Abstract
By using a fragment-assembly strategy and bioisosteric-replacement principle, a series of novel piperazine derivatives were designed, synthesized, and evaluated for their cellular target-effector fusion activities and in vitro antiviral activities against HIV-1. Preliminary structure-activity relationships (SARs) of target compounds were concluded in this study, and five compounds were found to exhibited medium to potent CCR5 fusion activities with IC50 values in low micromolar level. Among evaluated compounds, 23 h was found to be a CCR5 antagonist with an IC50 value of 6.29 µM and an anti-HIV-1 inhibitor with an IC50 value of 0.44 µM.
Introduction
AIDS, one of the leading threats for human health worldwide, is a disease of the human immune system caused by the human immunodeficiency virus (HIV). Although the highly active antiretroviral therapy (HAART) is an available option for AIDS treatment, many patients are suffered from incomplete efficacy, severe toxicity, and the eventual emergence of resistance [1]. Therefore, the development of potent antiretroviral agents with novel mechanism of action is of great interests in the field of medicinal chemistry and drug discovery.
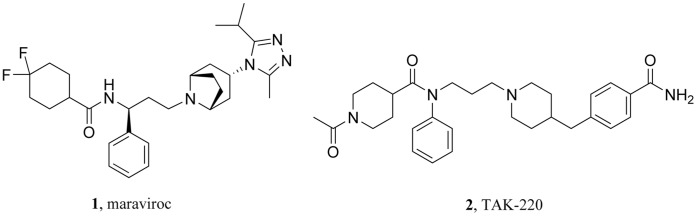
C-C Chemokine receptor 5 (CCR5), a G protein-coupled receptor (GPCR) for the β-chemokines MIP-1α, MIP-1β, and RANTES [2] and a primary co-receptor with CD4 for macrophage-tropic (M-tropic or R5) HIV-1 viruses [3] has been identified as a new target for HIV-1 epidemic prevention and treatment. Efforts devoted to the development of CCR5 antagonists have resulted in the discovery of the first marketed small-molecule inhibitor against CCR5, maraviroc (UK-427,857, 1, Figure 1) [4]. Besides, several other promising molecules are currently under clinical trials as potential anti-HIV agents, such as the Takeda disclosed compound TAK-220 (2) [5], [6].
Figure 1. Representative structures of small-molecule CCR5 antagonists.
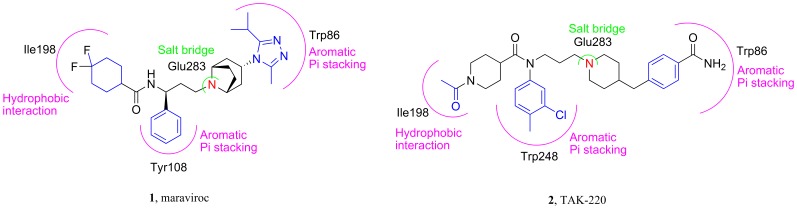
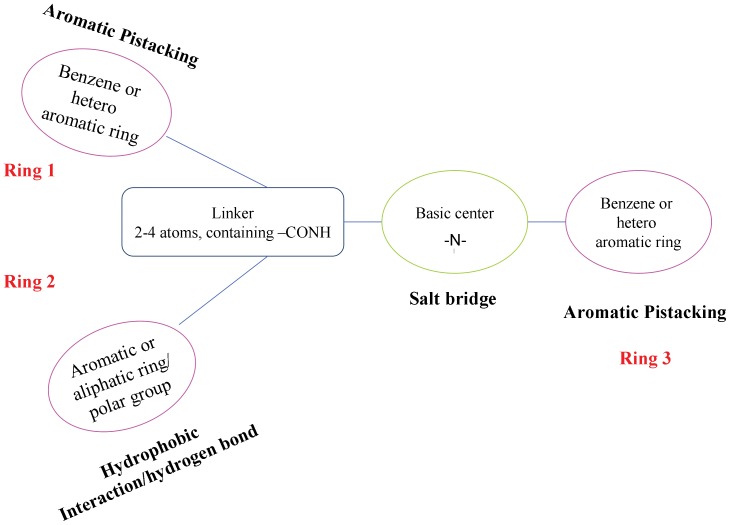
Although reported CCR5 antagonists are of different structures, the presence of one basic nitrogen center that tends to form strong salt-bridge interaction with the Glu283 residue of CCR5 receptor was found to be one of the most important features for CCR5 antagonists. A hydrophobic interaction involving the Ile198 residue was found for both maraviroc and TAK-220, together with a T-shaped π-π stacking interaction involving the Trp86 residue (Figure 2) [7]. Thus, a ‘Y shape’ pharmacophore model that contains one basic center, three hydrophobic domains, and an amide linker (Figure 3) was proposed in this study.
Figure 2. The binding models for maraviroc and TAK-220.
Figure 3. Proposed ‘Y shape’pharmacophore model of CCR5 antagonists.
Results and Discussion
Chemistry
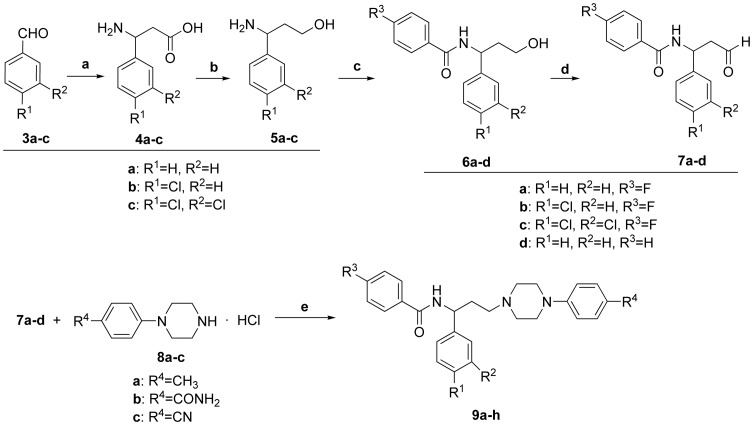
Target compounds listed in Tables 1 , 2 , and 3 were synthesized as outlined in Figure 4 , 5 , and 6 . As shown in Figure 4 , benzaldehydes 3a–c were reacted with malonic acid and ammonium acetate to give β-amino acids 4a–c, which was reduced to γ-amino alcohols 5a–c with the presence of LiAlH4 [11]. Acylation of 5a–c with corresponding benzoyl chlorides afforded amides 6a–d. Compounds 6a–d were then subjected to a Swern oxidation to give aldehydes 7a–d, whose following reductive amination with substituted phenylpiperazine hydrochlorides 8a–c [12] afforded target compounds 9a–h.
Table 1. Effects of different substituents R1–R4 and link between ring 2 and 3 on CCR5 fusion activity.
| Compd | R1 | R2 | R3 | R4 | inhibition rate (10 µM) a,b | IC50 (µM) a,b |
| 9a | H | H | H | CH3 | 47% | – |
| 9b | H | H | F | CH3 | 35% | – |
| 9c | H | H | F | F | 41% | – |
| 9d | H | H | H | F | 26% | – |
| 9e | H | H | F | CN | 0.64 | |
| 9f | Cl | H | F | CN | 3.70 | |
| 9g | Cl | Cl | F | CN | 10.01 | |
| 9h | H | H | F | CONH2 | 20% | – |
| 13a | inactive | – | ||||
| 13b | 22% | – | ||||
| 13c | inactive | – | ||||
| 13d | 25% | – | ||||
| 17 | inactive | – | ||||
Mean value of at least two experiments.
DMSO as a negative control.
Table 2. Modification of the p-fluorophenyl moiety of 9e.
| Compd | R5 | inhibition rate (10 µM)a,b | IC50 (µM)a,b |
| 23a | phenyl- | 2.44 | |
| 23b | 4-chlorophenyl- | 2.87 | |
| 23c | 4-trifluoromethylphenyl- | 10.00 | |
| 23d | 4-isopropylphenyl- | 20% | – |
| 23e | 1-acetylpiperidin-4-yl - | inactive | – |
| 23f | 1-benzoylpiperidin-4-yl- | 15% | – |
| 23g | furan-2-yl | inactive | – |
| 23h | cyclohexyl- | 6.29 |
Mean value of at least two experiments.
DMSO as a negative control.
Table 3. Antiviral activity of 9e–g, 23a–c, and 23h.
| Compd | inhibition rate (10 µM) a,b | IC50 (µM) a,b |
| 9e | 23.8% | – |
| 9f | inactive | – |
| 9g | 2.9% | – |
| 23a | 28.9% | – |
| 23b | 29.9% | – |
| 23c | 24.6% | – |
| 23h | 66.5% | 0.44 |
| Maraviroc | 96.8% | 0.0011 |
Mean value of two experiments.
DMSO as a negative control.
Figure 4. Synthesis of target piperazine derivatives 9a–h.
Reagents and conditions: a) CH3COONH4, CH2(COOH)2, C2H5OH, reflux, 12h; b) LiAlH4, THF, 65°C, 3h; c) corresponding benzoyl chlorides, Et3N, CH2Cl2, 0°C, 4h; d) (COCl)2, DMSO, CH2Cl2, −78°C, 2h; e) NaBH(OAc)3, Et3N, CH2Cl2, rt, 8h.
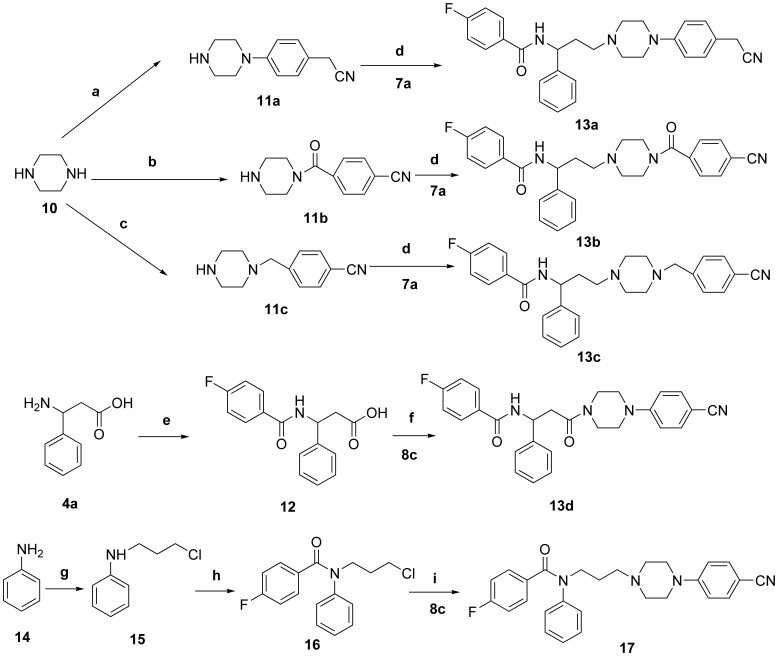
Figure 5. Synthesis of target piperazine derivatives 13a–d and 17.
Reagents and conditions: a) 2-(4-chlorophenyl)acetonitrile, K2CO3, DMSO, reflux, 18h; b) 4-cyanobenzoyl chloride, Et3N, CH2Cl2, rt, 3h; c) 4-(chloromethyl)benzonitrile, THF, reflux, 2.5h; d) NaBH(OAc)3, CH2Cl2, rt, 12h; e) 4-fluorobenzoyl chloride, Et3N, CH2Cl2, rt, 3h; f) EDC.HCl, CH2Cl2, rt, 8h; g) 1-bromo-3-chloropropane, KI, CH3CN, MWI, 15min; h) 4-fluorobenzoyl chloride, Et3N, CH2Cl2, 0°C, 5h; i) KI, K2CO3, CH3CN, reflux, 24h.
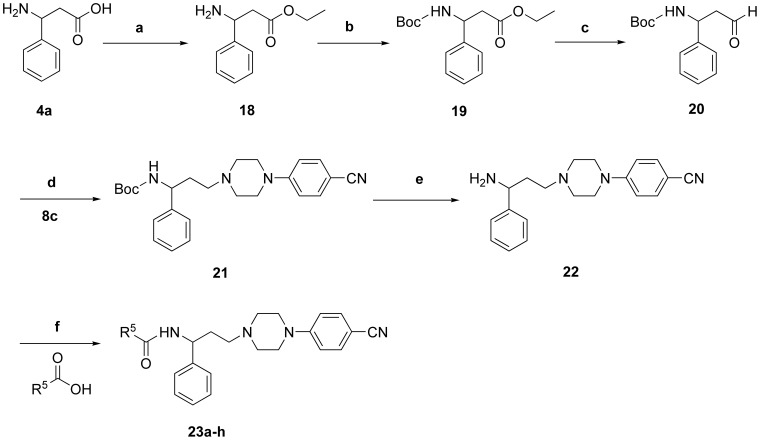
Figure 6. Synthesis of target piperazine derivatives 23a–h.
Reagents and conditions: a) SOCl2, EtOH, 0°C, 0.5h, rt, 3h, reflux, 1h; b) (Boc)2O, NaOH, H2O, Dioxane, rt, 12h; c) DIBAL-H, CH2Cl2, −78°C, 3h; d) NaBH(OAc)3, CH2Cl2, rt, 8h; e) 6NHCl, EtOAc, rt, 3h; f) EDC.HCl, CH2Cl2, rt, 6h.
Based on the ‘Y shape’ pharmacophore model, a series of 1,3-diamine compounds that combined the structural features of maraviroc and TAK-220 (1 and 2) were designed by using a fragment- assembly strategy and bioisosteric-replacement principle. Hydrophobic aromatic rings with a variety of other functional groups were introduced to study structure-activity relationship of target compounds. Although some piperazine-based compounds have been reported before, the structures of compounds involved here are different from that of the reported papers [8]–[10]. Herein, we report the design, synthesis, and biological evaluation of the novel piperazine derivatives 9a–h, 13a–d, 17, and 23a–h, with a goal to discover novel compounds as potential CCR5 antagonists for HIV treatment.
Synthesis of 13a–d and 17 is depicted in Figure 5 . Reaction of piperazine with 2-(4-chlorophenyl)acetonitrile afforded N-arylation compound 11a, which was submitted to reductive amination with aldehyde 7a in the presence of sodium triacetoxyborohydride to provide 13a. Compounds 11b–c were achieved by treatment with 4-cyanobenzoyl chloride/4-(chloromethyl)benzonitrile and piperazine, respectively. Compounds 13b–c were prepared from the corresponding intermediates 11b–c following the same procedure as described for the preparation of 13a from 11a. Target compound 13d was obtained by condensation mono-substituted piperazine 8c with β-amino acid 12, which was prepared by the acylation of β-phenylalanine 4a with 4-fluorobenzoyl chloride. Target compound 17 was obtained as follows. Reaction of aniline 14 with 1-bromo-3-chloropropane under microwave irradiation afforded compound 15. Acylation of 15 with 4-fluorobenzoyl chloride gave amide 16, whose reaction with 4-substituted piperazine 8c afforded target compound 17.
Target compounds 23a–h were synthesized through procedures as illustrated in Figure 6 . Compound 18 was prepared by esterification of 4a, and then protected by BOC group to get ester 19, which was reduced to the required aldehyde 20 using DIBAL-H, and then reductive amination with the compound 8c furnished the key BOC-protected intermediate 21. A BOC-deprotection step gave precursor 22, whose acylation with corresponding carboxylic acids afforded target compounds 23a–h.
Biological Evaluation
A total of 21 novel piperazine derivatives 9a–h, 13a–d, 17, and 23a–h were screened for their inhibitory activity against cell-cell fusion between target cells expressing CD4/CCR5 and effector cells expressing the envelope protein of HIV-1, gp-120 (fusion assay) [13]. All synthesized analogs, which were firstly tested in MTT assay, showed no significant cytotoxicity against HEK 293 cells at a concentration of 50 µM.
Among the first set of compounds with three hydrophobic phenyl rings and varied substituents on phenyl rings (Table 1), compound 9e exhibited the most potent CCR5 fusion activity with an IC50 value of 0.64 µM. The data in Table 1 clearly showed limited tolerance toward different substitutions (R4) at p-position of hydrophobic aromatic ring, as only compounds with a cyano-substituent retained good activity (9e–9g, IC50∶0.64 to 10.01 µM). Replacement of the p-cyano group with methyl (9b), fluoro (9c), or amide (9h) resulted in a sharp loss of potency. Replacement of the 4-fluorophenyl ring (hydrophobic aromatic ring 1) of 9b and 9c with phenyl group (9a and 9d) did not significantly improve in vitro fusion potency. These results indicated that the presence of a cyano substituent at R4 position is favorable for CCR5 fusion activity.
Thus, a further optimization process was initiated with the introduction of a chlorine atom at R1 position or two chlorine atoms both at R1 and R2 positions of the phenyl ring 2 of 9e. Compound 9f with monochloro-substitution at R1 position showed 5-fold less potent activity than that of 9e. 3,4-Dichloro-substituted analog (9g) demonstrated more than 15-fold less potent fusion activity in comparison to 9e. These results suggested that introduction of a more hydrophobic function to the R1 and R2 positions is not favorable for fusion potency.
We then turned our attention to the link between ring 2 and ring 3 of compound 9e, modifying the two basic nitrogen-atoms in piperazine and 4-cyanophenyl group. Replacing the cyano group at R4 with an acetonitrile group provided compound 13a, which was found inactive on CCR5-mediated fusion assay at a concentration of 10 µM. Subsequent replacement of the 4-cyanophenyl group with 4-cyanobenzyl group afforded compound 13c, which was also inactive in fusion assay. A carbonyl group was introduced in the right (13b) or left (13d) side of the piperazine to reduce the basicity of the piperazine. Neither 13b nor 13d showed potent CCR5 fusion activity. Our observation suggested that 4-(piperazin-1-yl)benzonitrile is a favored scaffold for CCR5 antagonists. Interestingly, replacement of the phenyl ring 2 from the carbon atom to the nitrogen atom (17) resulted in a substantial loss in CCR5 fusion activity.
Modifications as shown in Table 2 were also made to explore SARs of target compounds. Replacement of the fluoro-substitution at phenyl ring in ring 1 with a hydrogen (23a, IC50 = 2.44 µM) or a chlorine atom (23b, IC50 = 2.87 µM) resulted in a 4- or 5-fold reduction in CCR5-mediated fusion activity. Conversion of the fluoro-substituent of 9e to a trifluoromethyl group (23c) led to a 15-fold loss in potency (IC50 = 10.00 µM). p-Isopropyl replacement (23d) was found to lead an even more sharp decrease in potency. These observations indicated that the electron-withdrawing group at the 4-position in ring 1 rather than the corresponding electron-donating group contribute significantly in fusion activity. Not as expected, introducing of N-acetyl-piperidin group, effective in compound 2, to R5 position (23e) resulted in a complete loss in fusion activity. So did benzoylpiperdine derivatives (23f). Compound 23g with a five-membered furan ring was found to be inactive in CCR5-mediated fusion assay. It was interesting that compound 23h containing an aliphatic six-membered ring instead of the aromatic ring was found to be well-tolerated (IC50 = 6.29 µM). As such, hydrogen- or halogen-substituted phenyl ring and cyclohexyl group were identified as the optimal substituents.
Fluorophenyl analogs 9e–g, phenyl, chlorophenyl, and trifluorophenyl analog 23a–c, and cyclohexyl analog 23 h were evaluated for their anti-HIV activity using a recombinant HIV-1 virus pseudotyped with the envelope proteins of the CCR5-tropic virus (HIV-1 single cycle antiviral assay) [14]. For overall estimation of the therapeutic potential of these novel CCR5 antagonists, maraviroc was used as a positive control. Results are shown in Tables 3 . Albeit the observation of potent activity in fusion assay, compound 9e showed weak anti-HIV activity at a concentration of 10 µM. Compound 23 h showed potent anti- HIV activity in comparison with the other tested piperazine derivatives.
Conclusions
A novel series of piperazine derivatives were synthesized by adopting a fragment-assembly strategy and the bioisosteric-replacement principle. Target compounds were evaluated for their CCR5-mediated fusion activity and cytotoxicity, which showed that five compounds displayed medium to potent CCR5 antagosnist activity. Compound 23 h was evaluated as a CC5 antagonist with an IC50 value of 6.29 µM and an antiviral agent with an IC50 value of 0.44 µM. The piperazine derivatives developed in this study, as well as the concluded SAR, might be useful for following optimization toward the development of novel CCR5 antagonists for HIV treatment.
Materials and Methods
Cell Cytotoxicity Assays
Cytotoxicities of target compounds were tested by using MTT (Sigma-Aldrich) assay. Briefly, 100 µl of HEK293 cell suspension (5000 cells/well) were dispensed in a 96-well plate, and then pre-incubated the plate for 24 hours at 37°C in 5% CO2. 10 µl of various concentrations of substances to be tested were added to the plate. After 7 hours incubation, 10 µl of CCK-8 solution were added to each well of the plate. The plate was incubated at 37°C for another 2 hours in the incubator, the optical absorbance was measured at 430 nm using a microplate reader.
Two Stable Cell Lines
Effector cell line: This cell line express HIV envelope protein gp160 and chimera protein Rn-Dn. Rn-Dn is consist of the N terminal of renilla luciferase and the N terminal of DnaE intein from Anacystis nidulans R2 PCC7942.
Target cell line: This cell line express chemokine receptor 5(CCR5), CD4 protein and chimera protein Dc-Rc. Dc-Rc is consist of the C teminal of renilla luceferase and the C teminal of NnaE intein from Anacystis nidulans R2 PCC7942.
Cell-cell Fusion Assay (CCF Assay)
The cell-cell fusion assay was performed as described previously [13].The effector cells were plated in 24 well white culture plates at 7.5×104 cell per well in DMEM supplemented with 10% FBS, 800 µg/mL G418. The target cells in the growth medium were then added to the plates at 7.5×104 cells/50 µL/well and incubated for 5 hours. At the end of coculture, 70 µL of renilla luceferase assay lysis was added into each well, and the cultures were gently shaken for 15 min. At the same time, add 20 µl of Renilla Luciferase Assay Reagent to the luminometer tube. Add 20 µl of cell lysate to the tube. Mix quickly by flicking the tube for 1 second. Place the tube in FB12 luminometer and initiate measurement. Luminescence was integrated over 1 second with a 2-second delay. When small molecule compounds needed to be added to the CCF assay system, the compounds were diluted manually in DMSO. Then, 10 µL of the diluted compounds was added to the effector cells just before the addition of target cells, thus making the final concentration of DMSO in the coculture 0.5%.
Plasmids
HIV-1 proviral indicator construct pNL-Luc-E- contains a full-length HIV-1 genome, in which env was replaced by firefly Luciferase coding sequence. pENV-Ad8 expresses R5-tropic envelope (AD8).
Viral Infectivity Assays
Single-cycle HIV-1 replication assays were performed as described previously [14]. In brief, 4×105 293T cells were co-transfected with 0.4 µg of pNL-Luc-E- and 0.4 µg of pENV-R5. After 48h, the supernatant containing pseudovirion was harvested by filtration through a 0.45 µm filter and the amount of viral capsid protein was measured by p24 antigen capture ELISA (Biomerieux). The resultant supernatant (10 µL) was used to infect SupT1 cells (1×105) in 96-well plates in the presence of testing compound at the concentration indicated. The SupT1 cells were lysed 48h post-infection and firefly luciferase activities were determined using a firefly Luciferase Assay System (Promega). Values were normalized to the control group treated with DMSO, and represented relative infectivity of each sample testing.
Supporting Information
Experimental protocols, NMR data (1H and 13C), Mass spectrometry data (MS and HRMS) and Melting points data of compounds are presented in File S1.
Supporting Information
Experimental protocols, NMR data (1H and 13C), Mass spectrometry data (MS and HRMS) and Melting points data of compounds.
(DOC)
Acknowledgments
We thank Jianyang Pan (Pharmaceutical Informatics Insistute, Zhejiang University) for performing NMR spectrometry (or Mass spectrometry) for structure elucidation.
Funding Statement
This study was supported by grants from the National Nature Science Foundation of China (number 81072515). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yerly S, Kaiser L, Race E, Bru JP, Clavel F, et al. (1999) Transmission of antiretroviral-drug- resistant HIV-1 variants. Lancet 354: 729–733. [DOI] [PubMed] [Google Scholar]
- 2. Combadiere C, Ahuja SK, Tiffany HL, Murphy PM (1996) Cloning and functional expression of CC CKR5, a human monocyte CC chemokine receptor selective for MIP-1β, MIP-1α, and RANTES. J. Leukocyte Biol 60: 147–152. [DOI] [PubMed] [Google Scholar]
- 3. Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, et al. (1996) Identification of a major co-receptor for primary isolates of HIV-1. Nature 381: 661–666. [DOI] [PubMed] [Google Scholar]
- 4. Rodger DM, Richard MN (2008) Reviews of antiinfective agents: maraviroc: the first of a new class of antiretroviral agents. Clin Infect Dis 47: 236–241. [DOI] [PubMed] [Google Scholar]
- 5. Takashima K, Miyake H, Kanzaki N, Tagawa Y, Wang X, et al. (2005) Highly potent inhibition of human immunodeficiency virus type 1 replication by TAK-220, an orally bioavailable small-molecule CCR5 antagonist. Antimicrob Agents Ch 49: 3474–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nishikawa M, Takashima K, Nishi T, Furuta RA, Kanzaki N, et al. (2005) Analysis of binding sites for the new small-molecule CCR5 antagonist TAK-220 on human CCR5. Antimicrob Agents Ch 49: 4708–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kondru R, Zhang J, Ji CH, Mirzadegan T, Rotstein D, et al. (2008) Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol Pharmacol 73: 789–800. [DOI] [PubMed] [Google Scholar]
- 8.Dong MX, Lu L, Li H, Wang X, Lu H, et al. (2012) Design, synthesis, and biological activity of novel 1,4-disubstituted piperidine/piperazine derivatives as CCR5 antagonist-based HIV-1 entry inhibitors. Bioorg Med Chem Lett 22; 3284–3286. [DOI] [PubMed]
- 9. Feng DZ, Song YL, Jiang XH, Chen L, Long YQ (2007) Forward- and reverse-synthesis of piperazinopiperidine amide analogs: a general access to structurally diverse 4-piperazinopiperidine-based CCR5 antagonists. Org Biomol Chem 5: 2690–2697. [DOI] [PubMed] [Google Scholar]
- 10. Jiang XH, Song YL, Long YQ (2004) Facile synthesis of 4-substituted-4-aminopiperidine derivatives, the key building block of piperazine-based CCR5 antagonists. Bioorg Med Chem Lett 14: 3675–3678. [DOI] [PubMed] [Google Scholar]
- 11. Torre O, Gotor-Fernández V, Gotor V (2006) Lipase-catalyzed resolution of chiral 1,3-amino alcohols: application in the asymmetric synthesis of (S)-dapoxetine. Tetrahedron: Asymmetry 17: 860–866. [Google Scholar]
- 12.Weng ZY, Gao YP, Zhang JK, Dong XW, Liu T (2011) Synthesis and biological evaluation of novel N-[3-(4-phenylpiperazin-1-yl)-propyl]-carboxamide derivatives. J Chem Res 43–46.
- 13. Chen LJ, Zhang YP, Li G, Huang HS, Zhou NM (2010) Functional characterization of a naturally occurring trans-splicing intein from Synechococcus elongatus in a mammalian cell system. Anal Biochem 407: 180–187. [DOI] [PubMed] [Google Scholar]
- 14. Coakley E, Petropoulos C, Whitcomb J (2005) Assessing chemokine co-receptor usage in HIV. Curr Opin Infect Dis 18: 9–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental protocols, NMR data (1H and 13C), Mass spectrometry data (MS and HRMS) and Melting points data of compounds.
(DOC)