Abstract
Background and methods
In order to characterize the expression pattern of SALL4, BMI-1 and ABCA3 genes in patients with myeloid leukemia and those who achieved complete remission (CR) after chemotherapy. Real-time PCR was used to determine the expression level of these genes in peripheral blood mononuclear cells from 24 patients with AML, eight patients with AML-CR, 13 patients with CML in the chronic phase (CML-CP), 12 patients with CML in blast crisis (CML-BC), 13 patients with CML-CR and 11 healthy individuals (HI).
Results
Overexpression of the BMI-1 gene was found in the AML, CML-CP and CML-BC groups as compared with HI group, while the BMI-1 expression level was lower in patients who achieved CR. In contrast, significantly increased SALL4 expression was only found in AML group, additionally, SALL4 expression was lower in the CML-CP and CML-CR groups compared with the HI group, while the SALL4 expression level in the CML-BC group was higher and significantly greater than that in the CML-CP and CML-CR groups. Moreover, a positive correlation between the expression of SALL4 and BMI-1 genes was found in samples from most groups. There was no significant difference of ABCA3 expression level in AML and CML-BC group in comparison with HI group. Interestingly, the ABCA3 expression level was significantly decreased in the CML-CP, AML-CR and CML-CR in comparison with the HI group. Moreover, the ABCA3 expression level in all of the CR groups was lower than that in their corresponding groups.
Conclusions
These results describe the altered SALL4, ABCA3 and BMI-1 expression pattern in different phases of myeloid leukemia, which may relate to the development and progression to different diseases. SALL4 expression was strongly correlated with BMI-1 in most of the myeloid leukemia patient groups, providing a potential link between SALL4 and BMI-1 in leukemogenesis.
Keywords: SALL4 gene, BMI-1 gene, Real-time PCR, AML, CML
Background
The altered expression of genes, such as WT1, SCL, and Notch1, that play crucial roles in the regulation of hematopoietic progenitor cell proliferation is frequently found in leukemia [1-7]. Increasing data show that the genes involved in hematopoietic stem/progenitor cell (HSPC) proliferation change their expression pattern during leukemogenesis [8].
SALL4 (sal-like protein 4), a SALL gene family member that is a newly identified zinc-finger transcription factor, was originally cloned based on its sequence homology to Drosophila spalt (sal) [9-12]. Alternative splicing generates two variant forms of human SALL4 mRNA, SALL4A and SALL4B, and each has a different tissue distribution [9,13]. Recently, SALL4 has been shown to play an important role in maintaining ES cell (ESC) pluripotency and self-renewal properties. SALL4 is involved in the self-renewal of leukemic initiation and HSPC [14]. Moreover, recent data have shown that SALL4 plays an essential role in myeloid leukemogenesis. SALL4 is constitutively expressed in human leukemia cell lines and primary acute myeloid leukemia (AML) cells [9,13]. Transgenic mice that ubiquitously overexpress SALL4B exhibit myelodysplastic syndrome (MDS)-like symptoms and subsequently develop transplantable AML [9,13], while SALL4 knockdown in leukemia cell lines triggers apoptosis [15].
BMI-1 is a member of the polycomb group of proteins, and it was initially identified in Drosophila as a repressor of homeotic genes [9,16-18]. The BMI-1 gene was initially isolated as an oncogene that cooperates with c-myc in retroviral-induced B and T cell leukemia [19,20]. In humans, BMI-1 is highly expressed in purified HSCs, and its expression declines with differentiation [9,21], and it plays an essential role in regulating adult, self-renewing HSPC and leukemia stem cells [9,21-27]. Knockout of the BMI-1 gene in mice results in the progressive loss of all hematopoietic lineages [9,25]. BMI-1 expression appears to be important for the accumulation of leukemic cells. Interestingly, inhibiting tumor stem cell self renewal after BMI-1 deletion can prevent leukemic recurrence. Recently, BMI-1 expression has been used as an important marker for predicting MDS development and the progression to AML [9,28]. BMI-1 overexpression was also observed in a significant number of nasopharyngeal carcinoma tumors that correlated with advanced tumor progression, invasive stage and poor prognosis [19,29].
BMI-1 was recently demonstrated to be a direct SALL4 target gene. The induction of SALL4 expression is associated with increased levels of histone H3–K4 and H3–K79 methylation in the BMI-1 promoter, indicating a novel connection between SALL4 and polycomb group proteins in leukemogenesis and a mechanism whereby aberrant SALL4 expression can directly alter BMI-1 expression [9].
Moreover, SALL4 expression was higher in drug resistant primary acute myeloid leukemic patients than those from drug-responsive cases. In addition, SALL4 expression was enriched in the SP when compared to the non-SP counterpart. Recently, it is reported that SALL4 could promote the expression of the ABC transporter genes, such as ATP binding cassette transporter A3 (ABCA3), suggesting that SALL4 can contribute to the SP phenotype by regulating the expression of ABCA3 and ABCG2[15].
ABCA3 is a member of the ATP-binding cassette (ABC) family of transport proteins and is required for perinatal respiratory adaptation. Mutations in ABCA3 resulted in fatal neonatal lung disease [30,31]. ABCA3 is highly expressed in AML and ALL patient samples and its expression is associated with unfavorable clinical treatment outcome. Furthermore, the expression of ABCA3 is enriched in leukemic SP cells and has been linked to multidrug resistance by facilitating lysosomal sequestration of drugs in AML primary cells and cell lines [15,32-35]. RNAi specific for ABCA3 led to a decrease of ABCA3 expression in T-ALL cell line such as CCRF-CEM and Jurkat cells. Consequently, a significant sensitization of cells to cytostatic drugs was achieved [35]. Moreover, both pharmacological blockade and the silencing of ABCA3 enhanced susceptibility of target B-cell lymphoma cells to anti-CD20 antibody-mediated lysis. Mechanisms of cancer cell resistance to drugs and antibodies are linked in an ABCA3-dependent pathway of exosome secretion [36].
Little is known about the expression pattern of the SALL4, ABCA3 and BMI-1 genes in patients with myeloid leukemia and patients that achieved complete remission after chemotherapy. In this study, we determined the expression characteristics of the SALL4, ABCA3 and BMI-1 genes in de novo AML and CML and complete remission samples.
Results
The high amplification efficiency of the BMI-1 and SALL4 genes was consistent with that of the β2M reference gene. The PCR products from all of the genes of interest were confirmed using 2.5% agarose gel electrophoresis followed by sequencing (data not shown). The BMI-1 and SALL4 genes were detected in all of the PBMC samples from the healthy individuals and those with myeloid leukemia.
Higher expression of BMI-1 in AML and CML
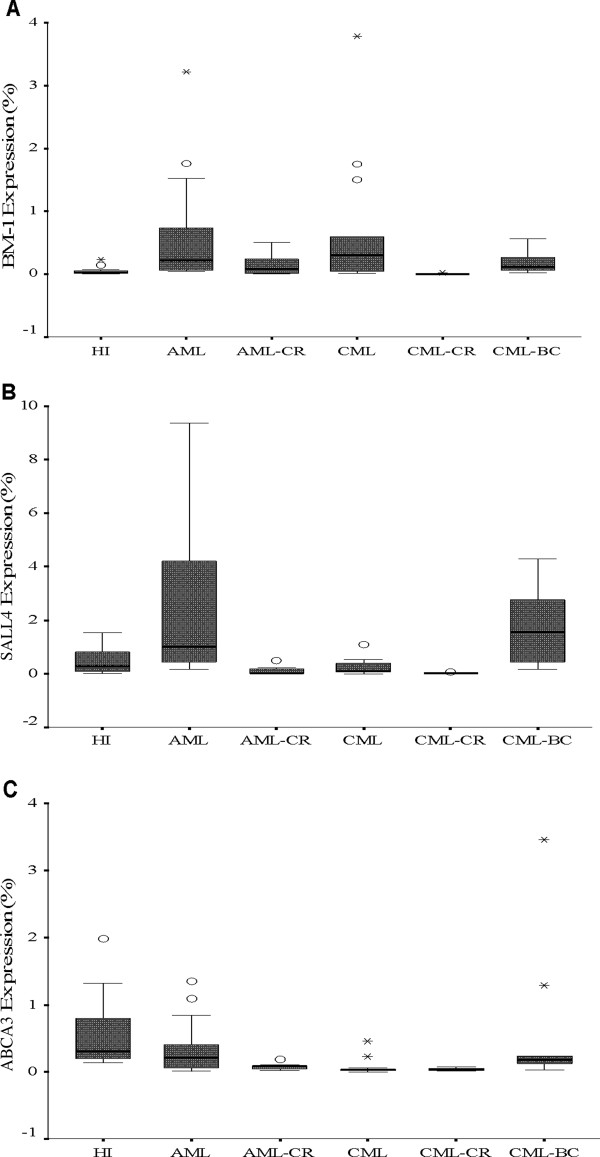
BMI-1 overexpression was found in the de novo AML (median: 0.303, p <0.001), CML-CP (median: 0.295, p=0.006), and CML-BC (median: 0.109, p=0.01) groups in comparison with the HI group (median: 0.027); the BMI-1 expression level in the AML-CR group (median: 0.078) was not significantly different compared with the HI group (p=0.322). Interestingly, the BMI-1 expression level in the CML-CR group (median: 0.003) was significantly lower than that in the HI (p< 0.0001) and CML groups (p< 0.0001).
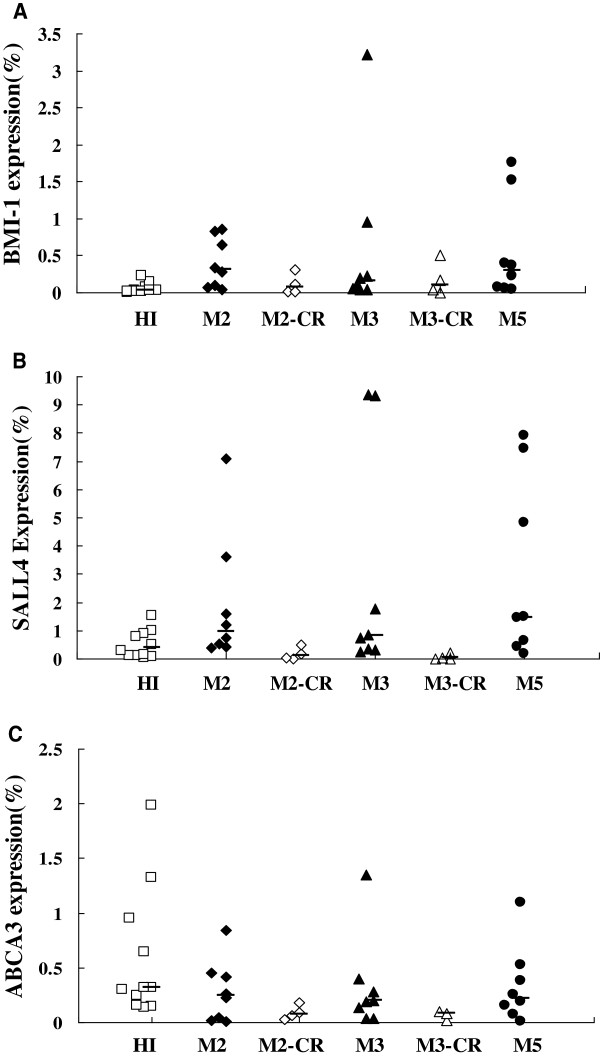
We next analyzed BMI-1 expression in different AML subtypes, including M2 (median: 0.400), M3 (median: 0.156) and M5 (median: 0.295), and all had a significantly higher expression level compared to the HI group (p=0.003, p=0.01 and p=0.004, respectively), while the BMI-1 expression level in the M2-CR (median: 0.070) and M3-CR (median: 0.099) groups was not significantly different in comparison to that of the HI group (p=0.514 and p=0.361, respectively) (Figures 1 and 2).
Figure 1.
The BMI-1 (A), SALL4 (B) and ABCA3 (C) expression levels in the different myeloid leukemia groups and healthy individuals.
Figure 2.
The BMI-1 (A), SALL4 (B) and ABCA3 (C) expression levels in the different AML subtypes and healthy individuals.
Different expression pattern of SALL4 in AML and CML
The expression pattern appeared different for SALL4 in the different myeloid leukemia in comparison with the HI group (median: 0.394). While the SALL4 expression level was increased in the AML group (median: 1.051; p=0.009), CML-BC group (median: 1.563; p=0.016) and the different AML subtypes M2 (median: 0.974; p=0.039), M3 (median: 0.799; p=0.083) and M5 (median: 1.465; p=0.026), it was lower in the AML-CR (median: 0.026; p=0.026), M2-CR (median: 0.105; p=0.151) and M3-CR (median: 0.023; p=0.037) groups. Interestingly, the level of SALL4 expression in the CML-CP (median: 0.093; p=0.213) and CML-CR groups (median: 0.025; p<0.0001) was lower in comparison with the HI group, and the increased SALL4 expression level in the CML-BC group (median: 1.563) was significantly higher than that in the CML-CP (p=0.001) and CML-CR (p<0.0001) groups. The SALL4 expression level in all of the CR groups was lower than that in their corresponding groups i.e., AML vs. AML-CR (p< 0.0001), M2 vs. M2-CR (p=0.017), M3 vs. M3-CR (p=0.007) and CML vs. CML-CR (p=0.011) (Figures 1 and 2).
Low expression of ABCA3 in myeloid leukemia
The expression level of ABCA3 seemed low in the different myeloid leukemia in comparison with the HI group. There was no significant difference of ABCA3 expression level in AML (median: 0.211; p=0.136), CML-BC (median: 0.174; p=0.097) and the different AML subtypes M2 (median: 0.242; p=0.215), M3 (median: 0.195; p = 0.186) and M5 (median: 0.221; p=0.364) in comparison with the HI group (median: 0.313). While the ABCA3 expression level was significantly decreased in the CML-CP (median: 0.025; p <0.0001), AML-CR (median: 0.078; p=0.0011) and CML-CR (median: 0.037; p <0.0001) in comparison with the HI group. Moreover, the ABCA3 expression level in all of the CR groups was lower than that in their corresponding groups i.e., AML vs. AML-CR (p=0.042) (Figures 1 and 2).
Correlation of relative expression of BMI-1, SALL4 and ABCA3 in myeloid leukemia
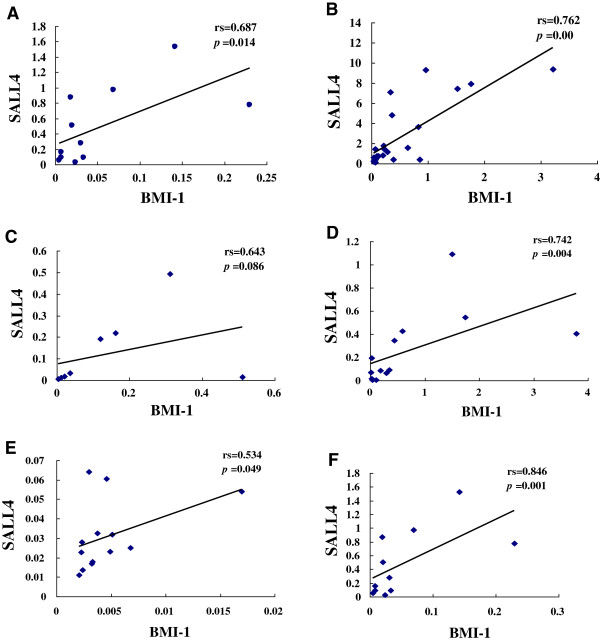
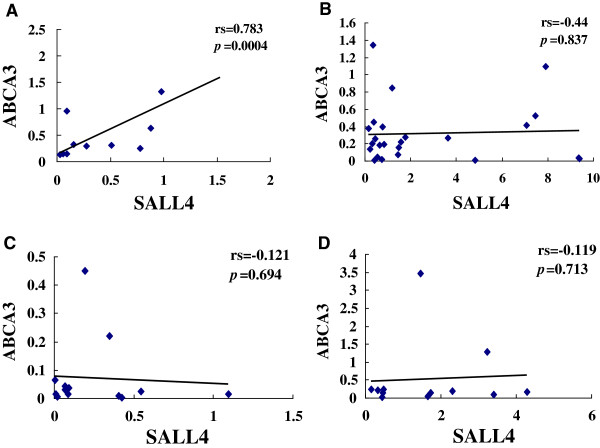
Correlation analysis of the relative expression levels of BMI-1 and SALL4,SALL4 and ABCA3 was performed using Spearman’s rank correlation analysis of the HI, AML and CML groups. A positive expression correlation level for BMI-1 and SALL4 genes was found in the HI (rs=0.687, p=0.014), AML (rs=0.762, p< 0.0001), M3 (rs=0.994, p< 0.0001), CML-CP (rs=0.742, p=0.004), CML-BC (rs=1=0.846, p=0.001), M2-CR (rs=1, p < 0.0001) and CML-CR (rs=0.534, p=0.049) groups. However, there was no significant correlation for both genes in the M2 (rs=0.381, p=0.352), M5 (rs=0.643, p=0.086), M3-CR (rs=0.40, p= 0.060) and AML-CR (rs=0.643, p=0.086) groups (Figure 3). A similar result with a positive expression correlation level for genes SALL4 and ABCA3 was found in the HI (rs=0.783, p=0.004), while there was no significant correlation between the expression levels of both genes in all myeloid leukemia groups (Figure 4).
Figure 3.
Correlation analysis of the relative BMI-1 and SALL4 expression levels in healthy individuals (A), AML (B), AML-CR (C), CML (D), CML-CR (E) and CML-BC (F) groups. The data of M2, M3, M3-CR, M2-CR, and M5 groups were not shown.
Figure 4.
Correlation analysis of the relative SALL4 and ABCA3 expression levels in healthy individuals (A), AML (B), CML (C) and the CML-BC (D) groups.
Discussion
BMI-1 and SALL4 are stem cell genes that modulate stem cell pluripotency and play a role in leukemogenesis. Dysregulated expression of both genes may have a cooperative effect in leukemogenesis [37]. Patients with RA and RARS who have a higher percentage of BMI-1+ cells showed disease progression to RAEB, suggesting that BMI-1 is a novel molecular marker that predicts the progression and prognosis of MDS [28]. In this study, we analyzed BMI-1 and SALL4 expression in primary AML and CML at diagnosis and those in complete remission.
It has been shown that BMI-1 overexpression occurs in a variety of cancers including several types of leukemias and lymphomas [38]. In this study, BMI-1 was found to be overexpressed in AML and chronic phase CML patient groups; and its expression level was lower in patients who achieved complete remission. Similar results were reported by Sawa, M et al. who found that moderate to high BMI-1 expression was detected in AML patients, and the AML-M0 subtype showed higher relative expression of the BMI-1 transcript [39]. In addition, Merkerova, M et al. demonstrated that BMI-1 and its significantly higher BMI-1 transcript level in CML cells seem to play a secondary role in CML transformation [40]. Our results also indicate that a decreased BMI-1 expression level is associated with complete disease remission. Interestingly, the BMI-1 expression level in the CML-BC group appeared to be low in comparison with the de novo CML group, although the difference was not significant. Further investigation is needed using a larger patient cohort to extend our findings. Preliminary results indicate that BMI-1 may have potential as a therapeutic target for myeloid leukemia. It has been reported that BMI-1 depletion by RNA interference leads to reduced U937 cell growth and proliferation and increased apoptosis [41], and an antisense BMI-1 gene can inhibit the growth of K562 cells and upregulate p16 expression in K562 cells [42].
Using immunohistochemistry and real-time PCR, SALL4 was demonstrated to be constitutively expressed in human primary acute myeloid leukemia [13]. In this study, we found that SALL4 was overexpressed in different primary AML subtypes, and its expression was lower in the AML-CR patient group. These results are similar to the findings of Jeong, HW et al. who showed that AML patients who responded to treatment had decreasing SALL4 expression throughout the course of treatment, while AML patients with disease relapse or drug resistance had increasing SALL4 expression, which was correlated with disease progression [15]. Interestingly, unlike the SALL4 expression characteristics in AML, the SALL4 expression level in the CML-CP and CML-CR groups was lower. Moreover, the SALL4 expression level in patients with chronic phase CML was significantly lower than that in the CML-CR group. There is no direct evidence demonstrating the SALL4 expression level in CML-CP and comparing the expression feature to healthy individuals; however, Lu and colleagues have found that the SALL4 protein was overexpressed in CML samples in blast crisis but not those in chronic phase by FACS [37]. Our results also demonstrated that SALL4 expression was higher in the CML-BC group in comparison with the CML-CP and CML-CR groups; however, there was no significant difference in comparison with the HI group. Is it possible that SALL4 is preferentially expressed in leukemic blasts? These results are similar to a report by Cui W et al. who demonstrated that only precursor B-cell lymphoblastic leukemias/lymphomas and AML had detectable SALL4 in neoplastic tissues [43]. The different SALL4 expression patterns in AML and CML suggest that these two disease entities may have different biological characteristics and/or mechanisms of leukemogenesis, at least for the association between SALL4 and pathogenesis. However, there are not reports comparing the data of SALL4 expression level in CML-CP to healthy individuals, it is difficult to evaluate the significance of this finding. Recently, research from Zhu et al. showed that hematopoietic transcription factor PU.1 expression was significantly lower in newly diagnosed APL patient samples as compared to normal hematopoietic cells, which may relate to the expression level of PML-RARα, and they found that suppression of PU.1 expression occurred concurrently with PML-RARα expression, the authors suggested that low PU.1 expression in APL patients is required for disease initiation and progression [44]. This finding might provide a direction in farther analysis the correlation of SALL4 with BCR-ABL in the pathogenesis of CML and to address this question.
In principle, the BMI-1 and SALL4 gene expression level should be positively correlated in stem cells [9]. Little is known about the expression pattern and differences in the SALL4 and BMI-1 genes in patients with AML and CML. In this study, we analyzed the correlation between the relative expression levels of BMI-1 and SALL4. A positive expression level correlation was found for both genes in HI, AML, chronic phase CML, CML-BC and CML-CR patient groups; however, there was no significant correlation between these genes in patients with AML-CR, leaving their role in this group an open question. These results indicate that a positively correlated expression pattern is a common feature in patients with myeloid leukemia and healthy individuals, and both genes may cooperate during cell proliferation and differentiation.
Based on the different expression features of SALL4 in AML and CML, we further analyzed its regulating gene ABCA3, which is a member of the ATP-binding cassette (ABC) family of transport proteins [30,31]. Unlike the description by Wult, Norwood and Steinbach groups, who showed that ABCA3 is highly expressed in acute meyloid leukemia samples and is associated with unfavorable clinical treatment outcome [24,33,45], in the present study, lower expression level of ABCA3 was found not only in AML but also in CML groups, especially in CML-CP and CR groups. Moreover, the expression level of ABCA3 lost the correlation with SALL4 expression in leukemia patients. To determine whether these results relate to favorable clinical outcome, further investigation is needed. Additionally, detection of ABCA2, ABCB2 and ABCC10, which were found overexpressed in childhood AML, may be worthy to build the gene regulation network in proliferation of myeloid leukemia cells.
In conclusion, we determined the expression characteristics of the SALL4, ABCA3 and BMI-1 genes in different phases of AML and CML. Further studies will be needed to determine whether BMI-1 and SALL4 are novel therapeutic targets for leukemic stem/initiation cells in primary myeloid leukemia.
Methods
Samples
Twenty-four newly diagnosed and untreated patients with AML, eight cases with AML in complete remission (AML-CR), thirteen newly diagnosed and untreated patients with CML in chronic phase, 13 cases with CML-CR, and 12 cases with CML in blast crisis (CML-BC),were recruited, the details of the samples was listed in Table 1. The diagnoses of all patients were based on cytomorphology, immunohistochemistry, and cytoimmunological and cytogenetic analysis. Peripheral blood mononuclear cells (PBMCs) from 11 healthy individuals (HI) served as controls. Peripheral blood was collected by heparin anticoagulation, and PBMCs were separated using the Ficoll-Hypaque gradient centrifugation method. All procedures were conducted in accordance with the guidelines of the Medical Ethics committees of the health bureau of Guangdong Province, China.
Table 1.
The details of samples used in study
| Diagnosis | Subtype |
Numbers |
Age (year) |
|||
|---|---|---|---|---|---|---|
| Total | Male | Female | Range | Median | ||
| AML |
|
24 |
12 |
12 |
6-69 |
32 |
| M2 |
8 |
4 |
4 |
16-61 |
42.5 |
|
| M3 |
8 |
3 |
5 |
24-52 |
30 |
|
| M5 |
8 |
6 |
2 |
6-69 |
30.5 |
|
| AML-CR |
|
8 |
4 |
4 |
16-61 |
32 |
| M2-CR |
4 |
3 |
1 |
39-61 |
47 |
|
| M3-CR |
4 |
1 |
3 |
16-25 |
20 |
|
| CML-CP |
|
13 |
11 |
2 |
13-64 |
38 |
| CML-BP |
|
12 |
6 |
6 |
23-66 |
42.5 |
| CML-CR |
|
13 |
7 |
6 |
15-55 |
32 |
| HI | 11 | 5 | 6 | 24-57 | 36 | |
RNA extraction and cDNA synthesis
RNA was extracted using the Trizol kit (Invitrogen, Carlsbad, CA, USA) and then reverse-transcribed into first-strand cDNA using random hexamer primers and the Superscript II reverse transcriptase Kit (Invitrogen) according to the manufacturer’s instructions.
Real-time quantitative reverse transcription–polymerase chain reaction (qRT–PCR)
The expression levels of BMI-1, SALL4, ABCA3 and the β2-microglobulin (β2-MG) reference gene were determined by SYBR Green I real-time PCR. Briefly, PCR was performed in a 25 μL total volume containing 1 μL of cDNA, 9 μL of 2.5× SYBR Green mix (Tiangen, Beijing, China), and 10 μmol/L primer pairs. After initial denaturation at 95°C for 2 min, 45 cycles consisting of the following procedure were performed using an MJ Research DNA Engine Opticon 2 PCR cycler (BIO-RAD, USA): 15 s at 95°C, 40 s at 64°C for β2M and BMI-1, 60°C for SALL4, and 63°C for ABCA3. The relative amounts of the genes of interest and the β2-MG reference gene were measured in two independent assays. The data are presented as the relative expression of the genes of interest relative to the internal control gene as determined by the 2(−∆∆CT) method [1-3,5,46]. Additionally, the specific amplification of the PCR products was analyzed by melting curve analysis and agarose gel electrophoresis. The primers used for real-time PCR for all gene amplifications were synthesized by Shanghai Biological Engineering Technology Services Co., Ltd. (Table 2). RT–PCR, for the BMI-1, SALL4 and ABCA3 genes, was performed using the same primers described above, and the PCR products were sent to Shanghai Invitrogen Biotechnology Co. for DNA sequence analysis.
Table 2.
Primer sequences used for real-time PCR
| Primer | Sequence | Accession No | PCR product size |
|---|---|---|---|
| SALL4-f |
5’-TGCAGCAGTTGGTGGAGAAC-3’ |
NM_020436.3 |
68 bp |
| SALL4-r |
5’-TCGGTGGCAAATGAGACATTC-3’ |
||
| BMI-1-f |
5’-TAAGCATTGGGCCATAGT-3’ |
NM_005180.8 |
140 bp |
| BMI-1-r |
5’-ATTCTTTCCGTTGGTTGA-3’ |
||
| ABCA3-f |
5’-CTCCGAGAAGGACTTTGAGG-3’ |
NM_001089.2 |
144 bp |
| ABCA3-r |
5’-TCCGTGTGTAACTGAACCGT-3’ |
||
| β2M-f |
5’-TACACTGAATTCACCCCCAC-3’ |
J00105 | 144 bp |
| β2M-r | 5’-CATCCAATCCAAATGCGGCA-3’ |
Statistical analyses
Differences in mRNA expression between two groups were analyzed using the Mann–Whitney U test. Data are presented as median. Spearman’s rank correlation analysis was used to analyze the SALL4, BMI-1 and ABCA3 mRNA levels in different samples using the SPSS 11.5 statistical software. Differences were considered statistically significant at P < 0.05.
Competing interests
The authors declare that they have no potential conflicts of interest.
Authors’ contributions
YQL and YPM contributed to concept development and study design. QS, SCL, SHC and YM performed the real-time PCR. JYH, LJY, BL, XLW, JCY were responsible for collection of clinical data. YQL and QS coordinated the study and helped drafting the manuscript. All authors read and approved the final manuscript.
Contributor Information
Qi Shen, Email: tshenq@jnu.edu.cn.
Sichu Liu, Email: 76194436@qq.com.
Junyan Hu, Email: 306006187@qq.com.
Shaohua Chen, Email: 452319674@qq.com.
Lijian Yang, Email: jnyanglijian@163.com.
Bo Li, Email: 420562894@qq.com.
Xiuli Wu, Email: 378246347@qq.com.
Yu Ma, Email: 316667210@qq.com.
Jianchang Yang, Email: jnyanglijian@163.com.
Yupo Ma, Email: yupo.ma@stonybrook.edu.
Yangqiu Li, Email: yangqiuli@hotmail.com.
Acknowledgements
This work was supported by Grants from National Natural Science Foundation of China (no. 81270604), the Fundamental Research Funds for the Central Universities (No. 21612116) and in part through NIH grant R01HL087948 (Y.M.).
References
- Stams WAG, den Boer ML, Beverloo HB, Meijerink JPP, Stigter RL, van Wering ER, Janka-Schaub GE, Slater R, Pieters R. Sensitivity to L-asparaginase is not associated with expression levels of asparagine synthetase in t(12;21)+ pediatric ALL. Blood. 2003;101(7):2743–2747. doi: 10.1182/blood-2002-08-2446. [DOI] [PubMed] [Google Scholar]
- Miglino M, Colombo N, Pica G, Grasso R, Clavio M, Bergamaschi M, Ballerini F, Ghiso A, Ghiggi C, Mitscheunig L, Beltrami G, Cagnetta A, Vignolo L, Lucchetti MV, Aquino S, Pierri I, Sessarego M, Carella AM, Gobbi M. WT1 overexpression at diagnosis may predict favorable outcome in patients with de novo non-M3 acute myeloid leukemia. Leuk Lymphoma. 2011;52(10):1961–1969. doi: 10.3109/10428194.2011.585673. [DOI] [PubMed] [Google Scholar]
- Cardoso BA, de Almeida SF, Laranjeira AB, Carmo-Fonseca M, Yunes JA, Coffer PJ, Barata JT. TAL1/SCL is downregulated upon histone deacetylase inhibition in T-cell acute lymphoblastic leukemia cells. Leukemia. 2011;25(10):1578–1586. doi: 10.1038/leu.2011.140. [DOI] [PubMed] [Google Scholar]
- Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011;223(2):262–273. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Zheng H, Wang C, Yang L, Chen S, Li B, Zhou Y, Tan H, Li Y. Mutations increased overexpression of Notch1 in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 2012;12:13. doi: 10.1186/1475-2867-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S. Current findings for recurring mutations in acute myeloid leukemia. J Hematol Oncol. 2011;4:36. doi: 10.1186/1756-8722-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braoudaki M, Papathanassiou C, Katsibardi K, Tourkadoni N, Karamolegou K, Tzortzatou-Stathopoulou F. The frequency of NPM1 mutations in childhood acute myeloid leukemia. J Hemotol Oncol. 2010;3:41. doi: 10.1186/1756-8722-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong AS, Stephens N, Weber G, Li Y, Savani BN, Eniafe R, Keyvanfar K, Kurlander R, Rezvani K, Barrett AJ. Improved outcome following allogeneic stem cell transplantation in chronic myeloid leukemia is associated with higher expression of BMI-1 and immune responses to BMI-1 protein. Leukemia. 2011;25(4):629–637. doi: 10.1038/leu.2010.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC, Chai L, Liu F, Fink LM, Lin P, Silberstein LE, Amin HM, Ward DC, Ma YP. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci USA. 2007;104(25):10494–10499. doi: 10.1073/pnas.0704001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhase J, Hausmann S, Stojmenovic G, Dixkens C. Bink Schulz-Schaeffer W, Altmann M, Engel W: SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics. 1999;62(2):216–222. doi: 10.1006/geno.1999.6005. [DOI] [PubMed] [Google Scholar]
- Al-Baradie R, Yamada K, St Hilaire C, Chan WM, Andrews C, McIntosh N, Nakano M, Martonyi EJ, Raymond WR, Okumura S, Okihiro MM, Engle EC. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am J Hum Genet. 2002;71(5):1195–1199. doi: 10.1086/343821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Aguila JR, Alipio Z, Lai R, Fink LM, Ma Y. Enhanced self-renewal of hematopoietic stem/progenitor cells mediated by the stem cell gene Sall4. J Hematol Oncol. 2011;4:38. doi: 10.1186/1756-8722-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YP, Cui W, Yang JC, Qu J, Di CH, Amin HM, Lai R, Ritz J, Krause DS, Chai L. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006;108(8):2726–2735. doi: 10.1182/blood-2006-02-001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster JA, Stupnikov MR, Ma G, Liao WB, Lai R, Ma YP, Aguila JR. Expansion of hematopoietic stem cells for transplantation: current perspectives. Exp Hematol Oncol. 2012;1:12. doi: 10.1186/2162-3619-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HW, Cui W, Yang YY, Lu JY, He J, Li AL, Song D, Guo Y, Liu BH, Chai L. SALL4, a stem cell factor, affects the side population by regulation of the ATP-binding cassette drug transport genes. PLoS One. 2011;6(4):e18372. doi: 10.1371/journal.pone.0018372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Paro R. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 1995;14(2):5660–5671. doi: 10.1002/j.1460-2075.1995.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- Franke A, DeCamillis M, Zink D, Cheng N, Brock HW, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11(8):2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hu D, Yang G, Zhou JY, Yang CF, Gao Y, Zhu ZY. Down-regulation of BMI-1 cooperates with artemisinin on growth inhibition of nasopharyngeal carcinoma cells. J Cell Bochem. 2011;112(7):1938–1948. doi: 10.1002/jcb.23114. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65(5):753–763. doi: 10.1016/0092-8674(91)90383-A. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Raaphorst FM. Self-renewal of hematopoietic and leukemic stem cells: a central role for the Polycomb-group gene Bmi-1. Trends Immunol. 2003;24(10):522–524. doi: 10.1016/S1471-4906(03)00241-2. [DOI] [PubMed] [Google Scholar]
- Dick JE. Stem cells: Self-renewal writ in blood. Nature. 2003;423(6937):231–233. doi: 10.1038/423231a. [DOI] [PubMed] [Google Scholar]
- Ohta H, Sawada A, Kim JY, Tokimasa S, Nishiguchi S, Humphries RK, Hara J, Takihara Y. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J Exp Med. 2002;195(6):759–770. doi: 10.1084/jem.20011911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423(6937):255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, Bwrns A. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8(7):757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, Ema H, Kamijo T, Katoh-Fukui Y, Koseki H, van Lohuizen M, Nakauchi H. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21(6):843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Mihara K, Chowdhury M, Nakaju N, Hidani S, Ihara A, Hyodo H, Yasunaga S, Takihara Y, Kimura A. Bmi-1 is useful as a novel molecular marker for predicting progression of myelodysplastic syndrome and patient prognosis. Blood. 2006;107(1):305–308. doi: 10.1182/blood-2005-06-2393. [DOI] [PubMed] [Google Scholar]
- Song LB, Zeng MS, Liao WT, Zhang L, Mo HY, Liu WL, Shao JY, Wu QL, Li MZ, Xia YF, Fu LW, Huang WL, Dimri GP, Band V, Zeng YX. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res. 2006;66(12):6225–6232. doi: 10.1158/0008-5472.CAN-06-0094. [DOI] [PubMed] [Google Scholar]
- Stahlman MT, Besnard V, Wert SE, Weaver TE, Dingle S, Xu Y, von Zychlin K, Olson SJ, Whitsett JA. Expression of ABCA3 in developing lung and other tissues. J Histochem Cytochem. 2007;55(1):71–83. doi: 10.1369/jhc.6A6962.2006. [DOI] [PubMed] [Google Scholar]
- Saugstad OD, Hansen TW, Rønnestad A, Nakstad B, Tølløfsrud PA, Reinholt F, Hamvas A, Coles FS, Dean M, Wert SE, Whitsett JA, Nogee LM. Novel mutations in the gene encoding ATP binding cassette protein member A3 (ABCA3) resulting in fatal neonatal lung disease. Acta Paediatr. 2007;96(2):185–190. doi: 10.1111/j.1651-2227.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- Chapuy B, Koch R, Radunski U, Corsham S, Cheong N, Inagaki N, Ban N, Wenzel D, Reinhardt D, Zapf A, Schweyer S, Kosari F, Klapper W, Truemper L, Wulf GG. Intracellular ABC transporter A3 confers multidrug resistance in leukemia cells by lysosomal drug sequestration. Leukemia. 2008;22(8):1576–1586. doi: 10.1038/leu.2008.103. [DOI] [PubMed] [Google Scholar]
- Wulf GG, Modlich S, Inagaki N, Reinhardt D, Schroers R, Griesinger F, Trümper L. ABC transporter ABCA3 is expressed in acute myeloid leukemia blast cells and participates in vesicular transport. Haematologica. 2004;89(11):1395–1397. [PubMed] [Google Scholar]
- Norwood K, Wang RY, Hirschmann-Jax C, Andreeff M, Brenner MK, Goodell MA, Wulf GG. An in vivo propagated human acute myeloid leukemia expressing ABCA3. Leuk Res. 2004;28(3):295–299. doi: 10.1016/j.leukres.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Efferth T, Gillet JP, Sauerbrey A, Zintl F, Bertholet V, de Longueville F, Remacle J, Steinbach D. Expression profiling of ATP-binding cassette transporters in childhood T-cell acute lymphoblastic leukemia. Mol Cancer Ther. 2006;5(8):1986–1994. doi: 10.1158/1535-7163.MCT-06-0086. [DOI] [PubMed] [Google Scholar]
- Aung T, Chapuy B, Vogel D, Wenzel D, Oppermann M, Lahmann M, Weinhage T, Menck K, Hupfeld T, Koch R, Trümper L, Wulf GG. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc Natl Acad Sci USA. 2011;108(37):15336–15341. doi: 10.1073/pnas.1102855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Ma Y, Kong N, Alipio Z, Gao C, Krause DS, Silberstein LE, Chai L. Dissecting the role of SALL4, a newly identified stem cell factor, in chronic myelogenous leukemia. Leukemia. 2011;25(7):1211–1213. doi: 10.1038/leu.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beà S, Tort F, Pinyol M, Puig X, Hernández L, Hernández S, Fernandez PL, van Lohuizen M, Colomer D, Campo E. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 2001;61(6):2409–2412. [PubMed] [Google Scholar]
- Sawa M, Yamamoto K, Yokozawa T, Kiyoi H, Hishida A, Kajiguchi T, Seto M, Kohno A, Kitamura K, Itoh Y, Asou N, Hamajima N, Emi N, Naoe T. BMI-1 is highly expressed in M0-subtype acute myeloid leukemia. Int J Hematol. 2005;82(1):42–47. doi: 10.1532/IJH97.05013. [DOI] [PubMed] [Google Scholar]
- Merkerova M, Bruchova H, Kracmarova A, Klamova H, Brdicka R. Bmi-1 over-expression plays a secondary role in chronic myeloid leukemia transformation. Leuk Lymphoma. 2007;48(4):793–801. doi: 10.1080/10428190601186002. [DOI] [PubMed] [Google Scholar]
- Zhu W, Huang L, Xu XJ, Qian H, Xu WR. Anti-proliferation effect of BMI-1 in U937 cells with siRNA. Int J Mol Med. 2010;25(6):889–895. [PubMed] [Google Scholar]
- Meng XX, Liu WH, Liu DD, Zhao XY, Su BL. Construction of antisense Bmi-1 expression plasmid and its inhibitory effect on K562 cells proliferation. Chin Med J. 2005;118(16):1346–1350. [PubMed] [Google Scholar]
- Cui W, Kong NR, Ma YP, Amin HM, Lai R, Chai L. Differential expression of the novel oncogene, SALL4, in lymphoma, plasma cell myeloma, and acute lymphoblastic leukemia. Mod Pathol. 2006;19(12):1585–1592. doi: 10.1038/modpathol.3800694. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhang H, Qian M, Zhao X, Yang W, Wang P, Zhang J, Wang K. The significance of low PU.1 expression in patients with acute promyelocytic leukaemia. J Hematol Oncol. 2012;5:22. doi: 10.1186/1756-8722-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach D, Gillet JP, Sauerbrey A, Gruhn B, Dawczynski K, Bertholet V, de Longueville F, Zintl F, Remacle J, Efferth T. ABCA3 as a possible cause of drug resistance in childhood acute myeloid leukemia. Clin Cancer Res. 2006;12(14 Pt 1):4357–4363. doi: 10.1158/1078-0432.CCR-05-2587. [DOI] [PubMed] [Google Scholar]
- Huang X, Chen S, Shen Q, Yang L, Li B, Zhong L, Geng S, Du X, Li Y. Analysis of the expression pattern of the BCL11B gene and its relatives in patients with T-cell acute lymphoblastic leukemia. J Hematol Oncol. 2010;3:44. doi: 10.1186/1756-8722-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]