Summary
Bacillus cereus strains elaborate pili on their surface using a mechanism of sortase-mediated crosslinking of major and minor pilus components. Here we used a combination of electron microscopy and atomic force microscopy to visualize these structures. Pili occur as single, double or higher order assemblies of filaments formed from monomers of the major pilin, BcpA, capped by the minor pilin, BcpB. Previous studies demonstrated that within assembled pili, four domains of BcpA – CNA1, CNA2, XNA, and CNA3 – each acquire intramolecular lysine-asparagine isopeptide bonds formed via catalytic glutamic acid or aspartic acid residues. Here we showed that mutants unable to form the intramolecular isopeptide bonds in the CNA2 or CNA3 domains retain the ability to form pilus bundles. A mutant lacking the CNA1 isopeptide bond assembled deformed pilin subunits that failed to associate as bundles. X-ray crystallography revealed that the BcpA variant Asp312Ala, lacking an aspartyl catalyst, did not generate the isopeptide bond within the jelly-roll structure of XNA. The Asp312Ala mutant was also unable to form bundles and promoted the assembly of deformed pili. Thus, structural integrity of the CNA1 and XNA domains are determinants for the association of pili into higher order bundle structures and determine native pilus structure.
Introduction
Many Gram-positive bacteria assemble proteinaceous filaments called pili or fimbriae on their surface (Yanagawa & Honda, 1976). Pili are implicated in bacterial adhesion to and colonization of host tissues, escape from innate immune responses and the establishment of bacterial infections (Telford et al., 2006, Hendrickx et al., 2011). Pilus assembly is catalyzed by pilus-specific sortases that cleave the C-terminal sorting signals of pilin precursors (Ton-That & Schneewind, 2003). Sortases generate acyl-enzyme intermediates (Ton-That et al., 1999), which are relieved by the nucleophilic attack of the ε-amino group of lysine (K) in the YPKN pilin motif, thereby forming intermolecular isopeptide bonds between the major pilin subunits (Budzik et al., 2008a). Immuno-electron microscopy techniques identified major and minor pilin subunits, which contribute specific functions during fimbrial assembly or the pathogenesis of infectious diseases (Ton-That & Schneewind, 2003, Hendrickx et al., 2011).
Some Gram-positive bacteria assemble pili from one major and one minor pilin subunit (Budzik et al., 2007). In these species, the minor subunit is deposited at the fimbrial tip, while the major pilins represent both their shaft and cell wall anchor subunits (Budzik et al., 2007). This distribution of pilins is accomplished through a unique property of their sorting signals. For example, the sorting signal of BcpB, the minor subunit of Bacillus cereus pili, is cleaved by pilus-specific sortase (SrtD) (Budzik et al., 2009a). The resulting acyl-intermediate is relieved by the nucleophilic attack of the major subunit, BcpA (Budzik et al., 2009a). Similarly, the sorting signal of BcpA is cleaved by pilus-specific sortase and polymerized with other BcpA subunits (Budzik et al., 2008a). However, the sorting signal of BcpA may also be cut by sortase A (Budzik et al., 2008b). Acyl-intermediates of sortase A are relieved by the nucleophilic attack of the amino group of m-diaminopimelic acid within the peptidoglycan cell wall peptides of B. cereus (Budzik et al., 2008b). Sortase A generates a covalent bond linking assembled pili to the cell wall envelope (Budzik et al., 2008b). Bacteria that assemble pili from three different pilins accomplish this with a similar reaction sequence (Oh et al., 2008). The minor (tip) and major pilins are cleaved by pilus-specific sortase (Ton-That & Schneewind, 2003). A unique feature of these pili is a minor pilin that is located at the base of the structure and functions as the cell wall anchoring unit (Mandlik et al., 2008a). This minor pilin contributes an ε-amino group of lysine to the formation of an intermolecular isopeptide bond with the major pilin subunit (Mandlik et al., 2008a). Its sorting signal is cleaved by sortase A to enable the anchoring of pili to the cell wall envelope (Swaminathan et al., 2007, Mandlik et al., 2008b).
CnaB domains with reverse immunoglobulin-like (Ig) domains and intra-molecular isopeptide bonds are key structural elements of pilin proteins (Kang et al., 2007). Intramolecular isopeptide bonds, first identified in the capsid protein subunits of bacteriophage HK97 (Duda et al., 1995, Duda, 1998), are formed between lysine-asparagine residues by a mechanism requiring the catalytic property of the side chain carboxyl group of either glutamyl or aspartyl (Kang et al., 2007). Although the intramolecular isopeptide bonds of CnaB domains in minor and major pilins are not required for sortase-mediated polymerization, assembled pili lacking these bonds are quickly degraded by extracellular proteases (Budzik et al., 2009b). Pilin proteins harbor additional structural elements. For example, RrgA, the adhesin of pneumococcal pili (Nelson et al., 2007), consists of four domains (Izore et al., 2010). The structure of one of these, D3, resembles the structure of the A3 domain of von Willebrand factor, a hemostasis factor that mediates association between platelets and collagen (Izore et al., 2010). The major pilin of S. pneumoniae, RrgB, is composed of four domains, designated D1–D4 (Spraggon et al., 2010, Paterson & Baker, 2011, El Mortaji et al., 2012). Each of these domains assembles into a β-sandwich structure with intramolecular isopeptide bonds (El Mortaji et al., 2012). Fitting of the RrgB fragment crystal structure into the overall structure of assembled RrgB pili, as revealed by high resolution transmission electron microscopy, suggests that the pilin subunits are stacked head to tail during sortase-mediated assembly (Hilleringmann et al., 2009, Spraggon et al., 2010).
The major pilin of B. cereus is also endowed with four structural units: CNA1, CNA2, XNA, and CNA3 (Budzik et al., 2009b). Within fully assembled pili, each of these domains harbors an intramolecular isopeptide bond (Budzik et al., 2009b). Recombinant BcpA forms the isopeptide bonds within CNA2, XNA, and CNA3, but not the isopeptide bond of CNA1. As a result, the CNA1 domain of the recombinant pilin can be degraded by proteases similar to the first CnaB domain of RrgB (Budzik et al., 2009b, Kang et al., 2011). Negative staining transmission electron microscopy (TEM) of S. pneumoniae TIGR4 pili (comprised of RrgB) revealed a beaded structure of potentially overlapping stacked RrgB subunits within the pilus (Hilleringmann et al., 2009). Detailed information regarding the native structure of other Gram-positive pili is still lacking. Here we report that B. cereus employs sortase polymerization of BcpA to generate pilus bundles and characterized the determinants that are required for bundle formation and pilus structure.
Results
Bacillus cereus pili assemble into bundles
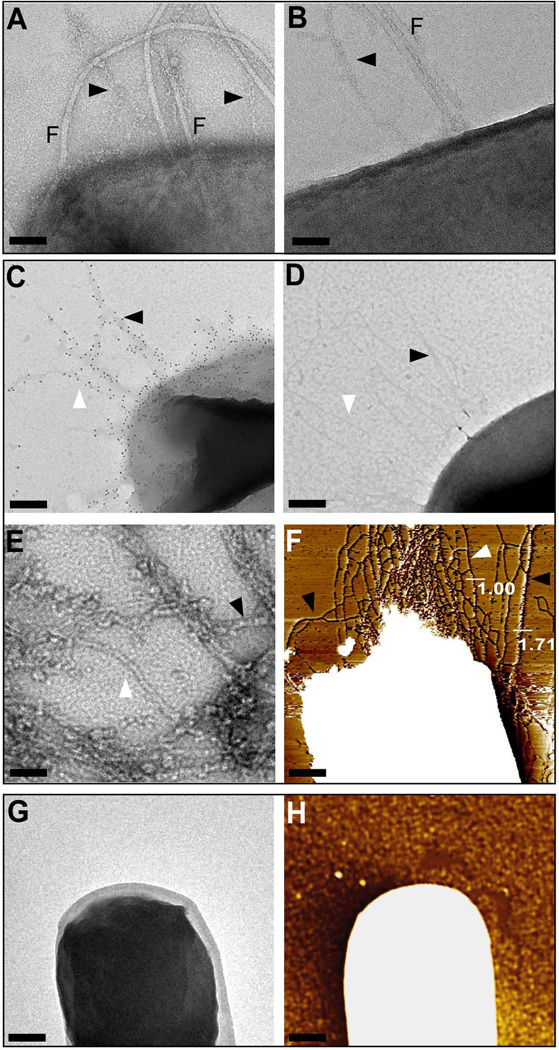
Negative-staining transmission electron microscopy (TEM) of B. cereus ATCC 14579 cells revealed bacilli that produce pili with varying diameter (Fig. 1A, B). In addition to pili, 10 to 15 flagella were present on the surface of each bacterial cell. Compared to pili, flagella are more electron dense and larger in diameter. To analyze pilus bundles without flagella, genes for pilus assembly were expressed in Bacillus anthracis Sterne (pJB12) via the IPTG-inducible expression of the bcpA-srtD-bcpB operon (Budzik et al., 2007). TEM and immunogold labelling with rabbit anti-BcpA and Goat anti-rabbit IgG 10 nm gold conjugate revealed pili on the surface of B. anthracis (pJB12); as observed with B. cereus, immunogold-labeled pili of B. anthracis (pJB12) varied in diameter (Fig. 1C). As a control, incubation of B. anthracis Sterne (pJB12) with 10 nm gold conjugate alone did not generate immunogold labeling; negatively stained pili appeared as filaments that extended up to 1.5 µm, forming bundles with variable diameter (Fig. 1D). To examine whether pilus bundles formed when pili are released from bacterial surfaces, pJB12 was transformed into sortase A mutant B. anthracis (srtA) (Gaspar et al., 2005). Following IPTG-induction of cultured bacilli, bacteria were sedimented by centrifugation and purified pili from the culture medium were analyzed by TEM. These experiments revealed wild-type BcpA pili with variable diameter (Fig. 1E). This result suggests that the release of pili from the bacterial surface does not prevent their association into bundles with variable diameter. To address whether bundle formation of pili occurred in the absence of specific antibody and to confirm TEM results, B. anthracis Sterne (pJB12) cells were analyzed by atomic force microscopy (AFM). Figure 1F shows a representative image of pilus bundles with variable diameter on the surface of bacilli. Height measurements along multiple pili revealed filaments with uniform diameter as well as assemblies of pili with larger diameters. As negative control, B. anthracis cells containing empty vector (pLM5) and analyzed by TEM and AFM did not produce pili (Fig. 1G, H). From this we infer that BcpA pili associate to form filamentous bundles on the bacterial surface (Fig. 1D, F).
Figure 1.
Assembly of pilus bundles on the surface of Bacilli. (A + B) Transmission electron micrographs of negatively stained wild-type B. cereus ATCC 14579. In both panels, B. cereus cells display surface exposed flagella (F) and pili (arrows) that associate as bundles. Bar = 50 nm. (C) Immuno transmission electron micrograph (TEM) of B. anthracis Sterne (pJB12). Pili were labeled using rabbit anti-BcpA antibodies followed by goat anti-rabbit IgG-10 nm gold conjugate to reveal fibers with variable diameter (bar = 100 nm). (D) B. anthracis Sterne (pJB12) was incubated with goat anti-rabbit IgG-10 nm gold conjugate alone. (E) Negative staining TEM micrograph of pili released B. anthracis Sterne srtA (pJB12) cultures. Single pili (white arrowhead), two-filament bundles (black arrowhead) pili, and higher order bundle assemblies were observed (bar = 20 nm). (F) Flattened atomic force microscopy (AFM) image of B. anthracis Sterne (pJB12) cells that elaborate pili with varying diameters as indicated by height measurements in nanometer (Bar = 100 nm). (G + H) Negative control, B. anthracis Sterne (pLM5) cells analyzed by negative staining TEM and AFM, respectively, did not display pili or pilus bundles at the surface of the cell (Bar = 100 nm).
Isopeptide bonds required for the bundle formation of BcpA pili
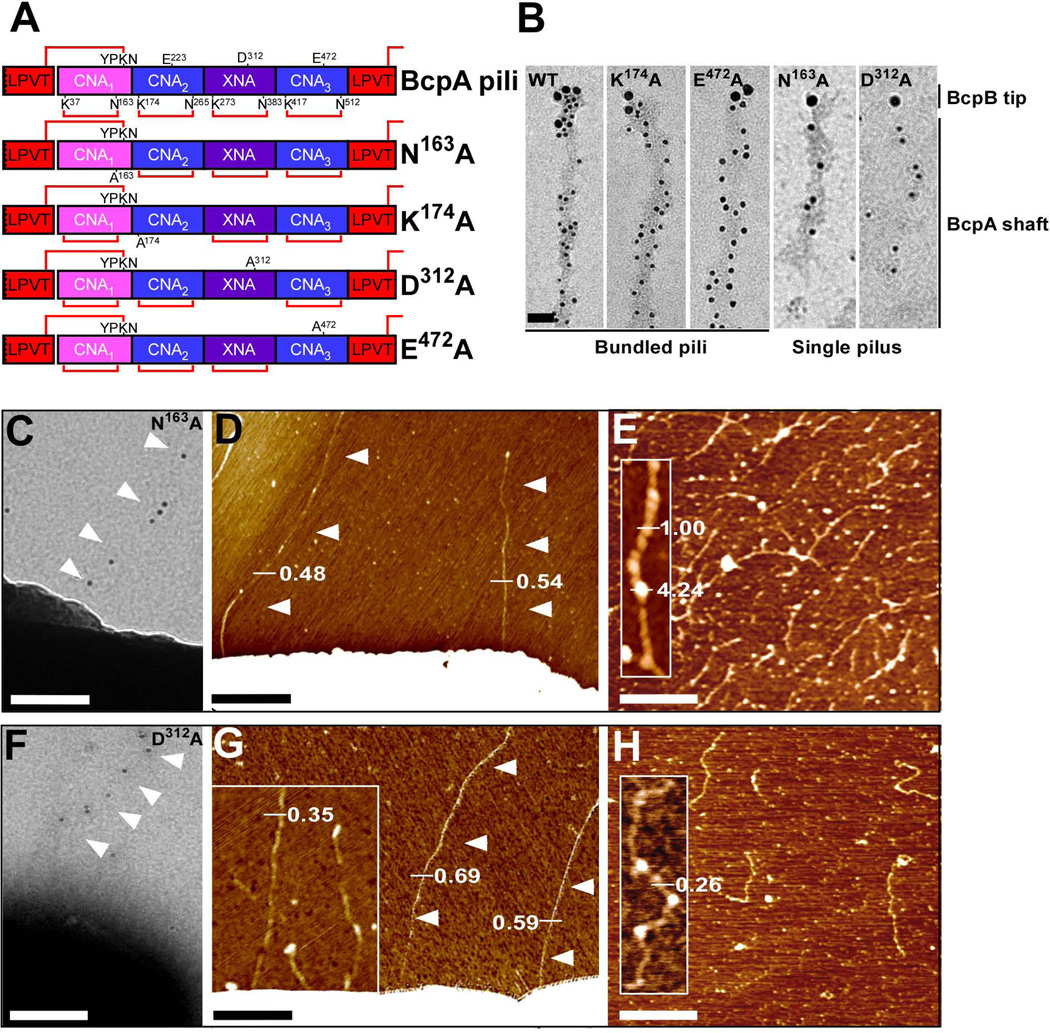
Within assembled pili, each subunit of the major pilin, BcpA, acquires four intramolecular isopeptide bonds, one in each of its four domains: CNA1, CNA2, XNA and CNA3 (Budzik et al., 2009b). Previous work generated plasmid encoded bcpA variants with single amino acid substitutions that abrogate the formation of the corresponding intramolecular isopeptide bond: N163A on pJB109 (CNA1), K174A on pJB61 (CNA2), D312A on pJB236 (XNA), and E472A on pJB107 (CNA3) (Fig. 2A). To analyze whether these internal isopeptide bonds contribute to the formation of pilus bundles and influence overall pilus morphology, plasmids pJB109, pJB61, pJB236 and pJB107 were transformed into B. anthracis Sterne or its srtA variant. Immuno TEM of pili released by srtA variants revealed that the BcpA K174A and E472A mutants assembled pilus bundles with BcpB at the tip, similar to wild-type BcpA pili (Fig. 2B). In contrast, N163A and D312A mutant pili appeared as thin, single-pilus filaments with BcpB at their tip (Fig. 2B). Similar results were obtained when pili were analyzed on the surface of B. anthracis (Fig. 2C, F as compared to Fig. 1C), suggesting that these isopeptide bonds within the major pilin BcpA may contribute to overall pilus morphology and bundle formation. As observed for wild-type BcpA, the K174A and E472A mutants continued to assemble bundled pili with BcpB at the tip (data not shown). AFM on whole Bacilli that produce either N163A or D312A pili demonstrated the presence of single pili not exceeding a pilus diameter of 1.0 nm. AFM analysis of these mutant N163A and D312A pili released and purified from B. anthracis Sterne ∆srtA revealed single pili with both unique morphology as compared to wild-type pili (Fig. 2E, H). The N163A mutant pili did not associate into bundles and harbored bulbous deformations in the CNA1 domain of their pilus structure. D312A pili also did not form bundles, likely due to instability of the XNA domain, which is described in greater detail below.
Figure 2.
Contribution of intramolecular isopeptide bonds towards pilus bundle formation. (A) Schematic representation of wild-type and mutant (N163A, K174A, D312A, and E472A) BcpA pilin that lack the CNA1, CNA2, XNA, or CNA3 intramolecular isopeptide bonds, respectively. The lysine (K) and asparagine (N) residues involved in intramolecular isopeptide bond formation (red lines) are identified. The glutamic acid (E) and aspartic acid (D) residues implicated in catalysis of the intramolecular isopeptide bonds are also identified. (B) Immuno TEM micrographs of wild-type, N163A, K174A, D312A, and E472A pili released and purified from B. anthracis Sterne srtA variants harboring pJB12, pJB109, pJB61, pJB236, or pJB107, respectively. Pili were labeled with anti-BcpB antibodies and anti-IgG-15 nm gold conjugate, fixed, and subsequently labeled with anti-BcpA and anti-IgG-10 nm gold conjugate (bar = 50 nm). (C + F) Immuno TEM micrographs in which mutant N163A and D312A pili were labeled with anti-BcpA serum followed by goat anti-rabbit IgG-10 nm gold conjugate and revealed thin pili on the surface of B. anthracis Sterne (pJB109 and pJB236) cells (bar = 100 nm). (D + G) AFM analysis of B. anthracis Sterne (pJB109 and pJB236) cells producing N163A and D312A pili revealed only single pili as indicated by height measurements in nanometer. (E + H) AFM analysis on N163A and D312A pili released and purified from B. anthracis Sterne srtA (pJB109 and pJB236) showed single filaments with unique structure. Insets, close-up of single filaments.
Pili formed by BcpA K174A and E472A mutants resemble wild-type pili
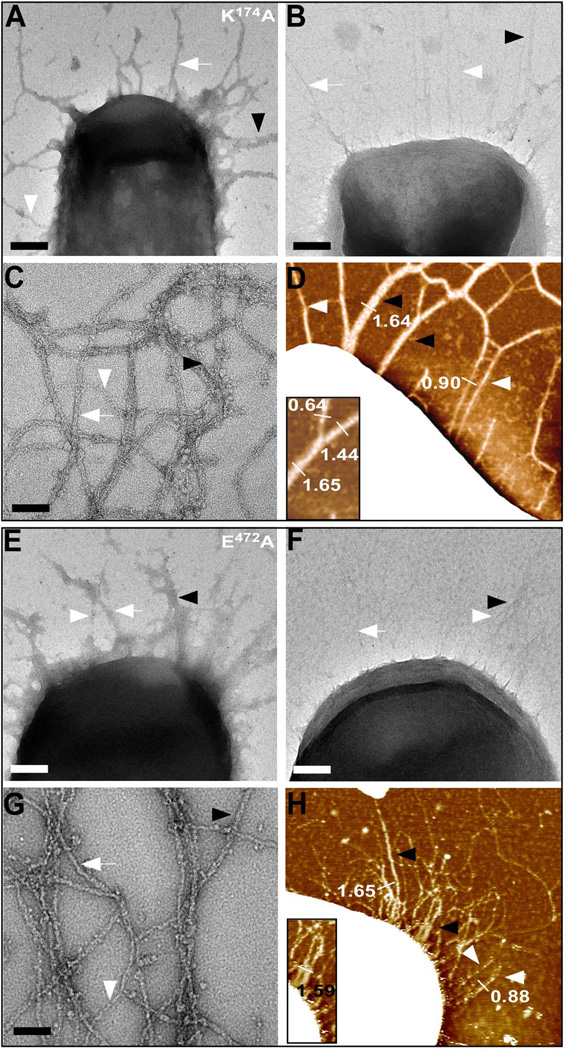
AFM and TEM were performed on B. anthracis Sterne expressing K174A or E472A bcpA mutants. Immuno TEM with anti-BcpA and Goat anti-rabbit 10 nm gold conjugate revealed pili with variable diameter (Fig. 3A, E). Addition of 10 nm gold conjugate without primary antibodies also identified pili with variable diameter (Fig. 3B, F). Negative staining TEM was performed on K174A or E472A pili released by B. anthracis srtA cells, confirming the formation of single, double, and higher order pilus filament assemblies (Fig 4C, G). Similar results were obtained with AFM (Fig. 3D, H). Height measurements along filaments confirmed the presence of pili with varying diameter and comparable overall morphology. Thus, the intramolecular isopeptide bonds of CNA2 and CNA3 are dispensable for pilus bundle formation and structure.
Figure 3.
Structure of BcpA pili lacking the CNA2 and CNA3 isopeptide bonds. (A, E) Immuno TEM of B. anthracis Sterne (pJB61 or pJB107) in which K174A and E472A pili are labeled using rabbit anti-BcpA followed by goat anti-rabbit IgG-10 nm gold conjugate. (B, F) Similar as panels (A, E), however cells were incubated with goat anti-rabbit IgG-10 nm gold conjugate alone (bar = 100 nm). (C+G) High resolution negative-staining TEM micrographs of pili released and purified from B. anthracis Sterne srtA (pJB61 or pJB107). Single pili, double pili, and higher order assemblies of pili are observed (bar = 50 nm). (D+H) Flattened AFM images of B. anthracis Sterne (pJB61 or pJB107) elaborating either K174A or E472A pili on their surface with varying diameters (bar = 100 nm). Insets, close-ups of double filaments.
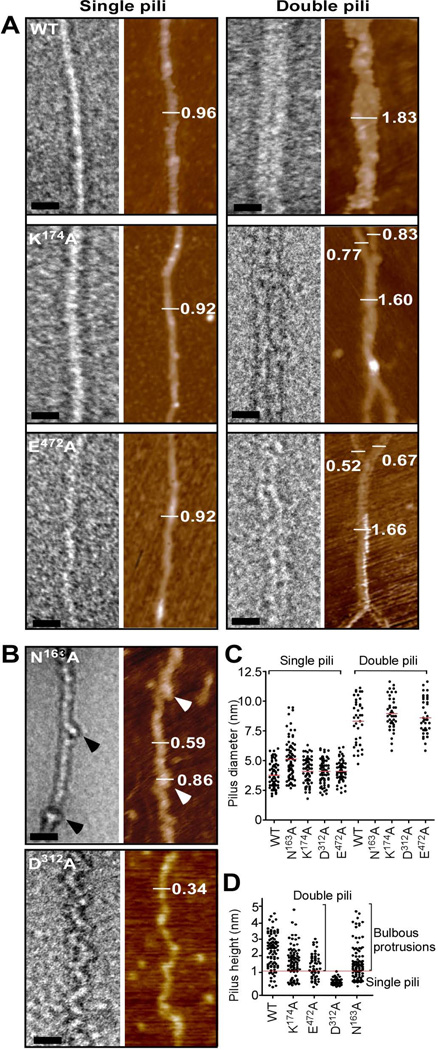
Figure 4.
Electron microscopy and atomic force microscopy of wild-type and mutant pili. (A) A combination of TEM and AFM analysis of wild-type BcpA (WT), K174A, and E472A pili. TEM images were obtained of single and double filament pili negatively stained with 2% uranyl acetate (bar = 20 nm) and by AFM (mocha color). Single pili assembled from wild-type (WT), K174A and E472A mutant BcpA assume a straight morphology, while double filaments represent a close association of two pili that may intertwine. The visualization of single and double filament pili by TEM was confirmed by AFM as indicated by height measurements in nanometer. (B) Pili assembled from N163A variants harbor bulbous protrusions (arrowheads) in regular intervals along an otherwise straight filament. Mutant D312A pili appear as filaments with zig-zag or undulating deformations. Both pilus variants were only observed as single filaments. (C) Quantification of pilus diameter from TEM micrographs of pilus extracts of at least 2 independent experiments. Each dot represents a measurement of the diameter of one pilus. Multiple measurements were taken along >20 pili. (D) Quantification of pilus height on flattened AFM images of pilus extracts of at least 2 independent experiments. Each dot represents a height measurement of one pilus and multiple measurements were taken along >20 pili.
AFM and TEM analysis of wild-type and mutant BcpA pili
Wild-type or mutant BcpA pili that had been released by B. anthracis srtA mutants were analyzed by TEM and AFM to gain insights into pilus morphology. Both techniques were used to exclude potential differences in pilus morphology due to variation in sample preparation (Fig. 4). Analysis of >20 filaments of wild-type, K174A and E472A mutant pili revealed comparable morphology; pili appeared as rigid straight filaments with either single, double or higher order assemblies (higher order assemblies are not shown). Pili derived from wild-type, K174A or E472A variants formed filaments that assembled into double pilus assemblies (Fig. 4A). The overall morphology of single B. cereus pili assumes a molecular thread, while double filaments occur as a close association of two that may intertwine. In contrast, N163A and D312A mutant pili were only detected as single filaments with irregularities in their structure (Fig. 4B). The N163A substitution in the CNA1 domain caused bulbous protrusions in intervals along the otherwise straight filaments of mutant BcpA pili. Further, the N163A substitution also abrogated the formation of pilus bundles. Pili harboring the D312A substitution do not form straight filaments and instead assume an undulating structure. D312A pili do not associate as filament bundles, as occurs for wild-type, K174A or E472A pili (Fig. 4A). Individual BcpA monomers could be distinguished within D312A pili.
To quantify pilus bundling, diameters of pili were determined from multiple measurements along pilus shafts of single filaments and double filaments from TEM micrographs (Fig. 4C). The average diameter of wild-type, N163A, K174A, D312A, and E472A pili was 3.8, 5.1, 4.1, 4.1 and 4.1 nm, respectively. The diameter of N163A pili is slightly larger, likely due to the observed bulbous protrusions along the filaments. The average diameter of double filament bundles was 8.4, 9.0 and 8.6 nm for wild-type BcpA pili, K174A and E472A pili, respectively. Double filament bundles were not detected for N163A and D312A mutant BcpA pili. Of note, the diameter of wild-type pili (3.8 nm) measured by electron microscopy was slightly larger than the observed width of BcpA in the rBcpA* crystal structure (3 nm, PDB code = 3PKT) (Budzik et al., 2009b). The observed increase in diameter may be due to the uranyl acetate staining of pilus filaments. In addition, multiple AFM height measurements were performed along >20 pili. Quantification of pilus height on flattened AFM scans confirmed that wild-type, K174A and E472A mutant pili occur as single, double, and higher order filaments (Fig. 4D). The average pilus height of single filaments of wild-type, K174A and E472A mutant pili was 0.68, 0.54 and 0.67 nm, respectively. Thus, the structure of K174A and E472A pilus filaments was not significantly altered as compared to wild-type pili. Mutant N163A pili had an increased pilus height owing to the structural irregularities. Filaments derived from D312A mutant BcpA displayed an average height of 0.45 nm; these pili appeared only as single 'collapsed' filaments.
X-ray crystallography and characterization of rBcpA* (D312A)
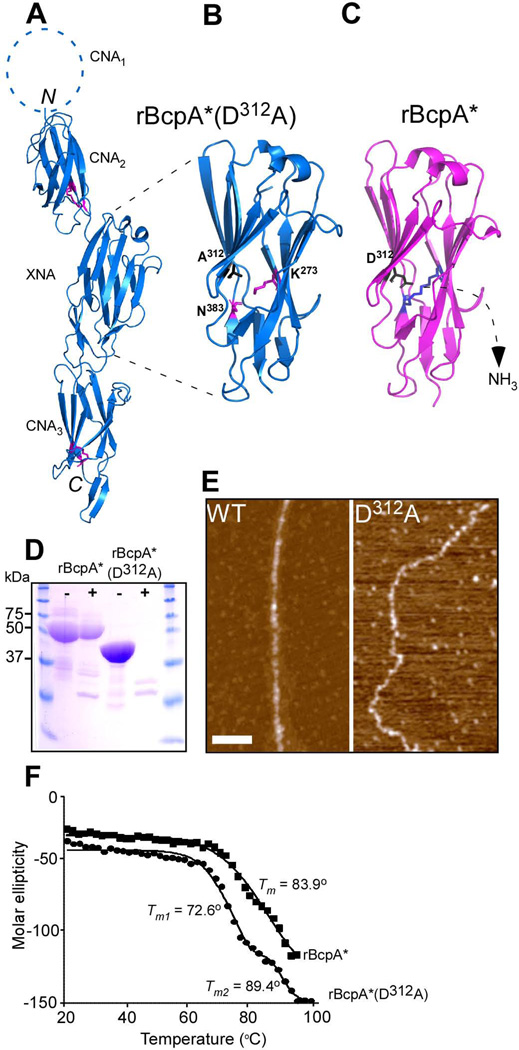
We sought to examine whether the D312A substitution abolishes intramolecular isopeptide bond formation and alters the jelly-roll (CnaA) fold of the XNA domain within BcpA. Therefore, GST-BcpA* (D312A), i.e. a fusion of glutathione S-transferase (GST) to truncated BcpA encompassing the CNA2-XNA-CNA3 domains (and lacking the CNA1 domain), was purified by affinity chromatography, GST cleaved off, and rBcpA* (D312A) purified by size exclusion chromatography. The protein was crystallized and its structure solved by X-ray crystallography and molecular replacement in the space group P21 to a resolution of 2.24Å (PDB code = 3RKP). Data collection and refinement statistics are represented in Table 1. Comparison of the rBcpA* (D312A) structure with that of the previously determined rBcpA* structure revealed a high degree of similarity (Fig. 5A and Fig. S1). The three domains of rBcpA* (D312A) and rBcpA* adopt β-sandwich structures and their CNA2 and CNA3 domains assume a reverse Ig-fold containing K174-N265 and K417-N512 isopeptide bonds. The XNA domain in rBcpA* (D312A) and rBcpA* adopts a jelly-roll fold. In rBcpA*, the isopeptide bond between K273-N383 covalently joins the first and the penultimate β-strands within XNA, which are oriented in an antiparallel manner. This isopeptide bond is absent in the rBcpA*(D312A) variant, as demonstrated by a discontinuity of the electron density between the K273 and N383 residues (Fig. 5B). The K273-N383 isopeptide bond is present in the rBcpA* structure (Fig. 5C), which is formed after release of a NH3 molecule.
Table 1.
Data Collection and Refinement Statistics
| Data Collection and Crystal Parametersa | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 68.9, 68.1, 97.3 |
| α, β, γ (°) | 90, 107.7, 90 |
| Wavelength (Å) | 1.0332 |
| Resolution (Å) | 50–2.24 (2.28–2.24)b |
| Rmerge (%)c | 6.5 (30.5) |
| I/σI | 17.0 (2.7) |
| Multiplicity | 3.5 (2.5) |
| Refinement | |
| Resolution (Å) | 38.8–2.24 (2.30–2.24) |
| Rwork/Rfree (%)d | 21.0/25.5 (31.1/42.3) |
| Rmsd bond lengths (Å) | 0.002 |
| Rmsd bond angles (°) | 0.579 |
PDB ID= 3RKP
Values for the highest resolution shell are shown in parentheses.
Rmerge= Σ|Ihkl - <Ihkl>|/ ΣIhkl, where I is the observed intensity for reflection hkl, and <I> is the mean intensity.
Rwork= Σ||Fo(hkl)|-|Fc(hkl)||/Σ|Fo(hkl)|; Rfree is calculated in the same way with 5% of reflections excluded from refinement.
Figure 5.
Crystal structure of rBcpA*(D312A). (A) The crystal structure of rBcpA*(D312A) was solved using molecular replacement. The CNA2, XNA, and CNA3 domains are shown as blue ribbons, the structure of the CNA1 domain is not included in rBcpA*(D312A). The Lys and Asn residues forming the intramolecular isopeptide bonds in the CNA2 and CNA3 domains are shown as black sticks. The intramolecular isopeptide bond in the XNA domain is not formed. The CNA2 and CNA3 domains assume reverse Ig-like folds and the XNA domain a jellyroll fold. (B+C) Close up of the XNA domain of rBcpA*(D312A) (blue) and rBcpA* (magenta), in which the A312 and the catalytic residue D312 are in black and the K273 and N383 residues in magenta and blue and presented as sticks, respectively. In rBcpA*(D312A), the K273-N383 intramolecular isopeptide bond between the two penultimate β-strands is not formed, as the catalyst D312 was replaced with alanine. Formation of the XNA isopeptide bond of rBcpA* (blue sticks) is accompanied by the loss of ammonium. (D) Treatment of rBcpA* and rBcpA*(D312A) with (+) and without (-) trypsin protease revealed that the absence of the XNA isopeptide bond in rBcpA*(D312A) renders the protein sensitive to protease; rBcpA* is protease resistant. (E) AFM images of wild-type (WT) and mutant D312A pili showing the morphological difference between these two types of pili. (F) Thermal melt spectroscopy analysis as a function of temperature demonstrated that rBcpA*(D312A) is less stable than rBcpA*.
In addition to the absence of the XNA isopeptide bond, we further characterized rBcpA*(D312A) in comparison with rBcpA*, to explain the zig-zag-like phenotype of D312A pili (Fig. 5E). The rBcpA* and rBcpA*(D312A) proteins were subjected to protease treatment, which revealed that the D312A mutation renders the recombinant protein protease sensitive as it is was degraded, while rBcpA* seemed to be unaffected by trypsin digestion (Fig. 5D). Notably, the rBcpA*(D312A) protein migrated faster through reducing SDS-PAGE. Thermal melt spectroscopy analysis demonstrated that the rBcpA*(D312A) protein is thermally less stable than the rBcpA* protein (Fig. 5F). The rBcpA* protein denatured by a two-state unfolding mechanism with a transition temperature (Tm) of 83.9°C without a stable intermediate. In contrast, the mutant rBcpA*(D312A) protein followed two transitions; the first between ~60°C and ~80°C (Tm1 of 72.6°C) and the second between ~80°C and 98°C (Tm2 of 89.4 °C). The rBcpA*(D312A) protein denatures 11.3 °C sooner than the rBcpA* protein. Taken together, these data show that the XNA isopeptide bond provides for protease resistance, thermal stability and determines pilus integrity.
Discussion
Pili of Gram-positive bacteria are surface-associated macromolecular assemblies of covalently linked major pilin subunits, forming a pilus shaft, and a minor pilin positioned at the fiber tip or at the pilus base (Ton-That & Schneewind, 2003). Sortase-assembled pili have been described in important human pathogens such as Actinomyces naeslundii (Mishra et al., 2007), B. cereus (Budzik et al., 2007), Corynebacterium diphtheriae (Ton-That & Schneewind, 2003), Enterococcus faecalis (Nallapareddy et al., 2006), Enterococcus faecium (Hendrickx et al., 2008), Streptococcus agalactiae (Lauer et al., 2005), Streptococcus pneumoniae (Barocchi et al., 2006) and Streptococcus pyogenes (Mora et al., 2005) or non-pathogenic microbes such as Lactobacillus rhamnosus (Kankainen et al., 2009). For several of these bacteria it has been shown that pili enable adherence, which is thought to be a key step for colonization of the host and the pathogenesis of bacterial infections (Mandlik et al., 2007, Edwards et al., 2008, Hendrickx et al., 2011).
Earlier work used TEM and AFM to demonstrate that pili of S. pneumoniae wrap around one another and associate into web-like structures (Falker et al., 2008). High resolution TEM of S. pneumoniae pili revealed that that the major pilin, RrgB, is stacked head-to-tail along the shaft of the filament, while the minor pilins, RrgA and RrgC, decorate the tip and base of the pilus, respectively (Hilleringmann et al., 2009). TEM and single particle image reconstruction revealed the three-dimensional structure of two neighboring RrgB subunits within the assembled S. pneumoniae pilus, which could accommodate the crystal structure derived from the recombinant D2-D4 domains of RrgB (Spraggon et al., 2010). Full length RrgB is comprised of four domains, D1-D4; X-ray crystallography of recombinant full-length RrgB revealed that the D2-D4 domains each assume a CnaB-type fold stabilized by Asn-Lys isopeptide bonds with catalytic glutamic acid residues (Paterson & Baker, 2011). The D1 domain of recombinant full length RrgB carrying the C-terminal IPQTG motif (for sortase cleavage and pilus assembly) harbors also an intramolecular isopeptide bond (El Mortaji et al., 2012). Even without sortase-mediated assembly, full length recombinant RrgB associates during crystallization head-to-tail with other RrgB molecules by docking the IPQTG motif from one subunit into the D1 domain of another (El Mortaji et al., 2012). Truncated RrgB, lacking the C-terminal sorting signal, is however unstable: its D1 domain does not form the intramolecular isopeptide bond and remains sensitive to protease degradation (Paterson & Baker, 2011). The D1 domain encompasses the YPKN pilin motif (Ton-That & Schneewind, 2003), whose lysine (K) residue is essential for sortase-mediated pilus polymerization and located in direct proximity to the asparagine residue (N) involved in the CNA1 intramolecular isopeptide bond of B. cereus BcpA pili (Budzik et al., 2008a). Of note, it is not yet known whether the intramolecular isopeptide bonds of RrgB (D1-D4) are also assembled in S. pneumoniae pili (Paterson & Baker, 2011, El Mortaji et al., 2012). Considering the similarity between the D1 domains of S. pneumoniae RrgB and B. cereus BcpA, it seems likely that D1 domains form their intramolecular isopeptide bonds in vivo only after the sortase-mediated assembly of the intermolecular isopeptide bond has been completed (Budzik et al., 2008a, Budzik et al., 2009b).
Here we used AFM and TEM to examine B. cereus pili, which were found to be associated into bundles of two or more filaments of polymerized BcpA with BcpB at the tip. Bundle formation of B. cereus pili appears to be distinct from the web-like pattern of RrgB pili on the surface of S. pneumoniae (Falker et al., 2008). RrgB pili are characterized by somewhat irregular associations and disassociations between neighboring filaments (Falker et al., 2008). In contrast, B. cereus pili associate into bundles of neighboring filaments that often span the entire length of these fiber structures, which may occur through electrostatic interactions between pili after simultaneous polymerization in close proximity. Previous work determined that BcpA is comprised of four domains, CNA1-CNA2-XNA-CNA3, with CnaB- (CNA1–3) or CnaA-type (XNA) Ig-like folds (Budzik et al., 2009b). Three of these domains (CNA2-XNA-CNA3) generate intramolecular (Asn-Lys) isopeptide bonds to stabilize their Ig-like fold even in non-polymerized BcpA; in contrast, the CNA1 domain assembles its intramolecular amide bond only after BcpA subunits have been polymerized into filaments by pilus-specific sortase (SrtD). Mutants unable to generate any one of these intramolecular isopeptide bonds were analyzed for pilus bundle formation and their contribution to pilus morphology. The CNA2 and CNA3 variants formed pilus bundles that were indistinguishable from wild-type pili, albeit that the variant filaments display increased sensitivity to extracellular proteases (Budzik et al., 2009b). CNA1 variant BcpA pili display a deformity of the pilin subunit structure that also prevented pilus bundle formation. Another variant (D312A), which failed to generate the intramolecular isopeptide bond within XNA, was also defective in pilus bundle formation. Within assembled pili, the D312A variant did not produce the bulbous protrusions of the CNA1 mutant, however the mutation caused filaments to assume undulating deformations throughout their entire length instead of the straight fiber structure observed for wild-type pili. When analyzed by X-ray crystallography, the D312A variant assumed the same jelly-roll fold as wild-type BcpA, however, it lacks the XNA isopeptide bond. This bond appears to be critical for protease resistance, thermal stability, pilus integrity and bundling.
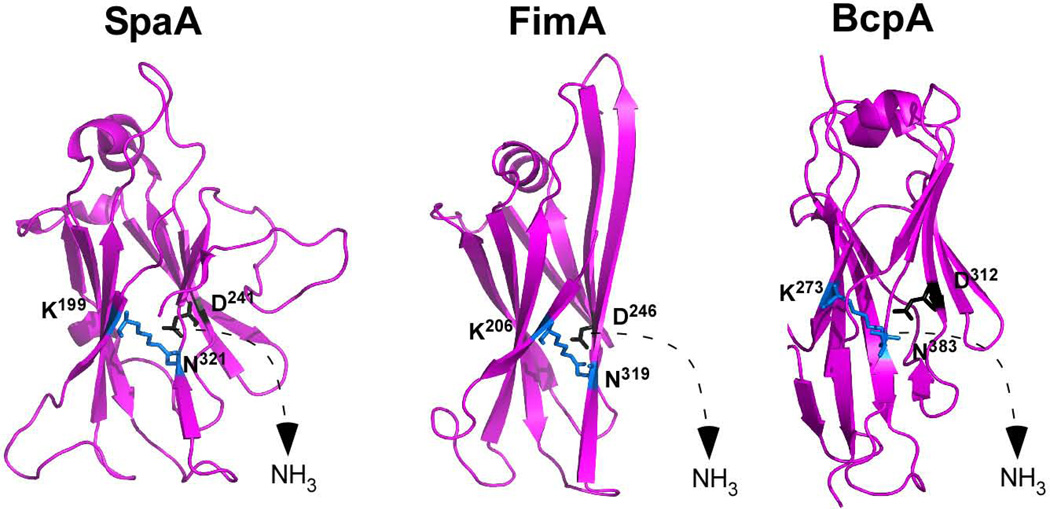
We entertained the possibility that the XNA domain of BcpA represents a common feature of pilin proteins that promote bundle formation. Analysis of the structure of the major pilins SpaA of C. diphtheriae (Kang et al., 2009) as well as FimA of A. naeslundii (Mishra et al., 2011) revealed jelly-roll structure domains (CnaA-type Ig-like fold) with intramolecular Asn-Lys isopeptide bonds (Fig. 6). Of note, in each of the three pilin proteins (BcpA, FimA and SpaA), an aspartyl residue, not glutamyl, is positioned for autocatalytic isopeptide bond formation (Fig. 6). Future work will need to determine whether C. diphtheriae and A. naeslundii pili form bundles in a manner requiring their jelly-roll domains and autocatalytic aspartyl residues.
Figure 6.
Pilin proteins harboring jelly-roll domains and autocatalytic aspartyl residues. Ribbon model for the X-ray crystallographic structures of three major pilin proteins from three Gram-positive microbes - Corynebacterium diphtheriae (SpaA), Actinomyces naeslundii (FimA) and Bacillus cereus (BcpA) - that harbor jelly-roll domains with intramolecular asparagines-lysine (K-N) isopeptide bonds requiring aspartyl (D) residues for autocatalysis.
The formation of pilus bundles is a general phenomenon that has been described for Gram-negative bacteria, for example the type IV pili (Tfp) of Neisseria gonorrhoeae (Swanson, 1973) and the bundle-forming pilus (BFP) of pathogenic Escherichia coli (Donnenberg et al., 1992). In these organisms, bundled pili were characterized as virulence factors that promote bacterial attachment to epithelial cells, biofilm formation, and twitching motility (Biais et al., 2008, Winther-Larsen et al., 2001, Bieber et al., 1998). Whether such varied functions apply also for Gram-positive bacterial pili remains to be elucidated.
Materials and Methods
Bacterial strains, culture conditions, and plasmids
B. cereus ATCC 14579 was propagated on Luria Bertani (LB) plates without antibiotics. B. anthracis Sterne and B. anthracis Sterne srtA::ermC (harboring pJB12, pJB61, pJB107, pJB109 or pJB236) and E. coli BL21(DE3) (harboring pAH16) strains were grown in LB broth or on LB agar plates at 30°C and 37°C, respectively. Kanamycin was added at a final concentration of 50 µg ml−1 for E. coli or 20 µg ml−1 for B. anthracis plasmid maintenance. Isopropyl-β-D-thio-galactoside (IPTG) was added at a final concentration of 1.0 mM for induction. The pJB12 plasmid is a pLM5 derivative containing bcpA-srtD-bcpB under control of the Pspac promoter for expression of wild-type BcpA pili of B. cereus in B. anthracis Sterne (Budzik et al., 2007). Plasmid pJB61, pJB107, pJB109 and pJB236 are pJB12 derivatives, containing D174A, E472A, N163A and D312A substitutions that abrogate the formation of intramolecular isopeptide bonds in the CNA2, CNA3 and XNA domains, respectively (Budzik et al., 2009b). The pAH16 plasmid is a pGEX2TK derivative (Novagene) for expression of GST-tagged recombinant BcpA*(D312A) (consisting of CNA2-XNA-CNA3 lacking the N-terminal signal sequence, the CNA1 domain and C-terminal LPXTG sorting signal, and containing strategically placed methionines and the D312A mutation) under control of an IPTG-inducible T7 promotor. The pAH16 plasmid was generated by quick change mutagenesis using primers 5'-CAAAAAATATGTGGTAACAGCTACTTTGGATAATCGTTTAG-3' and 5'-CTAAACGATTATCCAAAGTAGCTGTTACCACATATTTTTTG-3' with pJB215 plasmid as a template (Budzik et al., 2008a). The PCR reaction was performed with Accuprime Pfx polymerase (Invitrogen). The PCR product treated with DpnI and transformed to E. coli DH5α and BL21(DE3).
Isolation of B. cereus pili
B. anthracis Sterne ∆srtA::ermC harboring plasmid pJB12, pJB61, pJB107 or pJB236 was grown for 16–24 hrs at 30°C on LB agar supplemented with 20 µg/ml kanamycin and 1.0 mM IPTG. In these strains, pili are not linked to the cell wall (Budzik et al., 2007). Bacteria were harvested from agar plates and inoculated into 1.0 ml of MQ H2O to an OD660 of ~1.0, vortexed for 30 seconds, and cells removed by centrifugation at 6,000 ×g, followed by an additional centrifugation step. Pili were either dialyzed against 50 mM Tris-HCl pH 7.5, 150 mM NaCl buffer or ultrapure ddH2O for 16–24 hrs at 4°C in dialysis cassettes (Thermo scientific, pore 20 kDa) prior to TEM or AFM.
Transmission electron microscopy
Purified pili (dialyzed against 50 mM Tris-HCl, pH 7.5, 150 mM NaCl or water) from at least two different isolations were allowed to bind to glow discharged, carbon coated (Edwards Auto 306 Evaporator) copper grids (400 mesh) and were subsequently negatively stained using 2% uranyl acetate (Electron Microscopy Services). Images were recorded using a TeCNAi F30 (Philips/FEI) transmission electron microscope (Field emission gun, 300-kV accelerating voltage, with a magnification of 100,000 to 200,000×) and a high performance CCD camera with a 4k × 4k resolution. Images were acquired using the Gatan DigitalMicrograph software, contrast adjusted and processed using Adobe Photoshop (Adobe, San Jose, CA, USA).
Immuno-electron microscopy was performed as described previously with some modifications (Hendrickx et al., 2008). In brief, nickel grids (mesh Formvar-carbon coated) were incubated for 30 min with carbon side on a drop of 1.109 CFU ml−1 of B. cereus cells to detect surface-exposed pili. Grids were washed 3 times for 5 min on drops of 0.02 M glycine in PBS and subsequently blocked for 30 min on drops of 1% BSA in PBS (PBSb). Pili were labelled for 1 hr on drops of 1:100 diluted specific anti-BcpA rabbit immune sera in PBSb. Grids were washed 4 times for 2 min on drops of 0.1% BSA in PBS. Pilus-antibody complexes were labeled by incubation for 20 min on drops with 1:55 diluted Goat anti-Rabbit IgG-gold label (10 nm) in PBSb. Grids were washed four times for 2 min on drops of PBS, fixed by incubation on drops of 1% glutaraldehyde in PBS, and washed again 8 times for 2 min on drops of H2O. For double labelling experiments, the labeling was repeated similarly with 1:200 diluted anti-BcpB immune serum after fixing with 2% glutaraldehyde in PBS for 10 min, and a 1:55 diluted Goat anti-Rabbit IgG gold label (5 nm). Bacteria were stained by incubation of the grids for 5 min on drops containing 1% uranyl acetate (pH 4) and subsequently air dried for 10 min.
Quantification of pilus diameter
The diameter of pili released and purified from B. anthracis Sterne srtA::ermC was determined from TEM micrographs. Sites along pili were randomly chosen and the number of pixels (diameter) measured using the measuring tool in Adobe Photoshop. The number of pixels were converted to thickness in nanometer based on the number of pixels corresponding to the length of the scale bar (in nm) of the micrograph and plotted in Graphpad Prism 5.0 to perform statistical analysis (Student's t test, unpaired, 2-tailed) on the differences in pilus diameter.
Atomic force microscopy
Fixed pili or bacilli in H2O were spotted onto a freshly cleaved mica disk (Ted Pella, Inc.) for 5 min at room temperature, excess H2O was removed, and the sample was subsequently air dried for 15 minutes. Tapping mode images in air were collected on an Asylum Research MFP-3D-BIO atomic force microscope with Olympus AC240TS cantilevers (~70 kHz resonant frequency, 10 nm nominal tip radius, and 2 N/m spring constant). Images with a resolution of at least 512 × 512 pixels were acquired with typical line scan rates of 0.4 to 1 Hz. Images were processed by flattening and/or plane fitting with the Asylum MFP-3D software to remove scanning artifacts when required.
Quantification of pilus height by atomic force microscopy
AFM samples were prepared as described above and pilus height was determined from multiple height measurements along >20 pili. The line section tool in the Asylum MFP-3D software was used to measure heights along a cross-section of individual pili at randomly chosen sites. The maximum height at the top of the (roughly Gaussian) cross-section minus the height of the nearby surface was used as a measurement for pilus height.
Expression and purification of rBcpA* (D312A) for crystallization
E. coli BL21 (pAH16) was grown in 1.0 L of LB media to OD600 0.8, induced with 1 mM IPTG for 2 hours, and cells harvested. E. coli cells were lysed in a French Pressure cell in 15 ml of lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl). The lysate was clarified by centrifugation at 32,572 ×g for 15 minutes, and GST-rBcpA*(D312A) was purified by two rounds of GST-affinity chromatography. GST-rBcpA*(D312A) was applied to 1.5 ml of GST bead slurry (GE Healthcare) pre-equilibrated with lysis buffer and the beads were washed with 25 ml of lysis buffer. GST-rBcpA*(D312A) was eluted with 20 mM reduced glutathione prepared in lysis buffer, pH 8.0. The eluate was diluted to 50 ml of PBS, reapplied to new GST beads, which were washed with 25 ml of PBS. The GST tag was removed by cleavage with 55 U of thrombin (GE) in PBS at 22 °C for 16 hours with rotation. Flow-through containing rBcpA*(D312A) was concentrated with an Amicon Ultra–15 centrifugal filter device (Millipore) and subjected to gel filtration chromatography (Superose 12 column; GE Health Sciences) using lysis buffer. Gel filtration fractions were pooled and concentrated with an Ultrafree–0.5 centrifugal filter device (Millipore). Concentrated rBcpA* (D312A) was filtered with a Millex syringe filter unit (0.22 µm pore size, Millipore) and purity was analyzed by SDS-PAGE followed by Coomassie staining.
Crystallization and structure determination
Crystal trials were performed at a concentration of 13 mg/ml of rBcpA*(D312A) as determined by OD260 nm measurement. Crystals of rBcpA*(D312A) were grown at room temperature with a reservoir solution of 0.1 M HEPES (pH 7.5) and 20% PEG 10,000 using the hanging drop vapor diffusion method. Crystals were soaked in a cryoprotectant solution composed of the reservoir solution and 15% glycerol before freezing in liquid nitrogen. Data were collected at beamline 23-ID-D (GM/CA-CAT) at the Advanced Photon Source at Argonne National Laboratory. Data were processed using HKL2000 (Otwinowski & Minor, 1997) and phases determined by molecular replacement with Phaser (McCoy et al., 2007) using rBcpA* (PDB ID 3KPT) as the search model. The structure was refined with PHENIX (Adams et al., 2002) and deposited in the RCSB PDB with ID 3RKP. Figures were prepared with PyMOL (DeLano, 2002). Data collection and refinement statistics can be found in Table 1.
Trypsin cleavage of rBcpA
The recombinant rBcpA* and rBcpA*(D312A) proteins were purified as described above and incubated with 2.68 µg of sequencing grade modified trypsin (Promega) overnight at 37°C for 19 hours and subjected to reduced SDS-PAGE analysis.
Thermal melt circular dichroism spectroscopy
Purified recombinant rBcpA* and rBcpA*(D312A) proteins were dialyzed to PBS at 4°C overnight. Circular dichroism signals were measured on a AVIV 202 CD spectrometer employing a 0.1 mm path length quartz cuvette containing 300 µl of 40 µM recombinant protein. The circular dichroism signal at 222 nm was measured as a function of temperature, ranging from 20 to 98°C with 2°C intervals. Reversibility of the protein denaturation was checked visually and by cooling the sample. The data obtained in millidegree units were corrected for background signal and converted to molar ellipticity [⍰]. The melting temperature (Tm) was calculated from the inflection point of the curve after fitting the melting curve using the Boltzmann equation.
Supplementary Material
Acknowledgements
We are grateful to Dr. Robert Josephs, Dr. Jotham Austin and Yimei Chen of the Electron Microscopy facility and Dr. Elena Solomaha of the Biophysics Core Facility of the University of Chicago for experimental assistance. We thank Dr. Khaled Aly for critical reading of this manuscript. This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (NIAID), Infectious Diseases Branch (AI38997). C.H. and O.S. acknowledge membership within and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIH Award 1-U54-AI-057153).
References
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, Dahlberg S, Fernebro J, Moschioni M, Masignani V, Hultenby K, Taddei AR, Beiter K, Wartha F, von Euler A, Covacci A, Holden DW, Normark S, Rappuoli R, Henriques-Normark B. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biais N, Ladoux B, Higashi D, So M, Sheetz M. Cooperative retraction of bundled type IV pili enables nanonewton force generation. PLoS Biol. 2008;6:e87. doi: 10.1371/journal.pbio.0060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- Budzik JM, Marraffini LA, Schneewind O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 2007;66:495–510. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- Budzik JM, Marraffini LA, Souda P, Whitelegge JP, Faull KF, Schneewind O. Amide bonds assemble pili on the surface of bacilli. Proc. Nat. Acad. Sci. USA. 2008a;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Oh SY, Schneewind O. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J. Biol. Chem. 2008b;283:36676–36686. doi: 10.1074/jbc.M806796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Oh SY, Schneewind O. Sortase D forms the covalent bond that links BcpB to the tip of Bacillus cereus pili. J. Biol. Chem. 2009a;284:12989–12997. doi: 10.1074/jbc.M900927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Poor CB, Faull KF, Whitelegge JP, He C, Schneewind O. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc. Natl. Acad. Sci. USA. 2009b;106:19992–19997. doi: 10.1073/pnas.0910887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- Donnenberg MS, Giron JA, Nataro JP, Kaper JB. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- Duda RL. Protein chainmail: catenated protein in viral capsids. Cell. 1998;94:55–60. doi: 10.1016/s0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J. Mol. Biol. 1995;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Edwards AM, Manetti AG, Falugi F, Zingaretti C, Capo S, Buccato S, Bensi G, Telford JL, Margarit I, Grandi G. Scavenger receptor gp340 aggregates group A streptococci by binding pili. Mol. Microbiol. 2008;68:1378–1394. doi: 10.1111/j.1365-2958.2008.06220.x. [DOI] [PubMed] [Google Scholar]
- El Mortaji L, Contreras-Martel C, Moschioni M, Ferlenghi I, Manzano C, Vernet T, Dessen A, Di Guilmi AM. The full length Streptococcus pneumoniae major pilin RrgB crystallizes in a fiber-like structure, which presents the D1 isopeptide bond and provides details on the mechanism of pilus polymerization. Biochem. J. 2012;441:833–841. doi: 10.1042/BJ20111397. [DOI] [PubMed] [Google Scholar]
- Falker S, Nelson AL, Morfeldt E, Jonas K, Hultenby K, Ries J, Melefors O, Normark S, Henriques-Normark B. Sortase-mediated assembly and surface topology of adhesive pneumococcal pili. Mol. Microbiol. 2008;70:595–607. doi: 10.1111/j.1365-2958.2008.06396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar AH, Marraffini LA, Glass EM, DeBord KL, Ton-That H, Schneewind O. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J. Bacteriol. 2005;187:4646–4655. doi: 10.1128/JB.187.13.4646-4655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx AP, Bonten MJ, van Luit-Asbroek M, Schapendonk CM, Kragten AH, Willems RJ. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology. 2008;154:3212–3223. doi: 10.1099/mic.0.2008/020891-0. [DOI] [PubMed] [Google Scholar]
- Hendrickx AP, Budzik JM, Oh SY, Schneewind O. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- Hilleringmann M, Ringler P, Müller SA, De Angelis G, Rappuoli R, Ferlenghi I, Engel A. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 2009;28:3921–3930. doi: 10.1038/emboj.2009.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izore T, Contreras-Martel C, El Mortaji L, Manzano C, Terrasse R, Vernet T, Di Guilmi AM, Dessen A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus penumoniae. Structure. 2010;18:106–115. doi: 10.1016/j.str.2009.10.019. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Proft T, Baker EN. Crystal structure of Spy0129, a Streptococcus pyogenes class B sortase involved in pilus assembly. PLoS One. 2011;6:e15969. doi: 10.1371/journal.pone.0015969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. USA. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hämäläinen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjärvi T, Auvinen P, de Vos WM. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. USA. 2009;106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Genome analysis reveals pili in Group B streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc. Natl. Acad. Sci. USA. 2008a;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008b;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phasercrystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Das A, Cisar JO, Ton-That H. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J. Bacteriol. 2007;189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Devarajan B, Reardon ME, Dwivedi P, Krishnan V, Cisar JO, Das A, Narayana SV, Ton-That H. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol. Microbiol. 2011;81:1205–1220. doi: 10.1111/j.1365-2958.2011.07745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, Maggi T, Taddei AR, Grandi G, Telford JL. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpää J, Garsin DA, Höök M, Erlandsen SL, Murray BE. Endocarditis of biofilm-assoctaed pili of Enterococcus faecalis. J. Clin. Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AL, Ries J, Bagnoli F, Dahlberg S, Fälker S, Rounioja S, Tschöp J, Morfeldt E, Ferlenghi I, Hilleringmann M, Holden D, Rappuoli R, Normark S, Barocchi M, Henriques-Normark B. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 2007;66:329–340. doi: 10.1111/j.1365-2958.2007.05908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S-Y, Budzik JM, Schneewind O. Sortases make pili from three ingredients. Proc. Nat. Acad. Sci. USA. 2008;105:14147–14152. doi: 10.1073/pnas.0807334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paterson NG, Baker EN. Structure of the full-lengt major pilin from Streptococcus pneumoniae : implications for isopeptide bond formation in gram-positive bacterial pili. PLoS One. 2011;6:e22095. doi: 10.1371/journal.pone.0022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraggon G, Koesema E, Scarselli M, Malito E, Biagini M, Norais N, Emolo C, Barocchi MA, Giusti F, Hilleringmann M, Rappuoli R, Lesley S, Covacci A, Masignani V, Ferlenghi I. Supramolecular organization of the repetitive backbone unit of the Streptococcus penumoniae pilus. PLoS One. 2010;5:e10919. doi: 10.1371/journal.pone.0010919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J. Exp. Med. 1973;137:571–589. doi: 10.1084/jem.137.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford JL, Barocchi M, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc. Natl. Acad. Sci. USA. 2001;98:15276–15281. doi: 10.1073/pnas.261574998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa R, Honda E. Presence of pili in species of human and animal parasites and pathogens of the genus Corynebacterium. Infect. Immun. 1976;13:1293–1295. doi: 10.1128/iai.13.4.1293-1295.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.