates Peridinium foliaceum and Peridinium balticum (Pyrrhophyta). J Eukaryot Microbiol 44: 314–320. [DOI] [PubMed] [Google Scholar]
- 9. Horiguchi T, Takano Y (2006) Serial replacement of a diatom endosymbiont in the marine dinoflagellate Peridinium quinquecorne (Peridiniales, Dinophyceae). Phycol Res 54: 193–200. [Google Scholar]
- 10. Horiguchi T (2006) Algae and their chloroplasts with particularreference to the dinoflagellates. Paleontol Res 10: 299–309. [Google Scholar]
- 11. Horiguchi T, Pienaar RN (1991) Ultrustructure of a marine dinoflagellate, Peridinium quinquecorne Abé (Peridiniales) from South Africa with particular reference to its crysophyte endosymbiont. Bot Mar 34: 123–131. [Google Scholar]
- 12. Park MG, Park JS, Kim M, Yih W (2008) Plastid dynamics during survival of Dinophysis caudata without its ciliate prey. J Phycol 44: 1154–1163. [DOI] [PubMed] [Google Scholar]
- 13. Minnhagen S, Carvalho WF, Salomon PS, Janson S (2008) Chloroplast DNA content in Dinophysis (Dinophyceae) from different cell cycle stages is consistent with kleptoplasty. Environ Microbiol 10: 2411–2417. [DOI] [PubMed] [Google Scholar]
- 14. Nagai S, Nishitani G, Tomaru Y, Sakiyama S, Kamiyama T (2008) Predation by the toxic dinoflagellate Dinophysis fortii on the ciliate Myrionecta rubra and observation of sequestration of ciliate chloroplasts. J Phycol 44: 909–922. [DOI] [PubMed] [Google Scholar]
- 15. Garcia-Cuetos L, Moestrup Ø, Hansen PJ, Daugbjerg N (2010) The toxic dinoflagellate Dinophysis acuminate harbors permanent chloroplasts of cryptomonad origin, not kleptochloroplasts. Harmful Algae 9: 25–38. [Google Scholar]
- 16.Larsen J (1992) Endocytobiotic Consortia with Dinoflagellate Hosts. In Reisser W, editor. Algae and Symbiosis. Bristol: Biopress Limited. 427–442.
- 17. Kim E, Archibald JM (2010) Plastid evolution: gene transfer and the maintenance of ‘stolen’ organelles. BMC Biology 8: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnepf E (1992) From Parasitism to Symbiosis: the Dinoflagellate Example. In Reisser W, editor. Algae and Symbiosis. Bristol: Biopress Limited. 699–710.
- 19. Wisecaver JH, Hackett JD (2010) Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata . BMC Genomics 11: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilcox LW, Wedemeyer CJ (1984) Gymnodinium acidotum Nygaard (Pyrrophyta), a dinoflagellate with an endosymbiotic cryptomonad. J Phycol 20: 236–242. [Google Scholar]
- 21. Farmer MA, Roberts KR (1990) Organelle loss in the endosymbiont of Gymnodinium acidotum (Dinophyceae). Protoplasma 153: 178–185. [Google Scholar]
- 22. Fields SD, Rhodes RG (1991) Ingestion and retention of Chroomonas spp. (Cryptophyceae) by Gymnodinium acidotum (Dinophyceae). J Phycol 27: 525–529. [Google Scholar]
- 23. Yamaguchi H, Nakayama T, Kai A, Inouye I (2011) Taxonomy and Phylogeny of a New Kleptoplastidal Dinoflagellate, Gymnodinium myriopyrenoides sp. nov. (Gymnodiniales, Dinophyceae), and its Cryptophyte Symbiont. Protist 162: 650–667. [DOI] [PubMed] [Google Scholar]
- 24. Schnepf E, Winter S, Mollenhauer D (1989) Gymnodinium aeruginosum (Dinophyta): a blue-green dinofladellate with a vestigial, anucleate, cryptophycean endosymbiont. Plant Syst Evol 164: 75–91. [Google Scholar]
- 25. Hu H, Yu M, Zhang X (1980) Discovery of phycobilin in Gymnodinium cyaneum Hu sp. nov. and its phylogenetic significance. Kexue Tongbao 25: 882–884. [Google Scholar]
- 26. Hongjun H (1983) On Gymnodinium cyaneum Hu. Chin J Oceanol Limnol 1: 198–199. [Google Scholar]
- 27. Shi Z, Wei Y, Hu H (1983) An observation on the nucleus and nuclear substance of Gymnodinium eucyaneum Hu. Haiyang Yu Huzhao 14: 161–167. [Google Scholar]
- 28. Zhang S, Yu M, Li S (1989) Preliminary observation on the sexual reproduction of Gymnodinium eucyaneum Hu (Dinophyceae). ACTA Hydrobiol Sin 13: 289–291. [Google Scholar]
- 29. Chen B, Liu Q, Zhang X (1990) Ultrastructure and evolutionary relationship of the photosynsesis organelle in Gymnodinium eucyaneum Hu. J Chin Electron Micro Soc 3: 42. [Google Scholar]
- 30. Kofoid CA, Swezy O (1921) The Free-living Unarmored Dinoflagellata. Mem Univ California 5: 1–562. [Google Scholar]
- 31. Nygaard G (1949) Hydrobiological studies on some Danish ponds and lakes. Kongel Danske Videnskab Selskab Biolog Skrifter 7: 263. [Google Scholar]
- 32. Barsanti L, Evangelista V, Passarelli V, Frassanito AM, Coltelli P, et al. (2009) Microspectrophotometry as a method to identify kleptoplastids in the naked freshwater dinoflagellate Gymnodinium acidotum . J Phycol 45: 1304–1309. [DOI] [PubMed] [Google Scholar]
- 33. Daugbjerg N, Hansen G, Larsen J, Moestrup Ø (2000) Phylogeny of the major genera of dinoflagellates based on ultrastructure and partial LSU rDNA sequence data, including the erection of three new genera of unarmoured dinoflagellates. Phycologia 39: 302–317. [Google Scholar]
- 34. Flø_Jøgensen MF, Murray S, Daugbjerg N (2004a) A new genus of athecate interstitial dinoflagellates, Togula gen. nov., previously encompassed within Amphidinium sensu lato: Inferred from light and electron microscopy and phylogenetic analyses of partial large subunit ribosomal DNA sequences. Phycol Res 52: 284–299. [Google Scholar]
- 35. Flø Jøgensen MF, Murray S, Daugbjerg N (2004b) Amphidinium revised. I. redifinition of Amphidinium (Dinophyceae) based on cladistic and molecular phylogenetic analyses. J Phycol 40: 351–365. [Google Scholar]
- 36. Hansen G, Moestrup Ø (2005) Flagellar apparatus and nuclear chambers of the green dinoflagellate Gymnodinium chlorophorum. 53: 169–181. [Google Scholar]
- 37. Novarino G (2003) A companion to the identification of cryptomonad flagellates (Cryptophyceae = Cryptomonadea). Hydrobiologia 502: 225–270. [Google Scholar]
- 38. Hoef-Emden K (2008) Molecular phylogeny of phycocyanin-containing cryptophytes: evolution of biliprotein and geographical distribution. J Phycol 44: 985–993. [DOI] [PubMed] [Google Scholar]
- 39. Lewitus AJ, Glasgow Jr HB, Burkholder M (1999) Kleptoplastidy in the toxic dinoflagellate Pfiesteria piscicida (Dinophyceae). J Phycol 35: 303–312. [Google Scholar]
- 40. Schnepf E, Elbrähter M (1992) Nutritional strategies in dinoflagellates: a review with emphasis on cell biological aspects. Eur J Protistol 28: 3–24. [DOI] [PubMed] [Google Scholar]
- 41. Park MG, Park JS, Kim M, Yih W (2008) Plastid dynamics during survival of Dinophysis caudata without its ciliate prey. J Phycol 44: 1154–1163. [DOI] [PubMed] [Google Scholar]
- 42. Nagai S, Nitshitani G, Tomaru Y, Sakiyama S, Kamiyama T (2008) Predation by the toxic dinoflagellate Dinophysis fortii on the ciliate Myrionecta rubra and observation of sequestration of ciliate chloroplasts. J Phycol 44: 909–922. [DOI] [PubMed] [Google Scholar]
- 43. Skovgaard A (1998) Role of chloroplast retention in a marine dinoflagellate. Aquat Microb Ecol 15: 293–301. [Google Scholar]
- 44. Brett SJ, Perasso L, Wetherbee R (1994) Structure and Development of the Cryptomonad Periplast – a Review. Protoplasma 181: 106–122. [Google Scholar]
- 45.Kugrens P, Clay BL (2003) Cryptomonads. In: Wehr JD, Sheath RG, editors. Freshwater Algae of North America: Ecology and Classification. San Diego: Academic Press. 740.
- 46. Gillot MA, Gibbs SP (1980) The cryptomonad nucleomorph: Its ultrastructure and evolutionary significance. J Phycol 16: 558–568. [Google Scholar]
- 47. Maier UG, Sitte P (1994) Cryptomonad evolution – insights into a eucyte within a eucyte. Endocytobiosis Cell Res 10: 129–135. [Google Scholar]
- 48.Greenwood AD (1974) The Cryptophyta in relation to phylogeny and photosynthesis. In: Eighth International Congress on Electron Microscopy. Canberra: Australian Academy of Science. 566–567.
- 49. Greenwood AD, Griffiths HB, Santore UJ (1977) Chloroplasts and cell compartments in Cryptophyceae. Br Phycol J 12: 119. [Google Scholar]
- 50. Maier UG (1992) The 4 genomes of the alga Pyrenomonas salina (Cryptophyta). BioSystems 28: 69–73. [DOI] [PubMed] [Google Scholar]
- 51. Sitte P, Maier UG (1992) Evolution of complex plastids from eukaryotic endosymbionts. Endocytobiosis Cell Res 8: 223–225. [Google Scholar]
- 52. Douglas S, Zauner S, Fraunholz M, Baeton M, Penny S, et al. (2001) The highly reduced genome of an enslaved algal nucleus. Nature 410: 1091–1096. [DOI] [PubMed] [Google Scholar]
- 53. Yoon HS, Hackett JD, Van Dolah FM, Nosenko T, Lidie KL, et al. (2005) Tertiary endosymbiosis driven genome evolution in dinoflagellate algae. Mol Biol Evol 22: 1299–1308. [DOI] [PubMed] [Google Scholar]
- 54.Hoshaw RW, Rosowski JR (1973) Methods for microscopic algae. In: Stein JR, editor. Handbook of Phycological Methods. Culture methods and growth measurements. London: Cambridge University Press. 54–66.
- 55. Nichols HW, Bold HC (1965) Trichosarcina polymorpha gen et sp. nov. J Phycol 1: 34–38. [Google Scholar]
- 56. Doyle JJ, Dickson EE (1987) Preservation of plant samples for DNA restriction endonuclease analysis. Taxon 36: 715–722. [Google Scholar]
- 57. Scholin CA, Herzog M, Sogin M, Anderson DM (1994) identification of group-and strain-specific genetic markers for globally distributed alexandrium (dinophyceae). ii. sequence analysis of a fragment of the LSU rRNA gene. J Phycol 30: 999–1011. [Google Scholar]
- 58. Thompson JD, Gibson TJ, Plewniak F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic acids res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- 60. Hoef-Emden K, Marin B, Melkonian M (2002) Nuclear and nucleomorph SSU rDNA phylogeny in the Cryptophyta and the evolution of cryptophyte diversity. J Mol Evol 55: 161–179. [DOI] [PubMed] [Google Scholar]
- 61. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 62.Swofford DL (1998) PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4.0. Sunderland, MA: Sinauer Associates.
- 63. Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 64. Posada D, Crandall KA (1998) Modeltest: Testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]