Abstract
Identification of broadly neutralizing antibodies against influenza A viruses has raised hopes for the development of monoclonal antibody-based immunotherapy and ‘universal’ vaccines for influenza. However, a significant part of the annual flu burden is caused by two cocirculating, antigenically distinct lineages of influenza B viruses. Here we report human monoclonal antibodies, CR8033, CR8071 and CR9114, which protect mice against lethal challenge from both lineages. Antibodies CR8033 and CR8071 recognize distinct conserved epitopes in the head region of the influenza B hemagglutinin (HA), whereas CR9114 binds a conserved epitope in the HA stem and protects against lethal challenge with influenza A and B viruses. These antibodies may inform on development of monoclonal antibody-based treatments and a universal flu vaccine for all influenza A and B viruses.
Influenza viruses continue to cause significant morbidity and mortality worldwide (1). Because current vaccines are typically only effective against the specific viral strains used for vaccination and closely related viruses (2), and increasing resistance reduces the effectiveness of the available antiviral drugs (3), an urgent need remains for innovative new treatments, both prophylactic and therapeutic (4). To this end, we and others have previously described human monoclonal antibodies (mAbs) that neutralize a wide spectrum of influenza A viruses by binding to highly conserved epitopes in the stem region of hemagglutinin (HA), the major viral surface glycoprotein (5-9). To date, influenza B viruses have received less attention, as they are largely restricted to humans and thus lack the large animal reservoirs that are key to the emergence of pandemic influenza A viruses (10).
Although the morbidity and mortality rates attributable to influenza B are lower than H3N2 viruses, they are higher than H1N1 viruses (11). While influenza B viruses are classified as a single influenza type, two antigenically and genetically distinct lineages co-circulate (12), represented by the prototype viruses B/Victoria/2/1987 (Victoria lineage) and B/Yamagata/16/1988 (Yamagata lineage) (13). Vaccine manufacturers have therefore recently initiated clinical evaluation of quadrivalent vaccines that include strains from each influenza B lineage, H1N1 and H3N2 (14). Given that influenza B viruses are the major cause of seasonal influenza epidemics every two to four years leading to substantial absenteeism, hospitalization, and death (11), mAbs with broad neutralizing activity (bnAbs) against influenza B viruses have significant clinical potential.
Combinatorial display libraries, constructed from human B cells of volunteers recently vaccinated with the seasonal influenza vaccine (9), were panned using soluble recombinant HA from various influenza A and B viruses, and phages were subsequently screened for binding to HAs of both influenza B lineages (15). We recovered three immunoglobulins (IgGs) that bound HAs from both lineages, CR8033 (VH3-9, Vκ3-20) and CR8071 (VH1-18, Vλ1-47) (Fig. 1, A and B), as well as CR9114, a VH1-69 antibody, which additionally binds influenza A viruses from both group 1 and group 2 (Fig. 1A and fig. S1). Importantly, CR8033 and CR8071 neutralized representative viruses from either lineage (Fig. 1C), whereas polyclonal sheep sera did not (table S1). CR8033 showed hemagglutination-inhibition (HI) activity against the Yamagata lineage, but not against the Victoria lineage. Thus, while CR8033 likely neutralizes Yamagata strains by blocking receptor binding, it appears to neutralize Victoria strains by another mechanism. In contrast, CR8071 showed no HI activity against either lineage. Although CR9114 neutralized all influenza A viruses tested, it did not show in vitro neutralizing activity against influenza B viruses at the tested concentrations (Fig. 1C). Since recent work indicated that the protective efficacy of broadly neutralizing influenza antibody FI6 is substantially dependent on antibody effector functions (5), we evaluated the protective efficacy of all three mAbs against B/Florida/4/2006 (Yamagata) and B/Malaysia/2506/2004 (Victoria) infections in mice.
Fig. 1. In vitro binding and neutralizing activity of CR8033, CR8071 and CR9114.
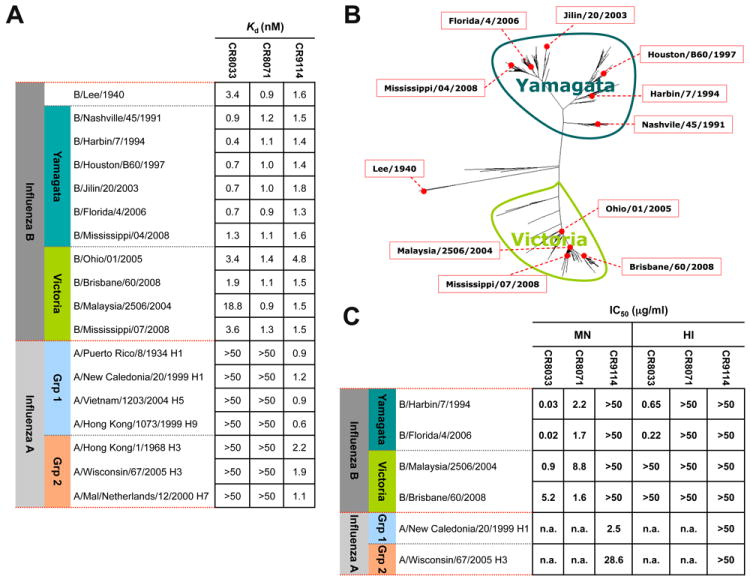
(A) Relative binding potency (Kd) for binding of CR8033, CR8071 and CR9114 to a wide range of influenza A and B hemagglutinins (HAs) as determined using a fluorescence-based plate assay. The HAs were selected to cover the two lineages and main phylogenetic branches of influenza B, as well as the major subtypes of influenza A that have infected humans. Values are from one representative of three independent experiments and reported in nM. (B) Dendrogram of all non-redundant, full-length influenza B HA sequences in the National Center for Biotechnology Information Flu database (85). The positions in the phylogenetic tree of the influenza B HAs used in these studies are indicated. (C) Fifty percent inhibitory concentrations (IC50, μg/ml) of CR8033, CR8071 and CR9114 against representative strains from the Yamagata and Victoria lineages of influenza B and the H1N1 and H3N2 subtypes of influenza A as determined by means of microneutralization (MN) and hemagglutination inhibition (HI) assays. Values are representative of three independent experiments and reported in micrograms per milliliter.n.a. = not applicable.
Doses as low as 0.6 mg/kg and 0.2 mg/kg of CR8033 fully protected mice from lethality upon challenge with B/Florida/4/2006 and B/Malaysia/2506/2004, respectively, and lower doses still resulted in increased survival and reduced weight loss (Fig. 2A and fig. S2A). Although CR8071 is somewhat less potent in vivo than CR8033 (Fig. 2B and fig. S2B), the difference is less marked than expected based on the microneutralization assay, indicating that in vitro neutralization is not fully predictive of in vivo potency. Despite the apparent lack of in vitro neutralizing activity, 15 mg/kg and ≥5 mg/kg of CR9114 fully protected mice from lethality following challenge with B/Florida/4/2006 and B/Malaysia/2506/2004, respectively, with significant protection against the latter virus with 1.7 and 0.6 mg/kg (Fig. 2C and fig. S2C). Similarly, 1.7 and 5 mg/kg CR9114 protected mice against challenge with lethal doses of influenza A H1N1 and H3N2 viruses, respectively (Fig. 2D and fig. S2D).
Fig. 2. In vivo efficacy of CR8033, CR8071 and CR9114.
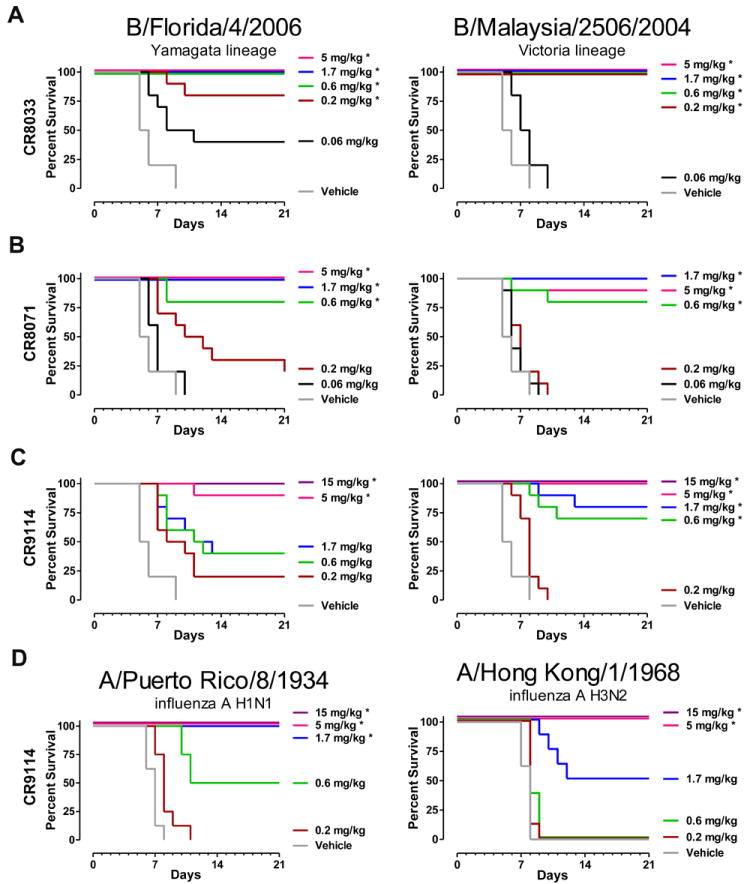
Efficacy of CR8033 (A), CR8071 (B), and CR9114 (C), against lethal challenge with mouse-adapted B/Florida/4/2006 (left panels), or B/Malaysia/2506/2004 (right panels) virus, and (D) of CR9114 against mouse-adapted A/Puerto Rico/8/1934 (left panel), or A/Hong Kong/1/1968 virus (right panel). Survival curves of mice (10 animals per group in A, B and C, 8 per group in D) treated with the indicated doses of CR8033, CR8071, or CR9114 are shown, or vehicle control 24 hours before challenge by intranasal inoculation (at day 0). Asterisks indicate significant improvements in survival proportions at day 21, compared to vehicle control (p<0.05).
CR8033, CR8071, and CR9114 do not compete with each other for HA binding, suggesting they recognize different epitopes (fig. S3). However, CR9114 competes with CR6261 and CR8020, (fig. S4), suggesting it binds an HA stem epitope. To identify their epitopes and understand how they achieve broad neutralization, we obtained x-ray and EM structures for CR8033, CR8071 and CR9114 (Fig. 3) in complex with representative HAs of influenza A and B viruses.
Fig. 3. CR8033, CR8071 and CR9114 bind to distinct epitopes on influenza B HA and conservation of the CR9114 neutralizing epitope on influenza A and B.
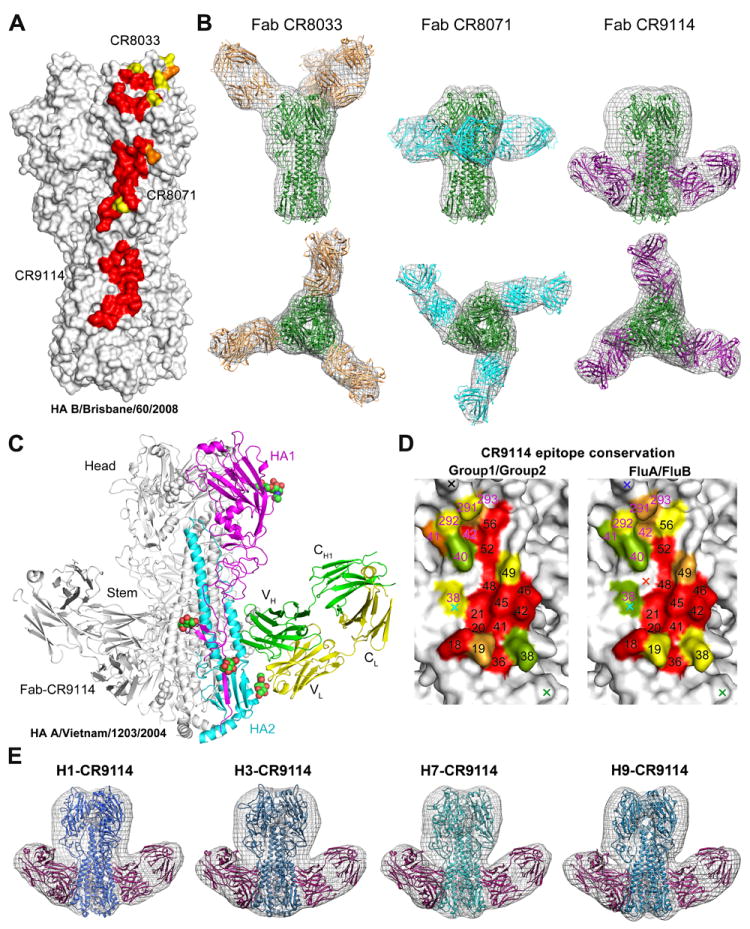
(A) Surface representation illustrating the neutralizing epitopes on HA B/Brisbane/60/2008 of CR8033, CR8071 and CR9114 as determined from the EM structures in (B) and the crystal structures of CR9114 in Fig 3C and CR8071 in fig. S8. The EM density maps allow unambiguous fits of known structures with good correspondence with the CR9114 epitope defined by both x-ray crystallography and EM, despite differences in resolution. The structure is colored by conservation of contact residues across all available influenza B virus sequences. Red = over 98% conserved; orange = 75-98% conserved; yellow = 50-75% conserved (B) Negative stain EM reconstructions (gray mesh) of CR8033 (left), CR8071 (middle) and CR9114 (right) in complex with B/Florida/4/2006. Side (top) and overhead (bottom) views show the fits of the individual crystal structures of the Fabs and flu B HA to the EM density. (C) Crystal structure of CR9114 in complex with H5 HA (group 1). One HA/Fab protomer of the trimeric complex is colored with HA1 in magenta, HA2 in cyan, Fab heavy chain in green, Fab light chain in yellow, and N-linked glycans in colored balls representing their atom type. The other two protomers are colored in gray. (D) Conservation of CR9114 contact residues across all 16 influenza A subtypes (left) and between influenza A and B viruses (right). Red, orange, yellow correspond to coloring used in Fig. 3A with green = 25-50% conserved. Carbohydrates positions are represented by a cross colored in black (group 1), cyan (group 2), green (group 1+2), orange (influenza B), and blue (influenza A and B). Residue numbers are shown with HA1 in magenta and HA2 in black. (E) Illustration of cross-reactivity of CR9114 for influenza A H1, H3, H7 and H9 subtypes using negative stain EM (from left to right). Single particle reconstructions of negatively stained CR9114 Fabs bound to HA trimers from four major subtypes of influenza A that have infected humans (SC1918/H1, group 1; HK68/H3, group 2; Neth03/H7, group 2; and Wisc66/H9, group 1). The HA trimers are colored in different shades of blue. The Fabs of CR9114 (three per trimer) are colored in purple (the third Fab, at the back, has been omitted for clarity). Broadly neutralizing CR9114 binds in a structurally similar manner to the stem region across all subtypes, groups and classes of HA. Additional density in the HA not accounted for by the protein likely corresponds to glycosylation.
We generated a 3D reconstruction of the HA ectodomain of B/Florida/4/2006 in complex with Fab CR8033 (Fig. 3B) using negative stain electron microscopy (EM). Crystal structures of Fab CR8033 at 1.9 Å and influenza B/Brisbane/60/2008 HA ectodomain at 3.45 Å provided atomic models for fitting into the EM reconstruction (fig. S5 and tables S2-S5). Three CR8033 Fabs bind the HA trimer on an epitope overlapping the receptor binding pocket and surrounding antigenic sites. The EM model suggests that almost all contacts are made by VH using all three HCDRs loops and framework 3 (fig. S6) (16). While the receptor binding pocket of influenza B HA is well conserved, variability in surrounding residues (fig. S7) may account for differences in CR8033 binding and neutralization of the two influenza B lineages.
EM reconstructions were also performed on Fab CR8071 in complex with both B/Florida/4/2006 (Yamagata) and B/Malaysia/2506/2004 HAs (Victoria) (Fig. 3B and fig. S8D), along with crystal structures of Fab CR8071 with a B/Florida/4/2006 HA1 construct (2.7 Å) and CR8059, of which CR8071 is a stabilized variant (see SOM), with B/Brisbane/60/2008 HA (5.65 Å) (fig. S8A-C). CR8071 binds the vestigial esterase domain at the base of the HA head distant from the receptor-binding site and in an orientation (Fig. 3B) consistent with the observed lack of HI. The epitope is highly conserved in influenza B HA with 17 of 19 residues >98 % conserved.
Crystal structures were determined for Fab CR9114 with HAs from a highly pathogenic H5N1 virus (A/Vietnam/1203/2004 (H5N1); Viet04/H5, 1.7 Å) (Fig. 3C), as well as H3 (5.25 Å) and H7 (5.75 Å) (fig. S9). EM studies further illustrated the CR9114 cross reactivity with influenza A H1, H3, H7, and H9 subtypes, and influenza B (Fig. 3E). CR9114 binds the HA stem (fig. S10), recognizing an epitope nearly identical to that of CR6261 (17), using the same HCDR loops (1, 2, 3) and FR3 with no light-chain contacts. Recently, human antibody FI6 was identified that broadly neutralizes influenza A viruses by targeting approximately the same epitope (5). However, FI6 uses a different V-gene (VH3-30) and its light chain (fig. S11), and is rotated by ~90°compared to VH1-69 antibodies CR6261 and CR9114. Thus, FI6 represents an alternative solution to cross-group neutralization of influenza A, but not influenza B viruses.
The CR9114 epitope is highly conserved in essentially all influenza A subtypes and influenza B (Fig. 3D, tables S6 and S7). At least three obstacles must be overcome for an antibody to bind to a similar epitope across influenza A and B. First, most group 1 HAs have HA2 Thr49, whereas most group 2 HAs have a larger Asn (fig. S10 and table S6) (6, 18) that can be accommodated by CR9114. Second, polymorphisms at HA2 position 111, His (group 1), Thr/Ala (group 2) or Glu (influenza B) result in subtly different conformations of HA2 Trp21, which affect its ability to make favorable interactions with hot spot residue HCDR2 Phe54 in CR6261 (fig. S10) (19). The more favorable orientation and approach of Phe54 and apparent plasticity of the CR9114 combining site (20) (fig. S12) may allow CR9114 to make high affinity interactions (including a hydrogen bond) with the various conformations of Trp21 in group 1 and group 2 HAs. Third, the predominant conformation of a conserved glycan at HA1 Asn38 in H3, H7, H10, and H15 in group 2, and at HA1 Asn332 in influenza B, would obscure the epitope surface (figs. S10 and S11) (21-23). Thus, this glycan and Asn side chain must be able to adopt a permissive alternative conformation as observed in the CR9114-H3 crystal structure (24). Thus, displacement of the HA1 38 glycan appears essential for CR9114 binding to several influenza A subtypes, and a similarly positioned glycan at HA1 332 in influenza B must also be avoided (fig. S11). Such glycan flexibility at HA1 38 was also observed in the FI6 structure with H3 HA but, in this case, the glycan at 332 in flu B may be more difficult to avoid (25).
CR9114 and CR6261 both utilize the VH1-69 germline Vgene and have relatively small numbers of somatic mutations compared to other bnAbs (26, 27). However, only five mutations are shared (fig. S13) and, hence, they are not simple variants of one another. Aside from the heavy-chain only binding, both antibodies are fairly conventional, in contrast to many bnAbs to other viruses, such as HIV-1, that have unusual features, such as extensive hypermutation, long indels in one or more CDRs, tyrosine sulfation, or domain swapping (26, 27). Consequently, antibodies like CR9114 are likely to be readily generated and may be present in the repertoire of many individuals, suggesting that an appropriate vaccination strategy may be able to selectively amplify such cross-reactive clones and trigger a bnAb response (4, 28, 29).
Despite CR8033 and CR8071 binding to the more variable HA globular head, but consistent with the apparent high level of epitope conservation, escape variants could only be generated after extensive passaging. For B/Florida/4/2006 virus, no CR8033 escape variants were generated in 20 passages, while 15 passages were required to generate Lys38Glu and Tyr40His mutants with reduced CR8071 susceptibility. For B/Malaysia/2506/2004 virus, 15 passages were required to generate a Pro161Gln reduced susceptibility mutant for CR8033, while 20 rounds of passaging did not result in any CR8071 escape variants. Lys38 and Pro161 are highly conserved (99.4% and 100% respectively) in all 494 (unique) full-length influenza B HA sequences in the NCBI database. Although Tyr40 is only 29.1% conserved, all other strains have His40, including B/Harbin/7/94, B/Malaysia/2506/04 and B/Brisbane/60/08, which are effectively neutralized by CR8071 (Fig. 1C). In contrast, the Lys38Glu mutation has not been observed in any influenza B isolates. Because CR9114 does not neutralize influenza B viruses in vitro, no influenza B escape variants could be generated (30).
The identification of conserved neutralizing epitopes in the influenza B HA globular head parallels the recent identification of such epitopes in influenza A viruses (31, 32) and establishes that both the HA stem and head regions contains broadly protective epitopes. Whereas the stem-binding CR9114 blocks the HA pH-induced conformational changes associated with membrane fusion (Fig. 4, A and B; and figs. S14 and S15) (33), this mechanism is not applicable to CR8033 and CR8071 that bind the head region (Fig. 4A). Indeed, whereas CR8033 has HI activity and blocks viral infection when pre-incubated with B/Florida/4/2006 virus (Fig. 1C), it had no such effect on B/Malaysia/2506/2004 virus (Fig. 4B). Furthermore, CR8071 did not prevent infection of cells by either virus (Fig. 4B) (34).
Fig. 4. Neutralization mechanisms of CR8033, CR8071 and CR9114.
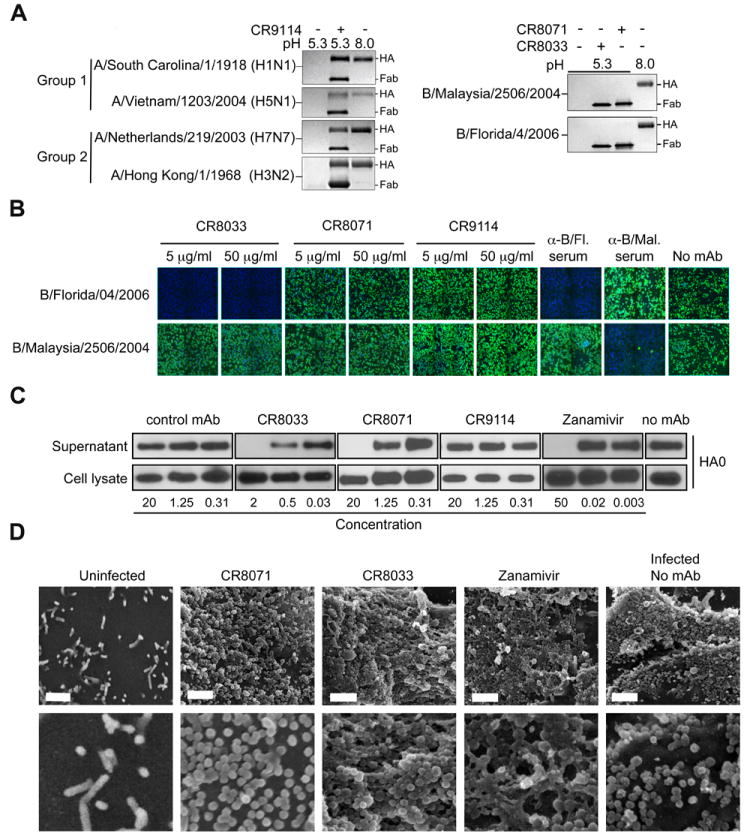
(A) CR9114 protects HAs of group 1 (A/South Carolina/1/1918 (H1N1) and A/Vietnam/1203/2004 (H5N1)) and group 2 (A/Netherlands/219/2003 (H7N7) and A/Hong Kong/1/1968 (H3N2)) influenza A viruses from the pH-induced protease sensitivity associated with membrane fusion. Exposure to low pH converts the HAs to the post-fusion state, rendering them sensitive to trypsin digestion (lane 1 vs. 3), but CR9114 prevents this conversion, retaining the HA in the protease-resistant, pre-fusion form (lane 2). CR8033 and CR8070 do not prevent this conversion at low pH (right panel) (N=4). (B) Expression of influenza NP in monolayers of MDCK cells 16-18 hrs after inoculation with B/Florida/4/2006 or B/Malaysia/2506/2004 viruses that were pre-incubated with CR8033, CR8071, CR9114, or polyclonal sheep sera directed against B/Florida/4/2006 or B/Malaysia/2506/2004, as indicated. NP expression is determined by immunofluorescence. Representative images of three independent experiments are shown. (C) Immunoblots of uncleaved HA (HA0) detected in the lysate and supernatant of MDCK cells infected with B/Florida/4/2006 virus and subsequently (from 3 to 20 hours post infection) incubated with different concentrations of antibodies or zanamivir as indicated. HA0 was detected using rabbit serum against B/Jiangsu/10/03 (Yamagata lineage). Concentrations are in μg/ml and μM for antibodies and zanamivir, respectively. Results from one representative of two independent experiments are shown. (D) SEM images of the surface of MDCK cells infected with B/Florida/4/2006 virus and subsequently (from 3 to 20 hours post infection) incubated with CR8071 (10 μg/ml), CR8033 (2.5 μg/ml), or zanamivir (60 μM). Representative images of three independent experiments are shown. Scale bar 1 μm.
Interestingly, these antibodies appear to prevent propagation of viruses without preventing entry and genome replication (Figs. 1C and 4B). Thus, we assessed the effect of CR8033 and CR8071 on formation of viral progeny from infected cells. Whereas HA was readily detected in both lysates and supernatants of MDCK cell cultures that were first infected and subsequently incubated with a non-binding control antibody or CR9114, no HA could be detected in the supernatants upon incubation with CR8033 or CR8071, despite abundant HA in the lysate (Fig. 4C). The effects of CR8033 and CR8071 then resemble that of neuraminidase (NA) inhibitor zanamivir, which interferes with release of progeny virions from infected cells (35). Indeed, the dense aggregation of virions on the surface of infected cells, particularly in the presence of antibody CR8033, closely resembles that observed in zanamivir-treated cells as examined by scanning electron microscopy (Fig. 4D).
The identification and characterization of monoclonal antibodies with broad neutralizing activity against influenza B viruses, together with previously described broadly neutralizing antibodies against group 1 and group 2 influenza A viruses (6, 7, 9) brings a universal therapy a step closer, and may serve as guides for design of broadly protective vaccines. In particular, a vaccine that elicits antibodies targeting the CR9114 epitope may provide the ultimate goal of protection against all influenza A and influenza B viruses.
Supplementary Material
Acknowledgments
We thank J. Juraszek, B. Brandenburg, M. Koldijk, K. Hegmans, J. Meijer, N. Hafkemeier, A. Apetri, J. Dulos, L. Dekking, E. Vietsch, H. Tien, D. Marciano, the staff of the APS GM/CA-CAT 23ID-B and 23ID-D, SSRL BL11-1, SSRL BL7-1 and ALS, X. Dai, R. Stanfield, W. Yu, J.P. Julien, and R.N. Kirchdoerfer, and D. Brown of Ottawa University, Canada for the mouse-adapted A/Hong Kong/1/68 strain, Crystallization experiments were carried out on the Rigaku Crystalmation system at the JCSG, which is supported by the NIGMS Protein Structure Initiative U54 GM094586. The electron microscopy and image analysis was conducted by R.K., J.H.L., and Z.M. at the National Resource for Automated Molecular Microscopy, which is supported by NIH through the National Center for Research Resources’ P41 program (RR017573). This project has been funded in part by the Area of Excellence Scheme of the University Grants Committee, Hong Kong (grant AoE/M-12/06); a predoctoral fellowship from the Achievement Rewards for College Scientists Foundation (D.C.E.); grant GM080209 from the NIH Molecular Evolution Training Program (D.C.E.); a Saper Aude Postdoc grant from The Danish Council for Independent Research, Natural Sciences (N.S.L) and the Skaggs Institute (I.A.W.). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy (DOE), Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Lightsource (SSRL) Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by NIH, National Center for Research Resources, Biomedical Technology Program (P41RR001209), and the National Institute of General Medical Sciences. The GM/CA CAT 23-ID-B beamline has been funded in whole or in part with federal funds from National Cancer Institute (Y1-CO-1020) and NIGMS (Y1-GM-1104). Use of the Advanced Photon Source (APS) was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or the NIH. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Coordinates and structure factors are deposited in the Protein Data Bank (PDB code 4FQH for CR9114 Fab, 4FQI for CR9114 Fab-A/H5 HA, 4FQJ for CR8071 Fab-B/Florida HA1, 4FQK for CR8059 Fab-B/Brisbane HA, 4FQL for CR8033 Fab, 4FQM for B/Brisbane HA, 4FQY for CR9114 Fab-A/H3 HA and 4FQV for CR9114 Fab-A/H7 HA). Image reconstructions have been deposited at the EMDB under accession numbers EMD-2143 (CR8033/Florida), EMD-2144 (CR8071/Florida), EMD-2145 (CR8071/Malaysia), EMD-2146 (CR9114/Florida), EMD-2147 (CR9114/H1), EMD-2148 (CR9114/H3), EMD-2149 (CR9114/H9), EMD-2150 (CR9114/H7). Nucleotide sequences for the CR8033, CR8059 (CR8071) and CR9114 variable regions have been deposited in GenBank (accession numbers JX213635 (CR8033, VH), JX213636 (CR8033, Vκ), JX213637 (CR8059, VH), JX213638 (CR8059, Vλ), JX213639 (CR9114, VH) and JX213640 (CR9114, Vλ)). Patent applications relating to antibodies CR8033, CR8059, CR8071 and CR9114 have been filed (CR8033, CR8059 and CR8071 are claimed in EP 12158525.1 and US 61/608,414. CR9114 is claimed in EP 11173953.8 and US 61/572,417). Sharing will be subject to standard material transfer agreements.
This is publication 21741 from The Scripps Research Institute.
Footnotes
Supplementary Materials:
Materials and Methods
Figures S1-S18
Tables S1-S8
References (36-62)
References and Notes
- 1.WHO. Fact sheet 211: Influenza. 2009 [Google Scholar]
- 2.Salzberg S. The contents of the syringe. Nature. 2008;454:160–161. doi: 10.1038/454160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowen AC, Palese P. Influenza virus transmission: basic science and implications for the use of antiviral drugs during a pandemic. Infect Disord Drug Targets. 2007;7:318–328. doi: 10.2174/187152607783018736. [DOI] [PubMed] [Google Scholar]
- 4.Wang TT, Palese P. Universal epitopes of influenza virus hemagglutinins? Nature Struct Biol. 2009;16:233–234. doi: 10.1038/nsmb.1574. [DOI] [PubMed] [Google Scholar]
- 5.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 6.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature Struct Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies crossprotective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. Influenza B virus in seals. Science. 2000;288:1051–1053. doi: 10.1126/science.288.5468.1051. [DOI] [PubMed] [Google Scholar]
- 11.Thompson WW, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita M, Krystal M, Fitch WM, Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology. 1988;163:112–122. doi: 10.1016/0042-6822(88)90238-3. [DOI] [PubMed] [Google Scholar]
- 13.Rota PA, et al. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- 14.Ambrose CS, Levin MJ. The rationale for quadrivalent influenza vaccines. Hum Vaccin Immunother. 2012;8:81–86. doi: 10.4161/hv.8.1.17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and Methods are available as supporting material on Science Online.
- 16.We have previously shown (our previous control analyses of CR6261 and other known HA-Fab complexes) that because we have high-resolution crystal structures of HAs and antibodies that the fit to the density is excellent and generally unambiguous. Nevertheless, the lower resolution of these data does not permit as precise a definition of the Ab epitope.
- 17.This epitope consists of residues from the N- and C-terminal regions of HA1 (38, 40-42, 291-293), and the N-terminal portion of HA2 (18-21, 36, 38, 41, 42, 45, 46, 48, 49, 52, 53, 56), including helix A. CR9114 buries a total of ~1293 Å2 at the interface with HA (654 Å2 for HA and 639 Å2 for Fab).
- 18.Thr49 is buried by CR6261, and affinity of CR6261 for the SC1918/H1 HA is reduced over 100-fold by a Thr49Asn mutation designed to mimic the group 2 sequence. Further, CR6261 binding to H12 HA (group 1) is undetectable under our assay conditions (Kd>~10 uM), likely due to a Thr49Gln substitution. In contrast, CR9114 binds the SC1918/H1 Thr49Asn mutant and H12 HA (Thr49Gln) with high affinity, since the conformation of its HCDR1 loop allows the larger Asn and Gln side chains to be accommodated.
- 19.A His111Leu mutation that mimics the group 2 Trp21 conformation on a group 1 background drastically reduces CR6261 binding and correlates with virus escape from neutralization (9).
- 20.HCDRs 1, 2, and 3 adopt significantly different conformations in the unbound CR9114 and the CR9114-H5 complex crystal structures, suggesting that these loops may be quite flexible in solution (fig. S12) and allow the antibody to adapt to the subtle variation seen in the largely conserved epitope in HA proteins from different virus subtypes and types.
- 21.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology. 2003;309:209–218. doi: 10.1016/s0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Cheng F, Lu M, Tian X, Ma J. Crystal structure of unliganded influenza B virus hemagglutinin. J Virol. 2008;82:3011–3020. doi: 10.1128/JVI.02477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoSPathog. 2010;6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.This alternate glycan configuration is supported by the observation of additional electron density near Asn38 in the CR8020-H3 crystal structure, indicating at least two accessible glycan conformations (7). Although not modeled explicitly, the second conformation would move the glycan away from the epitope and facilitate antibody binding
- 25.Although isolate-specific variation in residues proximal to the epitope may also influence interaction of FI6 with influenza B, one possible problem is Tyr100C at the tip of HCDR3 that makes an H-bond with Thr318 (Group1/2) in the hydrophobic groove at the junction between helix A and HA1 (fig. S10). In influenza B, the equivalent residue is a glycosylated Asn332, which will likely lead to a steric clash with Tyr100C of FI6. CR9114 does not interact with Thr318 and the orientation of the HCDR2 loop allows CR9114 to accommodate larger residue at this position (figs. S10 and S11).
- 26.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 29.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.No escape variants were generated; however, two substitutions previously described to affect neutralization of group 2 and group 1 influenza A viruses by CR8020 and CR6261, respectively also affected CR9114 binding. Asp19Asn affects H3 and H7 viruses and reduces H3 binding (~30-fold increase in Kd). In addition, all influenza B viruses have Ala19, which may account in part for the somewhat lower affinity for influenza B. In contrast, several group 1 subtypes with Asp19Asn or Asp19Ala mutations are neutralized by CR9114 and/or bound with high affinity, presumably because other substitutions offset the detrimental effect of substitutions at position 19. The Asp19Asn variant is rare in most subtypes, suggesting that most viruses will be sensitive to CR9114 (table S7) (7). The other Ile45Phe escape variant is naturally present in essentially all human H2N2 viruses that circulated from 1957 to 1968 and Phe45Ile substitution in a human H2 HA restores high affinity binding for both CR6261 and CR9114 (table S8). In contrast, Phe45 occurs in only 3 of 8550 sequences from avian H2N2 isolates and the remaining 15 subtypes.
- 31.Krause JC, et al. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Why influenza B viruses are not neutralized in the same way by CR9114 remains currently unknown.
- 34.In contrast, polyclonal sheep sera directed against B/Florida/4/2006 and B/Malaysia/2506/2004 did prevent initial infection, but not of virus from the opposite lineage.
- 35.Gubareva LV, Kaiser L, Hayden FG. Influenza virus neuraminidase inhibitors. Lancet. 2000;355:827–835. doi: 10.1016/S0140-6736(99)11433-8. [DOI] [PubMed] [Google Scholar]
- 36.Kramer RA, et al. The human antibody repertoire specific for rabies virus glycoprotein as selected from immune libraries. Eur J Immunol. 2005;35:2131–2145. doi: 10.1002/eji.200526134. [DOI] [PubMed] [Google Scholar]
- 37.Kramer RA, et al. A novel helper phage that improves phage display selection efficiency by preventing the amplification of phages without recombinant protein. Nucleic Acids Res. 2003;31:e59. doi: 10.1093/nar/gng058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown EG, Liu H, Kit LC, Baird S, Nesrallah M. Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: identification of functional themes. Proc Natl Acad Sci U S A. 2001;98:6883–6888. doi: 10.1073/pnas.111165798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 40.Stevens J, et al. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 41.Xu R, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koefoed K, et al. Rational identification of an optimal antibody mixture for targeting the epidermal growth factor receptor. MAbs. 2011;3:584–595. doi: 10.4161/mabs.3.6.17955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown PH, Cronan JE, Grotli M, Beckett D. The biotin repressor: modulation of allostery by corepressor analogs. J Mol Biol. 2004;337:857–869. doi: 10.1016/j.jmb.2004.01.041. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W. Xds Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 50.Sheriff S, Hendrickson WA, Smith JL. Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol. 1987;197:273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- 51.Connolly M. Analytical molecular surface calculation. J Appl Crystallogr. 1983;16:548–558. [Google Scholar]
- 52.Abhinandan KR, Martin AC. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol Immunol. 2008;45:3832–3839. doi: 10.1016/j.molimm.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 57.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera - A Visualization System for Exploratory Research And Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Bao Y, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss MS, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J Appl Crystallog. 1997;30:203–205. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


