Abstract
Single crystal X-ray diffraction show that Zr(IV) forms an octa-coordinated complex with 4 bidentate hydroxamates whose solution structures were investigated utilizing density functional theory at the level of B3LYP/DGDZVP. Stability constants obtained by potentiometry were in accordance with the tendency observed when radiolabeling with 89Zr.
The positron-emitter 89Zr exhibits favorable physical characteristics for applications in nuclear imaging, especially when associated with a cancer targeting antibody. Its 78.4 h half-life matches the biodistribution kinetics of full antibodies, allowing for the achievement of high tumor-to-background signal ratio a few days after administration. Recent pre-clinical and clinical studies have shown the high potential of this radionuclide,1 and the relatively low cost and simplicity of production makes 89Zr a serious challenger to 124I (T1/2 = 100.2 h), a more costly and extensively investigated β−-emitter.2
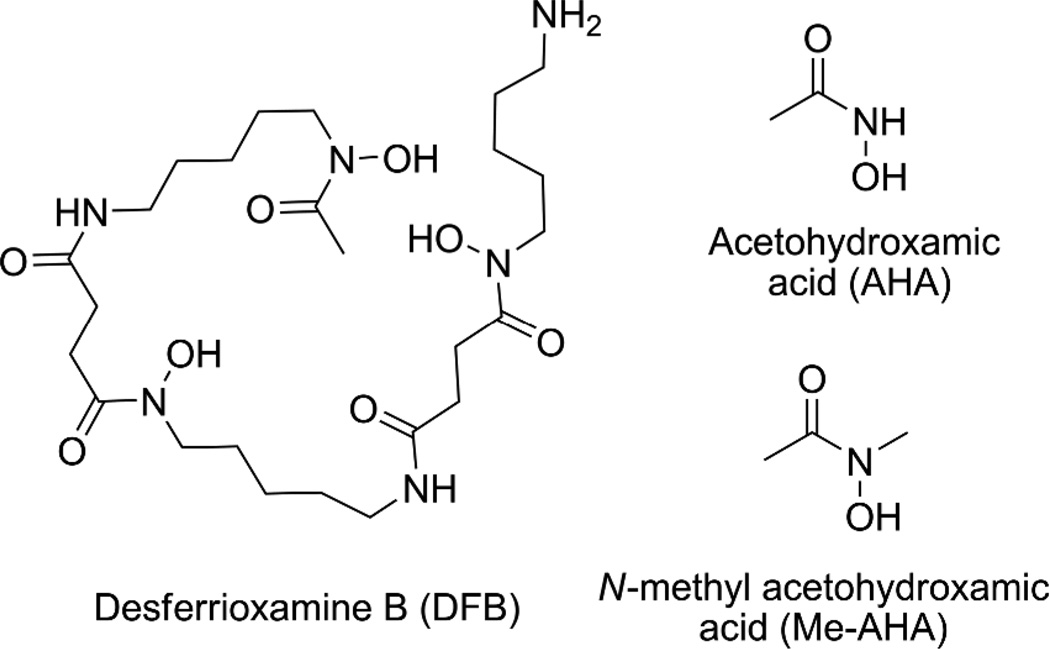
Currently, antibody radiolabeling with 89Zr is performed via the Desferrioxamine B chelator (DFB), a siderophore bearing 3 hydroxamate units coordinated to the metal (Fig. 1).3 However, while use of DFB has allowed radiolabeling of antibodies with 89Zr in proof-of-concept studies, concerns remain regarding the in vivo stability of this complex. This instability has been observed in several animal studies with uptake of 89Zr in bone when release from the chelate occurs.4
Figure 1.
Compounds discussed in this work.
While several works have focused on the modification of the linkage between the DFB and the antibody,3,5 no attempts to improve the chelator itself have been reported. Yet, with only 3 hydroxamate donor groups, DFB does not appear to be ideal to saturate the coordination sphere of Zr4+. In contrast, a chelator with 4 hydroxamates that would form an octa-coordinated complex with this cation would reasonably be expected to be more appropriate for this application.
The medium sized and highly charged Zr4+ (ionic radius = 0.84 Å)6 is a hard cation exhibiting a strong affinity for anionic oxygen donors. Particularly, hydroxamates have been known as good ligands for this cation for decades.7 However, there is a dearth of data concerning the complexation chemistry of Zr4+ with hydroxamates from the structural and thermodynamic point of view. To our knowledge, no X-ray structures have been reported and stability constants of the previously investigated complexes are either incomplete or unknown.
In this study, we aim to provide new data regarding this specific type of complex that should assist in the rational design of hydroxamate-based chelating agents for 89Zr4+. We initiated this work with 2 simple hydroxamic acids: acetohydroxamic acid (AHA) and N-methyl acetohydroxamic acid (Me-AHA) (Fig. 1). The presence of a methyl at the carbonyl and nitrogen positions makes Me-AHA the simplest and the most electronically similar ligand to binding sites provided by DFB. The desired complexes were obtained by ligand exchange from Zr(IV) acetylacetonate in the presence of excess of hydroxamic acid (4.1 eq.) in refluxing methanol. After isolation, crystallization of Zr(Me-AHA)4 was successful from dichloromethane/nitrobenzene (5/1), allowing for the determination of the crystal structure by X-ray diffraction.
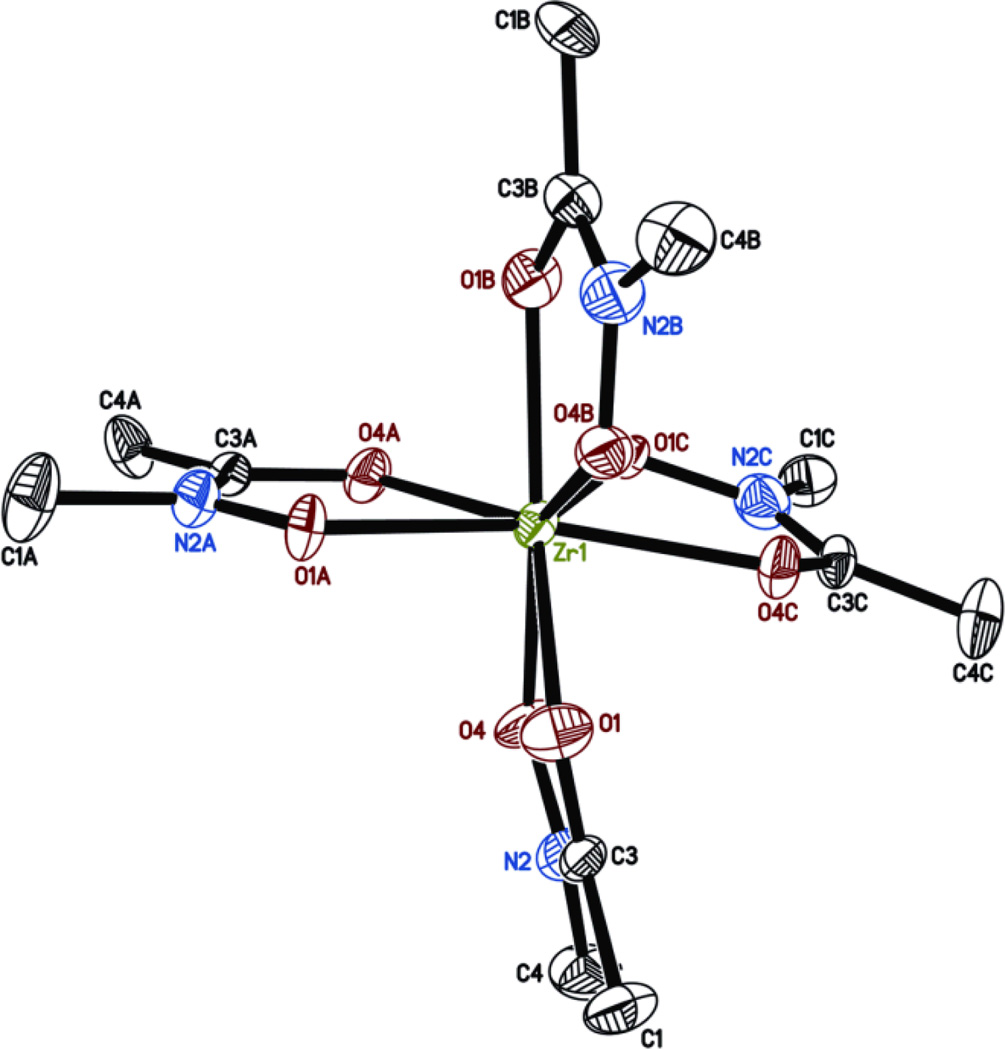
Single-crystal x-ray diffraction data were collected at 150 K using MoKα radiation and a Brüker APEX 2 CCD area detector. The final anisotropic full matrix least-squares refinement on F2 with 363 variables converged at R1 = 3.48% for the observed data. To our knowledge, this is the first time that the X-ray structure of a Zr complex with this class of ligands is reported. The X-ray structure shows that the metal is octa-coordinated by 4 hydroxamates via their oxygen atoms (Fig.2). The coordination by each pair of oxygen exhibits a relatively high symmetry with Zr-O distances of about 2.2 Å. However, analysis of the X-ray data suggests that the ligands are disordered such that the N and C atoms may swap positions. As a result, the averaged structure obtained exhibits some non-ideal bond lengths (e.g. N-CH3 (1.485 Å) > C-CH3 (1.464 Å) in fragment A). Attempts to resolve this disorder by refining the occupancy of the N and C atoms with the methyl substituent did not provide noticeable improvement, probably due to a disorder affecting actually all the ligands.
Figure 2.
X-ray structure of Zr(Me-AHA)4 obtained from the 0.572 × 0.258 × 0.097 mm3 crystal. Displacement ellipsoids are shown at the 50% level, hydrogen atoms and water molecules have been omitted for clarity. Space group Cc, unit cell dimensions: a = 17.8295(12), b = 14.3111(14), c = 13.2223(8) Å, and β = 129.061(2)°. Selected bond lengths [Å] and angles [°]: Zr-O1 2.233(4), Zr-O4 2.196(3), Zr-O1A 2.215(3), Zr-O4A 2.172(5), Zr-O1B 2.189(4), Zr-O4B 2.163(5), Zr-O1C 2.178(4), Zr-O4C 2.199(3), O1-Zr-O4 69.6(1), O1A-Zr-O4A 70.4(1), O1B-Zr-O4B 68.4(1), O1C-Zr-O4C 69.3(1).‡
Consequently, in order to refine the bond lengths, and also to better understand the bonding nature of Zr4+ with the hydroxamate ligands in solution, quantum chemical calculations at the B3LYP/DGDZVP level8 in the reaction field of water were carried out from this X-ray structure. The resulting calculated complex closely resembles the X-ray structure with a heavy atom root mean square deviation of 0.28 Å (Fig. S1 in ESI†). Whereas the calculated average bond distance of the Zr–ON (2.199 Å) is shorter than the Zr–OC (2.324 Å), these two respective average distances are very comparable in the X-ray structure (2.183 and 2.204 Å). In a similar hydroxamate-based structure of tetrakis(N-benzoyl-N-phenylhydroxylaminato)hafnium(IV) or Hf(NBPHA)4 reported by Tranqui et al,9 the average distance of Hf–ON (2.116 Å) is shorter than Hf–OC (2.258 Å). This strongly suggests that the Zr–ON bond distance is shorter than Zr–OC as observed in the calculated structure. Also, the discordance between the C-CH3 and the N-CH3 bond lengths observed for the X-ray structure was resolved in the calculated structure (1.509 Å and 1.454 Å for all C-CH3 and N-CH3, respectively). Another quantum chemical calculation after replacement of Hf by Zr and NBPHA by Me-AHA in the X-ray structure of Hf(NBPHA)4 resulted in a conformer 0.7 kcal/mol less stable than the one calculated from the X-ray structure (Fig. S2†), suggesting the probable co-existence of these two conformers of Zr(Me-AHA)4 in solution. Both calculated coordinates are given in Tables S1 and S2†.
The NMR study of both Zr(Me-AHA)4 and free ligand Me-AHA was also carried out in order to probe their dynamics in solution. The experimental 1H and 13C NMR spectra of the free ligand (Fig. S4†) show the presence of Z and E rotamers of Me-AHA at room temperature due to the hindered rotation with respect to the C–N amide bond. Quantum chemical calculation at the B3LYP/6-31G* level in the reaction field of CHCl3 indicated that the Z form is more stable than E by 0.4 kcal/mol (Structures and coordinates in Fig. S3, Tables S3 and S4†). This was confirmed by the presence of a slight excess of the Z form in the experimental 1H spectrum, with a ratio of 52/48 for the integration of the N-CH3 split signal. Based on the calculated energetics together with the experimental NMR data, both rotamers were fully characterized. For example, the N-methyl carbon of the E form syn to oxygen (δ 36.14) is better shielded than the anti carbon of the Z form (δ 36.89). This assignment is consistent with the previous 13C NMR studies, which have shown that alkyl carbon atoms syn to amide oxygen are better shielded than the corresponding anti carbons.10 In line with these 13C assignments, the two methyl peaks at δ 3.24 and 3.37 are assigned to the E and Z of the N-CH3 respectively (Tables S4 and S5†). In contrast, all the 1H and 13C doublets associated with the Z and E rotamers of Me-AHA appear as a singlet in Zr(Me-AHA)4 since only the Z rotamer can form a bidentate complex with Zr4+ (Fig. S5†). This demonstrates the formation of Zr(Me-AHA)4 in solution that is consistent with the present X-ray structure. Additionally, these single peaks also strongly suggest that the two calculated conformers of Zr(Me-AHA)4 derived from the X-ray structures, undergo rapid inter-conversion at room temperature that cannot be detected at the NMR time scale. As a result, the 4 methyl groups appear as equivalent.
To further explore the Zr affinity for hydroxamates, we assessed the complexation by potentiometric titrations in aqueous solutions (0.10 M ionic strength in KNO3 at 25°C). At first, the acid-base properties of AHA and Me-AHA were investigated. The calculated protonation constants, pKa = 9.33 and 8.75 obtained respectively for AHA and Me-AHA were in good agreement with the literature data.11 Next, we turned our attention to the complexation of Zr(IV). Based on the structure of the complex determined in the X-ray study, we performed potentiometric titrations for a 1:4 (M:L) adduct. Thus, metal ratio of approximately 0.25 equivalent of Zr(IV) (relative to the ligand amount) were used during complexation titrations, and the titration curves obtained were used jointly to refine the complete model of stability constants for all the complex species. However, different potential adducts from 1:4 to 1:1 metal-to-ligand complexation modes and the corresponding complex species have been considered. The data used for Zr hydroxides were taken from the literature and especially from the potentiometric study reported by Veyland et al.12 For both ligands, species of 1:4 stoichiometry were predominant in equilibrium on the studied pH scale (pH = 2–12). A selection of calculated overall (log β) and stepwise (log K) stability constants is available in Table 1. These values are in good agreement with the literature data for Zr(IV) hydroxamates complexes investigated by other experimental methodologies,7,13 and are also in the same order of magnitude than those observed with Fe3+, known to form strong complexes with hydroxamic acids in various natural biological processes.11 Speciation diagrams calculated from the complexation constants are available in ESI (Fig. S6†). Interestingly, log K values decrease in the order ZrL > ZrL2 > ZrL3, to finally reach a maximum for the ZrL4 species. Overall, these results illustrate the preference of these ligands to form 1:4 complexes with adequate metal concentrations and the stronger affinity of Me-AHA for Zr(IV) with higher log β values compared to AHA.
Table 1.
Overall and stepwise stability constants of the Zr(IV) complexes of AHA and Me-AHA (25.0 °C, I = 0.1 M in KNO3).
| AHA | Me-AHA | |||
|---|---|---|---|---|
| Speciesa | Log β | Log Kb | Log β | Log Kb |
| ZrL | 12.01 | 12.01(3) | 13.21 | 13.21(2) |
| ZrL2 | 24.00 | 11.99(2) | 25.22 | 12.01(2) |
| ZrL3 | 29.69 | 5.69(5) | 28.66 | 3.44(5) |
| ZrL4 | 45.07 | 15.38(3) | 45.98 | 17.32(1) |
Charges are omitted for clarity.
Values in parentheses are standard deviations in the last significant digit.
The superiority of Me-AHA was confirmed by a complexation study with the radionuclide 89Zr. AHA and Me-AHA were incubated for 30 min at 50 °C with a 89Zr oxalate solution (3.33 to 3.7 MBq) at pHs varying from 4 to 10. The resulting mixtures were analyzed by ITLC-SG using a 50 mM EDTA solution as eluant. DFB was also included in the study for comparison. It was by far the best ligand with > 99 % of the activity complexed (activity remaining at the bottom of the TLC) for pHs ranging from 5 to 9, due to the pre-organization of the hydroxamates, while the best results with the single hydroxamates were obtained at pH 7, with about 20 % and only 3.5 % activity bound to Me-AHA and AHA, respectively. The rest of the activity was spread along the TLC up to the top, and corresponded to either uncomplexed 89Zr, or progressively transchelated 89Zr from the weaker hydroxamate ligands to the stronger chelator EDTA during the elution (Fig S7†). Even though it was not possible to determine the radiochemical yield of complex actually formed, this method of analysis proved Me-AHA to be better than AHA as observed in the potentiometry study described above. The higher localized charge on the oxygen atom at the nitrogen position provided by the methyl group in Me-AHA is probably the main reason of a stronger affinity for the metal, similarly to what has been reported for Fe(III) with this class of ligands.14
In conclusion, the X-ray and potentiometric data presented in this study open new perspectives for the synthesis of chelators for 89Zr. First, the fact that Zr(IV) forms octa-coordinated complexes with the single hydroxamates strongly supports the hypothesis of the accessibility to a chelator better than DFB, if appropriately assembled from 4 of these subunit ligands. Secondly, it was observed that the coordination geometry of Zr(IV) with hydroxamate can be investigated at the B3LYP/DGDZVP level. This will allow for the design of hydroxamates-bearing chelators with an adequate pre-organization for the Zr(IV) cation. Lastly, the ability to measure stability constants of the investigated complexes by potentiometry as reported here for the first time allows for the rationalization of the stabilities observed for different ligands, and also provides a method for the exploration of alternative ligands to hydroxamates (i.e. catecholates and hydroxypyridinones) for the improved complexation of 89Zr. Research through the different axes of study opened here to improve the stability of the antibody radiolabeling with 89Zr is warranted.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and the Center for Information Technology. The quantum chemical study utilized PC/LINUX clusters at the Center for Molecular Modeling of the NIH (http://cit.nih.gov). X-ray crystallographic studies were supported by NIDA through Interagency Agreement #Y1-DA1101 with the Naval Research Laboratory (NRL).
Footnotes
† Electronic Supplementary Information (ESI) available: Details of syntheses, NMR study, crystallographic study, quantum chemical calculation, potentiometry and radiochemistry as well as crystallographic data in CIF format. See DOI: 10.1039/b000000x/
Atomic coordinates for Zr(Me-AHA)4 have been deposited at the Cambridge Crystallographic Data Centre (deposition number 902586).
Notes and references
- 1.Zhang Y, Hong H, Cai W. Current Radiopharm. 2011;4:131. doi: 10.2174/1874471011104020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chacko A-M, Divgi CR. Med. Chem. 2011;7:395–412. doi: 10.2174/157340611796799221. [DOI] [PubMed] [Google Scholar]
- 3.Perk LR, Vosjan MJWD, Visser GWM, Budde M, Jurek P, Kiefer GE, van Dongen GAMS. Eur. J. Nucl. Med. Mol. Imaging. 2010;37:250. doi: 10.1007/s00259-009-1263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nayak TK, Garmestani K, Milenic DE, Brechbiel MW. J. Nucl. Med. 2012;53:113. doi: 10.2967/jnumed.111.094169. [DOI] [PMC free article] [PubMed] [Google Scholar]; van Rij CM, Sharkey RM, Goldenberg DM, Frielink C, Molkenboer JDM, Franssen GM, van Weerden WM, Oyen WJG, Boerman OC. J. Nucl. Med. 2011;52:1601. doi: 10.2967/jnumed.110.086520. [DOI] [PubMed] [Google Scholar]; Holland JP, Divilov V, Bander NH, Smith-Jones PM, Larson SM, Lewis JS. J. Nucl. Med. 2010;51:1293. doi: 10.2967/jnumed.110.076174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verel I, Visser GWM, Boellaard R, Walsum MS, Snow GB, van Dongen GAMS. J. Nucl. Med. 2003;44:1271–1281. [PubMed] [Google Scholar]; Tinianow JN, Gill HS, Ogasawara A, Flores JE, Vanderbilt AN, Luis E, Vandlen R, Darwish M, Junutula JR, Williams S-P, Marik J. Nucl. Med. Biol. 2010;37:289–297. doi: 10.1016/j.nucmedbio.2009.11.010. [DOI] [PubMed] [Google Scholar]; Zeglis BM, Mohindra P, Weissmann GI, Divilov V, Hilderbrand SA, Weissleder R, Lewis JS. Bioconjugate Chem. 2011;22:2048–2059. doi: 10.1021/bc200288d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shannon RD. Acta Crystallogr. A. 1976;32:751–767. [Google Scholar]
- 7.Baroncelli F, Grossi G. J. Inorg. Nucl. Chem. 1965;27:1085–1092. [Google Scholar]
- 8.Frisch MJ, et al. Gaussian 09, revision A.02. Wallingford CT: Gaussian Inc.; 2009. [Google Scholar]
- 9.Tranqui D, Laugier J, Boyer P, Vulliet P. Acta Crystallogr. B. 1978;34:767–773. [Google Scholar]
- 10.Lee Y-S, Siméon FG, Briard E, Pike VW. ACS Chem. Neurosci. 2012;3:325–335. doi: 10.1021/cn3000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Codd R. Coord. Chem. Rev. 2008;252:1387–1408. [Google Scholar]
- 12.Veyland A, Dupont L, Pierrard J, Rimbault J, Aplincourt M. Eur. J. Inorg. Chem. 1998;1998:1765–1770. [Google Scholar]
- 13.Matteson BS, Tkac P, Paulenova A. Separ. Sci. Technol. 2010;45:1733–1742. [Google Scholar]
- 14.Brink CP, Crumbliss AL. Inorg. Chem. 1984;23:4708–4718. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.