Abstract
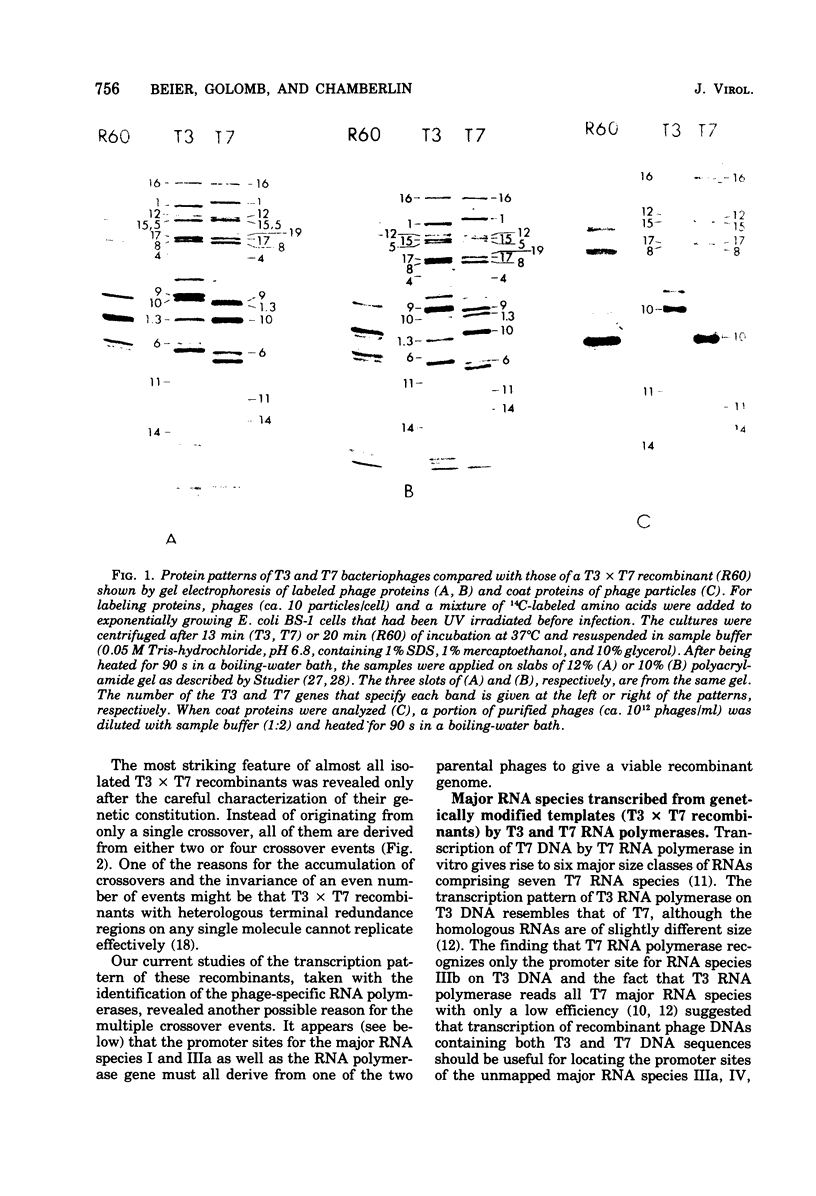
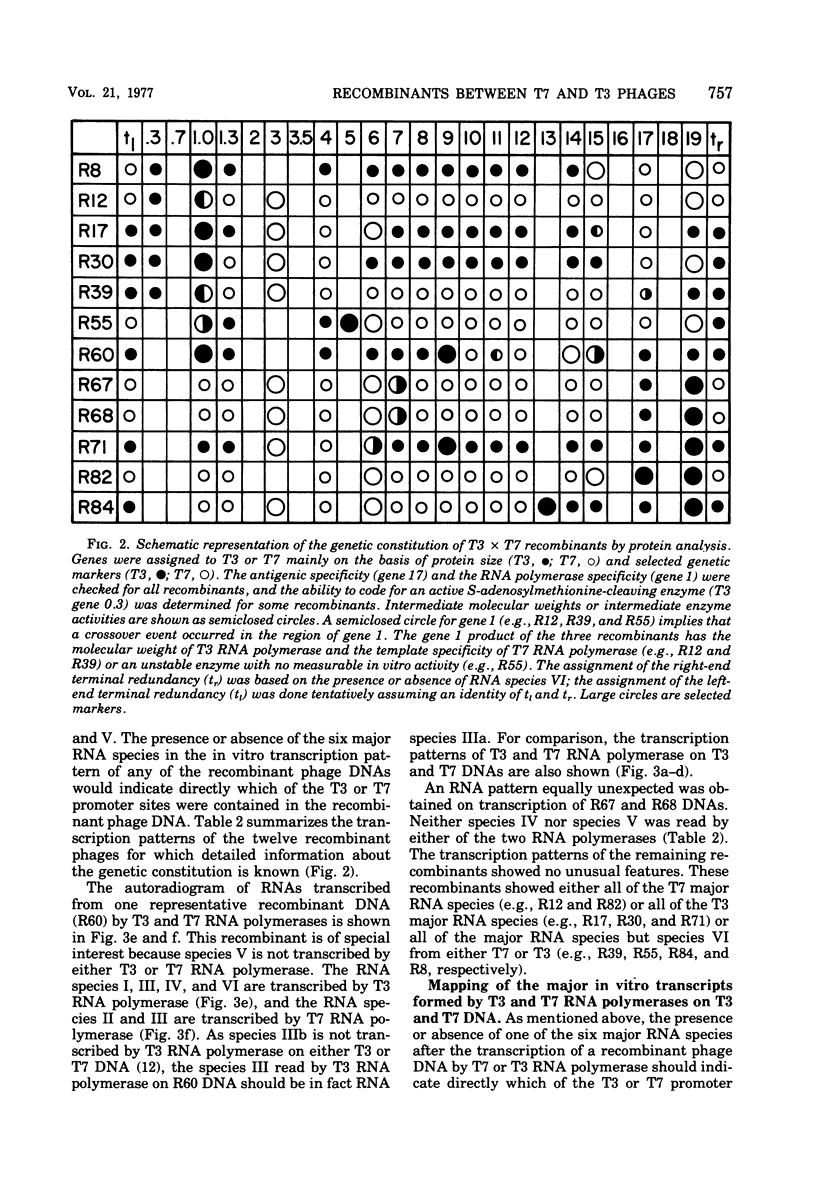
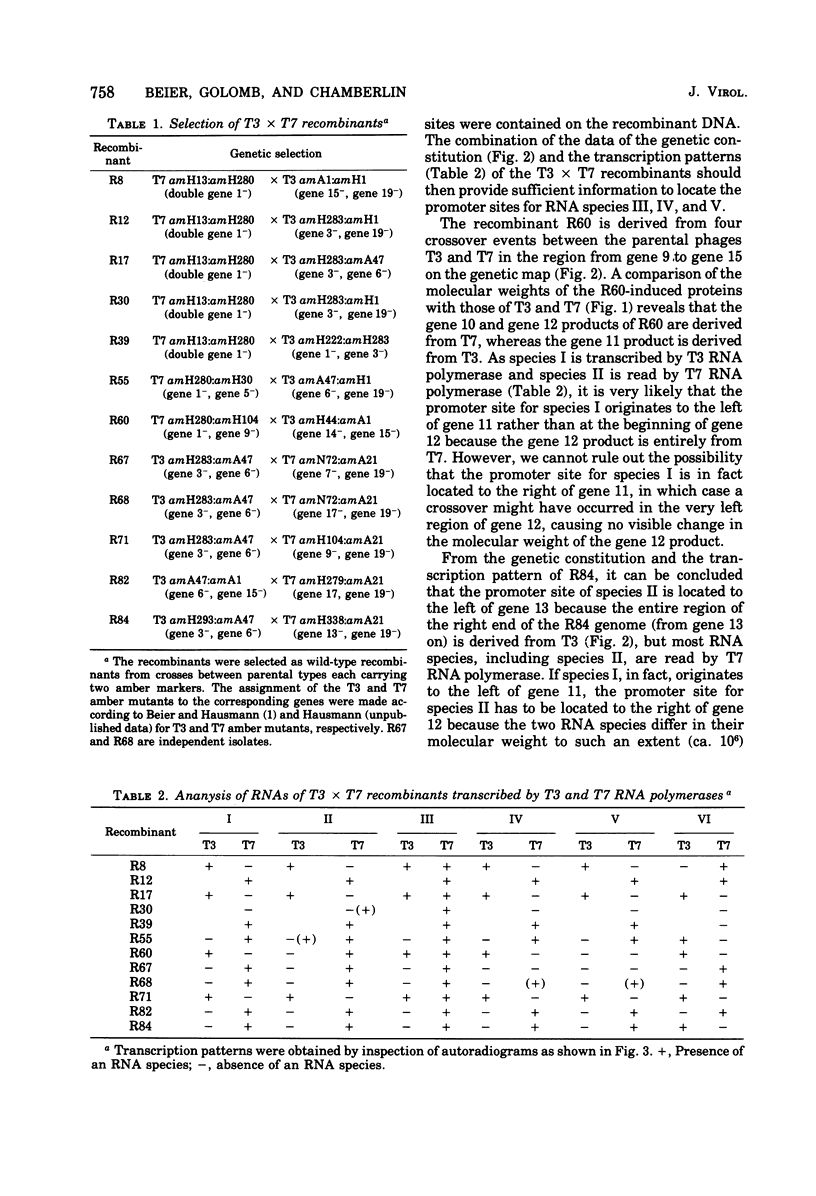
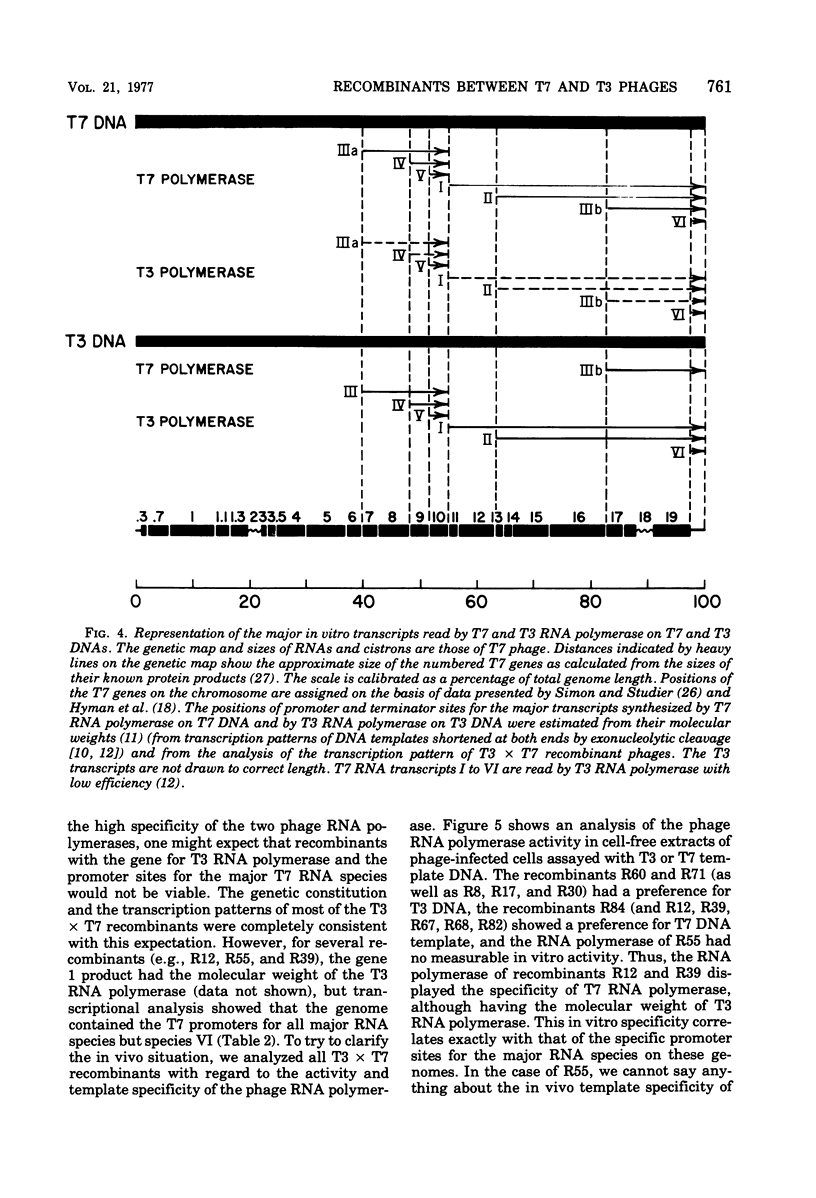
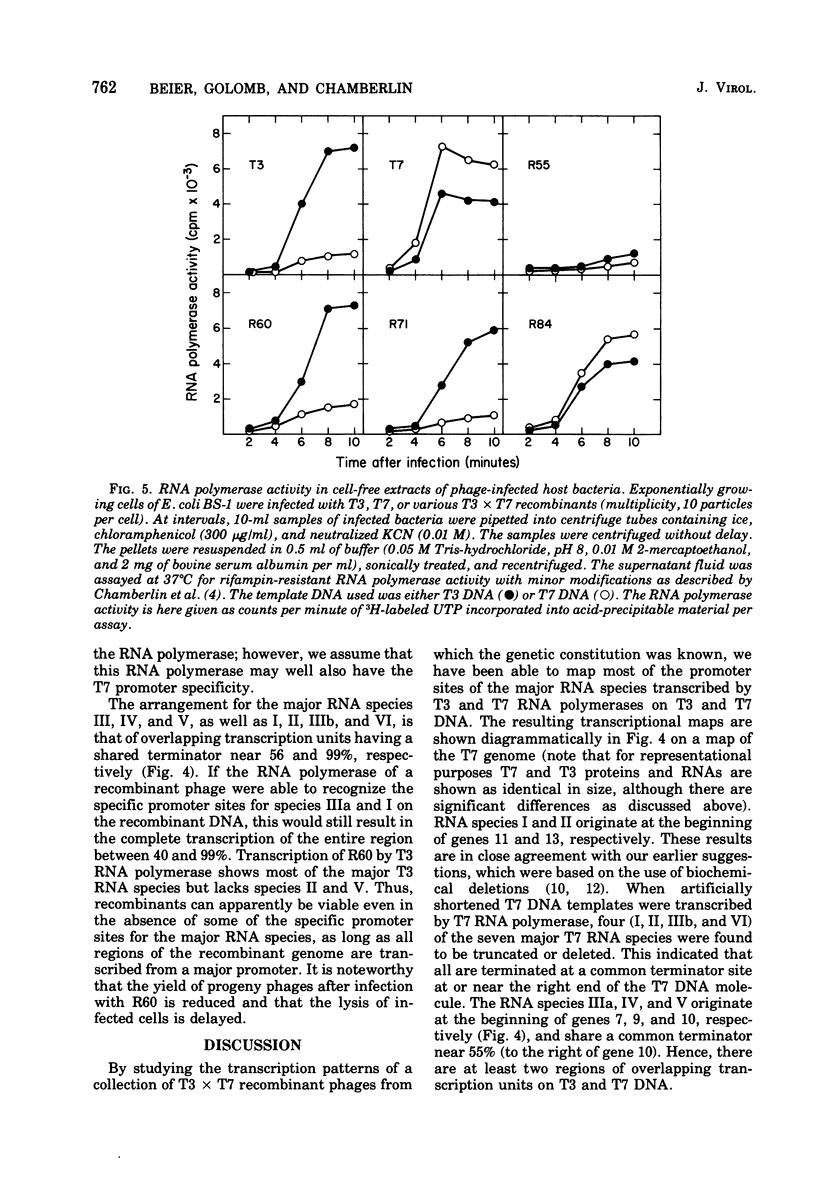
A variety of T3 X T7 recombinants were isolated from crosses between T3 and T7 parental phages carrying amber markers in various genes (gene 1 to gene 19). The genetic constitution of these recombinants was determined by reference to the selected markers and also directly by analysis of the proteins translated from the T3 X T7 recombinants in vivo. Although T3 and TM phages are closely related, most T3 and T7 proteins differ slightly in size, and hence the genetic origin of a gene can be determined by protein analysis. The major transcripts read by T3 and T7 RNA polymerases from T3 X T7 recombinant phage DNAs vary, depending on which regions of the T3 or T7 chromosome are present. T7 RNA polymerase is unable to utilize major promoter sites employed by T3 polymerase at an appreciable rate, and the converse is also true. Hence the transcriptional pattern for a recombinant phage DNA obtained with the T3 or T7 polymerase allows a determination of the identity of the different promoter sites on the genome. The transcriptional analysis of T3 X T7 recombinant DNAs together with earlier observations has been used to map the promoter sites for five out of seven major T3 and T7 RNA species on the genetic maps of T3 and T7. The promoter sites for the T7 and T3 RNA species IIIa, IV, and V originate at the beginning of genes 7, 9, and 10, respectively; the promoter sites for the T7 and T3 RNA species I and II are located to the left of gene 11 and gene 13, respectively. No T3 X T7 recombinants were found for which the specificity of the phage RNA polymerase was not correlated with the corresponding promoter sites for species IIIa and I (the transcription of which covers 60% of the genome). That means that the RNA polymerase specified by the recombinant genome is able to read all of the information encoded in sequences read normally from major promoters by the enzyme on the wild-type phage genome. This suggests that the in vitro specificity for promoter site selection by the phage polymerases is also maintained in vivo.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beier H., Hausmann R. Genetic map of bacteriophage T3. J Virol. 1973 Aug;12(2):417–419. doi: 10.1128/jvi.12.2.417-419.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H., Hausmann R. T3 times T7 phage crosses leading to recombinant RNA polymerases. Nature. 1974 Oct 11;251(5475):538–540. doi: 10.1038/251538a0. [DOI] [PubMed] [Google Scholar]
- Bradley C., Ling O. P., Egan J. B. Isolation of phage P2-186 intervarietal hybrids and 186 insertion mutants. Mol Gen Genet. 1975 Sep 29;140(2):123–135. doi: 10.1007/BF00329780. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Couturier M., Dambly C., Thomas R. Control of development in temperate bacteriophages. V. Sequential activation of the viral functions. Mol Gen Genet. 1973 Feb 2;120(3):231–252. doi: 10.1007/BF00267155. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., McAllister W. T., Bautz E. K. In vitro transcription of T3 DNA by Escherichia coli and T3 polymerases. Virology. 1972 Apr;48(1):112–125. doi: 10.1016/0042-6822(72)90119-5. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter M., Hausmann R., Gold M., Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. X. Bacteriophage T3-induced S-adenosylmethionine cleavage. J Biol Chem. 1966 May 10;241(9):1995–2006. [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. J. T7- and T3-specific RNA polymerases: characterization and mapping of the in vitro transcripts read from T3 DNA. J Virol. 1977 Feb;21(2):743–752. doi: 10.1128/jvi.21.2.743-752.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. A preliminary map of the major transcription units read by T7 RNA polymerase on the T7 and T3 bacteriophage chromosomes. Proc Natl Acad Sci U S A. 1974 Mar;71(3):760–764. doi: 10.1073/pnas.71.3.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J Biol Chem. 1974 May 10;249(9):2858–2863. [PubMed] [Google Scholar]
- Hausmann R., Gomez B. Amber mutants of bacteriophages T3 and T7 defective in phage-directed deoxyribonucleic acid synthesis. J Virol. 1967 Aug;1(4):779–792. doi: 10.1128/jvi.1.4.779-792.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann R. Synthesis of an S-adenosylmethionine-cleaving enzyme in T3-infected Escherichia coli and its disturbance by co-infection with enzymatically incompetent bacteriophage. J Virol. 1967 Feb;1(1):57–63. doi: 10.1128/jvi.1.1.57-63.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker S., Botstein D. An early regulatory gene of Salmonella phage P22 analogous to gene N of coliphage lambda. Virology. 1975 Dec;68(2):510–524. doi: 10.1016/0042-6822(75)90291-3. [DOI] [PubMed] [Google Scholar]
- Hyman R. W., Brunovskis I., Summers W. C. A biochemical comparison of the related bacteriophages T7, phiI, phiII, W31, H, and T3. Virology. 1974 Jan;57(1):189–206. doi: 10.1016/0042-6822(74)90120-2. [DOI] [PubMed] [Google Scholar]
- Hyman R. W., Brunovskis I., Summers W. C. DNA base sequence homology between coliphages T7 and phiII and between T3 and phiII as determined by heteroduplex mapping in the electron microscope. J Mol Biol. 1973 Jun 25;77(2):189–196. doi: 10.1016/0022-2836(73)90330-6. [DOI] [PubMed] [Google Scholar]
- Issinger O. G., Beier H., Hausmann R. In vivo and in vitro "phenotypic mixing" with amber mutants of phages T3 and T7. Mol Gen Genet. 1973 Mar 27;122(1):81–88. doi: 10.1007/BF00337976. [DOI] [PubMed] [Google Scholar]
- Niles E. G., Condit R. C. Translational Mapping of Bacteriophage T7 RNAs synthesized in vitro by purified T7 RNA polymerase. J Mol Biol. 1975 Oct 15;98(1):57–67. doi: 10.1016/s0022-2836(75)80101-x. [DOI] [PubMed] [Google Scholar]
- Niles E. G., Conlon S. W., Summers W. C. Purification and physical characterization of T7 RNA polymerase from T7-infected Escherichia coli B. Biochemistry. 1974 Sep 10;13(19):3904–3912. doi: 10.1021/bi00716a014. [DOI] [PubMed] [Google Scholar]
- Pachl C. A., Young E. T. Detection of polycistronic and overlapping bacteriophage T7 late transcripts by in vitro translation. Proc Natl Acad Sci U S A. 1976 Feb;73(2):312–316. doi: 10.1073/pnas.73.2.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M., Herrlich P. DNA-directed enzyme synthesis in vitro. Curr Top Microbiol Immunol. 1974;65:59–132. doi: 10.1007/978-3-642-65875-4_3. [DOI] [PubMed] [Google Scholar]
- Simon M. N., Studier F. W. Physical mapping of the early region of bacteriophage T7 DNA. J Mol Biol. 1973 Sep 15;79(2):249–265. doi: 10.1016/0022-2836(73)90004-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Bacteriophage T7. Science. 1972 Apr 28;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J Mol Biol. 1975 May 15;94(2):283–295. doi: 10.1016/0022-2836(75)90083-2. [DOI] [PubMed] [Google Scholar]
- Watson J. D. Origin of concatemeric T7 DNA. Nat New Biol. 1972 Oct 18;239(94):197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]