Abstract
Nuclear factor erythroid 2-related factor 2 (Nrf2, NM 006164, 605 AA) is essential for the antioxidant responsive element (ARE)-mediated expression of a group of detoxifying antioxidant genes that detoxify carcinogens and protect against oxidative stress. Several proteins have been identified as Nrf2-interacting molecules. In this study, we found that the overexpression of RAC3/AIB-1/SRC-3, a nuclear co-regulator and oncogene frequently amplified in human breast cancers, induced heme oxygenase-1 (HO-1) through Nrf2 transactivation in HeLa cells. Next, we determined the interaction between RAC3 and Nrf2 proteins using a co-immunoprecipitation assay (co-IP) and fluorescence resonance energy transfer (FRET) analysis. The results showed that RAC3 bound directly to the Nrf2 protein in the nucleus. Subsequently, we identified the interacting domains of Nrf2 and RAC3 using a GST pull-down assay. The results showed that both the N-terminal RAC3-pasB and C-terminal RAC3-R3B3 domains were tightly bound to the Neh4 and Neh5 transactivation domains. Furthermore, chromatin immunoprecipitation (ChIP) showed that RAC3 bound tightly to the ARE enhancer region of the HO-1 promoter via Nrf2 binding. These data suggest that Nrf2 activation is modulated and directly controlled through interactions with the RAC3 protein in HeLa cells.
Keywords: Nrf2, RAC3/SRC-3, HO-1
Introduction
Mammalian cells have developed efficient machinery to maintain the redox balance against the extracellular environment and/or intracellular oxidative electrophilic stresses. Defense mechanisms against such oxidative stress include the expression of detoxifying enzymes, including phase II drug-metabolizing detoxifying enzymes, such as glutathione S-transferase (GST), NAD(P)H:quinone oxidoreductase 1 (NQO1), and UDP-glucuronosyltransferase (UGT) (Chan et al., 2001; Morse & Stoner, 1993; Zhang et al., 1992); antioxidant enzymes, such as HO-1 (Alam et al., 1999) and γ-glutamylcystein synthetase (γGCS) (McMahon et al., 2001); and multidrug transporters, such as multidrug-resistance-associated protein 1 or 2 (MRP1/2) (Hayashi et al., 2003; Vollrath et al., 2006).
These genes are generally controlled and regulated by nuclear factor erythroid 2-related factor 2 (Nrf2) (Lee et al., 2003; McMahon et al., 2001; Vollrath et al., 2006), a transcription factor belonging to the cap’n’collar (Cnc) family. Nrf2 possesses a highly conserved basic leucine zipper (bZIP) region (Kobayashi & Yamamoto, 2005) with a cis-acting enhancer element called an antioxidant-responsive element/electrophile-response element (ARE/EpRE, 5′-(A/G)TGACNNNGC (A/G)-3′) at the 5′-flanking region (Kong et al., 2001; Li & Jaiswal, 1993; Rushmore & Pickett, 1990).
Under normal conditions, Nrf2 forms a dimer with Kelch-like ECH-associated protein 1 (Keap1), an inhibitory protein of Nrf2, in the cytoplasm. However, under oxidative stress conditions, the Nrf2-Keap1 complex is disrupted, allowing Nrf2 to translocate to the nucleus and transactivate detoxifying genes through binding to the ARE enhancer. Nrf2 consists of several functional domains (Neh1 to Neh6), and the cysteine-rich Keap1 interacts with the Neh2 domain (Kobayashi et al., 2002; Motohashi & Yamamoto, 2004).
A pivotal role for Nrf2 in managing oxidative or electrophilic stress has been shown in studies using the Nrf2-knockout mouse. In the absence of the Nrf2 gene, the induction of detoxifying enzymes against oxidative stress is dramatically abolished, and Nrf2-knockout mice are more sensitive to oxidative stress-induced lung injury and more susceptible to carcinogens (Chan & Kan, 1999; Enomoto et al., 2001; Lee et al., 2004).
Several mechanisms regarding Nrf2 transactivation have been reported. Nrf2 transactivation activity can be triggered by kinases, such as the mitogen-activated protein kinases (MAPKs) (Nguyen et al., 2003), protein kinase C (PKC) (Huang et al., 2002), and phosphoinositide-3-kinase (PI3K) (Nakaso et al., 2003). In addition, among the Maf basic leucine zipper (bZIP) transcription regulators, small Mafs (MafF, MafK, and MafG) play an important role in Nrf2 transactivation. These proteins contain leucine zipper motifs, which allow for heterodimerization with Nrf2 and binding to the ARE DNA sequence region (Blank, 2008). CREB-binding protein (CBP), a nuclear coactivator, has been implicated in Nrf2 signaling through direct binding to the two transactivation domains (Neh4 and Neh5) of Nrf2 (Katoh et al., 2001). Furthermore, a member of the p160 protein cofactor family, ARE-binding protein-1 (ARE-BP-1), has previously been speculated to be an Nrf2/ARE-binding protein (Zhu & Fahl, 2001).
Receptor-associated coactivator 3 (RAC3), also called steroid receptor coactivator (SRC-3/ACTR/AIB-1/pCIP/TRAM-l), is a member of the p160 family, which includes SRC-1 (NCoA-1) and SRC-2 (TIF2/GRIP1/NCoA2), that plays a pivotal role in cell growth (Wang et al., 2000; Zhou et al., 2003) and mammary gland development (Xu et al., 2000). This coactivator family interacts with not only steroid hormone receptors, such as estrogen receptor (ER), progesterone receptor, and thyroid receptor (Han et al., 2006; Takeshita et al., 1997; Ying et al., 2005) but also several transcription factors, including activator protein-1 (AP-1), nuclear factor-kappa B (NF-κB), and signal transducer and activator of transcription (STAT) (Arimura et al., 2004; Lee et al., 1998; Werbajh et al., 2000). Furthermore, Nrf2 recruits other nuclear co-regulators and chromatin modification factors, such as CREB-binding protein (CBP/p300), p300/CBP-associated factor (p/CAF), protein arginine methyltransferase 1 (PRMT1), and coactivator-associated arginine methyltransferase 1 (CARM1), which initiates the transcription of target genes (Chen et al., 1999; Chen et al., 1997; Koh et al., 2001; McKenna et al., 1999; Yao et al., 1996).
RAC3 shares common structural domains like other family members: the N-terminal basic helix-loop-helix (bHLH) domain; the Per/ARNT/Sim (Pas) domain; the transcriptional activation domains for interaction with CBP/p300, PRMT1, and CARM1; the LXXLL motifs (where L is leucine and X is any amino acid); and the polyglutamate (poly Q) domain. In general, the bHLH domain is considered to be involved in interactions with many transcription factors, whereas the Pas domains (PasA and PasB) are involved in potential protein–protein interactions (Leo & Chen, 2000).
RAC3 was first identified as a gene that was highly expressed and amplified in many breast and ovarian cancer cell lines (Anzick et al., 1997). Recent data suggest that RAC3 can play an important role in cell growth in breast and prostate cancer cell lines (Yan et al., 2006; Zhou et al., 2003). Furthermore, in the transgenic mouse model, overexpressed RAC3 caused mammary hypertrophy, hyperplasia, and abnormal post-weaning involution and showed high malignant tumor incidence, potentially through the stimulation of the PI3K/AKT/mTOR signaling pathway (Torres-Arzayus et al., 2004). In contrast, RAC3-knockout mice showed a lower incidence of ras-related mammary gland tumorigenesis (Kuang et al., 2004) and exhibited growth retardation and an abnormal adult body size (Wang et al., 2000; Xu et al., 2000). Based on these reports, unlike other p160 coactivators, RAC3 may potentially play a role in cell growth and tumorigenesis.
Previously, Zhu and Fahl (2001) proposed that ARE-binding protein-1 was a member of the p160 family, but the identity of this molecule was still unknown. However, based on recent data from our lab, we suggested that RAC3 might be the p160 family member that directly interacted with Nrf2 because Gal4-luciferase activity was induced by co-transfection with Nrf2 (1-370) and RAC3 in HepG2 cells. However, it is still not clear how RAC3 modulates Nrf2 signaling because the Gal4-luciferase reporter system merely informs about the binding interaction between two molecules, and the functional domains of the two proteins have not verified (Lin et al., 2006). If RAC3 interacts with and regulates Nrf2 signaling, it would be interesting to determine how oncogenic RAC3 can regulate the cytoprotective Nrf2 protein. Therefore, we investigated the role of RAC3 in Nrf2 signaling and investigated the functional domains of Nrf2 and RAC3 in HeLa cells.
Results
The expression of heme oxygenase-1 (HO-1) was increased by RAC3
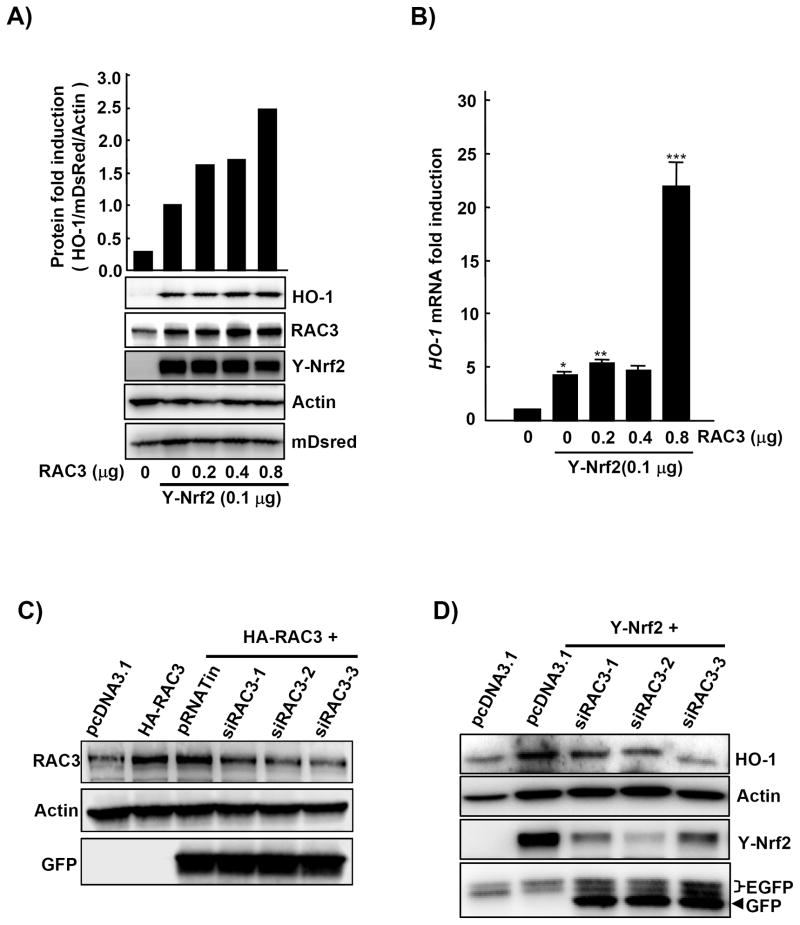
To determine the effects of RAC3 on the expression of HO-1, an Nrf2 target protein, HeLa cells were transfected with Nrf2 and various concentrations of HA-RAC3 plasmids. First, we measured the protein and mRNA levels of HO-1. The results showed that transfection with HA-RAC3 increased the HO-1 protein (Fig. 1A) and mRNA levels (Fig. 1B) in a dose-dependent manner in the presence of a YFP-Nrf2 construct.
Fig. 1. HO-1 protein and mRNA were induced by transient transfection with Nrf2 and RAC3 and decreased by siRAC3.
A) HeLa cells were transfected with YFP-Nrf2 and HA-RAC3 constructs for 24 h. The construct concentrations are indicated in the materials and methods. The cell lysates (20 μg) were subjected to western blotting to determine HO-1 protein levels. pcDNA3.1 was used to confirm equal transfections. To determine the transfection efficiency, pDsredmono (10 ng) was transfected into each well. The relative fold induction of HO-1 was analyzed using densitometry analysis (upper bar). B) To determine the effect of RAC3 on HO-1 mRNA expression, cDNA from (A) was subjected to qPCR analysis. C) For RAC3 silencing, HeLa cells in 6-well plates were transfected with three different siRAC3 constructs (2 μg; siRAC3-I, -II, and -III) and HA-RAC3 (1 μg) constructs for 24 h. The cells were lysed with RIPA buffer and subjected to western blot analysis. To determine equal plasmid transfection, GFP expression from pRNATin was measured. D) To determine the effects of the siRAC3 constructs on HO-1 expression, HeLa cells were transfected with YFP-Nrf2 (0.8 μg) and three different siRAC3s (0.7 μg) for 24 h. The EGFP protein from the pEGFP empty vector (0.2 μg) was used to confirm equal transfection. GFP expressed by the siRAC3 construct was also used to confirm equal transfection among the siRAC3 constructs. pcDNA3.1 was used to maintain equal transfections for the transfection study. *, p<0.001; **, p<0.02; ***, p<0.0001
To support these results, we initially used three different siRAC3 constructs to silence the expression of the RAC3 protein. Western blot analysis indicated that of the three siRAC3s, siRAC3-III strongly decreased the expression level of RAC3. To confirm equal amounts of siRAC3 transfection, GFP (constitutively expressed from the vector) was measured using an anti-GFP antibody (Fig. 1C). Next, the different siRAC3 constructs were transfected into the HeLa cells in the presence of a YFP-Nrf2 construct to determine the inhibition of HO-1 induction. As expected, both HO-1 and YFP-Nrf2 proteins were inhibited by siRAC3s (Fig. 1D).
Nrf2 directly binds RAC3
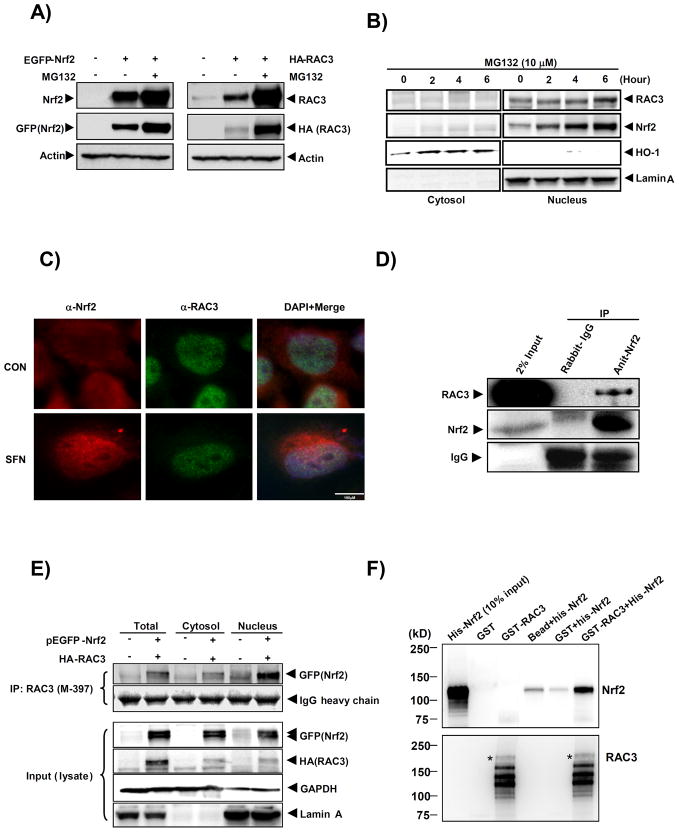
Before determining the binding interaction between Nrf2 and RAC3 proteins, the level of overexpressed EGFP-Nrf2 and HA-RAC3 proteins were first measured after transfection with pEGFP-Nrf2 and pSG5-RAC3, respectively, in HeLa cells. The cells were treated with a proteasome inhibitor (MG132) after transfection to prevent protein degradation and enhance the accumulation of the proteins. Western blot analysis showed that MG132 stabilized the overexpressed EGFP-Nrf2 and HA-RAC3 proteins, as detected using anti-Nrf2 (C-20) and anti-GFP (B-2) or anti-RAC3 (M397) and anti-HA (Zymed) antibodies for EGFP-Nrf2 and RAC3 proteins, respectively (Fig. 2A).
Fig. 2. Nrf2 binds directly to the RAC3 protein.
A) Prior to the co-IP study with Nrf2 and RAC3, the two constructs were verified by determining EGFP-Nrf2 and HA-RAC3 expression levels in HeLa cells. The cells were transiently transfected with GFP-Nrf2 (3 μg) or HA-RAC3 (3 μg) using jetPEI reagent (Polyplus-Transfection) for 24 h followed by treatment with MG132, a proteasome inhibitor, for 6 h. The protein samples (20 μg) were subjected to western blot analysis for Nrf2 and RAC3 using anti-GFP or anti-Nrf2 (C-20) and anti-HA or anti-RAC3 (M-397) antibodies. Actin expression was detected to confirm equal loading. B) To determine the subcellular localization of RAC3 and Nrf2 in HeLa cells, MG132 (10 μM) was administered for different times, and the cytosolic and nuclear fractions were isolated using M-PER buffer (Pierce). The fractionated samples (20 μg) were subjected to western blot analysis to measure endogenous protein levels using specific antibodies as indicated. Lamin A was used as the positive control for the nuclear fraction. C) The cellular co-localization of endogenous Nrf2 and RAC3 in HeLa cells was visualized using immunofluorescence microscopy after DL-sulforaphane (SFN, 20 μM) treatment for 16 h. Anti-Nrf2 (Epitomics, California, Burlingame, USA) and an Alexa Fluor 594 secondary antibody was used to visualize Nrf2. Anti-RAC3 (E-11) and an Alexa Fluor 488 secondary antibody was used to visualize RAC3. Magnification, 100X. D) To confirm the binding interaction between Nrf2 and RAC3, whole-cell lysates from MCF7 cells were subjected to the IP of endogenous Nrf2 using an anti-Nrf2 antibody followed by western blotting against endogenous RAC3 using an anti-RAC3 antibody. E) To determine whether Nrf2 could bind to the RAC3 protein, HeLa cells in 6-well plates were transfected with EGFP-Nrf2 (2 μg) and HA-RAC3 (2 μg) constructs for 24 h, and a co-IP assay was performed. A total of 200 μg of protein from the different fractions were immunoprecipitated using anti-RAC3 (M-397) and blotted for EGFP-Nrf2 using an anti-GFP antibody using western blot analysis. The co-IP method is described in the Materials and Methods. GAPDH and Lamin A were used as the controls for the cytosolic and nuclear fractions, respectively. The IgG heavy chain was used to confirm equal bead loading. F) To determine whether Nrf2 could directly bind to RAC3, purified His-Nrf2 and GST-RAC3 expressed in a bacterial expression system were co-incubated, and GST-RAC3 was pulled down using GSH beads in vitro. The protein-bead complexes were subjected to western blot analysis using an anti-Nrf2 (C-20) antibody. The detailed procedures are described in the Materials and Methods. Asterisks indicate the predicted size of the GST-RAC3 protein.
To examine the subcellular distribution of endogenous Nrf2, RAC3, and HO-1 proteins in HeLa cells, the cells were treated with MG132 (10 μM) for different times, and the cells were fractionated into the cytosolic and nuclear fractions. Western blot analysis showed that the majority of endogenous Nrf2 and RAC3 proteins was localized in the nucleus (Fig. 2B).
To visualize the localization of Nrf2 and RAC3 proteins, the cells were treated with sulforaphane. Figure 2C shows that the Nrf2 protein partially translocated into the nucleus and co-localized with the RAC3 protein after treatment with an Nrf2 activator, DL-sulforaphane.
To investigate whether Nrf2 bound to RAC3 in HeLa cells, the cells were transiently co-transfected with EGFP-Nrf2 and HA-RAC3 constructs. After co-transfection, whole-cell, cytosolic, and nuclear extracts were subjected to a co-IP assay using an anti-RAC3 (M-397) antibody followed by western blotting against EGFP-Nrf2 (using anti-GFP). The results indicated that GFP-Nrf2 and HA-RAC3 were tightly bound (Fig. 2E). In addition, we confirmed the endogenous binding interactions between Nrf2 and RAC3 using MCF7 cell lysates (Fig. 2D).
To provide additional evidence for the binding of these two proteins, an in vitro GST pull-down experiment was conducted using human GST-RAC3 (full length) and His-Nrf2 (full length) proteins, which were overexpressed in and purified from bacteria. The GST-RAC3 protein was first purified using glutathione (GSH) sepharose beads and then incubated with purified His-Nrf2. Then, GSH-protein-bead complexes were subjected to western blot analysis against His-Nrf2 using an anti-Nrf2 antibody (C-20). The results strongly suggested that the RAC3 protein could directly bind to Nrf2 (Fig. 2F).
Construction of GST-RAC3, His-Nrf2, and their fragments for a GST pull-down assay
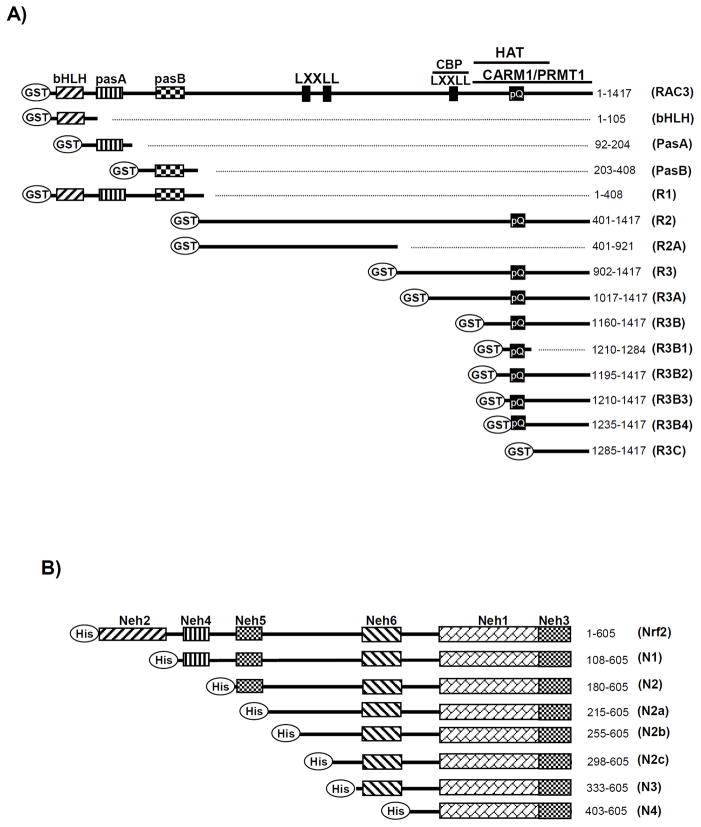
To identify the interacting domains of RAC3 and Nrf2 using a GST pull-down assay, differently sized cDNAs from RAC3 and Nrf2 were subcloned into pGEX2T/4T3 and pET28b(+), respectively (Fig. 3A and B).
Fig. 3. A schematic diagram showing the different structures of the fragmented domains of His-Nrf2 and GST-RAC3 used in the GST pull-down assay.
A) RAC3 and its fragments were subcloned into the bacterial expression vector pGEX2T or pGEX4T to introduce the N-terminal GST-tag. B) Nrf2 and its fragments were subcloned into the bacterial expression vector pET28b(+) to introduce the N-terminal His-tag. The different sizes of the segments are indicated in the figure based on the amino acid (aa) number from the full-length Nrf2 and RAC3. The Nrf2 and RAC3 domains have been previously reported (UniProt Q16236 and Q9Y6Q9).
N-terminal pasB and C-terminal RAC3 (R3B3) regions bind to Nrf2
In previous studies using the Nrf2/ARE luciferase assay, co-IP and GST pull-down assays, we concluded that Nrf2 activation could be modulated by direct binding to the RAC3 protein. We investigated the possible binding region of RAC3 to the Nrf2 protein. Differently sized purified GST-RAC3 polypeptides were incubated with His-Nrf2 protein followed by a GST pull-down assay. The constructs of the different GST-RAC3 fragments in the pGEX2T or pGEX4T vector are shown in Fig. 3A.
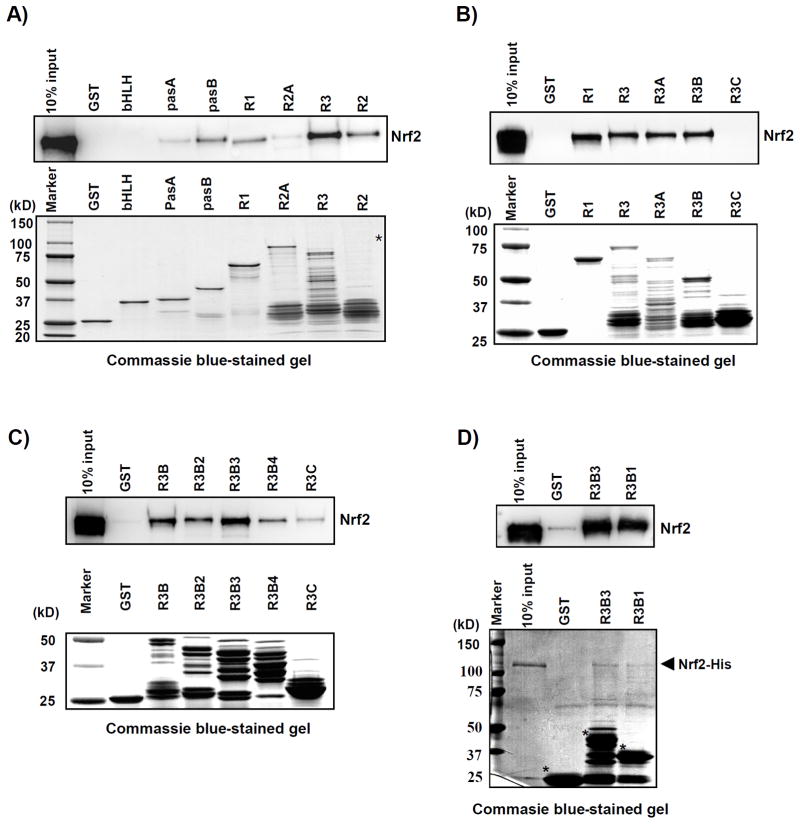
First, we conducted a GST pull-down assay by incubating His-Nrf2 and different GST-RAC3 fragments (bHLH, pasA, pasB, R1, R2, R2A, and R3; Fig. 3A). The samples were immunoblotted against Nrf2 using an anti-Nrf2 (C-20) antibody. The results indicated that His-Nrf2 strongly interacted with pasB and R3 of the GST-RAC3 protein (Fig. 4A). R1 and R2 also showed some interaction with Nrf2 (Fig. 4A).
Fig. 4. The RAC3 pasB and R3B3 domains bind to Nrf2.
A) The purified GST-RAC3 fragments were incubated with His-Nrf2 protein followed by a GST pull-down assay. The N-terminal fragments (bHLH, pasA, pasB, and R1), the central fragment (R2A), and the C-terminal fragments (R2 and R3) were incubated with His-Nrf2, followed by a GST pull-down assay. The protein-bead complex samples were analyzed using a western blot to detect His-Nrf2 using anti-Nrf2 (C-20). Of the GST-RAC3 segments, pasB and R3 bound strongly to the His-Nrf2 protein. B) To identify the Nrf2-binding domain in the R3 region of RAC3, N-terminal-deleted fragments of R3 (R3A, R3B, and R3C) were incubated with His-Nrf2, followed by a GST pull-down assay. Immunoblotting against His-Nr2 was the same as above. The R3B fragment of GST-RAC3 bound strongly to the His-Nrf2 protein. C) To identify the Nrf2-binding domain in the R3B region of RAC3 based on the previous results in (B), N-terminal-deleted fragments of R3 (R3B2, R3B3, and R3B4) were incubated with His-Nrf2, followed by a GST pull-down assay and western blotting against His-Nr2. The R3B3 segment of GST-RAC3 bound strongly to the His-Nrf2 protein. D) R3B3 and the C-terminal-deleted R3B1 fragment were incubated with His-Nrf2 to verify the binding region of RAC3. The GST pull-down assay was the same as in (A). Equal volumes of purified His-Nrf2 were applied to the reactions, and the same amount of the GST-RAC3 fragments in the reaction was analyzed using an SDS-PAGE gel, which was stained with Coomassie brilliant blue, as shown at the bottom of each experiment. The asterisk indicates the expected size of the proteins. Degraded GST-RAC3 fragments are also shown in Coomassie blue-stained gels.
To more specifically determine the regions of R3 that were involved in His-Nrf2 binding, R3 was dissected into three parts, R3A, R3B, and R3C (Fig. 3A), and used in a GST pull-down assay, as described above. The results showed that R3A and R3B interacted with His-Nrf2, whereas the short C-terminal protein (R3C) did not (Fig. 4B).
To investigate the binding of the R3B fragment to Nrf2, the N-terminus of R3B was deleted to construct R3B fragments: R3B1, R3B2, R3B3, and R3B4 (Fig. 3A). The GST pull-down assay showed that R3B3 bound strongly to His-Nrf2, whereas the binding of R3B4 to HisNrf2 was significantly reduced (Fig. 4C).
Next, we compared the ability of R3B1 and R3B3 to bind to His-Nrf2. The results showed that R3B1 had a lower binding affinity for His-Nrf2 compared to R3B3 (Fig. 4D). These results suggest that the pasB (92–204) and R3B3 (1210–1417) regions of the RAC3 protein play critical roles in Nrf2 activation.
The PasB and R3B3 regions of RAC3 bind to the N-terminal transactivation domain Neh5 of Nrf2
We determined the critical regions of RAC3 responsible for binding to the full-length Nrf2 protein. However, the region of Nrf2 that binds to the RAC3 protein has not been elucidated. We next investigated the Nrf2 region that binds to the RAC3 protein. Prior to the GST pull-down assay, various His-Nrf2 fragments (Fig. 3B) were subcloned into the pET28b(+) vector for bacterial expression.
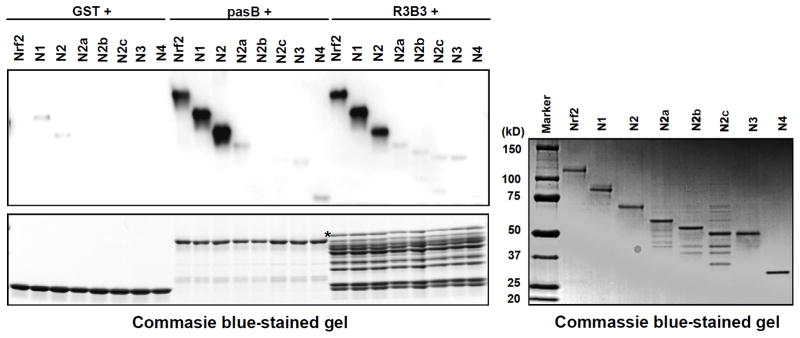
The purified His-Nrf2 protein and the differently sized fragments (N1, N2, N2-a, N2-b, N2-c, N3, and N4; Fig. 3B) were incubated with pasB or R3B3 fragments (from above) conjugated to GSH-beads. After incubation, the samples were immunoblotted against Nrf2 using anti-Nrf2 (C-20). Because the Nrf2 antibody (C-20) recognizes the C-terminal epitope of human Nrf2, all Nrf2 fragments were detected in the immunoblotting analysis. The results showed that both pasB and R3B3 of RAC3 bound strongly to N1 and N2 of Nrf2 but not to the other fragments (Fig. 5).
Fig. 5. The pasB and R3B3 regions of RAC3 bind strongly to N2 containing the Neh5 domain of Nrf2.
To identify the domain of Nrf2 that bound to the pasB and R3B3 regions of RAC3, GST-pasB and GST-R3B3 were incubated with His-Nrf2 and its fragments. The GST pull-down procedures were identical to the previous experiments. An equal amount of purified His-Nrf2 and its fragments were subjected to SDS-PAGE and stained with Coomassie brilliant blue, as shown in the right panel in the figure. In addition, the amount of GST, GST-pasA, and GST-R3B3 segments used in the pull-down assay was subjected to SDS-PAGE and stained with Coomassie brilliant blue, as shown in the bottom of the left panel. An asterisk indicates the expected size of the proteins.
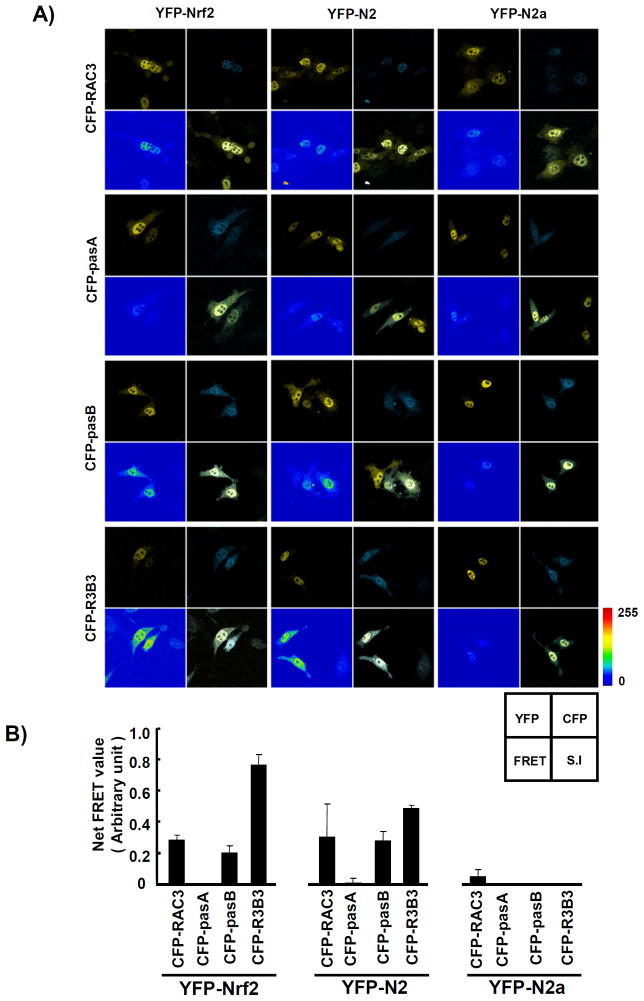
To corroborate the results from the GST pull-down study, we conducted a fluorescence resonance energy transfer (FRET) assay to verify the interactions between Nrf2 and RAC3. This assay was designed to identify molecules that were located within 10 nm of each other. Recently, cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) pairs have been used in FRET assays to investigate the functional interactions between two molecules. If, for example, a CFP-tagged protein is closely localized with or bound to a YFP-tagged protein, energy (457 nm) given to the CFP-tagged protein can transfer to the YFP-tagged protein, resulting in yellow fluorescence (545–600 nm). Based on this rationale, we constructed ECFP-RAC3 and its fragments (ECFP-pasA, ECFP-pasB, and ECFP-R3B3) and EYFP-Nrf2 and its fragments (EYFP-N2 and EYFP-N2a) and co-transfected them into HeLa cells in different combinations. The FRET results showed that the net FRET signal was strong in the pairs with ECFP-R3B3 and EYFP-Nrf2 or EYFP-N2 (Fig. 6A, B).
Fig. 6. Fluorescence resonance energy transfer (FRET) signals were strong between the pasB and R3B3 regions in RAC3 and N2 containing the Neh5 domain of Nrf2.
To analyze the potential interaction between Nrf2 and RAC3 proteins in HeLa cells, FRET signals were measured using the Zeiss LSM510 laser scanning confocal microscope (Zeiss, Thornwood, New York, USA). The cells plated in glass-bottom dishes were co-transfected with EYFP-Nrf2 or its fragments (EYFP-N2 and EYFP-N2a) and ECFP-RAC3 or its fragments (ECFP-pasB and ECFP-R3B3) in different combinations using jetPEI transfection reagent for 24 h. The procedures for the FRET assay are described in the Materials and Methods. The fluorescent channels are shown in the figure. The intensity of the FRET signal is indicated by a rainbow of colors. The net FRET values were measured using densitometry analysis of the three different FRET signals (bottom).
The RAC3/Nrf2 complex binds to ARE regions in heme oxygenase-1
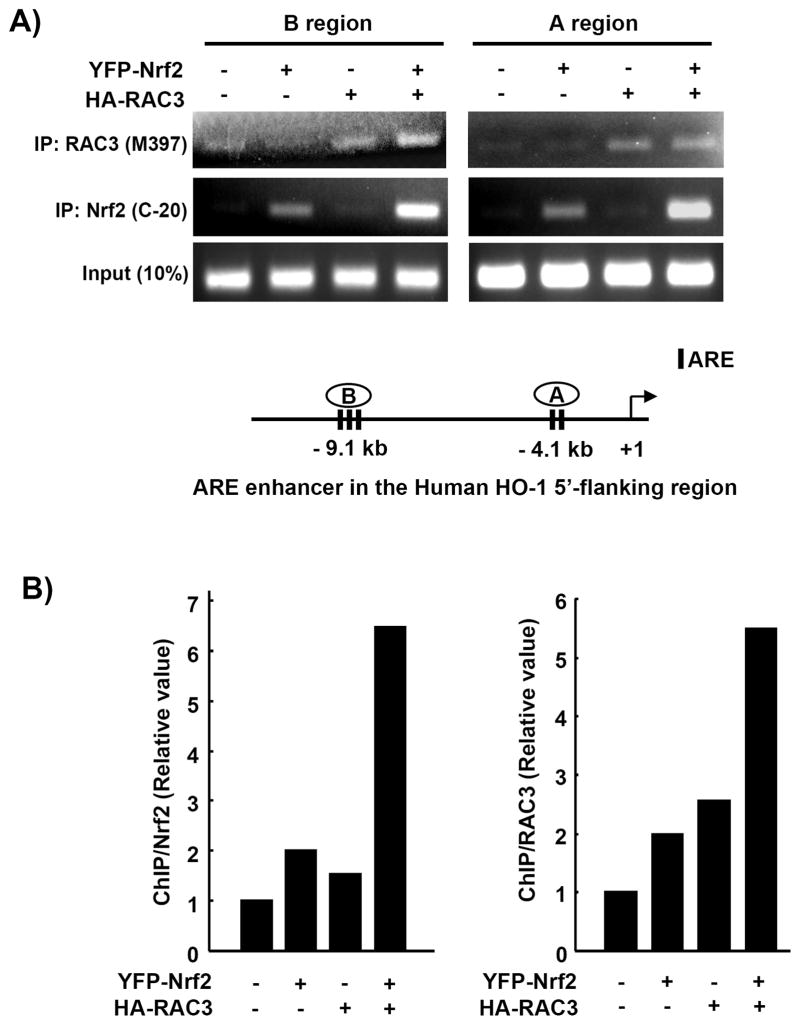
Based on the above results, we suggest that RAC3 can regulate Nrf2 signaling via direct binding of its N-terminal pasB domain and C-terminal R3B3 region to the Neh4 and/or Neh5 domain of Nrf2. Therefore, RAC3 may be detectable on the Nrf2-bound ARE DNA sequence. To test this hypothesis, we conducted a ChIP assay to elucidate whether the RAC3 protein was detectable on the ARE sequences bound to Nrf2. HeLa cells were transfected with the EYFP-Nrf2 and/or HA-RAC3 constructs, and the total cell lysates were subjected to a ChIP assay against the ARE enhancers in the HO-1 promoter.
As described previously, the HO-1 5′-flanking region contains ARE enhancer region A at −4.1 kb and region B at −9.1 kb. These two regions were amplified using PCR and qPCR after immunoprecipitation using anti-RAC3 (M-397) or anti-Nrf2 (C-20) antibodies. The PCR and qPCR results showed that both ARE regions were strongly amplified in the EYFP-Nrf2 plus HA-RAC3 transfection group from two different sets of antibody-treated groups (Fig. 7A, B). These results suggest that RAC3 can bind to Nrf2, which binds to the ARE sequence in the 5′-flanking region of human HO-1.
Fig. 7. RAC3 and Nrf2 bind to the ARE enhancers in the 5′-flanking region of the heme oxygenase-1 (HO-1) promoter.
A) To determine whether RAC3 could bind to Nrf2/ARE enhancers in HO-1, a ChIP assay was performed. HeLa cells were plated in 100-mm culture dishes and transfected with EYFP-Nrf2 (3.5 μg) and HA-RAC3 (3.5 μg) constructs using the jetPEI reagent for 22 h. The procedures for the ChIP assay are described in the Materials and Methods. The ChIP DNA samples using anti-Nrf2 (C-20) or anti-RAC3 (M-397) were subjected to PCR to amplify the two ARE regions [−4.1 kb (region A) and −9.1 kb (region B)] using specific primers. Non-IP samples were used as input controls. B) ARE region A was also quantified using qPCR.
Discussion
Nrf2 is a key transcription factor in the regulation of ARE-mediated gene expression in response to oxidative and electrophilic insults. Although numerous papers have been published on the mechanism of the Nrf2-mediated induction of phase II detoxifying enzymes and antioxidant enzymes, the exact mechanisms of Nrf2 signaling are still not clear.
Previously, several interacting molecules, such as Keap1, CBP/p300, and small Maf proteins (MafF, MafG, and MafK), were identified as potential regulators of Nrf2 signaling; however, it is possible that other unknown partners could interact with Nrf2 in the regulation of the antioxidant response. In this report, we demonstrated that the nuclear co-regulator RAC3, a member of the p160 cofactor family, interacted with Nrf2 through binding between two regions of RAC3 and the Neh5 domain of Nrf2 in HeLa cells.
From a previous report by Zhu and Fahl (2001), we assumed that ARE-binding protein-1 (ARE-BP-1) could be a p160 coactivator because a MafK antibody ablated the binding of ARE-binding protein-1 in a gel-shift assay. In addition, SRC-1 and SRC-2 antibodies have been used to ablate binding between ARE-BP-1 and the ARE sequence. However, it was shown that the antibodies had no effect on that binding. Indeed, it remains to be determined whether RAC3 is involved in binding to the ARE sequence.
A recent publication from our lab elucidated the interaction between RAC3 and Nrf2 using a Gal4-luciferase reporter system in HepG2 cells (Lin et al., 2006). The results revealed that the Gal4-Nrf2 (1-370) chimera containing two transactivation domains (TADs), Neh4 and Neh5, interacted with RAC3, resulting in the induction of luciferase activity. However, the enhancement in the transactivation activity of Nrf2 by RAC3 was not explained because the Gal4-luciferase system was designed to identify a binding partner that could be either a transcriptional activator or suppressor. To determine the functional role of RAC3 in Nrf2 signaling, it is necessary to verify the induction of a Nrf2-induced target protein, such as heme oxygenase-1. Therefore, we investigated whether RAC3 was a coactivator or corepressor of Nrf2 signaling.
First, we evaluated the role of RAC3 in the Nrf2 transactivation signal by examining Nrf2/ARE-luciferase activity. If this assay system was appropriate for the functional study of Nrf2 partners (coactivator or corepressor), the expression of the Nrf2-mediated target protein HO-1 would be consistent with results of the western blot and RT-PCR analysis. However, as shown in Fig. 1, the Nrf2/ARE luciferase assay results conflicted with the western blotting and RT-PCR results. We postulate that the short size of the ARE luciferase gene in a reporter vector may not be sufficient for the Nrf2-partner interaction study, and co-transfection (RAC3 and Nrf2 constructs) may not be a good tool because the overexpression of one gene can be hindered by the overexpression of the other gene. Therefore, we excluded the results of the Nrf2/ARE luciferase assay.
In this study, based on western blot analysis and RT-PCR, we showed that the overexpression of RAC3 increased the HO-1 protein and mRNA levels in HeLa cells. We also investigated the interaction between the Nrf2 and RAC3 proteins. Because endogenous RAC3 expression and stability is low in HeLa cells, we used an overexpression system by transfecting Nrf2 and RAC3 plasmids into HeLa cells. We then confirmed the interactions between endogenous Nrf2 and RAC3 using MCF7 cells, which overexpress RAC3.
Furthermore, we identified the possible interaction domains of the two molecules. From the GST pull-down assay shown in Fig. 4, the interaction domains between RAC3 and Nrf2 were somewhat surprising. The data suggest that R3B3 has a stronger binding affinity for Nrf2 than the pasB domain of RAC3 (Fig. 3A, B). Twenty amino acids, 1210MQPQQGFLNAQMVAQRSRELLS1231, might be indispensable for the binding of RAC3 to the Nrf2 protein because the removal of this segment caused a significant loss of binding ability to Nrf2 (compare R3B3 to R3B4; Fig. 3A and Fig. 4C). Figures 4 and 5 show that the N-terminal pasB and C-terminal R3B3 regions of RAC3 bind to both Nrf2-N1 and Nrf2-N2 but not to the Nrf2-N2a segments. The results suggest that the Neh5 domain of Nrf2 may be indispensable for RAC3 binding. However, the possibility of the Neh4 domain binding to RAC3 remains because N1 binds strongly to R3B3 than N2. To elucidate whether the Neh4 domain binds to both the pasB and R3B3 regions of RAC3, different mutational methods are needed. It is possible that RAC3 require both the Neh4 and Neh5 domains of Nrf2 for the modulation of signaling. Katoh et al. (2001) showed that both the Neh4 and Neh5 domains of Nrf2 could cooperatively bind to CBP, resulting in a synergistic increase in Nrf2 transactivation. In addition, Shen et al. (2004) showed that the Nrf2 transactivation domain containing Neh4 and Neh5 could bind to CBP, resulting in the activation of Nrf2, using the Gal4-luciferase system.
RAC3 shares conserved domains with other p160 cofactor family members (McKenna & O’Malley, 2002). The N-terminus contains a basic helix-loop-helix (bHLH) domain and Per/Arnt/Sim (pasA/B) domains for nuclear localization (Li et al., 2007) and protein–protein interactions (McKenna & O’Malley, 2002), respectively. The middle region contains an α-helical LXXLL motif that can bind to steroid receptors (Heery et al., 1997). The C-terminus contains HAT domains for acetyl-transferase activity, a CBP-binding domain and a CARMI/PRMT1 interaction domain (Leo & Chen, 2000), which also overlaps with the R3B3 region of RAC3 (Fig. 3A). Based on this information, it may be speculated that the pasB domain of RAC3 may bind to Nrf2 for recognition and dimerization, which is followed by R3B3 (which we named the Nrf2-binding domain, NBD) binding to Nrf2.
Numerous reports have shown that RAC3 is expressed in not only breast cancer cells (Kuang et al., 2004; Torres-Arzayus et al., 2004) but also other types of cancers, such as prostate cancer (Gnanapragasam et al., 2001), esophageal squamous cell carcinomas (Xu et al., 2007), pancreatic adenocarcinoma (Henke et al., 2004), hepatocellular carcinoma (Wang et al., 2002), colorectal carcinoma (Xie et al., 2005), and gastric cancer (Sakakura et al., 2000). Therefore, the oncogenic RAC3 protein may play an important role in tumorigenesis.
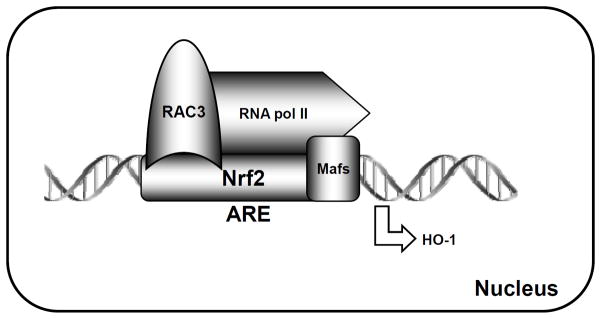
In summary, we show that RAC3 binds directly to Nrf2 in the nucleus to stimulate Nrf2 activity in HeLa cells (Fig. 8). Furthermore, we elucidated the interacting regions between Nrf2 and RAC3. The results indicate that the pasB and R3B3 regions of RAC3 interact with the Neh4 and Neh5 transactivation domains of Nrf2. Therefore, understanding RAC3 and Nrf2 molecular mechanisms may be useful for cancer or chemoprevention research.
Fig. 8.
Cartoon showing a possible interaction between Nrf2 and RAC3 in the ARE enhancer region of the HO-1 promoter (upper) through direct interactions between specific domains (lower).
Materials and Methods
Materials
Minimum essential medium (MEM), fetal bovine serum (FBS), and penicillin/streptomycin antibiotic mixtures were obtained from Invitrogen (Carlsbad, CA, USA). The anti-Nrf2 (C-20, H-300), anti-RAC3 (M-397), anti-GFP (B-2), anti-Dsred, anti-GAPDH, anti-lamin A, anti-actin, and secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, California, USA). Anti-HA was obtained from Zymed Laboratories (San Francisco, California, USA). ECL femto signal substrate and M-PER mammalian cell lysis buffer were obtained from Pierce Biotech Inc. The transfection reagent jetPEI was purchased from Polyplus-Transfection (Bioparc, Illkirch, France). The polypropylene columns were from Qiagen, and covered glass-bottomed culture dishes were purchased from MatTek Corporation (Ashland, Massachusetts, USA). The polyvinylidene difluoride (PVDF) membranes were from Millipore, and sulforaphane (SFN) and MG132 were purchased from Alexis (San Diego, California, USA). The protease inhibitor cocktail was obtained from Roche Molecular Biochemicals. The mammalian expression vectors pEGFP-C1, pDsredmono-C1, pEYFP-C1, and pECFP-C1 were obtained from Clontech. The pET28b(+) and pGEX4T3 vectors were from Novagen and GE Healthcare Life Sciences, respectively. The pRNATin-H1.2/Hygro vector for the siRAC3 constructs was purchased from Genscript (Piscataway, New Jersey, USA). All other chemicals were of analytical grade or the highest grade available.
Cell culture
HeLa and MCF7 cells (from ATCC) were maintained at 37°C in a humidified atmosphere of 5% CO2/95% air in MEM or RPMI 1640 medium, respectively, supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 50 U/ml of penicillin/streptomycin (Gibco BRL, Grand Island, New York). The cells were grown to 60–80% confluence and trypsinized with 0.05% trypsin containing 2 mM EDTA.
Construction of plasmids
Full-length wild-type human Nrf2 cDNA (NM 006164, 605 AA) was purchased from the I.M.A.G.E. consortium (Open Biosystems, Huntsville, Alabama, USA). To express Nrf2 in HeLa cells, Nrf2 cDNA was subcloned into the pEGFP-C1 vector (Clontech), resulting in EGFP-Nrf2. For GST pull-down studies, Nrf2 cDNA and its fragments were subcloned into the pET28b (+) vector (Novagen) for the expression of the His-Nrf2 fusion protein in BL21 Star™ (DE3) E. coli (Invitrogen). The wild-type Nrf2 and its fragments were amplified and subcloned into the vector using XhoI-BamHI digestion. The cloned plasmids were designated as follows: His-Nrf21-605, His-N1108-605, His-N2180-605, His-N2a215-605, His-N2b255-605, His-N2c298-605, His-N3333-605, and His-N4403-605. For FRET analysis, Nrf21-605, N1108-605, N2180-605, and N2a215-605 were subcloned into the pEYFP-C1 vector (Clontech).
For the RAC3 plasmids, pSG5-RAC3-HA, pGEX2T-bHLH, pGEX2T-pasA, pGEX2T-pasB and pGEX2T-R1 were used (Wu et al., 2001). To prepare different fragments of GST-RAC3 proteins, different fragments of RAC3 were amplified and sub-cloned into the pEGX2T (for full-length RAC3) and pGEX4T3 (for segmented RAC3) vectors using BamHI/BglII-EcoRI/MfeI and BamHI-XhoI digestion, respectively. The cloned plasmids were designated as follows: GST-RAC31-1417, GST-bHLH1-105, GST-PasA92-204, GST-PasB203-408, GST-R11-408, GST-R2401-1417, GST-R2A401-1417, GST-R3902-1417, GST-B3A1017-1417, GST-R3B1160-1417, GST-R3B11210-1284, GST-R3B21195-1417, GST-B3B31210-1417, GST-R3B41235-1417, and GST-R3C1285-1417.
For FRET analysis, CFP-RAC3 and its fragments were subcloned into the pECFP-C1 vector using KpnI-BamHI/BglII (for CFP-RAC3) or XhoI-BamHI (for CFP-pasA, CFP-pasB, and CFP-R3B3) digestion. Figure 3 shows each proposed protein designated as amino acid lengths.
For RAC3-silencing experiments, three different small interfering RNAs (siRNAs) were designed against RAC3 using Genscript siRNA Target Finder (Genscript, New Jersey, USA), and synthesized DNA duplexes were inserted into the pRNATin-H1.2/Hygro vector. The sequences of each siRAC3 construct were as follows: pRNATin-siRAC3-I, 5′-GAT CCC GTA TGT CTG TCC ATA TAA TCC TTT GAT ATC CGA GGA TTA TAT GGA CAG ACA TAT TTT TTC CAA A-3′; pRNATin-siRAC3-II, 5′-GAT CCC GTT TGT TAC AGG ATT TCG GAA GTT GAT ATC CGC TTC CGA AAT CCT GTA ACA AAT TTT TTC CAA A-3′; and pRNATin-siRAC3-III, 5′-GAT CCC GTC ATA GGT TCC ATT CTG CCG GTT GAT ATC CGC CGG CAG AAT GGA ACC TAT GAT TTT TTC CAA A-3′. The duplexes were directly ligated into the BamHI and XhoI restriction enzyme sites of the pRNAT-CMV3.2/neo/cGFP vector.
Fusion protein purification and the GST pull-down assay
For the purification of GST-RAC3 and its fragments, pGEX2T- or pGEX4T-based RAC3 constructs were transformed into BL21 Star™ (DE3) E. coli (Invitrogen) and cultured in 5 ml of liquid LB media containing ampicillin (50 μg/ml) overnight at 37°C in an orbital shaker. Then, 50 ml of LB media was added and incubated until the OD600 reached 0.6. These cultures were incubated for an additional 4 h after 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce GST-RAC3 proteins. The cultured bacteria were then centrifuged at 5,000×g for 3 min at 4°C. The pellets were suspended and sonicated in phosphate-buffered saline (PBS) containing 10 mM EDTA and protease inhibitor cocktail (Roche) on ice. After bacterial lysis, the lysates were centrifuged at 12,000 rpm for 5 min at 4°C. The supernatants were then applied to the polypropylene column (Qiagen), loaded with 1 ml of 50% slurry glutathione (GSH) sepharose beads (GE Healthcare Life Sciences) and incubated for 5 min at 4°C on a rotator. After incubation, the beads were washed 3 times with 20 volumes of PBS (containing 0.1% Triton X-100) in the cold room. After washing, the beads were suspended in 1 ml of incubation buffer (25 mM Tris-phosphate (pH 7.8), 2 mM DTT, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol and 1% Trion X-100) and stored at 4°C before the GST pull-down assay. The same volume of purified GST and GST-RAC3 proteins was subjected to SDS-PAGE and stained with Coomassie brilliant blue to verify the purity of proteins and to determine the bead volumes for GST pull-down assays.
For the purification of His-Nrf2 and its fragments, pET28b(+)-based Nrf2 constructs were transformed into BL21 Star™ (DE3) E. coli and cultured in 5 ml LB media containing 50 μg/ml kanamycin overnight at 37°C in an orbital shaker. Bacteria in 50 ml of LB media were incubated until the OD600 reached 0.6. IPTG was then added for 1 h to induce the six histidine (His6)-tagged fusion protein (His-Nrf2). The rest of the procedure was the same as described above. His-Nrf2 proteins were purified using a Ni-NTA agarose system according to the manufacturer’s instructions (Qiagen).
To study the binding interactions between RAC3 and Nrf2 proteins, purified GST-RAC3-beads and His-Nrf2 protein samples were mixed in 500 μl incubation buffer (25 mM Tris-phosphate (pH 7.8), 2 mM DTT, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol and 1% Trion X-100) and incubated overnight at 4°C. After incubation, the beads were vigorously washed 5 times with 10 volumes of washing buffer (PBST, PBS with 0.1% Tween 20, pH 7.2). The beads were then mixed with 200 μl of 2x SDS buffer, boiled for 5 min and subjected to SDS-PAGE and western blot analysis. To confirm equal protein loading, the same samples were subjected to SDS-PAGE, stained with Coomassie brilliant blue, and blotted with antibodies against the C-terminus or His-tag of Nrf2.
Western blot analyses
HeLa cells were plated in 100-mm culture dishes at ~2.0 × 106 cells/plate for 16 h prior to plasmid transfection. After transfection, the cells were scraped and lysed with RIPA lysis buffer (150 mM NaCl, 0.5% Triton X-100, 50 mM Tris-HCl, pH 7.4, 25 mM NaF, 20 mM EGTA, 1 mM DTT, 1 mM Na3VO4, and protease inhibitor cocktail) for 30 min on ice, followed by centrifugation at 14,800 × g for 15 min. The protein concentration of the supernatant was measured using the BCA reagent. The protein (20 μg) was electrophoresed on a 4–15% gradient tris-HCl gel (Biorad) and electrotransferred to a polyvinylidene difluoride (PVDF) membrane in tris-glycine buffer (pH 8.4) containing 20% methanol. The membrane was blocked with 5% fat-free dry milk in PBST buffer for 1 h. The membranes were probed with primary antibodies and horseradish peroxidase-conjugated secondary antibodies using standard western blotting procedures. The proteins were visualized using the femto signal chemiluminescent substrate (Pierce) using an image analyzer (Biorad).
Quantitative real-time PCR (qPCR) analysis
Complementary DNA (cDNA) from HeLa cells was synthesized from 1 μg total mRNA using the SuperScript II reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA). qPCR was performed using the Applied Biosystems Prism 7900HT Sequence Detection System. All reactions were performed in a 10 μl volume consisting of 25 ng cDNA, 300 nM of each primer, and 5 μl of SYBER Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) at 95°C for 10 min and 40 cycles of 95°C for 3 sec and 60°C for 30 sec. The primers used for the amplification were as follows: HO-1 (105 bp), 5′-GCC AGC AAC AAA GTG CAA G-3′ and 5′-GAG TGT AAG GAC CCA TCG GA-3′ and β-actin (500 bp), 5′-GCA CAG AGC CTC GCC TT-3′ and 5′-GTT GTC GAC GAC GAG CG-3′. The levels of cDNA were quantified using the comparative threshold cycle method with β-actin as the reference.
Transient transfections
To analyze the ARE luciferase activity, HeLa cells were plated in 6-well plates at ~4.0 × 105 cells/well and cultured for 16 h prior to transfection. The cells were transfected with pSG5-RAC3-HA, pEGFP-Nrf2, and pRNATin-siRAC3s constructs for 20–24 h. The amount of the plasmids used in the different experiments is indicated in the respective figures and legends. Transfection was performed according to the manufacturer’s instructions (Bioparc, Illkirch, France). In brief, plasmid mixtures in 100 μl of NaCl (150 mM) and jetPEI transfection reagent in 100 μl of NaCl (150 mM) were mixed and incubated for 25 min at room temperature. The ratio between plasmid and jetPEI was 1:4. After incubation, the cells were subjected to Western blotting or RT-PCR analysis.
Immunocytochemistry
HeLa cells were plated on cover slips and treated with DMSO (0.1%) or sulforaphane (20 mM) for 16 h. The cells were washed once with PBS and fixed with fresh 4% paraformaldehyde for 10 min. The cells were then washed 3 times with PBS containing 0.1% Triton X-100 (PBST) and incubated with 3% BSA-PBST blocking solution for 2 h. The cells were incubated with primary antibodies (1:100) in 3% BSA-PBST in a cold room overnight, washed with a PBST solution 3 times and incubated with secondary antibodies for 1 h. The cells were then washed 3 times with PBST and incubated with DAPI solution for nuclear staining. After mounting, cell images were obtained using florescence microscopy.
Co-immunoprecipitation (IP)
HeLa cells were plated in 100-mm dishes at ~2.0 × 106 cells/plate and cultured for 16 h prior to transfection. After cell stabilization, the cells were transfected with 2 μg of pEGFP-Nrf2 and/or 2 μg of pSG5-RAC3-HA for 24 h using jetPEI transfection reagent. The total amount of plasmids was adjusted using the pcDNA3.1 control vector. After transfection, the cells were lysed in M-PER mammalian lysis buffer (Pierce) containing protease inhibitors. M-PER lysis buffer was designed for whole-cell lysis of certain cell lines; however, this buffer only disrupts cellular membranes in HeLa cells (data not shown). Therefore, M-PER can be used to fractionate the cells into cytosolic and nuclear fractions, which was exploited in this study. In brief, the cells were first lysed to release the cellular membranes using M-PER lysis buffer for 10 min on ice. Then, the samples were centrifuged at 12,000 rpm for 1 min at 4°C to obtain the nuclear fraction. The pellets were gently washed once with the same buffer and then resuspended in the same buffer. The nuclear pellets in the buffer were then sonicated and centrifuged at 12,000 rpm for 10 min at 4°C. To obtain whole-cell lysates, the cells were sonicated in the M-PER buffer to suspend the proteins. The protein concentration of the fractionated samples was measured using the BCA reagent. A total of 200 μg of protein samples were precleared with 50 μl of TrueBlot™ anti-rabbit IgG IP beads (eBioscience, San Diego, California, USA) for 30 min. Then, the supernatants were incubated with anti-RAC3 (2 μg, M-397) for 1 h at 4°C followed by the pull-down step of adding TrueBlot™ anti-rabbit IgG IP beads to the samples for 1 h. After the reaction, the beads were washed 3 times with 500 ml of M-PER buffer at 4°C. Then, the beads were mixed with 200 μl of 2x SDS sample buffer and boiled for 5 min. The samples were subjected to western blot analysis and blotted against GFP-Nrf2 using an anti-GFP antibody. To pull down the His-tagged Nrf2 protein from HeLa cells, Ni-NTA agarose beads were used instead of Ig IP beads.
For co-IP using MCF7 cells, the cells were cultured in three 100-mm dishes. The cells were scraped and pooled, and whole cells were lysed with 1 ml of M-PERT (M-PER containing 0.2% Triton X-100). Each 400-ml fraction of cell lysate was incubated with rabbit IgG or anti-Nrf2 antibody for 2 h in a cold room. Then, 50 ml of protein A/G agarose beads was added and incubated for 1 h. The beads were washed with the same lysis buffer 3 times and subjected to western blotting.
Fluorescence resonance energy transfer (FRET) analysis
HeLa cells were cultured in glass-bottomed dishes (MatTek, Ashland, Massachusetts, USA) and co-transfected with pECFP-RAC3 or its fragments (pECFP-pasA, -pasB, or -R3B3) and pEYFP-Nrf2 or its fragments (pEYFP-N1, -N2, or -N2a). After 24 h, the cells were examined under a Zeiss LSM510 laser scanning confocal microscope (Zeiss, Thornwood, New York) with a 63x objective. We used a sensitized emission method for the FRET assay (Trinkle-Mulcahy et al., 2001). Three channels were used to detect the donor (ECFP, excitation at 457 nm and emission at 475–525 nm), acceptor (EYFP, excitation at 543 nm and emission at 475–525 nm), and FRET (excitation at 457 nm and emission at 545–600 nm) signals. The FRET signal was corrected by subtracting the crosstalk from each single ECFP or EYFP signal, according to Trinkle-Mulcahy’s method (Trinkle-Mulcahy et al., 2001). The net FRET was calculated according to the following formula:
In our system, α and β were measured as 20% and 21%, respectively. The data were analyzed using LSM510 SP2 software (Zeiss, version 3.2).
Chromatin immunoprecipitation (ChIP) Assay
HeLa cells were plated in 100-mm culture dishes at 50% confluence overnight. Then, the cells were transfected with pEYFP-Nrf2 (3.5 μg) and pSG5-RAC3-HA (3.5 μg) using jetPEI reagent for 22 h. pcDNA3.1 was used as a control. The transfection protocol was identical to the method described above for the ARE reporter assay. After transfection, the cells were washed twice with cold PBS and fixed with 10 ml of 1% formaldehyde for 10 min at room temperature. To halt cross-linking, 500 μl of glycine (1.25 M) was added to the plates for 5 min, and cells were washed briefly with cold PBS. The cells then were scraped in ice-cold PBS and centrifuged at 12,000 rpm for 1 min at 4°C. After the cells were harvested and suspended in 500 μl of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.1), and protease inhibitor cocktail), the samples were sonicated using the Bioruper system (Cosmo bio, Japan) for 15 min (30 sec ON/30 sec OFF mode) in an ice-water bath and centrifuged at 12,000 rpm for 5 min at 4°C. Each sample (150 μl) was mixed with 1.35 ml of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl, and protease inhibitor cocktail). Then, 60 μl of TrueBlot™ anti-rabbit IgG IP beads (eBioscience, San Diego, CA) was added to the samples and incubated for 1 h at 4°C in a rotary shaker. After the preclearing step, the samples were centrifuged at 2,000 rpm for 1 min at 4°C, and the supernatants were transferred to new tubes. The samples in the new tubes were mixed with 15 μl of anti-Nrf2 (H-300) or anti-RAC3 (M397) antibodies and incubated for 6 h in a cold room. To pull down the DNA-protein complex, 60 μl of TrueBlot™ anti-rabbit IgG IP beads was added and incubated for 1 h after primary antibody incubation. After incubation, the samples were sequentially washed once with the following washing buffers for 15 min: low-salt washing buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, and 150 mM NaCl), high-salt washing buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, and 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.1), and TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0.). Then, the samples were eluted with 200 μl of freshly prepared 2x elution buffer (1% SDS and 0.1 M NaHCO3). For reverse cross-linking, 16 μl of 5 M NaCl was added to the samples and incubated for 8 h at 65°C followed by DNA purification using the Qiagen PCR purification kit. The DNA samples were eluted with 50 μl of ultrapure water (Invitrogen) and run on a DNA electrophoresis gel to confirm that the DNA sizes were between 200 and 1,000 base pairs (bp) after reverse cross-linking. PCR was performed using 5 μl of ChIP DNA samples using the following primers for the 2 different regions of AREs: A region: 5′-GGC GCC TTG GGA ATG CTG AGT CGCG-3′ and 5′-CAC TTC CTC CTG CC TAC CAT TAA AGC -3′; B region: 5′-CCC TGC TGA GTA ATC CTT TCC CGA-3′ and 5′-ATG TCC CGA CTC CAG ACT CCA -3′. The expected PCR products were 200 bp and 242 bp, respectively. PCR was performed using HotstarTaq Master Mix (Qiagen) under the following reaction conditions: an initial activation step at 95°C for 15 min; 34 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 40 sec; and a final extension at 72°C for 1 min. In addition, real-time PCR was performed using 1 μl of ChIP DNA samples with the same primers used above for region A. The relative fold DNA binding levels were determined using means of the 2−ΔCT method. The ChIP assay was repeated in two independent experiments.
Statistics
Statistical analyses were performed using the Student’s t-test using Sigma plot software (Ver. 7). The results are presented as the mean ±S.D. (n=3), and P values <0.05 were considered significant.
Acknowledgments
We thank Dr. Jianjie Ma (Department of Physiology and Biophysics, University of Medicine and Dentistry of New Jersey, Newark, New Jersey, USA) for help with confocal microscopy. This study was supported by NIH grant R01 NIH R01-CA-94828.
ABBREVIATIONS
- Nrf2
nuclear factor erythroid 2-related factor 2
- RAC3
receptor-associated coactivator 3
- AIB-1
amplified in breast cancer-1 protein
- SRC-3
steroid receptor coactivator protein-3
- NCoA-3
nuclear receptor coactivator-3
- TRAM-1
thyroid hormone receptor activator molecule-1
- ACTR
activator of retinoid and thyroid receptors
- pCIP
CBP-interacting protein
- Neh
Nrf2-ECH homology
- TAD
transactivation domain
- CARM1
coactivator-associated arginine methyltransferase 1
- PRMT1
protein arginine methyltransferase 1
- p/CAF
p300/CBP-associated factor
- GST
glutathione S-transferase
- NQO1
NAD(P)H:quinone oxidoreductase 1
- UGT
UDP-glucuronosyltransferase
- γGCS
γ-glutamylcysteine synthetase
- ARE/EpRE
antioxidant responsive element/electrophile response element
- Keap1
Kelch-like ECH-associated protein 1
- HO-1
heme oxygenase-1
- MAPK
mitogen-activated protein kinase
References
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. J Biol Chem. 1999;274:26071–8. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. Science. 1997;277:965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Arimura A, vn Peer M, Schroder AJ, Rothman PB. J Biol Chem. 2004;279:31105–12. doi: 10.1074/jbc.M404428200. [DOI] [PubMed] [Google Scholar]
- Blank V. J Mol Biol. 2008;376:913–25. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- Chan K, Han XD, Kan YW. Proc Natl Acad Sci U S A. 2001;98:4611–6. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Kan YW. Proc Natl Acad Sci U S A. 1999;96:12731–6. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Cell. 1997;90:569–80. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O’Connor T, Harada T, Yamamoto M. Toxicol Sci. 2001;59:169–77. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- Gnanapragasam VJ, Leung HY, Pulimood AS, Neal DE, Robson CN. Br J Cancer. 2001;85:1928–36. doi: 10.1054/bjoc.2001.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, DeMayo FJ, Xu J, Tsai SY, Tsai MJ, O’Malley BW. Mol Endocrinol. 2006;20:45–55. doi: 10.1210/me.2005-0310. [DOI] [PubMed] [Google Scholar]
- Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. Biochem Biophys Res Commun. 2003;310:824–9. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Henke RT, Haddad BR, Kim SE, Rone JD, Mani A, Jessup JM, Wellstein A, Maitra A, Riegel AT. Clin Cancer Res. 2004;10:6134–42. doi: 10.1158/1078-0432.CCR-04-0561. [DOI] [PubMed] [Google Scholar]
- Huang HC, Nguyen T, Pickett CB. J Biol Chem. 2002;277:42769–74. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. Genes Cells. 2001;6:857–68. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Genes Cells. 2002;7:807–20. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Yamamoto M. Antioxid Redox Signal. 2005;7:385–94. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- Koh SS, Chen D, Lee YH, Stallcup MR. J Biol Chem. 2001;276:1089–98. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- Kong AN, Owuor E, Yu R, Hebbar V, Chen C, Hu R, Mandlekar S. Drug Metab Rev. 2001;33:255–71. doi: 10.1081/dmr-120000652. [DOI] [PubMed] [Google Scholar]
- Kuang SQ, Liao L, Zhang H, Lee AV, O’Malley BW, Xu J. Cancer Res. 2004;64:1875–85. doi: 10.1158/0008-5472.can-03-3745. [DOI] [PubMed] [Google Scholar]
- Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. J Biol Chem. 2003;278:12029–38. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- Lee JM, Chan K, Kan YW, Johnson JA. Proc Natl Acad Sci U S A. 2004;101:9751–6. doi: 10.1073/pnas.0403620101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Kim HJ, Na SY, Kim TS, Choi HS, Im SY, Lee JW. J Biol Chem. 1998;273:16651–4. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- Leo C, Chen JD. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- Li C, Wu RC, Amazit L, Tsai SY, Tsai MJ, O’Malley BW. Mol Cell Biol. 2007;27:1296–308. doi: 10.1128/MCB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jaiswal AK. J Biol Chem. 1993;268:21454. [PubMed] [Google Scholar]
- Lin W, Shen G, Yuan X, Jain MR, Yu S, Zhang A, Chen JD, Kong AN. J Biochem Mol Biol. 2006;39:304–10. doi: 10.5483/bmbrep.2006.39.3.304. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O’Malley BW. Endocr Rev. 1999;20:321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O’Malley BW. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. Cancer Res. 2001;61:3299–307. [PubMed] [Google Scholar]
- Morse MA, Stoner GD. Carcinogenesis. 1993;14:1737–46. doi: 10.1093/carcin/14.9.1737. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Trends Mol Med. 2004;10:549–57. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Nakaso K, Yano H, Fukuhara Y, Takeshima T, Wada-Isoe K, Nakashima K. FEBS Lett. 2003;546:181–4. doi: 10.1016/s0014-5793(03)00517-9. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. J Biol Chem. 2003;278:4536–41. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- Rushmore TH, Pickett CB. J Biol Chem. 1990;265:14648–53. [PubMed] [Google Scholar]
- Sakakura C, Hagiwara A, Yasuoka R, Fujita Y, Nakanishi M, Masuda K, Kimura A, Nakamura Y, Inazawa J, Abe T, Yamagishi H. Int J Cancer. 2000;89:217–23. [PubMed] [Google Scholar]
- Shen G, Hebbar V, Nair S, Xu C, Li W, Lin W, Keum YS, Han J, Gallo MA, Kong AN. J Biol Chem. 2004;279:23052–60. doi: 10.1074/jbc.M401368200. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. J Biol Chem. 1997;272:27629–34. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- Torres-Arzayus MI, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, Sellers WR, Brown M. Cancer Cell. 2004;6:263–74. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Sleeman JE, Lamond AI. J Cell Sci. 2001;114:4219–28. doi: 10.1242/jcs.114.23.4219. [DOI] [PubMed] [Google Scholar]
- Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu MC, Sham JS, Zhang W, Wu WQ, Guan XY. Cancer. 2002;95:2346–52. doi: 10.1002/cncr.10963. [DOI] [PubMed] [Google Scholar]
- Wang Z, Rose DW, Hermanson O, Liu F, Herman T, Wu W, Szeto D, Gleiberman A, Krones A, Pratt K, Rosenfeld R, Glass CK, Rosenfeld MG. Proc Natl Acad Sci U S A. 2000;97:13549–54. doi: 10.1073/pnas.260463097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbajh S, Nojek I, Lanz R, Costas MA. FEBS Lett. 2000;485:195–9. doi: 10.1016/s0014-5793(00)02223-7. [DOI] [PubMed] [Google Scholar]
- Wu X, Li H, Chen JD. J Biol Chem. 2001;276:23962–8. doi: 10.1074/jbc.M101041200. [DOI] [PubMed] [Google Scholar]
- Xie D, Sham JS, Zeng WF, Lin HL, Bi J, Che LH, Hu L, Zeng YX, Guan XY. Hum Pathol. 2005;36:777–83. doi: 10.1016/j.humpath.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Xu FP, Xie D, Wen JM, Wu HX, Liu YD, Bi J, Lv ZL, Zeng YX, Guan XY. Cancer Lett. 2007;245:69–74. doi: 10.1016/j.canlet.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O’Malley BW. Proc Natl Acad Sci U S A. 2000;97:6379–84. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Yu CT, Ozen M, Ittmann M, Tsai SY, Tsai MJ. Cancer Res. 2006;66:11039–46. doi: 10.1158/0008-5472.CAN-06-2442. [DOI] [PubMed] [Google Scholar]
- Yao TP, Ku G, Zhou N, Scully R, Livingston DM. Proc Natl Acad Sci U S A. 1996;93:10626–31. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Furuya F, Willingham MC, Xu J, O’Malley BW, Cheng SY. Mol Cell Biol. 2005;25:7687–95. doi: 10.1128/MCB.25.17.7687-7695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Talalay P, Cho CG, Posner GH. Proc Natl Acad Sci U S A. 1992;89:2399–403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Hashimoto Y, Kwak I, Tsai SY, Tsai MJ. Mol Cell Biol. 2003;23:7742–55. doi: 10.1128/MCB.23.21.7742-7755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Fahl WE. Biochem Biophys Res Commun. 2001;289:212–9. doi: 10.1006/bbrc.2001.5944. [DOI] [PubMed] [Google Scholar]