Summary
We narrowed chromosome 15q21-23 linkage to plasma high density lipoprotein cholesterol (HDL-C) levels in atherogenic dyslipidemic Turkish families by fine mapping, then focused on glucuronic acid epimerase (GLCE), a heparan sulfate proteoglycan (HSPG) biosynthesis enzyme. HSPGs participate in lipid metabolism along with apolipoprotein (apo) E. Of 31 SNPs in the GLCE locus, nine analyzed by haplotype were associated with plasma HDL-C and triglyceride levels (permuted p = 0.006 and 0.013, respectively) in families. Of five tagging GLCE SNPs in two cohorts of unrelated subjects, three (rs16952868, rs11631403, rs3865014) were associated with triglyceride and HDL-C levels in males (non-permuted p < 0.05). The association was stronger in APOE 2/3 subjects (apoE2 has reduced binding to HSPGs) and reached multiple-testing significance (p < 0.05) in both males and females (n = 2612). Similar results were obtained in the second cohort (n = 1164). Interestingly, at the GLCE locus, bounded by recombination hotspots, Turks had a minor allele frequency of SNPs resembling Chinese more than European ancestry; adjoining regions on chromosome 15 resembled the European pattern. Studies of glce+/–apoe–/– mice fed a chow or high-fat diet supported a role for GLCE in lipid metabolism. Thus, SNPs in GLCE are associated with triglyceride and HDL-C levels in Turks, and mouse studies support a role for glce in lipid metabolism.
Introduction
Atherogenic dyslipidemia, characterized by low levels of plasma high density lipoprotein cholesterol (HDL-C) and elevated levels of triglycerides, is a risk factor for coronary artery disease (Gordon et al., 1989; Jacobs et al., 1990), the leading cause of death worldwide (Murray & Lopez, 1997). Plasma HDL-C and triglyceride levels are controlled by genetic factors, although heritability estimates vary widely (Heller et al., 1993; Knoblauch et al., 2004; Mahaney et al., 1995; Yu et al., 2005). In a recent scan for atherogenic dyslipidemia, the multinational Genetic Epidemiology of Metabolic Syndrome study (Wyszynski et al., 2005) found significant linkage to plasma HDL-C levels on chromosome 15q21-23 in Turkish families (Yu et al., 2005). Previous genome-wide scans showed significant linkage of this region not only to plasma HDLC (Almasy et al., 1999; Feitosa et al., 2007; Yang et al., 2010) but also triglyceride levels (Austin et al., 2003; Burgess-Herbert et al., 2009; Klimes et al., 2003; Li et al., 2009; Rollins et al., 2006; Suto et al., 1999). Similar linkage was found for plasma HDL-C and triglyceride levels in the orthologous region in mice and rats (Burgess-Herbert et al., 2009; Klimes et al., 2003; Rollins et al., 2006; Suto et al., 1999).
The linkage peak on chromosome 15 is >20 cM wide (Yu et al., 2005) and includes the gene encoding hepatic lipase (LIPC), which has important functions in lipid metabolism (Mahley et al., 2008). Recently, we analyzed LIPC variants in detail and found several single nucleotide polymorphisms (SNPs) that were significantly associated with plasma HDL-C levels in a Turkish population (Hodoğlugil et al., 2010). Likewise, recent genome-wide association studies have shown the importance of variants at the LIPC locus (Kathiresan et al., 2008; Willer et al., 2008). Turks, whether living in Turkey or abroad, have very low plasma HDL-C and slightly elevated triglyceride levels (Bersot et al., 1999; Mahley et al., 1995; Mahley et al., 2005; Onat et al., 1992; Porsch-Oezçueruemez et al., 1999; Tezcan et al., 2003). However, we suspected that variations in LIPC might not fully explain the linkage peak in Turkish families and sought additional gene(s) that might contribute to the peak.
Glucuronic acid epimerase (GLCE) is another candidate gene in the 15q21-23 region. GLCE encodes an enzyme that is critically important for the biosynthesis of heparan sulfate proteoglycans (HSPGs) (Bishop et al., 2007; Bishop et al., 2008). HSPGs play a major role in clearing triglyceride-rich lipoprotein particles from the plasma (Mahley & Ji, 1999; Mahley & Huang, 2007; Williams et al., 1992; Williams & Fuki, 1997), a process that involves apolipoprotein (apo) E (Mahley & Ji, 1999; Mahley & Huang, 2007). A common isoform, apoE2 (determined by APOE SNP rs429358, which has a frequency of ~8% in Turks), has reduced (50−90%) HSPG-binding activity (Mahley & Ji, 1999). However, only 10% of APOE2/2 homozygotes have type III hyperlipoproteinemia, suggesting that additional genetic, hormonal, or environmental factors are required to precipitate overt hyperlipidemia (Mahley et al., 1999; Utermann, 1985). A defect in the synthesis or function of GLCE could be such a factor.
HSPG biosynthesis involves N-deacetylase/N-sulfotransferase (ndst1) and heparan sulfate 2-O-sulfotransferase (hs2st) (Bishop et al., 2007; Bishop et al., 2008). In mice, conditional knockout of ndst1and hs2st in the liver results in accumulation of atherogenic triglyceride-rich particles in plasma (MacArthur et al., 2007; Stanford et al., 2010). Variants in the gene encoding N-acetylgalactosaminyltransferase 2 (GALNT2), another HSPG synthesis enzyme (Bishop et al., 2007; Bishop et al., 2008), were reported to be significantly associated with HDL-C levels (Kathiresan et al., 2008; Willer et al., 2008). Glce–/– knockout mice die neonatally, but hemizygous mice are viable (Li et al., 2003). However, no lipid metabolism studies have been reported in glce+/– mice.
In this study we sought to determine whether GLCE contributes to the linkage peak in the Turkish families and, if so, whether SNPs in GLCE were associated with plasma triglyceride and HDL-C levels in Turkish people. To answer the first question, we did fine mapping of the chromosome 15q21-23 linkage peak in 40 Turkish families, initially with 11 microsatellite markers and then with SNP markers. Next, we sequenced all exons of GLCE in 32 subjects from these families. To answer the second question, we selected SNPs from GLCE sequence results and the HapMap database and genotyped them in a family-based cohort and two separate cohorts of unrelated individuals from the Turkish Heart Study, a large, cross-sectional epidemiological survey of the Turkish population (Mahley et al., 1995; Mahley et al., 2005). In addition, we evaluated the effects of the interaction of GLCE SNPs and the APOE ε2 allele on triglyceride and HDL-C levels. We also examined plasma lipid and lipoprotein levels in heterozygous glce+/– mice given a standard chow or high-fat diet.
Materials and Methods
Study Population and Biochemical Analyses
The study population included 40 three-generation families with atherogenic dyslipidemia and two separate cohorts of unrelated individuals from Turkey. Families were recruited for the Genetic Epidemiology of Metabolic Syndrome study (Wyszynski et al., 2005). Unrelated individuals with complete lipid and demographic biodata were randomly selected from Turkish Heart Study participants. The first cohort (n = 2612) included subjects whose samples were collected between 1990 and 1995 (Mahley et al., 1995). The second cohort (n = 1164) included subjects whose samples were collected between 2000 and 2003 (Wyszynski et al., 2005) and was used mainly to verify results obtained in the first cohort. Detailed biodata and blood samples were collected from each subject after an overnight fast. Plasma lipids were measured as described (Mahley et al., 1995). Subjects who were taking lipid-lowering medication, had a history of diabetes mellitus, or had a plasma triglyceride level >800 mg/dl were excluded from the unrelated cohorts. The protocols were approved by the Committee on Human Research of the University of California, San Francisco, and were in accordance with the Helsinki Declaration.
Fine Mapping with Microsatellite and SNPs Markers
A genome-wide linkage scan using approximately 400 microsatellite markers was previously conducted in the Turkish families with atherogenic dyslipidemia (Yu et al., 2005). An additional 11 microsatellite markers on chromosome 15 from 51 to 75 cM were genotyped in these families using methods previously described (Yu et al., 2005) for fine mapping. Further fine mapping (± 1 Mb of D15S983 locus) was conducted with SNP markers. The HapMart tool (http://hapmap.ncbi.nlm.nih.gov/biomart/martview/) with a HapMap Centre d'Etude du Polymorphisme Humaine from Utah population (CEU) population (The International HapMap Consortium, 2007) was used to choose SNPs between 65.5 and 67.5 Mb. Briefly, 26 nonsynonymous, 5′ UTR and 3′ UTR SNPs with a minor allele frequency ≥10 % were retrieved. Nine of the 26 SNPs were further selected with the Tagger algorithm (r2 > 0.4) and genotyped in the families using TaqMan assays (ABI, Applied Biosystems, Foster City, CA). The average genotyping call rate exceeded 97%, and the estimated genotyping error rate, determined by analysis of duplicate samples, was about 1%.
Detection and Selection of GLCE Polymorphisms
Oligonucleotide primers were designed to amplify across the GLCE promoter (up to 1000 bases from exon 1) and all five exons, including intron/exon splicing boundaries and the 3′ UTR (up to 600 bases from the last exon). Additionally, two regions (Chr15:67,273,454 − 67,274,053 and 67,275,478 − 67,276,084) were selected for sequencing because of the possibility of spliced expressed sequence tags (UCSC Genome Browser, http://genome.ucsc.edu; human, March 2006 assembly). DNA from 32 family subjects was sequenced to identify polymorphisms in GLCE. DNA sequences were aligned and analyzed with Sequencher DNA analysis software (GeneCodes, Ann Arbor, MI). Since we did not sequence all of the GLCE, we selected additional SNPs using HapMap (http://www.hapmap.org) CEU and Chinese Han from Beijing (CHB) populations in which the frequency and linkage disequilibrium (LD) information among SNPs were available. Selected SNPs were genotyped with TaqMan assays.
Genetic Structure of the Turkish Cohort
Our previous studies suggested that Turks more closely resemble CEU than other HapMap populations with respect to allele frequency (Hodoğlugil et al., 2005; Hodoğlugil et al., 2006; Hodoğlugil et al., 2010). In preliminary analyses of SNPs in the chromosome 15: 67.0−67.5 Mb region (where GLCE is located), the minor allele frequencies of some SNPs were more similar to those of Chinese in the HapMap reference panel (CHB) and the Human Genome Diversity Panel (HGDP) (Li et al., 2008) than to CEU. Therefore, we examined the population substructure of the Turkish cohort by comparing high-density SNP chip data for a subset of the Turkish samples with available HGDP data (http://hagsc.org/hgdp/files.html) (Li et al., 2008). Recently, 1043 HGDP samples were genotyped with Illumina HumanHap650K BeadChips (Illumina, San Diego, CA), and genotype data were made available (Li et al., 2008).
We genotyped 64 unrelated Turkish samples (including one duplicate pair) from the 1990–1995 cohort using Infinium Human 610-quad BeadChip assays (Illumina, San Diego, CA) according to the manufacturer's specifications. All samples had call rates >98%. The rate of concordance between the duplicate samples was >99.99%. SNPs were filtered out if they contained replication errors or if their call rates across the 64 samples were <95%. SNPs that deviated from Hardy-Weinberg equilibrium (p < 0.001, n = 590) were also excluded. These filtering and exclusion criteria resulted in 573,782 high-quality SNPs. HGDP genotype data from unrelated individuals (Rosenberg, 2006) were combined with our filtered 63-sample set and resulted in high-quality genotypes for 535,045 SNPs.
To assess population substructure from the high-density genetic marker data, we used a Bayesian clustering algorithm implemented in the STRUCTURE software package (version 2.2) (Falush et al., 2003). Default parameter settings of 30,000 replicates and 30,000 burn-in cycles were used. Seven parental populations (K = 7) were assumed in this analysis. Because STRUCTURE has a large memory demand, a reduced set of 6,858 SNPs was selected. For maximum representation of the genome, SNPs in LD were first pruned with PLINK (Purcell et al., 2007), which removes one of a pair of SNPs if r2 >0.3 in 50-SNP windows, repeats this process for every pair, and then shifts the window 5 SNPs forward and repeats the procedure again. Next, high Fst SNPs [CEU vs. CHB + JPT (Japanese from Tokyo) >0.25] were selected. SNPs were thinned if adjacent SNPs were <0.1 cM apart and were filled with SNPs 0.25 > Fst> 0.20 if they were >1 cM apart. Only autosomal SNPs were used. Pairwise HapMap Fst and mapping (cM) data for individual SNPs were provided by Stephen Schaffner (Broad Institute of MIT and Harvard) and Tara Matise (Rutgers University), respectively.
Animal Studies
Glce+/– (also known as hsepi) mice (Li et al., 2003) were provided by J.-P. Li (University of Uppsala, Sweden). Apoe–/– mice were purchased from Jackson Laboratory (Bar Harbor, ME). Both glce+/– and apoe–/– mice were on a C57BL/6 background. Male glce+/– mice were bred with female apoe–/– mice to produce glce+/–/apoe–/– mice. Age-matched 10- to 12-week-old male mice were used for all experiments. Mice were housed in a pathogen-free barrier facility (12-h light/12-h dark cycle) and fed standard rodent chow (12% calories from fat, 5053 PicoLab, Purina). For high-fat experiments, mice were fed either TD.88137 (42% calories from fat, 0.2% cholesterol, Harlan Teklad) or TD02028 (43% calories from fat, 1.3% cholesterol, 0.5% cholic acid, Harlan Teklad) diets for 4−6 weeks. All experiments were approved by the Committee on Animal Research of the University of California, San Francisco.
Plasma Lipid Analyses
Blood was collected from the retro-orbital sinus after a 4−5 h fast. EDTA (10 mM) was used as an anticoagulant and phenylmethylsulfonyl fluoride (Sigma) (1 mM) was used as a protease inhibitor. Plasma was obtained by centrifugation at 14,000 rpm (microcentrifuge) for 15 min at 4°C. Total cholesterol and triglycerides were measured in total plasma by an enzymatic colorimetric method (Roche/Hitachi).
mRNA Expression
Total RNA from the liver was isolated with RNeasy kits and treated with DNase (Qiagen). Complementary cDNA was synthesized from RNA (5 μg) with Superscript II reverse transcriptase and random hexamers (Invitrogen). Quantitative PCR was performed with a 7900HT PCR system and the data were analyzed by the delta-delta Ct method as recommended by the manufacturer (Applied Biosystems). Beta-glucuronidase expression was used for internal normalization.
Statistical Analysis
Genotype data in the families were tested for Mendelian errors using Haploview 4.1 (Barrett et al., 2005). Inconsistencies were resolved by re-genotyping or eliminating all genotypes for particular markers in the families where the errors were noted. Fine mapping within the previously reported linkage region for HDL-C on chromosome 15q21-23 was performed by single-locus and multi-locus linkage analysis using the variance component method implemented in SOLAR (version 2.13) (Almasy & Blangero, 1998), as previously described (Yu et al., 2005). Marker locations were fixed using the deCode genetic map.
Association of single SNP markers or haplotypes with plasma HDL-C and triglyceride levels was evaluated in the family cohort using the transmission-disequilibrium test (TDT) (Spielman et al., 1993) implemented in Haploview. TDT compares the observed and expected transmission of alleles (or haplotypes) from heterozygous parents to affected offspring (low HDL-C or high triglyceride values). For these analyses, plasma HDL-C and triglyceride levels in family members were classified as high or low. Cut-off values for plasma HDL-C were ≤35 mg/dl and ≤40 mg/dl for affected males and females and ≥40 mg/dl and ≥45 mg/dl for unaffected (high HDL-C) males and females, respectively. Cut-off values for plasma triglyceride levels were ≥80th percentile (age and sex matched) for affected and ≤50th percentile for unaffected subjects (low triglyceride) (Wyszynski et al., 2005). Multiple-test correction for SNPs or haplotypes was conducted by the permutation test (50,000 permutations) in Haploview. Quantitative TDT (QTDT) in SOLAR was used to determine SNP associations with plasma triglycerides and HDL-C levels treated as quantitative traits.
In the unrelated individuals, Hardy-Weinberg equilibrium was tested for each SNP using Haploview. Log-transformed values of triglyceride and HDL-C levels were used for statistical comparisons to correct for departure from normality; untransformed mean values are reported. PLINK (version 1.06) was used to evaluate association of SNPs with plasma HDL-C or triglyceride values in an additive genotypic model including covariates for age, body mass index, smoking (number of cigarettes/day), and alcohol consumption (nondrinkers, 1–5 drinks/week, >5 drinks/week). Corrections for multiple testing were conducted with a permutation test (50,000 permutations) in PLINK. Values for p or permutated p were considered as significant (p < 0.05) and borderline significant (p < 0.1 and p >0.05). Covariate adjusted mean values were calculated by SPSS 10.0. Interaction between pairs of SNPs was evaluated using regression models including terms for genotypes for the two SNPs and an interaction term (snp1*snp2). Males and females were analyzed separately because plasma HDL-C and triglyceride levels showed gender-specific differences.
Data from mouse experiments were evaluated by t-test. p < 0.05 was considered significant.
Results
Population Characteristics
The demographic and biochemical characteristics of the atherogenic dyslipidemic families and the 1990–1995 and 2000–2003 cohorts are shown in Table 1. In all groups and in men and women, the plasma HDL-C levels were low, and plasma triglyceride levels and total cholesterol/HDL-C ratios were elevated.
Table 1.
Demographic and biochemical characteristics of the study subjects according to gender
| Families |
1990–1995 Cohort |
2000–2003 Cohort |
||||||
|---|---|---|---|---|---|---|---|---|
| Males (n = 278) | Females (n = 347) | Males (n = 1549) | Females (n = 1063) | p | Males (n = 474) | Females (n = 690) | p | |
| |
|
|
||||||
| Age (yrs) | 41.6 ± 16.5 | 41.5 ± 16.6 | 42.1 ± 13.2 | 42.2 ± 14.9 | NS | 44.1 ± 13.1 | 44.4 ± 13.9 | NS |
| Body mass index (kg/m2) | 26.1 ± 4.6 | 27.6 ± 5.4 | 26.1 ± 3.9 | 26.6 ± 5.4 | <0.05 | 28.1 ± 3.8 | 29.9 ± 5.4 | <0.01 |
| HDL-C (mg/dl) | 35.8 ± 9.9 | 44.7 ± 12.4 | 35.7 ± 7.5 | 41.2 ± 9 | <0.001 | 38.7 ± 8.5 | 47.0 ± 9.4 | <0.001 |
| Total cholesterol (mg/dl) | 181 ± 45 | 180 ± 42 | 183 ± 44 | 183 ± 42 | NS | 182 ± 37 | 184 ± 43 | NS |
| Total cholesterol/HDL-C ratio | 5.4 ± 1.9 | 4.3 ± 1.6 | 5.8 ± 2.9 | 4.5 ± 1.4 | <0.05 | 5.0 ± 1.6 | 4.0 ± 1.9 | <0.05 |
| LDL-C (mg/dl) | 110 ± 37 | 108 ± 35 | 126 ± 41 | 116 ± 39 | <0.001 | 112 ± 33 | 113 ± 35 | NS |
| Triglycerides (mg/dl) | 184 ± 140 | 139 ± 110 | 141 ± 89 | 111 ± 76 | <0.01 | 155 ± 105 | 118 ± 67 | <0.01 |
| Systolic blood pressure (mm Hg) | 131 ± 21 | 136 ± 28 | 125 ± 23 | 122 ± 21 | NS | 132 ± 20 | 133 ± 22 | NS |
| Diastolic blood pressure (mm Hg) | 81 ± 12 | 85 ± 14 | 82 ± 14 | 81 ± 13 | NS | 84 ± 12 | 85 ± 13 | NS |
| Consumption of alcohol (%)a | 31.3 | 4.6 | 29.8 | 5.6 | <0.001 | 36.7 | 9.6 | <0.001 |
| Cigarette smoking (%)b | 71.9 | 22.2 | 56.6 | 24.0 | <0.001 | 67.2 | 25.2 | <0.001 |
Data from atherogenic dyslipidemic families were collected between 2000 and 2003. Unrelated samples were collected between 1990 and 1995 and between 2000 and 2003.
LDL-C, low density lipoprotein cholesterol. Values are mean ± SD or percentages. Means were compared by t-test, and percentages were analyzed by chi-square test. P: males vs. females. The p value was determined for males versus females?????
One or more drinks per week.
One or more cigarettes per day.
Narrowing the Chromosome 15q21-23 Peak with Fine Mapping
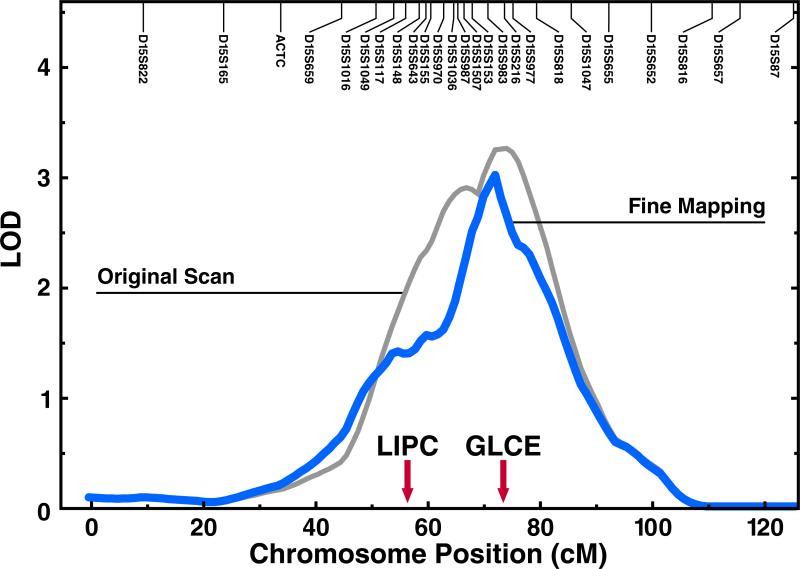
Analysis of the combined set of previously genotyped and the additional 11 microsatellite markers in the families substantially narrowed the linkage peak in the original study (Yu et al., 2005) from a 95% confidence interval (CI) of 20−25 cM to an interval of approximately 15 cM without changing the peak location at 71 cM (LOD = 2.91, Fig. 1). The marker nearest to the linkage peak is D15S983. Of note, GLCE is located almost precisely at the linkage peak, whereas LIPC is more than 10 cM outside the 95% CI for the linkage peak. Further fine mapping (± 1 Mb of D15S983) was performed with SNP markers and nine SNPs were selected with the Tagger algorithm implemented in Haploview. TDT analysis of the nine SNP markers located within 65.5–67.5 Mb identified two nominally significant results in the families (rs3865014: p = 0.023 for triglyceride status and p = 0.022 for HDL-C status; rs6545: p = 0.033 for triglyceride status, Table 2). Since these two SNPs are located in GLCE, we decided to focus on GLCE and the surrounding locus. Rs3865014 is a non-synonymous SNP (Val/Ile) and rs6545 is located in the 3′ UTR.
Figure 1.
Fine mapping of chromosome 15 in atherogenic dyslipidemic Turkish families. An additional 11microsatellite markers (located at 51–75 cM) were used for fine mapping in these families. The original scan and a new scan with additional markers are shown. The deCode genetic map was utilized.
Table 2.
Transmission-disequilibrium test analysis of Chr15:67,025K..67,483K region SNPs and haplotypes with atherogenic dyslipidemia in 40 Turkish families
| Triglycerides | HDL-C | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | SNP | Position | Allelesa | Frequencyb | Overtransmitted allele | Tc | Uc | T/Uc | p | Permuted pd | Tc | Uc | T/Uc | p | Permuted pd |
| 1 | rs3743093 | 67,025,498 | A/G | 0.474 | A | 20 | 12 | 1.67 | NS | 28 | 25 | 1.12 | NS | ||
| 2 | rs12907196 | 67,115,279 | C/T | 0.482 | T | 20 | 12 | 1.67 | NS | 23 | 19 | 1.21 | NS | ||
| 3 | rs11072067 | 67,199,022 | C/T | 0.368 | T | 22 | 11 | 2.00 | 0.056 | NS | 41 | 28 | 1.46 | NS | NS |
| 4 | rs7177250 | 67,225,596 | T/A | 0.114 | – | 6 | 6 | 1.00 | NS | 14 | 12 | 1.17 | NS | ||
| 5 | rs17358457 | 67,230,768 | T/C | 0.088 | C | 5 | 4 | 1.25 | NS | 11 | 10 | 1.10 | NS | ||
| 6 | rs16952868 | 67,233,927 | A/C | 0.483 | C | 25 | 13 | 1.92 | 0.052 | NS | 48 | 35 | 1.37 | NS | NS |
| 7 | rs11631403 | 67,236,313 | C/T | 0.369 | T | 21 | 9 | 2.33 | 0.029 | 0.075 | 39 | 24 | 1.63 | 0.059 | NS |
| 8 | rs1553251 | 67,236,900 | C/T | 0.369 | T | 21 | 9 | 2.33 | 0.029 | 0.075 | 39 | 24 | 1.63 | 0.059 | NS |
| 9 | rs7176782 | 67,264,433 | A/G | 0.422 | G | 23 | 11 | 2.09 | 0.040 | NS | 44 | 33 | 1.33 | NS | NS |
| 10 | rs4777126 | 67,294,513 | C/T | 0.11 | C | 7 | 4 | 1.75 | NS | 14 | 9 | 1.56 | NS | ||
| 11 | rs11629932 | 67,323,011 | C/T | 0.431 | T | 25 | 13 | 1.92 | 0.052 | NS | 47 | 35 | 1.34 | NS | NS |
| 12 | rs17360351 | 67,329,100 | A/G | 0.119 | G | 17 | 7 | 2.43 | 0.041 | NS | 26 | 12 | 2.17 | 0.023 | 0.094 |
| 13 | rs3865014 | 67,348,571 | A/G | 0.308 | G | 20 | 8 | 2.50 | 0.023 | 0.062 | 36 | 19 | 1.89 | 0.022 | 0.076 |
| 14 | rs6545 | 67,351,396 | T/C | 0.407 | C | 25 | 12 | 2.08 | 0.033 | 0.087 | 45 | 31 | 1.45 | NS | NS |
| 15 | rs2047822 | 67,351,961 | C/G | 0.106 | C | 6 | 5 | 1.20 | NS | 13 | 12 | 1.08 | NS | ||
| 16 | rs905508 | 67,358,721 | C/T | 0.151 | T | 16 | 9 | 1.78 | NS | 22 | 17 | 1.29 | NS | ||
| 17 | rs7171293 | 67,367,474 | G/A | 0.071 | G | 5 | 3 | 1.67 | NS | 10 | 8 | 1.25 | NS | ||
| 18 | rs12593391 | 67,379,320 | C/T | 0.442 | T | 24 | 14 | 1.71 | NS | 45 | 36 | 1.25 | NS | ||
| 19 | rs10851802 | 67,383,494 | G/A | 0.312 | A | 23 | 9 | 2.56 | 0.013 | 38 | 25 | 1.52 | NS | ||
| 20 | rs12441411 | 67,384,261 | A/G | 0.032 | A | 5 | 3 | 1.67 | NS | 6 | 6 | 1.00 | NS | ||
| 21 | rs7173953 | 67,385,179 | A/T | 0.373 | A | 22 | 11 | 2.00 | 0.056 | 42 | 31 | 1.35 | NS | ||
| 22 | rs7173355 | 67,385,566 | C/T | 0.422 | T | 23 | 14 | 1.64 | NS | 47 | 35 | 1.34 | NS | ||
| 23 | rs870335 | 67,392,196 | G/T | 0.39 | T | 24 | 11 | 2.18 | 0.028 | 47 | 33 | 1.42 | NS | ||
| 24 | rs8034725 | 67,393,781 | T/C | 0.03 | T | 7 | 2 | 3.50 | NS | 9 | 6 | 1.50 | NS | ||
| 25 | rs16953133 | 67,404,769 | C/T | 0.028 | C | 6 | 2 | 3.00 | NS | 8 | 6 | 1.33 | NS | ||
| 26 | rs7182765 | 67,410,893 | C/G | 0.054 | C | 5 | 1 | 5.00 | NS | 8 | 6 | 1.33 | NS | ||
| 27 | rs4505253 | 67,415,937 | T/C | 0.25 | C | 18 | 11 | 1.64 | NS | 34 | 18 | 1.89 | 0.027 | ||
| 28 | rs12900857 | 67,436,279 | A/C | 0.05 | A | 6 | 2 | 3.00 | NS | 9 | 7 | 1.29 | NS | ||
| 29 | rs7172559 | 67,464,551 | C/T | 0.011 | – | 1 | 1 | 1.00 | NS | 2 | 1 | 2.00 | NS | ||
| 30 | rs1394415 | 67,483,302 | C/T | 0.036 | C | 2 | 1 | 2.00 | NS | 5 | 3 | 1.67 | NS | ||
| 31 | rs3743096 | 67,483,365 | G/T | 0.017 | – | 0 | 0 | 0.00 | NS | 1 | 1 | 1.00 | NS | ||
| SNPs of 3,6-9,11-14 | Haplotype | Frequencyb | Tc | Uc | T/Uc | p | Permuted pd | Tc | Uc | T/Uc | p | Permuted pd | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CACCACAAT | 0.461 | 16 | 26 | 0.62 | NS | NS | 37 | 49 | 0.76 | NS | NS | ||||
| TCTTGTAGC | 0.131 | 6 | 3 | 2.00 | NS | NS | 10 | 11 | 0.91 | NS | NS | ||||
| CCCCGTAAC | 0.108 | 6 | 5 | 1.20 | NS | NS | 12 | 13 | 0.92 | NS | NS | ||||
| TCTTGTGGC | 0.094 | 15 | 5 | 3.00 | 0.025 | 0.013 | 25 | 8 | 3.13 | 0.003 | 0.006 | ||||
| TCTTACAAT | 0.052 | 1 | 2 | 0.50 | NS | NS | 4 | 4 | 1.00 | NS | NS | ||||
| CCTTGTAGC | 0.051 | 0 | 1 | 0.00 | NS | NS | 2 | 3 | 0.67 | NS | NS | ||||
| TACCACAAT | 0.026 | 0 | 0 | 0.00 | NS | NS | 0 | 0 | 0.00 | NS | NS | ||||
| TCTTGCAAT | 0.019 | 1 | 1 | 1.00 | NS | NS | 4 | 4 | 1.00 | NS | NS | ||||
Haplotypes consisting of markers (n = 9) in GLCE locus are shown in bold and other significant or borderline significant SNPs (n = 4) outside the haplotype block are shown in italic. NS, not significant (p ≥ 0.1).
Common/rare in Turkish families in forward strand (UCSC genome browser on the human March 2006 assembly).
Frequency of rare allele in parental chromosomes.
Transmitted and untransmitted chromosomes and the transmission ratio (T/U).
Permutation p values were calculated from 50,000 random iterations of the genotype data in Haploview and corrected for multiple testing.
Analysis of GLCE SNPs in Families
Seven known SNPs in the GLCE locus (rs3865014, rs3205721, rs16953674, rs17360351, rs6545, rs2047822, rs11633143) with minor allele frequencies between 9% and 35% and a novel coding variant (Met/Val, A/G at 67,384,254) were identified by sequencing DNA from 32 subjects. This coding variant was observed in only one heterozygous subject (frequency, 1.6%) and was not further studied. There was complete LD (r2 = 1) between SNP pairs rs3865014 and rs3205721, and among rs6545, rs16953674 and rs11633143 SNPs. After eliminating three redundant SNPs, we selected four SNPs for genotyping in atherogenic dyslipidemic families (rs3865014, rs6545, rs17360351, and rs2047822 corresponding to SNPs 11−14 in Table 2). No variants were observed in the two regions sequenced for possibly spliced mRNA.
In addition to these four SNPs, 27 additional HapMap SNPs were selected to cover GLCE and the surrounding regions (Table 2). Since the minor allele frequencies of some SNPs in the GLCE region in Turkish families were more similar to those of CHB than CEU, we utilized both HapMap populations in the selection process. The additional SNPs were chosen based on LD with rs3865014 and rs6545 SNPs (r2 < 0.8), coverage of the GLCE locus, and minor allele frequency (>5%). SNPs located in the recombination hotspots were also selected to define haplotype blocks surrounding GLCE. Thirteen of the 31 SNPs showed significant (p < 0.05) or borderline significant (p < 0.06) results for triglyceride or HDL-C status, of which nine are located within the coding, promoter, or 5′ UTR regions of GLCE (shown in bold, Table 2). However, permuted p values for the nine SNPs were either not significant (p > 0.1) or borderline (p < 0.1) significant. Analysis of haplotypes comprised of these nine SNPs showed that one haplotype (frequency, 9.4%) was associated with both triglyceride and HDL-C status in the nominal and permuted tests. Analysis of pairwise LD (r2) among the 13 significant SNPs revealed that the block of nine SNPs forming a haplotype block is separated from the remaining four SNPs (rs10851802, rs7173953, rs870335, rs4505253, Table 2 in italics) by multiple recombination hotspots and (Fig. 2 and Table S1). Inclusion of one or more of these four SNPs in the haplotype block weakened the significance of the results, suggesting that the association signal is more likely due to a variant in GLCE than in the region downstream from GLCE. Analysis of the quantitative traits revealed notable results for plasma triglyceride levels with GLCE SNPs rs3865014 (p = 0.018) and rs11631403 (p = 0.064).
Figure 2.
Recombination rate and LD structure in the chromosome 15: 67.0−67.5 Mb locus in Turkish families. Top panel shows recombination rate. Middle panel shows the LD (r2) plot from Haploview for 31 SNPs . Darker diamonds represent regions of high pairwise r2. The black arrows indicate the size and the direction of expression of GLCE and PAQR5 (progestin and adipoQ receptor family member V). Red dotted lines show the location of recombination hotspots. The bottom panel shows the nine-SNP haplotype block.
Genetic Structure of the Turkish Cohort
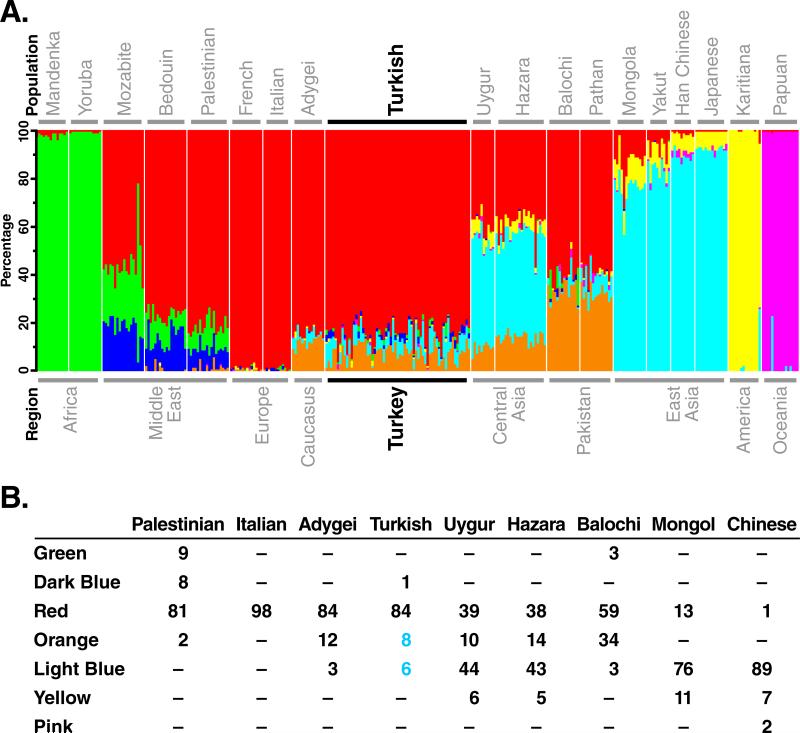
Analysis of HGDP data with 6858 SNP markers genotyped in 260 unrelated individuals using the STRUCTURE software package (version 2.2) (Falush et al., 2003) revealed that individuals from the same geographic region or predefined population nearly always shared similar parental ancestry components, as previously shown (Li et al., 2008; Rosenberg et al., 2002). A similar analysis including Turks was performed to compare with other populations. The genetic structure of the Turkish samples revealed three major parental ancestries (Fig. 3A). The percentage ancestry was calculated as an average across 63 Turks as described under Materials and Methods. In Turks, the largest portion (red), about 84%, was also present as the major ancestry in European populations and to a lesser extent in the Middle Eastern, Central Asian, and Pakistani populations. About 8% of ancestry (orange) in the Turkish population was also present in Central Asian and Pakistani populations. About 6% of Turkish ancestry (light blue) was also present to a significant extent in Central Asian and to a major extent in East Asian populations. Results from the Caucasus (Adygei population) were very similar to the Turks. The average ancestry for selected populations is shown for comparison (Fig. 3B).
Figure 3.
STRUCTURE analysis of global dataset with 6,858 SNP markers genotyped in 323 subjects. (A) Representative populations from each geographic region were selected from HGDP data (Li et al., 2008), and Turkish samples were included in the analysis. Populations are labeled above the figure, with their geographic affiliations below. Each individual is represented by a thin vertical line, which is partitioned into K colored segments (K = 7). Colors represent the inferred ancestry from parental populations. White lines separate individuals of different populations. Seven other STRUCTURE runs produced very similar individual ancestry estimates. (B) The average ancestry estimates for selected populations are shown for comparison.
Further analysis of the chromosome 15:67.0−67.5 Mb region revealed that the minor allele frequencies in Turks are very similar to those of European–HGDP populations (French, Sardinian, Orcadian, Italian, Tuscan, and Basque) and European descendants (CEU) for SNPs outside the recombination hotspots (67,162−67,170 K and 67,195−67,205 K, and 67,358−67,362 K and 67,383−67,386 K, Fig. 4A). In contrast, in the area between the recombination hotspots, which includes GLCE, the minor allele frequencies in Turks were more similar to the Chinese−HGDP (Han and Han from North China) and Chinese−Hapmap (CHB) populations (Fig. 4B).
Figure 4.
Allele frequency plot of the chromosome 15: 67.0−67.5 Mb locus. (A) Minor allele frequencies in Turks vs. European (CEU and HGDP). (B) Minor allele frequencies in Turks vs. Chinese (CHB and HGDP). Blue arrows indicate the location of GLCE and PAQR5 (progestin and adipoQ receptor family member V) with respect to SNPs. Red arrows show recombination hotspots.
Association of GLCE SNPs with Plasma Triglyceride and HDL-C Levels in Unrelated Turks
The nine SNPs that showed significant results at the GLCE locus in the families (Table 2) were genotyped in a small cohort of unrelated Turkish subjects (n = 260) to assess the LD structure. Five SNPs (rs16952868, rs11631403, rs17360351, rs3865014, and rs6545) were selected by Tagger (r2 > 0.8, Table 3) and genotyped in the 1990–1995 and 2000–2003 cohorts.
Table 3.
Pair-wise LD coefficients (r2, up-right and D′, bottom-left) between GLCE SNPs in the 1990–1995 Turkish cohort
| r2/D′ | rs11072067 | rs16952868 | rs11631403 | rs1553251 | rs7176782 | rs11629932 | rs17360351 | rs3865014 | rs6545 |
|---|---|---|---|---|---|---|---|---|---|
| rs11072067 | 0.28 | 0.81 | 0.70 | 0.16 | 0.21 | 0.30 | 0.61 | 0.17 | |
| rs16952868 | 0.85 | 0.72 | 0.43 | 0.72 | 0.29 | 0.12 | 0.27 | 0.72 | |
| rs11631403 | 0.90 | 1.00 | 0.86 | 0.32 | 0.30 | 0.25 | 0.56 | 0.30 | |
| rs1553251 | 1.00 | 0.89 | 0.93 | 0.23 | 0.30 | 0.31 | 0.70 | 0.21 | |
| rs7176782 | 0.62 | 0.89 | 0.74 | 0.62 | 0.81 | 0.13 | 0.30 | 0.83 | |
| rs11629932 | 0.53 | 0.92 | 0.62 | 0.66 | 0.80 | 0.14 | 0.48 | 0.85 | |
| rs17360351 | 0.82 | 0.84 | 0.89 | 1.00 | 0.84 | 0.92 | 0.45 | 0.18 | |
| rs3865014 | 0.78 | 0.85 | 0.90 | 1.00 | 0.85 | 0.85 | 1.00 | 0.41 | |
| rs6545 | 0.56 | 1.00 | 0.64 | 0.53 | 1.00 | 0.95 | 0.87 | 0.88 |
Selected SNPs (Tagger algorithm in Haploview, r2 > 0.8) are shown in bold.
Three SNPs (rs16952868, rs11631403 and rs3865014) were significantly associated with plasma triglyceride and HDL-C levels in males in the 1990–1995 cohort, but not in females (Tables 4 and 5). However, after multiple-test correction, these results were no longer significant. Since the apoE2 isoform has reduced HSPG binding activity, and apoE3 (the most common isoform) and apoE4 have normal and similar binding activities (Mahley & Ji, 1999), genotype data were stratified by common APOE genotypes (apo2/3 vs. apo3/3 + apo3/4) to determine whether the significance would increase. ApoE2/2, apoE4/4, and apoE2/4 individuals (total ~2.5%) were excluded from the analysis because many of these subjects had extreme values for plasma triglyceride levels. Among apoE2/3 subjects, rs16952868 and rs3865014 were significantly or borderline significantly associated with plasma triglyceride and HDL-C levels in males (p and permutated p values), and rs3865014 and rs11631403 were significantly associated with plasma triglyceride levels in females, whereas rs3865014 was significantly associated with plasma HDL-C in females, even after multiple-test correction (Tables 4 and 5). Mean lipid values for subjects according to APOE genotype are presented in Table S2. The non-synonymous coding SNP rs3865014 is a logical candidate for a functional SNP, but valine at this position in the amino acid sequence is not conserved among species, and SIFT (Ng & Henikoff, 2002) and PolyPhen (Sunyaev et al., 2001) did not predict any function for this amino acid change.
Table 4.
GLCE SNPs and adjusted plasma triglyceride (mg/dl) levels in the 1990–1995 cohort
| Males | Females | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | AA | AB | BB | p | Permuted p | AA | AB | BB | p | Permuted p | |
| |
|
||||||||||
| rs16952868 | 135 ± 81 (504) | 143 ± 92 (752) | 146 ± 95 (285) | 0.037 | 0.098 | 105 ± 62 (337) | 115 ± 83 (502) | 113 ± 78 (219) | NS | NS | |
| rs11631403 | 136 ± 83 (689) | 144 ± 90 (684) | 151 ± 109 (168) | 0.027 | 0.092 | 106 ± 61 (451) | 114 ± 85 (467) | 119 ± 84 (143) | 0.087 | NS | |
| rs17360351 | 139 ± 86 (1177) | 148 ± 94 (336) | 159 ± 137 (28) | 0.061 | NS | 109 ± 69 (792) | 118 ± 94 (251) | 120 ± 86 (17) | NS | NS | |
| rs3865014 | 137 ± 82 (824) | 144 ± 96 (609) | 153 ± 93 (105) | 0.037 | NS | 109 ± 75 (562) | 112 ± 72 (415) | 122 ± 96 (85) | NS | NS | |
| rs6545 | 138 ± 83 (626) | 142 ± 93 (689) | 148 ± 92 (214) | NS | NS | 110 ± 79 (440) | 108 ± 61 (468) | 122 ± 104 (147) | NS | NS | |
| GLCE | APOE | AA | AB | BB | p | Permuted p | AA | AB | BB | p | Permuted p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
||||||||||
| rs16952868 | 3/3 + 3/4 | 136 ± 79 (432) | 140 ± 86 (645) | 143 ± 96 (243) | NS | NS | 103 ± 67 (281) | 107 ± 85 (425) | 109 ± 80 (183) | NS | NS |
| 2/3 | 130 ± 82 (66) | 153 ± 99 (75) | 154 ± 85 (40) | 0.021 | 0.042 | 92 ± 42 (44) | 111 ± 80 (66) | 123 ± 87 (28) | 0.027 | 0.074 | |
| rs11631403 | 3/3 + 3/4 | 136 ± 82 (588) | 139 ± 85 (583) | 144 ± 100 (146) | NS | NS | 103 ± 62 (380) | 107 ± 86 (393) | 111 ± 87 (121) | NS | NS |
| 2/3 | 136 ± 88 (80) | 150 ± 105 (77) | 152 ± 97 (27) | 0.044 | NS | 93 ± 45 (54) | 113 ± 81 (60) | 126 ± 94 (20) | 0.016 | 0.048 | |
| rs17360351 | 3/3 + 3/4 | 138 ± 87 (1004) | 145 ± 95 (288) | 146 ± 109 (26) | NS | NS | 104 ± 72 (665) | 111 ± 89 (212) | 114 ± 92 (15) | NS | NS |
| 2/3 | 143 ± 101 (134) | 145 ± 112 (46) | 149 ± 87 (3) | NS | NS | 106 ± 55 (99) | 118 ± 102 (34) | 133 ± 21 (2) | NS | NS | |
| rs3865014 | 3/3 + 3/4 | 137 ± 82 (706) | 139 ± 95 (520) | 145 ± 96 (92) | NS | NS | 106 ± 75 (470) | 105 ± 74 (348) | 117 ± 103 (71) | NS | NS |
| 2/3 | 134 ± 99 (91) | 151 ± 89 (72) | 163 ± 64 (18) | 0.021 | 0.043 | 95 ± 67 (71) | 120 ± 81 (57) | 122 ± 64 (11) | 0.014 | 0.046 | |
| rs6545 | 3/3 + 3/4 | 137 ± 83 (534) | 139 ± 91 (591) | 142 ± 97 (181) | NS | NS | 102 ± 80 (368) | 111 ± 67 (395) | 112 ± 99 (125) | NS | NS |
| 2/3 | 138 ± 98 (75) | 144 ± 96 (78) | 150 ± 77 (30) | 0.073 | NS | 102 ± 71 (59) | 107 ± 50 (62) | 125 ± 95 (18) | 0.043 | NS | |
All means ± SD were calculated by analysis of covariance using general linear models and were adjusted for age, body mass index, smoking, and alcohol consumption. Total number of subjects: 1549 males, 1063 females.
Number of subjects for each group is shown in parentheses. p values were calculated using an additive genotypic model in PLINK. Permutation Permuted???? p values were calculated from 50,000 random iterations in PLINK.
A, common allele; B, rare allele. NS, not significant (p ≥ 0.1). Significant (<0.05) results are shown in bold.
Table 5.
GLCE SNPs and adjusted plasma HDL-C (mg/dl) levels in the 1990–1995 cohort
| Males | Females | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | AA | AB | BB | p | Permuted p | AA | AB | BB | p | Permuted p | |
| |
|
||||||||||
| rs16952868 | 36.4 ± 7.9 (504) | 35.4 ± 7.3 (752) | 35.3 ± 7.5 (285) | 0.029 | 0.091 | 41.1 ± 8.8 (337) | 41.4 ± 9.6 (502) | 40.9 ± 9.4 (219) | NS | NS | |
| rs11631403 | 36.1 ± 7.8 (689) | 35.5 ± 7.4 (684) | 35.0 ± 6.8 (168) | 0.046 | NS | 41.0 ± 8.8 (451) | 41.7 ± 9.6 (467) | 40.1 ± 9.8 (143) | NS | NS | |
| rs17360351 | 35.9 ± 7.6 (1177) | 35.4 ± 7.4 (336) | 35.0 ± 6.1 (28) | 0.300 | NS | 41.3 ± 9.1 (792) | 41.0 ± 10.1 (251) | 39.0 ± 7.5 (17) | NS | NS | |
| rs3865014 | 36.1 ± 7.9 (824) | 35.5 ± 7.2 (609) | 34.5 ± 6.7 (105) | 0.038 | 0.098 | 40.9 ± 8.6 (562) | 41.8 ± 10.1 (415) | 39.1 ± 9.7 (85) | NS | NS | |
| rs6545 | 36.1 ± 7.8 (626) | 35.6 ± 7.4 (689) | 35.2 ± 7.3 (214) | 0.092 | NS | 40.9 ± 8.6 (440) | 41.8 ± 9.9 (468) | 40.2 ± 9.6 (147) | NS | NS | |
| GLCE | APOE | AA | AB | BB | p | Permuted p | AA | AB | BB | p | Permuted p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
||||||||||
| rs16952868 | 3/3 + 3/4 | 36.6 ± 7.9 (432) | 36 ± 7.4 (645) | 35.8 ± 7.4 (243) | NS | NS | 41.3 ± 8.9 (281) | 42.1 ± 9.6 (425) | 41.4 ± 9.5 (183) | NS | NS |
| 2/3 | 37.7 ± 9.6 (66) | 36.1 ± 6.8 (75) | 34.7 ± 6.7 (40) | 0.015 | 0.043 | 43.9 ± 9.5 (44) | 41.6 ± 9.1 (66) | 40.8 ± 8.9 (28) | NS | NS | |
| rs11631403 | 3/3 + 3/4 | 36.4 ± 8.0 (588) | 35.9 ± 7.4 (583) | 35.5 ± 6.8 (146) | NS | NS | 41.3 ± 9.0 (380) | 42.5 ± 9.7 (393) | 41.2 ± 9.8 (121) | NS | NS |
| 2/3 | 38 ± 9.0 (80) | 36.1 ± 7.1 (77) | 35.7 ± 6.9 (27) | 0.075 | NS | 42.9 ± 8.8 (54) | 42.2 ± 9.5 (60) | 40 ± 9.1 (20) | NS | NS | |
| rs17360351 | 3/3 + 3/4 | 36.2 ± 8.0 (1005) | 35.8 ± 7.5 (288) | 35.1 ± 7.1 (26) | NS | NS | 41.7 ± 9.1 (665) | 42.2 ± 10.1 (212) | 40.5 ± 6.8 (15) | NS | NS |
| 2/3 | 36.9 ± 8.3 (134) | 35.9 ± 7.8 (46) | 36.8 ± 9.1 (3) | NS | NS | 42.7 ± 9.4 (99) | 41.6 ± 8.3 (34) | 39.1 ± 16.3 (2) | NS | NS | |
| rs3865014 | 3/3 + 3/4 | 36.4 ± 8.1 (706) | 35.8 ± 7.4 (520) | 35.1 ± 7.4 (92) | NS | NS | 41.3 ± 8.9 (470) | 42.5 ± 10.1 (348) | 40.6 ± 9.7 (71) | NS | NS |
| 2/3 | 37.5 ± 8.9 (93) | 36.6 ± 7.3 (72) | 33.9 ± 6.5 (18) | 0.027 | 0.069 | 42.9 ± 8.4 (71) | 41.9 ± 9.9 (57) | 39.8 ± 9.2 (11) | 0.013 | 0.041 | |
| rs6545 | 3/3 + 3/4 | 36.5 ± 7.8 (534) | 35.8 ± 7.6 (591) | 35.7 ± 7.4 (181) | NS | NS | 41.4 ± 8.7 (368) | 42.1 ± 10.2 (395) | 41.6 ± 9.8 (125) | NS | NS |
| 2/3 | 37.3 ± 9.4 (75) | 36.9 ± 7.2 (78) | 34.6 ± 6.2 (30) | 0.085 | NS | 43.1 ± 9.1 (59) | 42.2 ± 9.6 (62) | 39.6 ± 8.6 (18) | NS | NS | |
All means ± SD were calculated by analysis of covariance using general linear models and were adjusted for age, body mass index, smoking, and alcohol consumption. Total number of subjects: 1549 males, 1063 females.
Number of subjects for each group is shown in parentheses. p values were calculated using an additive genotypic model in PLINK. Permutation Permuted??? p values were calculated from 50,000 random iterations in PLINK.
A, common allele; B, rare allele. NS, not significant (p ≥ 0.1). Significant (<0.05) results are shown in bold.
Similar results were obtained in the 2000−2003 cohort for plasma triglyceride (Table 6) and HDL-C levels (Table 7). Three SNPs (rs16952868, rs11631403, and rs3865014) were significantly associated with triglyceride or HDL-C levels, but not after multiple-test correction. However, after stratification by APOE genotype, significant associations were observed, even after multiple-test correction, for rs16952868 and rs3865014 with triglycerides and for rs3865014 with HDL-C levels in males; and for rs11631403 and rs3865014 with triglycerides and for rs11631403 with HDL-C levels in females in the APOE2/3 group (Tables 6 and 7).
Table 6.
GLCE SNPs and adjusted plasma triglyceride (mg/dl) levels in the 2000–2003 cohort
| Males | Females | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GLCE SNPs | AA | AB | BB | p | Permuted p | AA | AB | BB | p | Permuted p | |
| |
|
||||||||||
| rs16952868 | 148 ± 89 (153) | 157 ± 101 (228) | 160 ± 104 (87) | 0.043 | NS | 112 ± 66 (217) | 122 ± 88 (323) | 120 ± 83 (141) | NS | NS | |
| rs11631403 | 150 ± 91 (209) | 158 ± 99 (208) | 166 ± 120 (51) | 0.031 | 0.098 | 113 ± 65 (290) | 121 ± 90 (301) | 129 ± 89 (92) | 0.028 | 0.085 | |
| rs17360351 | 153 ± 95 (357) | 163 ± 103 (102) | 168 ± 151 (8) | 0.081 | NS | 116 ± 73 (510) | 125 ± 100 (162) | 128 ± 91 (11) | NS | NS | |
| rs3865014 | 151 ± 90 (250) | 158 ± 106 (185) | 168 ± 102 (32) | 0.041 | NS | 112 ± 80 (362) | 119 ± 77 (267) | 136 ± 102 (55) | 0.021 | 0.076 | |
| rs6545 | 152 ± 91 (190) | 156 ± 102 (209) | 163 ± 101 (65) | NS | NS | 117 ± 84 (283) | 115 ± 65 (301) | 130 ± 111 (95) | NS | NS | |
| GLCE | APOE | AA | AB | BB | p | Permuted p | AA | AB | BB | p | Permuted p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
||||||||||
| rs16952868 | 3/3 + 3/4 | 149 ± 90 (131) | 152 ± 94 (195) | 155 ± 103 (75) | NS | NS | 108 ± 77 (188) | 115 ± 90 (281) | 112 ± 82 (123) | NS | NS |
| 2/3 | 141 ± 91 (19) | 162 ± 101 (22) | 166 ± 92 (12) | 0.017 | 0.045 | 100 ± 55 (23) | 119 ± 81 (33) | 120 ± 95 (15) | 0.077 | NS | |
| rs11631403 | 3/3 + 3/4 | 149 ± 93 (180) | 151 ± 95 (178) | 157 ± 104 (44) | NS | NS | 109 ± 72 (250) | 112 ± 92 (261) | 116 ± 89 (80) | NS | NS |
| 2/3 | 143 ± 92 (23) | 163 ± 98 (22) | 164 ± 99 (6) | 0.041 | NS | 98 ± 52 (28) | 115 ± 84 (34) | 139 ± 101 (10) | 0.018 | 0.048 | |
| rs17360351 | 3/3 + 3/4 | 151 ± 92 (306) | 157 ± 102 (87) | 159 ± 119 (7) | NS | NS | 110 ± 85 (442) | 117 ± 100 (142) | 120 ± 101 (10) | NS | NS |
| 2/3 | 155 ± 102 (38) | 159 ± 102 (12) | 164 (1) | NS | NS | 109 ± 61 (51) | 121 ± 88 (18) | 141 (1) | NS | NS | |
| rs3865014 | 3/3 + 3/4 | 149 ± 90 (210) | 155 ± 96 (164) | 158 ± 108 (30) | NS | NS | 111 ± 84 (311) | 114 ± 82 (231) | 123 ± 102 (50) | NS | NS |
| 2/3 | 147 ± 89 (27) | 166 ± 98 (22) | 178 ± 72 (4) | 0.021 | 0.047 | 99 ± 73 (37) | 126 ± 82 (27) | 128 ± 71 (7) | 0.023 | 0.048 | |
| rs6545 | 3/3 + 3/4 | 151 ± 95 (163) | 153 ± 102 (180) | 158 ± 103 (56) | NS | NS | 112 ± 85 (243) | 111 ± 73 (266) | 117 ± 101 (82) | NS | NS |
| 2/3 | 150 ± 94 (21) | 159 ± 101 (21) | 164 ± 71 (10) | 0.078 | NS | 107 ± 77 (28) | 115 ± 62 (30) | 126 ±101 (10) | 0.039 | NS | |
All means ± SD were calculated by analysis of covariance using general linear models and were adjusted for age, body mass index, smoking, and alcohol consumption. Total number of subjects: 474 males, 690 females.
Number of subjects for each group is shown in parentheses. p values were calculated using an additive genotypic model in PLINK. Permutation p values were calculated from 50,000 random iterations in PLINK.
A, common allele; B, rare allele. NS, not significant (p ≥ 0.1). Significant (<0.05) results are shown in bold.
Table 7.
GLCE SNPs and adjusted plasma HDL-C (mg/dl) levels in the 2000–2003 cohort
| Males | Females | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GLCE SNPs | AA | AB | BB | p | Permuted p | AA | AB | BB | p | Permuted p | |
| |
|
||||||||||
| rs16952868 | 39.2 ± 8.6 (153) | 38.4 ± 7.9 (228) | 38.7 ± 8.1 (87) | NS | NS | 46.9 ± 9.9 (217) | 47.2 ± 11.0 (323) | 46.7 ± 10.7 (141) | NS | NS | |
| rs11631403 | 39.2 ± 8.5 (209) | 38.5 ± 8.0 (208) | 37.9 ± 7.4 (51) | 0.028 | 0.094 | 46.8 ± 9.8 (290) | 47.6 ± 11.0 (301) | 45.7 ± 11.2 (92) | 0.091 | NS | |
| rs17360351 | 38.8 ± 8.2 (357) | 38.4 ± 8.0 (102) | 38.3 ± 6.6 (8) | NS | NS | 47.1 ± 10.4 (510) | 46.8 ± 11.5 (162) | 44.5 ± 8.6 (11) | NS | NS | |
| rs3865014 | 39.1 ± 8.6 (250) | 38.5 ± 7.8 (185) | 37.4 ± 7.3 (32) | 0.039 | NS | 46.7 ± 9.8 (362) | 47.7 ± 11.5 (267) | 44.6 ± 11.1 (55) | NS | NS | |
| rs6545 | 39.1 ± 8.5 (190) | 38.6 ± 8.1 (209) | 38.2 ± 7.9 (65) | NS | NS | 46.7 ± 9.8 (283) | 47.7 ± 11.3 (301) | 45.9 ± 11.0 (95) | NS | NS | |
| GLCE | APOE | AA | AB | BB | p | Permuted p | AA | AB | BB | p | Permuted p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
||||||||||
| rs16952868 | 3/3 + 3/4 | 39.4 ± 8.2 (131) | 38.9 ± 8.5 (195) | 38.5 ± 8.1 (75) | NS | NS | 46.8 ± 9.8 (188) | 48 ± 10.6 (281) | 48.1 ± 10.9 (123) | NS | NS |
| 2/3 | 40.9 ± 10.1 (19) | 39.3 ± 7.4 (22) | 39.5 ± 7.0 (12) | 0.085 | NS | 49.4 ± 10.8 (23) | 48.3 ± 10.2 (33) | 48.1 ± 10.4 (15) | NS | NS | |
| rs11631403 | 3/3 + 3/4 | 39.5 ± 8.5 (180) | 38.8 ± 8 (178) | 38.4 ± 7.9 (44) | NS | NS | 47.4 ± 9.8 (250) | 48.1 ± 10.9 (261) | 47 ± 11.2 (80) | NS | NS |
| 2/3 | 41.1 ± 9.4 (23) | 39.1 ± 7.5 (22) | 38.6 ± 7.1 (6) | 0.043 | NS | 49.5 ± 9.9 (28) | 48.4 ± 10.1 (34) | 46.2 ± 10.0 (10) | 0.021 | 0.048 | |
| rs17360351 | 3/3 + 3/4 | 39.2 ± 8.8 (306) | 38.7 ± 8.3 (87) | 38.2 ± 7.0 (7) | NS | NS | 46.9 ± 10.3 (442) | 47.7 ± 11.5 (142) | 45.5 ± 7.7 (10) | NS | NS |
| 2/3 | 39.6 ± 8.2 (38) | 38.9 ± 8.2 (12) | 39.3 (1) | NS | NS | 48.9 ± 10.9 (51) | 47.5 ± 9.4 (18) | 42.8 (1) | NS | NS | |
| rs3865014 | 3/3 + 3/4 | 39.4 ± 8.7 (210) | 38.7 ± 8 (164) | 38.1 ± 7.6 (30) | NS | NS | 47.5 ± 9.9 (311) | 48.1 ± 11.5 (231) | 46.6 ± 11 (50) | NS | NS |
| 2/3 | 40.5 ± 8.2 (27) | 39.8 ± 7.5 (22) | 36.8 ± 7.1 (4) | 0.017 | 0.044 | 49.6 ± 9.3 (37) | 47.8 ± 11.1 (27) | 46.1 ± 9.2 (7) | 0.036 | 0.091 | |
| rs6545 | 3/3 + 3/4 | 39.4 ± 8.8 (163) | 38.8 ± 8.3 (180) | 38.7 ± 8.2 (56) | NS | NS | 47 ± 9.8 (243) | 48.4 ± 11.3 (266) | 47.1 ± 10.9 (82) | NS | NS |
| 2/3 | 40.1 ± 9.1 (21) | 40.1 ± 7.7 (21) | 39.1 ± 6.7 (10) | NS | NS | 48.8 ± 10.6 (28) | 48.6 ± 10.8 (30) | 47.7 ± 9.8 (10) | NS | NS | |
All means ± SD were calculated by analysis of covariance using general linear models and were adjusted for age, body mass index, smoking, and alcohol consumption. Total number of subjects: 474 males, 690 females.
Number of subjects for each group is shown in parentheses. p values were calculated using an additive genotypic model in PLINK. Permutation p values were calculated from 50,000 random iterations in PLINK.
A, common allele; B, rare allele. NS, not significant (p ≥ 0.1). Significant (<0.05) results are shown in bold.
Because LIPC variants are significantly associated with plasma HDL-C levels in this unrelated Turkish cohort (Hodoğlugil et al., 2010), we evaluated the interaction between GLCE (rs16952868, rs11631403, and rs3865014) and LIPC SNPs. However, no significant interaction was observed for either HDL-C or triglyceride levels between SNPs of these two genes (data not shown).
Plasma Lipid and Lipoprotein Analysis of glce+/– Mice
On a chow or high fat diet (TD.88137 or TD.02028, Harlan Teklad) glce+/– heterozygous mice did not have altered plasma triglyceride and total cholesterol levels compared to wildtype mice (glce+/+). However, glce+/– heterozygous mice on apoe knockout (apoe–/–) background had a significant elevation in both plasma triglyceride and total cholesterol levels on high fat diet compared to apoe knockout mice (Table 8).
Table 8.
Plasma triglyceride and total cholesterol levels in groups of mice under different dietary conditions
| Triglyceride (mg/dl) |
Total cholesterol (mg/dl) |
||||||
|---|---|---|---|---|---|---|---|
| Chow | High fat_1 (TD88137) | High fat_2 (TD02028) | Chow | High fat_1 (TD88137) | High fat_2 (TD02028) | ||
| apoe +/+ | glce+/+ (wildtype) | 49 ± 9 (9) | 50 ± 15 (7) | 46 ± 11 (15) | 65 ± 6 (9) | 171 ± 34 (7) | 182 ± 31 (15) |
| glce +/– | 55 ± 23 (18) | 48 ± 10 (15) | 42 ± 14 (12) | 76 ± 22 (18) | 168 ± 48 (15) | 179 ± 21 (12) | |
| p (glce+/+ vs. glce+/–) | NS | NS | NS | NS | NS | NS | |
| apoe –/– | glce +/+ | 108 ± 23 (41) | 99 ± 23 (29) | 86 ± 19 (25) | 399 ± 70 (41) | 1068 ± 276 (29) | 1695 ± 323 (25) |
| glce +/– | 119 ± 28 (25) | 121 ± 28 (18) | 107 ± 35 (19) | 428 ± 103 (25) | 1302 ± 237 (18) | 1996 ± 507 (19) | |
| p (glce+/+ vs. glce+/–) | NS | <0.005 | <0.01 | NS | <0.005 | <0.02 | |
The number of subjects for each group is shown in parentheses.
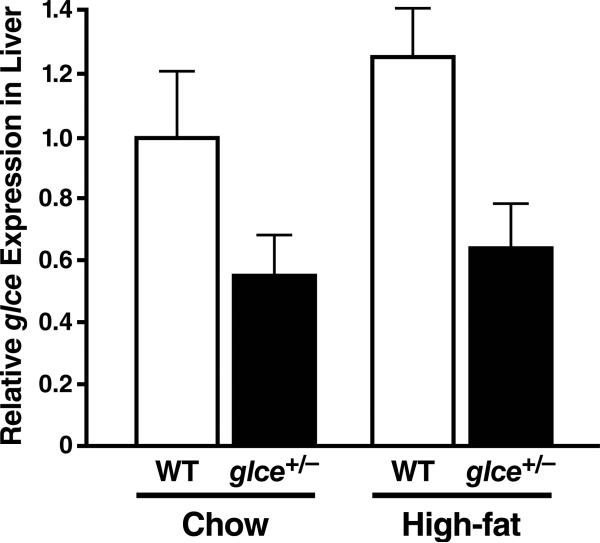
Glce mRNA expression was measured in liver samples of wildtype and glce+/– mice on a chow or high-fat diet (TD.02028). Glce mRNA expression was ~50% lower in glce+/– mice and was not increased by a high-fat diet (Fig. 5). There was no clear difference in the weights of wildtype and glce+/– mice on the different diets (data not shown).
Figure 5.
Glce expression in mouse liver. Quantitative PCR was performed with cDNA. Results were analyzed with the delta-delta Ct method. Data were normalized to expression in the wildtype (WT) liver, and gusb expression was used for internal normalization. Values are mean ± SD (n = 4 per group) of triplicate measurements and two different experiments.
Discussion
Previously, using a linkage analysis approach, we mapped a locus for HDL-C on chromosome 15q21-23 in a group of Turkish families (Yu et al., 2005). LIPC is located under this linkage peak and thus emerged as a strong candidate gene. By fine mapping with additional genetic markers, we substantially narrowed the region under the linkage peak. Because LIPC is not located under the more narrowly defined linkage peak, we evaluated association of HDL-C and triglyceride levels in the 40 Turkish families with a set of SNPs tagging all of the haplotype blocks in a one-half megabase region under the linkage peak. Nominal significant results for both traits were obtained with several SNPs, primarily in the coding and promoter regions of GLCE. Haplotype analysis of these SNPs showed that one particular haplotype (composed of rare alleles) was associated with both traits in permutation tests in families. Three GLCE SNPs were associated with plasma triglyceride and HDL-C levels in two cohorts of unrelated Turkish subjects. The association was noticeably stronger in the subset of individuals with the APOE2/3 genotype. Plasma lipid analyses of glce+/– mice on the apoe–/– background also support the involvement of glce in lipid metabolism, because these mice had higher total cholesterol and triglycerides when fed a high-fat diet.
GLCE plays a significant role in the biosynthesis of HSPGs, which are composed of a core protein and one or more heparan sulfate glycosaminoglycan chains. Several enzymes are involved in HSPG synthesis, including GLCE, which converts D-glucuronic acid to L-iduronic acid (Bishop et al., 2007; Bishop et al., 2008). This epimerization process gives more flexibility to the HS chain and promotes ligand apposition by increasing conformational flexibility (Casu & Lindahl, 2001). All identified protein-binding heparan sulfate epitopes contain L-iduronic acid, suggesting the importance of iduronic acid (and GLCE) in the diverse functions of heparan sulfate chains. With their high negative charge, heparan sulfate chains bind to a wide range of proteins, including apoE- and apoB-containing lipoproteins, hepatic lipase, and lipoprotein lipases; therefore, they have many important functions in the clearance of atherogenic triglyceride-rich lipoproteins (Williams et al., 1992; Williams & Fuki, 1997). These particles can be cleared by HSPG acting alone, by the HSPG/low density lipoprotein receptor–related protein complex, or by the low density lipoprotein receptor in the liver. HSPGs in the space of Disse sequester triglyceride-rich lipoproteins, thereby facilitating interactions with cell-surface receptors. ApoE is a critical ligand in this process (for a review, see Mahley et al., 1999; Mahley & Ji, 1999; Mahley & Huang, 2007).
Several GLCE SNPs and a haplotype constructed with minor alleles of nine SNPs in this gene were associated with elevated triglyceride levels and decreased HDL-C in Turkish families. Three of these SNPs (rs16952868, rs11631403, and rs3865014) were nominally associated with these traits in two independent, large, unrelated Turkish cohorts (3776 individuals). These associations were stronger and limited to APOE2/3 individuals. These observations are consistent with the idea that impaired binding of apoE2 to hepatic lipoprotein receptors and HSPGs exacerbates the impact of GLCE variants on lipoprotein clearance and strengthen the conclusion that GLCE plays a role in modulating lipoprotein metabolism.
Apoe–/–/glce+/– mice fed a high-fat diet had higher plasma cholesterol and triglyceride levels than apoe–/– mice. Targeted disruption of glce results in neonatal lethality; however, no overt abnormalities were observed in glce+/– mice (Li et al., 2003). To elicit an effect of glce+/– on lipoprotein metabolism, a high-fat diet was required to impair lipoprotein clearance. In the future, conditional knockout of glce in the liver will be required to determine the full spectrum of effects of glce hepatic expression.
Other enzymes in the HSPG synthetic pathway also modulate lipoprotein metabolism (Bishop et al., 2007; Bishop et al., 2008). Conditional knockout of ndst1 (MacArthur et al., 2007) and hs2st (Stanford et al., 2010) in mice increases the level of triglyceride-rich lipoproteins. In humans, GALNT2 polymorphisms are significantly associated with plasma HDL-C levels (Willer et al., 2008). This observation was replicated in another genome-wide association study (Kathiresan et al., 2008).
The importance of the human chromosome 15q21-23 in lipoprotein metabolism has also been suggested by studies of orthologous regions in mice and rats. Quantitative trait analysis revealed a linkage to triglyceride levels in rats (Klimes et al., 2003) and to plasma triglycerides (Suto et al., 1999) and HDL-C levels (Burgess-Herbert et al., 2009) in mice. The nearest markers for triglycerides (D9Mit163) (Suto et al., 1999) and HDL-C (rs4227694) (Burgess-Herbert et al., 2009) are only 0.6 Mb and 3.8 Mb away from the mouse glce locus, respectively. Additional support for the involvement of glce in lipid metabolism comes from two sources. One is Jackson Laboratory's mouse phenome database (http://phenome.jax.org) (Blake et al., 2009; Svenson et al., 2007), where several phenotypic measurements, including plasma lipid measurements, for different strains are deposited. The other source is the Perlegen mouse haplotype viewer (http://mouse.perlegen.com/mouse/), where over 8 million SNPs and haplotypes for 16 commonly used strains of inbred laboratory mice are stored (Frazer et al., 2007). Using these two sources, we grouped mouse strains according to their glce haplotypes and analyzed their lipid data (DiPetrillo et al., 2005; Guo et al., 2007; Wang et al., 2004). Mice with different glce haplotypes had different plasma triglyceride and total cholesterol values (Table S3).
Since Turks have lower HDL-C than other populations and borderline high triglycerides (Bersot et al., 1999; Mahley et al., 1995; Mahley et al., 2005; Onat et al., 1992; Porsch-Oezçueruemez et al., 1999; Tezcan et al., 2003), we compared the ancestry admixture of our subjects with data from HapMap and HGDP for other populations, paying specific attention to the 67.0–67.5 Mb region encompassing GLCE on chromosome 15. The Anatolian peninsula (present day Turkey) has served as a junction connecting the Middle East, Europe, and Central Asia, and thus has been subject to major population movements (Findley, 2005). The Turks, mainly Oghuz tribes, arrived in Turkey from Central Asia between the 11th and 13th centuries A.D. (Grousset, 1970; Güvenç, 1993). There was also admixture of the Indo-European-speaking people of Central Asia with nomadic people (including Turkic populations) from the northern steppes of Central and East Asia (Frye, 1996). The results of our population substructure analyses are consistent with these admixture events. About 40% of the ancestry estimates were indistinguishable among Europeans, Turks, and Central Asians (Fig. 3, red color). However, the subjects in our Turkish cohort have about 14% (8% Central Asian and 6% East Asian) of the ancestry estimates found in Central and East Asian populations as well; these ancestry estimates were absent in European populations (Fig. 3, orange and blue colors).
GLCE has not previously been genetically associated with lipid traits. There are well-powered examples of association studies that have not been replicated in populations of non-European descent (Hiura et al., 2009; Ken-Dror et al., 2010; Klos et al., 2005; Nakayama et al., 2009), and this may partly be explained by genetic uniqueness among various populations. GLCE is bounded by recombination hotspots (Fig. 2) and displays population differences with respect to SNP allele frequencies. Plotting the minor allele frequencies of SNPs across the chromosome 15 67.0−67.5 Mb region demonstrated a divergence of those frequencies in Turks and Europeans within the GLCE locus (Fig. 4A). In this locus, the Turkish pattern more closely resembles the Chinese (Fig. 4B). Furthermore, the frequency of the significant nine-SNP GLCE haplotype that was linked to high triglyceride and low HDL-C levels differs among various populations (9.4% in Turks, 5.4% in CEU, and 13.4% in CHB). This difference further suggests that admixture in Turks may have functional consequences in lipid metabolism.
Since the original linkage peak included LIPC and GLCE and LIPC variants are associated with HDL-C levels (Hodoğlugil et al., 2010; Kathiresan et al., 2008; Willer et al., 2008) and LIPC is involved in triglyceride clearance (Mahley & Huang, 2007; Mahley et al., 2008), we considered the potential for interaction between LIPC and GLCE on HDL-C and triglyceride levels. However, no interaction was observed between LIPC and GLCE SNPs on either triglyceride or HDL-C levels. Thus, the effects of these loci on lipid levels are probably independent.
Because the GLCE SNPs evaluated in this study are highly correlated, a parsimonious interpretation is that a single common variant in LD with these SNPs is the effector of HDL-C and triglyceride levels. However, since rare variants can have a major effect on plasma HDL-C levels (Cohen et al., 2004; Cohen et al., 2006) and simulation studies suggest that uncommon or rare genetic variants can easily create synthetic associations that are credited to common variants (Dickson et al., 2010), a thorough analysis and comparison of the GLCE genomic sequence in Turks and individuals from other populations is warranted.
In summary, we showed that several SNPs of GLCE are associated with plasma triglyceride and HDL-C levels in atherogenic dyslipidemic Turkish families and unrelated Turkish individuals. This association is stronger among those with the APOE2/3 genotype. Studies of glce+/– mice on an apoe–/– background supported the role of GLCE in lipid metabolism. Although fine genetic mapping of this region and association studies support a role for GLCE and LIPC (Hodoğlugil et al., 2010) in the modulation of HDL-C and triglyceride levels, other genes within the 15q21-23 region may also affect lipid levels in Turks.
Supplementary Material
Acknowledgments
DNA samples were provided through the Turkish Heart Study and the Genetic Epidemiology of Metabolic Syndrome study, a multinational collaborative project supported by GlaxoSmithKline. The authors acknowledge the support of GlaxoSmithKline scientists and project leaders, especially Vincent Mooser, M.D., and Dawn M. Waterworth, Ph.D. The authors thank Stephen Schaffner (BROAD Institute of MIT and Harvard) and Tara Matise (Rutgers University) for providing pairwise HapMap Fst and mapping (cM) data for individual SNPs, respectively. The authors also thank Dr. Vivian G. Cheung, (University of Pennsylvania) for valuable input and critical reading of the manuscript. In addition, the authors are indebted to their associates at the American Hospital, Istanbul, especially Drs. K. Erhan Palaoğlu, Oryal Gökdemir, Sinan Özbayrakcı and Kerem Özer and Guy Pépin, Sibel Tanir, Judy Dawson-Pépin, and Linda L. Mahley. The authors thank Sylvia Richmond for manuscript preparation and Mimi Zeiger and Stephen Ordway for editorial assistance. The authors acknowledge the generous support of the American Hospital, especially Mr. George Rountree, and the J. David Gladstone Institutes. This work was supported in part by grant R01 HL71027 from the National Institutes of Health.
Footnotes
Conflict of Interest
The Authors have no conflict of interest to disclose.
References
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Hixson JE, Rainwater DL, Cole S, Williams JT, Mahaney MC, VandeBerg JL, Stern MP, MacCluer JW, Blangero J. Human pedigree-based quantitative-trait–locus mapping: Localization of two genes influencing HDL-cholesterol metabolism. Am J Hum Genet. 1999;64:1686–1693. doi: 10.1086/302425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MA, Edwards KL, Monks SA, Koprowicz KM, Brunzell JD, Motulsky AG, Mahaney MC, Hixson JE. Genome-wide scan for quantitative trait loci influencing LDL size and plasma triglyceride in familial hypertriglyceridemia. J Lipid Res. 2003;44:2161–2168. doi: 10.1194/jlr.M300272-JLR200. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bersot TP, Vega GL, Grundy SM, Palaoğlu KE, Atagündüz P, Özbayrakçi S, Gökdemir O, Mahley RW. Elevated hepatic lipase activity and low levels of high density lipoprotein in a normotriglyceridemic, nonobese Turkish population. J Lipid Res. 1999;40:432–438. [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Stanford KI, Esko JD. Heparan sulfate proteoglycans and triglyceride-rich lipoprotein metabolism. Curr Opin Lipidol. 2008;19:307–313. doi: 10.1097/MOL.0b013e3282feec2d. [DOI] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE, Mouse Genome Database Group The Mouse Genome Database genotypes::phenotypes. Nucleic Acids Res. 2009;37:D712–D719. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Herbert SL, Tsaih S-W, Stylianou IM, Walsh K, Cox AJ, Paigen B. An experimental assessment of in silico Haplotype association mapping in laboratory mice. BMC Genet. 2009;10:81. doi: 10.1186/1471-2156-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casu B, Lindahl U. Structure and biological interactions of heparin and heparan sulfate. Adv Carbohydr Chem Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci USA. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPetrillo K, Wang X, Stylianou IM, Paigen B. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 2005;21:683–692. doi: 10.1016/j.tig.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitosa MF, Province MA, Heiss G, Arnett DK, Myers RH, Pankow JS, Hopkins PN, Borecki IB. Evidence of QTL on 15q21 for high-density lipoprotein cholesterol: The National Heart, Lung, and Blood Institute Family Heart Study (NHLBI FHS). Atherosclerosis. 2007;190:232–237. doi: 10.1016/j.atherosclerosis.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Findley CV. Islam and empire from the Seljuks through the Mongols. The Turks in World History. Oxford University Press; New York: 2005. pp. 56–92. [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. others. [DOI] [PubMed] [Google Scholar]
- Frye RN. The Heritage of Central Asia From Antiquity to the Turkish Expansion. Marcus Wiener Publishers; Princeton: 1996. The present is born. pp. 233–239. [Google Scholar]
- Gordon DJ, Probstfield JL, Garrison RJ, Neaton JD, Castelli WP, Knoke JD, Jacobs DR, Jr., Bangdiwala S, Tyroler HA. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation. 1989;79:8–15. doi: 10.1161/01.cir.79.1.8. [DOI] [PubMed] [Google Scholar]
- Grousset R. The Empire of the Steppes A History of Central Asia. Rutgers University Press; New Brunswick: 1970. The Turks and Islam to the thirteenth century. pp. 141–170. [Google Scholar]
- Guo Y, Lu P, Farrell E, Zhang X, Weller P, Monshouwer M, Wang J, Liao G, Zhang Z, Hu S. In silico and in vitro pharmacogenetic analysis in mice. Proc Natl Acad Sci USA. 2007;104:17735–17740. doi: 10.1073/pnas.0700724104. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güvenç B. Türk Kimliği Kültür Tarihinin Kaynakları (in Turkish) (Turkish Identity Sources of Cultural History) Kültür Bakanlığı; Ankara: 1993. Türklerin Kimliği: Kim Bu Türkler? (Identity of Turks: Who are the Turks?). pp. 19–52. [Google Scholar]
- Heller DA, de Faire U, Pedersen NL, Dahlén G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–1156. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- Hiura Y, Shen C-S, Kokubo Y, Okamura T, Morisaki T, Tomoike H, Yoshida T, Sakamoto H, Goto Y, Nonogi H. Identification of genetic markers associated with high-density lipoprotein-cholesterol by genome-wide screening in a Japanese population—the Suita Study. Circ J. 2009;73:1119–1126. doi: 10.1253/circj.cj-08-1101. others. [DOI] [PubMed] [Google Scholar]
- Hodoğlugil U, Williamson DW, Huang Y, Mahley RW. Common polymorphisms of ATP binding cassette transporter A1, including a functional promoter polymorphism, associated with plasma high density lipoprotein cholesterol levels in Turks. Atherosclerosis. 2005;183:199–212. doi: 10.1016/j.atherosclerosis.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Hodoğlugil U, Tanyolaç S, Williamson DW, Huang Y, Mahley RW. Apolipoprotein A-V: A potential modulator of plasma triglyceride levels in Turks. J Lipid Res. 2006;47:144–153. doi: 10.1194/jlr.M500343-JLR200. [DOI] [PubMed] [Google Scholar]
- Hodoğlugil U, Williamson DW, Mahley RW. Polymorphisms in the hepatic lipase gene affect plasma HDL-cholesterol levels in a Turkish population. J Lipid Res. 2010;51:422–430. doi: 10.1194/jlr.P001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs DR, Jr., Mebane IL, Bangdiwala SI, Criqui MH, Tyroler HA. High density lipoprotein cholesterol as a predictor of cardiovascular disease mortality in men and women: The follow-up study of the Lipid Research Clinics Prevalence Study. Am J Epidemiol. 1990;131:32–47. doi: 10.1093/oxfordjournals.aje.a115483. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ken-Dror G, Goldbourt U, Dankner R. Different effects of apolipoprotein A5 SNPs and haplotypes on triglyceride concentration in three ethnic origins. J Hum Genet. 2010;55:300–307. doi: 10.1038/jhg.2010.27. [DOI] [PubMed] [Google Scholar]
- Klimes I, Weston K, Kovacs P, Gasperikova D, Jezova D, Kvetnansky R, Thompson JR, Sebokova E, Samani NJ. Mapping of genetic loci predisposing to hypertriglyceridaemia in the hereditary hypertriglyceridaemic rat: Analysis of genetic association with related traits of the insulin resistance syndrome. Diabetologia. 2003;46:352–358. doi: 10.1007/s00125-003-1035-6. [DOI] [PubMed] [Google Scholar]
- Klos KLE, Hamon S, Clark AG, Boerwinkle E, Liu K, Sing CF. APOA5 polymorphisms influence plasma triglycerides in young, healthy African Americans and whites of the CARDIA Study. J Lipid Res. 2005;46:564–570. doi: 10.1194/jlr.M400437-JLR200. [DOI] [PubMed] [Google Scholar]
- Knoblauch H, Bauerfeind A, Toliat MR, Becker C, Luganskaja T, Günther UP, Rohde K, Schuster H, Junghans C, Luft FC. Haplotypes and SNPs in 13 lipid-relevant genes explain most of the genetic variance in high-density lipoprotein and low-density lipoprotein cholesterol. Hum Mol Genet. 2004;13:993–1004. doi: 10.1093/hmg/ddh119. others. [DOI] [PubMed] [Google Scholar]
- Li J-P, Gong F, Hagner-McWhirter Å, Forsberg E, Åbrink M, Kisilevsky R, Zhang X, Lindahl U. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J Biol Chem. 2003;278:28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. others. [DOI] [PubMed] [Google Scholar]
- Li X, Monda KL, Göring HHH, Haack K, Cole SA, Diego VP, Almasy L, Laston S, Howard BV, Shara NM. Genome-wide linkage scan for plasma high density lipoprotein cholesterol, apolipoprotein A-1 and triglyceride variation among American Indian populations: The Strong Heart Family Study. J Med Genet. 2009;46:472–479. doi: 10.1136/jmg.2008.063891. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur JM, Bishop JR, Stanford KI, Wang L, Bensadoun A, Witztum JL, Esko JD. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. J Clin Invest. 2007;117:153–164. doi: 10.1172/JCI29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaney MC, Blangero J, Rainwater DL, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW, Hixson JE. A major locus influencing plasma high-density lipoprotein cholesterol levels in the San Antonio Family Heart Study: Segregation and linkage analyses. Arterioscler Thromb Vasc Biol. 1995;15:1730–1739. doi: 10.1161/01.atv.15.10.1730. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Palaoğlu KE, Atak Z, Dawson-Pepin J, Langlois A-M, Cheung V, Onat H, Fulks P, Mahley LL, Vakar F. Turkish Heart Study: Lipids, lipoproteins, and apolipoproteins. J Lipid Res. 1995;36:839–859. others. [PubMed] [Google Scholar]
- Mahley RW, Huang Y, Rall SC., Jr. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia): Questions, quandaries, and paradoxes. J Lipid Res. 1999;40:1933–1949. [PubMed] [Google Scholar]
- Mahley RW, Ji Z-S. Remnant lipoprotein metabolism: Key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40:1–16. [PubMed] [Google Scholar]
- Mahley RW, Can S, Özbayrakçı S, Bersot TP, Tanir S, Palaoğlu KE, Pépin GM. Modulation of high-density lipoproteins in a population in Istanbul, Turkey, with low levels of high-density lipoproteins. Am J Cardiol. 2005;96:547–555. doi: 10.1016/j.amjcard.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Huang Y. Atherogenic remnant lipoproteins: Role for proteoglycans in trapping, transferring, and internalizing. J Clin Invest. 2007;117:94–98. doi: 10.1172/JCI30889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, Bersot TP. Disorders of lipid metabolism. In: Kronenberg HM, Melmed S, Polonsky KS, Larsen PR, editors. Williams Textbook of Endocrinology. Saunders; Philadelphia: 2008. pp. 1589–1653. [Google Scholar]
- Murray CJL, Lopez AD. Mortality by cause for eight regions of the world: Global Burden of Disease study. Lancet. 1997;349:1269–1276. doi: 10.1016/S0140-6736(96)07493-4. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Bayasgalan T, Yamanaka K, Kumada M, Gotoh T, Utsumi N, Yanagisawa Y, Okayama M, Kajii E, Ishibashi S. Large scale replication analysis of loci associated with lipid concentrations in a Japanese population. J Med Genet. 2009;46:370–374. doi: 10.1136/jmg.2008.064063. others. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onat A, Surdum-Avcı G, Senocak M, Örnek E, Gözükara Y. Plasma lipids and their interrelationship in Turkish adults. J Epidemiol Community Health. 1992;46:470–476. doi: 10.1136/jech.46.5.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porsch-Oezçueruemez M, Bilgin Y, Wollny M, Gediz A, Arat A, Karatay E, Akinci A, Sinterhauf K, Koch H, Siegfried I. Prevalence of risk factors of coronary heart disease in Turks living in Germany: The Giessen Study. Atherosclerosis. 1999;144:185–198. doi: 10.1016/s0021-9150(99)00054-4. others. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins J, Chen Y, Paigen B, Wang X. In search of new targets for plasma high-density lipoprotein cholesterol levels: Promise of human–mouse comparative genomics. Trends Cardiovasc Med. 2006;16:220–234. doi: 10.1016/j.tcm.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Stanford KI, Wang L, Castagnola J, Song D, Bishop JR, Brown JR, Lawrence R, Bai X, Habuchi H, Tanaka M. Heparan sulfate 2-O-sulfotransferase is required for triglyceride-rich lipoprotein clearance. J Biol Chem. 2010;285:286–294. doi: 10.1074/jbc.M109.063701. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe III W, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Suto J, Matsuura S, Yamanaka H, Sekikawa K. Quantitative trait loci that regulate plasma lipid concentration in hereditary obese KK and KK-Ay mice. Biochim Biophys Acta. 1999;1453:385–395. doi: 10.1016/s0925-4439(99)00013-7. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Von Smith R, Magnani PA, Suetin HR, Paigen B, Naggert JK, Li R, Churchill GA, Peters LL. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol. 2007;102:2369–2378. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- Tezcan S, Altıntaş H, Sönmez R, Akinci A, Doğan B, Çakır B, Bilgin Y, Klör HU, Razum O. Cardiovascular risk factor levels in a lower middle-class community in Ankara, Turkey. Trop Med Int Health. 2003;8:660–667. doi: 10.1046/j.1365-3156.2003.01057.x. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermann G. Genetic polymorphism of apolipoprotein E – impact on plasma lipoprotein metabolism. In: Crepaldi G, Tiengo A, Baggio G, editors. Diabetes, Obesity and Hyperlipidemias. III Elsevier Science Publishers; Amsterdam: 1985. pp. 1–28. [Google Scholar]
- Wang X, Korstanje R, Higgins D, Paigen B. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Res. 2004;14:1767–1772. doi: 10.1101/gr.2668204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KJ, Fless GM, Petrie KA, Snyder ML, Brocia RW, Swenson TL. Mechanisms by which lipoprotein lipase alters cellular metabolism of lipoprotein(a), low density lipoprotein, and nascent lipoproteins. Roles for low density lipoprotein receptors and heparan sulfate proteoglycans. J Biol Chem. 1992;267:13284–13292. [PubMed] [Google Scholar]
- Williams KJ, Fuki IV. Cell-surface heparan sulfate proteoglycans: Dynamic molecules mediating ligand catabolism. Curr Opin Lipidol. 1997;8:253–262. doi: 10.1097/00041433-199710000-00003. [DOI] [PubMed] [Google Scholar]
- Wyszynski DF, Waterworth DM, Barter PJ, Cohen J, Kesäniemi YA, Mahley RW, McPherson R, Waeber G, Bersot TP, Sharma SS. Relation between atherogenic dyslipidemia and the Adult Treatment Program-III definition of metabolic syndrome (Genetic Epidemiology of Metabolic Syndrome Project). Am J Cardiol. 2005;95:194–198. doi: 10.1016/j.amjcard.2004.08.091. others. [DOI] [PubMed] [Google Scholar]
- Yang R, Li L, Seidelmann SB, Shen G-Q, Sharma S, Rao S, Abdullah KG, MacKinlay KG, Elston RC, Chen Q. A genome-wide linkage scan identifies multiple quantitative trait loci for HDL-cholesterol levels in families with premature CAD and MI. J Lipid Res. 2010;51:1442–1451. doi: 10.1194/jlr.M004325. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wyszynski DF, Waterworth DM, Wilton SD, Barter PJ, Kesäniemi YA, Mahley RW, McPherson R, Waeber G, Bersot TP. Multiple QTLs influencing triglyceride and HDL and total cholesterol levels identified in families with atherogenic dyslipidemia. J Lipid Res. 2005;46:2202–2213. doi: 10.1194/jlr.M500137-JLR200. others. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.