Abstract
The population genetic structure of sea bass (Dicentrarchus labrax) along a transect from the Atlantic Ocean (AO) to the Eastern Mediterranean (EM) Sea differs from that of most other marine taxa in this area. Three populations (AO, Western Mediterranean [WM], EM) are recognized today, which were originally two allopatric populations. How two ancestral genetic units have evolved into three distinct units has not been addressed yet. Therefore, to investigate mechanisms that lead to the emergence of the central WM population, its current status, and its connectivity with the two parental populations, we applied 20 nuclear loci that were either gene associated or gene independent. Results confirmed the existence of three distinct gene pools, with higher differentiation at two transitional areas, the Almeria-Oran Front (AOF) and of the Siculo-Tunisian Strait (STS), than within any population. Significant linkage disequilibrium and heterozygote excess indicated that the STS is probably another tension zone, as already described for the AOF. Neutrality tests fail to reveal marker loci that could be driven by selection within or among metapopulations, except for locus DLA0068. Collectively, results support that the central WM population arose by trapping two tensions zones at distinct geographic locations of limited connectivity. Population assignment further revealed that WM individuals were more introgressed than individuals from the other two metapopulations. This suggests that this population might result from hybrid swarming, and was or is still seeded by genes received through the filter of each tension zone.
Keywords: Hybrid swarm, introgression, microsatellite, minisatellite, sea bass, tension zones
Introduction
It is well established that, despite the apparent connectivity of marine habitats and the high dispersal capabilities of marine organisms, populations may nevertheless be organized into well-defined genetic units (Hellberg et al. 2002; Hauser and Carvalho 2008; Hellberg 2009; Sanford and Kelly 2011). If present-day processes can contribute to patterns of differentiation (Galindo et al. 2006; Gerlach et al. 2007; Selkoe et al. 2008, 2010), recognized phylogeographic barriers shared by a wide range of species represent the main barriers to gene flow in the marine environment (e.g., Benzie 1999; Barber et al. 2000; Patarnello et al. 2007; Galarza et al. 2009; Kelly and Palumbi 2010).
Nevertheless, nearly all marine organisms have sporadic opportunities to cross almost all physical barriers and to exchange genes between phylogeographically differentiated populations. Opportunities for any neutral or unconditionally favorable mutation to invade most of – or the whole – the species range frequently occur after secondary contacts, so gene pools should be homogenized on either side because minimal gene migration easily overcomes mutation and drift over generations. Genetic differentiation persisting across phylogeographic barriers probably indicates nonneutral limitations to gene flow, as exemplified by numerous marine hybrid zones worldwide (e.g., Gardner 1997; Strelkov et al. 2007; Hobbs et al. 2009; Gagnaire et al. 2011). The classical “porous” (or “semipermeable”) genome model states that differentiation would be wiped out from most genomic regions, except around loci conferring fitness advantage to one of the populations or, on the contrary, loci acquired during isolation and involved in endogenous incompatibilities of the Dobzhansky–Muller type (e.g., Barton and Hewitt 1985; Mallet 2005; Payseur 2010; Bierne et al. 2011). Such postzygotic barriers contribute to natural selection against hybrid F1, F2, and backcrosses (Nosil et al. 2005; Burton et al. 2006; Marshall et al. 2010), and may reveal costs of adaptation for immigrants (Hereford 2009). They, hence, contribute to the establishment of so-called tension zones reflecting the conflicting effects of dispersal of parental forms and selection against hybrids (e.g., Barton and Hewitt 1985). As the porous genome model implies wide heterogeneity in the distribution of differentiation along chromosomes, estimation of population divergence will depend on the loci investigated, their position relative to sites under selective pressure, and recombination rate (see, e.g., Bierne 2010; Payseur 2010). This leads to different introgression patterns among (classes of loci) across tension zones (e.g., Barton and Hewitt 1985). Conversely – but considerably less studied – new fit hybrid genotypes may arise from successfully recombinant individuals (Harrison 1993; Edmands et al. 2005; Hwang et al. 2011; Willett 2012). A new population (or taxon), designed as a hybrid swarm, can then emerge and successfully occupy a portion of the available habitat (Moore 1977; Arnold 1993; Jiggins and Mallet 2000).
The European sea bass Dicentrarchus labrax L. is a model for evolutionary studies among marine fishes, in both wild and cultured populations (Volckaert et al. 2008; Chatain and Chavanne 2009). Over its distribution area, the species is structured into three main metapopulations (Atlantic, Western, and Eastern Mediterranean [EM]), separated by well-defined barriers to gene flow (Benharrat et al. 1983; Naciri et al. 1999; Bahri-Sfar et al. 2000; Lemaire et al. 2005). Genetic homogeneity predominates in Atlantic and Western Mediterranean (WA) basins (e.g., Garcia de León et al. 1997; Naciri et al. 1999; Fritsch et al. 2007; Coscia and Mariani 2011), while the less well-studied EM basin is more structured (Bahri-Sfar et al. 2000; Castilho and Ciftci 2005). The genetic transitions over the range are each coincident with an oceanic front (the Almeria-Oran front [AOF] and the Siculo-Tunisian Strait [STS]). This may reflect tension zones that have been trapped there, with low dispersal rates and somewhat reduced population densities (Barton's trapping hypothesis; discussed in Bierne et al. 2011). Evidence that these transitional areas are tension zones in sea bass has however only been documented for the AOF by Lemaire et al. (2005). Authors demonstrated differential introgression at mtDNA and nuclear markers, but also among anonymous nuclear DNA markers (microsatellites) at the AOF. Lemaire et al. (2005) also reported significant linkage disequilibrium (LD) at population samples located close to the AOF, which are classical features of tension zones (Barton and Hewitt 1985).
All studies on sea bass have been based on a limited number of anonymous microsatellites of unknown genomic location. Therefore, the objectives of this study were to use distinct classes of nuclear markers that were either associated or independent of genes, in order to: (1) provide better estimates on the levels of differentiation at major transitional areas; (2) establish connectivity and introgression patterns for each group of markers or patterns shared by markers irrespective the group they belong to; and (3) improve knowledge on the origin of the central WM population of sea bass, in particular whether it is a hybrid swarm.
Materials and Methods
Samples and molecular analysis
Sixteen natural populations (N = 474) were sampled among the three phylogeographic units of D. labrax distribution area: Atlantic Ocean (AO), WM, and EM basins (Table 1). Most samples have been already analyzed in previous studies (Table 1). Nineteen microsatellite and one minisatellite markers were used, classified into two classes of markers: gene-associated loci (hereafter GAL) and gene-independent loci (GIL). A marker was classified as a GAL when it was situated either inside the span of an annotated gene or at <2 kilobases (kb) from one extremity of a coding sequence (CDS). The limit of 2 kb was retained because it generally encompasses the proximal promoter of most genes in vertebrates. Proximal promoters are known to be under functional constraints and, together with their closeness from CDS, mini- and microsatellite loci they contain may have greater probability to be influenced by selection (e.g., Li et al. 2004; Gemayel et al. 2010). A symmetrical size limit of 2 kb was also retained after the 3′-end of the CDS while – as far as we know – documented functional constraints are less reported on this side of genes. Those limits are, however, operating limits to classify GAL and GIL; they do not mean that loci located farther from CDS cannot be impacted by selection (and vice versa). The GAL comprises one mini- and three microsatellite markers that have been specifically designed for their association with growth hormone (GH: μGH and mGH), somatolactin (SL: μSL), and insulin-like growth factor (IGF-1: μIGF-1; Quéré et al. 2010; description and conditions herein) and seven microsatellite loci from two simple sequence repeat (SSR)-enriched genomic libraries (Tsigenopoulos et al. 2003; Chistiakov et al. 2004) that appeared to be close to CDS afterward. The GIL class contains nine loci originating from the same SSR-enriched genomic libraries than GAL (Table 2).
Table 1.
Samples used in this study
| Basin | Abbreviation | Population | Country | Number of individuals | Geographic location | Origin of sample | |
|---|---|---|---|---|---|---|---|
| AO (31) | BB | Bay of Biscay | France | 31 | 45°44′19″N | 1°25′11″O | Fritsch et al. (2007) |
| WM (365) | GOU | La Goulette | Tunisia | 30 | 36°50′06″N | 10°19′59″E | Guinand et al. (2008) |
| ICH | Ichkeul | Tunisia | 49 | 37°10′21″N | 9°39′55″E | Bahri-Sfar et al. (2000) | |
| MAR | Marsala | Italia | 24 | 37°48′09″N | 12°25′11″E | Naciri et al. (1999) | |
| ANN | Annaba | Algeria | 24 | 36°54′47″N | 7°46′36″E | Naciri et al. (1999) | |
| SBD | Sabaudia | Italia | 16 | 41°19′36″N | 13°59′30″E | Lemaire et al. (2000) | |
| FIUM | Fiumicino | Italia | 22 | 41°51′36″N | 12°70′02″E | Lemaire et al. (2000) | |
| OR | Etang de l'Or | France | 54 | 43°34′35″N | 4°01′40″E | This study | |
| PER | Pérols | France | 30 | 43°33′50″N | 3°58′10″E | This study | |
| LUN | Lunel | France | 30 | 43°36′05″N | 4°05′12″E | This study | |
| SET | Sète | France | 38 | 43°23′06″N | 3°42′19″E | Guinand et al. (2008) | |
| MUR | Murcia | Spain | 48 | 37°51′50″N | 0°43′31″O | Lemaire et al. (2005) | |
| EM (98) | SYR | Syria | Syria | 32 | 35°34′50″N | 35°31′36″E | This study |
| CYP | Cyprus | Cyprus | 15 | 34°33′03″N | 32°58′36″E | Boutet et al. (2008) | |
| SYRA | Syracuse | Italia | 26 | 37°47′07″N | 18°48′27″E | This study | |
| SEL | Selinunte | Italia | 25 | 36°50′53″N | 13°34′07″E | This study | |
AO, WM, and EM indicate the basin from where samples originate (AO: Atlantic Ocean; WM: Western Mediterranean; EM: Eastern Mediterranean). Numbers of individuals considered in each basin are given in brackets. References of published work that already used individual samples are reported.
Table 2.
Microsatellite loci used in this study, including size ranges, GenBank accession numbers, fluorochrome labeling, and PCR primers
| Locus | Category | Annotation | Motif | GenBank Accession | Size range (bp)1 | Fluorochrome | Primers (5′⇒3′) | |
|---|---|---|---|---|---|---|---|---|
| DLA0041 | GIL | – | (TC)8(AC)21AA(AC)3AA(AC)3 | DQ363864 | 167–201 | FAM | F | AAAAGGAACAGCCCTCCAC |
| R | AGCATTGTTCTTCTGAGTGACC | |||||||
| DLA0044 | GAL | Unknown | (TC)18TTTT(TC)6CTCC | DQ363867 | 105–129 | HEX | F | TCCGCTCCGCACCGAGTGAC |
| R | ACCGCCCAAGGGTTGGACTG | |||||||
| DLA0051 | GAL | MAPK3a | (GT)16 | DQ363874 | 149–181 | ROX | F | AGGTTCTTGGCCTGGGAATC |
| R | AGTGACAGCAGCCTCCAGAG | |||||||
| DLA0060 | GAL | Bestrophin 3b | (CA)12(TA)3AA(CA)2 | DQ363883 | 119–131 | FAM | F | GAGAGTTCATCCTGTTCGCTC |
| R | TGTAGTAATAATGCGCTCTGCAA | |||||||
| DLA0061 | GIL | – | (TG)14 | DQ363884 | 153–167 | FAM | F | AAAGGCCAGTGAAACTCATGT |
| R | CTCCCTGTCCATCTGTCCTC | |||||||
| DLA0066 | GIL* | – | (AG)22 | DQ363887 | 135–161 | PET | F | GTTGACCGGAGTCCTAGC |
| R | GGCCATATGTGTCTTGCTT | |||||||
| DLA0068 | GIL | – | (CA)7CGCACG(CA)3 | DQ363889 | 247–266 | NED | F | CAACACCTGTTCCTCTGAACC |
| R | GCATTAGCATTGATTGTCCTG | |||||||
| DLA0070 | GAL | Unknown | (AC)30 | DQ363891 | 126–155 | VIC | F | TCTGCTTGCATCTGTGGAAT |
| R | GCCATCTGGCTAGCTTCACT | |||||||
| DLA0073 | GIL* | – | (CT)36 | DQ363894 | 157–181 | NED | F | CATGACTTCATGTGCTAATGTCC |
| R | AGTTCAGAGCGGCAACTGT | |||||||
| DLA0075 | GIL | – | (CA)15 | DQ363896 | 180–186 | PET | F | CACATACACAAGCTTAACCC |
| R | GGCAGAGATGGGAAATAGACA | |||||||
| DLA0078 | GAL | MAN2A1c | (AG)29 | DQ363899 | 223–243 | VIC | F | AAGACTGGACCTCTGGAGACC |
| R | CACAAGGAACCGAGACAAGA | |||||||
| DLA0081 | GAL* | PPP2R5Ad | (CA)16 | DQ363902 | 202–222 | PET | F | GACGAAGACTTCAGACGAGCTAT |
| R | ATACCGAGCGACCATGTTG | |||||||
| DLA0086 | GIL | – | (AC)26 | DQ363907 | 191–205 | FAM | F | GCTAGAGGATTCATGTCGCTT |
| R | ACCTGGTGATTGGCAATTCT | |||||||
| DLA0089 | GAL | LLGL1e | (GT)15 | DQ363910 | 127–135 | NED | F | ACGAGTAATGAGGACCCA |
| R | GTCAAAACAGCCCACCTA | |||||||
| DLA0096 | GIL* | – | (TG)16 | EF471091 | 256–270 | ROX | F | AACTTAGTGAAGTAACTTGTGGCAA |
| R | TCGATGCATCTAGGACAGGA | |||||||
| DLA0097 | GIL | – | (GT)4GC(GT)2GC(GT)13 | EF471092 | 216–240 | HEX | F | GCTGCAGGAGTGTGAGAGG |
| R | GCGAGAGACTCGAGGAAGA |
Categories of loci to which markers belong to are reported, together with their functional annotation for GAL when available. Note that GAL developed by Quéré et al. (2010; μGH, mGH, μSL, and μIGF-1) associated with growth hormone (GH), somatolactin (SL), and insulin-like growth factor 1 (IGF-1) are not reported in this table. Loci marked with an asterisk indicated loci that did not amplified in the EM samples, and were analyzed only in the AO and WM samples. Alphabetical superscripts refer to selected references in which functional role of annotated GAL markers are reported.
Size range established before this study.
Locus that was not genotyped in the EM basin.
Markers from SSR libraries were amplified in two multiplexes PCR (Table 2). For the first multiplex, amplification conditions have been reported in Guinand et al. (2008). The second multiplex was optimized for a final volume of 10 μL with 10× Taq buffer, 30 mmol/L MgCl2, 2.7 mmol/L dNTP, 10 ng of genomic DNA and 4 μmol/L of each primer. Reactions were performed on a PTC-200 (MJ Research, St Bruno, Québec, Canada) as follows: an initial denaturation at 95°C for 3 min, 35 cycles at 94°C for 45 s, annealing at the 58°C for 45 s, and 72°C for 45 s, followed by a final elongation at 72°C for 10 min. Genotyping was performed on ABI PRISM 3130xl or 3700 DNA Analyser (Life Technologies, St Aubin, France), using 5′-labeled reverse primers and the GeneScan® TM-500 LIZ Size Standard (Life Technologies) as internal size standard.
Data analyses
After control for null alleles with MicroChecker (Van Oosterhout et al. 2004), all analyses – except neutrality tests – were first performed with individual samples (Fig. 1; Table 1), then with pooled samples corresponding to the three metapopulations. In order to provide descriptive summary statistics, the mean number of alleles (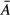 ), the mean unbiased expected gene diversity (
), the mean unbiased expected gene diversity (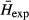 ; Nei 1987) were computed for each locus and each (meta)population. Significant within or among basins differences in
; Nei 1987) were computed for each locus and each (meta)population. Significant within or among basins differences in 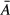 and
and 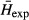 were tested using the nonparametric Mann–Whitney U-test implemented in the Microsoft Excel add-on, StatEL. Complementarily, genetic description of sample was assessed by computing LD. LD between pairs of loci was computed using GENETIX v4.05 (http://genetix.univ-montp2.fr/) in each sample, and tested using permutation tests (5000 permutations). Finally, deviations from Hardy–Weinberg expectations (HWE) within each original sample and within each metapopulation were investigated using
were tested using the nonparametric Mann–Whitney U-test implemented in the Microsoft Excel add-on, StatEL. Complementarily, genetic description of sample was assessed by computing LD. LD between pairs of loci was computed using GENETIX v4.05 (http://genetix.univ-montp2.fr/) in each sample, and tested using permutation tests (5000 permutations). Finally, deviations from Hardy–Weinberg expectations (HWE) within each original sample and within each metapopulation were investigated using 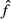 (Weir and Cockerham 1984), also using GENETIX. The null hypothesis (f = 0) of no significant departure from panmixia was tested by randomly permuting alleles (n = 1000) from the original matrix of genotypes.
(Weir and Cockerham 1984), also using GENETIX. The null hypothesis (f = 0) of no significant departure from panmixia was tested by randomly permuting alleles (n = 1000) from the original matrix of genotypes.
Figure 1.
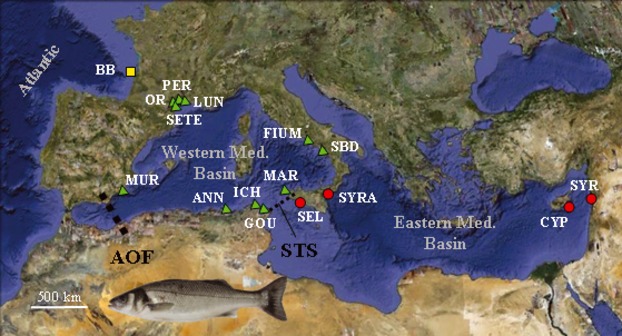
Map of sampling locations; samples are abbreviated as in Table 1. The colored symbols for samples belonging to each metapopulation will be used in other figures. Location of the Almeria-Oran Front and the Siculo-Tunisian Strait are reported.
Among-sample comparisons were assessed by estimating levels of population differentiation using 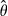 , an estimator of FST (Weir and Cockerham 1984). Estimates of
, an estimator of FST (Weir and Cockerham 1984). Estimates of 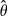 were independently computed for three data sets (i.e., the full set of 20 loci, then GAL and GIL sets). Their significance was tested according to the permutation procedure available in GENETIX and corrected for multiple tests according to Benjamini and Yukitieli (2001) false discovery rate (FDR; Narum 2006). Distance trees were inferred from Reynolds et al. (1983) coancestry genetic distance matrix [
were independently computed for three data sets (i.e., the full set of 20 loci, then GAL and GIL sets). Their significance was tested according to the permutation procedure available in GENETIX and corrected for multiple tests according to Benjamini and Yukitieli (2001) false discovery rate (FDR; Narum 2006). Distance trees were inferred from Reynolds et al. (1983) coancestry genetic distance matrix [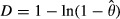 ] by the Neighbor procedure of the phylogenetic package Phylip 3.6 (Felsenstein 2005). Trees were visualized using Treeview (Page 1996). Following O'Reilly et al. (2004), we explored the relationship between
] by the Neighbor procedure of the phylogenetic package Phylip 3.6 (Felsenstein 2005). Trees were visualized using Treeview (Page 1996). Following O'Reilly et al. (2004), we explored the relationship between 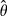 and Hexp to check for effects of size homoplasy. We specifically controlled whether GIL and GAL markers were differently affected. We further explored the possible role of selection in shaping differentiation in the full, GAL, and GIL data sets using neutrality tests developed by Beaumont and Nichols (1996; hereafter B&N and implemented in LOSITAN Antao et al. 2008, http://popgen.eu/soft/lositan/), by Vitalis et al. (2001; implemented in DETSEL Vitalis et al. 2003, http://www.genetix.univ-montp2.fr/detsel.html), and by Foll and Gaggiotti (2008; hereafter: F&G and implemented in BayeScan, http://www-leca.ujf-grenoble.fr/logiciels.htm). All tests were first performed among populations within each basin, then among the three basins except for DETSEL, specifically designed for pairwise comparisons then between each pair of basins. Outlier loci detected using several methods would be stronger candidate to the action of selection than loci detected once. The B&N and, when considered, the DETSEL tests were both performed according to P = 0.95 and P = 0.99 successively. F&G test considered a Log10(BF) of 1.3 and 2 (BF = Bayes Factor) corresponding to posterior probabilities of locus effects of 0.95 and 0.99, respectively.
and Hexp to check for effects of size homoplasy. We specifically controlled whether GIL and GAL markers were differently affected. We further explored the possible role of selection in shaping differentiation in the full, GAL, and GIL data sets using neutrality tests developed by Beaumont and Nichols (1996; hereafter B&N and implemented in LOSITAN Antao et al. 2008, http://popgen.eu/soft/lositan/), by Vitalis et al. (2001; implemented in DETSEL Vitalis et al. 2003, http://www.genetix.univ-montp2.fr/detsel.html), and by Foll and Gaggiotti (2008; hereafter: F&G and implemented in BayeScan, http://www-leca.ujf-grenoble.fr/logiciels.htm). All tests were first performed among populations within each basin, then among the three basins except for DETSEL, specifically designed for pairwise comparisons then between each pair of basins. Outlier loci detected using several methods would be stronger candidate to the action of selection than loci detected once. The B&N and, when considered, the DETSEL tests were both performed according to P = 0.95 and P = 0.99 successively. F&G test considered a Log10(BF) of 1.3 and 2 (BF = Bayes Factor) corresponding to posterior probabilities of locus effects of 0.95 and 0.99, respectively.
The program MIGRATE-n v3.0 (Beerli 2008) was used to infer M, the migration rate, among basins only (M = m/μ, where m is the immigration rate per generation). Contrary to most Fst-based statistics, MIGRATE-n provides estimation of asymmetric migrations among (meta)populations, indicating if a (meta)population is a net donor or net receiver of individuals over evolutionary times. It hence provides a picture how tension zones may function. These calculations used the Brownian mutation model and the mutation rate was considered equal for all loci (μ = 10−4) even for GAL. We used coalescent maximum likelihood (ML) based on Markov Chain Monte Carlo with Hastings Metropolis importance sampling to infer the various parameters (Beerli and Felsenstein 1999, 2001). Fst estimates among basins were used as initial parameters for the estimation of Θ and M in MIGRATE-n. For each locus, the ML was run for 10 short and five long chains with 50,000- and 10,000-recorded genealogies, respectively, after discarding the first 1000 genealogies (burn-in) for each chain. One of every 20 reconstructed genealogies was sampled for both the short and long chains. We used an adaptive heating scheme with four concurrent chains. Analyses were all performed in triplicates either on the full set of loci or on the GAL and GIL sets independently.
As a complement to estimates of population differentiation and asymmetric migration rates, we computed the probability of membership of individuals to each metapopulation (i.e., individuals should have higher probability of membership in the metapopulation they were sampled). Assignments of individuals to populations and associated probabilities were inferred with the software STRUCTURE v2.3 (Pritchard et al. 2000; http://pritch.bsd.uchicago.edu/structure.html) by setting K = 3 (i.e., three basins) after verification of the most likely number of independent population clusters was K = 3 using Evanno et al.'s (2005) ΔK method (details not reported). The software used a Monte Carlo Markov Chain (MCMC) Bayesian clustering method that maximizes the within-cluster Hardy–Weinberg and linkage equilibriums. The admixture model with noncorrelated allele frequencies was used for the full, GAL, and GIL data sets. A burn-in length of 50,000 iterations and subsequent 500,000 additional MCMC iterations were carried out. Individuals were assigned to clusters based on the highest probability of membership (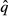 -value). Five replicates were independently performed, giving reproducible results. Average
-value). Five replicates were independently performed, giving reproducible results. Average 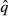 -values (±1 standard deviation) of individuals to each metapopulation were hence computed from those individual assignments.
-values (±1 standard deviation) of individuals to each metapopulation were hence computed from those individual assignments.
Analyses using MIGRATE-n and STRUCTURE were first performed using initial sample sizes of the three basins (Table 1), but, as sample sizes were unbalanced, we also performed analyses with 31 individuals in each basin (i.e., the number of individuals from the AO basin). Results were highly reproducible and qualitatively comparable to those obtained with the full set of individuals and will not be reported.
Results
Numerous amplifications failures were observed for fishes from EM populations, at several loci (GAL: DLA0081, mGH; GIL: DLA0066, DLA0073, DLA0096). This was probably due to loss of sequence homology at PCR priming sites. Those loci were discarded from some analyses, as relevant. Therefore, genetic comparisons were performed with 20 loci (11 GAL, nine GIL), except those involving EM populations, which were only based on 15 loci (nine GAL, six GIL).
Gene and allelic diversities
Allelic and gene diversities are reported in Table 3. There were no differences among populations within the WM or the EM basin for the whole set of loci, or for the GAL and GIL marker sets considered separately (details not reported), (20 or 15 loci, respectively). 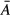 and
and 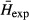 were not significantly different between the AO and WM metapopulations (P > 0.05 in each case; 20 loci). In contrast,
were not significantly different between the AO and WM metapopulations (P > 0.05 in each case; 20 loci). In contrast, 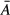 and
and 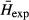 of the EM metapopulation were significantly lower than the other metapopulations for the 15 loci (P < 0.001). When considering the GAL and GIL marker sets independently,
of the EM metapopulation were significantly lower than the other metapopulations for the 15 loci (P < 0.001). When considering the GAL and GIL marker sets independently, 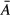 was significantly larger for GAL than for GIL in the WM metapopulation (Mann–Whitney U-test, P = 0.006). No such difference was observed between GAL and GIL in the AO and EM basins for
was significantly larger for GAL than for GIL in the WM metapopulation (Mann–Whitney U-test, P = 0.006). No such difference was observed between GAL and GIL in the AO and EM basins for 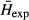 (P = 0.055 and P = 0.12, respectively).
(P = 0.055 and P = 0.12, respectively).
Table 3.
Gene and allelic diversity indices for each individual samples and metapopulations (AO, EM, WM) of sea bass considered in this study
| Multilocus | GAL | GIL | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Basin | Sample | Hexp | Ar | f | Hexp | Ar | f | Hexp | Ar | f |
| AO | GG | 0.77 | 8.70 | 0.01 | 0.78 | 9.33 | −0.01 | 0.76 | 7.75 | 0.03 |
| WM | MUR | 0.79 | 9.65 | 0.01 | 0.79 | 10.17 | 0.01 | 0.78 | 8.88 | 0.01 |
| OR | 0.77 | 9.20 | 0.00 | 0.76 | 9.48 | 0.04** | 0.79 | 8.73 | 0.06*** | |
| SET | 0.77 | 7.84 | 0.01 | 0.77 | 8.64 | −0.01 | 0.78 | 6.75 | 0.03 | |
| PER | 0.77 | 8.61 | −0.01 | 0.76 | 9.17 | −0.01 | 0.77 | 7.75 | 0.00 | |
| LUN | 0.78 | 8.90 | 0.02 | 0.78 | 8.58 | 0.00 | 0.79 | 9.38 | 0.05 | |
| FIUM | 0.75 | 7.65 | 0.00 | 0.74 | 8.25 | −0.04 | 0.76 | 6.70 | −0.02 | |
| SBD | 0.77 | 7.36 | −0.03 | 0.74 | 7.25 | −0.04 | 0.81 | 7.53 | −0.02 | |
| MAR | 0.76 | 8.13 | −0.01 | 0.76 | 8.06 | −0.02 | 0.76 | 8.26 | 0.01 | |
| ANN | 0.78 | 9.47 | 0.00 | 0.79 | 9.54 | −0.02 | 0.77 | 9.38 | 0.03 | |
| ICH | 0.74 | 8.12 | 0.02 | 0.73 | 7.83 | −0.01 | 0.76 | 8.48 | −0.07*** | |
| GOU | 0.77 | 8.54 | −0.03** | 0.76 | 8.83 | −0.03** | 0.77 | 8.13 | −0.01 | |
| WM metapop. | 0.77 | 8.49 | 0.01 | 0.76 | 8.71 | −0.01 | 0.78 | 8.18 | 0.01 | |
| EM | SEL | 0.68 | 5.24 | −0.07 | 0.69 | 6.10 | −0.08* | 0.66 | 4.19 | −0.05 |
| SYRA | 0.60 | 3.93 | −0.22*** | 0.61 | 4.20 | −0.24*** | 0.56 | 3.25 | −0.18 | |
| CYP | 0.61 | 4.13 | −0.06 | 0.67 | 4.48 | −0.06 | 0.57 | 3.62 | −0.07 | |
| SYR | 0.66 | 5.21 | −0.08 | 0.67 | 5.82 | −0.06 | 0.62 | 4.03 | −0.06 | |
| EM metapop. | 0.64 | 4.62 | −0.07*** | 0.67 | 5.15 | −0.07*** | 0.61 | 3.76 | −0.06 | |
AO, Atlantic Ocean; EM, Eastern Mediterranean; WM, Western Mediterranean; GAL, gene-associated loci; GIL, gene-independent loci.
Multilocus estimates of f are also provided. *P < 0.05; **P < 0.01; ***P < 0.001.
Hardy–Weinberg equilibrium and linkage disequilibrium
After corrections for multiple tests, no significant departure from HWE was detected for any individual samples belonging to the AO and WM basins, whatever data were considered (not reported). HWE was also respected after pooling the WM samples into a single metapopulation (all markers: 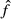 = 0.01, NS; GAL:
= 0.01, NS; GAL: 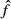 = −0.01, NS; GIL:
= −0.01, NS; GIL: 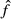 = 0.01, NS; Table 3). For the EM populations, there was a significant heterozygote excess over the 15 loci in the SYRA (Syracuse) population (
= 0.01, NS; Table 3). For the EM populations, there was a significant heterozygote excess over the 15 loci in the SYRA (Syracuse) population (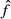 = −0.22; P < 0.001), while HWE was respected in other individual samples. This induced a significant heterozygote excess in the EM metapopulation (
= −0.22; P < 0.001), while HWE was respected in other individual samples. This induced a significant heterozygote excess in the EM metapopulation (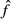 = −0.07; P < 0.001). It was mostly due to GAL markers in the SYRA and SEL (Selinunte) populations (
= −0.07; P < 0.001). It was mostly due to GAL markers in the SYRA and SEL (Selinunte) populations (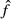 = −0.24, P < 0.001;
= −0.24, P < 0.001; 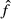 = −0.08, P < 0.05, respectively). No significant departure from HWE was detected in the EM metapopulation for the multilocus GIL marker set (
= −0.08, P < 0.05, respectively). No significant departure from HWE was detected in the EM metapopulation for the multilocus GIL marker set (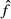 = −0.06; NS; Table 3).
= −0.06; NS; Table 3).
Because larger sample sizes prevent detection of spurious genotypic associations among loci, analysis of LD was primarily performed at the metapopulation level for Mediterranean samples (i.e., AO consists of a single sample). After correction for multiple tests, there were only three significant cases of LD in the WM metapopulation. In the EM metapopulation, 12 significant cases of LD were found, that involved only four loci: two GIL and two GAL markers (DLA0068, DLA0086, DLA0070 and DLA0089).
Relationships between 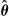 and gene diversity
and gene diversity
Relationship between 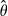 and Hexp within WM and EM revealed no significant negative correlations (WM[20 loci]: R2 = 0.062, NS; EM[15 loci]: R2 = 0.129, NS; Fig. S1). There were no significant correlations by pairwise metapopulation comparison, AO–WM, AO–EM and WM–EM, nor comparison among the three metapopulations (AO–WM[20 loci]: R2 = 0.158, NS; AO–EM[15 loci]: R2 = 0.052, NS; WM–EM[15 loci]: R2 = 0.043, NS; three metapopulations[15 loci]: R2 = 0.028, NS). When present, homoplasy did not significantly affect data and these results legitimize further interpretation in terms of differential gene flow and/or selection.
and Hexp within WM and EM revealed no significant negative correlations (WM[20 loci]: R2 = 0.062, NS; EM[15 loci]: R2 = 0.129, NS; Fig. S1). There were no significant correlations by pairwise metapopulation comparison, AO–WM, AO–EM and WM–EM, nor comparison among the three metapopulations (AO–WM[20 loci]: R2 = 0.158, NS; AO–EM[15 loci]: R2 = 0.052, NS; WM–EM[15 loci]: R2 = 0.043, NS; three metapopulations[15 loci]: R2 = 0.028, NS). When present, homoplasy did not significantly affect data and these results legitimize further interpretation in terms of differential gene flow and/or selection.
Genetic differentiation within the WM and EM metapopulations
Within the WM basin, no significant differentiation was detected (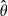 = 0.004, NS). There was no differentiation for the GIL or the GAL data separately (
= 0.004, NS). There was no differentiation for the GIL or the GAL data separately (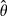 = 0.001, NS;
= 0.001, NS; 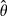 = 0.004, NS, respectively). Within the EM basin, there was significant differentiation only for the GAL data (
= 0.004, NS, respectively). Within the EM basin, there was significant differentiation only for the GAL data (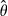 = 0.049, P < 0.01), but not for the full or GIL data (
= 0.049, P < 0.01), but not for the full or GIL data (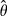 = 0.021, NS;
= 0.021, NS;  = −0.006, NS, respectively).
= −0.006, NS, respectively).
Genetic differentiation among basins
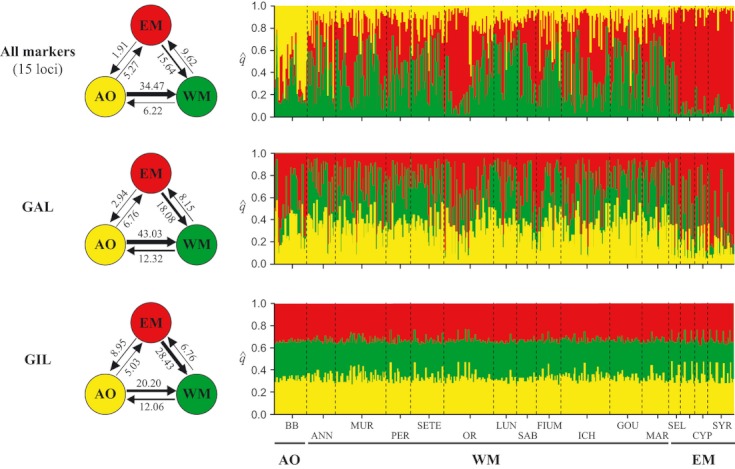
Reynolds' distance trees based on the 15 locus data clearly partitioned individual samples into three basins (Fig. 2). Levels of differentiation were generally significant and dependent on the pairwise basin comparison and markers used (GAL or GIL; Fig. 2). The differentiation between AO and WM was greater than between WM and EM, whatever data considered (Fig. 2).
Figure 2.
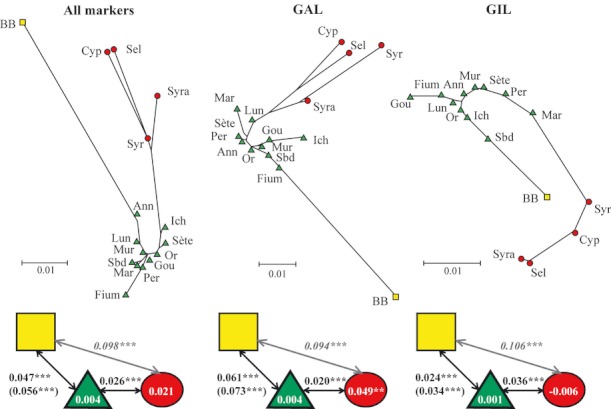
Reynold's distance trees for each marker set used in this study (all markers [15 loci]: GAL [9 loci] and GIL [6 loci]). Figures located at the bottom of each tree summarized patterns of genetic differentiation among samples within metapopulation (no estimate for Atlantic Ocean [AO] as one unique sample was considered in this metapopulation; Table 1), and among metapopulations as estimated by  for each marker set. In the case of the AO–Western Mediterranean,
for each marker set. In the case of the AO–Western Mediterranean, 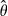 values are also reported in brackets for the 20 loci marker set, then for the set of 11 and 9 GAL and GIL marker sets, respectively. Samples are abbreviated as in Table 1 and colored symbols as in Fig. 1. **P < 0.01, ***P < 0.001.
values are also reported in brackets for the 20 loci marker set, then for the set of 11 and 9 GAL and GIL marker sets, respectively. Samples are abbreviated as in Table 1 and colored symbols as in Fig. 1. **P < 0.01, ***P < 0.001.
The individual values of 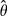 for each locus in the AO–WM and the WM–EM comparisons are reported in Figure 3A,B, respectively. Between AO and WM (20 loci used), five loci had
for each locus in the AO–WM and the WM–EM comparisons are reported in Figure 3A,B, respectively. Between AO and WM (20 loci used), five loci had 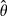 > 10%, including locus mGH where
> 10%, including locus mGH where 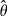 ≍ 25%. The multilocus differentiation estimator
≍ 25%. The multilocus differentiation estimator 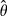 was 0.056 (P < 0.001) with significant differentiation at nine markers (GAL: six markers; GIL: three markers, Fig. 3A). The GAL data had higher values of
was 0.056 (P < 0.001) with significant differentiation at nine markers (GAL: six markers; GIL: three markers, Fig. 3A). The GAL data had higher values of 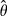 than the GIL data (
than the GIL data (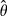 = 0.073 vs.
= 0.073 vs. 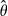 = 0.034; both P < 0.001; Fig. 2). Using 15 loci, levels of genetic differentiation were significant between WM and EM (
= 0.034; both P < 0.001; Fig. 2). Using 15 loci, levels of genetic differentiation were significant between WM and EM (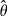 = 0.026; P < 0.001), with significant individual estimates of
= 0.026; P < 0.001), with significant individual estimates of 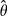 at nine of the 15 loci (GAL: four markers; GIL: five markers; Fig. 3B). Contrary to the AO–WM comparison, differentiation at the GIL markers was greater than at the GAL markers in the WM–EM comparison (respectively,
at nine of the 15 loci (GAL: four markers; GIL: five markers; Fig. 3B). Contrary to the AO–WM comparison, differentiation at the GIL markers was greater than at the GAL markers in the WM–EM comparison (respectively, 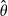 = 0.036 and
= 0.036 and 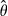 = 0.020, both P < 0.001; Fig. 2).
= 0.020, both P < 0.001; Fig. 2).
Figure 3.
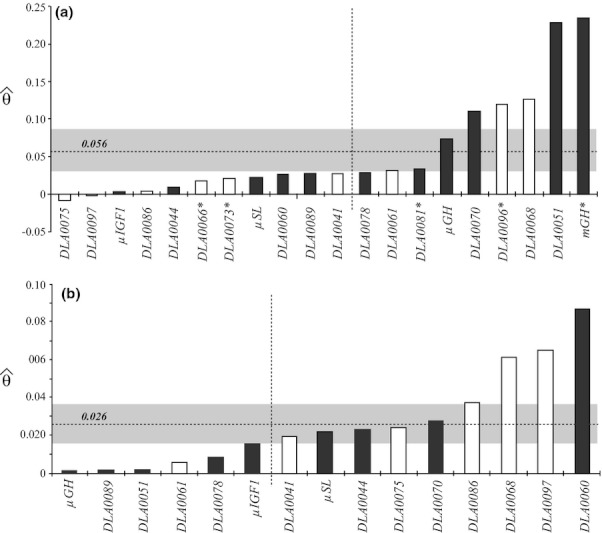
Estimates of  observed at each individual locus for (a) the Atlantic Ocean–Western Mediterranean (WM) comparison (20 loci), and (b) the WM–Eastern Mediterranean (EM) comparison (15 loci). Loci are arranged according to increased values Of
observed at each individual locus for (a) the Atlantic Ocean–Western Mediterranean (WM) comparison (20 loci), and (b) the WM–Eastern Mediterranean (EM) comparison (15 loci). Loci are arranged according to increased values Of  in each comparison. Horizontal lines indicate mean values of
in each comparison. Horizontal lines indicate mean values of  observed in each comparison (shaded area = 95% CI). Vertical lines indicate which individual loci were found significant (on the right) compared to those that were not (on the left) for each comparison. Open and black boxes indicate GIL and GAL, respectively. Asterisks indicate loci absent in the WM–EM comparison.
observed in each comparison (shaded area = 95% CI). Vertical lines indicate which individual loci were found significant (on the right) compared to those that were not (on the left) for each comparison. Open and black boxes indicate GIL and GAL, respectively. Asterisks indicate loci absent in the WM–EM comparison.
Patterns of genetic differentiation
There was wide variability in spatial patterns of allelic frequency changes (Fig. 4), including cases where allele frequencies were (1) almost identical in all metapopulations, reflecting either high gene flow, shared ancestral polymorphisms, or general selective sweeps at these loci (e.g., DLA0089, μGH, resulting in nonsignificant  values in Fig. 3B), (2) identical in two adjacent metapopulations (AO–WM or WM–EM) then marking a sharp break at one of the transition zones (e.g., DLA0051, DLA0097), or (3) in an apparent east–west gradient of allele frequencies (e.g., DLA0068). Those latter two classes of loci naturally coincided with loci achieving higher levels of differentiation (Fig. 3). Locus DLA0060 appeared as a particular case with almost identical allele frequencies for the metapopulations located at each side of the distribution but drastic change in frequency for the most frequent alleles in the central metapopulation.
values in Fig. 3B), (2) identical in two adjacent metapopulations (AO–WM or WM–EM) then marking a sharp break at one of the transition zones (e.g., DLA0051, DLA0097), or (3) in an apparent east–west gradient of allele frequencies (e.g., DLA0068). Those latter two classes of loci naturally coincided with loci achieving higher levels of differentiation (Fig. 3). Locus DLA0060 appeared as a particular case with almost identical allele frequencies for the metapopulations located at each side of the distribution but drastic change in frequency for the most frequent alleles in the central metapopulation.
Figure 4.
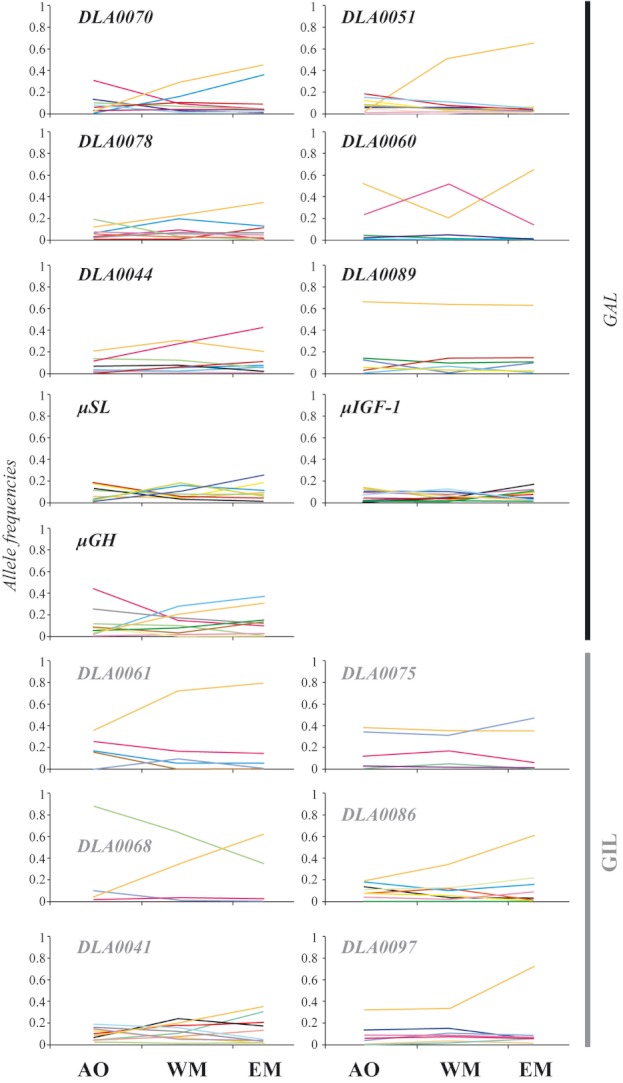
Variation of allele frequencies observed at each individual locus among sea bass metapopulations. For each marker, colored lines represent variation for a single allele. Because of size of the panels associated to loci, all alleles might not be visible at each locus. Metapopulations are abbreviated as in Table 1.
Population assignment and migration
Analyses regarding assignment were initially performed for the 15 markers in all metapopulations, then only using GAL (n = 9; DLA0044, DLA0051, DLA0060, DLA0070, DLA0078, DLA0089, μGH, μIGF1, μSL) or GIL (n = 6; DLA0041, DLA0061, DLA0068, DLA0075, DLA0086, DLA0097; Fig. 5). Unfortunately, some loci with a dramatically high  between AO and WM could not be considered because they failed to amplify in EM (e.g., mGH; Fig. 3A). This lowered the probability of assignment for AO and WM individuals. Nevertheless, better mean average assignment to their population of origin was achieved for AO and EM individuals (mean
between AO and WM could not be considered because they failed to amplify in EM (e.g., mGH; Fig. 3A). This lowered the probability of assignment for AO and WM individuals. Nevertheless, better mean average assignment to their population of origin was achieved for AO and EM individuals (mean  -value = 0.548 ± 0.156 and 0.868 ± 0.151, respectively) than for the WM individuals (
-value = 0.548 ± 0.156 and 0.868 ± 0.151, respectively) than for the WM individuals (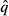 = 0.326 ± 0.206; WM individuals were in average better assigned to the EM cluster:
= 0.326 ± 0.206; WM individuals were in average better assigned to the EM cluster: 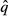 = 0.487 ± 0.291 when using all loci). This suggested higher introgression in the WM compared with the other basins (Fig. 5), and correlated well with lower all-marker genetic differentiation estimated at the STS compared to the AOF (Fig. 2). When analyzing the GAL, only EM individuals were better assigned on average to their actual population of origin (
= 0.487 ± 0.291 when using all loci). This suggested higher introgression in the WM compared with the other basins (Fig. 5), and correlated well with lower all-marker genetic differentiation estimated at the STS compared to the AOF (Fig. 2). When analyzing the GAL, only EM individuals were better assigned on average to their actual population of origin (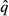 = 0.596 ± 0.223), while correct assignment was far lower for other metapopulations (AO:
= 0.596 ± 0.223), while correct assignment was far lower for other metapopulations (AO: 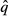 = 0.417 ± 0.117; WM:
= 0.417 ± 0.117; WM: 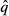 = 0.370 ± 0.134). When the GIL was used and whatever the metapopulation considered, probabilities of correct assignment did not differ significantly from
= 0.370 ± 0.134). When the GIL was used and whatever the metapopulation considered, probabilities of correct assignment did not differ significantly from 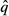 = 0.33 (details not reported). It has to be emphasized that results for GIL in this analysis did not indicate lower intrinsic relevance of GIL to assign individual, nor increased gene flow for this class of loci, but were primarily due to the low number of GIL in this particular analysis (n = 6) with only one locus demonstrating genetic differentiation among each pair of basin (DLA0068; Fig. 3). At the scale of the three metapopulations, it resulted that those six loci collectively had significant but low average
= 0.33 (details not reported). It has to be emphasized that results for GIL in this analysis did not indicate lower intrinsic relevance of GIL to assign individual, nor increased gene flow for this class of loci, but were primarily due to the low number of GIL in this particular analysis (n = 6) with only one locus demonstrating genetic differentiation among each pair of basin (DLA0068; Fig. 3). At the scale of the three metapopulations, it resulted that those six loci collectively had significant but low average 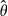 = 0.031 (P < 0.001), resulting in poor ability to successful individual assignment. Using simulated data sets in a five deme system, Latch et al. (2006) reported an average value of θ = 0.030 as a lower bound to consistently assign individuals to a subpopulation (i.e., with a probability distinct from equiprobability). Empirical
= 0.031 (P < 0.001), resulting in poor ability to successful individual assignment. Using simulated data sets in a five deme system, Latch et al. (2006) reported an average value of θ = 0.030 as a lower bound to consistently assign individuals to a subpopulation (i.e., with a probability distinct from equiprobability). Empirical 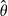 observed in sea bass is very close from this lower bound, easily explaining poor assignment capacities with this data set.
observed in sea bass is very close from this lower bound, easily explaining poor assignment capacities with this data set.
Figure 5.
Summary of results obtained by MIGRATE-n regarding the estimation of number of migrants among sea bass metapopulations for each set of markers considered in this study (15 loci data set; left panel), and STRUCTURE regarding probability of membership of each individual to one of the K = 3 sea bass metapopulations considered in this study. Thickness of arrows is broadly proportional to estimated number of migrants. Samples and metapopulations are abbreviated as in Table 1.
Individual WM samples located close to the STS (e.g., GOU, MAR; Fig. 1) present a compound allele pattern slightly closer to that of individuals from the EM samples than to WM samples located further west (e.g., MUR) for all assignments, either performed over the full data set or with the random subsets of 31 individuals. Despite this, WM individuals from western and eastern WM samples were not assigned significantly differently from EM samples over five replicates, whatever the number and class of markers considered (details not reported).
Maximum-likelihood estimates of migrations using MIGRATE-n revealed asymmetric exchange from AO to WM with a approximately threefold to fivefold difference in number of successful migrants when using GAL and all markers (GAL: MAO→WM = 43.03 vs. MWM→AO = 12.32; all markers: MAO→WM = 34.47 vs. MWM→AO = 6.22; Fig. 5). This asymmetry of migration between AO and WM was inferior to two for the GIL data set (Fig. 5). Between WM and EM, the asymmetry was also estimated with a higher number of alleles from EM migrating toward WM. The extent of the asymmetrical exchange was however lower when considering all markers (MEM→WM = 15.64 vs. MWM→EM = 9.62), or the GIL and GAL data sets separately (Fig. 5). Results of MIGRATE-n therefore indicate greater introgression in WM than in other metapopulations. That is, WM received more alleles by migration than they provided to the other two metapopulations, suggesting it may represent a hybrid swarm separated by two tension zones at the AOF and STS where migration of genes are asymmetrically filtered. It was however expected that lower estimates of genetic differentiation should translate in higher estimated number of migrants at the STS. This point has to be investigated further.
Neutrality tests
Neutrality tests were initially performed on individual samples within the WM and EM metapopulations, respectively. DETSEL was not used at this scale because of too many, not fully independent pairwise comparisons. In WM, no congruence between the B&N or the F&G tests was detected among potentially selected loci when using P = 0.95 (or Log10(BF) = 1.3). No locus was significant for a more stringent probability or BF (not reported). Indeed, B&N indicated that three loci were outliers and hence could potentially be under selection (μSL, DLA0066 and DLA0070) in WM, while F&G detected eight different loci (DLA0044, DLA0068, DLA0081, DLA0089, DLA0096, μIGF-1, μGH, and mGH). In EM, F&G did not detected potentially selected loci whatever the Log10(BF) considered, while B&N detected three loci at P = 0.95 (μSL, μGH, and μIGF-1; none at P = 0.99). It must be noted that those loci demonstrated the highest levels of differentiation among EM samples (details not shown). As results of neutrality tests failed to converge, the reality of nonneutrality within metapopulations is open to question.
Results were slightly more conclusive when performing neutrality tests among metapopulations. In the AO–WM comparison, the three tests did not provide similar results, but all indicated locus DLA0068 (GIL) as a possible outlier (Table 4). For the WM–EM comparison, B&N and DetSel detected the DLA0060 (GAL), DLA0068, and DLA0097 (GIL) loci as outliers for a minimal probability level of P = 0.95. F&G also detected three outlier loci for Log10 (BF) > 1.3: DLA0068, μIGF-1 and μSL (Table 4). Locus DLA0068 was detected by all three tests and was the locus with the sharpest pattern of graded variation across metapopulations (Fig. 4).
Table 4.
Summary of loci found as being potentially selected at the metapopulation level in each outlier detection selective test used in this study (B&N: Beaumont and Nichols 1996; DetSel: Vitalis et al. 2001; F&G: Foll and Gaggiotti 2008)
| Neutrality Test | ||||
|---|---|---|---|---|
| Locus | B&N | DetSel | F&G | |
| A: AO vs. WM | DLA0051 | 0.993 | >0.99 | – |
| DLA0060 | – | >0.95 | – | |
| DLA0061 | – | >0.95 | 0.992 | |
| DLA0068 | 0.973 | >0.99 | 1.0 | |
| DLA0070 | 0.995 | >0.95 | – | |
| DLA0075 | 0.999 | – | 0.976 | |
| DLA0096 | 0.958 | >0.99 | – | |
| DLA0097 | 0.999 | – | 0.952 | |
| mGH | 0.978 | >0.99 | – | |
| μIGF1 | 0.997 | – | 1.0 | |
| μSL | 0.967 | – | 0.983 | |
| B: WM vs. EM | DLA0060 | 0.993 | >0.95 | – |
| DLA0068 | 0.964 | >0.99 | 0.982 | |
| DLA0097 | 0.996 | >0.99 | – | |
| μIGF1 | – | – | 1.0 | |
| μSL | – | – | 0.997 | |
AO, Atlantic Ocean; EM, Eastern Mediterranean; WM, Western Mediterranean.
Locus DLA0068 found as an outlier in each test is indicated in bold.
Discussion
Genetic differentiation between Atlantic and Mediterranean populations of marine organisms is usually interpreted as the signature of secondary contact between two formerly allopatric lineages, occurring since the last glacial maximum (Borsa et al. 1997; Patarnello et al. 2007; but see, e.g., Grant 2005; Limborg et al. 2012). Accordingly, support for two distinct mtDNA clades, or nuclear differentiation, between Atlantic and Mediterranean populations at the AOF is numerous in marine organisms (Patarnello et al. 2007). When differentiation at the AOF is not demonstrated (e.g., Bargelloni et al. 2003), differentiation is generally detected in another area of the Mediterranean, including the STS and some other locations (e.g., south of Peloponnesus; Patarnello et al. 2007). The originality of sea bass resides in their partition into three metapopulations, separated by two areas of reduced genetic connectivity: the AOF and the STS (Naciri et al. 1999; Bahri-Sfar et al. 2000; Lemaire et al. 2005). To our knowledge, reports of two locations of reduced connectivity within the Mediterranean are pretty scarce, apart from the Black Sea (but see Rolland et al. 2007; Zulliger et al. 2009; Limborg et al. 2012). In sea bass, the hypothesis of a purely physical barrier shaping variation at the AO–WM transition has been seriously reassessed by Lemaire et al. (2005), who demonstrated a tension zone at this location. By using nuclear markers (GAL and GIL), we expanded on the study by Lemaire et al. (2005) to dissect the sea bass' patterns of genetic variation further.
Patterns of genetic differentiation and support for a tension zone at the Siculo-Tunisian strait
This study did not change the perception of sea bass genetic structure within each basin at nuclear loci (i.e., no differentiation inside the WM basin but some differentiation within the EM basin; Garcia de León et al. 1997; Naciri et al. 1999; Bahri-Sfar et al. 2000; Castilho and Ciftci 2005), nor the existence of three metapopulations. Nevertheless, estimates of differentiation among successive pairs of basins were larger than previously estimated with fewer microsatellite loci. The differentiation between the AO and WM basin reported in this study ( [20 loci] = 5.6%) was more than twice that provided by Naciri et al. (1999) over six loci (
[20 loci] = 5.6%) was more than twice that provided by Naciri et al. (1999) over six loci ( ≍ 2.1%). Estimates of differentiation at the WM–EM transition zone were also higher (
≍ 2.1%). Estimates of differentiation at the WM–EM transition zone were also higher (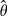 [15 loci] ≍ 2.6%) than those reported by Bahri-Sfar et al. (2000) (
[15 loci] ≍ 2.6%) than those reported by Bahri-Sfar et al. (2000) (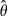 ≍ 1.4%; six loci). Results hence confirmed a stronger nuclear genetic barrier at the AOF than at the STS. This pattern was also demonstrated for mtDNA in sea bass (Lemaire et al. 2005; R. Rondon, E. Desmarais, F. Bonhomme and B. Guinand, unpubl. results), but also in sprat (Limborg et al. 2012). Using far less nuclear markers, Rolland et al. (2007) suggested on the contrary that differentiation should be stronger at the STS compared with the AOF for the common sole (Solea solea).
≍ 1.4%; six loci). Results hence confirmed a stronger nuclear genetic barrier at the AOF than at the STS. This pattern was also demonstrated for mtDNA in sea bass (Lemaire et al. 2005; R. Rondon, E. Desmarais, F. Bonhomme and B. Guinand, unpubl. results), but also in sprat (Limborg et al. 2012). Using far less nuclear markers, Rolland et al. (2007) suggested on the contrary that differentiation should be stronger at the STS compared with the AOF for the common sole (Solea solea).
GAL and GIL markers may both be involved in overall differentiation and/or differentiation among adjacent metapopulations, but the extent of differentiation depended on the particular comparison. Indeed, GAL that are located closer from or even within genes consistently demonstrated in average reduced genetic connectivity compared GIL at the AO–WM transition (20 loci; 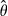 GAL = 7.3%;
GAL = 7.3%; 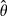 GIL = 3.4%). However, a reverse relationship was found at the WM–EM transition (15 loci;
GIL = 3.4%). However, a reverse relationship was found at the WM–EM transition (15 loci; 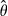 GAL = 2.0%;
GAL = 2.0%; 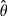 GIL = 3.6%). This second observation hence partly conflicts with those reported in other studies in which gene-associated markers had, on average, better discriminatory power than independent markers (e.g., Vasemägi et al. 2005; Yatabe et al. 2007; Shikano et al. 2010; Vilas et al. 2010).
GIL = 3.6%). This second observation hence partly conflicts with those reported in other studies in which gene-associated markers had, on average, better discriminatory power than independent markers (e.g., Vasemägi et al. 2005; Yatabe et al. 2007; Shikano et al. 2010; Vilas et al. 2010).
Besides differentiation, it is also interesting to note that significant LD and heterozygote excess was concentrated in a few samples. Heterozygote excess was especially marked for GAL at the WM–EM transition on both sides of Sicily: for the SEL and GOU samples, and at the Messina canal for the SYRA sample. LD only occurred in the SEL and SYRA samples from the EM basin. This observation is unlikely to be due to low sizes of SYRA and SEL samples, as others with similar sizes did not demonstrate any heterozygote excess and very few cases of LD. The GOU, SEL, and SYRA samples are all located at the WM–EM transition, suggesting that heterozygote excess and/or LD may be the signature of hybridization among parental pools at the STS, as already demonstrated by Lemaire et al. (2005) at the AOF. There, significant LD was only found in the sample closest to the AOF. In this study, we report a nearly identical situation at the STS (but note that the MAR sample also located close to the front did not show significant LD and/or heterozygote excess, in line with previous results by Naciri et al. (1999) and Bahri-Sfar et al. (2000) who also used this sample). Results are in agreement with the definition of a tension zone rather than with strict isolation by distance or parapatric differentiation. By theoretical modeling, Irwin (2012) has shown that genetically differentiated haploid clades arising from local adaptation along an environmental gradient can arise in a parapatric fashion under certain conditions and persist in a metapopulation even in the presence of moderate gene flow. Nevertheless, extension to multilocus diploid cases in high gene flow species has not been done, so it seems reasonable to stick to the less costly hypothesis of historical differentiation followed by secondary contact. If there was a single reason for this more parsimonious explanation, this would be at least because paleobiogeography indicates that favorable ecological zones have indeed been fragmented several time during the glacial maximums, notwithstanding other theoretical considerations about all the conditions required for the progressive building up of multilocus differentiation in presence of gene flow in highly dispersing species. We hence can reasonably be confident that the encounter of differentiated genetic backgrounds in the sea bass is a likely possibility as demonstrated for other marine organisms with large dispersal capabilities (e.g., bivalve mollusks, Nikula et al. 2008).
Spatial patterns support tension zones, but rarely selection at target loci
In addition to LD and/or heterozygote excess, spatial patterns of allele frequency changes are in accordance with the prediction of a tension zone at both the AO–WM and WM–EM transitions. The theory predicts that allele frequency changes are more or less pronounced depending on the marker investigated, with steeper transitions for loci closer to “isolation genes” (Barton and Hewitt 1985). In this study, some markers present no change in allele frequency, reflecting either one unconstrained high neutral gene flow, a selective sweep of one uniformly advantageous allele or the inheritance of an ancestral state where allele frequencies were already homogeneous. On the other hand, some loci show relatively sharp changes at AO–WM transition as already shown for mtDNA (Lemaire et al. 2005), or at the WM–EM transition, which has been less extensively investigated (Bahri-Sfar et al. 2000). A vicariant event that produced at least two separate Atlantic and Mediterranean metapopulations of sea bass during one of the quaternary climatic oscillations will have led to the emergence of isolation genes. These endogenous barriers influenced many other loci by indirect (hitchhiking) selection at genomic scales, depending on their strength and the amount of recombination (Barton and Hewitt 1985; Piálek and Barton 1997; Slatkin and Wiehe 1998). According to general hybrid zone models, we postulate that barriers have been trapped at various locations which correspond to areas of reduced density and/or environmental change, such as the AOF and STS. A rationale of this phenomenon is provided in Bierne et al. (2011), including a review of marine examples (see also Luttikhuizen et al. 2012).
Locus DLA0060 (GAL; located in intron one of the bestrophin three gene) presents a distinct pattern of spatial allele frequency compared to other loci, with the two most frequent alleles at higher frequencies in WM than in the marginal AO and EM metapopulations In the present case, we cannot distinguish whether locus DLA0060 was linked to an “endogenous” locus whose variation in allele frequency reflects genomic incompatibilities, or to an “exogenous” locus whose variation reflects local adaptation, both fitting with the coupling hypothesis of Bierne et al. (2011).
In hitchhiking models of genetic variation, direct selection on markers is not needed (Piálek and Barton 1997; Bierne et al. 2003). In sea bass, neutrality tests were unable to identify loci with clear footprint of selection, except DLA0068 (GIL). This locus presents a steeper gradient of allele frequencies than other loci, with a nearly fixed allele in the AO sample and an alternative one that predominates in the EM sample. This marker was not clearly associated with a gene. It is situated at approximately 4 kb from the 3′ end of a Retinal G Protein Coupled Receptor gene (RGR) on one side and approximately 6 kb from the 5′ end of the CDS of a solute carrier gene (SLC18A3 also known as VAChT, e.g., Weihe et al. 1996; Karila et al. 1998) on the other. We cannot therefore reach firm conclusions about its functional importance.
A hybrid swarm in the Western Mediterranean?
The WM contains strongly introgressed individuals, but also receives migrants from other basins. Taken together, this indicates that the WM metapopulation may have a dual origin from both AO and EM nuclear genomes, which have introgressed into each other and led to the emergence of a new reproductive unit, known as a hybrid swarm (Arnold 1993; Jiggins and Mallet 2000). The absence of LD among loci in each WM sample, except the GOU sample located close to the STS, also supports this hypothesis of a sea bass hybrid swarm. Interestingly, Schunter et al. (2011) recently reported a similar result in the comber Serranus cabrilla within the WM basin. In their case, combers from locations located between the two recognized fronts for this species (AOF and the Ibiza Channel) were significantly more introgressed than individuals from populations on the external sides of each front, although the presence of tension zones was not investigated. Our results on sea bass are also comparable to experimental studies by Edmands et al. (2005) on the copepod Tigriopus californicus. After approximately 15 generations of mixing, Edmands et al. (2005) found more hybrids than expected, together with heterozygote excess. These experimental results conform to the situation observed in wild sea bass near tension zones, as heterozygote excess and LD were recorded at the STS and AOF (this study and Lemaire et al. 2005, respectively). Overall, the results suggest that the WM metapopulation of sea bass could result from hybrid swarming, that is separated from parental forms by two distinct tension zones.
Hybrid swarms are poorly documented in marine organisms (McMillan et al. 1999; Roques et al. 2001; Roberts et al. 2010) by comparison with terrestrial or freshwater taxa (e.g., Arnold 1997; Nolte et al. 2009; Stemshorn et al. 2011). They may be restricted to narrow marine zones (estuaries; see Roberts et al. 2010) or extend over larger areas (Ayres et al. 2004; Strelkov et al. 2007; Nikula et al. 2008). Strelkov et al. (2007) analyzed patterns of genetic variation in a hybrid swarm of M. balthica that extends over hundreds of kilometers in the Norwegian, Barents, and Pechora seas. The samples within this swarm also showed a component of regional differentiation that was not observed in sea bass: within WM, no differentiation was detected and there was no significant indication of differential introgression with AO and EM individuals for samples located to the west or the east.
The random coupling of exo- and endogenous incompatibilities hypothesized by Bierne et al. (2011) can elicit emergence of distinct tension zones between successive fragmented patches of habitat. Using a two-locus stepping-stone model of gene flow with overdominance, Goodisman and Crozier (2001) also demonstrated that two spatially distinct tension zones can emerge between formerly isolated populations, giving rise to an additional central population. Goodisman and Crozier (2001) also demonstrated that LD accumulated at the two tension zones separating the three successive populations, while it disappeared in the central population, as observed in sea bass in the WM basin.
Although sea bass population structure and pattern of population differentiation conform to the expectations of some models, it is essential to acquire further data to investigate the hypothesis of a hybrid swarm in the WM. Such caution particularly derives from statistical considerations. This includes the discrepancy between estimated levels of differentiation and estimated number of migrants at the STS. It was expected that lower genetic differentiation should translate in higher number of estimated migrants, but this trend was not observed possibly casting doubt on this particular result. Furthermore, numerous methods for analysis of hybridization processes have emerged over recent years. They range from identification of F1 to FX hybrids and backcrosses (e.g., Anderson and Thompson 2002), to evaluating the fitness effects of alleles transferred across different genetic backgrounds (i.e., introgression; Gompert and Buerkle 2009), to genetic diffusion models (Stemshorn et al. 2011). Methods aim to analyze introgression (admixture) patterns together with the dynamics and history of introgression (Nolte and Tautz 2010; Lamaze et al. 2012). To derive consistent outputs, however, they require clear recognition of parental genotypes (Nolte et al. 2006, 2009; Gagnaire et al. 2011; Luttikhuizen et al. 2012). This is typically not observed in sea bass, where parental genotypes (AO, but also EM metapopulations) are both significantly introgressed, hence limiting the capacity for reliable inference. High throughput genome-wide genotyping will probably provide the statistical power to disentangle recent from old and shared from unshared polymorphism across transition zones in sea bass.
Acknowledgments
Authors wish to thank B. Chatain, M. Vandeputte (Ifremer, Palavas, France), D.M. Rand (Brown University, Providence, USA), and F. Volckaert (Katholieke Universiteit Leuven, Belgium) for constructive advices. M. Hassan and M. Arculeo kindly provided, respectively, the Syrian and Sicilian samples used in this study. We are also grateful to R. Reinhardt, H. Kuhl, M. Tine at MPI for Molecular Genetics, Berlin-Dahlem, for supplying unpublished data from their “European sea bass sequencing project,” and to D. McKenzie for English editing. Genetic data were produced at facilities of the HCMR and of the LabEx CeMEB (Centre Méditerranéen Environnement Biodiversité). This is publication ISEM2012-154 from the Institut des Sciences de l'Évolution de Montpellier (CNRS UMR 5554).
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Figure S1. Single-locus estimates of Fst as estimated by 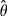 (mean ± 95% CI) vs. expected heterozygosity for (a) the 20 loci used for the Western Mediterranean samples, and (b) the 15 loci used for the Eastern Mediterranean samples. Relationship between
(mean ± 95% CI) vs. expected heterozygosity for (a) the 20 loci used for the Western Mediterranean samples, and (b) the 15 loci used for the Eastern Mediterranean samples. Relationship between 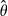 and Hexp revealed no significant negative correlations (Western Mediterranean: R2 = 0.062, NS; Eastern Mediterranean: R2 = 0.129, NS). Open and filled diamonds indicate GIL and GAL, respectively. Labels of individual loci are not reported.
and Hexp revealed no significant negative correlations (Western Mediterranean: R2 = 0.062, NS; Eastern Mediterranean: R2 = 0.129, NS). Open and filled diamonds indicate GIL and GAL, respectively. Labels of individual loci are not reported.
References
- Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J. Cytonuclear disequilibria in hybrid zones. Annu. Rev. Ecol. Syst. 1993;24:521–554. [Google Scholar]
- Arnold J. Natural hybridizaton and evolution. Oxford, U.K: Oxford Univ. Press; 1997. p. 232. [Google Scholar]
- Ayres DR, Smith DL, Zaremba K, Klorh S, Strong DR. Spread of exotic cordgrasses and hybrids (Spartina sp.) in the tidal marshes of San Francisco Bay, California, USA. Biol. Invasions. 2004;6:221–231. [Google Scholar]
- Bahri-Sfar L, Lemaire C, Ben Hassine OK, Bonhomme F. Fragmentation of seabass populations in the western and eastern Mediterranean as revealed by microsatellite polymorphism. Proc. R. Soc. B Biol. Sci. 2000;B267:929–935. doi: 10.1098/rspb.2000.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber PH, Palumbi SR, Erdmann MV, Moosa MK. A Marine Wallace's Line? Nature. 2000;406:692–693. doi: 10.1038/35021135. [DOI] [PubMed] [Google Scholar]
- Bargelloni L, Alarcon JA, Alvarez MC, Penzo E, Magoulas A, Reis C, et al. Discord in the family Sparidae (Teleostei): divergent phylogeographical patterns across the Atlantic-Mediterranean divide. J. Evol. Biol. 2003;16:1149–1158. doi: 10.1046/j.1420-9101.2003.00620.x. [DOI] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. B Biol. Sci. 1996;B263:1619–1626. [Google Scholar]
- Beerli P. 2008. MIGRATE documentation (version 3.0). Technical report. Available at http://popgen.sc.fsu.edu. Accessed May 23, 2010.
- Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl Acad. Sci. USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benharrat K, Pasteur N, Siau Y, Bouain A. Recherches Biologiques et Aquaculture. Vol. I. Journées d'études de Montpellier. Brest, France: CNEXO-BNDO; 1983. Polymorphisme biochimique de loups (Dicentrarchus labrax) originaires de quatre populations naturelles et d'un élevage; pp. 1–17. (in French) [Google Scholar]
- Benjamini Y, Yukitieli D. The control of false discovery rate under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- Benzie JAH. Genetic structure of coral reef organisms: ghosts of dispersal past? Am. Zool. 1999;39:131–145. [Google Scholar]
- Bierne N. The distinctive footprints of local hitchhiking in a varied environment and global hitchhiking in a subdivided population. Evolution. 2010;64:3254–3272. doi: 10.1111/j.1558-5646.2010.01050.x. [DOI] [PubMed] [Google Scholar]
- Bierne N, Daguin C, Bonhomme F, David P, Borsa P. Direct selection on allozymes is not required to explain heterogeneity among marker loci across a Mytilus hybrid zone. Mol. Ecol. 2003;12:2505–2510. doi: 10.1046/j.1365-294x.2003.01936.x. [DOI] [PubMed] [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol. Ecol. 2011;20:2044–2072. doi: 10.1111/j.1365-294X.2011.05080.x. [DOI] [PubMed] [Google Scholar]
- Borsa P, Naciri M, Bahri L, Chikhi L, Garcia de León FJ, Kotoulas G, et al. Zoogéographie infraspécifique de la mer Méditerranée: analyse des données génétiques populationnelles sur seize espèces Atlanto-Méditerranéennes (poissons et invertébrés) Vie Milieu. 1997;47:295–305. [Google Scholar]
- Boutet I, Quéré N, lecomte F, Agnèse JF, Guinand B. Putative transcription factor binding sites and polymorphisms in the proximal promoter of the PRL-A gene in percomorphs and European sea bass (Dicentrarchus labrax. Mar. Ecol. 2008;29:354–364. [Google Scholar]
- Burton RS, Ellison CK, Harrison JS. The sorry state of F2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am. Nat. 2006;168:S14–S24. doi: 10.1086/509046. [DOI] [PubMed] [Google Scholar]
- Castilho R, Ciftci Y. Genetic differentiation between close eastern Mediterranean Dicentrarchus labrax (L.) populations. J. Fish Biol. 2005;67:1746–1752. [Google Scholar]
- Chatain B, Chavanne H. La génétique du bar (Dicentrarchus labrax, L.) Cah. Agric. 2009;18:249–255. [Google Scholar]
- Chistiakov DA, Hellemans B, Tsigenopoulos CS, Law AS, Bartley N, Bertotto D, et al. Development and linkage relationships for new microsatellite markers of the sea bass (Dicentrarchus labrax L.) Anim. Genet. 2004;35:53–57. doi: 10.1046/j.1365-2052.2003.01076.x. [DOI] [PubMed] [Google Scholar]
- Coscia I, Mariani S. Phylogeography and population structure of European sea bass in the north-east Atlantic. Biol. J. Linn. Soc. 2011;104:364–377. [Google Scholar]
- Edmands S, Feaman HV, Harrison JS, Timmerman CC. Genetic consequences of many generations of hybridization between divergent copepod populations. J. Hered. 2005;96:114–123. doi: 10.1093/jhered/esi014. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package) version 3.6. Seattle, WA: Department of Genome Sciences, Univ. of Washington; 2005. [Google Scholar]
- Foll M, Gaggiotti O. A genome scan method to identify selected loci appropriate for both dominant and co-dominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch M, Morizur Y, Lambert E, Bonhomme F, Guinand B. Assessment of sea bass (Dicentrarchus labrax, L.) stock delimitation in the Bay of Biscay and the English Channel based on mark-recapture and genetic data. Fish. Res. 2007;83:123–132. [Google Scholar]
- Gagnaire PA, Minegishi Y, Zenboudji S, Valade P, Aoyama J, Berrebi P. Within-population structure highlighted by differential introgression across semipermeable barriers to gene flow in Anguilla marmorata. Evolution. 2011;65:3513–3525. doi: 10.1111/j.1558-5646.2011.01404.x. [DOI] [PubMed] [Google Scholar]
- Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, et al. The influence of oceanographic fronts and early-life history traits on connectivity among littoral fish species. Proc. Natl Acad. Sci. USA. 2009;106:1473–1478. doi: 10.1073/pnas.0806804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo HM, Olson DB, Palumbi SR. Seascape genetics: a coupled oceanographic-genetic model predicts population structure of Caribbean corals. Curr. Biol. 2006;16:1622–1626. doi: 10.1016/j.cub.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Garcia de León FJ, Chikhi L, Bonhomme F. Microsatellite polymorphism and population subdivision in natural populations of European sea bass Dicentrarchus labrax (Linnaeus, 1758) Mol. Ecol. 1997;6:51–62. [Google Scholar]
- García F, Zalba G, Páez G, Encío I, de Miguel C. Molecular cloning and characterization of the human p44 mitogen-activated protein kinase gene. Genomics. 1998;50:69–78. doi: 10.1006/geno.1998.5315. [DOI] [PubMed] [Google Scholar]
- Gardner JPA. Hybridization in the sea. Adv. Mar. Biol. 1997;31:1–78. [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- Gerlach G, Atema J, Kingsford M, Black KP, Miller-Sims V. Smelling home can prevent dispersal of reef fish larvae. Proc. Natl Acad. Sci. USA. 2007;104:858–863. doi: 10.1073/pnas.0606777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompert Z, Buerkle CA. A powerful regression-based method for admixture mapping of isolation across the genome of hybrids. Mol. Ecol. 2009;18:1207–1224. doi: 10.1111/j.1365-294X.2009.04098.x. [DOI] [PubMed] [Google Scholar]
- Goodisman MA, Crozier RH. Clines maintained by overdominant selection in hybrid zones. Hereditas. 2001;134:161–169. doi: 10.1111/j.1601-5223.2001.00161.x. [DOI] [PubMed] [Google Scholar]
- Grant WS. A second look at mitochondrial DNA variability in European anchovy (Engraulis encrasicolus): assessing models of population structure and the Black Sea isolation hypothesis. Genetica. 2005;125:293–309. doi: 10.1007/s10709-005-0717-z. [DOI] [PubMed] [Google Scholar]
- Guinand B, Dujardin E, Dufour V, Tsigenopoulos CS. Characterisation of genetic structure of Dicentrarchus labrax larvae in two nurseries of the Gulf of Lions (NW Mediterranean) Aquat. Living Resour. 2008;21:81–87. [Google Scholar]
- Harrison RG. Hybrid zones and the evolutionary process. New York: Oxford Univ. Press; 1993. p. 364. [Google Scholar]
- Hauser L, Carvalho GR. Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish Fish. 2008;9:333–362. [Google Scholar]
- Hellberg ME. Gene flow and isolation among populations of marine animals. Annu. Rev. Ecol. Evol. Syst. 2009;40:291–310. [Google Scholar]
- Hellberg ME, Burton RS, Neigel JE, Palumbi SR. Genetic assessment of marine population connectivity. Bull. Mar. Sci. 2002;70(Suppl):273–290. [Google Scholar]
- Hereford J. A quantitative survey of local adaptation and fitness trade-offs. Am. Nat. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Hobbs JPA, Frisch AJ, Allen GR, Van Herwerden Marine hybrid hotspot at Indo-Pacific biogeographic border. Biol. Lett. 2009;5:258–261. doi: 10.1098/rsbl.2008.0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang AS, Northrup SL, Alexander JK, Vo KT, Edmands S. Long-term experimental hybrid swarms between moderately incompatible Tigriopus californicus populations: hybrid inferiority in early generations yields to hybrid superiority in later generations. Conserv. Genet. 2011;12:895–902. [Google Scholar]
- Irwin DE. Local adaptation to smooth ecological gradients causes phylogeographical breaks and phenotypic clustering. Am. Nat. 2012;180:35–49. doi: 10.1086/666002. [DOI] [PubMed] [Google Scholar]
- Ishida N, Yoshioka S, Chiba Y, Takeuchi M, Kawakita M. Molecular cloning and functional expression of the human Golgi UDP-N-Acetylglucosamine transporter. J. Biochem. 1999;126:68–77. doi: 10.1093/oxfordjournals.jbchem.a022437. [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD, Mallet J. Bimodal hybrid zones and speciation. Trends Ecol. Evol. 2000;15:250–255. doi: 10.1016/s0169-5347(00)01873-5. [DOI] [PubMed] [Google Scholar]
- Karila P, Shahbazi F, Jensen J, Holmgren S. Projections and actions of tachykininergic, cholinergic, and serotonergic neurones in the intestine of the Atlantic cod. Cell Tissue Res. 1998;291:403–413. doi: 10.1007/s004410051010. [DOI] [PubMed] [Google Scholar]
- Kelly RP, Palumbi SR. Genetic structure among 50 species of the northeastern Pacific rocky intertidal community. PLoS ONE. 2010;5:e8594. doi: 10.1371/journal.pone.0008594. doi: 10.1371/journal.pone.0008594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultz D. Phylogenetic and functional classification of mitogen- and stress-activated protein kinases. J. Mol. Evol. 1998;46:571–588. doi: 10.1007/pl00006338. [DOI] [PubMed] [Google Scholar]
- Lamaze FC, Sauvage C, Marie A, Garant D, Bernatchez L. Dynamics of introgressive hybridization assessed by SNP population genomics of coding genes in stocked brook charr (Salvelinus fontinalis. Mol. Ecol. 2012;21:2877–2895. doi: 10.1111/j.1365-294X.2012.05579.x. [DOI] [PubMed] [Google Scholar]
- Latch EK, Dharmarajan G, Glaubitz JC, Rhodes OE., Jr Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv. Genet. 2006;7:295–302. [Google Scholar]
- Lemaire C, Allegrucci G, Naciri M, Bahri-Sfar L, Kara H, Bonhomme F. Do discrepancies between microsatellite and allozyme variation reveal differential selection between sea and lagoon in the sea bass (Dicentrarchus labrax)? Mol. Ecol. 2000;9:457–467. doi: 10.1046/j.1365-294x.2000.00884.x. [DOI] [PubMed] [Google Scholar]
- Lemaire C, Versini JJ, Bonhomme F. Maintenance of genetic differentiation across a transition zone in the sea: discordance between nuclear and cytoplasmic markers. J. Evol. Biol. 2005;18:70–80. doi: 10.1111/j.1420-9101.2004.00828.x. [DOI] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Limborg MT, Hanel R, Debes PV, Ring AK, André C, Tsigenopoulos CS, et al. Imprints from genetic drift and mutation imply relative divergence times across marine transition zones in a pan-European small pelagic fish (Sprattus sprattus. Heredity. 2012;102:96–107. doi: 10.1038/hdy.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttikhuizen PC, Dren J, Peijnenburg KTCA, Johannesson HC, van der Veer K. Genetic architecture in a marine hybrid zone: comparing outlier detection and genomic clines analysis in the bivalve Macoma balthica. Mol. Ecol. 2012;21:3048–3061. doi: 10.1111/j.1365-294X.2012.05586.x. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Marshall DJ, Monro K, Bode M, Keough MJ, Swearer S. Phenotype–environment mismatches reduce connectivity in the sea. Ecol. Lett. 2010;13:128–140. doi: 10.1111/j.1461-0248.2009.01408.x. [DOI] [PubMed] [Google Scholar]
- Matchkov VV, Larsen P, Bouzinova EV, Rojek A, Boedtkjer DMB, Golubinskaya V, et al. Bestrophin-3 (Vitelliform Macular Dystrophy 2-Like 3 Protein) is essential for the cGMP-dependent calcium-activated chloride conductance in vascular smooth muscle cells. Circ. Res. 2008;103:864–872. doi: 10.1161/CIRCRESAHA.108.178517. [DOI] [PubMed] [Google Scholar]
- McMillan WO, Weigt LA, Palumbi SR. Color pattern evolution, assortative mating, and genetic differentiation in brightly colored butterflyfishes (Chaetodontidae) Evolution. 1999;53:247–260. doi: 10.1111/j.1558-5646.1999.tb05350.x. [DOI] [PubMed] [Google Scholar]
- Moore WS. An evaluation of narrow hybrid zones in vertebrates. Q. Rev. Biol. 1977;52:263–278. [Google Scholar]
- Naciri M, Lemaire C, Borsa P, Bonhomme F. Genetic study of the Atlantic/Mediterranean transition in seabass (Dicentrarchus labrax. J. Hered. 1999;90:591–596. [Google Scholar]
- Narum SR. Beyond Bonferroni: less conservative analyses for conservation genetics. Conserv. Genet. 2006;7:783–787. [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia Univ. Press; 1987. p. 512. [Google Scholar]
- Nikula R, Strelkov P, Vaïnölä R. A broad transition zone between an inner Baltic hybrid swarm and a pure North Sea subspecies of Macoma balthica (Mollusca, Bivalvia) Mol. Ecol. 2008;17:1505–1522. doi: 10.1111/j.1365-294X.2007.03688.x. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Tautz D. Understanding the onset of hybrid speciation. Trends Genet. 2010;26:54–58. doi: 10.1016/j.tig.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Freyhof J, Tautz D. When invaders meet locally adapted types: rapid moulding of hybrid zones between sculpins (Cottus, Pisces) in the Rhine system. Mol. Ecol. 2006;15:1983–1993. doi: 10.1111/j.1365-294X.2006.02906.x. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Gompert Z, Buerkle CA. Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Mol. Ecol. 2009;18:2615–2627. doi: 10.1111/j.1365-294X.2009.04208.x. [DOI] [PubMed] [Google Scholar]
- Nosil P, Vines TH, Funk DJ. Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution. 2005;59:705–719. [PubMed] [Google Scholar]
- O'Driscoll KE, Hatton WJ, Burkin HR, Leblanc N, Britton FC. Expression, localization, and functional properties of Bestrophin 3 channel isolated from mouse heart. Am. J. Physiol. Cell Physiol. 2008;295:C1610–C1624. doi: 10.1152/ajpcell.00461.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly PTO, Canino MF, Bailey KM, Bentzen P. Inverse relationship between FST and microsatellite polymorphism in the marine fish, walleye Pollock (Theragra chalcogramma): implications for resolving weak population structure. Mol. Ecol. 2004;13:1799–1814. doi: 10.1111/j.1365-294X.2004.02214.x. [DOI] [PubMed] [Google Scholar]
- Page RDM. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Patarnello T, Volckaert FAJM, Castilho R. Pillars of Hercules: is the Atlantic–Mediterranean transition a phylogeographical break? Mol. Ecol. 2007;16:4426–4444. doi: 10.1111/j.1365-294X.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- Payseur BA. Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Mol. Ecol. Resour. 2010;10:806–820. doi: 10.1111/j.1755-0998.2010.02883.x. [DOI] [PubMed] [Google Scholar]
- Piálek J, Barton NH. The spread of an advantageous allele across a barrier: the effects of random drift and selection against heterozygotes. Genetics. 1997;145:493–504. doi: 10.1093/genetics/145.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéré N, Guinand B, Kuhl H, Reinhardt R, Bonhomme F, Desmarais E. Genomic sequences and genetic differentiation at associated tandem repeat markers in growth hormone, somatolactin and insulin-like growth factor-1 genes of the sea bass, Dicentrarchus labrax. Aquat. Living Resour. 2010;23:285–296. [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient, basis for a short term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DG, Gray CA, West RJ, Ayre DJ. Marine genetic swamping: hybrids replace an obligately estuarine fish. Mol. Ecol. 2010;19:508–520. doi: 10.1111/j.1365-294X.2009.04501.x. [DOI] [PubMed] [Google Scholar]
- Rolland JL, Bonhomme F, Lagardère F, Hassan M, Guinand B. Population structure of the common sole (Solea solea) in the Northeastern Atlantic and the Mediterranean Sea: revisiting the divide with EPIC markers. Mar. Biol. 2007;151:327–341. [Google Scholar]
- Roques S, Sévigny JM, Bernatchez L. Evidence for broadscale introgressive hybridization between two redfish (genus Sebastes) in the North-west Atlantic: a rare marine example. Mol. Ecol. 2001;10:149–165. doi: 10.1046/j.1365-294x.2001.01195.x. [DOI] [PubMed] [Google Scholar]
- Sanford E, Kelly MW. Local adaptation in the sea. Ann. Rev. Mar. Sci. 2011;3:509–535. doi: 10.1146/annurev-marine-120709-142756. [DOI] [PubMed] [Google Scholar]
- Schunter C, Carreras-Carbonell J, Macpherson E, Tintoré J, Vidal-Vijande E, Pascual A, et al. Matching genetics with oceanography: directional gene flow in a Mediterranean fish species. Mol. Ecol. 2011;20:5167–5181. doi: 10.1111/j.1365-294X.2011.05355.x. [DOI] [PubMed] [Google Scholar]
- Selkoe KA, Henzler CM, Gaines SD. Seascape genetics and the spatial ecology of marine populations. Fish Fish. 2008;9:363–377. [Google Scholar]
- Selkoe KA, Watson JR, White C, Ben Horin T, Iacchei M, Mitarai S, et al. Taking the chaos out of genetic patchiness: seascape genetics reveals ecological and oceanographic drivers of genetic patterns in three temperate reef species. Mol. Ecol. 2010;19:3708–3726. doi: 10.1111/j.1365-294X.2010.04658.x. [DOI] [PubMed] [Google Scholar]
- Shikano T, Ramadevi J, Merilä J. Identification of local and habitat-dependent selection: scanning functionally important genes in nine-spined sticklebacks (Pungitius pungitius. Mol. Biol. Evol. 2010;27:2775–2789. doi: 10.1093/molbev/msq167. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Wiehe T. Genetic hitch-hiking in a subdivided population. Genet. Res. 1998;71:155–160. doi: 10.1017/s001667239800319x. [DOI] [PubMed] [Google Scholar]
- Stemshorn KC, Reed FA, Nolte AW, Tautz D. Rapid formation of distinct hybrid lineages after secondary contact of two fish species (Cottus sp.) Mol. Ecol. 2011;20:1475–1491. doi: 10.1111/j.1365-294X.2010.04997.x. [DOI] [PubMed] [Google Scholar]
- Strelkov P, Nikula R, Väinölä R. Macoma balthica in the White and Barents Seas: properties of a widespread marine hybrid swarm (Mollusca: Bivalvia) Mol. Ecol. 2007;16:4110–4127. doi: 10.1111/j.1365-294X.2007.03463.x. [DOI] [PubMed] [Google Scholar]
- Tsigenopoulos CS, Hellemans B, Chistiakov DA, Libertini A, Kotoulas G, Volckaert F. Eleven new microsatellites of the sea bass (Dicentrarchus labrax L.) Mol. Ecol. Notes. 2003;3:352–354. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Willis DPM, Shipley PF. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. [Google Scholar]
- Vasemägi A, Nilsson J, Primmer CR. Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic salmon (Salmo salar L.) Mol. Biol. Evol. 2005;22:1067–1076. doi: 10.1093/molbev/msi093. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V. Lethal giant puzzle of Lgl. Dev. Neurosci. 2006;28:13–24. doi: 10.1159/000090749. [DOI] [PubMed] [Google Scholar]
- Vilas R, Bouza C, Vera M, Millán A, Martinez P. Variation in anonymous and EST-microsatellites suggest adaptive population divergence in turbot. Mar. Ecol. Prog. Ser. 2010;420:231–239. [Google Scholar]
- Vitalis R, Dawson K, Boursot P. Interpretation of variation across marker loci as evidence of selection. Genetics. 2001;158:1811–1823. doi: 10.1093/genetics/158.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis R, Dawson K, Boursot P, Belkhir K. DetSel 1.0: a computer program to detect markers responding to selection. J. Hered. 2003;94:429–431. doi: 10.1093/jhered/esg083. [DOI] [PubMed] [Google Scholar]
- Volckaert FAM, Batargias C, Canario A, Chatziplis D, Chistiakhov D, Haley C. European sea bass. In: Kocher TD, Cole C, et al., editors. Genome mapping and genomics in animals. Vol. 2: Genome mapping and genomics in fishes and aquatic animals. Berlin: Springer-Verlag; 2008. pp. 117–133. [Google Scholar]
- Weihe E, Tao-Cheng JH, Schaefer MKH, Erickson JD, Eiden LE. Visualization of the vesicular acetylcholine transporter in cholinergic nerve terminals and its targeting to a specific population of small synaptic vesicles. Proc. Natl Acad. Sci. USA. 1996;93:3547–3552. doi: 10.1073/pnas.93.8.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Willett CS. Hybrid breakdown weakens under thermal stress in population crosses of the copepod Tigriopus californicus. J. Hered. 2012;103:103–114. doi: 10.1093/jhered/esr109. [DOI] [PubMed] [Google Scholar]
- Wodarz A. Tumor suppressors: linking cell polarity and growth control. Curr. Biol. 2000;10:R624–R626. doi: 10.1016/s0960-9822(00)00658-8. [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 2007;175:1883–1893. doi: 10.1534/genetics.106.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulliger DE, Tanner S, Ruch M, Rubi G. Genetic structure of the high dispersal Atlanto-Mediterreanean sea star Astropecten aranciacus revealed by mitochondrial DNA sequences and microsatellite loci. Mar. Biol. 2009;156:597–610. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


