Abstract
Water diffusion magnetic resonance imaging (dMRI) is a powerful tool for studying biological tissue microarchitectures in vivo. Recently, there has been increased effort to develop quantitative dMRI methods to probe both length scale and orientation information in diffusion media. Diffusion spectrum imaging (DSI) is one such approach that aims to resolve such information based on the three‐dimensional diffusion propagator at each voxel. However, in practice, only the orientation component of the propagator function is preserved when deriving the orientation distribution function. Here, we demonstrate how a straightforward extension of the linear spherical deconvolution (SD) model can be used to probe tissue orientation structures over a range (or “spectrum”) of length scales with minimal assumptions on the underlying microarchitecture. Using high b‐value Cartesian q‐space data on a rat brain tissue sample, we demonstrate how this “restriction spectrum imaging” (RSI) model allows for separating the volume fraction and orientation distribution of hindered and restricted diffusion, which we argue stems primarily from diffusion in the extraneurite and intraneurite water compartment, respectively. Moreover, we demonstrate how empirical RSI estimates of the neurite orientation distribution and volume fraction capture important additional structure not afforded by traditional DSI or fixed‐scale SD‐like reconstructions, particularly in gray matter. We conclude that incorporating length scale information in geometric models of diffusion offers promise for advancing state‐of‐the‐art dMRI methods beyond white matter into gray matter structures while allowing more detailed quantitative characterization of water compartmentalization and histoarchitecture of healthy and diseased tissue. Hum Brain Mapp, 2013. © 2012 Wiley Periodicals, Inc.
Keywords: diffusion MRI, DTI, HARDI, DSI, FOD, fiber orientation, tractography, histology, cerebellum, cerebral cortex, striatum, basal ganglia, rat
INTRODUCTION
Owing to its exquisitely sensitive contrast mechanism, water diffusion magnetic resonance imaging (dMRI) is a powerful technique for studying the microstructural and physiological properties of biological tissue in vivo [Le Bihan et al., 1986]. At normal brain temperatures, the diffusion coefficient of water in the CNS is about 1 μm2/ms. During typical diffusion times of 20–80 ms water molecules probe lengths scales on the order of 5–20 μm, making dMRI signals uniquely sensitive to a wide range of microstructural information at the cellular and subcellular level. Over the past decade, a number of important methodological advances have been made to probe both length scale and orientation information from biological tissue samples [for a review see Yablonskiy and Sukstanskii, 2010].
Q‐space imaging (QSI) is one such technique that utilizes data collected over multiple q‐values (defined as q = γδG/2π where γ is the gyromagnetic ratio of hydrogen, and δ and G are the diffusion gradient duration and magnitude, respectively) under the so‐called narrow gradient pulse regime (δ<<Δ where Δ is the time between the leading edges of the gradient pulses) [Callaghan et al., 1990; Cory and Garroway, 1990] to probe length scale information in terms of the statistical probability of water displacements along a single (1D) dimension [Assaf et al., 2000; Cohen and Assaf, 2002]. One of the major contributions of QSI has been the identification and characterization of separable hindered and restricted diffusion pools in nerve tissue samples, with the length scale of the restricted pool offering information on the size of the compartment [Assaf and Cohen, 2000]. Length scale information of separable diffusion pools can also be probed in terms of the apparent diffusion coefficient (ADC) of water using multiexponential signal models for data collected with multiple b‐values (b∼q 2Δ) [Mulkern et al., 1999]. Similar to QSI, these multiexponential signal models which assume a Gaussian mixture model for the displacement distribution routinely show evidence for water compartmentation in biological tissue when measured over an extended b‐value range. Typically, at least two Gaussian diffusion pools (exponentials) are observed, one with a high ADC, corresponding to coarse scale (“fast”) diffusion, and one with a small ADC, corresponding to fine scale (“slow”) diffusion [Mulkern et al., 1999]. While many have attributed the “fast” and “slow” components to hindered and restricted diffusion in the extracellular (ECS) and intracellular space (ICS) of tissue respectively [Mulkern et al., 2009], this theory is not without controversy, particularly due to paradoxically reversed estimates of their partial volume fractions [Mulkern et al., 2009].
There is also a large body of literature describing the use of multidirectional dMRI acquisitions at a fixed b‐value (or q‐value) for studying the geometric organization of white matter tissue in vivo [Basser et al., 1994b; Behrens et al., 2003; Jansons and Alexander, 2003; Tournier et al., 2004; Tuch, 2004; Anderson, 2005; Alexander, 2005b; Hess et al., 2006; Ozarslan et al., 2006; Kaden et al., 2007]. The most popular of these methods is diffusion tensor imaging (DTI), which uses a set of six or more images with noncollinear diffusion directions, plus one or more images with no diffusion weighting (b = 0) to estimate the apparent diffusion tensor in three‐dimensional (3D) space at each voxel [Basser et al., 1994a). The eigensystem of the tensor is commonly used to quantify both the degree of anisotropy and the principal directions of the diffusion process [Basser and Pierpaoli, 1996], with the later forming the basis for white‐matter fiber‐tracking studies [Basser et al., 2000]. However, the well‐known limitation of the tensor model in describing diffusion in nonhomogenous media [Beaulieu, 2002] has led to the development of numerous high‐angular resolution diffusion imaging (HARDI) techniques to resolve complex (e.g., crossing or bending) fiber orientations within voxels [Alexander et al., 2002; Frank, 2002; Tournier et al., 2004; Tuch, 2004; Anderson, 2005; Alexander, 2005b; Hess et al., 2006; Ozarslan et al., 2006; Jian and Vemuri, 2007; Kaden et al., 2007]. In contrast to DTI, these HARDI methods require a large number of diffusion directions (>6) to be collected at a fixed diffusion weighting, and include techniques such as spherical harmonic modeling of the ADC [Frank, 2002], Q‐ball numerical approximation of the water diffusion orientation distribution (dODF) [Tuch, 2004], and spherical deconvolution (SD) reconstruction of the fiber orientation distribution (FOD) [Tournier et al., 2004; Alexander, 2005a; Dell'Acqua et al., 2007; Jian and Vemuri, 2007; Kaden et al., 2007]. However, despite their successful application in resolving complex white‐matter fiber orientations in vivo, these HARDI methods are generally limited to studying only the geometric aspect of the diffusion process, and are agnostic to length scale information (e.g., fast/slow hindered/restricted diffusion) due to the adherence of a single diffusion weighting factor with fixed diffusion time.
In recent years, there has been increased interest in developing alternative dMRI methods to probe both length scale and orientation information from multidirectional diffusion acquisitions with multiple diffusion weightings. Wedeen et al. introduced a nonparametric technique called diffusion spectrum imaging (DSI), which generalizes the QSI method to a 3D Cartesian sampling of q‐space to obtain the 3D water displacement probability density function or diffusion propagator [Kärger and Heink, 1983] at each voxel [Wedeen et al., 2005]. However, it is common practice in DSI to integrate the propagator in the radial direction to yield the dODF, which while capturing the orientation structure of the diffusion function, removes all scale information inherent in the propagator itself. In addition, DSI adopts the classic q‐space formalism to obtain the propagator through Fourier Transform, but it does so without satisfying the narrow gradient pulse requirement, which challenges the interpretation of the propagator and dODF [Basser, 2002]. Assaf et al. proposed a parametric multicompartmental hindered and restricted model of diffusion (CHARMED) for white matter, which was later extended to measure axon diameter distributions in a technique called AxCaliber [Assaf et al., 2008; Barazany et al., 2009]. This composite framework makes use of a more efficient concentric shell sampling of q‐space (sometimes referred to as a multishell HARDI acquisition), where each shell measurement is differentially weighted by diffusion at different length scales. Alexander et al. used a simplified version of the CHARMED model to measure an axon diameter index in vivo [Alexander et al., 2010]. Jespersen et al. introduced a parametric multicompartmental model of the cytoarchitecture that uses a distribution of cylinders to model fine scale diffusion in axons and dendrites (collectively called neurites), and a tensor model for coarse scale diffusion elsewhere (cell bodies, glia, ECS) [Jespersen et al., 2007; Jespersen et al., 2010]. In this model, a multishell HARDI acquisition is harnessed to disambiguate the intraneurite versus extraneurite water fraction yielding estimates of the neurite volume fraction and orientation distribution [Jespersen et al., 2007; Jespersen et al., 2010]. However, an important limitation of many of these hybrid diffusion methods is that they often require complex and time‐consuming nonlinear optimization of the model parameters.
In this study, we show how a rather straightforward extension of the SD model for HARDI acquisitions can be used to probe the orientation structure of tissue microstructures over a range (or “spectrum”) of length scales with minimal assumptions on the underlying microstructure and while preserving an efficient linear implementation. Using high b‐value Cartesian q‐space data collected on a fixed rat brain, we show how this linear analysis approach, which we call “restriction spectrum imaging” (RSI), can be used to separate the volume fraction and orientation structure of fine and coarse scale diffusion processes in rat brain tissue, which we believe stems from restricted and hindered diffusion in the intraneurite and extraneurite water compartment, respectively. We support this hypothesis using both theoretical and empirical evidence, including a diverse set of histological material from corresponding brain regions. We further demonstrate how the resultant neurite orientation distribution provides additional structure beyond that which can be gleaned from traditional DSI and fixed‐scale SD‐like reconstructions.
MATERIALS AND METHODS
Sample Preparation and Image Acquisition
The diffusion data and histological materials used in this study were the same as used in a previous report [Leergaard et al., 2010]. Briefly, an adult Sprague Dawley® rat (Charles River Laboratories International, Wilmington, MA) was deeply anesthesized (ketamine hydrochloride 50 mg/kg, and sodium pentobarbital 12 mg/kg, i.p.) and transcardially perfused with 4% paraformaldehyde. The brain was extracted and immersed in contrast enhancing Magnevist® solution (Bayer HealthCare Pharmaceuticals) for ∼2 weeks [D'Arceuil and de Crespigny, 2007; Leergaard et al., 2010]. For image acquisition, the brain was immobilized in a molded plastic holder and placed in a sealable custom‐built plastic chamber filled with perfluorocarbon liquid (Fomblin® LC/8, Solvay Solexis, Thorofare, NJ) to fixate the tissue. High‐resolution diffusion images were collected using a 4.7T Bruker BioSpec Avance scanner (Bruker Instruments, Freemont, CA) featuring a 40‐cm warm bore diameter and equipped with a 3‐cm solenoid receiver coil. Data were acquired using a single‐shot pulsed gradient spin echo (PGSE) echo planar imaging sequence with Cartesian q‐space sampling and the following pulse sequence parameters: TR/TE = 650/49 ms, Δ/δ = 23/12 ms, 515 q‐space vectors [Wedeen et al., 2005], |G|max = 380 mT/m, b max = 30452 s/mm2, matrix = 64 × 64 × 128, voxel size = 265 μm isotropic, total imaging time ∼12 h. Following tomographic imaging, the brain was coronally sectioned at 50 μm using a freezing microtome (Microm HM450, Microm Gmbh, Waldorf, Germany). Every fourth section was stained for myelin using a standard procedure modified from Woelche [Woelche, 1942].
Supplementary histological material was derived from another Sprague Dawley rat (Scanbur, Norway) that was sacrificed as described above. This brain was sectioned sagitally (at 50 μm), and selected sections stained for myelin. High‐resolution mosaic images of the histological sections were obtained through UPlanApo 20/0.70 and 40/0.85 dry objectives using a motorized Olympus BX52 microscope running the Neurolucida 7.0 software (Virtual Slice module, MBF Bioscience, Williston, VT) or a slide scanner (Mirax Scan, Carl Zeiss MicroImaging, Jena, Germany). Additional histological reference images were downloaded from the BrainMaps.org website (http://www.brainmaps.org) and the Rodent Brain Workbench (http://www.rbwb.org). This concerned images showing 40‐μm thick coronal sections from an adult Sprague Dawley rat stained for potassium channel interacting protein (KChlP1) using the K55/7 monoclonal antibody (NeuroMab; http://neuromab.ucdavis.edu/), and images showing the distribution of axonal plexuses anterogradely labeled with Phaseolus vulgaris leucoagglutinin [http://www.rbwb.org; Whole Brain Connectivity Atlas; case R606; see also Zakiewicz et al., 2011].
DSI Reconstruction
DSI is based on the Fourier relationship between the diffusion signal S(q,Δ) and the 3D propagator P(R,Δ) [Kärger and Heink, 1983]
| (1) |
where R is the net displacement of water molecules during the diffusion time Δ, and q = γδG/2π is the q‐space diffusion wave vector. To obtain P(R, Δ), the signal values were filled into 3D Cartesian coordinate space consisting of a 17 × 17 × 17 voxel grid according to their respective position in q‐space. Then the inverse Fourier Transform was applied directly to the gridded data. Before the Fourier inversion, a 3D Hanning window h(r) = 0.5 · (1 + cos (2π r/17)) was used to filter the data at high |q| to reduce truncation artifacts (Gibbs ringing) in the reconstructed propagator [Wedeen et al., 2005]. Finally, the orientation distribution function in the direction of the unit vector x was obtained by evaluating the integral
| (2) |
To evaluate this integral, we used a third‐order tessellation of the sphere (642 vertices) to define each point x, and Sinc interpolation along 20 equally spaced points between 0 and R max.
Restriction Spectrum Imaging
Spherical deconvolution
In traditional SD, the diffusion signal magnitude in each voxel s(q) can be written as
| (3) |
where f(·) is the fiber orientation distribution (FOD), s 0 is the signal measured with no diffusion weighting [i.e., s 0 ≡ s(q = 0)], and R(·,x) is the signal attenuation to a single “fiber” with orientation given by the unit vector x. To obtain the FOD, a popular approach uses an axially symmetric Gaussian model for the fiber response function [Anderson, 2005]
| (4) |
where b = |q|2 (Δ − δ/3) is the diffusion weighting factor (or b‐value), α is the measurement angle relative to the fiber axis (i.e., α = |r · x|, where r = q/|q| is a unit vector oriented along q), and D L and D T are the longitudinal and transverse diffusivities of the response function, respectively, with D L > D T. When these diffusivities are known a priori (or can be estimated from the data and held fixed) the convolution in Eq. (3) can be implemented as a linear system of equations. Given N q‐space measurements {q 1, q 2,…,q N} and M desired FOD reconstruction points on the sphere {x1,x2,…,xM}, Eq. (3) can be written
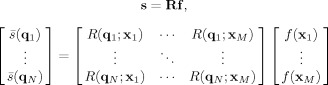 |
(5) |
where s = s/s 0 denotes the normalized signal. However, rather than treating each of the M values of f(·) as unknown parameters, the FOD is often parameterized using a set of even order spherical harmonics (SH) Y (·) with order l = 0,2,…,L and degree m = −l,…,0,…,l,
| (6) |
where P = (L + 2)(L + 1)/2 is the total number of SH basis functions in the series,
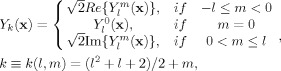 |
(7) |
is the kth SH basis function, and {β1,β2,…,βP} are the unknown real‐valued parameters (weights) to be estimated. This particular basis in Eq. (7) ensures that the recovered FOD is both real and symmetric [Descoteaux et al., 2007]. Substituting Eqs. (6) and (7) into (5) yields the parameterized SD signal model
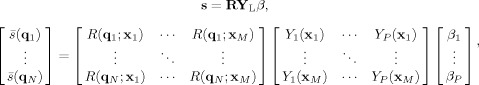 |
(8) |
where the subscript L is used to make the SH expansion order of Y explicit.
RSI model
The two main assumptions of the SD model in Eq. (8) are that (1) the orientation structure of the FOD lies within the subspace spanned by the spherical harmonic basis vectors in Y L, which places an intrinsic upper limit on the angular resolution of the FOD estimate [White and Dale, 2009], and (2) the tissue architecture can be described by a linear mixture of (nonexchanging) cylindrical fiber elements with identical diffusion characteristics. In other words, the diffusion length scale is presumed fixed for all fibers within the voxel. To do away with this assumption, we implement a straightforward extension to the deconvolution model that allows for a mixture of cylindrically symmetric Gaussian kernel functions with different D T and fixed (constant) D L. Thus, Eq. (5) is rewritten as
| (9) |
where D < D < … < D < D L are the transverse diffusivities for each of the J total FODs (Fig. 1). The distribution of FODs f 1,f 2,…,f J, which we call the “FOD spectrum”, models the orientation structure of the diffusion process at length scales given by D < D < … < D . Once again, if the diffusivities are known a priori, the multiscale model retains its efficient linear implementation and can be written
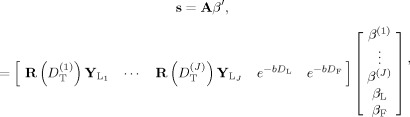 |
(10) |
where A is now a large
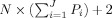 matrix, where P
i = (L
i + 2)(L
i + 1)/2 is the total number of SH for the ith FOD. To model the partial volume fraction of isotropic diffusion in both brain and nonbrain voxels, we included two terms e
and e
to our forward model in Eq. (10), where the longitudinal diffusivity D
L is the longitudinal diffusivity estimated from white matter, and D
F is the “free” water diffusivity estimated from the intraventricular space of our tissue sample (as described in the following “Estimation” section section). Note that unlike traditional SD, the RSI model in Eq. (10) requires multiple b‐values and diffusion gradient directions to separate scale and geometric information.
matrix, where P
i = (L
i + 2)(L
i + 1)/2 is the total number of SH for the ith FOD. To model the partial volume fraction of isotropic diffusion in both brain and nonbrain voxels, we included two terms e
and e
to our forward model in Eq. (10), where the longitudinal diffusivity D
L is the longitudinal diffusivity estimated from white matter, and D
F is the “free” water diffusivity estimated from the intraventricular space of our tissue sample (as described in the following “Estimation” section section). Note that unlike traditional SD, the RSI model in Eq. (10) requires multiple b‐values and diffusion gradient directions to separate scale and geometric information.
Figure 1.
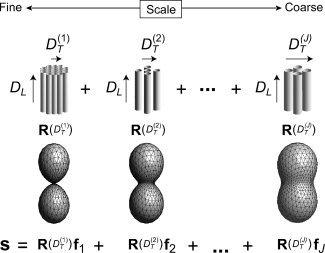
Restriction spectrum model. The oriented component of the diffusion signal is written a linear mixture (spectrum) of cylindrically symmetric Gaussian response functions R with different fixed transverse diffusivities D T and unknown volume fraction and orientation distribution (f).
Estimation
To fit the RSI model in Eq. (10) we first derived an estimate of D
L by fitting a tensor model to the diffusion data in a small region of the corpus callosum known to have a high density of uniformly oriented white‐matter fibers. In this region, we estimated D
L to be ≈0.34 μm2/ms. This value of D
L was also used to estimate the theoretical compartment size diameter for restricted diffusion using Monte Carlo simulations (see “Compartment Size Diameter Simulation” section). Next, we derived an estimate of D
F by fitting the same tensor model to voxels comprising the intraventricular space of our tissue sample. In this region, we estimated D
F to be ≈2.0 μm2/ms. Finally, with D
L and D
F now fixed, we let D
T take on J = 12 linearly spaced values between 0 and 0.9·D
L, and used a fourth‐order (L = 4) SH expansion for each FOD, and a third‐order tessellation of the sphere to define the M = 642 FOD reconstruction points {x1,x2,…,xM}. Given the large number of unknown parameters for this particular parameterization (182 total) we sought a regularized solution for
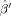 to prevent overfitting
to prevent overfitting
| (11) |
where α is the Tikhonov regularization factor. To optimize the regularization level, we selected the α which minimized the Bayesian Information Criterion (BIC) [Schwarz, 1978] over a large region of our tissue specimen, which included both gray and white matters (not shown). The BIC is defined as
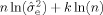 , where n is the number of measurements,
, where n is the number of measurements,
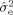 is the error variance, and k = trace(A
†
A) is the effective number of model parameters. The BIC can be interpreted as managing the tradeoff between goodness‐of‐fit on the one hand, by penalizing models with large residual error (i.e., nln(
is the error variance, and k = trace(A
†
A) is the effective number of model parameters. The BIC can be interpreted as managing the tradeoff between goodness‐of‐fit on the one hand, by penalizing models with large residual error (i.e., nln(
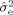 ) term), and reducing model complexity on the other, by penalizing models with a large number of free parameters [i.e., kln(n) term]. At the optimum regularization level (minimum BIC), the effective number of model parameters was reduced to approximately k ≈ 20.
) term), and reducing model complexity on the other, by penalizing models with a large number of free parameters [i.e., kln(n) term]. At the optimum regularization level (minimum BIC), the effective number of model parameters was reduced to approximately k ≈ 20.
Compartment Size Diameter Simulation
The theoretical compartment size diameter for restricted diffusion was estimated using Monte Carlo simulations consisting of a population of “spins” undergoing random‐walk diffusion within a single impermeable cylinder with known diameter. The exact diameter (d) varied for each simulation experiment and ranged between 0.1 and 100 μm. Spins were initially randomly distributed within the cylinder and allowed to diffuse with an intrinsic diffusion coefficient of D L. Spins that encountered a boundary were reflected off the cylinder wall while preserving their unreflected diffusion path length [Hall and Alexander, 2009]. The accumulated spin phase was used to generate synthetic diffusion signals for our Cartesian q‐space acquisition parameters G, Δ, and δ (see “Sample Preparation and Image Acquisition” section). For each cylinder diameter (simulation experiment), the logarithm of the signal measured perpendicular to the long axis of the cylinder was plotted as a function of b‐value to obtain an estimate of D T. This yielded a plot of D T versus d after compiling across all simulation experiments. This curve was then interpolated into for each D T of our RSI model to yield the theoretical cylinder diameter for restricted diffusion at each scale.
RESULTS AND DISCUSSION
General Observations and the Neurite Hypothesis
The goal of this study was to investigate whether a rather straightforward extension of traditional fixed‐scale SD model for HARDI acquisitions could be used to probe the orientation structure of our tissue sample at multiple length scales in a manner that reflects the underlying biology. The results of our RSI analysis is illustrated in Figure 2 for a single horizontal slice taken at the level of the dorsal striatum, hippocampus, and tectum. We found that the majority of the diffusion signal in our fixed tissue sample occurred at the fine (Fig. 2, left hand side; red frame) and coarse scale (Fig. 2, right hand side, blue frame), with little or no signal at intermediate scales. The fine scale diffusion processes were characterized by transverse diffusivities significantly smaller than the longitudinal diffusivity (D T/D L < 0.1), whereas the coarse scale processes were characterized by transverse diffusivities ∼60–90% of the longitudinal diffusivity (0.6 < D T/D L < 0.9). This apparent bimodal separation of fine and coarse scale diffusion is consistent with in vivo biexponential models of “slow” and “fast” diffusion, respectively [Niendorf et al., 1996; Mulkern et al., 1999; Maier and Mulkern, 2008]. Yet, unlike these biexponential models, our RSI model allows for additional quantification of the orientation structure at each scale through analysis of the FOD spectrum. In Figure 2, we plot the FOD spectrum for two representative voxels in white (Fig. 2, row 2) and gray (Fig. 2, row 3) matter. Interestingly, in many gray‐matter voxels and some white‐matter voxels, we found dissimilar orientation structure at the fine and coarse scale (see Fig. 2, row 4). However, in all voxels, we noted similar orientation structure across neighboring scales, suggesting some degree of local blurring of model parameters. In the Appendix (“RSI Model Resolution” section), we quantify the degree of blurring using the model resolution matrix, and show that, in our fixed tissue sample, our optimally regularized RSI model allows for resolving approximately three length scales at the fine, intermediate, and coarse scale level. This means that each of the 12 scales in Figure 2 cannot be considered independent, and that the lack of signal contribution at the intermediate scale is a property of the data and not the model. It should be noted that the intrinsic “scale resolution” of the RSI model is mainly due to the acquisition protocol and not the model itself or Tikhonov regularization level. Rather, the Tikhonov regularization level had a larger impact on the smoothness of the individual FODs of the spectrum (data not shown).
Figure 2.
RSI analysis for a single horizontal slice taken at the level of the dorsal striatum, hippocampus, and tectum. The diffusion length scale is shown increasing from left to right. Volume fraction maps (top row) are derived from the zeroth‐ordered SH coefficients normalized to sum to one (negative weights set to zero). These maps provide an estimate of the fractional contribution of diffusion at each scale to the total signal, where dark voxels indicate large contributions (volume fractions), and light voxels indicate little or no contribution. Note the apparent bimodal separation of fine (left, red frame) and coarse (right, blue frame) scale diffusion processes, which is consistent with in vivo biexponential models of “slow” and “fast” diffusion, respectively. The FOD spectra for two representative voxels in white and gray matter are shown in rows 2, and 3, respectively. Note the differences in orientation structure at the fine and coarse scale, particularly in gray matter (row 3). All FODs are displayed by subtracting the minimum (which highlights the oriented component of the FOD) and scaling their amplitudes by their relative volume fraction. Values for the geometric tortuosity (λg) were computed directly from the RSI model scale, while the compartment size diameters (d) were estimated using Monte Carlo simulations (see “Compartment Size Diameter Simulation section”). Together, the geometric tortuosity and theoretical compartment size diameter support the hypothesis that the fine‐ and coarse‐scale diffusion processes stem from restricted and hindered diffusion in the intraneurite and extraneurite water compartment, respectively (see “Theoretical and Empirical Support for the Neurite Hypothesis” section). In the “Histological Substrates for the r‐FOD and h‐FOD” section, we compare the restricted (r‐FOD) and hindered (h‐FOD) FOD against histology at the two scales illustrated with arrows. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
We were also interested in the physical nature of diffusion at the length scales probed by our RSI model. In biological tissue, there are two general modes of diffusion: hindered and restricted [Le Bihan, 1995]. Hindered diffusion relates to the increase in diffusion path length molecules must travel when diffusing around cellular obstructions and is classically described in terms of the “tortuosity”
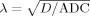 , which relates the measured, or apparent diffusion coefficient (ADC) with the free (aqueous) diffusion coefficient D of the molecule [Sykova and Nicholson, 2008]. Restricted diffusion, on the other hand, relates to the physical blockage of molecules trapped within cellular compartments. If the diffusion time Δ is long enough, the length scale of diffusion will vary dramatically depending on whether diffusion is hindered or restricted [Le Bihan, 1995]. To provide insight into this phenomenon in our data, we computed the geometric tortuosity of our model
, which relates the measured, or apparent diffusion coefficient (ADC) with the free (aqueous) diffusion coefficient D of the molecule [Sykova and Nicholson, 2008]. Restricted diffusion, on the other hand, relates to the physical blockage of molecules trapped within cellular compartments. If the diffusion time Δ is long enough, the length scale of diffusion will vary dramatically depending on whether diffusion is hindered or restricted [Le Bihan, 1995]. To provide insight into this phenomenon in our data, we computed the geometric tortuosity of our model
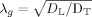 (assuming hindered diffusion) and compartment size diameter (assuming restricted diffusion, estimated using Monte‐Carlo simulations, see “Compartment Size Diameter Simulation” section) for each scale and plot these in Figure 2 (bottom rows). Based on these tortuosity and compartment size calculations, together with the nature of the FOD spectrum, we believe that the fine scale diffusion processes probed by our model has to be restricted and not hindered, and that the geometry of the restricted compartment is consistent with neurites (axons and dendrites), and possibly even long slender glial cell processes. Moreover, we believe that the coarse scale diffusion processes reflect water hindered within the extraneurite compartment, including large cell bodies, ECS, and glia. In the remainder of this article, we provide theoretical and experimental support for this neurite hypothesis. We begin by explaining the rationale for the neurite hypothesis using prior theoretical and empirical evidence, together with our own calculations for the geometric tortuosity and compartment size diameters. Then in the “Histological Substrates for the r‐FOD and h‐FOD section” we provide further empirical support for this hypothesis by comparing the restricted FOD (r‐FOD) and hindered FOD (h‐FOD) at two representative length scales (D
T/D
L = 0 and 0.82, respectively; cf. Fig. 2) against histological material from corresponding anatomical regions.
(assuming hindered diffusion) and compartment size diameter (assuming restricted diffusion, estimated using Monte‐Carlo simulations, see “Compartment Size Diameter Simulation” section) for each scale and plot these in Figure 2 (bottom rows). Based on these tortuosity and compartment size calculations, together with the nature of the FOD spectrum, we believe that the fine scale diffusion processes probed by our model has to be restricted and not hindered, and that the geometry of the restricted compartment is consistent with neurites (axons and dendrites), and possibly even long slender glial cell processes. Moreover, we believe that the coarse scale diffusion processes reflect water hindered within the extraneurite compartment, including large cell bodies, ECS, and glia. In the remainder of this article, we provide theoretical and experimental support for this neurite hypothesis. We begin by explaining the rationale for the neurite hypothesis using prior theoretical and empirical evidence, together with our own calculations for the geometric tortuosity and compartment size diameters. Then in the “Histological Substrates for the r‐FOD and h‐FOD section” we provide further empirical support for this hypothesis by comparing the restricted FOD (r‐FOD) and hindered FOD (h‐FOD) at two representative length scales (D
T/D
L = 0 and 0.82, respectively; cf. Fig. 2) against histological material from corresponding anatomical regions.
Theoretical and Empirical Support for the Neurite Hypothesis
The main source of water restriction in biological tissues comes from cell membranes, and the vast majority of oriented cylindrical compartments in the brain relate to neuronal extensions, or neurites (axons and dendrites). In the rat brain, the diameter of unmyelinated and myelinated subcortical axons range between 0.02 and 3.0 μm, with a mean of 0.2–0.6 μm [Partadiredja et al., 2003; Barazany et al., 2009], which is consistent with our diameter calculations at the fine scale (see the values for d on the bottom of Fig. 2, left). If we assume that the fine‐scale diffusion is hindered and not restricted by neurites, the geometric tortuosity at this scale would have to be greater than 2 (see the values for λg on the bottom of Fig. 2, left). This is quite certainly not the case. We know from previous reports that tortuosities greater than 2 can only be achieved under extreme pathophysiological states such as severe brain ischemia [Nicholson and Sykova, 1998; Chen and Nicholson, 2000]. In fact, the maximum geometric tortuosity introduced by various packed cellular objects in the brain ECS has empirically been estimated to be no greater than 1.225 [Tao et al., 2005]. Even in an ideal simulated environment consisting of a bundle of cylindrical fibers organized in the most compact way, the transverse ADC (D
T) cannot exceed (2/π)2 or 0.4 times the longitudinal ADC (D
L), which corresponds to a geometric tortuosity maximum of λg = π/2 = 1.57 [Le Bihan, 1995]. We therefore argue that all water contributing to the fine scales of our model has to be restricted and not hindered. As neurites fit well with the predicted geometry of the restricting compartment (i.e., cylinders with diameters < 3–4 μm), as opposed to say large spherical cell bodies, we argue that the source of restriction primarily stems from neurites, possibly with a small contribution from elongated cylindrical glial processes. In addition, we argue that all hindered water (with tortuosities less than 1.57) must contribute to the coarse length scales probed by our model. However, the source of this hindered water is less certain, and likely includes not only water in the ECS but also cell bodies and glia. For one, the short diffusion time of our experiment (Δ = 23 ms), together with the fact that the tissue was fixed with paraformaldehyde (which decreases the ADC) limits our ability to probe any restricted compartment (spherical or cylindrical) greater than about
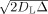 ≈ 4 μm; meaning that any water restricted within compartments greater than 4 μm would be indistinguishable from hindered water. Thus, it's likely that water trapped within many cell bodies (with diameters on the order of 5–15 μm), which may otherwise appear restricted at long diffusion times, would appear hindered in our short diffusion time experiment. Second, while some elongated glial processes may contribute to the restricted volume fraction (as mentioned above), the majority of intraglial water should contribute to the hindered fraction for two reasons. First, the permeability of glia cell membranes are orders of magnitude greater than that of neurons [Solenov et al., 2004], raising the likelihood of increased exchange with the hindered ECS compartment. Second, as we demonstrate in Figure 3, many glia, and in particular protoplasmic astrocytes, which take up as much as 10‐20% of the neuropil volume fraction [Reichenbach A, Wolburg H, 2005] have far more “spongiform” morphologies than neurons [Oberheim et al., 2008; Wilhelmsson et al, 2006], which may allow even restricted water in the glial compartment to travel fast, effectively hindered paths during the experimental diffusion time.
≈ 4 μm; meaning that any water restricted within compartments greater than 4 μm would be indistinguishable from hindered water. Thus, it's likely that water trapped within many cell bodies (with diameters on the order of 5–15 μm), which may otherwise appear restricted at long diffusion times, would appear hindered in our short diffusion time experiment. Second, while some elongated glial processes may contribute to the restricted volume fraction (as mentioned above), the majority of intraglial water should contribute to the hindered fraction for two reasons. First, the permeability of glia cell membranes are orders of magnitude greater than that of neurons [Solenov et al., 2004], raising the likelihood of increased exchange with the hindered ECS compartment. Second, as we demonstrate in Figure 3, many glia, and in particular protoplasmic astrocytes, which take up as much as 10‐20% of the neuropil volume fraction [Reichenbach A, Wolburg H, 2005] have far more “spongiform” morphologies than neurons [Oberheim et al., 2008; Wilhelmsson et al, 2006], which may allow even restricted water in the glial compartment to travel fast, effectively hindered paths during the experimental diffusion time.
Figure 3.
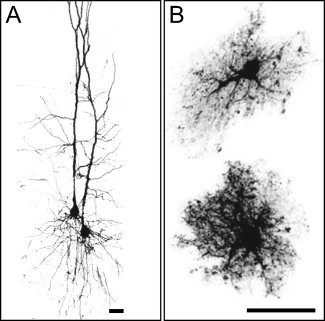
Comparison of neuronal and glial cell morphologies in the rat cerebral cortex. (A) Confocal image of two‐layer‐V pyramidal neurons [modified from Kalisman et al., 2005, with permission]. (B) Maximum projections of dye‐filled, nonreactive astrocytes in layer IV [reprinted from Wilhelmsson et al., 2006 with permission]. In contrast to pyramidal cell dendrites, astrocytes have a “spongiform” appearance with numerous, highly tortuous, and permeable processes that are not well modeled by piecewise cylindrical elements. As a reference, the maximum length scale probed by our experiment is on the order of
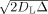 ≈ 4μm. Scale bars, 25 μm
≈ 4μm. Scale bars, 25 μm
In summary, given the aforementioned empirical and theoretical evidence, we surmise that our RSI model has separated the intraneuritic restricted water from extraneuritic hindered water in our fixed tissue sample. Note, this neurite‐specific assignment is fundamentally different from the common (albeit controversial) view that fine (“slow”) and coarse (“fast”) scale diffusion stems from water in the ICS and ECS of tissue [see Mulkern et al., 2009 for a review). Also note that this neurite assignment fits well with model presented by Jespersen for measuring the neurite density and orientation distribution [Jespersen et al., 2007]. In the next section, we provide further empirical support for this hypothesis, by comparing RSI estimates of the r‐FOD and h‐FOD with histological material taken from corresponding anatomical regions.
Histological Substrates for the r‐FOD and h‐FOD
If the neurite hypothesis holds true, then the volume fraction and orientation distribution of neurites should be reflected in the volume fraction and orientation distribution of the r‐FOD, while the h‐FOD should reflect the orientation distribution of hindered extraneuritic water. To investigate the anatomical substrate of the r‐FOD and h‐FOD, we compared their orientation distribution and volume fraction against the histoarchitecture in selected brain regions (the striatum, globus pallidus, cerebral cortex, and cerebellum) known to have complex but characteristic tissue architectures.
Example 1: Striatum and globus pallidus
In both the striatum and globus pallidus, the tissue architecture is characterized by the dense bundles of penetrating corticofugal axons, and by a relatively complex architecture with topographically organized axonal terminal fields [Brown et al., 1998; Alloway et al., 1999; Gerfen, 2005], neurons and glial cells. The penetrating corticofugal axons appear dark in the myelin stain (Figs. 4B,E and 5G), while the complex network of dendritic arbors and axonal terminal fields can be visualized using voltage‐gated potassium channel stains (KChlP1, Fig. 5F,I) and axonal tracing techniques (Fig. 6D), respectively. In the striatum and globus pallidus, the dissociation between the h‐FOD and r‐FOD is pronounced (Figs. 4A,C and D,F and 5E,H).
Figure 4.
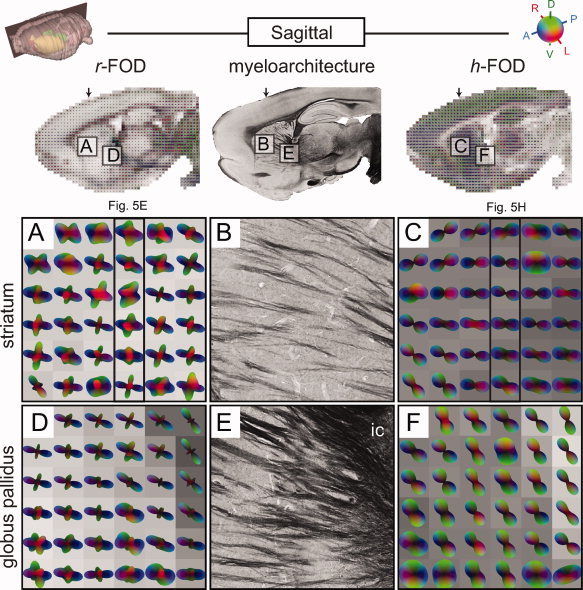
Sagittal comparison of RSI and histology in the striatum and globus pallidus. Color‐coded RSI reconstructions of the r‐FOD (left column; A, D) and h‐FOD (right column; C, F) are shown together with myelin stained histological sections (middle column; B, E) from the same specimen. All RSI distributions were scaled to fit within the voxel boundary, while the h‐FOD was further normalized by subtracting the minimum to highlight its preferred orientation. RSI volume fractions are shown as voxel grayscale intensities (dark corresponds to increased volume fraction). The upper left inset indicates position of the sagittal slice. Two regions are compared, one in the dorsolateral striatum (A–C) and one in the globus pallidus (D–F). Arrows on top of the inset images and frames in A and C indicate the position of the coronal slices shown in Figure 5E,H, respectively. The oblique orientation of the elongated dark, myelin stained fibers in B and E correspond well to the anterioposterior (blue) component of the r‐FOD, but less well with the elongated h‐FOD. The r‐FOD volume fractions (grayscale darkening from left to right in D) fit well with the density of myelin labeled fibers in E. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 5.
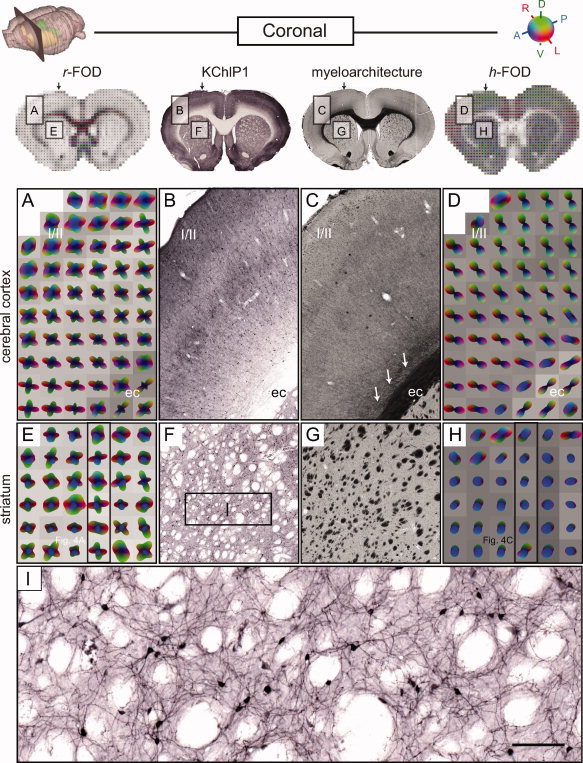
Coronal comparison of RSI and histology in the cerebral cortex and striatum. Presentation as in Figure 4. In addition to the myelin stained histological sections (middle column; C, G) from the same specimen, potassium channel (KChlP1) stains (middle column; B, F, I) are also included from a similar specimen (from http://www.brainmaps.org; see Sample Preparation and Image Acquisition section). Two regions are compared, one in the parietal cortex (A–D) and one in the dorsolateral striatum (E–I). Arrows on top of the inset images and frames in E and H indicate the position of the sagittal slices shown in Figure 4A,C, respectively. In the cortex, the primary orientation of the r‐FOD and h‐FOD are aligned radial to the cortical surface, while the r‐FOD displays additional tangential orientations, which are most prominent in superficial layers I/II. In the striatum, the through‐plane (anterioposterior, blue) r‐FOD and h‐FOD orientations reflect the cross‐sectioned myelin stained fibers passing through the striatum (dark dots in E, light dots in F). Here, again the r‐FOD displays substantial additional in‐plane structure (red and green peaks), which we argue reflects the complex geometry of corticostriatal terminal plexuses (Fig. 6D) and vast networks of dendritic arbors in the region (I). Scale bar 50 μm.
Figure 6.
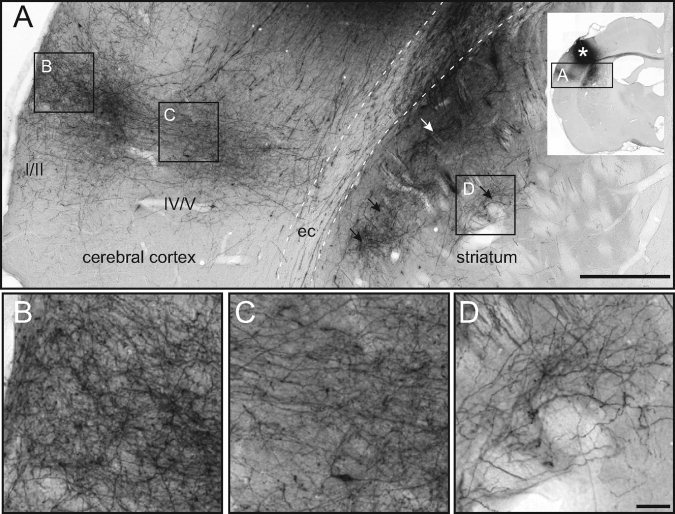
Axonal architecture in the cerebral cortex and dorsal striatum. Histological images [from the Whole Brain Connectivity Atlas at http://www.rbwb.org, case R606; Zakiewicz et al., 2011] showing specific corticocortical and corticostriatal axonal projections, anterogradely labeled by axonal tracer injection in the primary somatosensory cortex (darkly stained region indicated by asterisk in inset, see “Sample Preparation and Image Acquisition” section). (A) Overview of the secondary somatosensory cortex and dorsal striatum showing the distribution of distinct, darkly labeled axonal plexuses. Note the columnar distribution of labeled fibers in the cerebral cortex, with profuse arborizations in layers I–III and V, and laminar distribution of striatal fiber plexuses (arrowheads) extending in parallel with the overlying external capsule (ec). Frames indicate the position of panels B–D. (B–D) Image magnifications showing detailed fiber architectures in cerebral cortex (B, layer I/II; C, layer IV/V) and dorsal striatum (D). Note the perpendicular fiber orientations in the deeper part of the cortex (C) and striatum (D), which match the r‐FOD in Figure 5A,E, respectively. Also, the more complex fiber orientations in the superficial cortex (B) fit well with the more pronounced tangential r‐FOD peaks in Figure 5A. Scale bars, 500 and 50 μm.
For example, in the anterior dorsal striatum, the r‐FOD features three peaks, of which the anterioposterior component corresponds nicely with the through‐plane orientations of penetrating corticofugal axons (compare blue peaks in Fig. 4A with B), while the more complex, crossing orientations appear to reflect the more random organization of corticostriatal terminal plexuses [Brown et al., 1998; Alloway et al., 1999; Veinante and Deschenes, 2003], dendritic arbors, and cellular processes in the region (compare red and green peaks of the r‐FOD in Fig. 5E with the histoarchitecture in Figs. 5F,I and 6D).
In the globus pallidus, a similar pattern of r‐FOD orientations is observed, with prominent peaks well aligned with the oblique axial fibers penetrating the region (compare blue peaks in Fig. 4D with E), and less prominent, dorsoventrally (green) and mediolaterally (red) oriented peaks. While some of the smaller amplitude green and red peaks may be due to ringing artifacts introduced during reconstruction (see Appendix 5.2 for discussion and simulation of these artifacts), the larger amplitude peaks cannot be explained based on ringing artifacts alone, and are consistent with the orientation of pallidal dendrites and striatopallidal axonal terminations in the region [Gerfen, 2005; Sadek et al., 2007].
By contrast, the h‐FOD in the globus pallidus and dorsal striatum has a broader disk‐like shape, with oblique dorsoventral orientation in the globus pallidus (Fig. 4F), and horizontal orientation in the striatum (Figs. 4C and 5H). Recall, the h‐FOD was normalized by subtracting the minimum to highlight their preferred orientation, which turns spherical FODs into disk‐like distributions. Interestingly, in none of these regions is the primary orientation of the h‐FOD aligned consistently with the penetrating myelinated fibers (compare Fig. 4B with C and E with F). While we lack data to interpret this observation conclusively, we surmise that the primary orientation of the h‐FOD in this region reflects the combined hindrance of extraneurite water by both penetrating corticofugal and striatofugal axonal projections, and the highly oriented striatal [Brown et al., 1998; Alloway et al., 1999; Veinante and Deschenes, 2003] and pallidal neurites [Sadek et al., 2007]. This can explain why the h‐FOD appears to align somewhere in between the primary (blue) and secondary and tertiary (green and red) peaks of the r‐FOD (Fig. 4B,C,E,F). This also supports the existing notion that in most regions (excluding uniformly oriented dense white‐matter fiber tracts) the fiber architecture is less well characterized by the fast hindered water fraction compared to the slow restricted fraction [Ronen et al., 2003; Assaf and Basser, 2005].
Example 2: Cerebral cortex
The cerebral cortex features a characteristic laminar and columnar organization with radially and tangentially oriented neurites [Lorente de No, 1938; Szentagothai, 1975; Mountcastle, 1997]. Here, the h‐FOD is aligned radially to the cortical surface (Fig. 5D), consistent with the orientation of pyramidal cell axons and dendrites, as well as most corticoefferent and corticoafferent fibers [Deschenes et al., 1998] (Figs. 5B and 6B,C). By contrast, the r‐FOD not only reflects the same radial orientations, but also displays prominent orientations tangential to the cortical surface (Fig. 5A), with the largest tangential amplitude occurring in the superficial cortical layers I/II. Given their large amplitude relative to the main (radial) peak, is it unlikely that the tangential peaks of the r‐FOD are caused solely by ringing artifacts (again, see Appendix 5.2 for a detailed discussion and simulation of these artifacts), rather they likely reflect the well‐known horizontal orientation of cortical neurites, see e.g., [Kristt, 1978; Cowan and Wilson, 1994; Veinante and Deschenes, 2003]. The complex, characteristic radial and tangential organization of cortical neurites is demonstrated in Figure 6A–C.
In the external capsule (ec) below the cortical gray matter, both the h‐FOD and r‐FOD are elongated parallel with mediolaterally oriented fibers (Fig. 5A–D), while the r‐FOD displays additional perpendicular orientations (Fig. 5A), which appear to reflect corticoefferent fibers passing through the external capsule toward the underlying striatum [Cowan and Wilson, 1994; Veinante and Deschenes, 2003] and the fibers peeling off the ec into the cortex (arrows in Fig 5C).
To lend further support for the notion that the fine (restricted) scales probed by the RSI model reflect water restricted within neurites, we qualitatively compared RSI maps of the total restricted volume fraction (summing scales D T/D L = 0, 0.08, 0.16, 0.25, Fig. 2, red frame) with the myelin and KChlP1 stained sections (Fig. 7). We found that the RSI maps were indeed largely consistent with the expected neurite volume fraction within gray and white matter as the model predicts. RSI estimates of the neurite volume fraction in the cerebral cortex varied from ∼18–31%, with the highest fraction occurring in layers I/II followed by layer V. This pattern is consistent with the high density of apical dendrites and horizontally oriented neurites in layers I/II [Zhou and Hablitz, 1996] (see, also Fig. 6B), as well as numerous dendritic arbors and profuse plexuses of thalamocortical afferents in layer V [Deschenes et al., 1998] (see Fig. 6C). Histological substrates for this pattern are seen in the KChlP1 stain (Fig. 5B, Fig. 7), and the axonal tracer data (Fig. 6B,C). In the underlying white‐matter (ec, and corpus callosum, cc) RSI estimates of the total restricted volume fraction increased dramatically to ∼42–84%, consistent with the presence of densely packed white‐matter fibers in this region, which appear dark red on the myelin stain (Fig. 7). Taken together, these results compare favorably with the expected neurite volume fraction in gray and white matter, and indicate that the restricted volume fraction is a likely surrogate measure of the neurite volume fraction.
Figure 7.
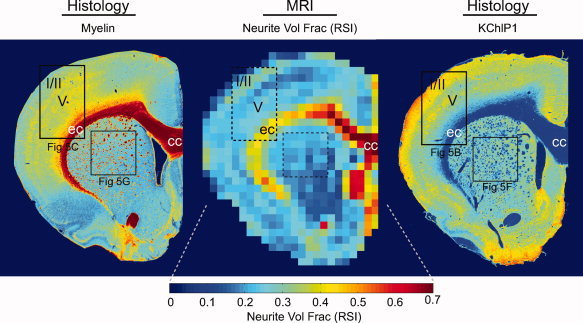
Coronal comparison of RSI estimates of (restricted) neurite volume fraction with histological measures of fiber architecture. The coronal section is the same as in Figure 5 (frames indicate corresponding panels). As expected, the RSI neurite volume fraction is consistent with the combination of both myelinated axons (left panel) and unmyelinated dendrites (right panel). Note also the increased neurite volume fraction in superficial layers of the cerebral cortex (I/II), which are also clearly visible in the potassium channel (KChlP1) stain (right panel). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Example 3: Cerebellum
We finally explored the anatomical substrates of the r‐FOD and h‐FOD in the cerebellum (Fig. 8), which features a well‐known stereotypic architecture, see e.g., [Voogd, 1995]. Our analysis focused on the cerebellar vermis in a horizontal slice at the level of lobules 3, 5, and 8, where the cerebellar folia are mediolaterally oriented, and orientation structure is easier to interpret in relation to the employed slice planes. The cerebellar white matter, granule cell layer, and molecular layers (mls) were all readily identified based on the r‐FOD and h‐FOD volume fractions and orientations (Fig. 8). The narrow Purkinje cell layer could not be distinguished due to partial voluming.
Figure 8.
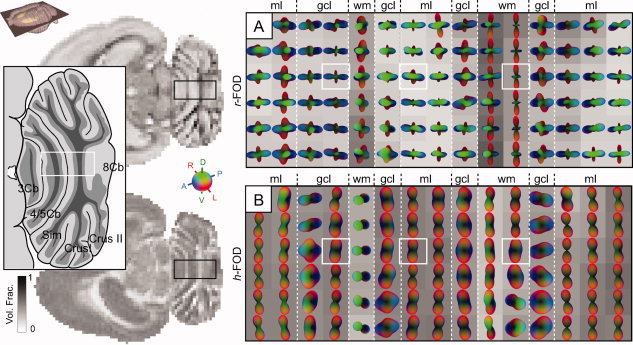
Axial RSI analysis in the cerebellum. Color‐coded RSI reconstructions of the (A) r‐FOD and (B) h‐FOD are shown for a horizontal cerebellar slice taken at level of lobules 3, 5, and 8, corresponding to a level ∼4.6 mm below bregma, illustrated in the inset atlas diagram redrawn from Figure 201 in Paxinos and Watson 2007 [Paxinos and Watson, 2007]. All distributions were scaled to fit within the voxel boundary, while the h‐FOD was further normalized by subtracting the minimum to highlight its preferred orientation. RSI volume fractions are shown as voxel grayscale intensities (dark corresponds to increased volume fraction). The cerebellar white matter and cortical layers are differentiated by distinct differences in r‐FOD and h‐FOD orientation and volume fraction. White boxes correspond to voxels plotted in Figure 10. Cb, cerebellar lobule, CrusI, crus 1 of the ansiform lobule; CrusII, crus 2 of the ansiform lobule; gcl, granule cell layer; ml, molecular layer; Sim, simple lobule; wm, white matter. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In white matter (wm), the cerebellar afferent and efferent fibers largely follow the foliated structure of the cerebellum, but also form more complex (crossing) orientations in some regions, see e.g., [Wu et al., 1999]. As observed in the earlier examples, the preferred orientation of white‐matter fibers was clearly reflected in both r‐FOD and h‐FOD, while the r‐FOD also showed additional secondary and tertiary orientations (Fig. 8), which are consistent with crossing afferent and efferent fibers in this region [Wu et al., 1999], but may also reflect partial voluming with neighboring layers.
The granule cell layer (gcl) is characterized by the presence of numerous small granule cells, and further also smaller numbers of other cell types. Radially (translobularly) oriented mossy fiber and climbing fiber penetrate the gcl and ascend together with granule cell axons through the Purkinje cell layer into the overlying ml [Voogd, 1995]. In this layer, the r‐FOD showed prominent radial (blue and green) peaks (Fig. 8), consistent with the orientation of mossy and climbing fibers, and less prominent mediolaterally (red) oriented peaks (Fig. 8), which may be associated with the different cellular extensions in the gcl. The h‐FOD, on the other hand, tended to be more variably elongated in both translobular or parlobular directions (Fig. 8).
The ml contains several categories of radially (translobularly) oriented neurites (including Purkinje cell dendrites, ascending granule cell axons, dendrites of Golgi‐ and stellate cells, and glial extensions [Voogd, 1995]. These orientations are clearly visible as blue and green peaks in the r‐FOD (Fig. 8). Another characteristic feature of the ml is the presence of numerous, mediolaterally (parlobularly) oriented parallel fibers, which fit well with the prominent mediolateral (red) peaks of the r‐FOD (Fig. 8). The h‐FOD, on the other hand, illustrates a single peak pointed in the direction of the parallel fibers (Fig. 8), which makes sense given the high packing density of parallel fibers in this region compared with other neurites in the ml (e.g., Purkinje cell dendrites and ascending granule cell axons), leading to increased tortuosity of extraneurite water in the orthogonal direction.
Taken together, we found that the r‐FOD demonstrates the stereotypic organization of cerebellar neurites in each layer, while the h‐FOD tends to be more indicative only of the densely packed white‐matter and parallel fibers, which provides further support for the notion that the coarse scale hindered fraction of diffusion is less well characteristic of the underlying fiber architecture compared with the fine scale restricted fraction.
Comparison With SD and DSI
We have demonstrated how the r‐FOD and h‐FOD reflect different aspects of the neurite architecture in both white and gray matter, but we were also interested in how these multiscale RSI measures compared with traditional fixed‐scale SD and DSI. To address this issue, we computed fixed‐scale estimates of the FOD (FS‐FOD) using a single deconvolution kernel (D T/D L = 0.08), which can be viewed as an extension of the traditional SD for HARDI acquisitions to arbitrary q‐space data [Leergaard et al. 2010], and DSI estimates of the water diffusion orientation distribution function (DSI‐dODF) (see Materials and Methods), and compared them with the r‐FOD and h‐FOD in the cerebral cortex and cerebellum.
In the cerebral cortex, we found that the FS‐FOD and DSI‐dODF reflected mainly the radial orientation of cortical neurites in contrast to the r‐FOD, which was able to resolve both the radial and tangential organization of cortical neurites (Fig. 9). It is also interesting to note the qualitative similarity between the h‐FOD and the DSI‐dODF (Fig. 9), which were both normalized by subtracting the minimum to highlight their preferred orientation (the r‐FOD and FS‐FOD were not normalized as their minima was effectively zero). This similarity is not surprising given the DSI‐dODF reflects the overall statistical likelihood of water diffusion in a given direction, which is heavily weighted by the volume fraction of “fast” hindered diffusion along that direction and less so by the “slow” restricted fraction.
Figure 9.
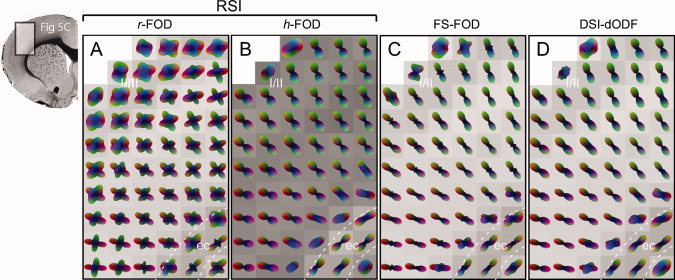
Comparison of RSI with SD and DSI in the cerebral cortex. Color‐coded RSI reconstructions of the (A) r‐FOD and (B) h‐FOD are compared against (C) SD reconstructions of the fixed‐scale FOD (FS‐FOD) and (D) DSI reconstructions of the dODF (DSI‐dODF). The region sampled is the same as in Figure 5A–D. All distributions were scaled to fit within the voxel boundary, while both the h‐FOD and DSI‐dODF were further normalized by subtracting the minimum to highlight their preferred orientation. RSI volume fractions are shown as voxel grayscale intensities (dark corresponds to increased volume fraction). The FS‐FOD and DSI‐dODF are plotted on the r‐FOD volume fraction map. In contrast to the r‐FOD, both the FS‐FOD and DSI‐dODF demonstrate little evidence for the numerous horizontally oriented neurites tangential to the cortical surface (see Fig. 6B,C), and have similar orientation structure to the h‐FOD. I/II, cerebral cortex layer I/II; ec, external capsule. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 10 compares the r‐FOD and h‐FOD versus the FS‐FOD and DSI‐dODF in three representative voxels from different layers of the cerebellum. In white matter (Fig. 10, bottom row), the main (red) peak of the r‐FOD and h‐FOD were well aligned with the main peak of the FS‐FOD and DSI‐dODF, which follow the cerebellar afferent and efferent fibers in this region. Note that in these voxels, the secondary and tertiary peaks of the r‐FOD and FS‐FOD may be due in part to ringing artifacts introduced during reconstruction, as their amplitudes are quite small compared to the main peak (see Appendix 5.2 for a discussion). Small spurious peaks in the r‐FOD, FS‐FOD, and DSI‐dODF may also due to numerical sampling errors associated with nonuniform (Cartesian) sampling of q‐space.
Figure 10.
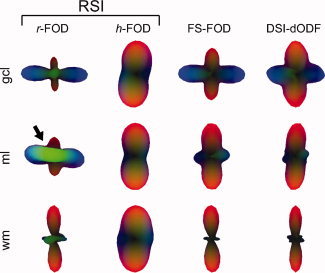
Comparison of RSI with SD and DSI in the cerebellum. Color‐coded RSI reconstructions of the r‐FOD and h‐FOD are compared against the FS‐FOD and DSI‐dODF. The sampled voxels are shown as white boxes in Figure 8. In the molecular layer (ml), the r‐FOD shows additional radially (translobularly) oriented green peaks (arrow) that are consistent with the orientation of Purkinje cell dendrites and ascending granule cell axons in the region. These peaks are not evident in either the FS‐FOD or DSI‐dODF. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In the granule cell layer (gcl, Fig. 10 top row), the FS‐FOD and DSI‐dODF both demonstrated prominent mediolateral (red) and radial (blue) peaks consistent with the orientation of mossy fiber and climbing fiber in the region. These same peaks were also present in the RSI r‐FOD and h‐FOD. Yet additional information can be gleaned from RSI. For example, the strong mediolateral (red) orientation of the h‐FOD suggests mediolaterally oriented neurites may have higher packing densities compared with radially oriented neurites, leading to higher tortuosities of extraneurite water in the orthogonal direction.
The ability to separate hindered and restricted diffusion may also increase the sensitivity of RSI to resolve neurite orientations in voxels with a high partial volume fraction of hindered diffusion. For example, in the ml (Fig. 10 middle row), the FS‐FOD, DSI‐dODF, and RSI h‐FOD again demonstrated prominent mediolateral (red) peaks consistent with the orientation of parallel fibers in the ml. However, the r‐FOD showed strong additional radially (translobularly) oriented blue and green peaks, which are consistent with the known orientation of Purkinje cell dendrites, ascending granule cell axons, dendrites of Golgi‐ and stellate cells, and glial extensions in this region [Voogd, 1995]. These results were generally consistent across other cerebellar voxels (see Fig. 8). Taken together, the r‐FOD seems to capture additional anatomical structure not afforded by fixed‐scale SD‐like reconstruction or DSI.
Advantages and Disadvantages
The RSI analysis method presented in this study is similar in spirit to the earlier model of neurite architectures proposed by Jespersen and coworkers [Jespersen et al. 2007; Jespersen et al. 2010]. Both use a distribution of nonexchanging cylinders to model the tissue microstructure and spherical harmonics to describe their orientation distribution. Both also harness multiple b‐value data to separate scale information at the hindered and restricted level. Similar to RSI, Jespersen's model also showed good agreement with the neurite volume fraction across various gray‐ and white‐matter regions [Jespersen et al. 2010]. The major difference between the two methods is that our RSI model does not explicitly fit for the intracylindrical and extracylindrical diffusivities (which requires nonlinear optimization of model parameters), but rather assumes the diffusivities can take on a broad range (or spectrum) of values when fitting the data. This allows RSI to preserve a linear implementation, which greatly simplifies estimation and decreases computation time significantly. The spectrum of independent scales and orientation distributions further allows RSI to model the tissue microstructure with minimal a priori assumptions on the number of hindered and restricted water compartments, their respective geometries, and partial volume fractions. This is in contrast to other biophysical models of diffusion in white matter, which often impose some form of geometric dependence between the hindered and restricted compartment to infer on nonlinear parameters describing the axon diameter distribution [Assaf et al., 2008; Alexander et al., 2010]. While it may be reasonable to equate the geometry of the hindered and restricted compartment in white matter, as axons provide the major source of both diffusion hindrance and restriction in this region, in gray matter, the physical relationship between the two compartments is more complex. For example, in the current study, and in Jespersen's model, the geometries of the hindered and restricted compartments are modeled independently to account for the fact that there are numerous additional structures in the extraneurite compartment (cell bodies, glia, ECS) that can alter the tortuosity of the hindered compartment besides neurites alone. Also, the tortuosity is known to fall off rapidly with increasing ECS volume fraction α [Chen and Nicholson, 2000; Kume‐Kick et al, 2002] and therefore it's likely only populations of tightly packed neurites (i.e., small α) will produce measurable increases in tortuosity in gray‐matter voxels, while sparsely packed neurites will not. This may explain why in the current study not all peaks in the r‐FOD were demonstrated in the h‐FOD and helps validate the use of independent geometries for the hindered and restricted compartment. With that said, the disadvantage of using a relatively unconstrained mixture model for the tissue architecture is the risk of overfitting and blurring of model parameters, as well as reduced inferential power. In the current study, we mitigated the risk of overfitting using carefully chosen model regularization techniques in conjunction with the Bayesian information criterion (BIC) to optimize the regularization level. We also quantified the extent and nature of model blurring in the optimally regularized model using the resolution matrix (as described in Appendix 5.1). Finally, we gained inferential power in the current study using the combination of simulations and theoretical (tortuosity) models of hindered and restricted diffusion in known geometries to relate our observations to the underlying histoarchitecture in various anatomical regions.
Some care must be taken when extrapolating our findings in a fixed tissue sample to the in vivo situation. Formalin fixation is known to reduce the mean diffusivity in gray and white matter by about 64% and 80%, respectively, but has little effect on the fractional anisotropy (FA) [D'Arceuil et al., 2007]. Given the FA is preserved, we would not expect the fixation process to alter the orientation distribution of the r‐FOD or h‐FOD, nor the overall pattern of the length scale spectrum per se (i.e., separation of hindered and restricted diffusion), as both are based on measures of the relative diffusivity D T/D L. However, as the fixation process has different effects on gray and white matter [D'Arceuil et al., 2007] and we assumed a constant D L across both regions, one should be cautious when interpreting the absolute volume fraction of hindered and restricted diffusion at a given length scale.
CONCLUSIONS
In this work, we presented an efficient linear reconstruction and modeling framework for multidimensional (multidirection and multi b‐value) diffusion data called RSI that allows for resolving both length scale and orientation information of biological tissue microstructures. Our model is based on a straightforward extension of the linear SD HARDI reconstruction method, by including a range (or spectrum) of Gaussian deconvolution kernels. We demonstrate in high b‐value Cartesian q‐space data how RSI analysis can be used to separate fine and coarse scale diffusion processes, which we argue stems mainly from restricted and hindered diffusion in the intraneurite and extraneurite water compartment, respectively. We support this hypothesis both theoretically and empirically, and demonstrate the correspondence of the restricted volume fraction and corresponding orientation distribution (r‐FOD) with the underlying neurite volume fraction and three‐dimensional histoarchitecture of white‐ and gray‐matter structures. We further demonstrate how RSI reconstructions of the r‐FOD captures additional anatomical structure beyond that which can be gleaned from traditional fixed‐scale SD and DSI analyses, particularly in gray‐matter regions. Future work is needed to understand the conditions and experimental protocols required to further probe hindered and restricted diffusion length scales in greater detail, which may include using multidiffusion time protocols to explore the diameter distribution of various cellular compartments, similar to the AxCaliber method for quantifying axon diameter distributions in vivo [Barazany et al., 2009; Assaf et al., 2008].
We conclude that incorporating diffusion length scale information in geometric models of biological diffusion will be of upmost importance for advancing state‐of‐the‐art dMRI methods beyond quantifying white‐matter orientations, to providing a more detailed quantitative characterization of tissue microstructures in health and disease. While future work will be required to test the application of this method on clinical scanners, we anticipate the general multiscale linear mixture model framework of RSI will find important clinical applications in probing salient microstructural features of normal and pathological tissue in vivo.
RSI Model Resolution
The concept of the model resolution matrix in linear estimation problems is a well established and important way to characterize the bias in linear inverse problems [Aster et al., 2005]. The basic idea is to see how well a particular inverse solution
 matches the original model parameters β through the expression
matches the original model parameters β through the expression
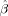 =A
†
A β, where A
† is the regularized inverse matrix, and A
†
A is the model resolution matrix. In practice, the model resolution matrix is commonly used in two different ways. The first is to examine the diagonal elements of A
†
A for their deviation from unity. A value of one would indicate perfect resolution, i.e., where
=A
†
A β, where A
† is the regularized inverse matrix, and A
†
A is the model resolution matrix. In practice, the model resolution matrix is commonly used in two different ways. The first is to examine the diagonal elements of A
†
A for their deviation from unity. A value of one would indicate perfect resolution, i.e., where
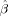 =Iβ, with I being the identity matrix. In this way, the trace of A
†
A can be used to indicate the intrinsic dimensionality of the problem, by quantifying the total number of resolvable parameters. The second is to multiply A
†
A by a particular “test model” βtest to see how well the true (test) model would be resolved using the regularized inverse matrix. Often the test models are column vectors of all zeros, except for a single element equal to one, which when multiplied on the left hand side by A
†
A equals the corresponding column of A
†
A. Thus, the columns of A
†
A, or “resolution kernels,” describe how well the true parameters are recovered or blurred by the regularized inverse matrix. Similarly the rows of A
†
A can be viewed as quantifying how sensitive a particular parameter estimate is to the true model.
=Iβ, with I being the identity matrix. In this way, the trace of A
†
A can be used to indicate the intrinsic dimensionality of the problem, by quantifying the total number of resolvable parameters. The second is to multiply A
†
A by a particular “test model” βtest to see how well the true (test) model would be resolved using the regularized inverse matrix. Often the test models are column vectors of all zeros, except for a single element equal to one, which when multiplied on the left hand side by A
†
A equals the corresponding column of A
†
A. Thus, the columns of A
†
A, or “resolution kernels,” describe how well the true parameters are recovered or blurred by the regularized inverse matrix. Similarly the rows of A
†
A can be viewed as quantifying how sensitive a particular parameter estimate is to the true model.
In this section, we use the model resolution matrix to quantify the blurring and intrinsic dimensionality of our optimally regularized RSI model. To do this, we first projected the regularized model resolution matrix A † A (cf. Eqs. 10,11) onto the FOD surface via Y L A † A Y , where Y L is a large block diagonal matrix of SH vectors. In this way, the model resolution can be visualized directly on the FOD surface, rather than in spherical harmonic space, which is more difficult to interpret. The resultant matrix was then row normalized (i.e., the same axis limits were used for each row element) and plotted in Figure A1. For simplicity, and without loss of generality, we plot only the columns of the resolution matrix that corresponds to unit fibers pointed along the horizontal axis (i.e., “test models” were horizontal unit fibers), and ignore the two isotropic terms. Each row of the matrix can be interpreted as the sensitivity of the recovered (estimated) FOD at that scale to FODs at all other scales. Thus, perfect resolution would result in a diagonal matrix of cigar‐like FODs.
Figure A1.
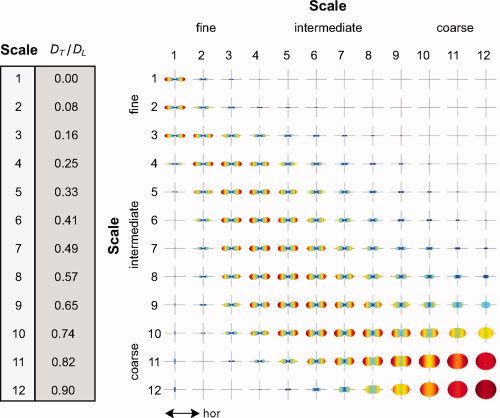
RSI model resolution matrix. Each row illustrates the sensitivity of the recovered (estimated) FOD at that scale (row number) to FODs at all other scales (column number). Perfect resolution would result in a diagonal matrix of “cigar‐like” FODs along the horizontal axis (labeled). All FODs are shown unnormalized (without subtracting the minimum) with negative values set to zero. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
As expected, we found that FODs at the fine and coarse scale were separable, despite significant blurring across neighboring scales. For example, rows 1–2 have little or no contribution from columns 10–12, and rows 10–12 have little to no contribution from columns 1–2. This separability fits well with our empirical observations (see Fig. 2). Also, the isotropic FODs in the bottom right‐hand corner demonstrate how estimates of coarse scale hindered FODs have higher angular uncertainly compared with the restricted FODs in the upper left, which also fits well with our empirical observations. As mentioned above, the resolution matrix in Figure A1 quantitatively demonstrates how information is blurred across neighboring scales using our optimally regularized model and q‐space acquisition parameters (i.e., G, Δ, and δ). The model resolution matrix can also be used to optimize the experimental acquisition parameters offline to achieve finer scale resolutions. For example, it may be possible to use multidiffusion time protocols to resolve restricted length scales in more detail, which would result in diagonal terms in the upper left‐hand corner of Figure A1. One can also use the trace of the resolution matrix to quantify the effective number of resolvable scales, which we calculated in our q‐space data to be just under 3 (2.67).
Reconstruction Bias (Ringing Artifacts)
An additional source of bias well‐known to all deconvolution‐based HARDI methods using spherical harmonic basis functions are ringing artifacts (spurious sidelobes) introduced during reconstruction. These artifactual sidelobes result from truncation of the spherical harmonic series, leading to a form of Gibbs ringing on the surface. For a 4th order (L = 4) harmonic expansion, there will be exactly L/2 − 1 = 1 sidelobes that occurs at 90° to the main peak [Hess et al., 2006]. Often, these artifactual peaks can be mitigated with appropriate model regularization techniques such as Tikhonov regularization (used in this study) or Laplace‐Beltrami regularization while balancing the tradeoff between angular resolution on the one hand and suppression of ringing artifacts on the other.
To test the extent to which our regularized RSI model was prone to ringing artifacts, we performed an additional Monte Carlo simulation study. Briefly, we generated synthetic diffusion signals using a simplified RSI model consisting of a single fiber system with two diffusion scales, one restricted (D T/D L = 0), and one hindered (D T/D L = 0.82) with various volume fractions. For each Monte Carlo run, we added random Rician noise to the synthetic signal at a given signal‐to‐noise (SNR) ratio with respect to the signal power at b = 0. Finally, we used the fully regularized RSI model (all 12 scales as implemented in this study) to estimate the model parameters. We then repeated the experiment for various SNR levels and hindered and restricted volume fractions.
The results of our Monte Carlo simulations are shown in Figure A2. Note that we only plot RSI estimates of the r‐FOD, as the h‐FOD was found to be largely insensitive to these ringing artifacts. Not surprisingly we found that the ringing artifacts increased both as the SNR decreased and as the volume fraction of hindered diffusion increased (Fig. A2). This is consistent with prior simulation studies showing both the SNR dependence of these artifacts [Tournier et al., 2008] and their sensitivity to isotropic “background” diffusion [Dell'acqua et al., 2010]. However, we also found that the amplitude of these ringing artifacts was generally very small compared to the main peak, even at very low SNR (Fig. A2). Thus, given the SNR of our q‐space data was in the range of ≈29.8 and ≈55.1 for white and gray matter, respectively, we would not expect the amplitude of the ringing artifacts to be greater than about 7% of the main peak, even in regions with a high partial volume fraction of hindered diffusion, such as cortical gray matter (˜70–80%). Taken together with the fact that many of our r‐FOD peaks were not always at 90° to each other (see Fig. 5E), we argue that it is unlikely that many of the secondary and tertiary orientations of the r‐FOD presented in this study can be explained on the basis of ringing artifacts alone, but rather likely reflects to a large extent the underlying neurite architecture of the region.
Figure A2.
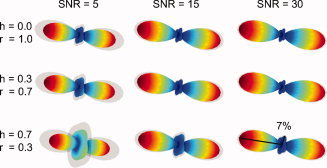
RSI Monte Carlo simulations. The mean r‐FOD (color‐coded surface) and mean plus standard deviation (gray opaque surface) over 100 Monte Carlo runs are shown for various SNR levels and hindered (h) and restricted (r) volume fractions. The amplitude of the 90° ringing artifact increases both with decreasing SNR and increasing hindered volume fraction. The SNR of our q‐space data in white and gray matter was ≈29.8 and ≈55.1, respectively. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
REFERENCES
- Alexander D ( 2005a): Maximum entropy spherical deconvolution for diffusion MRI In: Christensen G, Sonka M, editors. Information Processing in Medical Imaging: Springer; Berlin/Heidelberg: pp 27–57. [DOI] [PubMed] [Google Scholar]
- Alexander DC ( 2005b): Multiple‐fiber reconstruction algorithms for diffusion MRI. Ann NY Acad Sci 1064: 113–133. [DOI] [PubMed] [Google Scholar]
- Alexander DC, Barker GJ, Arridge SR ( 2002): Detection and modeling of non‐Gaussian apparent diffusion coefficient profiles in human brain data. Magn Reson Med 48: 331–340. [DOI] [PubMed] [Google Scholar]
- Alexander DC, Hubbard PL, Hall MG, Moore EA, Ptito M, Parker GJ, Dyrby TB ( 2010): Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage 52: 1374–1389. [DOI] [PubMed] [Google Scholar]
- Alloway KD, Crist J, Mutic JJ, Roy SA ( 1999): Corticostriatal projections from rat barrel cortex have an anisotropic organization that correlates with vibrissal whisking behavior. J Neurosci 19: 10908–10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AW ( 2005): Measurement of fiber orientation distributions using high angular resolution diffusion imaging. Magn Reson Med 54: 1194–1206. [DOI] [PubMed] [Google Scholar]
- Assaf Y, Basser PJ ( 2005): Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. Neuroimage 27: 48–58. [DOI] [PubMed] [Google Scholar]
- Assaf Y, Blumenfeld‐Katzir T, Yovel Y, Basser PJ ( 2008): AxCaliber: A method for measuring axon diameter distribution from diffusion MRI. Magn Reson Med 59: 1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y, Cohen Y ( 2000): Assignment of the water slow‐diffusing component in the central nervous system using q‐space diffusion MRS: Implications for fiber tract imaging. Magn Reson Med 43: 191–199. [DOI] [PubMed] [Google Scholar]
- Aster RC, Borchers B, Thurber CH ( 2005): Parameter Estimation and Inverse Problems. Dmowska R, Holton JR, Rossby HT, editors: Elsevier Academic Press; pp 62‐64. [Google Scholar]
- Barazany D, Basser PJ, Assaf Y ( 2009): In vivo measurement of axon diameter distribution in the corpus callosum of rat brain. Brain 132( Pt 5): 1210–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ ( 2002): Relationships between diffusion tensor and q‐space MRI. Magn Reson Med 47: 392–397. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D ( 1994a): Estimation of the effective self‐diffusion tensor from the NMR spin echo. J Magn Reson B 103: 247–254. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D ( 1994b): MR diffusion tensor spectroscopy and imaging. Biophys J 66: 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A ( 2000): In vivo fiber tractography using DT‐MRI data. Magn Reson Med 44: 625–632. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C ( 1996): Microstructural and physiological features of tissues elucidated by quantitative‐diffusion‐tensor MRI. J Magn Reson B 111: 209–219. [DOI] [PubMed] [Google Scholar]
- Beaulieu C ( 2002): The basis of anisotropic water diffusion in the nervous system – a technical review. NMR Biomed. 15: 435–455. [DOI] [PubMed] [Google Scholar]
- Behrens TE, Woolrich MW, Jenkinson M, Johansen‐Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM ( 2003): Characterization and propagation of uncertainty in diffusion‐weighted MR imaging. Magn Reson Med 50: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Brown LL, Smith DM, Goldbloom LM ( 1998): Organizing principles of cortical integration in the rat neostriatum: Corticostriate map of the body surface is an ordered lattice of curved laminae and radial points. J Comp Neurol 392: 468–488. [PubMed] [Google Scholar]
- Chen KC, Nicholson C ( 2000): Changes in brain cell shape create residual extracellular space volume and explain tortuosity behavior during osmotic challenge. Proc Natl Acad Sci USA 97: 8306–8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan RL, Wilson CJ ( 1994): Spontaneous firing patterns and axonal projections of single corticostriatal neurons in the rat medial agranular cortex. J Neurophysiol 71: 17–32. [DOI] [PubMed] [Google Scholar]
- D'Arceuil H, de Crespigny A ( 2007): The effects of brain tissue decomposition on diffusion tensor imaging and tractography. Neuroimage 36: 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arceuil HE, Westmoreland S, de Crespigny AJ ( 2007): An approach to high resolution diffusion tensor imaging in fixed primate brain. Neuroimage 35: 553–565. [DOI] [PubMed] [Google Scholar]
- Dell'Acqua F, Rizzo G, Scifo P, Clarke RA, Scotti G, Fazio F ( 2007): A model‐based deconvolution approach to solve fiber crossing in diffusion‐weighted MR imaging. IEEE Trans Biomed Eng 54: 462–472. [DOI] [PubMed] [Google Scholar]
- Dell'acqua F, Scifo P, Rizzo G, Catani M, Simmons A, Scotti G, Fazio F ( 2010): A modified damped Richardson‐Lucy algorithm to reduce isotropic background effects in spherical deconvolution. Neuroimage 49: 1446–1458. [DOI] [PubMed] [Google Scholar]
- Deschenes M, Veinante P, Zhang ZW ( 1998): The organization of corticothalamic projections: Reciprocity versus parity. Brain Res Brain Res Rev 28: 286–308. [DOI] [PubMed] [Google Scholar]
- Descoteaux M, Angelino E, Fitzgibbons S, Deriche R ( 2007): Regularized, fast, and robust analytical Q‐ball imaging. Magn Reson Med 58: 497–510. [DOI] [PubMed] [Google Scholar]
- Frank LR ( 2002): Characterization of anisotropy in high angular resolution diffusion‐weighted MRI. Magn Reson Med 47: 1083–1099. [DOI] [PubMed] [Google Scholar]
- Gerfen CR ( 2005): Basal Ganglia In: Paxinos G, editor. The Rat Nervous System, 3rd ed San Diego, CA: Elsevier Academic Press; pp 455‐508. [Google Scholar]
- Hall MG, Alexander DC ( 2009): Convergence and parameter choice for Monte‐Carlo simulations of diffusion MRI. IEEE Trans Med Imaging 28: 1354–1364. [DOI] [PubMed] [Google Scholar]
- Hess CP, Mukherjee P, Han ET, Xu D, Vigneron DB ( 2006): Q‐ball reconstruction of multimodal fiber orientations using the spherical harmonic basis. Magn Reson Med 56: 104–117. [DOI] [PubMed] [Google Scholar]
- Jansons KM, Alexander DC ( 2003): Persistent angular structure: New insights from diffusion MRI data. Inf Process Med Imaging 18: 672–683. [DOI] [PubMed] [Google Scholar]
- Jespersen SN, Bjarkam CR, Nyengaard JR, Chakravarty MM, Hansen B, Vosegaard T, Østergaard L, Yablonskiy D, Nielsen NC, Vestergaard‐Poulsen P ( 2010): Neurite density from magnetic resonance diffusion measurements at ultrahigh field: Comparison with light microscopy and electron microscopy. Neuroimage 49: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen SN, Kroenke CD, Østergaard L, Ackerman JJ, Yablonskiy DA ( 2007): Modeling dendrite density from magnetic resonance diffusion measurements. Neuroimage 34: 1473–1486. [DOI] [PubMed] [Google Scholar]
- Jian B, Vemuri BC ( 2007): A unified computational framework for deconvolution to reconstruct multiple fibers from diffusion weighted MRI. IEEE Trans Med Imaging 26: 1464–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaden E, Knosche TR, Anwander A ( 2007): Parametric spherical deconvolution: Inferring anatomical connectivity using diffusion MR imaging. Neuroimage 37: 474–488. [DOI] [PubMed] [Google Scholar]
- Kalisman N, Silberberg G, Markram H ( 2005): The neocortical microcircuit as a tabula rasa. Proc Natl Acad Sci USA 102: 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärger J, Heink W ( 1983): The propagator representation of molecular transport in microporous crystallites. J Magn Reson 51: 1–7. [Google Scholar]
- Kristt DA ( 1978): Neuronal differentiation in somatosensory cortex of the rat. I. Relationship to synaptogenesis in the first postnatal week. Brain Res 150: 467–486. [DOI] [PubMed] [Google Scholar]
- Kume‐Kick, J , Mazel, T , Vorisek, I , Hrabetova, S , Tau, L , Nicholson, C. 2002: Independence of extracellular tortuosity and volume fraction during osmotic challenge in rat neocortex. J Physiol 542: 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan, D , Breton, E , Lallemand, D , Grenier, P , Cabanis, E , Laval‐Jeantet, M ( 1986): MR imaging of intravoxel incoherent motions: Application to diffusion and perfusion in neurologic disorders. Radiology 161: 401–407. [DOI] [PubMed] [Google Scholar]
- Le Bihan, D ( 1995): Molecular diffusion, tissue microdynamics and microstructure. NMR Biomed 8: 375–386. [DOI] [PubMed] [Google Scholar]
- Leergaard TB, White NS, de Crespigny A, Bolstad I, D'Arceuil H, Bjaalie JG, Dale AM ( 2010): Quantitative histological validation of diffusion MRI fiber orientation distributions in the rat brain. PLoS One 5: e8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente de No R (1938): The cerebral cortex: Architecture, intracortical connections and motor projections In Fulton JF, editor. Physiology of the Nervous System. London: Oxford University Press; pp 291–321. [Google Scholar]
- Maier SE, Mulkern RV ( 2008): Biexponential analysis of diffusion‐related signal decay in normal human cortical and deep gray matter. Magn Reson Imaging 26: 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountcastle VB ( 1997): The columnar organization of the neocortex. Brain 120: 701–722. [DOI] [PubMed] [Google Scholar]
- Mulkern RV, Gudbjartsson H, Westin CF, Zengingonul HP, Gartner W, Guttmann CR, Robertson RL, Kyriakos W, Schwartz R, Holtzman D, Jolesz FA, Maier SE, ( 1999): Multi‐component apparent diffusion coefficients in human brain. NMR Biomed 12: 51–62. [DOI] [PubMed] [Google Scholar]
- Mulkern RV, Haker SJ, Maier SE ( 2009): On high b diffusion imaging in the human brain: Ruminations and experimental insights. Magn Reson Imaging 27: 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson C, Sykova E ( 1998): Extracellular space structure revealed by diffusion analysis. Trends Neurosci 21: 207–215. [DOI] [PubMed] [Google Scholar]
- Niendorf T, Dijkhuizen RM, Norris DG, van Lookeren Campagne M, Nicolay K ( 1996): Biexponential diffusion attenuation in various states of brain tissue: Implications for diffusion‐weighted imaging. Magn Reson Med 36: 847–857. [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Tian GF, Han X, Peng W, Takano T, Ransom B, Nedergaard M ( 2008): Loss of astrocytic domain organization in the epileptic brain. J Neurosci 28: 3264–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozarslan E, Shepherd TM, Vemuri BC, Blackband SJ, Mareci TH ( 2006): Resolution of complex tissue microarchitecture using the diffusion orientation transform (DOT). Neuroimage 31: 1086–1103. [DOI] [PubMed] [Google Scholar]
- Partadiredja G, Miller R, Oorschot DE ( 2003): The number, size, and type of axons in rat subcortical white matter on left and right sides: A stereological, ultrastructural study. J Neurocytol 32: 1165–1179. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. 2007. The rat brain in stereotaxic coordinates. 6th Edition Amsterdam: Elsevier; 1223 p. [Google Scholar]
- Rhodes KJ, Carroll KI, Sung MA, Doliveira LC, Monaghan MM, Burke SL, Strassle BW, Buchwalder L, Menegola M, Cao J, An WF, Trimmer JS ( 2004): KchIPs and Kv4 alpha subunits as integral components of A‐type potassium channels in mammalian brain. J Neurosci 24: 7903–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach A, Wolburg H ( 2005): Astrocytes and ependymal glia In Kettenmann H, Ransom BR, editors. Neuroglia. New York: Oxford University Press; p 23 [Google Scholar]
- Ronen I, Kim KH, Garwood M, Ugurbil K, Kim DS ( 2003): Conventional DTI vs. slow and fast diffusion tensors in cat visual cortex. Magn Reson Med 49: 785–790. [DOI] [PubMed] [Google Scholar]
- Sadek AR, Magill PJ, Bolam JP ( 2007): A single‐cell analysis of intrinsic connectivity in the rat globus pallidus. J Neurosci 27: 6352–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G ( 1978): Estimating the dimension of a model. Ann Stat 6: 461–464. [Google Scholar]
- Solenov E, Watanabe H, Manley GT, Verkman AS ( 2004): Sevenfold‐reduced osmotic water permeability in primary astrocyte cultures from AQP‐4‐deficient mice, measured by a fluorescence quenching method. Am J Physiol Cell Physiol 286: C426–C432. [DOI] [PubMed] [Google Scholar]
- Syková E, Nicholson C ( 2008): Diffusion in brain extracellular space. Physiol Rev 88: 1277–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentagothai J ( 1975): The ‘module‐concept’ in cerebral cortex architecture. Brain Res 95: 475–496. [DOI] [PubMed] [Google Scholar]
- Tao A, Tao L, Nicholson C ( 2005): Cell cavities increase tortuosity in brain extracellular space. J Theor Biol 234: 525–536. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Gadian DG, Connelly A ( 2004): Direct estimation of the fiber orientation density function from diffusion‐weighted MRI data using spherical deconvolution. Neuroimage 23: 1176–1185. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Yeh CH, Calamante F, Cho KH, Connelly A, Lin CP ( 2008): Resolving crossing fibres using constrained spherical deconvolution: Validation using diffusion‐weighted imaging phantom data. Neuroimage 42: 617–625. [DOI] [PubMed] [Google Scholar]
- Tuch DS ( 2004): Q‐ball imaging. Magn Reson Med 52: 1358–1372. [DOI] [PubMed] [Google Scholar]
- Veinante P, Deschenes M ( 2003): Single‐cell study of motor cortex projections to the barrel field in rats. J Comp Neurol 464: 98–103. [DOI] [PubMed] [Google Scholar]
- Voogd J ( 1995): Cerebellum In: Paxinos G, editor. The Rat Nervous System. San Diego: Academic Press; pp 309–352. [Google Scholar]
- Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM ( 2005): Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med 54: 1377–1386. [DOI] [PubMed] [Google Scholar]
- White NS, Dale AM ( 2009): Optimal diffusion MRI acquisition for fiber orientation density estimation: An analytic approach. Hum Brain Mapp Nov 30( 11): 3696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson U, Bushong EA, Price DL, Smarr BL, Phung V, Terada M, Ellisman MH, Pekny M ( 2006): Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proc Natl Acad Sci USA 103: 17513–17518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelche M ( 1942): Eine neue Methode der Markscheidenfarbung. J Psychol Neurol 51: 199–202. [Google Scholar]
- Wu HS, Sugihara I, Shinoda Y ( 1999): Projection patterns of single mossy fibers originating from the lateral reticular nucleus in the rat cerebellar cortex and nuclei. J Comp Neurol 411: 97–118. [DOI] [PubMed] [Google Scholar]
- Yablonskiy DA, Sukstanskii AL ( 2010): Theoretical models of the diffusion weighted MR signal. NMR Biomed 23: 661–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakiewicz IM, van Dongen YC, Leergaard TB, Bjaalie JG ( 2011) Workflow and Atlas System for Brain‐Wide Mapping of Axonal Connectivity in Rat. PLoS One. 6(8): e22669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FM, Hablitz JJ ( 1996): Morphological properties of intracellularly labeled layer I neurons in rat neocortex. J Comp Neurol 376: 198–213. [DOI] [PubMed] [Google Scholar]


