Abstract
Human leukocyte antigen (HLA) B*27 and B*57 are associated with protection against HIV-1 disease progression, yet most persons expressing these alleles are unable to control HIV-1. Here we show that HLA-B*27-restricted CD8+ T cells in controllers and progressors differ in their ability to inhibit virus replication through targeting of the immunodominant Gag epitope. This is associated with distinct TCR clonotypes, characterized by superior control of HIV-1 replication in vitro, greater cross-reactivity against epitope variants, and enhanced perforin delivery. Clonotype-specific differences in antiviral efficacy were also observed for an immunodominant HLA-B*57 restricted response in controllers and progressors. Thus, the efficacy of protective alleles is modulated by specific TCR clonotypes selected in natural infection, providing a functional explanation for divergent HIV-1 outcomes.
A subset of HIV-1-infected persons, here termed elite controllers, are distinguished by their ability to maintain a state of apparent durable control of HIV-1 replication without the need for antiviral therapy 1,2. Viral control is linked to expression of certain human leukocyte antigen (HLA) class I alleles 3-5, particularly B*57, B*27, and B*5801, suggesting an immunologic basis related to CD8+ T cell function. Indeed, a recent genome-wide association study (GWAS) implicated the nature of the HLA-viral peptide interaction as the major genetic factor modulating durable control of HIV-1 infection in the absence of antiretroviral therapy 6. However, the mechanistic basis for the association remains unclear, and it is also unclear why the majority of persons with so-called ‘protective HLA alleles’ actually develop progressive disease.
A number of studies have attempted to define quantitative and qualitative differences in CD8+ T cell responses that may associate with different outcomes in terms of immune control of viremia. Simple quantitative measures have shown little correlation with viral control 7,8, suggesting that qualitative features of CD8+ T cells may modulate efficacy. Factors potentially modulating protective HLA-associated CD8+ T cell responses include, among others, polyfunctionality 9, antigen sensitivity or functional avidity 10,11 , proliferative capacity 12, loading of lytic granules 13, ex vivo expression of perforin 14, specific targeting of conserved regions 15,16, immunoregulatory mechanisms 17-19, concurrent responses to multiple epitopes restricted by different HLA alleles 20, CD8+ T cell-associated mutations that impair viral fitness 21,22 and immune escape 23. Numerous studies also suggest that properties of the T cell receptor (TCR)-peptide-MHC interaction may be involved 24,25. However, the extent to which any of these factors influences the antiviral efficacy of the human immune response, as reflected by in vivo viral load, remains unclear, in part due to lack of direct comparison of epitope-specific CD8+ T cell responses in controllers and progressors, sequence diversity within targeted epitopes and immune escape, and the potential confounding effect of targeting of multiple epitopes through diverse HLA alleles.
To address these limitations, we focused on HIV-1-infected persons expressing HLA-B*2705, a situation in which the immune response is largely if not exclusively mediated by targeting of a single Gag epitope (KK10, KRWIILGLNK, aa 263 - 272) 23. From a large, well pedigreed cohort 26, we specifically selected five controllers and five progressors expressing HLA-B*2705, for whom the circulating viruses and cellular proviruses all harbored wild-type sequences within the dominant KK10 epitope targeted through this allele at the time of analysis. This allowed for comparative assessment of adaptive CD8+ T cell responses in persons in whom the dominant CD8+ T cell response is to a single epitope in Gag, in a setting in which the viral load, and by inference the degree of CD8+ T cell mediated control, and not HLA allele or sequence variation within the targeted viral epitope, were the primary variables. We performed a detailed analysis of the epitope-specific CD8+ T cell responses and then extended these to include a dominant B*57-restricted epitope. These data indicate that HLA-B*27- and HLA-B*57-restricted CD8+ T cells targeting the same epitopes in elite controllers and progressors are clearly differentiated based on potency and cross-reactivity of TCR recognition of HIV-1 and viral variants, which is in turn related to specific TCR clonotypes that are selected in natural infection.
Results
Quantitative measures of KK10-specific T cells
Previous studies have shown that persons expressing HLA-B*2705 generate an immunodominant response to an epitope in Gag p24 termed KK10 (KRWIILGLNK, aa 263-272), targeting of which is critical to long-term control in these persons 23,27. Although escape from this response leads to accelerated disease progression, variable viral loads and rates of CD4+ T cell decline are already observed before escape occurs 23. Given the importance of the KK10-specific CD8+ T cell response to disease control, we reasoned that identification of both controllers and progressors with wild-type KK10 sequence would afford the opportunity to define characteristics of effective and ineffective CD8+ T cell responses, independent of any confounding effects of immune escape.
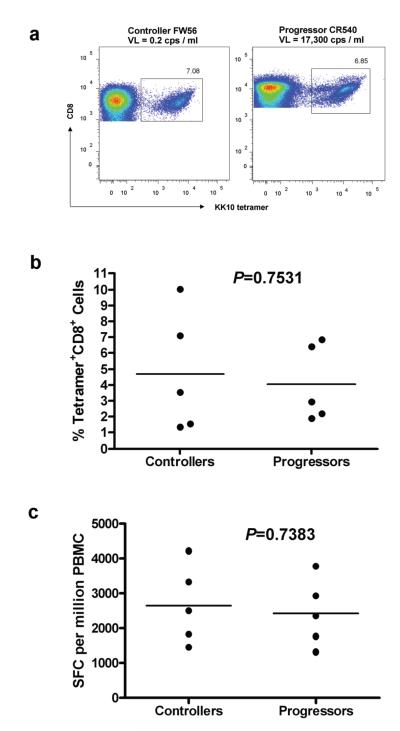
We recruited 5 HIV-1 elite controllers and 5 HIV-1 progressors for detailed studies, all expressing HLA-B*2705 and having autologous virus containing the wild-type KK10 epitope in plasma HIV-1 RNA and cellular HIV-1 DNA (Table 1). These subjects were selected from a larger population including individuals in whom variants within this epitope were present in vivo. Viral loads in controllers were all <50 RNA copies per ml plasma, and ranged as low as 0.2 RNA copies per ml plasma in those for whom more sensitive testing was performed. Progressors ranged from 4,073 to 22,094 RNA copies per ml plasma (Table 1). The dominance of the KK10-specific responses in these subjects was confirmed by fine mapping with HLA-restricted optimal peptides in interferon-γ (IFN-γ) ELISPOT assays (data not shown). Strong KK10-specific CD8+ T cell responses as defined by tetramer analysis were detected in both the controllers and progressors expressing wild-type KK10 (Fig. 1a). Comparing the controllers with the progressors, there was no significant difference in magnitude of HLA-B*27-KK10 tetramer staining (p = 0.7531; Fig. 1b) or IFN-γ production (p = 0.7383; Fig. 1c) despite substantial differences in plasma viremia between the two groups.
Table 1.
Study subjects of HIV controllers (EC) and progressors (CP) and clonal analysis of HLA-B*2705-restricted KK10-specific CD8+ T cell populations
| Subject HLA |
KK10 epitope sequence (Gag 263-272) in plasma virus (above) and PBMC provirus (below) |
% tetramer+ cells in bulk CD8+ T cells |
pVL (copies/ml) |
CD4 count (cells/ul) |
TCR β chain CDR3 region |
Clonotypic frequency |
|---|---|---|---|---|---|---|
| EC CTR22 | KRWIILGLNK | 3.53 | <50 | 337 | Vβ 20-CSARDRTRANYGYT-J1.2 | 22/25 |
| A*0201/0201 | KRWIILGLNK | Vβ 27-CASSGGRRAF-J1.1 | 1/25 | |||
| B*2705/5101 | Vβ 5.1-CASSSPDRTYGYT-J1.2 | 1/25 | ||||
| C*0102/1402 | Vβ 21-CASTNRGSEQY-J2.7 | 1/25 | ||||
| EC CTR40 | KRWIILGLNK | 1.56 | <50 | 1415 | Vβ 7.2-CASSLSGRWSTDTQY-J2.3 | 11/21 |
| A*0201/0201 | KRWIILGLNK | Vβ 7.2-CASSLEGRYSNQPQH-J1.5 | 4/21 | |||
| B*2705/5701 | Vβ 7.6-CASSLGTGKVEGYT-J1.2 | 1/21 | ||||
| C*0102/0602 | Vβ 7.9-CASSPEGPRAIEQF-J2.1 | 2/21 | ||||
| Vβ 15-CATSRELTGGPSYEQY-J2.7 | 1/21 | |||||
| Vβ 7.8-CASSQARASHLI-J1.4 | 1/21 | |||||
| Vβ 9-CASSEDRDTEAF-J1.1 | 1/21 | |||||
| EC CTR203 | KRWIILGLNK | 9.99 | 1.1 | 411 | Vβ 25.1-CASSEADFEAF-J1.1 | 13/37 |
| A*2601/6801 | KRWIILGLNK | Vβ 18-CASSPGQFSHEQY-J2.7 | 11/37 | |||
| B*0702/2705 | Vβ 27-CASSARTGELF-J2.2 | 2/37 | ||||
| C*0702/0202 | Vβ 20.1-CSARDGGEQY-J2.7 | 6/37 | ||||
| Vβ 9-CASSPLGNSGNTIY-J1.3 | 2/37 | |||||
| Vβ 7.9-CASSLDRLEQF-J2.1 | 3/37 | |||||
| EC FW56 | KRWIILGLNK | 7.08 | 0.2 | 937 | Vβ 4.3-CASRPGLASNEQF-J2.1 | 16/31 |
| A*0201/0301 | KRWIILGLNK | Vβ 6.5-CASRPGQGATEAF-J1.1 | 10/31 | |||
| B*1501/2705 | Vβ 20.1-CSARDRGTREVADNYGYT-J1.2 | 3/31 | ||||
| C*0304/0102 | Vβ 15-CATSETGTTLEQY-J2.7 | 2/31 | ||||
| EC 13587 | KRWIILGLNK | 1.35 | <50 | 994 | Vβ 7.9-CASSRDSNEQF-J2.1 | 15/27 |
| A*0206/3201 | NA | Vβ 20.1-CSAREGLAGVLYEQY-J2.7 | 1/27 | |||
| B*1524/2705 | Vβ 28-CASSSSGGAGDTQY-J2.3 | 1/27 | ||||
| C*0202/0304 | Vβ 20.1-CSAPTTEVAGSTDTQY-J2.3 | 1/27 | ||||
| Vβ 5.4-CASSLTNLGEQY-J2.7 | 3/27 | |||||
| Vβ 27-CASSRTTGELF-J2.2 | 6/27 | |||||
| CP CR540 | KRWIILGLNK | 6.85 | 17300 | 505 | Vβ 2-CASSAGPGQYGNTIY-J1.3 | 16/34 |
| A*0201/0201 | KRWIILGLNK | Vβ 5.6-CASGGGTVYEQY-J2.7 | 8/34 | |||
| B*2705/4402 | Vβ 21-CASTNRGSEQY-J2.7 | 6/34 | ||||
| C*0102/0501 | Vβ 4.3-CASSPGTNAYEQY-J2.7 | 4/34 | ||||
| CP CR420 | KRWIILGLNK | 2.93 | 13900 | 582 | Vβ 20.1-CSAREGVEGYT-J1.2 | 21/30 |
| A*0201/1101 | KRWIILGLNK | Vβ 27-CASSGGRRAF-J1.1 | 6/30 | |||
| B*2705/4402 | Vβ 4.3-CASSQGSGSGNTIY-J1.3 | 1/30 | ||||
| C*0102/0501 | Vβ 4.3-CASSQVLRGVYGYT-J1.2 | 1/30 | ||||
| Vβ 5.4-CASSLLAGGTDTQY-J2.3 | 1/30 | |||||
| CP CR338 | KRWIILGLNK | 2.2 | 6800 | 870 | Vβ 27-CASSPRTGELF-J2.2 | 16/29 |
| A*0101/0201 | KRWIILGLNK | Vβ 27-CASSQRTGELF-J2.2 | 3/29 | |||
| B*0702/2705 | Vβ 27-CASSRATGELF-J2.2 | 4/29 | ||||
| C*0102/0702 | Vβ 5.1-CASSLEGGANL-J1.2 | 1/29 | ||||
| Vβ 6.4-CASSVVRGNPNEQF-J2.1 | 5/29 | |||||
| CP 8222 | KRWIILGLNK | 1.9 | 4073 | 269 | Vβ 27-CASSGSNLEAF-J1.1 | 21/30 |
| A*0201/2902 | KRWIILGLNK | Vβ 7.9-CASSPLGVRAYEQY-J2.7 | 3/30 | |||
| B*2705/4403 | Vβ 4.3-CASSPGTSTYEQY-J2.7 | 6/30 | ||||
| C*0102/1601 | ||||||
| CP FEN33 | KRWIILGLNK | 6.4 | 22094 | 603 | Vβ 7.9-CASSLAGGDSYEQY-J2.7 | 21/23 |
| A*0201/1101 | KRWIILGLNK | Vβ 27-CASSGGVFYGYT-J1.2 | 1/23 | |||
| B*2705/5101 | Vβ 19-CATLGGFPDGYT-J1.2 | 1/23 |
Fig. 1. Quantification of KK10-specific CD8+ T cell responses.
(a) Scatterplot of the percentage of HLA-B*27-KK10-specific CD8+ T cells in a controller (FW56) and a progressor (CR540) determined by flow cytometry and staining with HLA class I tetramers. (b) No significant difference between the controllers (n = 5) and progressors (n = 5) in terms of the percentage of KK10 tetramer-positive cells in bulk CD8+ T cells. (c) KK10-specific CD8+ T cell responses in PBMC were assessed directly ex vivo in an IFN-γ ELISPOT assay following KK10 peptide stimulation. There were no significant differences in response magnitude (calculated as spot forming cells (SFC) per million PBMC) between the controllers (n = 5) and progressors (n = 5). Statistical comparisons were made using the Mann-Whitney test.
Since TCR is a key structure that defines antigen recognition, we next evaluated whether differences in TCR usage might be associated with differential ability to control viremia, due to TCR clonotypes with heterogeneous antiviral potential. KK10 tetramer positive cells were sorted and subjected to TCR sequencing. Consistent with the findings in other studies 28,29, there was striking diversity of clonotype recruitment in all KK10-specific CD8+ T cell populations, and despite dominance of single clonotypes in each person, we did not observe preferential usage of a particular TCRBV or CDR3 motif among the various KK10-specific CD8+ T cell clonotypes (Table 1). Indeed, of the clonotypes identified, only two clonotypes were the same in two different subjects (CTR22 and CR420: TCRBV27-CASSGGRRAF/J1-1, as well as CTR22 and CR540: TCRBV21-CASTNRGSEQY/J2-7), and only one subject (CR420) possessed a TCRBV4-3/J1-3 clonotype similar to that recently reported in persons expressing HLA-B*27 in whom viral loads varied between 1,880 and 202,590 28,30,31.
These data indicate that KK10-specific CD8+ T cells are quantitatively similar but demonstrate marked heterogeneity in TCR usage among persons targeting a genetically identical epitope through a genetically identical HLA allele, in whom we observe marked differences in viral load.
Functional characteristics of KK10-specific T cells
A number of reports have suggested qualitative features of CD8+ T cells that are associated with viral control. One such measure is antigen sensitivity (often termed ‘functional avidity’) 10,11,31, and HLA-B*27 is characterized by high sensitivity T cell responses 28. However, whether antigen sensitivity varies with viral load in persons expressing HLA-B*27 and wild-type epitope has not been determined. We next assessed antigen sensitivity in each of these subjects, by examining IFN-γ ELISPOT responses at limiting KK10 peptide concentrations. There was no difference in SD50 between controllers and progressors (p = 0.4678; Fig. 2a). These results, in which all responses were detected in the presence of wild-type KK10 and therefore not confounded by potentially cross-reactive responses induced by mutations within the epitope, are consistent with previous reports showing that the majority of clones specific for KK10 are of similar antigen sensitivity 10.
Fig. 2. Functional characteristics of KK10-specific CD8+ T cells.
(a) Functional avidity of KK10-specific CD8+ T cells as measured by peptide titration of PBMC in the IFN-γ ELISPOT assay. (b) Expression of IFN-γ, TNF, IL-2, MIP-1β and CD107a were measured in KK10-specific CD8+ T cells from the five HLA-B*2705-positive controllers (EC) and five progressors (CP) following PBMC stimulation with KK10 peptide. The bars represent proportion of subpopulations of KK10-specific cells expressing different combinations of effector functions. The y-axis shows the mean percentage of all cells displaying a particular combination. Statistical comparisons were made using the Mann-Whitney test; * denotes p < 0.05. (c) Scatterplot of proliferation of tetramer-positive KK10-specific CD8+ T cells, as shown by CFSE-low cells, at day 7 following stimulation of bulk CD8+ T cells from a controller (FW56) and a progressor (CR540) with virally infected HLA-B*2705-encoding GXR cells. (d) No significant difference was observed in proliferative capacity of KK10-specific CD8+ T cells as measured by CFSE intensity by flow cytometry between the controllers (n = 5) and progressors (n = 5).
Polyfunctionality was next examined, including the capacity of ex vivo B*2705 KK10-specific CD8+ T cells to simultaneously produce the effector cytokines and chemokines IFN-γ, IL-2, TNF, and MIP-1β and to release cytotoxic factors by monitoring the expression of the degranulation marker CD107a upon KK10 peptide stimulation. Previous population studies have shown that the ability to produce 4 and 5 functions concurrently is associated with HIV-1 controllers 9, and although some epitope-specific responses have been evaluated in this manner 10,28,32, this has not been examined for HLA-B*27 restricted responses in persons known to express wild-type virus—in whom the inducing antigen is thus the same. When this analysis was done for the KK10 epitope (Fig. 2b), MIP-1β secreting cells dominated the responses in both progressors and controllers, consistent with previous findings 9. Pairwise comparisons revealed several subsets with significantly different functional profiles between the two groups. For example, cells dually expressing IFN-γ and MIP-1β were significantly higher in controllers (p = 0.031), whereas cells dually expressing IFN-γ and TNF were higher in progressors (p = 0.012). However, although cells with greater than three functions were enriched in controllers, this did not reach statistical significance (p = 0.1) and these cells made up only a small subset of the total KK10-specific CD8+ T cell response.
Previous studies have shown that chronic HIV-1 infection skews maturation of HIV-1-specific CD8+ T cells towards pre-terminally differentiated cells with poor cytotoxic activity 33. CD27 and CD45RA staining was used as described previously 34 to phenotypically distinguish four distinct subpopulations of KK10-specific cells. Comparable proportions of KK10-specific cells with central memory (KK10+CD27+CD45RA−), effector memory (KK10+CD27−CD45RA−), or terminally differentiated effector memory (KK10+CD27−CD45RA+) phenotype were detected in the controllers and progressors (Supplementary Fig. 1). Moreover, we observed that HIV-1 infected HLA-B*2705-encoding GXR cells stimulated proliferation to a comparable degree in both groups by giving the same signal (p = 0.2222; Fig. 2c and 2d).
Together, these data indicate that the superior control of wild-type viremia in this cohort of elite controllers and chronic progressors expressing HLA-B*27, all of whom were treatment naive, is not reflected in quantitative measures of KK10-specific CD8+ T cells, nor in qualitative assessment of their functional avidity, cytokine secretion, proliferative capacity, or differentiation phenotypes.
Virus neutralization by KK10-specific T cells
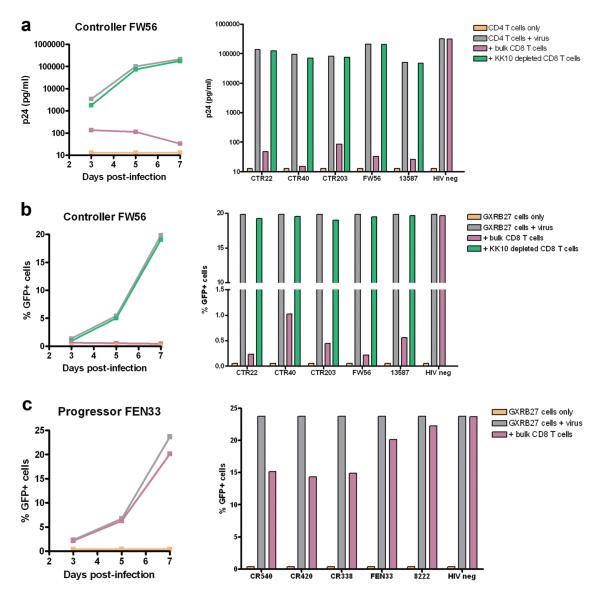
Having shown that the above measures of responses to the immunodominant KK10 Gag epitope did not differentiate these controllers from progressors, we next evaluated the functional ability of responses to this epitope to inhibit HIV-1 replication in vitro 35. We limited the initial analysis to the elite controllers, in whom outgrowth of autologous virus in CD4+ T cells is markedly delayed 36, allowing us to use controlled inocula of exogenous HIV-1 isolates to infect these cells and measure the ability of defined numbers of CD8+ T cells to inhibit virus replication. We included all 5 HLA-B*2705-positive elite controllers demonstrated to harbor wild-type KK-10 epitope sequences in plasma virus and PBMC provirus (Table 1) and assessed antiviral ability of bulk CD8+ T cells and KK10-specific cell-depleted CD8+ T cells to inhibit virus replication in autologous CD4+ T cells by measurement of p24 antigen production in the supernatant over 7 days 35. Addition of bulk CD8+ T cells to virally infected CD4+ T cells resulted in a 3 to 4 log reduction in p24 antigen production at day 7 in culture, whereas viral inhibition was reduced by greater than 90% using cells in which KK10-specific CD8+ T cells were depleted (Fig. 3a). This confirmed that the major immune control was mediated by the KK10 response in each of these subjects, and showed that all of the B*27-positive controllers were able to limit virus replication in vitro.
Fig. 3. Virus neutralization by ex vivo KK10-specific CD8+ T cells.
(a) The production of p24 antigen by autologous CD4+ T cells infected with NL4-3 wild-type virus were evaluated in the presence of bulk CD8+ T cells or KK10-specific cell-depleted CD8+ T cells over 7 days for all 5 controllers. Data are shown over 7 days for a representative subject (FW56, left) and for all 5 controllers at day 7 (right). Uninfected CD4+ T cells and virally infected CD4+ T cells were used as negative and positive controls, respectively. (b) The virus inhibition assay was performed using the HLA-B*2705-encoding GXR cell line and GFP reporter assay. The reporter cells were cultured in the presence of bulk CD8+ T cells or KK10-specific cell-depleted CD8+ T cells over 7 days. The proportions of GFP-positive cells were analyzed by flow cytometry as shown for an representative subject over 7 days (FW56, left) and for all 5 controllers at day 7 (right). (c) Virus replication was evaluated in HLA-B*2705-encoding GFP reporter GXR cells in the presence of bulk CD8+ T cells over 7 days for all 5 progressors. The proportion of GFP-positive cells was analyzed by flow cytometry as shown for an example over 7 days (FEN33, left) and for all 5 progressors at day 7 (right).
We next sought to examine the antiviral function of the CD8+ T cells from the progressors. However, outgrowth of autologous virus in HIV-1 progressors complicates the viral inhibition assay using autologous CD4+ T cells (data not shown). We therefore sought to validate an assay based on the use of HLA-B*2705-encoding GFP reporter CEM-derived GXR cells as target cells, which fluoresce green upon infection 37,38. When these target cells were used with bulk CD8+ T cells from the controllers, there was marked inhibition of replication; moreover, there was near complete loss of virus inhibition by depletion of KK10-specific cells from bulk CD8+ T cell population (Fig. 3b). In contrast, no inhibition was observed by bulk CD8+ T cells from HIV-1 negative individuals either using virally infected antologous CD4+ T cells or HLA-B*2705-encoding GFP reporter GXR cells. In addition, no inhibition was observed by bulk CD8+ T cells from the controllers using HLA-B*2705-negative GFP reporter GXR cells after infection of the same virus (data not shown).
Having shown that this assay provides evidence of active virus neutralization by ex vivo KK10-specific CD8+ T cells in HLA-B*2705-positive elite controllers, and that this assay is sensitive to KK10 epitope specificity and HLA-B*2705 expression, we next evaluated CD8+ T cells from the HIV-1 progressors. Infected GXR cells expressing HLA-B*2705 were inhibited by addition of CD8+ T cells from progressors (Fig. 3c), but to a dramatically lesser degree than had been seen with the controllers (p < 0.0001). This was observed despite the fact that there were no quantitative differences in KK10-specific cell numbers comparing the controllers to progressors, as shown by tetramer staining and IFN-γ ELISPOT assay.
We next extended these studies to determine the ability of KK10-specific CD8+ T cell responses in controllers and progressors to recognize viral escape variants known to arise in vivo 38. Recent computational studies suggest that protective HLA alleles are associated with enhanced cross-reactivity 25, but this has not been evaluated in the context of a single HLA class I allele and single epitope, comparing controllers and progressors. Recognition of HIV-1 and viral variants by KK10-specific CD8+ T cells in bulk CD8+ T cells from the controllers and progressors was analyzed by flow cytometry. This was done by evaluating the proportion of GFP-positive cells after infecting HLA-B*2705-encoding GFP reporter GXR cells. We chose the GFP reporter GXR cell assay system rather than autologous CD4+ T cells, again to overcome outgrowth of autologous virus from the chronic progressors and avoid potential variability of CD4+ T cell responses among study subjects.
An example of recognition of HIV-1 NL4-3 harboring KK10 epitope (KRWIILGLNK) and KK10 viral variants (Table 2) by KK10-specific CD8+ T cells from an HLA-B*2705-positive elite controller (FW56) and a chronic progressor (CR540) is shown in Fig. 4a. There was essentially complete inhibition of wild-type NL4-3 virus replication and broad recognition of viral variants by KK10-specific CD8+ T cells from controller FW56, as well as marked inhibition of the typical early L6M mutant, which was not detected in this individual. In contrast, inhibition of both of these targets by CD8+ T cells from progressor CR540 was present but minimal. Although peak infection with the other mutant viruses was less than with wild-type virus or L6M mutant, controller FW56 inhibited all variants, whereas CD8+ T cells from progressor CR540 were ineffective, even though both subjects had comparable KK10-specific effector cell proportions quantified by tetramer staining (Table 1) and proliferative capacity in response to HIV-1 infected HLA-B*2705-encoding GXR cells (Fig. 2c). The superiority in antiviral efficacy against NL4-3 wild-type virus and viral variants by HIV-1-specific CD8+ T cells from elite controllers was consistently observed when these detailed studies were extended to the 5 elite controllers and 5 chronic progressors (p < 0.0001; Fig. 4b). In addition, bulk CD8+ T cells from HIV-1 negative individuals (n = 12, including 4 HLA-B*2705-positive donors) did not inhibit virus replication in B*2705-encoding GFP reporter GXR cells (Fig. 4b). Similar results in terms of killing efficacy against the same virus-infected HLA-B*2705-encoding GXR cells were observed when we performed standard chromium release assays in the controllers and progressors (p < 0.0001; Fig. 4c).
Table 2.
HLA-B*2705-restricted KK10 and HLA-B*57-restricted TW10 epitope sequences in HIV-1 NL4-3 strain and its viral variants
| Viral isolate | Epitope amino acid |
|---|---|
| KK10 (residues 263 to 272) | KRWIILGLNK |
| NL4-3 KK10 WT | .......... |
| NL4-3 L268M | .....M.... |
| NL4-3 R264T | .T........ |
| NL4-3 R264T/L268M | .T...M.... |
| NL4-3 R264Q | .Q........ |
| NL4-3 R264Q/L268M | .Q...M.... |
| TW10 (residues 240 to 249) | TSTLQEQIGW |
| NL4-3 TW10 WT | .......... |
| NL4-3 T242N | ..N....... |
| NL4-3 G248A | ........A. |
| NL4-3 G248D | ........D. |
Fig. 4. Recognition of viral variants by KK10-specific CD8+ T cells.
(a) Inhibition of replication of NL4-3 wild-type virus and the designated NL4-3 variants was evaluated in HLA-B*2705-encoding GFP reporter GXR cells in the presence of ex vivo CD8+ T cells isolated from a controller (FW56) and a progressor (CR540) at an effector/target cell ratio of 1:1. Virus replication was calculated as the proportion of GFP-positive cells by flow cytometry at day 7 in culture. (b) Summary of data from pools of 5 controllers, 5 progressors, and 12 HIV-1 negative individuals demonstrating different antiviral efficacy for ex vivo CD8+ T cells from these groups. Significance was tested with a Mann-Whitney test; * denotes p < 0.0001. (c) The ability of ex vivo CD8+ T cells from controllers (CTR203 and FW56) and progressors (CR540 and FEN33) to kill live virally infected HLA-B*2705-encoding GFP reporter GXR cells was tested in the standard 4-h chromium release assay at an effector/target cell ratio of 10:1. Viable virally infected (GFP-positive) GXR cells were sorted by a FACS Aria cell-sorting instrument after infection for 5 days and used as target cells.
Together, these data indicate that HLA-B*2705-restricted CD8+ T cells targeting the KK10 epitope can be clearly differentiated between the elite controllers and chronic progressors based on potency and cross-reactivity of recognition of cells infected with wild-type HIV-1 and HIV-1 containing naturally arising mutations in the KK10 epitope. These cross-reactive neutralization data are consistent with the observation that the CDR3 sequences of KK10-specific clonotypes were significantly closer to germline in the controllers compared to the progressors (p = 0.001; Supplementary Fig. 2), which confers a greater ability to recognize epitope variants 39. These functional data are also consistent with computational modeling showing that thymic selection in the context of protective HLA alleles is more likely to generate a cross-reactive CD8+ T cell repertoire targeting mutants of viral epitopes, thereby contributing to improved control of a highly variable pathogen 25.
Antiviral efficacy of KK10-specific clonotypes
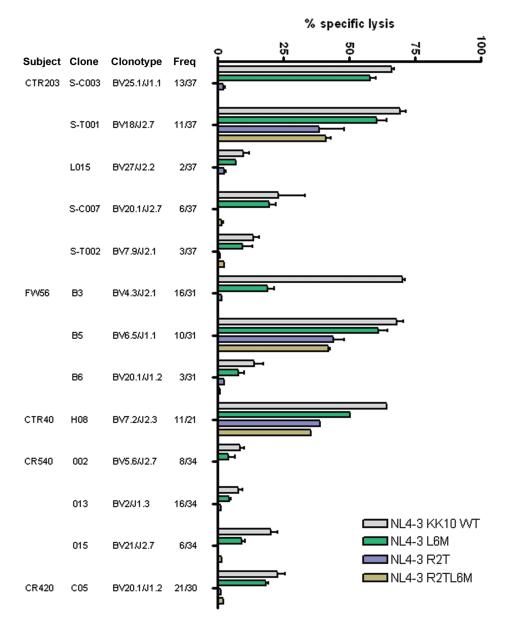
The above data indicate that there are differences in potency and cross-reactivity of recognition of wild-type HIV-1 and viral variants in the KK10-specific CD8+ T cell responses between controllers and progressors, suggesting that the fine specificity of the TCR might be modulating these effects. Given that we had observed different clonotypes in the tetramer-positive populations in these ten subjects (Table 1), we next sought to determine clonotypic antiviral efficacy. We therefore cloned HLA-B*2705 KK10-specific CD8+ T cells by limiting dilution from the KK10 tetramer-sorted cells from three elite controllers (CTR203, FW56 and CTR40) and two chronic progressors (CR540 and CR420), and then determined clonotypes by TCR sequencing (Table 3). In each of these individuals, the dominant clonotypes identified in vivo was cloned, and in 3 of the five subjects we were able to generate multiple clones. In addition, for multiple subjects we were able to establish clones for additional subdominant clonotypes.
Table 3.
Clonotypes of HLA-B*2705-restricted KK10-specific CD8+ T cell clones
| Clone | TCRBV | CDR3 | TCRBJ |
|---|---|---|---|
| CTR203 S-C003 | 25.1 | CASSEADFEAF | 1.1 |
| CTR203 S-T001 | 18 | CASSPGQFSHEQY | 2.7 |
| CTR203 L015 | 27 | CASSARTGELF | 2.2 |
| CTR203 S-C007 | 20.1 | CSARDGGEQY | 2.7 |
| CTR203 S-T002 | 7.9 | CASSLDRLEQF | 2.1 |
| FW56 B3 | 4.3 | CASRPGLASNEQF | 2.1 |
| FW56 B5 | 6.5 | CASRPGQGATEAF | 1.1 |
| FW56 B6 | 20.1 | CSARDRGTREVADNYGYT | 1.2 |
| CTR40 H08 | 7.2 | CASSLSGRWSTDTQY | 2.3 |
| CR540 002 | 5.6 | CASGGGTVYEQY | 2.7 |
| CR540 013 | 2 | CASSAGPGQYGNTIY | 1.3 |
| CR540 015 | 21 | CASTNRGSEQY | 2.7 |
| CR420 C0525.1 | 20.1 | CSAREGVEGYT1.1 | 1.2 |
We tested the ability of the clonotypic KK10-specific CD8+ T cells to kill virally infected target cells in a standard chromium release assay, and to inhibit virus replication, again using the HLA-B*2705-encoding GFP reporter GXR cells. For controller CTR203, we were able to establish clones representing 5 of the 6 clonotypes detected in peripheral blood. We observed marked variation in the ability of these clonotypes to recognize HIV-1 and viral variants (Fig. 5), ranging from broad recognition of the variants by the two most dominant CTR203 clonotypes, to weak and narrow recognition by all three CTR203 subdominant TCR variants. Extending this to clones from five subjects, the most effective clonotypes were the immunodominant in vivo clonotypes from the controllers, including the two co-dominant responses in subject CTR203, the two co-dominant responses in subject FW56 and the one dominant response in subject CTR40. These were significantly more potent at viral recognition than the immunodominant clones from the progressors (p = 0.005). Co-dominant clonotype TCRBV4-3 in controller FW56, which exhibited efficient recognition of wild-type virus and had less robust activity against the L6M variant and was unable to recognize any of the other variants, was observed at a low frequency in progressors CR540, CR420 and 8222 in the context of different CDR3 or J segments.
Fig. 5. Differential antiviral efficacy of B*27-KK10 specific clonotypes.
The ability of KK10-specific clonotypes to recognize NL4-3 wild-type and variant viruses was tested in the standard 4-h chromium release assay with virally infected HLA-B*2705-encoding GFP reporter GXR cells at an effector/target cell ratio of 1:1. Viable infected (GFP-positive) GXR cells were sorted by a FACS Aria cell-sorting instrument after infection for 5 days and used as target cells. Data are shown for three controllers (CTR203, FW56 and CTR40) and two progressors (CR540 and CR420).
Subdominant clonotypes from the elite controllers included TCRBV27/J2-2, TCRBV20-1/J2-7 and TCRBV20-1/J1-2, all of which were associated with inferior recognition of wild-type virus and the L6M variant and exhibited the least efficacy against the other viral variants. However, these less effective clonotypes were dominantly selected by progressors CR338, 8222 and CR420. They were rearranged with variant CDR3 or J segments, but were likewise less efficient at recognizing virally infected cells. And none of the subdominant clonotypes, whether from controllers or progressors, was able to efficiently recognize virally infected cells or to inhibit virus replication. Consistent findings in terms of the ability of individual clonotypes to inhibit virus replication were observed (Supplementary Fig. 3).
Together, these data indicate that differential antiviral efficacy of KK10-specific CD8+ T cells in HIV-1-infected persons is defined by dominance of clonotypes which confer distinct antiviral potential on CD8+ T cells.
Antiviral efficacy of B*57-TW10 specific clonotypes
The effect of individual TCR clonotypes on antiviral efficacy was further examined with HLA-B*5701-restricted TW10 (TSTLQEQIGW, Gag residues 240 to 249)-specific CD8+ T cell clones in the chromium assay with virally infected HLA-B*5701-encoding GFP reporter GXR cells (Fig. 6). HLA-B*5701-restricted TW10-specific CD8+ T cell clones were generated by limiting dilution from TW10 (TSTLQEQIGW) tetramer-sorted cells from HLA-B*57-positive elite controllers (CTR53, CR462) and a chronic progressor (CR555) and then clonotypically determined by TCR sequencing. Again, we observed that variant clonotypes possessed differential antiviral efficacy and that overall clones from the elite controllers were more potent and cross-reactive in recognition of HIV-1 and viral variants than clones from the progressor.
Fig. 6. Differential antiviral efficacy of B*57-TW10 specific clonotypes.
The ability of HLA-B*5701 TW10-specific CD8+ T cell clones generated from HLA-B*57 positive elite controllers (CTR53, CR462) and a chronic progressor (CR555) to recognize NL4-3 wild-type and variant viruses was tested in the standard 4-h chromium release assay with virally infected HLA-B*5701-encoding GFP reporter GXR cells at an effector/target cell ratio of 1:1. Viable infected (GFP-positive) GXR cells were sorted by a FACS Aria cell-sorting instrument after infection for 5 days and used as target cells.
Together, these data indicate that the difference of the same epitope-specific CD8+ T cell responses between the controllers and progressors in recognition of HIV-1 and viral variants is related to distinct TCR clonotypes that are selected in natural infection.
Lytic granule loading and delivery by clonotypes
The above data indicate clonotype-specific functional differences in antiviral function, offering the opportunity to define mechanisms that account for these phenotypes. We next determined the effect of TCR clonotype on lytic granule loading upon recognition of HIV-infected target cells, and the ability of these T cells to deliver perforin to infected target cells. Expression of perforin and granzyme B (GrB) by representative clonotypes was measured by flow cytometry 12,13. Upon culture with KK10 wild-type HIV-1-infected HLA-B*2705-expressing GXR cells for 3 days, dominant clonotypes from controllers efficiently expressed perforin. In contrast, subdominant clonotypes from controllers and all the clonotypes from progressors were significantly less efficient at expressing perforin (p < 0.0001; Fig. 7a). Similar results were observed for GrB expression (p < 0.0001; Supplementary Fig. 4).
Fig. 7. Differential perforin loading and delivery of clonotypes.
(a) Perforin expression of clonotypes was examined by flow cytometry after 3 days of culture with virally infected and uninfected HLA-B*2705-expressing GFP reporter GXR cells. Values indicate percentages of perforin secreting KK10-specific cells upon culture with virally infected target cells after subtracting background from effector cells incubated with uninfected target cells. (b) Differential interference contrast (DIC) images of effector cells with GFP-fluorescing target cells (DIC+GFP) are shown on the Left. Confocal microscope z-series were obtained. Projected serial confocal sections through conjugation between effector cells and HLA-B*2705-encoding GFP reporter GXR cells are shown (Green). F-actin was stained with phalloidin-Alexa Fluor 647 (Red). Perforin was stained with anti-perforin primary Ab followed by Alexa Fluor 568-conjugated secondary mAb (Purple). Merged overlays are on the Right. Clones were imaged 30 min after incubation with HIV-1 infected GXR cells. The dominant clonotype S-C003 (top) and subdominant clonotype S-C007 (bottom) from controller CTR203 are indicated. Scale bars are 3.0 μm. (c) Total intensity of perforin per T cell from representative dominant clonotypes (S-C003 and 013) and subdominant clonotypes (S-C007 and 015) from controller CTR203 and progressor CR540 are shown following exposure to HIV-1 infected GXR cells. (d) Intensity of perforin staining in GXR target cells following exposure to the clonotypes shown in (c). Data for (c) and (d) were obtained from two independent experiments. Error bars indicate the standard error of the mean (SEM).
We also examined effector-target cell conjugation and granule loading and delivery by confocal microscopy 40. Upon incubation with virally infected HLA-B*2705-expressing GXR cells for 30 minutes, more perforin in the inhibitory clonotypes was polarized to synapses and released into target cells, as compared to the non-inhibitory clonotypes (Fig. 7b). An extended quantitative analysis of perforin loading and delivery showed that significantly less perforin was delivered to synapses (p < 0.0001; Fig. 7c) and released into target cells (p < 0.0001; Fig. 7d) by subdominant clonotypes from controllers and all the clonotypes from progressors. As control, endogenous perforin was confirmed to be undetectable in virally infected GXR cells (Supplementary Fig. 5). Comparable basal levels of perforin were observed in inhibitory and non-inhibitory clonotypes without target cell stimulation (p = 0.96; Supplementary Fig. 6), indicating that the functionally effective clones rapidly upregulate perforin loading upon cognate antigen recognition.
These data indicate that TCR clonotypes that are associated with enhanced abillty to inhibit HIV-1 replication do so by rapid upregulation of lytic granules at the immunological synapes following target cell engagement, and delivery of these into the infected cell.
Discussion
Certain HLA B alleles are associated with enhanced control of viremia in HIV-1 infected persons 3-5, and this effect maps to specific host amino acids within the HLA-B peptide binding groove 6. However, the majority of persons with so-called protective alleles such as HLA B*27 and HLA-B*57 experience progressive infection 26. To address the basis for these differences in outcome, we compared CD8+ T cell responses to immunodominant epitopes in treatment-naive elite controllers and chronic progressors. In order to limit the number of potential confounding variables, all subjects harbored wild-type sequences of the respective T cell epitopes in plasma virus and PBMC provirus, and all shared the same HLA class I restricting allele. In this setting, in which contemporaneous immune escape is not a confounding issue, we find that CD8+ T cell responses from HIV-1 controllers are more potent at inhibiting HIV-1 replication than those in progressors targeting the same epitopes, and better able to cross-recognize HIV-1 viral variants that typically arise in vivo. Moreover, these effects are associated with a unique ability of the dominant TCR clonotypes to upregulate perforin and GrB, providing a mechanistic explanation for the divergent disease outcomes in persons with protective HLA alleles.
There have been numerous characteristics of CD8+ T cells that have been reported to be associated with enhanced control of viremia, including differences in polyfunctionality, proliferative capacity, and functional avidity of KK10-specific CD8+ T cells. In the carefully controlled comparative studies done here, in a small cohort of well pedigreed controllers and progresssors expressing protective alleles, none of these previously reported associations reached statistical significance. In contrast, functional ability of both bulk CD8+ T cells as well as epitope-specific TCR clonotypes to inhibit virus replication, cross recognize viral vaiants and upregulate perforin and GrB, which are likely the most important in vivo function of these cells, were highly significant. Overall, this study links the antiviral efficacy of the two most protective HLA class I alleles to CD8+ T cell clonotypes selected in HIV-1 natural infection to viral control in vivo, and demonstrates that TCR rearrangement modulates the effect of protective alleles on disease outcome.
This study is distinct from other reports of TCR clonotype usage by KK10-specific CD8+ T cells in persons expressing HLA-B*27, in that the subjects here were stratified by extremes of viral load. As in previous studies 28,29, we observed striking diversity of clonotype recruitment and CDR3 motif in KK10-specific CD8+ T cell populations, as well as a dominance of clonotypes in progressors that were unable to cross-recognize the L6M mutation. This mutation is known to occur early during the course of HIV-1 infection with little or no impact on peptide processing 27, HLA-B*27 binding 27, TCR recognition 41, or viral fitness 38, but it is an important intermediate mutation on the path to complete escape. Although such ineffective responses were detected at a clonal level in both controllers and progressors in our cohort, HLA-B*27 positive elite controllers possess dominant clonotypes that not only target the L6M mutant, but also other mutants including substitution at KK10 residue position 2 alone or in combination with the L6M mutation, which reduce peptide binding to HLA-B*27 and impair viral replication 38. Thus the TCR clonotypes in controllers are comprised of effective and ineffective clonotypes, whereas those in progressors are limited to less effective clonotypes.
In contrast to some other reports 42,43, we found no significant differences in the usage of public clonotypes between the two groups despite siginificant differences in plasma viremia and that public clonotypes do not appear to dominate among controllers 29. However, controllers used TCRB clonotypes with CDR3 sequences that were significantly more “germline-like” than those used by the progressors. The lower number of nucleotide additions in the germline-like CDR3 regions is a hallmark of clonotypes that are found at high frequecny in naive and memory T cell pools and that are also shared between multiple individuals 44. The mechanism underlying this advantage bestowed by germline-like CDR3 regions may be related to higher precursor frequency and (or) greater ability to recognize mutational variants of the epitope 39,42. Furthermore, the most effective clonotypes, in terms of viral inhibition and cytotoxic recognition of wild-type and variant viruses, were dominantly selected in vivo by the controllers but either absent or subdominantly selected in KK10-specific CD8+ T cell populations by the progressors. In contrast, the clonotypes associated with inferior recognition of wild-type virus and the least efficacy against the viral variants were dominantly selected by the progressors but subdominantly selected by the controllers.
Although this study clearly shows that TCR modulates the protective effect of HLA alleles, it has a number of limitations. The HLA-B*2705 studies were limited to only 5 controllers and 5 progressors, and a number of previously reported associations with viral control did not reach statistical significance in this small study group. Nevertheless, even with these small numbers the results are highly significant in showing greater cytotoxic killing and greater cross-reactivity by the dominant clonotypes in controllers compared to progressors. We were not able to generate CD8+ T cell clones representing all detectable TCR clonotypes in all persons, but in one subject we were able to test 5 of 6 clonotypes in vivo, representing 90% of the detectable TCR diversity in that subject. Moreover, all three controllers evaluated at the clonal level had dominant TCR clonotypes that were highly effective, whereas these were absent in both dominant and subdominant clones established in the progressors. Of note, the important role of TCR clonotypes in distinguishing virus control from lack of control likely was mediated by direct cytotoxicity of HIV-1-infected cells, with perforin upregulation noted within 30 minutes of cognate epitope recogntion and not requiring proliferation of CD8+ T cells. The data presented here contrast with the well documented differential proliferative capacity of CD8+ T cells between nonprogressors and progressors 12, likely due to the way in which proliferation was measured. In the current study, we used virally infected GXR cells expressing HLA-B*2705 as stimulator cells in culture with bulk CD8+ T cells and observed comparable proliferative capacity of HLA-B*27 KK10-specific CD8+ T cells between the controllers and progressors. This could be explained by the comparable level of CD4+ T cells (p = 0.4206) in the progrssors and controllers, properties of T regulatory cells in persons expressing protective HLA alleles 17, or cytokines produced by the cell line used for stimulation compensating for in vivo impaired CD4+ T cell helper function in progressors. Finally, whether these results can be extrapolated to other protective alleles and other epitopes will require additional study.
Taken together, our data indicate that TCR usage modulates viral inhibitory capacity and recognition of naturally arising HIV-1 variants, and thus modulates the effect of protective HLA alleles. The data suggest that TCR clonotypes that inhibit viral replication and confer cross-recognition of viral epitope variants that can eventually arise in vivo may be critical to long term control of viremia. Efforts to define the factors that contribute to junctional rearrangement of more effective TCR may be of critical importance for T cell vaccine design and therapeutic strategies for highly variable pathogens like HIV-1.
Materials and Methods
Study subjects
PBMC and plasma samples from HIV-1-infected individuals and HIV-1 negative persons were used for this study according to protocols approved by the Institutional Review Board of the Massachusetts General Hospital. Elite controllers were defined as having HIV-1 RNA below the level of detection for the respective available ultrasensitive assay (e.g., < 75 RNA copies/ml by bDNA or < 50 copies by ultrasenstive PCR) without antiretroviral therapy. Treatment-naive chronic progressors in the study had a median virus load of 12,833 copies/ml (4073 - 22,094 copies/ml). CD4+ T cell counts, viral loads and HLA types were determined as described 26. Characteristics of the study subjects are shown in Table 1.
Viruses and synthetic peptides
The CXCR4-utilizing HIV-1 laboratory strain NL4-3 was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (Bethesda, Maryland, USA). HIV-1 laboratory strain NL4-3 was also modified to express one or more mutations in Gag p24 as previously described 38,45. Peptides corresponding to described optimal HIV-1 epitopes and their variants (http://hiv-web.lanl.gov) were synthesized at the MGH Peptide Core Facility on an automated peptide synthesizer using F-moc technology.
Virus sequencing
Nested PCR for viral DNA or RNA was performed as previously described 46. PCR fragments were population sequenced to identify regions of sequence variation. All fragments were sequenced bi-directionally on an ABI 3730×l automated sequencer (Applied Biosystems, Foster City, CA).
ELISPOT assay
IFN-γ enzyme-linked immunospot (ELISPOT) assays were performed as described, using optimally defined epitopes and designated concentrations of peptide 8. Input cells ranged from 10,000 to 100,000 per well. To calculate the number of specific spot-forming cells (SFC), the number of spots in the negative control wells was subtracted from the number of spots in each experimental well. Responses were regarded as positive if they had at least 3 times the mean number of SFC in the 3 negative control wells; positive responses also had to be at least 50 SFC/106 PBMCs. The magnitude of epitope-specific response was calculated as SFC per million cells.
Generation of CD8+ T cell clones
PBMC were stained with fluorophore-labeled HLA tetramer refolded with epitopic HIV-1 peptides (ProImmune, Oxford, UK) and fluorophore-labeled anti-CD8 and anti-CD3 antibodies. Tetramer-positive, CD8-positive cells were sorted on a FACS Aria cell-sorting instrument (BD Biosciences) at 70 pounds per square inch (PSI) and single cells were placed into each well of 96-well plates, using irradiated allogeneic PBMC and CD3-specific mAb 12F6 as a stimulus for T cell proliferation 47. Developing epitope-specific clones were further tested in IFN-γ ELISPOT assays with optimal epitopes and with tetramer staining. Cloned CD8+ T cells were maintained by restimulation every 14 to 21 days with an anti-CD3 mAb and irradiated allogeneic PBMC in RPMI 1640 medium containing 50 U/ml of recombinant IL-2, as described 47.
TCR α and β chain sequencing
Tetramer-positive CD8-positive cells were sorted from PBMC or cloned CD8+ T cells and mRNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA). Anchored RT-PCR was then performed using a modified version of the SMART (switching mechanism at 5′ end of RNA transcript) procedure and a TCR α or β chain constant region 3′-primer to obtain PCR products containing the Vα or Vβ chain in addition to the CDR3 region, the Jα/β region and the beginning of the Cα/β region. RT-PCR and TCR α and β chain gene sequencing and analysis were performed as described previously 48.
Flow cytometry
Cell staining was performed as previously described 9. Briefly, cells were stained with indicated tetramer (ProImmune, Oxford, UK) for 20 minutes at room temperature. Following one wash with PBS containing 1% FCS, the cells were stained with surface antibodies. After 30 minutes at room temperature, the cells were washed and fixed using the Cytofix/Cytoperm kit (BD PharMingen) according to the manufacturer’s instructions. Following fixation, the cells were washed twice in the perm wash buffer and stained with antibodies against intracellular markers. Following staining, the cells were re-suspended in PBS containing 2% paraformaldehyde. The cells were acquired on a LSRII cytometer (BD Biosciences). Flow data were analyzed with the FlowJo software package (Treestar, Ashland, OR).
Proliferation assay
Primary CD8+ T cells were isolated from PBMC by negative selection (Dynabeads, Invitrogen) with the proportion of CD3+ CD8+ T cells >98% detected by flow cytometry. Cells were stained with 0.35 μM carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Breda, Netherlands) for 7 min at 37°C and then cultured with medium alone or with HIV-1 infected or uninfected HLA-B*27-encoding CEM-derived GXR cells for 7 days in RPMI 1640 medium in the absence of IL-2. After labeling with indicated tetramer (ProImmune, Oxford, UK), anti-CD8 and anti-CD3 antibodies, cells were fixed in 1% paraformaldehyde and analyzed on an LSRII flow cytometer (BD Biosciences).
Chromium release assay
HLA-B*27- or HLA-B*57-expressing green fluorescent protein (GFP) reporter CEM-derived GXR cells were constructed as described elsewhere 37,38 and infected with the designated HIV-1 strains or viral variants at the specified multiplicity of infection (MOI). On day 5 after infection, viable virally infected cells (which contain a plasmid encoding GFP driven by the HIV-1 long terminal repeat) were sorted on a FACS Aria cell-sorting instrument (BD Biosciences) and labeled with chromium for 1 h at 37°C. Bulk CD8+ T cells isolated from PBMC by negative selection (Dynabeads, Invitrogen) or CD8+ T cell clones were then added at the indicated effector-target ratios, and a standard 4-h chromium release assay was performed as previously described 49. Percent specific lysis was calculated as [(mean experimental cpm – mean spontaneous cpm)/(mean maximum cpm – mean spontaneous cpm)] × 100. Spontaneous and maximum releases were determined by incubating the labeled target cells with medium alone or 2% Triton X-100, respectively.
Viral inhibition assay
HLA-B*27- or HLA-B*57-expressing GFP reporter CEM-derived GXR cells were infected with the designated HIV-1 strains and viral variants at the specified MOI for 4 h at 37°C, then washed and cocultured with bulk CD8+ T cells isolated from PBMC by negative selection (Dynabeads, Invitrogen) or CD8+ T cell clones at the indicated effector-target cell ratios. The ability to recognize HIV-1 and viral variants by CD8+ T cells was analyzed by flow cytometry to evaluate the proportion of GFP-positive cells over 7 days in culture. To additionally address the relative antiviral efficacy of epitope-specific CD8+ T cell responses we measured the ability of bulk CD8+ T cells and epitope-specific cell-depleted CD8+ T cells to inhibit virus replication in autologous primary CD4+ T cells by analysis of p24 production, as described elsewhere 35. Briefly, primary CD4+ T cells were isolated from PBMC by negative selection (Dynabeads, Invitrogen). Greater than 98% of these primary cells coexpressed CD3 and CD4 by flow cytometric analysis. These CD4+ T cells were stimulated with CD3-CD8-bispecific monoclonal antibody 50 and infected at day 3 with the designated HIV-1 isolates at a MOI of 0.1, except as otherwise specified, for 4 h at 37°C. Virally infected cells were then washed and incubated in the absence or presence of effector cells at an effector-to-target cell ratio of 1:1 in RPMI 1640 medium in addition of IL-2 at 50 U/ml. At regular intervals, the cultures were fed by removing and replacing one-half of the culture supernatant with fresh medium. The removed supernatant was cryopreserved for subsequent p24 antigen quantitation by ELISA (Dupont, Boston, MA). Virus inhibition was calculated as follows: % inhibition = 100 x [1 – ([% GFP+ cells with effectors]/[% GFP+ cells without effectors])].
Effector-target cell conjugation, granule loading and delivery
CD8+ T cell clones were cultured with HIV-1 infected or uninfected HLA-B*27-expressing GFP reporter GXR cells for 30 minutes in RPMI 1640 medium in the absence of IL-2. Perforin staining in CD8+ T cell clones was as described 40. Briefly, cells were fixed with freshly prepared 4% paraformaldehyde for 15-30 minutes at room temperature and washed with PBS for 3 times. Cells were permeabilized in 0.5% Triton-X100 (Sigma) and 10% normal donkey serum (NDS; Jackson Immunoresearch, West Grove, PA) in PBS for 30 minutes at room temperature. Cells were stained with anti perforin monoclonal antibody mouse IgG2b for 60 minutes at room temperature. The primary antibody was diluted with 0.05% Triton-X100 and 3% NDS in PBS (1:333 dilution). After three washes in PBS, cells were incubated for 1 hour at room temperature with appropriate secondary antibodies in 0.05% Triton-X100 and 3% NDS in PBS. All secondary antibodies used were Alexa Fluor dyes conjugated (1:1000 dilution). F-actin was stained with phalloidin-Alexa Fluor 647 (1:50 dilution). Confocal images were collected on a Zeiss LSM510 Meta Confocal microscope using a plan apochromat 63×1.4 oil immersion objective. Differential interference contrast (DIC) images were collected simultaneously with the fluorescent images. Multi-track acquisition mode was used to avoid crosstalk between the different fluorophores. Images were analyzed with Imaris software (Bitplane).
Statistical analyses
An unpaired t-test with Welch’s correction and Mann-Whitney tests were performed using GraphPad Prism version 4.0a. All tests were two-tailed and p-values of p < 0.05 were considered significant.
Supplementary Material
Supplementary Fig. 1. Differentiation phenotypes of KK10-specific CD8+ T cells. (a) KK10-specific cells identified using HLA class I-peptide complexes (blue) are superimposed on the whole CD8+ T cell population (red). Four different phenotypes are shown: CD27+CD45RA− phenotype are central memory (CM) cells; CD27-CD45RA-phenotype are effector memory (EM) cells; CD27−CD45RA+ phenotype are terminally differentiated effector memory (TDEM) cells; and CD27+CD45RA+ phenotype are naïve cells. Plots are gated on CD3+CD8+ cells. (b) No significant difference was observed in the phenotypic profile of KK10-specific CD8+ T cells between the controllers (n = 5) and progressors (n = 5). Statistical comparisons were made using the Mann-Whitney test.
Supplementary Fig. 2. Germline analysis of TCRB genes. Sequencing reads were analyzed to determine TCRBV, TCRBJ and TCRBD usage, CDR3 sequence and length, and the number of nucleotides within the CDR3 contributed to by germline TCRBV, TCRBJ and TCRBD as well as by random nucleotide insertions. TCRBV, TCRBJ and TCRBD usage were determined by aligning the reads against human TCRB sequences. A “germline-like” index was calculated by subtracting the number of random nucleotide insertions within the CDR3 from the total length of the CDR3 and then dividing the product by the total length of the CDR3. This was performed in order to obtain a normalized value which could then be used to compare the amount of CDR3 that originates from germline sequences. Statistical tests comparing the germline-like index, CDR3 length, and the percentage of the CDR3s from germline TCRBV, TCRBJ and TCRBD between the controller and progressor groups were performed using an unpaired t-test with Welch’s correction.
Supplementary Fig. 3. Differential neutralization of HIV-1 by KK10-specific clonotypes. The ability of KK10-specific clonotypes to neutralize NL4-3 wild-type and variant viruses was evaluated in HLA-B*2705-encoding GFP reporter GXR cells at an effector/target cell ratio of 1:1 after 7 days of culture. Virus inhibition was calculated as follows: % inhibition = 100 x [1 – ([% GFP+ cells with effectors]/[% GFP+ cells without effectors])].
Supplemental Fig. 4. Differential expression of GrB by TCR clonotypes. Expression of GrB by clonotypes after 3 days of culture with virally infected and uninfected HLA-B*2705-expressing GFP reporter GXR cells. Values indicate percentages of GrB secreting KK10-specific cells upon culture with virally infected target cells after subtracting background from effector cells incubated with uninfected target cells.
Supplemental Fig. 5. GXR cells do not express endogenous perforin. Confocal imaging of GXR cell alone (a) and KK10-specific clone cell alone (b) imaged at ~30 min after addition to the Poly-L-lysine-coated coverslips. Fixed and permeabilized cells were incubated with Ab to perforin and then incubated with Alexa Fluor 647-conjugated secondary Abs (Red). Cells were mounted using prolong Gold with DAPI (Invitrogen). The scale bars are 3.0 μm.
Supplemental Fig. 6. No differences in basal level of perforin between KK10-specific clones alone. Quantitative measurement of total fluorescence intensity of perforin by confocal microscopy was performed. The total fluorescence intensity of perforin per cell within clones (inhibitory clonotype S-C003 and non-inhibitory clonotype S-C007 from subject CTR203) was calculated. The clones imaged on the poly-L-lysine coated coverslips. Fixed and permeabilized cells were incubated with Ab to perforin and then incubated with Alexa Fluor 647-conjugated secondary Abs. The total fluorescence intensity of perforin per cell in S-C003 and S-C007 clones are 3714 ± 485.9 (n = 20) and 3682 ± 325.1 (n = 27), respectively. Error bars indicate the standard error of the mean (SEM).
ACKNOWLEDGMENTS
We thank all study participants for their contributions. This work was supported by the Harvard University Center for AIDS Research (5 P30 AI060354-04), grants from the Bill and Melinda Gates Foundation (B.D.W and D.C.D.), the Doris Duke Charitable Foundation (B.D.W.), the NIH (B.D.W. AI030914 and T.M.A. AI074415), the Howard Hughes Medical Institute (B.D.W.), the Mark and Lisa Schwartz Foundation (B.D.W.), the Intramural Research Program and the Office of AIDS Research of the NIH (D.C.D. and S.D.), the New Investigator Award from the Canadian Institutes for Health Research (Z.L.B.), and the Canada Research Chair in Viral Pathogenesis and Immunity (M.A.B.).
Footnotes
AUTHOR CONTRIBUTIONS
H.C. was responsible for the overall conduct of the study, under the supervision of B.D.W.; H.C., Z.M.N., and B.D.W. contributed to the experimental design; H.C., Z.M.N., L.C.P., J.W.F., S.D., T.M., Z.L.B., K.T.C. and J.S. performed the experiments; M.A.B., A.S. and T.M.A. constructed HIV-1 variants and GXR cell lines; D.L. and D.E.K. performed the imaging expriments; T.D.C. and X.G.Y. helped TCR sequencing; F.P., A.P. and I.T. provided clinical samples; H.C., Z.M.N., and B.D.W. wrote the paper, and all authors contributed to revisions.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Migueles SA, Connors M. Long-term Nonprogressive Disease Among Untreated HIV-Infected Individuals. JAMA: The Journal of the American Medical Association. 2010;304:194–201. doi: 10.1001/jama.2010.925. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG, Walker BD. Human Immunodeficiency Virus Controllers: Mechanisms of Durable Virus Control in the Absence of Antiretroviral Therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Kaslow RA, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 4.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. PNAS. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrington M, O’Brien SJ. The Influence of HLA Genotype on AIDS*. Annual Review of Medicine. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 6.Pereyra F, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betts MR, et al. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75:11983–91. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Addo MM, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–92. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almeida JR, et al. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351–60. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett MS, Ng HL, Dagarag M, Ali A, Yang OO. Epitope-Dependent Avidity Thresholds for Cytotoxic T-Lymphocyte Clearance of Virus-Infected Cells. J. Virol. 2007;81:4973–4980. doi: 10.1128/JVI.02362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Migueles SA, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–8. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 13.Migueles SA, et al. Lytic Granule Loading of CD8+ T Cells Is Required for HIV-Infected Cell Elimination Associated with Immune Control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hersperger AR, et al. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 2010;6:e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahirel V, et al. Coordinate linkage of HIV evolution reveals regions of immunological vulnerability. Proceedings of the National Academy of Sciences. 2011;108:11530–11535. doi: 10.1073/pnas.1105315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiepiela P, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 17.Elahi S, et al. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med. 2011;17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufmann DE, et al. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat Immunol advanced online publication. 2007 doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- 19.Day CL, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–4. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 20.Perez CL, et al. Broadly Immunogenic HLA Class I Supertype-Restricted Elite CTL Epitopes Recognized in a Diverse Population Infected with Different HIV-1 Subtypes. The Journal of Immunology. 2008;180:5092–5100. doi: 10.4049/jimmunol.180.7.5092. [DOI] [PubMed] [Google Scholar]
- 21.Miura T, et al. HLA-B57/B*5801 Human Immunodeficiency Virus Type 1 Elite Controllers Select for Rare Gag Variants Associated with Reduced Viral Replication Capacity and Strong Cytotoxic T-Lymphotye Recognition. J. Virol. 2009;83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassen KG, et al. Elite Suppressor Derived HIV-1 Envelope Glycoproteins Exhibit Reduced Entry Efficiency and Kinetics. PLoS Pathog. 2009;5:e1000377. doi: 10.1371/journal.ppat.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulder PJ, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–7. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 24.Lee K-H, et al. The Immunological Synapse Balances T Cell Receptor Signaling and Degradation. Science. 2003;302:1218–1222. doi: 10.1126/science.1086507. [DOI] [PubMed] [Google Scholar]
- 25.Kosmrlj A, et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereyra F, et al. Genetic and Immunologic Heterogeneity among Persons Who Control HIV Infection in the Absence of Therapy. The Journal of Infectious Diseases. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 27.Goulder PJ, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–8. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 28.Almeida JR, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. The Journal of Experimental Medicine. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendoza D, et al. HLA B*5701+ Long-Term Nonprogressors/Elite Controllers are not Distinguished from Progressors by the Clonal Composition of HIV-Specific CD8+ T-Cells. J Virol. 2012 doi: 10.1128/JVI.06982-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Bockel DJ, et al. Persistent Survival of Prevalent Clonotypes within an Immunodominant HIV Gag-Specific CD8+ T Cell Response. The Journal of Immunology. 2011;186:359–371. doi: 10.4049/jimmunol.1001807. [DOI] [PubMed] [Google Scholar]
- 31.Iglesias MC, et al. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood. 2011;118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Day CL, et al. Proliferative capacity of epitope-specific CD8 T-cell responses is inversely related to viral load in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:434–8. doi: 10.1128/JVI.01754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Champagne P, et al. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410:106–11. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 34.Hamann D.r., et al. Phenotypic and Functional Separation of Memory and Effector Human CD8+ T Cells. The Journal of Experimental Medicine. 1997;186:1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, et al. Differential Neutralization of Human Immunodeficiency Virus (HIV) Replication in Autologous CD4 T Cells by HIV-Specific Cytotoxic T Lymphocytes. J. Virol. 2009;83:3138–3149. doi: 10.1128/JVI.02073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Julg B, et al. Infrequent Recovery of HIV from but Robust Exogenous Infection of Activated CD4+ T Cells in HIV Elite Controllers. Clinical Infectious Diseases. 2010;51:233–238. doi: 10.1086/653677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brockman MA, Tanzi GO, Walker BD, Allen TM. Use of a novel GFP reporter cell line to examine replication capacity of CXCR4- and CCR5-tropic HIV-1 by flow cytometry. J Virol Methods. 2006;131:134–42. doi: 10.1016/j.jviromet.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Schneidewind A, et al. Structural and Functional Constraints Limit Options for Cytotoxic T-Lymphocyte Escape in the Immunodominant HLA-B27-Restricted Epitope in Human Immunodeficiency Virus Type 1 Capsid. J. Virol. 2008;82:5594–5605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89:375–87. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 40.Liu D, et al. Integrin-dependent organization and bidirectional vesicular traffic at cytotoxic immune synapses. Immunity. 2009;31:99–109. doi: 10.1016/j.immuni.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart-Jones GBE, et al. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. European Journal of Immunology. 2005;35:341–351. doi: 10.1002/eji.200425724. [DOI] [PubMed] [Google Scholar]
- 42.Price DA, et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. The Journal of Experimental Medicine. 2009;206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong T, et al. HIV-specific cytotoxic T cells from long-term survivors select a unique T cell receptor. J Exp Med. 2004;200:1547–57. doi: 10.1084/jem.20032044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venturi V, et al. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J Immunol. 2011;186:4285–94. doi: 10.4049/jimmunol.1003898. [DOI] [PubMed] [Google Scholar]
- 45.Miura T, et al. Genetic characterization of human immunodeficiency virus type 1 in elite controllers: lack of gross genetic defects or common amino acid changes. J Virol. 2008;82:8422–30. doi: 10.1128/JVI.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen TM, et al. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol. 2004;78:7069–78. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker BD, et al. Long-term culture and fine specificity of human cytotoxic T-lymphocyte clones reactive with human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989;86:9514–8. doi: 10.1073/pnas.86.23.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varadarajan N, et al. A high-throughput single-cell analysis of human CD8+ T cell functions reveals discordance for cytokine secretion and cytolysis. The Journal of Clinical Investigation. 2011;121:4322–31. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang OO, et al. Impacts of avidity and specificity on the antiviral efficiency of HIV-1-specific CTL. J Immunol. 2003;171:3718–24. doi: 10.4049/jimmunol.171.7.3718. [DOI] [PubMed] [Google Scholar]
- 50.Wilson CC, et al. Ex vivo expansion of CD4 lymphocytes from human immunodeficiency virus type 1-infected persons in the presence of combination antiretroviral agents. J Infect Dis. 1995;172:88–96. doi: 10.1093/infdis/172.1.88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Differentiation phenotypes of KK10-specific CD8+ T cells. (a) KK10-specific cells identified using HLA class I-peptide complexes (blue) are superimposed on the whole CD8+ T cell population (red). Four different phenotypes are shown: CD27+CD45RA− phenotype are central memory (CM) cells; CD27-CD45RA-phenotype are effector memory (EM) cells; CD27−CD45RA+ phenotype are terminally differentiated effector memory (TDEM) cells; and CD27+CD45RA+ phenotype are naïve cells. Plots are gated on CD3+CD8+ cells. (b) No significant difference was observed in the phenotypic profile of KK10-specific CD8+ T cells between the controllers (n = 5) and progressors (n = 5). Statistical comparisons were made using the Mann-Whitney test.
Supplementary Fig. 2. Germline analysis of TCRB genes. Sequencing reads were analyzed to determine TCRBV, TCRBJ and TCRBD usage, CDR3 sequence and length, and the number of nucleotides within the CDR3 contributed to by germline TCRBV, TCRBJ and TCRBD as well as by random nucleotide insertions. TCRBV, TCRBJ and TCRBD usage were determined by aligning the reads against human TCRB sequences. A “germline-like” index was calculated by subtracting the number of random nucleotide insertions within the CDR3 from the total length of the CDR3 and then dividing the product by the total length of the CDR3. This was performed in order to obtain a normalized value which could then be used to compare the amount of CDR3 that originates from germline sequences. Statistical tests comparing the germline-like index, CDR3 length, and the percentage of the CDR3s from germline TCRBV, TCRBJ and TCRBD between the controller and progressor groups were performed using an unpaired t-test with Welch’s correction.
Supplementary Fig. 3. Differential neutralization of HIV-1 by KK10-specific clonotypes. The ability of KK10-specific clonotypes to neutralize NL4-3 wild-type and variant viruses was evaluated in HLA-B*2705-encoding GFP reporter GXR cells at an effector/target cell ratio of 1:1 after 7 days of culture. Virus inhibition was calculated as follows: % inhibition = 100 x [1 – ([% GFP+ cells with effectors]/[% GFP+ cells without effectors])].
Supplemental Fig. 4. Differential expression of GrB by TCR clonotypes. Expression of GrB by clonotypes after 3 days of culture with virally infected and uninfected HLA-B*2705-expressing GFP reporter GXR cells. Values indicate percentages of GrB secreting KK10-specific cells upon culture with virally infected target cells after subtracting background from effector cells incubated with uninfected target cells.
Supplemental Fig. 5. GXR cells do not express endogenous perforin. Confocal imaging of GXR cell alone (a) and KK10-specific clone cell alone (b) imaged at ~30 min after addition to the Poly-L-lysine-coated coverslips. Fixed and permeabilized cells were incubated with Ab to perforin and then incubated with Alexa Fluor 647-conjugated secondary Abs (Red). Cells were mounted using prolong Gold with DAPI (Invitrogen). The scale bars are 3.0 μm.
Supplemental Fig. 6. No differences in basal level of perforin between KK10-specific clones alone. Quantitative measurement of total fluorescence intensity of perforin by confocal microscopy was performed. The total fluorescence intensity of perforin per cell within clones (inhibitory clonotype S-C003 and non-inhibitory clonotype S-C007 from subject CTR203) was calculated. The clones imaged on the poly-L-lysine coated coverslips. Fixed and permeabilized cells were incubated with Ab to perforin and then incubated with Alexa Fluor 647-conjugated secondary Abs. The total fluorescence intensity of perforin per cell in S-C003 and S-C007 clones are 3714 ± 485.9 (n = 20) and 3682 ± 325.1 (n = 27), respectively. Error bars indicate the standard error of the mean (SEM).