Summary
The human pathogen Staphyloccocus aureus requires cell wall anchored surface proteins to cause disease. During cell division, surface proteins with YSIRK signal peptides are secreted into the cross wall, a layer of newly synthesized peptidoglycan between separating daughter cells. The molecular determinants for the trafficking of surface proteins are, however, still unknown. We screened mutants with non-redundant transposon insertions by fluorescence-activated cell sorting for reduced deposition of protein A (SpA) into the staphylococcal envelope. Three mutants, each of which harbored transposon insertions in genes for transmembrane proteins, displayed greatly reduced envelope abundance of SpA and surface proteins with YSIRK signal peptides. Characterization of the corresponding mutations identified three transmembrane proteins with abortive infectivity (ABI) domains, elements first described in lactococci for their role in phage exclusion. Mutations in genes for ABI domain proteins, designated spdA, spdB and spdC (surface protein display), diminish the expression of surface proteins with YSIRK signal peptides, but not of precursor proteins with conventional signal peptides. spdA, spdB and spdC mutants display an increase in the thickness of cross walls and in the relative abundance of staphylococci with cross walls, suggesting that spd mutations may represent a possible link between staphylococcal cell division and protein secretion.
Introduction
Staphylococcus aureus is the single most frequent cause of infectious disease mortality in the United States (Klevens et al., 2008). The manifestations of staphylococcal diseases vary greatly, ranging from minor skin infections to life threatening pneumonia, septicemia, and endocarditis (Lowy, 1998). Over the past thirty years, S. aureus strains have acquired resistance to many different antibiotics, most notably β-lactam compounds (MRSA, methicillin-resistant S. aureus)(DeLeo & Chambers, 2009). In the United States, MRSA strains are isolated in more than 60% of nosocomial infections and are also a frequent cause of community-acquired disease (DeLeo et al., 2010). Therapeutic strategies for MRSA bacteremia are based on vancomycin, the antibiotic of last resort (Lowy, 2003). Nevertheless, even for vancomycin-sensitive MRSA isolates, case fatalities approach 50% (Klevens et al., 2007). The recent emergence of vancomycin-resistant MRSA strains has been viewed as testimony for the possible return to a pre-antibiotic era. More specifically, it documents the pressing need of identifying new targets for S. aureus drug therapy (Weigel et al., 2003).
A central virulence strategy of S. aureus is its ability to secrete proteins that interact with the host (Morfeldt et al., 1988, Recsei et al., 1986). These secreted polypeptides are either released into the extracellular milieu or anchored within the bacterial cell wall envelope, a pathway that requires the activity of sortases (Novick, 2003, Marraffini et al., 2006). Sortase A is responsible for anchoring 19 substrates in S. aureus strain Newman through the recognition of a conserved LPXTG cell wall sorting signal (Mazmanian et al., 1999, Mazmanian et al., 2002). Sortase A-mediated cleavage of sorting signals immediately following the conserved threonine (T) results in the formation of amide bonds between the C-terminal end of the polypeptide chain and the pentaglycine crossbridge of lipid II (Ton-That et al., 1999, Ton-That et al., 2000, Perry et al., 2002). Transglycosylation and transpeptidation reactions lead to the incorporation of surface proteins via their lipid II moiety into the staphylococcal cell wall (Ton-That & Schneewind, 1999, Ton-That et al., 1998). Sortase A is required for staphylococcal virulence in all disease models examined thus far (Mazmanian et al., 2000, Weiss et al., 2004, Jonsson et al., 2002, Cheng et al., 2009, Kim et al., 2010). These defects stem from the phenotype that srtA variants are unable to anchor any one of 19-21 surface proteins in the bacterial envelope (Mazmanian et al., 2000). Among the many functions of sortase A-anchored proteins are hemeiron uptake (Mazmanian et al., 2003, Skaar et al., 2004), binding to host extracellular matrix components (Patti et al., 1994) or evasion of host innate immune responses (Silverman & Goodyear, 2006, Foster, 2005, Thammavongsa et al., 2009).
The distribution of one surface molecule, protein A (SpA), in the bacterial envelope has been investigated in detail. SpA is assembled in a ring-like structure encircling the spherical cells (DeDent et al., 2007). Its distribution is uneven, however, with increased abundance of SpA at cell division sites, i.e. the location where cross walls are formed (DeDent et al., 2007). Staphylococci divide perpendicular to their previous cell division plane and, at least during logarithmic growth, appear to be born unit size (Tzagoloff & Novick, 1977, Giesbrecht et al., 1998). The latter implies that peptidoglycan synthesis at the cross wall is ultimately responsible for the bulk of bacterial envelope expansion. Once completed, cross walls are split down the middle, enabling the separation of daughter cells and the display of 50% of the cell wall envelope on the staphylococcal surface. SpA is among a family of surface proteins that possess a conserved YSIRK-G/S sequence within signal peptides that direct this subset of surface proteins to the cross-wall of dividing staphylococci (Bae & Schneewind, 2003, DeDent et al., 2008). Streptococcus pyogenes M protein, which also harbors YSIRK-G/S within its signal peptide, is deposited at a similar site (Carlsson et al., 2006). Conversely, conventional signal peptide bearing surface proteins are deposited at the cell poles - SasF and PrtF in S. aureus and S. pyogenes, respectively (Carlsson et al., 2006, DeDent et al., 2008). While mutations directly to the conserved YSIRK-G/S sequence did not appear to affect the site of protein deposition, motif containing signal peptides are essential for the delivery of substrates to their final destination. For example, when the signal peptide of SasF (non-YSIRK) is replaced with that of ClfA (+YSIRK), SasF is now detected at the cross-wall of staphylococci (DeDent et al., 2008). Additionally, it has been demonstrated that mutations to the YSIRK-G/S sequence negatively affect the rate of signal peptide processing (Bae & Schneewind, 2003). Despite the critical importance of surface proteins for staphylococcal disease, very little is known about the factors and mechanisms of their trafficking. Through genome sequencing and in silico predictions, staphylococcal homologues of Escherichia coli or Bacillus subtilis secretion factors have been identified (e.g. SecYEG)(Sibbald et al., 2006). There are no obvious differences between known secretion genes for these genomes that can explain why staphylococci deposit some surface proteins at the cross wall, whereas bacilli apparently do not.
Here we identified three genes encoding transmembrane proteins with ABI (abortive infectivity) domains, designated spdA, spdB and spdC (for surface protein display), that are required for the abundant deposition of SpA at the cross wall. Mutations in each of these genes reduces the abundance of surface protein mRNA and the expression of the corresponding gene products. This phenotype is restricted to surface proteins with YSIRK-G/S signal peptides, and does not apply to surface proteins secreted via canonical signal peptides. Mutants with defects in ABI domain proteins accumulate cross walls, suggesting that the reduced expression of surface proteins may result from associated defects in cell division and/or separation.
Results
Screening staphylococcal mutants for defects in the display of protein A
Five immunoglobulin binding domains of staphylococcal protein A (SpA) capture the Fcγ portion of IgG on the staphylococcal surface (Jensen, 1958, Forsgren, 1968). Exploiting Cy5-labled IgG to measure the surface display of SpA, 1,996 non-redundant insertional mutants of S. aureus Newman (Bae et al., 2004) were analyzed by FACS for defects in surface protein trafficking. To account for non-specific cell lysis and release of SpA, only live staphylococci were gated for FACS interpretation. As a control, a mutant with an insertion in the sortase A gene (srtA), which encodes the transpeptidase responsible for linking surface proteins to the cell wall (Mazmanian et al., 2000), did not display SpA on the bacterial surface (data not shown). In addition to srtA, we identified three mutants with a four-fold or greater reduction in the surface display of SpA. These mutants harbored bursa aurealis insertions in nwmn1671, nwmn2215 and nwmn2236. Each of the three genes encodes for a transmembrane protein with a unique functional domain, termed abortive infectivity (ABI). Interestingly, the mutant with a transposon insertion in nwmn2236 had previously been described as being important for lysostaphin resistance (LyrA, lysostaphin resistance A)(Gründling et al., 2006). Lysostaphin is a glycyl-glycine endopeptidase that cleaves the pentaglycine crossbridges of the staphylococcal peptidoglycan (Schindler & Schuhardt, 1964, Browder et al., 1965). Of note, the synthesis of pentaglycine crossbridges in lyrA mutant strains is not affected by the transposon insertion (Gründling et al., 2006). Through sequence analysis of the S. aureus Newman genome (Baba et al., 2007), we identified a fourth ABI-containing gene, nwmn1939. A mutant with a transposon insertion in nwmn1939 was not affected for the surface display of SpA. Of note, transposon insertions with spdA, spdB, and spdC did not result in a growth defect or cell aggregation (Fig. S1, data not shown).
SpA surface display is diminished in spdABC mutants
The insertional lesions of the bursa aurealis mini-transposon in four genes encoding for ABI domain proteins (nwmn1671, nwmn1939, nwmn2215, and nwmn2236) were transduced via bacteriophage infection from the mutant strains into the chromosome of the wild-type parent, S. aureus Newman (Bae et al., 2006). Transductants were propagated on erythromycin containing tryptic soy agar and the mutational lesions in spdA (nwmn1671), spdB (nwmn2215), spdC (nwmn2236 or lyrA) as well as nwmn1939 were verified by DNA sequencing. To measure the surface display of SpA, staphylococci were stained with Cy5-labeled IgG and subjected to FACS analysis. Compared to S. aureus Newman, the spdA, spdB and spdC mutants displayed a four fold or greater reduction of Cy5-IgG fluorescence (Fig. 1B). The spd phenotype was not observed for transductants of the nwmn1939 lesion. SEJ2, a spa deletion variant (DeDent et al., 2008), was used as a negative control and did not generate surface fluorescence when incubated with Cy5 labeled IgG (Fig. 1B).
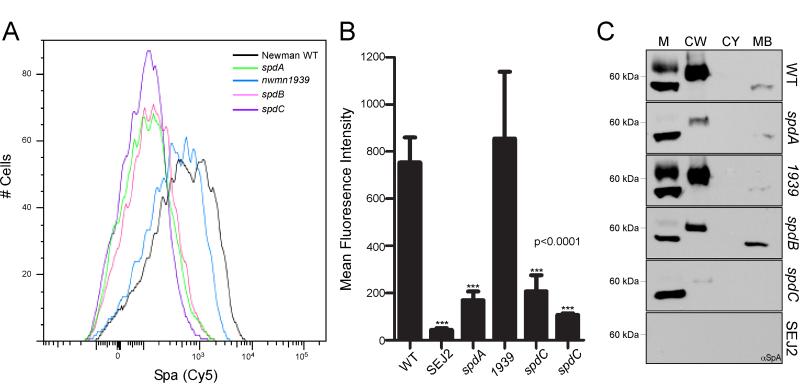
Figure 1. Mutations in Staphylococcus aureus spd genes affect the display of protein A on the surface of staphylococci.
(A) Wild-type S. aureus Newman and the indicated mutant strains were stained with Cy5 conjugated rabbit IgG and analyzed by FACS for the surface expression of SpA. A representative histogram is shown from two independent experiments performed in triplicate.
(B) The mean fluorescence intensity from experiments performed in A is plotted. Only single cell, live staphylococci are interrogated for these analyses. Error bars represent SEM. Statistical significance (P values) was analyzed with the student’s t-test.
(C) Wild-type S. aureus Newman, the spd mutants (surface protein display), and SEJ2 (Δspa) variants were fractionated into media (M), cell wall (CW), cytosol (CY) and membrane (MB) and immunoblotted for SpA to investigate its cellular distribution.
Protein A is synthesized in the bacterial cytoplasm as the P1 precursor with an N-terminal signal peptide and a C-terminal sorting signal (Schneewind et al., 1992). Upon initiation of P1 into the secretory pathway, the signal peptide is cleaved (Schneewind et al., 1992). The resulting P2 intermediate is recognized by sortase to cut the sorting signal and establish the amide bond between the C-terminal threonine and the pentaglycine crossbridge of lipid II (Schneewind et al., 1993, Navarre & Schneewind, 1994, Schneewind et al., 1995, Ton-That et al., 1997). The product of this reaction, designated the P3 precursor (Perry et al., 2002), is subsequently incorporated into cell wall peptidoglycan to generate the mature, anchored surface protein (Ton-That & Schneewind, 1999). To determine whether mutational lesions in spd genes caused SpA precursors to be missorted along this pathway, staphylococci were first fractionated into subcellular compartments and the distribution of protein A analyzed by immunoblotting (Fig. 1C). Briefly, cultures were centrifuged to sediment staphylococci. Proteins secreted into the culture medium (supernatant) were precipitated. The cell wall envelope of staphylococci was digested with lysostaphin. Resulting protoplasts were sedimented by centrifugation and cell wall proteins (supernatant) were precipitated. Following protoplast lysis, membranes were sedimented by ultracentrifugation and separated from the cytoplasm. Proteins in all cellular compartments were precipitated with TCA, washed in acetone and analyzed by immunoblot using monoclonal antibodies. As controls, mature protein A is found in the cell wall compartment of wild-type staphylococci, with small amounts of precursors detectable in the membrane fraction (Fig. 1C). During cell division, staphylococci release significant amounts of cell wall peptidoglycan into the culture medium, some of it linked to protein A. As expected, SpA immunoreactive signals could not be detected in the spa deletion variant SEJ2 (Fig. 1C). Mutants with insertions in spdA, spdB or spdC displayed the expected decrease in the abundance of protein A in the cell wall fraction (Fig. 1C). Nevertheless, the subcellular distribution of SpA was not significantly affected by the spdA, spdB, and spdC mutations (Fig. 1C). For example, we did not observe an abundance of SpA within the cytosolic fraction in the Spd mutant strains, indicative of a defect in the secretion of SpA precursors. Of note, the spdB mutant displayed an increased abundance of SpA in the membrane fraction, while the spdC mutant resulted in elevated amounts of SpA found in the media (Fig. 1C). Additionally, we do not observe the higher molecular weight band found in the media fraction from wild-type and nwmn1939 strains within the Spd mutant samples. This could indicate an alteration to the peptidoglycan profile of the mutant bacteria, which release only the faster migrating species of SpA into the media. Previous work was unable to discern such a difference between wild-type and spdC bacteria (Gründling et al., 2006). Neither the abundance of SpA nor its subcellular distribution was affected by the bursa aurealis insertion in nwmn1939 (Fig. 1C).
In addition to the analysis by immunoblot of the ABI mutants, we queried whether the deposition of SpA at the cell surface was affected. For example, it may be possible that SpA is secreted to the cross-wall of the mutant bacteria yet is unable to be distributed throughout the cell surface. Using both immuno-fluorescence and immuno-electron microscopy, a marked decrease in SpA signal intensity was observed for samples derived from spd mutant staphylococci that prevented us from identifying the subcellular location of protein A in mutant cells (Fig. 3BC, data not shown).
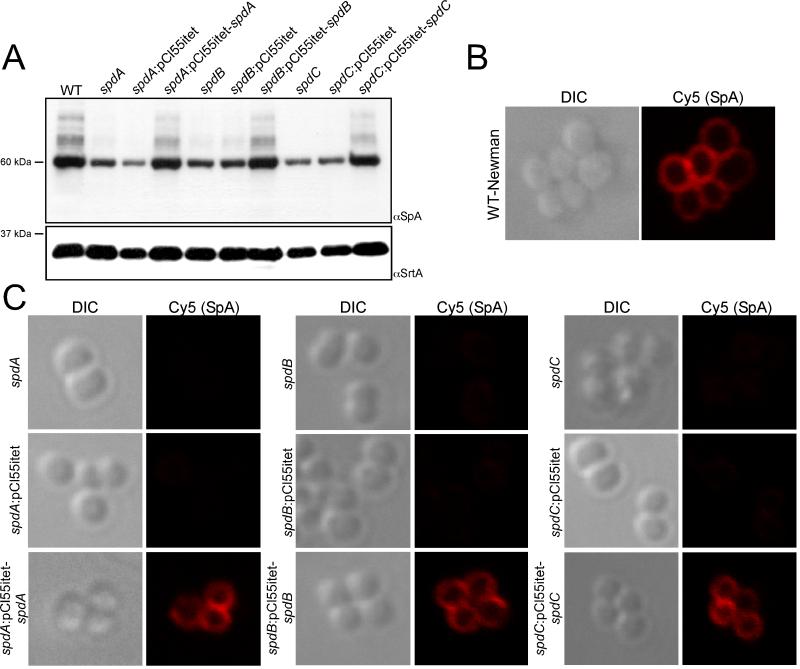
Figure 3. Complementation analysis of spd mutants.
(A) Cell wall extracts from the indicated strains were subjected to immunoblot to assess the abundance of SpA as well as sortase A (SrtA, loading control).
(B) Wild-type S. aureus Newman (WT) was grown to mid logarithmic phase, then fixed and stained with Cy5 conjugated rabbit IgG. Samples were analyzed by immunofluorescence for the presence of SpA at the cell surface. The left panel is the differential interference contrast (DIC) image and the fluoresent micrograph on the right panel.
(C) spd mutants with either pCL55-itet (vector control) or with their complementation vectors were stained and imaged as in B.
Domain architecture of ABI-domain containing S. aureus proteins
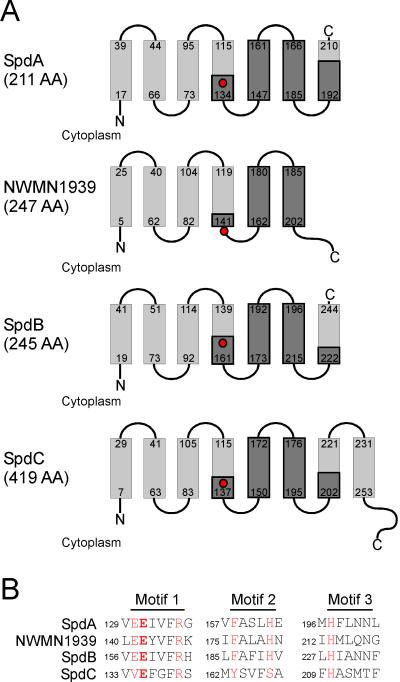
ABI domain containing transmembrane proteins were first characterized in lactococcal species for their role in the exclusion of bacteriophages (Chopin et al., 2005). Although 20 different ABI genes (abiA-U) are now identified, there is currently no mechanistic appreciation for their function in preventing phage infections (Chopin et al., 2005). Moreover, proteins with ABI domains have been found in both prokaryotic and eukaryotic organisms. Within eukaryotes, the ABI domain is part of the CAAX type II prenyl endopeptidase family (Pei & Grishin, 2001). CAAX proteases are located in the membrane of the endoplasmic reticulum and remove the C-terminal end of proteins that have been prenylated at the cysteine (C) residue of their CAAX motif (Pei & Grishin, 2001). This cleavage of prenylated proteins is critical for their subsequent targeting to membranes, as has been characterized for the Ras GTPase (Boyartchuk et al., 1997, Kim et al., 1999). ABI domains encompass three motifs. Motif 1 possesses a semi-variant glutamic acid and arginine. A second glutamic acid is predicted to be the catalytic center of these proteases, as a substitution of this residue abolishes function in RCE1, a yeast CAAX protease (Dolence et al., 2000). Motif 2 includes a conserved phenylalanine and histidine separated by three variable amino acids (Pei & Grishin, 2001). Finally, motif 3 consists of a structural element with a single conserved histidine residue. Although this has not yet been substantiated by crystallographic or NMR studies, the glutamic acid residue within motif 1 together with the histine residues of motifs 2 and 3 are thought to coordinate a catalytically essential zinc atom in the active site (Pei & Grishin, 2001). With the exception of SpdC, ABI-containing transmembrane proteins of S. aureus harbor all of the conserved residues listed above (Fig. 2AB); SpdC encodes for valine rather than the first glutamic acid of motif 1 and tyrosine and serine in the place of the conserved phenylalanine and histidine residues, respectively, of motif 2. However, the staphylococcal genome does not encode for geranyl-geranyl and farensyl protein transferases or proteins with a C-terminal CAAX box (Baba et al., 2007). The translation product of nwmn1939 differs from that of spdA, spdB, and spdC in that the conserved glutamic acid residue of motif 1 is situated in a loop between the predicted transmembrane segments four and five, but not within a hydrophobic transmembrane helix (Fig. 2A). This positional difference may explain the varying phenotypes between spdA, spdB, spdC, and nwmn1939.
Figure 2. spd mutations identify ABI-domain containing proteins of S. aureus.
(A) The architecture of all four ABI-domain containing proteins in S. aureus. Transmembrane spanning regions are indicated by boxes with the corresponding amino acids numbered. The conserved functional motifs are highlighted in dark grey with the predicted catalytic glutamic acid (E) indicated with a red circle.
(B) Amino acid alignment of the conserved residues from the three motifs of all ABI-domain containing S. aureus proteins. In red are residues with high sequence conservation among type II CAAX proteases, with the catalytic glutamic acid (E) residue in bold (motif 1). The glutamic acid of motif 1, histidines (H) of motif 2 and 3 are predicted to coordinate zinc.
Complementation analysis of spd mutants
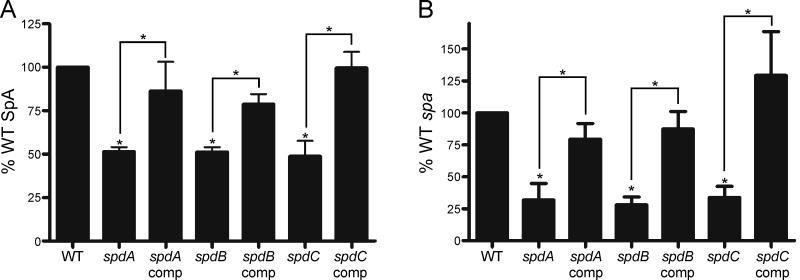
In addition to the transposon mutations of spdA, spdB, and spdC that are described above, we evaluated the phenotype of ten other bursa aurealis insertions in the same genes: one mutation each within spdA (NMTN 6341) and spdC (NMTN 9166), as well as eight within spdB (NMTN 10183, 11440, 6248, 6318, 9178, 4299, 3658, and 9064)(Bae et al., 2004). As before, transposons were transduced into wild-type strain Newman and assayed for SpA abundance by immunoblot. All ten mutations in spdA, spdB or spdC caused similar decreases in the abundance of SpA (data not shown). To further confirm the role of ABI-containing genes in the proper deposition of SpA on the cell surface, complementation analyses were performed. Each gene (spdA, spdB and spdC) was cloned into a vector for chromosomal integration and expressed from a constitutive promoter (Lee et al., 1991, Gründling & Schneewind, 2007). Complementation strains were then analyzed for the surface display of protein A using immunofluorescence microscopy and immunoblotting (Fig. 3). In all cases examined, expression of wild-type spd genes from chromosomal integration vectors in spdA, spdB or spdC mutant strains restored the surface display of SpA as well as the overall abundance of protein A to wild-type levels (Fig. 3ABC).
Protein A precursor processing in spd mutants
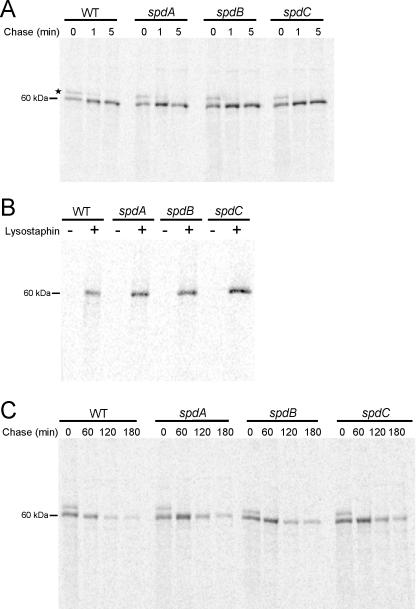
In order to ascertain the defect of SpA display in the spd mutants, we utilized radiolabeling to monitor the path of protein A secretion. To examine the rates of protein A signal peptide and sorting signal processing, staphylococcal proteins were pulse-labeled for 1 minute with [35S]methionine and [35S]cysteine. During the pulse (0 minute) or at 1 and 5 minute intervals following the addition of the chase (excess of unlabeled methionine/cysteine), aliquots of the culture were treated with TCA to quench surface protein processing within the sorting pathway. The staphylococcal envelope was removed with lysostaphin and released protein A was immunoprecipitated prior to SDS-PAGE and autoradiography. The P1 precursor (signal peptide and cell wall sorting signal present) is cleaved very rapidly to generate mature anchored surface protein, and this processing occurs at a speed that does not allow for the detection of the P2 precursor. The rate of precursor processing in spdA, spdB and spdC mutants was similar to that observed in wild-type staphylococci (Fig. 4A).
Figure 4. Pulse-chase analysis of surface protein processing in spd mutants.
(A) Wild-type and spd mutant staphylococci were examined for defects in signal peptide and sorting signal processing through pulse-chase. Samples were incubated with [35S]methionine/cysteine followed by the addition of excess non-labeled amino acids (chase). Aliquots of the reaction were harvested at the indicated times (time 0 is during the pulse). Proteins were immunoprecipitated with polyclonal SpA rabbit antisera, resolved by SDS-PAGE, and autoradiographed. Asterisk indicates the P1 precursor of SpA evident at time 0.
(B) spd mutants were tested for their ability to anchor surface proteins. The same strains as in A were briefly incubated with [35S]methionine/cysteine followed by a five minute chase. Samples were then split into two and incubated in the presence (+) or absence (-) of lysostaphin. Protein was immunoprecipitated and analyzed as in A.
(C) The stability/turnover of SpA was measured using an identical experiment as described in A, except allowing extended chase periods and processed as described above.
Discrete defects in surface protein anchoring can be detected by boiling samples in SDS. Without prior lysostaphin treatment of the staphylococcal cell wall, hot SDS cannot release anchored protein A molecules. In contrast, SDS treatment does solubilize protein A in mutants with defects in cell wall anchoring (Schneewind et al., 1992). Therefore, we examined the solubility of SpA in the presence or absence of lysotaphin treatment. As in wild-type staphylococci, hot SDS failed to solubilize protein A from the envelope of spdA, spdB and spdC mutants (Fig. 4B). Additionally, cell fractionation experiments revealed that pulse labeled protein A was found in the cell wall envelope, but not in the extracellular media (data not shown).
To examine the stability of SpA among wild-type and mutant bacteria, we subjected each strain to a brief pulse with [35S]methionine/cysteine followed by an extended chase over 180 minutes, analyzing cells in 60 minute intervals. Similar to previous experiments, SpA was immunoprecipitated and subjected to autoradiography. Compared to wild-type staphylococci, an increase in the degradation of SpA was not detected in spdA, spdB or spdC mutant cells (Fig. 4C). A similar experiment was performed in which cells were either mock treated or the cell wall envelope digested with lysostaphin following extended chase periods to measure the rates at which SpA was released from the staphylococcal surface over time. Again, no apparent differences between wild-type, spdA, spdB and spdC mutant strains were encountered (data not shown). From these combined experiments we can conclude that mutations within spdA, spdB, and spdC do not result in defects in SpA signal peptide processing, anchoring, or stability.
The decrease in SpA display is caused by a defect in gene expression
To discern whether reduced surface display of protein A was due to a defect in expression, the rate of SpA synthesis was addressed by pulse labeling staphylococci with [35S]methionine/cysteine. Briefly, staphylococci were labeled for 1 minute, treated with TCA and their envelope disintegrated with lysostaphin. Immunoprecipitated SpA was quantified by autoradiography and compared to the total amount of radioactivity in all TCA precipitated proteins of each sample. Compared to wild-type staphylococci, spdA, spdB and spdC mutants displayed a two-fold reduction in the rate of protein A synthesis (Fig. 5A). This defect was restored to at or near wild-type levels in spd complemented strains (Fig. 5A). As a control, similar rates of SpA synthesis were measured for wild-type and the nwmn1939 mutant cells (data not shown).
Figure 5. spd mutations affect the surface display of protein A by decreasing the abundance of its transcript.
(A) Wild-type (WT), spd mutant, and spd complemented staphylococci were subjected to [35S]methionine/cysteine labeling to determine the rate of SpA synthesis. Samples were pulsed for 60 seconds and immediately plunged into ice-cold TCA acid to quench cellular activity. SpA was immunoprecipitated and analyzed as in Figure 4. Samples were normalized to WT according to the total amount of radiolabeled proteins prior to immunoprecipitation of SpA. Band intensities of SpA were then compared to WT. Data is the composite of four independent experiments (three for complemented strains). Error bars represent SEM. Statistical significance was analyzed with the student’s t-test with a p-value of <0.01.
(B) The abundance of spa transcript was determined by quantitative PCR of cDNA derived from total cellular RNA from wild-type (WT) and each spd mutant with the corresponding complemented strain. RNA was normalized using 16s rRNA transcript abundance. The abundance of spa transcript from each mutant was compared to wild-type staphylococci. Data are the composite results of six independent experimental trials (three for complemented strains). Error bars represent SEM. Statistical significance was analyzed with the student’s t-test with a p-value of <0.01.
We next queried whether the reduced rate of SpA synthesis is caused by the diminished abundance of its mRNA within spd mutants. spa transcript was analyzed by quantitative PCR from cDNA that had been derived from the total RNA of each strain and was normalized compared to 16S ribosomal RNA abundance. We observed a four-fold reduction in spa transcript levels determined for spdA, spdB and spdC mutant cells as compared to the wild-type control (Fig. 5B). Again, spa levels were restored to at or near wild-type levels in the spd complemented strains (Fig. 5B). From these data, we presume that the reduced display of SpA on the staphylococcal surface is at least in part caused by the diminished abundance of its transcript in spd mutant strains.
spd mutations affect the display of surface proteins with YSIRK-G/S signal peptides
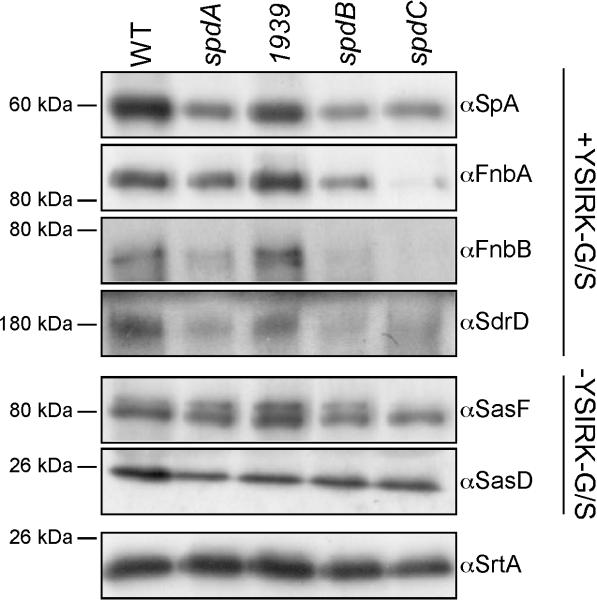
Previous work established that staphylococcal surface proteins are deposited at two distinct cellular locations, depending on the presence of a conserved YSIRK-G/S sequence within the signal peptide (DeDent et al., 2007, DeDent et al., 2008). SpA is among the 13 YSIRK-G/S containing surface proteins encoded in the genome of S. aureus Newman (DeDent et al., 2008). These substrates are delivered to the cross-wall of dividing staphylococci, whereas non-YSIRK signal peptide substrates are sent to the cell poles (DeDent et al., 2008). In addition to our analysis of SpA at the cell surface, we examined the effects of spd mutations on other surface proteins. Cell wall extracts of wild-type and spd mutant staphylococci were isolated and investigated by immunoblot. As shown in Figure 6, a consistent marked decrease in the abundance of the YSIRK/G-S signal peptide proteins SpA, FnbA, FnbB, and SdrD was observed in spdA, spdB and spdC mutants, but not in wild-type staphylococci or nwmn1939. In contrast, the surface display of SasF and SasD, which are endowed with a conventional signal peptide, were not significantly reduced in spd mutants (Fig. 6). Again, protein levels were restored to wild-type levels in complemented strains (data not shown). Together these data suggest a role for SpdA, SpdB and SpdC proteins in the display of proteins secreted via YSIRK-G/S signal peptides.
Figure 6. spd mutations affect the display of surface proteins with YSIRK-G/S signal peptides.
Cell wall extracts derived from wild-type (WT) or spd mutant staphylococci were subjected to immunoblotting with antibodies specific for SpA, FnbA, FnbB, SdrD, SasF, SasD, and SrtA. The presence or absence of a YSIRK-G/S motif in the signal peptides of these gene products is indicated. SrtA serves as a loading control.
Given the importance of spdA, spdB, and spdC with respect to YSIRK-G/S bearing precursors, we investigated the cellular distribution of the translated products using the fluorescent protein, mCherry. We created translational hybrids at the C-terminus of each Spd protein and queried its localization by immunofluorescence. Using BODIPY-Vancomycin to illuminate the cross-wall of the bacteria, we observed that only SpdC showed strong association with the cross-wall of dividing staphylococci (Fig. S2). SpdA and SpdB displayed staining throughout the cell membrane with some staining along the cross-wall, but with less intensity (Fig. S2). Of note, these data are derived from hybrids of truncated Spd proteins that are expressed from a multicopy vector. Future work is needed to determine precise location of SpdA, SpdB and SpdC in staphylococcal cells, which may be achieved by generated specific antibodies directed against each protein.
Spd mutants display an increase in the abundance and thickness of cross-walls
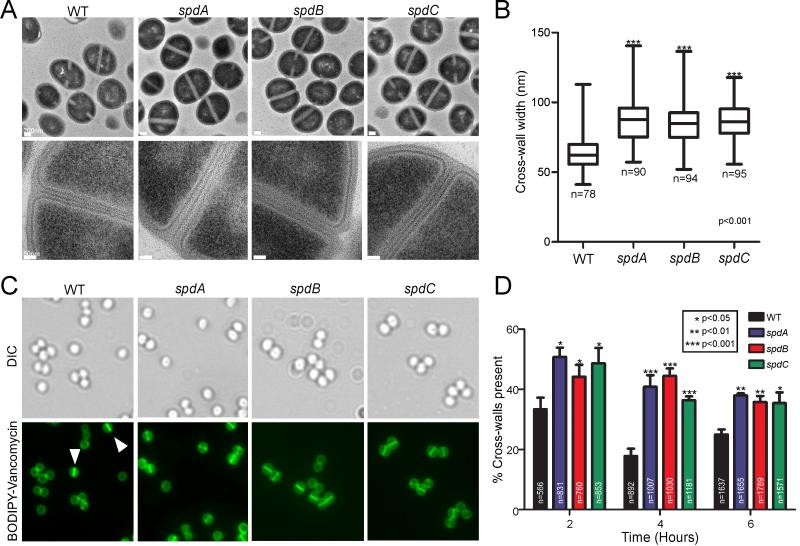
We next investigated whether the phenotype of reduced surface protein display in spd mutants could be explained by defects in cell division or separation. These processes involve the cross wall of staphylococci, which is also the site of deposition for surface protein precursors with YSIRK/G-S motifs. To address this, wild-type and mutant staphylococcal cells were examined via transmission electron microscopy using uranyl acetate stained thin-sectioned samples (Fig. 7A). As compared to wild-type staphylococci, spdA, spdB and spdC mutants displayed two discrete phenotypes. First, the diameter of cross walls in dividing cells appeared to be increased and, second, many more staphylococcal cells harbored cross walls, i.e. they were in the process of cell division (Fig. 7AC). To quantify these observations, we determined the thickness of cross-walls between strains by measuring the diameter of this structure in at least 75 dividing cells of each strain (Fig. 7B). In contrast to wild-type staphylococci where cross walls have a mean diameter of 63.6 nm, the diameter of the same structure in spdA, spdB and spdC mutants was determined as 87.5 nm, 83.9 nm, and 86.3 nm, respectively (Fig. 7B). We observed a decrease in the thickness of the cross-wall in spd complemented bacteria to wild-type levels (Fig. S3A).
Figure 7. Architecture of the cross-wall in wild-type and spd mutant staphylococci.
(A) Staphylococci were grown to mid-logarithmic growth and processed for electron microscopy as described in Materials and Methods. Top panels show dividing staphylococci and the bottom panel is an enlarged representative image of the cross-wall from each strain. Scale bars for the top panels represent 200 nm, for the bottom panels 50 nm.
(B) The width of the cross-walls from each strain was determined from the indicated number of bacteria (n) of each strain from images derived by electron microscopic analysis. The non-parametric Anova (Kruskal-Wallis) test and the Dunn’s multiple comparison test were used to assess significance of these observations.
(C) Staphylococci were grown as described in panel A, fixed and stained with BODIPY-vancomycin to discriminate the cross-wall of dividing bacteria. In each case, the cross walls are the most intensely stained sections of the cell wall envelope (arrowheads). The top panels are DIC micrographs and bottom panels represent the corresponding fluorescent microscopy images.
(D) Overnight cultures of wild-type and spd mutant staphylococci were diluted into fresh media and aliquots removed after 2, 4, and 6 hours of incubation. Cells were fixed and stained as in C. The percent amount of cross-walls was quantified and compared to the total number of cells per field of view. The number of cells counted is listed in each bar (n). Error bars represent the SEM. Statistical significance was examined with the student’s t-test (P values), comparing wild-type with spd mutant staphylococci at each time interval.
To measure the abundance of cross-walls formed by each strain, staphylococci were stained with BODIPY-vancomycin (DeDent et al., 2007). Vancomycin binds to D-Ala-D-Ala residues within peptidoglycan, which occurs with greater intensity at the cross-wall of S. aureus due to the increased synthesis of peptidoglycan (Walsh, 1993). Overnight cultures were subcultured in fresh media and cells were harvested at various times, stained with BODIPY-vancomycin, and viewed by fluorescence microscopy (Fig. 7C). By comparing the number of cross-walls formed relative to the total number of cells present, at two hours after dilution of cultures more than 50% of the spdA, spdB and spdC mutants were found to harbor cross walls as compared to 35% of wild-type cells (Fig. 7CD). Different stages of staphylococcal growth are associated with different amounts of cross walls. For example, four hours after the dilution of cultures, 40.8%, 44.4% and 36.4% of spdA, spdB, and spdC mutants harbored cross walls, compared to 17.9% for wild-type staphylococci (Fig. 7D). Again, restoration of the affected gene in each spd mutant significantly reduced the abundance of cross-walls (Fig. S3B). Additionally, we did not observe an overall increase in the thickness of the peptidoglycan cell wall in the Spd mutants relative to wild-type bacteria (Fig. S4). Together these data suggest that spd mutations impact the formation and separation of cross walls and may thereby also impact the secretion of surface proteins with YSIRK/G-S signal peptides into this subcellular structure.
Discussion
The cross wall of S. aureus and of other staphylococcal species is a subcellular structure that initially arises from cytokinesis, i.e. FtsZ-mediated separation of two daughter cells (Giesbrecht et al., 1998). The compartment between two newly formed cytoplasmic membranes becomes the site for de novo cell wall synthesis and assembly of proteins into the peptidoglycan scaffold, which is designated as the cross wall (Cole & Hahn, 1962). Earlier work showed that precursor proteins with YSIRK/G-S signal peptides traffic to the cross wall (Carlsson et al., 2006). In addition to sortase-anchored surface proteins, this includes lipases as well as the peptidoglycan hydrolase, LytN, an enzyme that may be responsible for cleaving the completed cross wall and separating the two daughter cells (Bae & Schneewind, 2003, Rosenstein & Gotz, 2000). In addition to protein secretion into the cross wall compartment, membrane proteins traffic to this site and contribute to the assembly of newly formed cell wall envelope. This includes peptidoglycan transpeptidases, transglycosylases, sortase A and many other proteins (Pinho & Errington, 2005, Pereira et al., 2007, Zapun et al., 2008, Raz & Fischetti, 2008). The molecular determinants responsible for directing these membrane proteins to the cross wall are still unknown; future work will need to map the composition of cell division and cell wall synthesis factors to these sites. Importantly, mutations that perturb factors contributing to cross wall formation and separation would be expected to alter its shape and construction period in relation to the staphylococcal cell division cycle. Finally, mutations that entirely abrogate the function of essential cross wall factors very likely cannot be isolated, as they would altogether abolish bacterial growth.
Here we sought to identify factors that contribute to the trafficking of surface proteins with YSIRK/G-S signal peptides. Using FACS analysis to monitor the abundance of protein A on the surface of staphylococci, we identified mutants that displayed four-fold or less of this surface protein. Three of these spd mutants (surface protein display) harbored a transposon insertion in membrane proteins that together belong to the family of proteins with ABI domains. The best characterized of these are the CAAX proteases of eukaryotic cells, which remove the C-terminal residues of polypeptides post-translationally modified with either geranyl-geranyl or farnesyl moieties (Reiss et al., 1990, Seabra et al., 1992). Prokaryotic organisms do not express proteins with a C-terminal CAAX box, i.e. the recognition motif for protein prenylation, and also lack the geranyl-geranyl or farensyl protein transferases of eukaryotic cells. While ABI domain proteins have been characterized as factors involved in lactococcal bacteriophage replication, the precise molecular mechanisms of action remain unknown (Chopin et al., 2005). S. aureus encodes four ABI domain proteins and mutations in three of these - spdA, spdB and spdC – affect the abundance of protein A and other YSIRK/G-S surface proteins as well as the abundance and the shape of cross walls. Mutations in the fourth ABI protein encoded by nwmn1939 do not trigger these phenotypes. Given the wide distribution of ABI motifs in prokaryotic and eukaryotic organisms, we do not observe any direct link of ABI proteins and its link to the secretion of YSIRK-G/S containing substrates.
The experiments in this report cannot distinguish between two broad possibilities. On one hand, SpdA, SpdB and SpdC may be directly involved in the trafficking of surface proteins with YSIRK/G-S signal peptides, for example by processing either catalytic factors or secretion substrates themselves. In this scenario, the defect in the secretion of proteins into the cross wall compartment may trigger changes in the shape and in the abundance of cells with cross walls. On the other hand, SpdA, SpdB and SpdC may contribute to cell division and mutations in these genes may slow the processes that lead to the formation of cross walls, thereby causing a secondary effect on the trafficking of surface proteins. Whatever mechanisms may underwrite the observed phenotypic defects of spd mutations, their consequence is the reduced expression of surface proteins, which was detected as the reduced abundance of transcript and translational products. Considering their primary membrane protein structure and putative protease activities, it seems unlikely that SpdA, SpdB and SpdC are directly involved in transcription or translation. Rather, the observed effect on surface protein expression may be indirect as occurs during the feedback inhibition of outer membrane proteins in Gram-negative bacteria with defects in protein assembly into the outer membrane surface organelle (Raivio & Silhavy, 1999). It is conceivable that Spd proteins act to cleave a transcription factor which in turn activates YSIRK-containing gene expression. Future work is required to elucidate the mechanism of action of these unique Spd proteins.
ABI proteins have been described as an altruistic bacterial module in which viral-infected cells cease cellular activity to prevent bacteriophage replication and subsequent further infection of the community (Chopin et al., 2005). These systems have also been implicated in self-immunity to bacteriocin with similarities to toxin-antitoxin modules (Kjos et al., 2010). It is conceivable that the Spd proteins in S. aureus may act to modify the cell surface of the bacterium to prevent infection by bacteriophages or serve to protect themselves from antimicrobials. Preliminary studies did not reveal significant differences in the sensitivity to Φ85 infection between wild-type and spd variants (data not shown). Mutations within spdC affect the resistance to lysostaphin, which led to the previous identification of this gene as lyrA (Gründling et al., 2006). However, this phenotype was not shared by the other three genes encoding ABI transmembrane proteins (data not shown). Together with the spd phenotypes that occur in three of the four genes, these observations suggest that ABI domain proteins may act independently of each other or fulfill only partial redundancy in function. Further investigations into the mechanisms by which Spd proteins control protein secretion will provide needed information about the function of this ubiquitous, yet largely uncharacterized family of proteins.
Experimental Procedures
Bacterial strains and culture conditions
S. aureus strains used were derived from strain Newman (Duthie & Lorenz, 1952). Plasmid DNA was first electroporated into RN4220 (Kreiswirth et al., 1983), isolated, and subsequently electroporated into strain Newman. All transposon mutants were transduced via Φ85 into Newman and SEJ2 (SpA deletion)(Bae et al., 2006). S. aureus was propagated in tryptic soy broth (TSB) or on tryptic soy agar plates with the appropriate antibiotics. Erythromycin and chloramphenicol were used at a 10 μg/mL concentration. Ampicillin was used for plasmid selection in E. coli at 100 μg/mL. E. coli was grown in LB broth or on agar plates containing antibiotics.
Primers and plasmids
To construct complementation vectors, the following primers were used:
spdA: 5′-gatcAGATCTtcaaactaatgctttttaggtaggtacaatg-3′ and 5′- gatcCCGCGGttatctccatattataaagagcagagctaacaag-3′ (BglII and SacII sites in uppercase, respectively).
spdB: 5′-gatcAGATCTgttccaatctgtcattaaagtattatatttaatcg-3′ and 5′- gatcCCGCGGctacgcataaatacttattactgatataacgaaaga-3′ (BglII and SacII sites in uppercase, respectively).
spdC: 5′-ggCCTAGGataaatttggttcaccttgcttgtacac-3′ and 5′-gatcCCGCGG gaggtactagcaagcgctttgttatta -3′ (AvrII and SacII sites in uppercase, respectively).
PCR products were amplified using Accuprime pfx (Invitrogen) from Newman genomic DNA, purified, and digested with the indicated restriction enzymes (NEB). The vector, pCL55-iTET (Gründling & Schneewind, 2007), was cut with either BglII and SacII (for spdA and B) or AvrII and SacII (spdC) and ligated with the digested PCR products. Transformants were analyzed by DNA sequencing to confirm correct sequence and subsequently transformed into S. aureus RN4220. Transformants were then transduced into the appropriate mutant strain and confirmed by PCR.
For mCherry hybrids to Spd proteins, we utilized SOE (splicing by overlapping extension) PCR to amplify spdA, B, and C, and mCherry with homologous extensions for two rounds of PCR amplification. All constructs contain a C-terminal truncation of the Spd protein fused to mCherry (C-terminal hybrid; depicted in Fig. S2). Constructs were designed to maintain mCherry within the bacterial cytosol while also including all transmembrane segments of Spd proteins. For spdA, primers 5′- gatcAGATCTtcaaactaatgctttttaggtaggtacaatg-3′ and 5′- ctcgcccttgctttttatataaattaggccaaaaatcaaacc-3′ were used. mCherry was amplified using primers 5′-atttatataaaaagcaagggcgaggaggataacatgg-3′ and 5′-gatcCCGCGGttacttgtacagctcgtccatgcc-3′. This yielded amino acids 1-186 of SpdA, which spans all transmembrane segments directly fused to mCherry with the first two amino acids absent. The SOE product contained a BglII (5′) and SacII (3′) restriction site.
For spdB, primers 5′-gatcAGATCTgttccaatctgtcattaaagtattatatttaatcg-3′ and 5′- ctcgcccttgctACCTAGGTTTCTTCCAGATTTTAAATAAGT-3′ were used. mCherry was amplified using primers 5′- AGAAACCTAGGTagcaagggcgaggaggataacatgg-3′ and 5′- gatcCCGCGGttacttgtacagctcgtccatgcc-3′. This yielded amino acids 1-222 of SpdB, which spans all transmembrane segments directly fused to mCherry with the first two amino acids absent. The SOE product contained a BglII and SacII restriction site.
For spdC, primers 5′-ggCCTAGGataaatttggttcaccttgcttgtacac-3′ and 5′- ctcgcccttgctTCGGATAATTAAGCTTAAACCAATG-3′ were used. mCherry was amplified using primers 5′- TTAATTATCCGAagcaagggcgaggaggataacatgg-3′ and 5′- gatcCCGCGGttacttgtacagctcgtccatgcc-3′. This yielded amino acids 1-253 of SpdC, which spans all transmembrane segments directly fused to mCherry with the first two amino acids absent. The SOE product contained an AvrII and SacII restriction site.
SOE PCR products were purified using the QIAquick PCR purification kit (Qiagen) and digested with the appropriate restriction enzymes and ligated with pMF312 digested with identical enzymes. Transformants were analyzed by DNA sequencing to confirm correct sequence and subsequently transformed into S. aureus RN4220. Transformants were then grown, plasmid DNA extracted, and electroporated into S. aureus strain Newman. Expression was then determined by immunofluorescence. pMF312 is a derivative of pOS1 (Schneewind et al., 1992) harboring the tetracycline inducible promoter, iTET, which was amplified from pCL55-iTET using primers 5′- GGGAATTCGAGCTCGGTACCTTGGTTA-3′ and 5′- CGCGTCGACGCATGCCTTAATTAAcgatGCGGCCGCTTCCCGCGGGTTTAAACAGATCTCCT AGGTCATTTGATATGCCTCCTC-3′. This added a NotI, PacI, and SphI restriction site to the multiple cloning site. The PCR product was ligated into pOS1 at the SmaI site. Expression from pMF312 is constitutive due to incomplete inhibition of transcription in the absence of anhydrotetracycline.
FACS, immunofluorescence, and electron microscopy
For FACS analysis, bacteria were grown with appropriate antibiotics to mid-logarithmic phase and added to a 96 well plate and centrifuged. Bacterial pellets were washed with PBS and fixed in 2.5% paraformaldehyde, 0.006% gluteraldehyde, 30 mM NaPO4 pH 7.4 for 20 minutes. Cells were then pelleted, washed with PBS, and blocked in 3% BSA in PBS. Staining was performed with Cy5 conjugated rabbit IgG (Jackson Immuno Research) at a 1:500 dilution and incubated for one hour in the dark. Cells were then washed three times with PBS and stained with Syto9 and propidium iodide (Invitrogen) to differentiate live versus dead cells. Cells were washed 3× with PBS and suspended in 4% formalin. Finally, cells were added to 500 μl of PBS and then subjected to analysis using the LSRII (BD Biosciences). Data were collected by first gating on staphylococci, followed by gating on Syto9 positive, propidium iodide negative cells. This population was then analyzed for the abundance of Cy5 reactive signal. Data are presented as either a representative histogram from two independent experiments performed in triplicate or from the average of the mean fluorescence intensity accumulated from two independent experiments performed in triplicate.
For SpA analysis by immunofluorescence, cells were grown to mid-logarithmic phase, centrifuged, washed, fixed, and blocked as described above. Cells were incubated with Cy-5 conjugated rabbit IgG for one hour, followed by washing in PBS. Cells were then applied to poly-lysine treated glass coverslips and subsequently mounted onto glass coverslips containing a drop of SlowFade anti-fading reagent (Invitrogen). Images were captured on a Leica TCS SP2 AOBS laser scanning confocal microscope with 100× oil objective using identical settings and exposure times.
Vancomycin staining was performed as described previously (DeDent et al., 2007). Slides were viewed on an Olympus AX-70 fluorescence microscope, and images were captured with a charge-coupled-device (CCD) camera. Total cells were counted and compared with the number of visible cross-walls formed through BODIPY-vancomycin staining. The number of cells counted per time point of each strain is listed within the figure. All experiments are representative of at least two independent trials. For electron microscopy, cells were grown as described above; staphylococci were sedimented and prepared as described previously (Gründling & Schneewind, 2007).
RNA isolation, cDNA synthesis, and quantitative PCR
Total cellular RNA was isolated from bacteria harvested during mid-logarithmic phase using TRIzol® reagent (Invitrogen). Cells were normalized by OD600 readings, sedimented and immediately placed on ice. Staphylococci were then incubated in TSM (50 mM Tris-HCl pH 7.5, 0.5 M sucrose, 10 mM MgCl2) containing 200 μg of lysostaphin (AMBI Products) at 4°C for 20 minutes. Cells were then centrifuged at 16,000 ×g for 2 minutes and suspended in 1 mL of TRIzol, following manufacturer’s protocol. Total RNA was then DNase treated (Invitrogen) and subjected to reverse transcription using the iScript™ cDNA synthesis kit (Biorad). cDNA was then diluted 10-fold and quantitative PCR was performed using SsoFast™ Evagreen® Supermix (Biorad) on a BioRad CFX96™ real-time PCR system according to manufacturer recommendations. To amplify spa primers 5′-GCGCAACGTAACGGCTTCATTC-3′ and 5′AGCCGTTACGTTGTTCTTCAGTTA-3′ were used. 16S rRNA was used as the housekeeping gene with which to normalize samples and primers 5′- ACGCCGTAAACGATGAGTGCTAAGTGT-3′ and 5′-GTGGTGTGACGGGCGGTGTG-3″ were used. Negative controls in which no reverse transcriptase was added to the cDNA synthesis reaction were performed for each sample set. Data collected for wild-type and Spd mutants are the composite results of 6 independent experimental trials, while complementing strains are the composite result of 3 independent experimental trials.
Cell fractionation and western immunoblot
For fractionation, exponentially growing staphylococci were first normalized according to their OD600 reading and 1 ml of bacterial suspension were centrifuged at 16,000 ×g for 5 minutes. The supernatants were removed (media fraction) and precipitated with trichloroacetic acid (TCA). The staphylococcal sediment was suspended in 1 ml of TSM containing 50 μg/ml of lysostaphin and incubated at 37°C for 10 minutes. Following incubation, protoplasts were centrifuged at 16,000 ×g for 5 minutes and the supernatant (cell wall fraction) was precipitated with TCA. The protoplast sediment was suspended in 1ml of membrane buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl) and subjected to five freeze-thaw cycles in a dry ice and ethanol bath. Samples were subjected to ultracentrifugation at 100,000 ×g for 30 minutes at 8°C. The supernatant (cytosolic fraction) was separated from the sediment, which was suspended in membrane buffer; both fractions were then precipitated with TCA. Proteins in TCA precipitates were sedimented by centrifugation at 16,000 ×g for 10 minutes. The sediment was washed with ice-cold acetone, dried and proteins solubilized in sample buffer.
To isolate the cell wall fraction, cells were treated as previously described and the supernatant collected following lysostaphin treatment. Sortase A was used as a loading control for cell wall protein abundance.
For immunoblot analysis, proteins were separated on a 10% SDS-PAGE, electro-transferred to PVDF membrane (Millipore), and residual binding sites on the filter blocked with 5% milk in PBS-0.1% Tween 20. For SpA detection, monoclonal antibodies (SPA-27; Sigma) were used. SrtA, SasD, SasF, FnbA, FnbB, and SdrD were detected using rabbit polyclonal antibodies previously raised against recombinant proteins. Secondary antibodies were HRP-conjugated (Cell Signaling Technology) and visualized using an ECL solution.
Radioactive labeling and quantification
For radiolabeling experiments, staphylococci were grown to mid-logarithmic phase and normalized according to their optical densities, sedimented at 7000 ×g for 10 minutes, and washed twice in MM4 medium (Schneewind et al., 1992). Cells were suspended in 1 ml of MM4 media and briefly starved prior to addition of [35S]methionine/cysteine (Perkin Elmer) for 90 seconds at 37°C. For rate of synthesis experiments, cells were labeled for 60 seconds, and immediately plunged into ice cold 5% TCA to quench all metabolic activity. For all other analyses, an excess of non-labeled amino acids were added to pulse-labeled staphylococci; at variable incubation times surface protein processing was quenched via the addition of ice-cold 5% TCA. Cells were washed with acetone, dried, and suspended in 1 mL of 0.5 M Tris pH 7.5 containing 50 μl of lysostaphin (from a 2 mg/ml stock). For experiments examining sortase anchoring, cells were radio-labeled for 90 seconds followed by a 5 minute chase. Samples were then split into two, with one sample treated with lysostaphin and the other mock treated. Lysostaphin treatment was performed at 37°C for 60 minutes, followed by TCA precipitation and acetone washing. The sediments were suspended in 50 μl of 4% SDS, 0.5 M Tris-HCl pH 8.0 and allowed to incubate for 30 minutes prior to boiling.
Solubilized proteins were then added to 1 ml of RIPA buffer (0.1% SDS, 0.5% deoxycholic acid, 1% Triton X100, 50 mM Tris-HCl pH 8.0, 150 mM NaCl) containing 1μl of rabbit polyclonal anti-SpA antibodies (Schneewind et al., 1992) and incubated overnight at 4°C with rotation. Protein A sepharose (50 μl of a 50% slurry, Sigma) was then added to this mixture to capture antibody-antigen complexes and incubated for an additional hour followed by washing 5× with RIPA buffer. Beads were dried, protein loading buffer was added, and samples were boiled for 10 minutes. Proteins were separated by 10% SDS-PAGE, gels dried onto Whatman 3M paper, and subjected to autoradiography for 24-72 hours and read on a phosphorimager (Storm; GE Healthcare). For rate of synthesis experiments, 10% of the total radio-labeled proteins (prior to immunoprecipitation) were subjected to liquid scintillation counts to normalize radioactivity to that of wild-type amounts. This was then compared to the band intensity of SpA derived from the immunoprecipitation. Data on spd mutants are representative of four independent experimental trials; data on the complementation of spd mutant phenotypes are comprised of three independent experimental trials.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge stimulating discussion and criticism of this manuscript by their laboratory colleagues. In particular, we would like to thank Dr. Antoni Hendrickx for his assistance with transmission electron microscopy. We would also like to thank Dr. Jess Leber and Kierstyn Schwartz for technical assistance and use of equipment for quantitative PCR work. This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (NIAID), Infectious Diseases Brach AI38897. M.B.F. was supported by a post-doctoral fellowship award from the National Institute of Allergy and Infectious Diseases (F32AI085709). O.S. and D.M.M. acknowledge membership within and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (GLRCE, National Institute of Allergy and Infectious Diseases Award 1-U54-AI-057153).
References
- Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes. J. Bacteriol. 2007;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 2006;62:1035–1047. doi: 10.1111/j.1365-2958.2006.05441.x. [DOI] [PubMed] [Google Scholar]
- Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA. 2004;101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Schneewind O. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J. Bacteriol. 2003;185:2910–2919. doi: 10.1128/JB.185.9.2910-2919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- Browder HP, Zygmunt WA, Young JR, Tavormina PA. Lysostaphin: enzymatic mode of action. Biochem. Biophys. Res. Com. 1965;19:383–389. doi: 10.1016/0006-291x(65)90473-0. [DOI] [PubMed] [Google Scholar]
- Carlsson F, Stalhammar-Carlemalm M, Flardh K, Sandin C, Carlemalm E, Lindahl G. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442:943–946. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:1–12. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin MC, Chopin A, Bidenko E. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Cole RM, Hahn JJ. Cell wall replication in Streptococcus pyogenes. Science. 1962;135:722–724. doi: 10.1126/science.135.3505.722. [DOI] [PubMed] [Google Scholar]
- DeDent AC, McAdow M, Schneewind O. Distribution of protein A on the surface of Staphylococcus aureus. J. Bacteriol. 2007;189:4473–4484. doi: 10.1128/JB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDent AC, Missiakas DM, Schneewind O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 2008;27:2656–2668. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR, Chambers HF. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolence JM, Steward LE, Dolence EK, Wong DH, Poulter CD. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry. 2000;39:4096–4104. doi: 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- Duthie ES, Lorenz LL. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- Forsgren A. Protein A from Staphylococcus aureus. VI. Reaction with subunits from guinea pig gamma-1- and gamma-2-globulin. J. Immunol. 1968;100:927–930. [PubMed] [Google Scholar]
- Foster TJ. Immune evasion by staphylococci. Nat. Rev. Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 1998;62:1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling A, Missiakas DM, Schneewind O. Staphylococcus aureus mutants with increased lysostaphin resistance. J. Bacteriol. 2006;188:6286–6297. doi: 10.1128/JB.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Nat. Acad. Sci. USA. 2007;104:8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. A normally occuring staphylococcus antibody in human serum. Acta Path. Microbiol. Scandin. 1958;44:421–428. doi: 10.1111/j.1600-0463.2007.apm_731a.x. [DOI] [PubMed] [Google Scholar]
- Jonsson IM, Mazamanian SK, Schneewind O, Vendrengh M, Bremell T, Tarkowski A. On the role of Staphylococcus aureus sortase and sortase-catalyzed surface protein anchoring in murine septic arthritis. J. Infect. Dis. 2002;185:1417–1424. doi: 10.1086/340503. [DOI] [PubMed] [Google Scholar]
- Kim E, Ambroziak P, Otto JC, Taylor B, Ashby M, Shannon K, Casey PJ, Young SG. Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J. Biol. Chem. 1999;274:8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- Kim HK, DeDent A, Cheng AG, McAdow M, Bagnoli F, Missiakas DM, Schneewind O. Antibodies that interfere with Staphylococcus aureus heme-iron transport protect mice against abscess formation and lethal challenge. Vaccine. 2010 doi: 10.1016/j.vaccine.2010.02.097. EPub: ahead of press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. The abi proteins and their involvement in bacteriocin self-immunity. J. Bacteriol. 2010;192:2068–2076. doi: 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens RM, Edwards JR, Gaynes RP, System NNIS. The impact of antimicrobial-resistant, health care-associated infections on mortality in the United States. Clin. Infect. Dis. 2008;47:927–930. doi: 10.1086/591698. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, A. B. C. s. A. M. Investigators Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Lee CY, Buranen SL, Ye Z-H. Construction of single-copy integration vectors for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- Lowy FD. Staphylococcus aureus infections. New Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 2003;111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, DeDent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase enzyme anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E, Janzon L, Arvidson S, Löfdahl S. Cloning of a chromosomal locus (exp) which regulates the expression of several exoprotein genes in Staphylococcus aureus Mol. Gen. Genet. 1988;211:435–440. doi: 10.1007/BF00425697. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- Patti JM, Allen BL, McGavin MJ, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 1994;48:89–115. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 2001;26:275–277. doi: 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- Pereira SFF, AHenriques AO, Pinho MG, de Lencastre H, Tomasz A. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 2007;189:3525–3531. doi: 10.1128/JB.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- Pinho MG, Errington J. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 2005;55:799–807. doi: 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- Raivio TL, Silhavy TJ. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 1999;2:159–165. doi: 10.1016/S1369-5274(99)80028-9. [DOI] [PubMed] [Google Scholar]
- Raz A, Fischetti VA. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc. Natl. Acad. Sci. USA. 2008;105:18549–18554. doi: 10.1073/pnas.0808301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick R. Regulation of exoprotein gene expression by agr. Mol. Gen. Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- Reiss Y, Goldstein JL, Seabra MC, Casey PJ, Brown MS. Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell. 1990;62:81–88. doi: 10.1016/0092-8674(90)90242-7. [DOI] [PubMed] [Google Scholar]
- Rosenstein R, Gotz F. Staphylococcal lipases:biochemical and molecular characterization. Biochimie. 2000;82:1005–1014. doi: 10.1016/s0300-9084(00)01180-9. [DOI] [PubMed] [Google Scholar]
- Schindler CA, Schuhardt VT. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. USA. 1964;51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Fowler A, Faull KF. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface protein of Gram-positive bacteria. EMBO. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- Seabra MC, Brown MS, Slaughter CA, Sudhof TC, Goldstein JL. Purification of component A of Rab geranylgeranyl transferase: possible identity with the choroideremia gene product. Cell. 1992;70:1049–1057. doi: 10.1016/0092-8674(92)90253-9. [DOI] [PubMed] [Google Scholar]
- Sibbald MJ, Ziebandt AK, Engelmann S, Hecker M, de Jong A, Harmsen HJ, Raangs GC, Stokroos I, Arends JP, Dubois JY, van Dijl JM. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 2006;70:755–788. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman GJ, Goodyear CS. Confounding B-cell defences: lessons from a staphylococcal superantigen. Nat. Rev. Immunol. 2006;6:465–475. doi: 10.1038/nri1853. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J. Exp. Med. 2009;206:2417–2427. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Faull KF, Schneewind O. Anchor structure of staphylococcal surface proteins. I. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 1997;272:22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Labischinski H, Berger-Bächi B, Schneewind O. Anchor structure of staphyococcal surface proteins. III. The role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall. J. Biol. Chem. 1998;273:29143–29149. doi: 10.1074/jbc.273.44.29143. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Mazmanian H, Faull KF, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. I. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J. Biol. Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J. Biol. Chem. 1999;274:24316–24320. doi: 10.1074/jbc.274.34.24316. [DOI] [PubMed] [Google Scholar]
- Tzagoloff H, Novick R. Geometry of cell division in Staphylococcus aureus. J. Bacteriol. 1977;129:343–350. doi: 10.1128/jb.129.1.343-350.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT. Vancomycin resistance: decoding the molecular logic. Science. 1993;261:308–309. doi: 10.1126/science.8392747. [DOI] [PubMed] [Google Scholar]
- Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- Weiss WJ, Lenoy E, Murphy T, Tardio L, Burgio P, Projan SJ, Schneewind O, Alksne L. Effect of srtA and srtB gene expression on the virulence of Staphylococcus aureus in animal infection. J. Antimicrob. Chemother. 2004;53:480–486. doi: 10.1093/jac/dkh078. [DOI] [PubMed] [Google Scholar]
- Zapun A, Vernet T, Pinho M. The different shapes of cocci. FEMS Microbiol. Rev. 2008;32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.