Abstract
Glutamatergic axons in the mammalian forebrain terminate predominantly onto dendritic spines. Long-term changes in the efficacy of these excitatory synapses are tightly coupled to changes in spine morphology. The reorganization of the actin cytoskeleton underlying this spine “morphing” involves numerous proteins that provide the machinery needed for adaptive cytoskeletal remodeling. Here we review recent literature addressing the chemical architecture of the spine, focusing mainly on actin-binding proteins (ABPs). Accumulating evidence suggests that ABPs are organized into functionally-distinct microdomains within the spine cytoplasm. This functional compartmentalization provides a structural basis for regulation of the spinoskeleton, offering a novel window into mechanisms underlying synaptic plasticity.
Excitatory signals in the mammalian forebrain are transmitted mainly by glutamatergic axons that terminate onto dendritic spines of pyramidal neurons. Most spines are tiny. A typical spine in CA1 hippocampus has a volume of ~0.02–0.03 fl [1], making them difficult to resolve with the limited resolution provided by the standard tools of live-cell imaging. Recent dramatic advances in super-resolution light microscopy [2, 3, 4, 5] can provide important insights into sub-spine dynamics. However, the outstanding spatial resolution provided by transmission electron microscopy (EM) makes it the primary tool of choice for studying the fine structure of the spine, though it is unsuitable for visualization of living cells.
Spine morphology
A variety of spine shapes can be distinguished, including thin, stuby, and mushroom-shaped (Fig. 1A, [6]). Mushroom-shaped spines are linked to their parent dendrite through a thin neck. It has been suggested that the spine neck might help to isolate the synapse from the shaft electrically (see review by [7]), though this idea remains controversial, and technical challenges make it difficult to address experimentally. In contrast, considerable evidence supports that the neck can provide a diffusion barrier for second messengers like Ca2+ [8, 9, 10, 11]. The biochemical compartmentalization thus achieved can isolate activity of synapses between neighboring spines, allowing independent modulation of single synapses [12, 13].
Figure 1. Appearance of dendritic spines in the electron microscope.
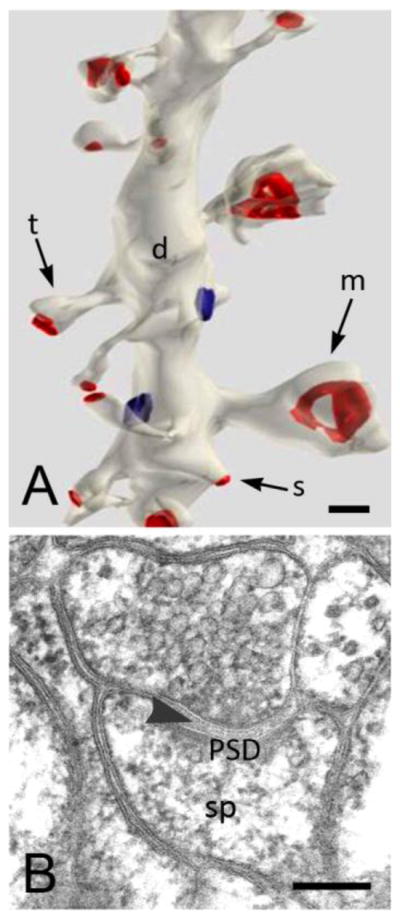
A. 3D reconstruction of serial thin sections (from stratum radiatum of rat CA1 shows stubby (s) spines on the same segment of a dendritic shaft as spines with thin (t) and mushroom (m) shapes. Synaptic contacts have been colorized in red. Image is from http://synapses.clm.utexas.edu/anatomy/compare/compare.stm, reprinted with kind permission from J. Spacek. B. Micrograph of a thin (70 nm) section shows a typical mushroom-shaped spine from the rat hippocampus. Note the presynaptic terminal with synaptic vesicles, the synaptic cleft (arrowhead) and the postsynaptic density (PSD). The filamentous material in the spine head (sp) represents the actin cytoskeleton. Scale bars: 200 nm
Spines proliferate rapidly in early postnatal life, but many of these early spines regress and disappear during maturation. Despite early suggestions that spines might be plastic or motile [14], by the latter part of the 20th century it was widely thought that spines in the mature brain were stable and fixed [see review by 15]. The introduction of new tools that allow direct visualization of spines in intact brain has led to the development of a more nuanced perspective. Large spines are generally stable [16, 17, 18]. In contrast, small spines are quite dynamic even in the adult; current evidence suggests that many of them are transient, and may either expand or disappear within a few days [16, 19, 20].
Extensive research in both hippocampus and neocortex documents a close relationship between spine size and synaptic efficacy [21, 22, 23, 24]. Spine volume itself predicts the size of the postsynaptic density (PSD), the number of AMPA receptors (AMPARs) at the synapse, and the magnitude of excitatory postsynaptic currents (EPSCs) in response to presynaptic glutamate release [25, 26, 27, 28, 29]. Moreover, stimuli that trigger long-term synaptic potentiation lead to spine growth and the insertion of additional AMPARs into the postsynaptic membrane [30, 31], whereas stimuli that trigger long-term depression lead to spine shrinkage and endocytosis of AMPARs [32, 33, 34].
The causal direction of this relationship between spine size and synaptic efficacy remains unclear, but the phenomenological evidence implies the presence of sophisticated biochemical machinery that links synaptic activity to the spine cytoskeleton (the “spinoskeleton”) [35, 36, 37, 38, 39, 40, 41]. Thus, a clearer understanding of the regulation of spine architecture may offer new insight into mechanisms underlying the control of synaptic processing. This line of inquiry may also have clinical implications, since several syndromes with prominent neuropsychiatric features are caused by genetic lesions directly linked to dysregulation of the actin cytoskeleton; these mutations can also lead to specific patterns of spine disruption [42, 43, 44, 45]. Considerable recent work suggests that the spatial organization of relevant proteins represents an important aspect of spinoskeletal regulation. Before reviewing these data, we will provide a brief overview of the functional organization of the spine.
Domains at the spine surface
When viewed with the electron microscope, the spine’s most prominent feature is the postsynaptic denisty (PSD), an electron-dense specialization extending beneath the plasma membrane, closely aligned with the presynaptic active zone across the 20–30 nm synaptic cleft (Fig. 1B). The PSD, which serves as the primary signaling domain of the spine, contains a complex matrix of receptors, scaffolding, and signal transduction molecules [46, 47]. Notwithstanding considerable variability, different synaptic proteins tend to segregate to distinct regions within the PSD. For example, the PSD exhibits a pronounced “laminar” organization: receptors are embedded in the plasma membrane, and adaptor proteins like PSD-95 concentrate near the exterior edge of the cytoplasm, whereas scaffolds like the Shank family lie deeper in the cytoplasm [2, 48]. Though less conspicuously, the PSD also seems to be organized in the tangential axis along the plasma membrane; for example, NMDA receptors generally lie in a central zone of the PSD, whereas AMPA receptors are more peripherally situated [49, 50, 51].
Immunogold electron microscopy has shown that a perisynaptic zone of the postsynaptic membrane is enriched in several signal-related proteins, including mGluR1, diacylglycerol lipase-α (an essential enzyme in endocannabinoid signaling at central synapses) and the glutamate transporter EAAC1 [52, 53, 54], though this zone is not morphologically distinctive. Beyond the perisynaptic region is a specialized trafficking zone for entry and exit of protein cargoes. This microdomain (originally identified with functional live-cell imaging) is not readily visualized even with electron microscopy, but its location has been verified using immunogold EM to demonstrate the organization of proteins associated with endo- and exocytosis [55, 56, 57]. It is generally assumed that the zone adjacent to the spine neck is also biochemically specialized, though little direct evidence is available. In summary, the presence of multiple functionally-distinct domains along the plasma membrane of the spine is now generally accepted [58], though the functional implications of this organization remain poorly-understood.
Compartments within the spine
Spines may also contain various endomembranous structures. Spines on cerebellar Purkinje cells are especially rich in smooth endoplasmic reticulum (SER) that contains high levels of the inositol trisphosphate receptor [59]. SER is less often associated with forebrain spines (the focus of this review), but large spines in the forebrain often contain a peculiar membrane-bounded structure, the spine apparatus (SA) [60, 61]. The SA, usually considered a specialized element of the SER [62], contains high levels of calcium [63], suggesting that it may help to regulate spinoplasmic free [Ca+2] [63, 64, 65, 66].
Though endosomes appear to play an important role in synaptic plasticity, they are rather uncommon in spines of the adult forebrain; a serial-section EM study detected endosomes in less than half of spines in CA1 stratum radiatum of the rat [67]. However, LTP-associated spine growth is accompanied by an increase of intraspinous vesicles, pointing to the importance of membrane trafficking events during synaptic plasticity [68]. During LTP, recycling endosomes may provide AMPA receptors to the synapse [69], while also supplying lipid membrane to the growing spine surface [70], thus linking morphological changes with synaptic potentiation. Conversely, the reduction of synaptic strength during LTD is accompanied by both internalization of synaptic AMPA receptors and spine shrinkage [71, 72].
Aside from these vesicular structures, the spine cytoplasm has traditionally been viewed as nondescript or amorphous, but contemporary work challenges this view. As outlined below, recent studies suggest the presence of multiple distinct domains within the cytoplasm.
The actin spinoskeleton
Spines are rich in actin [73], a highly-conserved globular protein of ~43 KDa molecular weight with ATPase activity. Extensive work in model systems demonstrates that actin cycles dynamically between soluble monomeric G-actin and polymerized ~8–9 nm diameter filaments of F-actin. These filaments can assemble into complex networks, which may undergo rapid extension and rearrangement, producing force and deformation of the plasma membrane [74, 75, 76]. Actin filaments also undergo “treadmilling,” in which filament length remains constant, while actin monomers add at the “barbed” end, and dissociate from the “pointed” end. Actin is essential for basic cellular processes, ranging from establishment of cell polarity and directional migration, to organelle trafficking and exo- and endocytosis [77].
At EM, the actin spinoskeleton typically appears as loose clumps of filamentous material [73, 78, 79]. Recent studies using high-resolution techniques like electron tomography (Fig. 2A–C) and metal shadowing (Fig. 2D) have elucidated important aspects of the structure of these complex actin networks [80, 81], but the functional organization of actin filaments within the spine remains unclear. Live-cell imaging experiments led to a “two-pool” model of spinoskeletal organization, which postulates that filaments are organized into a relatively stable core and a dynamic periphery [82, 83, 84]. Direct visualization of actin treadmilling has been demonstrated within spines using super-resolution live-cell imaging [3, 85, 86, 87, 88]. A distinct “enlargement pool” of filaments has also been described [89], and an actin pool that directly interacts with synaptic glutamate receptors within the postsynaptic density (PSD) has also been identified [36, 90]. Actin filaments can be seen to make direct contact with the PSD [81, 91, 92]. While actin has multiple functions at the PSD, perhaps its most fundamental role relates to stabilization of glutamate receptors [93]. NMDA receptors can be directly linked to actin filaments through the α-actinin (see below). The association of AMPA receptors to F-actin filaments is less direct, via a variety of scaffold proteins, including PSD-95 [94, 95], Abp1 [96], PICK1 [97], and neurabin [98].
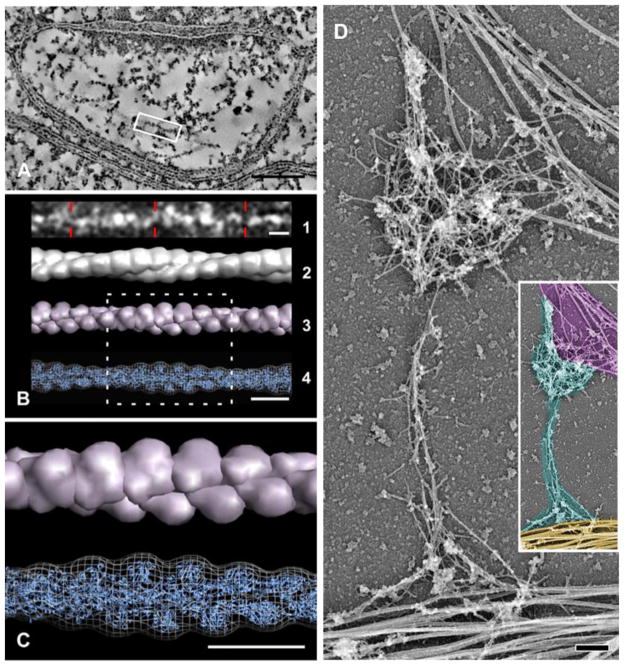
Figure 2. Appearance of F-actin in spines, demonstrated by high-resolution EM techniques.
Left panel shows electron tomography of a thin section from rat neocortex (images adapted with permission from [81]); right panel is from a platinum-shadowed replica of cultured hippocampal neurons ((image adapted with permission from [80]). A shows a virtual slice (~4 nm thick) through a spine (the PSD is at top). A volume containing a long, straight filament (highlighted by the white box) is shown in B1 (image inverted to highlight features). This filament displays helical periodicity corresponding closely to that predicted for F-actin. B2 shows a helically-averaged surface representation of the extracted density. B3 is a low-resolution representation of an atomic model of a canonical actin filament obtained from high-resolution electron microscopy analysis. B4 shows the fit of this atomic model (blue cartoon representation) into the symmetrized, extracted filament density (chicken wire representation). C is an enlargement of the boxed region in B. Scale bars: 100 nm for A, 10nm for B and C. D illustrates the cytoskeleton of a spine after membranes have been removed. The organization of filaments in the head and neck of a mushroom-shaped spine associated with dendrite (yellow pseudocolor, at bottom of inset) and with axon running along the head (magenta, top of inset) from 14 DIV neurons, treated with Triton X-100 to reveal internal features. Thick fibers in dendritic shaft represent microtubules. Actin filaments (cyan pseudocolor in inset) are the main cytoskeletal elements in the spine head and neck. Inset in D is a shrunk and pseudocolored version of panel D. Scale bar 200 nm.
Controlling the spinoskeleton
An elaborate cascade of proteins modulates F-actin remodeling; this protein network is crucial for regulating shape and motility in eukaryotic cells. The Rho family of small GTPases links receptors at the plasma membrane to mediators of actin remodeling (see [37, 38, 90, 99, 100] for information on their role in spines). These and other upstream regulatory proteins operate on actin via a cohort of functionally-distinct families of actin-binding proteins (ABPs) that catalyze the construction and reorganization of actin networks. Numerous ABPs have been identified in dendritic spines (Table 1). Intriguingly, these ABPs are targets of many of the same signaling pathways involved in long-term synaptic plasticity [101]. Furthermore, agents that interfere with actin remodeling also impair synaptic plasticity [102], confirming a long-suspected functional link between actin and synaptic efficacy [92, 103]
Table 1.
Functional families of actin-binding proteins in spines
| Function | Name (references) | |
|---|---|---|
| ABPs regulating F-actin assembly | Monomer sequestration | profilin [162, 164, 165, 184] |
| Depolymerization, severing | ADF/cofilin [30, 33, 137] | |
| Nucleation, branching | Arp2/3 complex [97, 149, 185]; WASP and SCAR/WAVE[142, 146]; formins [140]; cortactin [143] | |
| Capping | Capping protein (CapZ)[186] | |
| ABPs regulating network superstructure | Bundling | drebrin [174, 176, 177]; synaptopodin [60, 124, 125]; CaMKII [117, 118, 119, 187] |
| Cross-linking | α-actinin [108, 109, 112, 114, 188]; filamin [189]; spectrin [190] |
Regulation of the cytoskeleton requires tightly restricted spatial control of filament dynamics. Considering their functional specificity and their direct linkage to actin, defining the spatial organization of ABPs within the spine makes it possible to apply the static information available via EM to deduce aspects of the dynamic control of actin, at resolution not yet possible with live-cell imaging. This possibility is of special interest for investigating the sub-femtoliter volume of the dendritic spine. Evidence from immunogold electron microscopy that functionally-distinct ABPs concentrate in spatially-distinct domains within the spine cytoplasm points to a complex arrangement of multiple functional actin microdomains. Based on these data, a more quantitative and geometrically realistic view of spinoskeletal architecture is emerging, complementing functional LM studies. We here review these ABP-defined microdomains, focusing on a limited set of ABPs for which high-resolution ultrastructural data are available, to illustrate general principles underlying spinoskeletal design.
Linking the cytoskeleton to the plasma membrane
The actin cytoskeleton interacts with the plasma membrane via a family of intermediary proteins [104, 105]. α-actinin, an ABP of ~100 KDa MW that can crosslink filamentous actin in vitro [106], links the cytoskeleton to transmembrane proteins at cell-cell contact sites in a wide range of eukaryotic cells [107]. In neurons, α-actinin has been linked to the PSD both by proteomic screens and immunogold EM. The presence of α-actinin-1 and -2 isoforms in the brain has been shown directly [108, 109], and α-actinin-3 was demonstrated in proteomic studies [110]; the α-actinin-4 isoform is also implicated in neural function [111]. Immunogold electron microscopy shows that α-actinin is associated with glutamatergic synapses; in hippocampus it concentrates in the PSD [109] (Fig. 3), and is also associated with the spine apparatus (see below).
Figure 3. ABP content of spines, revealed by immunogold.
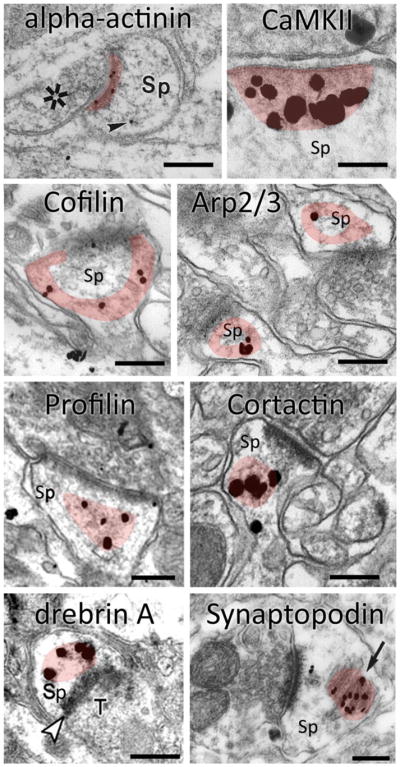
Hippocampal spines (Sp) labeled with immunogold for eight different actin-binding proteins. (Images adapted with permission from [109] (α-actinin), [116] CaMKII, [137] (cofilin), [149] (Arp2/3 complex),[163] (profilin), [157] (cortactin), [174] (drebrin) and [125] (synaptopodin). Shaded area represents the zone of concentration, for each protein. Scale bar: 200 nm.
α-actinin can bind directly to the GluN1 and GluN2B subunits of the N-methyl D-aspartate receptor (NMDAR) via its central spectrin repeats, providing a direct link between NMDARs and the actin spinoskeleton [112, 113, 114, 115]. The C-terminus of α-actinin can bind to the dodecameric holozyme CaMKII, and these two proteins may work cooperatively. LTP-inducing stimuli cause massive NMDAR-gated Ca2+ entry into the spine, recruiting CaMKII to the PSD, as has been demonstrated by several methods, including quantitative immunogold EM [116] (Fig. 3). Concurrently, by binding to calmodulin, the Ca2+ displaces α-actinin from the GluN1 subunit, allowing CaMKII to bind to the NMDA receptor [112]. Interestingly, the β subunit of CaMKII has been shown to bundle F-actin [117]. Thus, α-actinin and CaMKII may function together at the PSD to modify synaptic efficacy, while also acting on the actin cytoskeleton [118, 119, 120].
α-actinin can also interact with synaptopodin, a 74 kDa ABP originally described in podocytes of the kidney which can elongate and bundle actin filaments [121]. Synaptopodin’s α-actinin binding domain is crucial for its postsynaptic localization in neurons [60, 122, 123]. Both pre-embedding immunoperoxidase and postembedding immunogold methods demonstrate synaptopodin in a subset of spines of forebrain neurons (Fig. 3,[122]), concentrating in the spine neck and spine apparatus. This association with the SA is functionally significant: synaptopodin knock-out mice lack a typical SA [124]. Importantly, these knock-out mice also display impaired LTP [125]; moreover, synaptopodin has been implicated in memory formation in the behaving animal [124].
Turmoil under the surface: the shell domain
The spinoskeleton is continually changing in response to synaptic activity. A prerequisite for this remodeling is the destruction of preexisting architectural elements, to permit creation of a new network. This recycling task is performed by the ADF (Actin Depolymerizing Factor)/cofilin enzyme family, comprising three ~20 kDa proteins in vertebrates: ADF (also known as destrin), cofilin-1 (the major form in non-muscle tissue), and cofilin-2 (mainly in muscle). Proteins of the ADF/cofilin family enhance turnover of actin both by increasing the rate of depolymerization at filament ends, and by cutting long filaments to expose uncapped barbed ends, permitting new filament growth [126]. These proteins are enriched at sites of motility associated with rapid actin reorganization [127], as best documented at the leading edge and ruffling membranes of cultured fibroblasts [128]. An analogous enrichment is seen in corresponding regions of neuronal growth cones [129]. Here we focus on cofilin-1 (the major isoform in the brain), which we will refer to simply as “cofilin.”
Stimuli that trigger long-term depression (LTD) in hippocampal neurons also cause spine shrinkage. Considerable evidence implicates cofilin as an essential agent linking hippocampal LTD to structural plasticity [33, 130]. Cofilin’s enzymatic activity is tightly regulated by an interplay between (Lin-11/Isl-1/Mec-3)-domain-containing protein kinase (LIMK) and the phosphatase Slingshot [131, 132, 133]. The LIMK gene is deleted in Williams-Beuren syndrome, a disorder associated with severe mental retardation and visuo-spatial cognitive deficits [134]; thus, dysregulation of neuronal cofilin has clinical implications. Interestingly, cofilin-actin “rods,” which form under conditions of cellular stress, have been reported in postmortem studies of patients with Alzheimer’s disease [135, 136].
Immunogold EM labeling reveals that cofilin in hippocampal spines concentrates in the immediate vicinity of the plasma membrane (the “shell”), and is sparse in the central “core” of the spine (Fig. 3) [137]. Since cofilin is a prerequisite for rapid actin reorganization [138], these immunogold data point to the shell domain as the most dynamic part of the spinoskeleton. Cofilin concentrates near the extrasynaptic plasma membrane, so it is not surprising that it also lies in the PSD, where it may help to regulate the postsynaptic signaling scaffold. Cofilin has been reported to play an active role in LTP consolidation via modifications of both receptor number and spine size [30, 133, 139]. Phosphorylation inactivates cofilin, and spines enriched in phosphocofilin were reported to have unusually large synapses [35, 139]. Conversely, cofilin is dephosphorylated and activated by LTD inducing-stimuli, which lead to spine shrinkage. These observations are especially intriguing because spine volume is closely linked to synapse size, which in turn is closely linked to the number of AMPA receptors [26]. In summary, current evidence suggests a distinct shell microdomain that can remodel quickly in response to external signals.
Branches below the surface
Cytoskeletal remodeling requires extension and branching of actin filaments, and stabilization of these branches. A number of ABPs that serve these functions have been identified in model systems. The Arp2/3 complex, a multi-protein assembly of net molecular weight ~225 kDa, plays a central role in filament branching and membrane protrusion [140, 141]. The purified complex contains one copy each of seven subunits: two actin-related proteins (Arp2 and Arp3) whose tertiary structure resembles that of actin, and five additional subunits, ARPC1–5. Several upstream modulators, including the nucleation-promoting N-WASP (Neural-Wiskott-Aldrich syndrome protein), WAVE/Scar [142], cortactin [143], and the inhibitor PICK1, regulate the activity of the Arp2/3 complex [97]. Upon activation, Arp2/3 acts to initiate a new “daughter” actin filament, branching at ~70° from the side of an existing “mother” filament [144].
Activation of Arp2/3 in neurons promotes spine head expansion [145, 146, 147]. Inhibition of its activity by PICK1, a PDZ and BAR-domain containing protein that can bind to AMPA receptors, leads to defective neuronal architecture associated with a reduction in the number of mature dendritic spines [148]. The PICK1-Arp2/3 interaction is also necessary for spine shrinkage during LTD [97]. ImmunoEM indicates that the Arp2/3 complex concentrates in a restricted donut-shaped domain in the spine head: label is found away from the plasma membrane, but also avoids the center of the spine, lying mainly in a zone 20 – 100 nm from the plasma membrane on the “side” of the spine, away from the PSD (Fig. 3)[149]. In model systems, the Arp2/3 complex is spatially restricted within the extending lamellopodium during polarized extension [150]. The restricted Arp2/3 zone in the spinoskeleton may represent an analogous microdomain specialized for actin branching and nucleation [80, 144, 145, 147].
Deep in the spine: a stable but dynamic core
The central zone of the spine head is relatively stable, suggesting that the structure of the actin network in the core may be maintained for prolonged periods. Consistent with this notion, recent in vivo and vitro super-resolution imaging of spines [4, 85] indicates that the density of actin is higher in the center compared to the periphery. Since these studies imaged actin itself, the actin-related biochemical machinery could only be deduced indirectly. Using quantitative immunoelectron microscopy, three ABPs, cortactin, profilin and drebrin, have been found to localize to the central core, where they seem to play related but distinct roles in maintaining structural integrity.
Cortactin, an 80–85 kDa ABP implicated in nucleation, branching, and stabilization of actin filaments, can activate the Arp2/3 complex when phosphorylated [151, 152]. Studies in fibroblasts show that cortactin localizes to lamellopodia, filopodia, and membrane ruffles, where it colocalizes with F-actin and the Arp2/3 complex [153, 154]. In neurons, cortactin concentrates in spines [155]. Cortactin can directly bind to NMDA receptors through its SH3 domain [143, 156], and quantitative immunogold electron microscopic data show that cortactin is indeed found — though at modest levels — at the PSD. However, cortactin is at much higher levels within the spine core, 100–150 nm away from the PSD (Fig. 3) [157]. Multiple lines of evidence suggest that activation of NMDA receptors causes cortactin to move from the spine to the dendritic shaft [143, 155, 158]. Interestingly, this activity-driven translocation of synaptic cortactin is lost in the Fmr1 knock-out model of fragile X syndrome, in which stabilization of both actin filaments and LTP is impaired [44]. While this and other evidence suggests an important role for cortactin in both electrophysiologically-defined synaptic plasticity and behavioral models of learning, it remains unclear exactly how the loss of functional cortactin causes these deficiencies.
Profilins, a family of 14–16 kDa proteins, are essential for actin polymerization. By attaching G-actin to the barbed end of an F-actin filament, profilin mediates filament elongation [159]. Four isoforms of profilin have been identified, the products of four genes (Pfn1-Pfn4). The most ubiquitous isoform is profilin-1 [160]. Profilin-2 has two splice variants, 2a being primarily expressed in neurons [161], whereas expression of profilins 3 and 4 is largely restricted to kidney and testes [159]. In dendritic spines, profilin plays a role complimentary to cortactin [162]. Profilin-1 is present in most spiny neurons; immunogold analysis reveals that profilin-1 localizes to the spine core, though labeling is also seen in the PSD (Fig. 3) [163]. An elegant study in the lateral amygdala shows that fear conditioning drives profilin to the spine head, resulting in enlarged spines [164]. Profilin-2 has been demonstrated in spines of cultured hippocampal neurons expressing GFP-tagged protein. In contrast to cortactin, LTP-inducing activation of postsynaptic NMDA receptors recruits profilin-2 from the dendritic shaft into the spine core [162]. This recruitment stabilizes spine morphology; moreover, blocking its translocation from shaft to spine prevents formation of LTP. Nevertheless, profilin-1 and profilin-2 knock-out mice have normal LTP and LTD, and exhibit normal learning [165, 166], perhaps reflecting compensatory mechanisms. Interestingly, immunolabel for profilin is often visible in the spine neck [163], consistent with its ability to shuttle between the spine head and the dendritic shaft. The available evidence suggests that profilins are highly mobile, and can enter and leave the spine head in an activity-dependent manner.
Drebrins (developmentally regulated brain proteins) are a family of 95–100 kDa actin-binding proteins. Drebrin can bind to F-actin and inhibit its interactions with tropomyosin and α-actinin, leading to thick, curving bundles of actin filaments [167, 168, 169]. Two isoforms have been identified, products of alternative splicing from a single gene: the embryonic (drebrin E), which is found primarily in developing spines of the rat brain at postnatal day (PND) 7 [170, 171], and adult (drebrin A), which becomes common by PND21 [172]. While both isoforms are expressed in neurons, drebrin A is neuron-specific, localizing to the postsynaptic side of excitatory synapses [173]. Early in postnatal development, drebrin A and E localize to the spine shell [170, 173], suggesting that both isoforms play a role in spine maturation. However, drebrin A has been shown by immunoelectron microscopy to concentrate in the core region of mature spines (Fig. 3). Spines with large heads and PSDs contain higher levels of drebrin A than found in small spines [174]. This is particularly interesting in the view of a recent study in neurons showing that drebrin A changes the mechanical properties of actin filaments, rendering them resistant to depolymerization [175]. Thus, modulation of filament assemblies by drebrin A likely plays an important role in the activity-dependent spine expansion and stabilization associated with long-term synaptic potentiation [176, 177].
In summary, although the spine core is considered the stablest zone of the spinoskeleton [89], actin filaments in the core are nevertheless regulated by the coordinated interplay of cortactin, profilin, and drebrin, in an activity-dependent manner.
Conclusion and perspective
Actin filaments play a crucial role in regulating both spine morphology and synaptic plasticity. The architecture of the actin-based spinoskeleton is controlled by multiple families of ABPs. The data reviewed here suggest that these ABPs concentrate in distinct spatial domains, notwithstanding the rapid turnover of actin (Fig. 4). Since immunogold quantification relies on averaged values, it may fail to detect heterogeneity among spines [178]. However, this averaging attenuates underlying nonrandom distributions, so the true in vivo distribution of specific ABPs is likely to be even more restricted. These distinct ABP-defined microdomains imply a highly compartmentalized regulation of the spinoskeleton [179]. This conclusion is supported by observations from live-cell imaging, which also point to functionally discrete sub-spine actin domains [3, 89, 145, 180]. Taken together, these data suggest that the spinoskeleton is regulated by spatially-restricted ABP domains, which control the activity-dependent actin remodeling observed during synaptic plasticity.
Figure 4. ABP microdomains in the spinoskeleton.
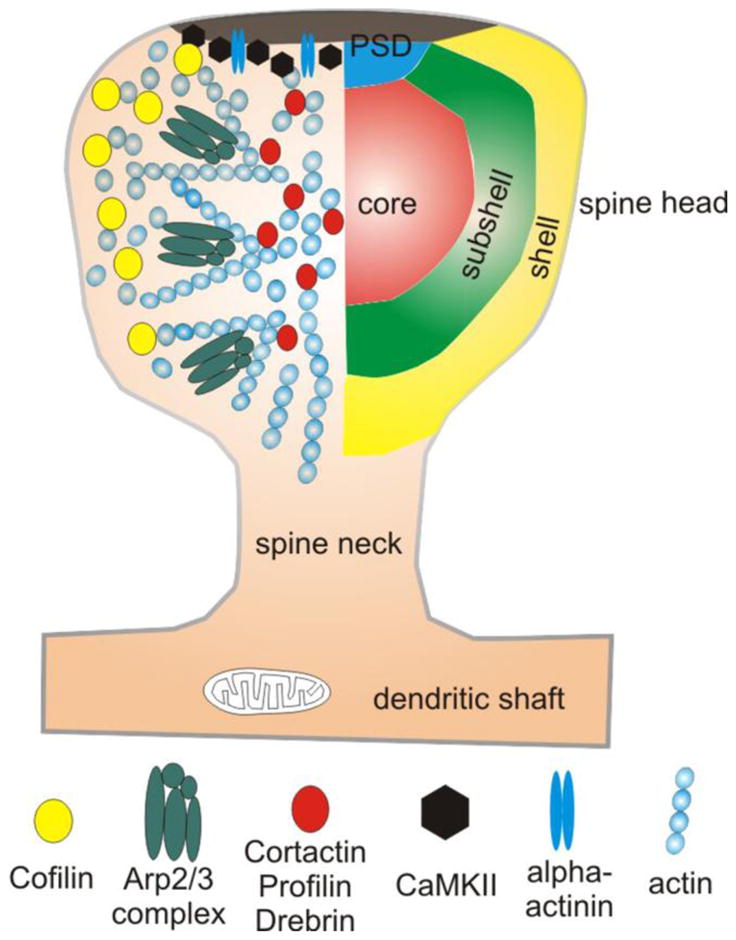
Proteomic studies of forebrain synapses have identified a sizable number of ABPs in the biochemically-defined PSD [110, 182, 183]. Some of these may be contaminants, but several, including α-actinin (blue double ovals) and CaMKII (black hexagons), have been demonstrated with immuno-EM to lie within the morphologically-defined PSD. Imaging studies reveal that actin (small blue circles) is more dynamic in the shell of the spine than in the center, suggesting a functional gradient of activity, from shell to core. Consistent with these data, cofilin (yellow circles) — a protein responsible for depolymerization of filaments — is heavily concentrated in this shell domain. The presence of a distinct “subshell” microdomain within the spinoplasm is suggested by the accumulation of the Arp2/3 complex (green composites), which mediates filament branching. The center (“core”) of the spine contains a relatively stable pool of actin. A heterogeneous pool of ABPs, including cortactin, profilin and drebrin (red circles), concentrate in this core microdomain.
While suggesting hitherto-unrecognized functional domains within the spine, this ABP-based view also raises a number of questions. For example, the mechanisms that target and hold ABPs in position within the spine are unclear: how can the spinoskeleton maintain a stable architecture in the face of rapid turnover of actin filaments? How dynamic are these cytoskeletal domains, and how much variability is there among different spines? It would also be useful to know how ABP domains are localized during neural development. Since spine morphology is closely linked to synaptic plasticity, and abnormalities in spine morphology have been linked to a variety of clinical disorders [42, 45, 181], it will be important to determine whether specific disease states are associated with specific pathological changes in ABP-defined microdomains. Gaining further insight into the organization of proteins that regulate actin in dendritic spines may provide important new clues to basic mechanisms underlying mental retardation and neuropsychiatric disease. This will require new tools and approaches, but these new tools cannot yet replace the resolution provided by the electron microscope.
Acknowledgments
The authors acknowledge grants from the Hungarian Scientific Research Fund (OTKA, grant #K83830 to B.L.R.) and from the National Institute of Health (#NS-39444 and #NS-35527 to R.J.W.). B.L.R. is also supported by the János Bólyai Research Fellowship from the Hungarian Academy of Sciences. We thank T. Svitkina for providing electron micrographs, and T. Blanpied, I. Ethell, P. Hotulainen, H. Kasai and S. Soderling for helpful comments.
Cited literature
- 1.Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9:2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dani A, Huang B, Bergan J, et al. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frost NA, Shroff H, Kong H, et al. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urban NT, Willig KI, Hell SW, et al. STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophys J. 2011;101:1277–1284. doi: 10.1016/j.bpj.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagerl UV, Willig KI, Hein B, et al. Live-cell imaging of dendritic spines by STED microscopy. Proc Natl Acad Sci U S A. 2008;105:18982–18987. doi: 10.1073/pnas.0810028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spacek J, Hartmann M. Three-dimensional analysis of dendritic spines. I. Quantitative observations related to dendritic spine and synaptic morphology in cerebral and cerebellar cortices. Anat Embryol (Berl) 1983;167:289–310. doi: 10.1007/BF00298517. [DOI] [PubMed] [Google Scholar]
- 7.Yuste R. Dendritic spines and distributed circuits. Neuron. 2011;71:772–781. doi: 10.1016/j.neuron.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller W, Connor JA. Dendritic spines as individual neuronal compartments for synaptic Ca2+ responses. Nature. 1991;354:73–76. doi: 10.1038/354073a0. [DOI] [PubMed] [Google Scholar]
- 9.Yuste R, Majewska A, Holthoff K. From form to function: calcium compartmentalization in dendritic spines. Nat Neurosci. 2000;3:653–659. doi: 10.1038/76609. [DOI] [PubMed] [Google Scholar]
- 10.Grunditz A, Holbro N, Tian L, et al. Spine neck plasticity controls postsynaptic calcium signals through electrical compartmentalization. J Neurosci. 2008;28:13457–13466. doi: 10.1523/JNEUROSCI.2702-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santamaria F, Wils S, De Schutter E, et al. Anomalous diffusion in Purkinje cell dendrites caused by spines. Neuron. 2006;52:635–648. doi: 10.1016/j.neuron.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuzaki M, Honkura N, Ellis-Davies GC, et al. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey CD, Svoboda K. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 2007;450:1195–1200. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fifkova E, Van Harreveld A. Long-lasting morphological changes in dendritic spines of dentate granular cells following stimulation of the entorhinal area. J Neurocytol. 1977;6:211–230. doi: 10.1007/BF01261506. [DOI] [PubMed] [Google Scholar]
- 15.Defelipe J. Brain plasticity and mental processes: Cajal again. Nat Rev Neurosci. 2006;7:811–817. doi: 10.1038/nrn2005. [DOI] [PubMed] [Google Scholar]
- 16.Holtmaat AJ, Trachtenberg JT, Wilbrecht L, et al. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Majewska AK, Newton JR, Sur M. Remodeling of synaptic structure in sensory cortical areas in vivo. Journal of Neuroscience. 2006;26:3021–3029. doi: 10.1523/JNEUROSCI.4454-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasumatsu N, Matsuzaki M, Miyazaki T, et al. Principles of long-term dynamics of dendritic spines. J Neurosci. 2008;28:13592–13608. doi: 10.1523/JNEUROSCI.0603-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen XW, Leischner U, Rochefort NL, et al. Functional mapping of single spines in cortical neurons in vivo. Nature. 2011;475:501–U597. doi: 10.1038/nature10193. [DOI] [PubMed] [Google Scholar]
- 20.Kasai H, Hayama T, Ishikawa M, et al. Learning rules and persistence of dendritic spines. Eur J Neurosci. 2010;32:241–249. doi: 10.1111/j.1460-9568.2010.07344.x. [DOI] [PubMed] [Google Scholar]
- 21.Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 22.Noguchi J, Nagaoka A, Watanabe S, et al. In vivo two-photon uncaging of glutamate revealing the structure-function relationships of dendritic spines in the neocortex of adult mice. J Physiol. 2011;589:2447–2457. doi: 10.1113/jphysiol.2011.207100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi Y, Majewska AK. Dendritic spine geometry: functional implication and regulation. Neuron. 2005;46:529–532. doi: 10.1016/j.neuron.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 25.Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annual Review of Neuroscience. 2008:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris KM, Weinberg RJ. Ultrastructure of synapses in the mammalian brain. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasai H, Fukuda M, Watanabe S, et al. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 2010;33:121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Matsuzaki M, Ellis-Davies GC, Nemoto T, et al. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001;4:1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nusser Z, Lujan R, Laube G, et al. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 1998;21:545–559. doi: 10.1016/s0896-6273(00)80565-6. [DOI] [PubMed] [Google Scholar]
- 30.Gu J, Lee CW, Fan Y, et al. ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat Neurosci. 2010;13:1208–1215. doi: 10.1038/nn.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto K, Nagai T, Miyawaki A, et al. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat Neurosci. 2004;7:1104–1112. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- 32.Bastrikova N, Gardner GA, Reece JM, et al. Synapse elimination accompanies functional plasticity in hippocampal neurons. Proc Natl Acad Sci U S A. 2008;105:3123–3127. doi: 10.1073/pnas.0800027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Snyder EM, Philpot BD, Huber KM, et al. Internalization of ionotropic glutamate receptors in response to mGluR activation. Nat Neurosci. 2001;4:1079–1085. doi: 10.1038/nn746. [DOI] [PubMed] [Google Scholar]
- 35.Chen LY, Rex CS, Casale MS, et al. Changes in synaptic morphology accompany actin signaling during LTP. J Neurosci. 2007;27:5363–5372. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerr JM, Blanpied TA. Subsynaptic AMPA receptor distribution is acutely regulated by actin-driven reorganization of the postsynaptic density. J Neurosci. 2012;32:658–673. doi: 10.1523/JNEUROSCI.2927-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tolias KF, Duman JG, Um K. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog Neurobiol. 2011;94:133–148. doi: 10.1016/j.pneurobio.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin B, Kramar EA, Bi X, et al. Theta stimulation polymerizes actin in dendritic spines of hippocampus. J Neurosci. 2005;25:2062–2069. doi: 10.1523/JNEUROSCI.4283-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zito K, Knott G, Shepherd GM, et al. Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron. 2004;44:321–334. doi: 10.1016/j.neuron.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 41.Ramachandran B, Frey JU. Interfering with the actin network and its effect on long-term potentiation and synaptic tagging in hippocampal CA1 neurons in slices in vitro. J Neurosci. 2009;29:12167–12173. doi: 10.1523/JNEUROSCI.2045-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penzes P, Cahill ME, Jones KA, et al. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai Y, Shaw CA, Dawson BC, et al. Protein interactome reveals converging molecular pathways among autism disorders. Sci Transl Med. 2011;3:86ra49. doi: 10.1126/scitranslmed.3002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seese RR, Babayan AH, Katz AM, et al. LTP induction translocates cortactin at distant synapses in wild-type but not FMR1 knock-out mice. J Neurosci. 2012;32:7403–7413. doi: 10.1523/JNEUROSCI.0968-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Brain Res Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- 46.Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: A more quantitative view. Annual Review of Biochemistry. 2007:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- 47.Okabe S. Molecular anatomy of the postsynaptic density. Mol Cell Neurosci. 2007;34:503–518. doi: 10.1016/j.mcn.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Valtschanoff JG, Weinberg RJ. Laminar organization of the NMDA receptor complex within the postsynaptic density. J Neurosci. 2001;21:1211–1217. doi: 10.1523/JNEUROSCI.21-04-01211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kharazia VN, Weinberg RJ. Tangential synaptic distribution of NMDA and AMPA receptors in rat neocortex. Neurosci Lett. 1997;238:41–44. doi: 10.1016/s0304-3940(97)00846-x. [DOI] [PubMed] [Google Scholar]
- 50.Racca C, Stephenson FA, Streit P, et al. NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. J Neurosci. 2000;20:2512–2522. doi: 10.1523/JNEUROSCI.20-07-02512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macgillavry HD, Kerr JM, Blanpied TA. Lateral organization of the postsynaptic density. Mol Cell Neurosci. 2011;48:321–331. doi: 10.1016/j.mcn.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baude A, Nusser Z, Roberts JD, et al. The metabotropic glutamate receptor (mGluR1 alpha) is concentrated at perisynaptic membrane of neuronal subpopulations as detected by immunogold reaction. Neuron. 1993;11:771–787. doi: 10.1016/0896-6273(93)90086-7. [DOI] [PubMed] [Google Scholar]
- 53.He Y, Janssen WG, Rothstein JD, et al. Differential synaptic localization of the glutamate transporter EAAC1 and glutamate receptor subunit GluR2 in the rat hippocampus. J Comp Neurol. 2000;418:255–269. [PubMed] [Google Scholar]
- 54.Katona I, Freund TF. Endocannabinoid signaling as a synaptic circuit breaker in neurological disease. Nat Med. 2008;14:923–930. doi: 10.1038/nm.f.1869. [DOI] [PubMed] [Google Scholar]
- 55.Blanpied TA, Scott DB, Ehlers MD. Dynamics and regulation of clathrin coats at specialized endocytic zones of dendrites and spines. Neuron. 2002;36:435–449. doi: 10.1016/s0896-6273(02)00979-0. [DOI] [PubMed] [Google Scholar]
- 56.Kennedy MJ, Davison IG, Robinson CG, et al. Syntaxin-4 defines a domain for activity-dependent exocytosis in dendritic spines. Cell. 2010;141:524–535. doi: 10.1016/j.cell.2010.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Racz B, Blanpied TA, Ehlers MD, et al. Lateral organization of endocytic machinery in dendritic spines. Nat Neurosci. 2004;7:917–918. doi: 10.1038/nn1303. [DOI] [PubMed] [Google Scholar]
- 58.Newpher TM, Ehlers MD. Spine microdomains for postsynaptic signaling and plasticity. Trends Cell Biol. 2009;19:218–227. doi: 10.1016/j.tcb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Fotuhi M, Sharp AH, Glatt CE, et al. Differential localization of phosphoinositide-linked metabotropic glutamate receptor (mGluR1) and the inositol 1,4,5-trisphosphate receptor in rat brain. J Neurosci. 1993;13:2001–2012. doi: 10.1523/JNEUROSCI.13-05-02001.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kremerskothen J, Plaas C, Kindler S, et al. Synaptopodin, a molecule involved in the formation of the dendritic spine apparatus, is a dual actin/alpha-actinin binding protein. J Neurochem. 2005;92:597–606. doi: 10.1111/j.1471-4159.2004.02888.x. [DOI] [PubMed] [Google Scholar]
- 61.Spacek J. Three-dimensional analysis of dendritic spines. II. Spine apparatus and other cytoplasmic components. Anat Embryol (Berl) 1985;171:235–243. doi: 10.1007/BF00341418. [DOI] [PubMed] [Google Scholar]
- 62.Segal M, Vlachos A, Korkotian E. The spine apparatus, synaptopodin, and dendritic spine plasticity. Neuroscientist. 2010;16:125–131. doi: 10.1177/1073858409355829. [DOI] [PubMed] [Google Scholar]
- 63.Fifkova E, Markham JA, Delay RJ. Calcium in the spine apparatus of dendritic spines in the dentate molecular layer. Brain Res. 1983;266:163–168. doi: 10.1016/0006-8993(83)91322-7. [DOI] [PubMed] [Google Scholar]
- 64.Rose CR, Konnerth A. Stores not just for storage. intracellular calcium release and synaptic plasticity. Neuron. 2001;31:519–522. doi: 10.1016/s0896-6273(01)00402-0. [DOI] [PubMed] [Google Scholar]
- 65.Korkotian E, Segal M. Fast confocal imaging of calcium released from stores in dendritic spines. Eur J Neurosci. 1998;10:2076–2084. doi: 10.1046/j.1460-9568.1998.00219.x. [DOI] [PubMed] [Google Scholar]
- 66.Vlachos A. Synaptopodin and the spine apparatus organelle-Regulators of different forms of synaptic plasticity? Ann Anat. 2012;194:317–320. doi: 10.1016/j.aanat.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 67.Cooney JR, Hurlburt JL, Selig DK, et al. Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J Neurosci. 2002;22:2215–2224. doi: 10.1523/JNEUROSCI.22-06-02215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toni N, Buchs PA, Nikonenko I, et al. Remodeling of synaptic membranes after induction of long-term potentiation. J Neurosci. 2001;21:6245–6251. doi: 10.1523/JNEUROSCI.21-16-06245.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park M, Penick EC, Edwards JG, et al. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- 70.Park M, Salgado JM, Ostroff L, et al. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 2006;52:817–830. doi: 10.1016/j.neuron.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim CH, Chung HJ, Lee HK, et al. Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc Natl Acad Sci U S A. 2001;98:11725–11730. doi: 10.1073/pnas.211132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 73.Fifkova E. Actin in the nervous system. Brain Res. 1985;356:187–215. [PubMed] [Google Scholar]
- 74.Jonas O, Duschl C. Force propagation and force generation in cells. Cytoskeleton (Hoboken) 2010;67:555–563. doi: 10.1002/cm.20466. [DOI] [PubMed] [Google Scholar]
- 75.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 76.Kessels MM, Schwintzer L, Schlobinski D, et al. Controlling actin cytoskeletal organization and dynamics during neuronal morphogenesis. Eur J Cell Biol. 2011;90:926–933. doi: 10.1016/j.ejcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 77.Anitei M, Hoflack B. Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat Cell Biol. 2012;14:11–19. doi: 10.1038/ncb2409. [DOI] [PubMed] [Google Scholar]
- 78.Fifkova E, Morales M. Actin matrix of dendritic spines, synaptic plasticity, and long-term potentiation. Int Rev Cytol. 1992;139:267–307. doi: 10.1016/s0074-7696(08)61414-x. [DOI] [PubMed] [Google Scholar]
- 79.Markham JA, Fifkova E. Actin filament organization within dendrites and dendritic spines during development. Brain Res. 1986;392:263–269. doi: 10.1016/0165-3806(86)90253-1. [DOI] [PubMed] [Google Scholar]
- 80.Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burette AC, Lesperance T, Crum J, et al. Electron tomographic analysis of synaptic ultrastructure. J Comp Neurol. 2012;520:2697–2711. doi: 10.1002/cne.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smart FM, Halpain S. Regulation of dendritic spine stability. Hippocampus. 2000;10:542–554. doi: 10.1002/1098-1063(2000)10:5<542::AID-HIPO4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 83.Matus A. Growth of dendritic spines: a continuing story. Curr Opin Neurobiol. 2005;15:67–72. doi: 10.1016/j.conb.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 84.Rao A, Craig AM. Signaling between the actin cytoskeleton and the postsynaptic density of dendritic spines. Hippocampus. 2000;10:527–541. doi: 10.1002/1098-1063(2000)10:5<527::AID-HIPO3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 85.Izeddin I, Specht CG, Lelek M, et al. Super-resolution dynamic imaging of dendritic spines using a low-affinity photoconvertible actin probe. PLoS One. 2011;6:e15611. doi: 10.1371/journal.pone.0015611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tatavarty V, Kim EJ, Rodionov V, et al. Investigating sub-spine actin dynamics in rat hippocampal neurons with super-resolution optical imaging. PLoS One. 2009;4:e7724. doi: 10.1371/journal.pone.0007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koskinen M, Bertling E, Hotulainen P. Methods to measure actin treadmilling rate in dendritic spines. Methods Enzymol. 2012;505:47–58. doi: 10.1016/B978-0-12-388448-0.00011-5. [DOI] [PubMed] [Google Scholar]
- 88.Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Curr Opin Neurobiol. 2011 doi: 10.1016/j.conb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Honkura N, Matsuzaki M, Noguchi J, et al. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 90.Fischer M, Kaech S, Wagner U, et al. Glutamate receptors regulate actin-based plasticity in dendritic spines. Nat Neurosci. 2000;3:887–894. doi: 10.1038/78791. [DOI] [PubMed] [Google Scholar]
- 91.Capani F, Martone ME, Deerinck TJ, et al. Selective localization of high concentrations of F-actin in subpopulations of dendritic spines in rat central nervous system: a three-dimensional electron microscopic study. J Comp Neurol. 2001;435:156–170. doi: 10.1002/cne.1199. [DOI] [PubMed] [Google Scholar]
- 92.Fifkova E, Delay RJ. Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J Cell Biol. 1982;95:345–350. doi: 10.1083/jcb.95.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Allison DW, Gelfand VI, Spector I, et al. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pak DT, Yang S, Rudolph-Correia S, et al. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- 95.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 96.Haeckel A, Ahuja R, Gundelfinger ED, et al. The actin-binding protein Abp1 controls dendritic spine morphology and is important for spine head and synapse formation. J Neurosci. 2008;28:10031–10044. doi: 10.1523/JNEUROSCI.0336-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakamura Y, Wood CL, Patton AP, et al. PICK1 inhibition of the Arp2/3 complex controls dendritic spine size and synaptic plasticity. Embo Journal. 2011;30:719–730. doi: 10.1038/emboj.2010.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Terry-Lorenzo RT, Roadcap DW, Otsuka T, et al. Neurabin/protein phosphatase-1 complex regulates dendritic spine morphogenesis and maturation. Mol Biol Cell. 2005;16:2349–2362. doi: 10.1091/mbc.E04-12-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carlisle HJ, Kennedy MB. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 100.Penzes P, Rafalovich I. Regulation of the actin cytoskeleton in dendritic spines. Adv Exp Med Biol. 2012;970:81–95. doi: 10.1007/978-3-7091-0932-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Penzes P, Cahill ME. Deconstructing signal transduction pathways that regulate the actin cytoskeleton in dendritic spines. Cytoskeleton (Hoboken) 2012 doi: 10.1002/cm.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fukazawa Y, Saitoh Y, Ozawa F, et al. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- 103.Fonseca R. Activity-dependent actin dynamics are required for the maintenance of long-term plasticity and for synaptic capture. Eur J Neurosci. 2012;35:195–206. doi: 10.1111/j.1460-9568.2011.07955.x. [DOI] [PubMed] [Google Scholar]
- 104.Ahmed S. Nanoscopy of cell architecture: The actin-membrane interface. Bioarchitecture. 2011;1:32–38. doi: 10.4161/bioa.1.1.14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chia PH, Patel MR, Shen K. NAB-1 instructs synapse assembly by linking adhesion molecules and F-actin to active zone proteins. Nat Neurosci. 2012;15:234–242. doi: 10.1038/nn.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sjoblom B, Salmazo A, Djinovic-Carugo K. Alpha-actinin structure and regulation. Cell Mol Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oikonomou KG, Zachou K, Dalekos GN. Alpha-actinin: a multidisciplinary protein with important role in B-cell driven autoimmunity. Autoimmun Rev. 2011;10:389–396. doi: 10.1016/j.autrev.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 108.Cabello N, Remelli R, Canela L, et al. Actin-binding protein alpha-actinin-1 interacts with the metabotropic glutamate receptor type 5b and modulates the cell surface expression and function of the receptor. J Biol Chem. 2007;282:12143–12153. doi: 10.1074/jbc.M608880200. [DOI] [PubMed] [Google Scholar]
- 109.Wyszynski M, Kharazia V, Shanghvi R, et al. Differential regional expression and ultrastructural localization of alpha-actinin-2, a putative NMDA receptor-anchoring protein, in rat brain. J Neurosci. 1998;18:1383–1392. doi: 10.1523/JNEUROSCI.18-04-01383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jordan BA, Fernholz BD, Boussac M, et al. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- 111.Walikonis RS, Oguni A, Khorosheva EM, et al. Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2+/calmodulin-dependent protein kinase II and (alpha)-actinin. J Neurosci. 2001;21:423–433. doi: 10.1523/JNEUROSCI.21-02-00423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Merrill MA, Malik Z, Akyol Z, et al. Displacement of alpha-actinin from the NMDA receptor NR1 C0 domain By Ca2+/calmodulin promotes CaMKII binding. Biochemistry. 2007;46:8485–8497. doi: 10.1021/bi0623025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wyszynski M, Lin J, Rao A, et al. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 114.Nakagawa T, Engler JA, Sheng M. The dynamic turnover and functional roles of alpha-actinin in dendritic spines. Neuropharmacology. 2004;47:734–745. doi: 10.1016/j.neuropharm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 115.Leonard AS, Bayer KU, Merrill MA, et al. Regulation of calcium/calmodulin-dependent protein kinase II docking to N-methyl-D-aspartate receptors by calcium/calmodulin and alpha-actinin. J Biol Chem. 2002;277:48441–48448. doi: 10.1074/jbc.M205164200. [DOI] [PubMed] [Google Scholar]
- 116.Otmakhov N, Tao-Cheng JH, Carpenter S, et al. Persistent accumulation of calcium/calmodulin-dependent protein kinase II in dendritic spines after induction of NMDA receptor-dependent chemical long-term potentiation. J Neurosci. 2004;24:9324–9331. doi: 10.1523/JNEUROSCI.2350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Okamoto K, Narayanan R, Lee SH, et al. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci U S A. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Borgesius NZ, Van Woerden GM, Buitendijk GHS, et al. Beta CaMKII plays a nonenzymatic role in hippocampal synaptic plasticity and learning by targeting alpha CaMKII to synapses. Journal of Neuroscience. 2011;31:10141–10148. doi: 10.1523/JNEUROSCI.5105-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin YC, Redmond L. CaMKII beta binding to stable F-actin in vivo regulates F-actin filament stability. Proc Natl Acad Sci U S A. 2008;105:15791–15796. doi: 10.1073/pnas.0804399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Okamoto K, Bosch M, Hayashi Y. The roles of CaMKII and F-actin in the structural plasticity of dendritic spines: a potential molecular identity of a synaptic tag? Physiology (Bethesda) 2009;24:357–366. doi: 10.1152/physiol.00029.2009. [DOI] [PubMed] [Google Scholar]
- 121.Asanuma K, Yanagida-Asanuma E, Faul C, et al. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol. 2006;8:485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- 122.Deller T, Merten T, Roth SU, et al. Actin-associated protein synaptopodin in the rat hippocampal formation: localization in the spine neck and close association with the spine apparatus of principal neurons. J Comp Neurol. 2000;418:164–181. doi: 10.1002/(sici)1096-9861(20000306)418:2<164::aid-cne4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 123.Deller T, Mundel P, Frotscher M. Potential role of synaptopodin in spine motility by coupling actin to the spine apparatus. Hippocampus. 2000;10:569–581. doi: 10.1002/1098-1063(2000)10:5<569::AID-HIPO7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 124.Vlachos A, Korkotian E, Schonfeld E, et al. Synaptopodin regulates plasticity of dendritic spines in hippocampal neurons. Journal of Neuroscience. 2009;29:1017–1033. doi: 10.1523/JNEUROSCI.5528-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Deller T, Korte M, Chabanis S, et al. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc Natl Acad Sci U S A. 2003;100:10494–10499. doi: 10.1073/pnas.1832384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bernstein BW, Bamburg JR. ADF/Cofilin: a functional node in cell biology. Trends in Cell Biology. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yonezawa N, Nishida E, Koyasu S, et al. Distribution among tissues and intracellular localization of cofilin, a 21kDa actin-binding protein. Cell Struct Funct. 1987;12:443–452. doi: 10.1247/csf.12.443. [DOI] [PubMed] [Google Scholar]
- 128.Cai L, Marshall TW, Uetrecht AC, et al. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Desmarais V, Ghosh M, Eddy R, et al. Cofilin takes the lead. Journal of Cell Science. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- 130.Wang XB, Yang Y, Zhou Q. Independent expression of synaptic and morphological plasticity associated with long-term depression. Journal of Neuroscience. 2007;27:12419–12429. doi: 10.1523/JNEUROSCI.2015-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Endo M, Ohashi K, Mizuno K. LIM kinase and slingshot are critical for neurite extension. Journal of Biological Chemistry. 2007;282:13692–13702. doi: 10.1074/jbc.M610873200. [DOI] [PubMed] [Google Scholar]
- 132.Takemura M, Mishima T, Wang Y, et al. Ca2+/calmodulin-dependent protein kinase IV-mediated LIM kinase activation is critical for calcium signal-induced neurite outgrowth. Journal of Biological Chemistry. 2009;284:28554–28562. doi: 10.1074/jbc.M109.006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yuen EY, Liu WH, Kafri T, et al. Regulation of AMPA receptor channels and synaptic plasticity by cofilin phosphatase Slingshot in cortical neurons. J Physiol-London. 2010;588:2361–2371. doi: 10.1113/jphysiol.2009.186353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gray V, Karmiloff-Smith A, Funnell E, et al. In-depth analysis of spatial cognition in Williams syndrome: A critical assessment of the role of the LIMK1 gene. Neuropsychologia. 2006;44:679–685. doi: 10.1016/j.neuropsychologia.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 135.Bamburg JR, Bernstein BW, Davis RC, et al. ADF/Cofilin-actin rods in neurodegenerative diseases. Curr Alzheimer Res. 2010;7:241–250. doi: 10.2174/156720510791050902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Davis RC, Maloney MT, Minamide LS, et al. Mapping cofilin-actin rods in stressed hippocampal slices and the role of cdc42 in amyloid-beta-induced rods. J Alzheimers Dis. 2009;18:35–50. doi: 10.3233/JAD-2009-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Racz B, Weinberg RJ. Spatial organization of cofilin in dendritic spines. Neuroscience. 2006;138:447–456. doi: 10.1016/j.neuroscience.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 138.Meng Y, Zhang Y, Tregoubov V, et al. Regulation of spine morphology and synaptic function by LIMK and the actin cytoskeleton. Rev Neurosci. 2003;14:233–240. doi: 10.1515/revneuro.2003.14.3.233. [DOI] [PubMed] [Google Scholar]
- 139.Fedulov V, Rex CS, Simmons DA, et al. Evidence that long-term potentiation occurs within individual hippocampal synapses during learning. J Neurosci. 2007;27:8031–8039. doi: 10.1523/JNEUROSCI.2003-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 141.Spillane M, Ketschek A, Jones SL, et al. The actin nucleating Arp2/3 complex contributes to the formation of axonal filopodia and branches through the regulation of actin patch precursors to filopodia. Dev Neurobiol. 2011 doi: 10.1002/dneu.20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soderling SH, Guire ES, Kaech S, et al. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci. 2007;27:355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hering H, Sheng M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci. 2003;23:11759–11769. doi: 10.1523/JNEUROSCI.23-37-11759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Goley ED, Rammohan A, Znameroski EA, et al. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc Natl Acad Sci U S A. 2010;107:8159–8164. doi: 10.1073/pnas.0911668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hotulainen P, Llano O, Smirnov S, et al. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. Journal of Cell Biology. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim Y, Sung JY, Ceglia I, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- 147.Wegner AM, Nebhan CA, Hu L, et al. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J Biol Chem. 2008;283:15912–15920. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rocca DL, Martin S, Jenkins EL, et al. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Racz B, Weinberg RJ. Organization of the Arp2/3 complex in hippocampal spines. Journal of Neuroscience. 2008;28:5654–5659. doi: 10.1523/JNEUROSCI.0756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shao D, Forge A, Munro PM, et al. Arp2/3 complex-mediated actin polymerisation occurs on specific pre-existing networks in cells and requires spatial restriction to sustain functional lamellipod extension. Cell Motil Cytoskeleton. 2006;63:395–414. doi: 10.1002/cm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weaver AM, Karginov AV, Kinley AW, et al. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- 152.Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Daly RJ. Cortactin signalling and dynamic actin networks. Biochem J. 2004;382:13–25. doi: 10.1042/BJ20040737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Weed SA, Karginov AV, Schafer DA, et al. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Iki J, Inoue A, Bito H, et al. Bi-directional regulation of postsynaptic cortactin distribution by BDNF and NMDA receptor activity. Eur J Neurosci. 2005;22:2985–2994. doi: 10.1111/j.1460-9568.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- 156.Naisbitt S, Kim E, Tu JC, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 157.Racz B, Weinberg RJ. The subcellular organization of cortactin in hippocampus. J Neurosci. 2004;24:10310–10317. doi: 10.1523/JNEUROSCI.2080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen YK, Hsueh YP. Cortactin-binding protein 2 modulates the mobility of cortactin and regulates dendritic spine formation and maintenance. J Neurosci. 2012;32:1043–1055. doi: 10.1523/JNEUROSCI.4405-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- 160.Schluter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- 161.Di Nardo A, Gareus R, Kwiatkowski D, et al. Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J Cell Sci. 2000;113(Pt 21):3795–3803. doi: 10.1242/jcs.113.21.3795. [DOI] [PubMed] [Google Scholar]
- 162.Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6:1194–1200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- 163.Neuhoff H, Sassoe-Pognetto M, Panzanelli P, et al. The actin-binding protein profilin I is localized at synaptic sites in an activity-regulated manner. Eur J Neurosci. 2005;21:15–25. doi: 10.1111/j.1460-9568.2004.03814.x. [DOI] [PubMed] [Google Scholar]
- 164.Lamprecht R, Farb CR, Rodrigues SM, et al. Fear conditioning drives profilin into amygdala dendritic spines. Nat Neurosci. 2006;9:481–483. doi: 10.1038/nn1672. [DOI] [PubMed] [Google Scholar]
- 165.Pilo Boyl P, Di Nardo A, Mulle C, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. Embo J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gorlich A, Zimmermann AM, Schober D, et al. Preserved morphology and physiology of excitatory synapses in profilin1-deficient mice. PLoS One. 2012;7:e30068. doi: 10.1371/journal.pone.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hayashi K, Ishikawa R, Kawai-Hirai R, et al. Domain analysis of the actin-binding and actin-remodeling activities of drebrin. Exp Cell Res. 1999;253:673–680. doi: 10.1006/excr.1999.4663. [DOI] [PubMed] [Google Scholar]
- 168.Ishikawa R, Hayashi K, Shirao T, et al. Drebrin, a development-associated brain protein from rat embryo, causes the dissociation of tropomyosin from actin filaments. J Biol Chem. 1994;269:29928–29933. [PubMed] [Google Scholar]
- 169.Ishikawa R, Katoh K, Takahashi A, et al. Drebrin attenuates the interaction between actin and myosin-V. Biochem Biophys Res Commun. 2007;359:398–401. doi: 10.1016/j.bbrc.2007.05.123. [DOI] [PubMed] [Google Scholar]
- 170.Asada H, Uyemura K, Shirao T. Actin-binding protein, drebrin, accumulates in submembranous regions in parallel with neuronal differentiation. J Neurosci Res. 1994;38:149–159. doi: 10.1002/jnr.490380205. [DOI] [PubMed] [Google Scholar]
- 171.Shirao T, Kojima N, Terada S, et al. Expression of three drebrin isoforms in the developing nervous system. Neurosci Res Suppl. 1990;13:S106–111. doi: 10.1016/0921-8696(90)90039-6. [DOI] [PubMed] [Google Scholar]
- 172.Hayashi K, Suzuki K, Shirao T. Rapid conversion of drebrin isoforms during synapse formation in primary culture of cortical neurons. Brain Res Dev Brain Res. 1998;111:137–141. doi: 10.1016/s0165-3806(98)00128-x. [DOI] [PubMed] [Google Scholar]
- 173.Aoki C, Sekino Y, Hanamura K, et al. Drebrin A is a postsynaptic protein that localizes in vivo to the submembranous surface of dendritic sites forming excitatory synapses. J Comp Neurol. 2005;483:383–402. doi: 10.1002/cne.20449. [DOI] [PubMed] [Google Scholar]
- 174.Kobayashi C, Aoki C, Kojima N, et al. Drebrin a content correlates with spine head size in the adult mouse cerebral cortex. J Comp Neurol. 2007;503:618–626. doi: 10.1002/cne.21408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sharma S, Grintsevich EE, Phillips ML, et al. Atomic force microscopy reveals drebrin induced remodeling of f-actin with subnanometer resolution. Nano Lett. 2011;11:825–827. doi: 10.1021/nl104159v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Biou V, Brinkhaus H, Malenka RC, et al. Interactions between drebrin and Ras regulate dendritic spine plasticity. Eur J Neurosci. 2008;27:2847–2859. doi: 10.1111/j.1460-9568.2008.06269.x. [DOI] [PubMed] [Google Scholar]
- 177.Takahashi H, Yamazaki H, Hanamura K, et al. Activity of the AMPA receptor regulates drebrin stabilization in dendritic spine morphogenesis. J Cell Sci. 2009;122:1211–1219. doi: 10.1242/jcs.043729. [DOI] [PubMed] [Google Scholar]
- 178.O’rourke NA, Weiler NC, Micheva KD, et al. Deep molecular diversity of mammalian synapses: why it matters and how to measure it. Nat Rev Neurosci. 2012;13:365–379. doi: 10.1038/nrn3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nicholson DA, Cahill ME, Tulisiak CT, et al. Spatially restricted actin-regulatory signaling contributes to synapse morphology. J Neurochem. 2012 doi: 10.1111/j.1471-4159.2012.07743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Frost NA, Kerr JM, Lu HE, et al. A network of networks: cytoskeletal control of compartmentalized function within dendritic spines. Curr Opin Neurobiol. 2010;20:578–587. doi: 10.1016/j.conb.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nadif Kasri N, Van Aelst L. Rho-linked genes and neurological disorders. Pflugers Arch. 2008;455:787–797. doi: 10.1007/s00424-007-0385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fernandez E, Collins MO, Uren RT, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Molecular Systems Biology. 2009;5 doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li KW, Hornshaw MP, Van Der Schors RC, et al. Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem. 2004;279:987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- 184.Michaelsen K, Murk K, Zagrebelsky M, et al. Fine-tuning of neuronal architecture requires two profilin isoforms. Proc Natl Acad Sci U S A. 2010;107:15780–15785. doi: 10.1073/pnas.1004406107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fan YJ, Tang X, Vitriol E, et al. Actin capping protein is required for dendritic spine development and synapse formation. Journal of Neuroscience. 2011;31:10228–10233. doi: 10.1523/JNEUROSCI.0115-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sanabria H, Swulius MT, Kolodziej SJ, et al. Beta CaMKII regulates actin assembly and structure. Journal of Biological Chemistry. 2009;284:9770–9780. doi: 10.1074/jbc.M809518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schulz TW, Nakagawa T, Licznerski P, et al. Actin/alpha-actinin-dependent transport of AMPA receptors in dendritic spines: role of the PDZ-LIM protein RIL. J Neurosci. 2004;24:8584–8594. doi: 10.1523/JNEUROSCI.2100-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bolduc FVBFV, Bell K, Rosenfelt C, et al. Fragile X mental retardation 1 and Filamin A interact genetically in Drosophila long-term memory. Front Neural Circuits. 2010;3 doi: 10.3389/neuro.04.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nestor MW, Cai X, Stone MR, et al. The actin binding domain of beta I-spectrin regulates the morphological and functional dynamics of dendritic spines. Plos One. 2011;6 doi: 10.1371/journal.pone.0016197. [DOI] [PMC free article] [PubMed] [Google Scholar]