Abstract
A comparative genomic analysis of the recently sequenced human body louse unicellular endosymbiont Candidatus Riesia pediculicola with a reduced genome (582 Kb), revealed that it is the only known organism that might have lost all post-transcriptional base and ribose modifications of the tRNA body, retaining only modifications of the anticodon-stem-loop essential for mRNA decoding. Such a minimal tRNA modification set was not observed in other insect symbionts or in parasitic unicellular bacteria, such as Mycoplasma genitalium (580 Kb), that have also evolved by considerably reducing their genomes. This could be an example of a minimal tRNA modification set required for life, a question that has been at the center of the field for many years, especially for understanding the emergence and evolution of the genetic code.
Keywords: tRNA, maturation, translation, modified nucleosides, comparative genomics
INTRODUCTION
As adapters between the mRNA and the elongating peptide, tRNAs are the central decoding molecules in translation. Their overall efficiency in protein synthesis depends both on the sequence/structure of the whole set of the tRNA repertoire and on modified nucleotides that are formed during the tRNA maturation process. Depending on the organism considered, a single functional tRNA isoacceptor may contain from 2 to 17 modified nucleosides [2]. These post-transcriptional modifications are required to maintain tRNA structure, insure correct mRNA decoding, optimize translation accuracy and efficiency, and/or regulate tRNA turn-over or its cellular localization (reviewed in: [3] and [4]).
Several studies have attempted to define a minimal, possibly ancestral tRNA modification set. By comparing the modification profiles in all available sequenced tRNAs from different kingdoms (Bacteria, Eucarya and Archaea, a total of about 500 tRNA in 1998), it was predicted that eight, possibly nine, modifications were present in the putative last universal common ancestor (LUCA or Cenancestor) [5, 6]. These modifications are the Ψ residues at positions 13, 38, 39, 55, Cm at position 34 [5] or Cm at position 32 [6], Q at position 34, t6A and m1G adjacent to the anticodon at position 37, and m1A at position 58 in the highly conserved sequence of the so-called TΨ-loop. Another study that combined comparative genomics and essentiality data predicted that LUCA harbored only three modifications [7]: Q34, Ψ13, and Ψ39. Finally Church and colleagues proposed that six modifications (k2C34, xs2U34 derivatives, I34, m1G37, t6A37, ms2i6A37) are required for the minimal bacterial translation set [8]. The discrepancies are due to inherent flaws in all the prediction methods used. Predictions based solely on gene essentiality can be misleading, as a dispensable tRNA modification can become essential if other modifications are missing [9, 10]. Moreover, ancient-primordial genes may have considerably diverged in different phyla of organisms, so that they are now unrecognizable by any sequence-relatedness algorithms [11]. Alternatively, distinct enzyme families can introduce the exact same modification (functional type of enzyme evolution [12, 13, 14]). These will also be missed with methods based on ortholog searches. Finally, several genes of unknown function predicted to be present in LUCA [7] have since turned out to be involved in tRNA modification [15–17] and had therefore been missed in previous searches.
The idea of defining an “absolute minimal” set when talking about tRNA modifications might be inherently flawed and probably elusive. First, parallel and convergent solutions are deployed by different organisms both for modifications involved in decoding (discussed below) and in maintaining tRNA structural integrity. For example, ribothymidine, (m5U54) that is critical for tRNA stability in bacteria [18, 19], is replaced by m1Ψ or Um in many Archaea [2, 20, 21]. Likewise, different modified uridines are used at the wobble position of tRNA to fulfill decoding requirements in different organisms [22]. Second, one cannot separate nucleoside modifications from the sequence context of a given tRNA repertoire as there is a clear co-evolution between the two sets. For example, the tilS gene responsible for the k2C (lysidine) modification at the wobble position 34 was lost in Mycoplasma mobile. This loss occurred with a concomitant change of the sequence of the minor tRNAIle that decodes AUA codons from a CAU to a UAU anticodon [23, 24], a cellular strategy that has been experimentally verified in B. subtilis [25]. Third, the G+C content at the third codon position conditions the use of modified bases at the wobble position of tRNA [26]. Lastly, the requirements for modifications are going to be extremely dependent on environmental and physiological factors and will hence vary from one organism to another, for example, halophiles are predicted to require less modifications (see discussion of [24]). It is therefore not a minimal tRNA modification set but a minimal set of organism specific functional constraints that needs to be defined. An efficient and biologically relevant method to tentatively identify these minimal sets of essential tRNA modification enzymes, possibly the most reluctant to be lost during cellular evolution, is to analyze organisms with reduced genomes, such as parasitic or symbiotic intracellular and extracellular bacteria.
tRNA modification sets in Mollicutes
Mollicutes are parasitic, small unicellular bacteria normally living within eukaryotic cells. They originated from gram-positive bacteria (phylum: Firmicutes) by considerably reducing their genomes [27]. The Mollicute with the smallest genome identified so far is Mycoplasma genitalium (580 kb encoding 480 predicted ORFs) [28]. When cultivated in extremely rich medium, several of these Mollicutes can grow as free-living organisms, albeit very poorly and thus are considered to have minimal genomes [29]. In agreement with gene economization strategies, all Mollicutes display a minimalist, non-redundant set of tRNAs (from 28 to 35 with distinct anticodons), that is sufficient to decode all sense codons corresponding to 20 canonical amino acids [24]. In this same study, we analyzed the presence or absence of genes coding for corresponding enzymes and predicted the tRNA modifications sets in 15 Mollicutes covering the four major clades (Spiroplasma, Pneumonia, Hominis and Phytoplasma). The genes were identified by homology with model systems such as Escherichia coli and Bacillus subtilis, and further validated from the knowledge of the modified nucleosides in the full set of 29 sequenced tRNAs of Mycoplasma capricolum [24]. The main conclusion was that only a few modification enzymes, all acting on nucleotides of the anticodon loop in tRNA (m1G37, t6A37 and cmnm5U34), seemed resistant to gene loss. However, all the Mollicutes analyzed retained additional genes coding for enzymes inserting modifications in the tRNA body. For example, TruB catalyzing the Ψ55 insertion and TrmB catalyzing the methylation of G47 (m7G47) are found in the majority of Mollicutes, and therefore resistant to loss. Inspection of 20 additional complete genome sequences of Mollicutes, made available since this study, does not fundamentally change the initial conclusion (S. Yokobori, H. Grosjean and S. Bessho, personal communication).
tRNA repertoires in insect bacterial symbionts
In the present work, we performed a similar computational analysis of 14 genomes of bacterial symbionts and endosymbionts of insects, covering Wolbachia (3 strains which infect arthropod species and some nematodes), Buchnera (6 strains, which infect aphids), Candidatus Blochmannia (2 strains, which infect bacteriocytes and ant ovaries), Baumannia cicadellinicola (infecting bacteriomes of sharpshooter leafhoppers), Wigglesworthia glossinidia (infecting the gut of the tsetse fly) and Candidatus Riesia pediculicola (infecting human body louse). All of these species are derived from gram-negative Proteobacteria, mainly gamma-proteobacteria and related to E. coli, with the exception of Wolbachia (an alpha-proteobacteria). Unlike most bacteria and Mollicutes, members of this group cannot live as free-living organisms and form an obligate relationship (intimate symbioses) with their eucaryal hosts. These symbionts are predominantly vertically transmitted along with their host, and thus extend the heritable genetic variation of the host cells [30–33].
The genome sizes of the set of organisms analyzed (Supplemental Table 1) varied from 416 kb with 371 predicted CoDing Sequences (CDSs) (Buchnera aphidicola str. CC) to 1,483 kb with 1586 CDSs (Wolbachia pipientis quinquefasciatus Mel) (numbers of CDS taken from the Rast server [34]). 557 CDSs have been predicted in C. R. pediculicola, but around 80 of these are very small (between 19 and 70 aa) with no homology to any known proteins. These types of small proteins are not found in the other insect symbiont genomes analyzed and might be overpredictions.
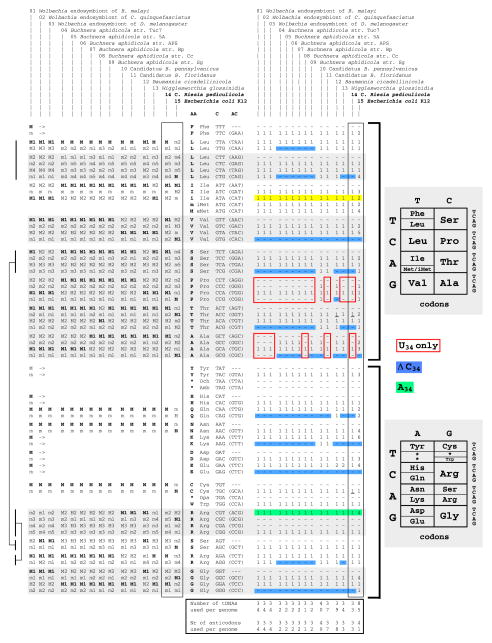
Figure 1 (right part) shows that all 14 symbionts analyzed harbor genes coding for a full set of tRNAs able to read all sense codons for the 20 canonical amino acids, indicating that no tRNAs from the host are needed. Like Mollicutes and at variance with bacteria with large genomes, these uncultivable symbionts display a quasi-non-redundant set of tRNAs, with each isoacceptor having a distinct anticodon (compare columns #1 through #14 with column #15 for E. coli). The total number of tRNAs varies from 31 for Buchnera aphidicola str Cc (#8) to 40 for C. Blochmannia pennsylvanicus (#10). These correspond to tRNA repertoires typically found in Bacteria and not in Eucarya and Archaea [22, 35]. For example, tRNA genes containing the wobble T34 and G34 are almost always present, while tRNA genes containing C34 are often absent (blue background in Figure 1). In both of the quartet boxes corresponding to Pro and Ala (boxed in red in Figure 1), only one tRNA gene harboring a wobble T34 is present. For the isoleucine triplet decoding box and the arginine quartet decoding box, the T34-containing tRNA genes are systematically replaced by a C34-containing tRNAIle and an A34-containing tRNAArg, respectively (indicated with yellow and green background in Figure 1). tRNA usage is usually correlated with codon usage, which in turn controls the efficiency of decoding [36]. By comparing the relative codon usage in each of the decoding box, it appears that G34-containing tRNAs more frequently read codons ending with the wobble U3 while U34-containing tRNAs mainly read codons ending with A3 (Watson-Crick base pairing). When the C34-containing tRNA isoacceptor is absent, U34 also reads codons ending with the wobble G3 (compare information about codon usage on the left part of Figure 1 with the identity of the wobble base in the tRNA, under the column symbol ‘AC’ for anticodon). This trend reflects the low average G+C content in ORFs of insect symbionts analyzed (from 23 to 35% compared to 52% in E. coli; Supplemental Table 1), particularly at the third position of codons (data not shown), and reflects the type of modified nucleotide present at the wobble position of tRNA. Non-redundancy of tRNA isoacceptor may affect cellular tRNA abundance, and hence the growth rate of the symbiont [37]. Finally, in contrast to Mycoplasma [24], UGA is a genuine stop codon in these insect symbiotic organisms, correlating with the presence of Release Factor 1 and Release Factor 2 (see the “tRNA modification E. coli” subsystem available on the Public SEED, http://pubseed.theseed.org/SubsysEditor.cgi).
Figure 1. Codon/anticodon/tDNA usage for the 20 canonical amino acids in the 14 symbiont genomes and E. coli.
The 15 genomes investigated are listed at the top. Full names are given in Supplementary Table SS1. Codon usage within each amino acid family decoding boxes is denoted by the letters on the left: “M” corresponds to most frequently used codon and “m” to the least used ones, with “M1” > “M2” > “m1” > “m2”, etc … to indicate decreasing frequency of codon usage. Rightwards arrows indicate a similar codon usage frequency among the 15 genomes. Details about codon usage in each of the 15 bacteria analyzed can be found in Supplemental Table SS2. These were obtained from automatic determination of all non-overlapping ORFs of 100 codons or more. Vertical bars at the left indicate the six codons of Leu, Ser and Arg respectively. The four columns in the center list the amino acids (indicated as “AA”, one- and three-letter code), the codon (“C”) and anticodon (“AC”) at DNA level. Anticodons never used are indicated as “---”. Numbers at the right indicate the number of tRNA genes harboring the respective anticodons found in each bacteria. Dash signs indicate absence of corresponding tRNA gene. tRNA gene search was performed with tRNAscan-SE [59], and the structure of each tRNA was carefully inspected for fit to the earlier defined bacterial-type tRNA cloverleaf structure [35]. Only three cases of tRNAs (underlined) with more nucleotide than expected (+1 nt in the D-loop) were found. None of the tRNA genes were found in plasmids. The key to the color code is: light gray background denotes four-codon family boxes encoding a single amino acid; yellow background for AUA codon read by the special Ile-tRNA (CAU with wobble C34 modified to k2C34 in mature tRNA, see text); green background for the unique A34-containing tRNAArg (I34 in mature tRNA, see text); red boxes correspond to ‘quartet’ decoding mode in which a single tRNA with T34 at the gene level reads the four codons; blue background denotes C-sparing strategy, the corresponding codon being read by a U34 –containing tRNA. The boxes to the right indicate the standard Genetic Code (split in two).
tRNA modifications sets in insect bacterial symbionts
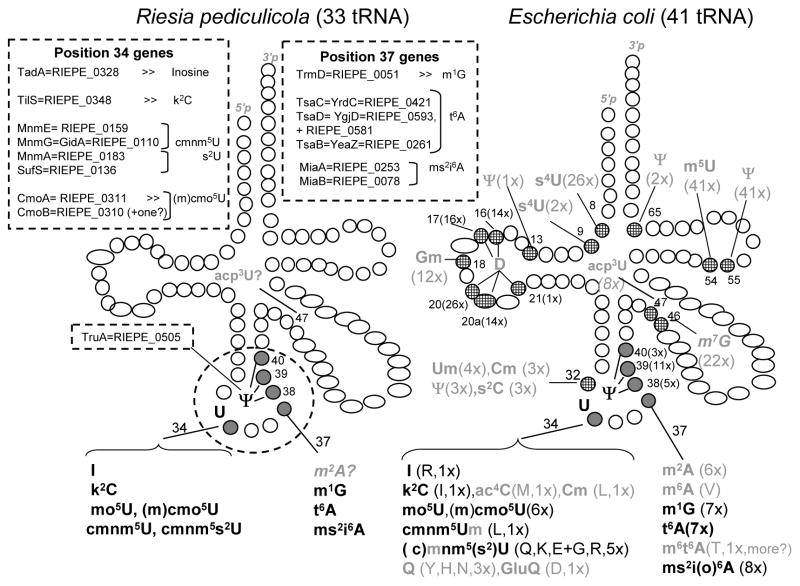
Genes coding for tRNA modification enzymes in the 14 genomes analyzed were identified by BLAST analysis against the genes found in E. coli (see [1] and Figure 2 legend). In E. coli, all but four of the fully matured isoacceptor tRNAs have been sequenced, and genes coding for most of the corresponding tRNA modification enzymes have been identified. Surprisingly, the recently sequenced human louse endosymbiont C. R. pediculicola [38] appears to have lost all modifications of the tRNA body, retaining only a few modifications of the anticodon loop and proximal stem (Figure 2): Ψ at position 38 and 39; I, k2C, xs2U derivatives and xo5U at position 34; and m1G, t6A, i6A and ms2i6A at position 37. As it is technically impossible to extract enough tRNA from the human louse symbiont to analyze the modifications profiles, we cannot rule out that additional or unknown modifications are present in this organism. For example the acp3U47 modification gene has not yet been identified in any organism, and could be present in C. R. pediculicola (Figure 2).
Figure 2. Prediction of the tRNA modifications present in C. R. pediculicola and comparison with E. coli.
The analysis of the modification genes present in the genome of C. R. pediculicola was performed using the SEED database. We constructed a subsystem containing all known E. coli tRNA modifications genes (see “tRNA modification E. coli” subsystem available on the Public SEED, http://pubseed.theseed.org/SubsysEditor.cgi) and extended it to C. R. pediculicola. A manual search of the genome (NC_014109, NC_013962) using BlastP and tBlastN [60] with the E. coli proteins from the “tRNA modification E. coli” subsystem as input was performed. The gene list used is also found in Table 1 of [61] with the addition of the gene encoding TsaA involved in m6t6A formation (T. Suzuki and V. de Crécy-lagard, personal communication) and TsaD/YgjD involved in t6A formation [16]. In E. coli, IscS and TusABCDE are required for thiol transfer [3], but no TusACDE homologs were found in C. R. pediculicola and SufS is the only IscS homolog in this organism. The m2A37 methylase encoding gene has not been identified in any organism. The same is true for the acp3U47 gene, hence the question marks. We previously predicted yfiF encodes the missing methylase [62], but this has not been experimentally validated. No yfiF homolog or no other methylase of unknown function could be identified in the C. R. pediculicola genome, making the presence of m2A37 in this organism unlikely. Finally, to make sure no other genes had been missed, all known tRNA modification genes from B. subtilis and S. cerevisiae were queried in C. R. pediculicola (using the subsystems “tRNA modification Bacteria”, “tRNA modification yeast cytoplasmic” and “tRNA modification yeast mitochondrial” [63]). The genes present in C. R. pediculicola are listed in the dashed boxes, with prediction of the resulting modification. Assuming that gene products in C. R. pediculicola exhibit the same specificity as the E. coli homologs, one can predict which modifications are found in the 33 tRNAs of the symbiont. They are all localized in the anticodon loop and proximal stem (indicated by numbered grey circles, the whole cluster of modified nucleotides being encircled by dashed line). Only acp3U, normally present at position 47, cannot be excluded because the gene coding for the corresponding enzyme is unknown. For the same reason, it is not certain if m2A37 is present. For comparison, the same tRNA cloverleaf is shown with all the modified nucleotides identified so far by sequencing the 37 fully mature E. coli tRNA, as indicated in Figure 1 (only 4 isoacceptor tRNA remain to be sequenced, see Figure 4). The modified nucleotides common to both bacteria are indicated in black, while the ones found only in E. coli are indicated in grey. In brackets, the number of isoacceptor tRNAs containing a given modification is indicated. When this number is low, the identity of the modified tRNA is also indicated using the one letter code for amino acid. Open circles correspond to positions in E. coli tRNAs where no modification has been found. This compilation was adapted from previously published data [2, 3]. Full names for the different acronyms used to define a given modified base can be found in the MODOMICS database [1].
An identical analysis was performed on the remaining 13 symbionts (#1 to #13, Figure 3). In some genomes, additional modifications were predicted to be present: s4U8, s4U9, D17, 20, 20a, Q34, m7G46 and Ψ55. However, all symbionts analyzed except C. R. pediculicola contain at least one modification outside the anticodon-stem loop (Figure 3).
Figure 3. Prediction of tRNA modifications present in insect symbionts.
A signature gene was chosen for every modification and the distribution of the genes analyzed in all genomes listed in Figure 1 by adding them to the “tRNA_modification_E._coli” subsystem on the Public SEED server. Only the genes that were found in at least one of the genomes analyzed other than E. coli are shown, with the exception of the ones responsible for m2A37 and acp3U47 modifications that have yet to be identified in E. coli. Grey boxes denote genes present in all genomes analyzed. Black boxes denote genes present in E. coli and in some of the symbiotic genomes. White boxes denote that a specific gene is missing in a specific organism.
Decoding strategy of synonymous codons in Candidatus R. pediculicola
Analysis of the sequences of both of the louse endosymbiont C. R. pediculicola and its host reveals that no eukaryotic genes, including putative tRNA modification enzymes, have been transferred to the insect bacterial genome, and that the genome reduction in C. R. pediculicola has not been associated with gene transfer to the host [38]. In the 14 proteobacterial symbionts analyzed, we are confident that the only genes coding for tRNAs and tRNA modification enzymes are those reported in Figures 1, 2 and 3 (except, see Figure 2 legend, for enzymes catalyzing acp3U47 and m2A37, for which the corresponding genes in E. coli are yet to be identified).
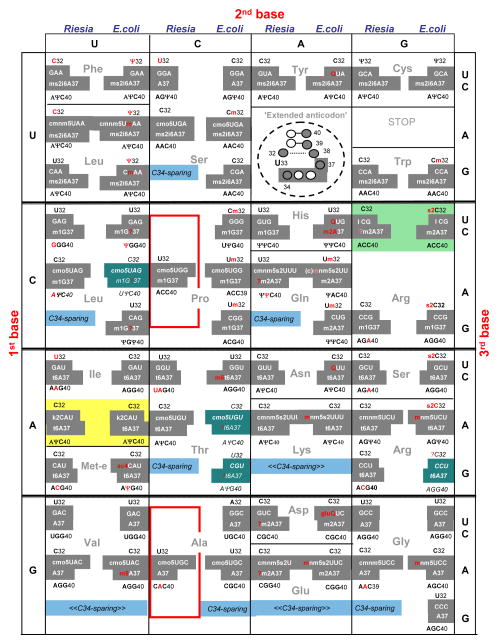
Beside the lack of some tRNA isoacceptors in the insect symbionts (discussed above and Figure 1), both nucleotide identities and post-transcriptional modifications are very similar when comparing tRNA isoacceptors from E. coli and C. R. pediculicola, attesting closely related and typical bacterial decoding strategies [22]. The differences between the two organisms are indicated in red in Figure 4. The main difference is the additional modification of several E. coli bases. These are 2′O-methylation of C32 (Cm) in tRNASer (*UGA) and tRNATrp (CCA) or U32 (Um) in tRNAs specific for Pro, His and Gln, as well as C34 (Cm) or U34 (Um) in E. coli tRNALeu (*UmAA) and tRNALeu (CmAA). Many complex modifications are predicted to be missing in C. R. pediculicola tRNAs: the acetyl group in elongator tRNAMet (ac4C34), the N6-methylations of A37 or t6A37 in tRNAThr (GGU/UGU), Q34 or GluQ34 in tRNA specific for Tyr, His, Asn and Asp, and the thio-group on C32 in several tRNA for Arg and Ser. The mnm5U34 modification found in some E. coli Gln, Lys, Glu, Arg and Gly tRNAs, is predicted to be cmnm5U34 in the corresponding C. R. pediculicola tRNAs, because of the presence of MnmE and MnmG and the absence of MnmC. This last enzyme normally catalyzes the stepwise decarboxylation of the ‘cmnm’ group attached to C5 of U34, followed by methylation of the resulting ‘nm’ group into the final product mnm5U34 [39]. An alternative mnm5U34 biosynthetic pathway using ammonium instead of glycine as a cofactor has been demonstrated in E. coli MnmE/MnmG mutants in vitro [40]. Therefore, it is possible that such an alternative ammonium mediated biosynthetic pathway leading to the final nm5U34 derivative is used in the insect symbiont (thus by-passing the formation of cmnm5U34). The cmo5U34 modification is found in E. coli tRNAs belonging to quartet decoding boxes for Leu (anticodon *UAG), Val (*UAC), Ser (*UGA), Pro (*UGG), Thr (*UGU) and Ala (*UGC). Its synthesis requires at least three enzymes, only two of which are known (CmoA-yecO and CmoB-yecP [41, 42]). The methylester of cmo5U at position 34 (mcmo5U not mentioned in any of the cases in Figure 4) is reported to be base labile and thus only cmo5U is usually detected during most analyses of modified nucleosides. In E. coli, tRNASer and tRNAAla, but not tRNAVal were reported to be substrates for the E. coli and Salmonella CmoA methyltransferase [3]. Remarkably, genes coding for CmoA-CmoB are found in C. R. pediculicola but absent in all the other 13 insect symbionts analyzed (Figure 3) as well as in Mycoplasma [24, 43]. This suggests that the cmo5U34 modification is dispensable, and cmoA/cmoB could be the next set of modification gene lost by C. R. pediculicola. Alternatively, the maintenance of cmo5U in several C. R. pediculicola tRNAs could result from subtle decoding constraints specific to that organism. Several studies exploring the function of (m)cmo5U derivatives versus ho5U modified U34 [3, 41, 42] concluded that the (m)cmo group added to the C5 atom of the wobble U base enhances the ability of the tRNA to pair with all four codons, a property that was also demonstrated for a non-modified wobble U-base [44]. These observations again suggest that U34 modification of tRNA belonging to quartet decoding boxes can be dispensable, however only in certain extended anticodon contexts [45].
Figure 4. Comparative decoding strategies of C. Riesia pediculicola and E. coli.
In the standard genetic code, each decoding box contains information about identity of nucleotides present in the anticodon loop and proximal stem, as illustrated in the decoding box corresponding to codons UAA/UAG (labeled in figure as “Extended anticodon”). Shown are the nucleotides at positions 32, the three anticodon bases (34–36) and nucleotide-37 (both in grey background) and the sequence of nucleotide 38–40. On the right side of each decoding box, is listed the information for E. coli isoacceptor tRNAs obtained from the tRNA data banks [2, 3]. On the left side of each decoding box, is listed the information for the homologous C. R. pediculicola (Riesia) isoacceptor tRNAs. The identities of the nucleotides were obtained directly from the tRNA gene analysis (this work, Figure 1), while the presence of modified nucleotides was deduced by combining knowledge from the analysis done in Figure 2 with the known modifications at identical positions in the corresponding E. coli tRNAs. The color code is as in Figure 1. In dark green background, are the only four mature tRNAs in E. coli that have not yet been sequenced, only the sequence of the corresponding genes are known. Differences between the two sets of bacterial isoacceptor species are highlighted by red letters. The exact chemical nature of the hypermodified m1G?37 in E. coli tRNALeu is not known [3], so only the m1G moiety was indicated for the insect symbiont tRNA. Also the presence of m2A37 in C. R. pediculicola is questionable (see Figure 2 legend) and indicated as ?m2A.
The situation is different in the cases of bacterial tRNAs belonging to the split/duet decoding boxes, such as tRNAs specific for Phe/Leu, His/Gln, Asn/Lys, Asp/Gly and Ser/Arg that depend strictly on the identity of modified nucleotides at wobble U base. E. coli, all of the insect symbionts analyzed in this work, and all of the Mollicutes analyzed earlier rely on xm5U and xm5s2U derivatives to allow accurate and efficient discrimination of the duet codons ending with a pyrimidine U or C. The other duet codons of the same decoding box ending with a purine A or G being efficiently read by a G34 or modified G34-containing tRNA (reviewed in: [22, 46]).
Other important, modified nucleotides conserved in C. R. pediculicola, and possibly essential in all bacteria, are the pseudouridines at positions 38, 39 and/or 40 and the modified purine at position 37 found in tRNAs harboring an anticodon ending with A36, G36 or U36, modified into (ms2)i6A37, m1G37 or m2A37 and (m6)t6A37, respectively (Figure 4). Removal of these modifications has been shown to have a detrimental effect on efficiency and accuracy of decoding (reviewed in [47, 48]). However, one cannot generalize the essentiality of these modifications to all tRNA sets, as the naturally occurring E. coli tRNASer(GGA) and/or tRNAs harboring an anticodon ending with A37 in most Mycoplasmas lack i6A37 or ms2i6A37 derivatives [24, 49]. It is clear from our analysis that the genes responsible for the insertion of Ψ38-40, m1G37, t6A37, cmnm5U34 and s2U34 remain resistant to loss. This suggests that these genes emerged early during cellular evolution, and, once fixed in the genome, became essential for the cell.
DISCUSSION
Both the insect symbionts and Mollicutes analyzed in our work are derived from bacteria with larger genomes (gram-negative Proteobacteria and gram-positive Firmicutes, respectively). During their evolutionary adaptation to their specific eukaryotic host cell, these organisms have massively lost genes, including genes coding for many isoacceptor tRNA and tRNA modification enzymes. With their minimal genomes, and unlike more specialized organelles, they are generally considered to correspond to the simplest living, autonomous organisms. We purposely did not include in our analysis genomes of insect symbionts with extremely reduced genomes (below 300 kb), such as Candidatus Carsonella ruddi, the endosymbiont of the psyllid Pachpsylla venusta (genome size of 160 kb with 183 CDSs [50]) and the very recently sequenced Candidatus Tremblaya princeps str PCVAL of the citrus mealybug Planococcus citri (genome size of 138kb, about 110 CDSs [51]). Both C. Carsonella ruddii and C. Tremblaya princeps have lost several enzymes required for self-replication, several ribosomal RNA, and many aminoacyl-tRNA synthetases. C. Tremblaya princeps has even lost most of its tRNA genes. These organisms must therefore rely on host proteins and tRNAs. They resemble organelles (mitochondria and plastids) [32, 52, 53], and cannot be used in our analysis as we cannot predict the presence of modifications from the presence of the corresponding genes in the endosymbiont.
The finding that C. R. pediculicola has lost all modifications of the tRNA body suggests that the structural and recognition roles of modifications outside the anticodon region (reviewed in [3] and [4]) are dispensable in the context of intracellular organisms with slow growth rates and probably with limited sets of nucleases genes and whertRNA degradation might be less of an issue [9]. Indeed, one can expect that protein synthesis might not be as accurate in Mollicutes and insect symbionts as in more sophisticated free-living bacteria. However, since these organisms are not in constant competition with other bacteria, they can certainly survive with a less efficient translation system. The positions of these parasites on the bacterial phylogenetic tree suggest that these are fast evolving bacteria with elevated mutation rates ([29] and several chapters of [54]). Proteins generated by an inaccurate translation system might provide an advantage to the parasite to evolve faster than other bacteria producing a more homogeneous proteome (discussed in [55–57]) and could be an advantage for fast adaptation to the host.
The conservation of genes coding for modification enzymes acting at the wobble position as well as the proximal anticodon bases (position 37–40), at least in organisms having a relatively low G+C content (below 35%, like Mollicutes and most insect symbionts), definitively pointed out the importance of these modifications for maintaining minimalist accuracy and efficacy in reading the genetic code based on 61 sense codons for 20 amino acids. Analyzing genomes of organisms having progressively reduced the size of their genomes allows for identification of the genes more resistant to loss. Hence, from an evolutionary perspective, Mollicutes and insect symbionts constitute excellent biological specimens to identify strategies developed during evolution for reading the genetic code with a minimal set of tRNAs and modification enzymes, a situation that could correspond to what might have occurred at an early stage of life, when the genetic code was just emerging [24, 58].
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01 GM70641) to VdC. HG hold a position of Emeritus Scientist at the Center of Molecular Genetic of the CNRS in Gif-sur-Yvette (France) in the laboratory of Dominique Fourmy. We thank David Reed and Bret Boyd for introducing us to the field of louse symbionts, Basma El Yacoubi for numerous and insightful discussions and Patrick C. Thiaville and Jennifer J. Thiaville for editing the manuscript.
Footnotes
Note added in proofs: It was recently found that the E. coli rlmN gene encodes the missing m2A37 methyltransferase (Eugenia Armengod, personal communication). RlmN homologs are present in most insect symbiont genomes, including C. R. pediculicola. A37 is therefore most certainly methylated into m2A37 in a few C. R. pediculicola tRNAs, which fits with our general conclusion above.
ABBREVIATIONS
Full names for the different acronyms used to define a given modified base can be found in [1].
References
- 1.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. Nucleic Acids Res. 2009;37:D118. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. Nucleic Acids Res. 2009;37:D159. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björk GR, Hagervall TG. Escherichia coli and Salmonella. In: Böck A, Curtis R, Kaper JB, Neidhardt FC, Nyström T, Squires CL, editors. Cellular and Molecular Biology. ASM. Press; Washington DC: 2005. http://www.ecosal.org Module 4.6.2. [Google Scholar]
- 4.Phizicky EM, Hopper AK. Genes & Dev. 2010;24:1832. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cermakian N, Cedergren R. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. ASM Press; Washington DC: 1998. p. 535. [Google Scholar]
- 6.Björk GR. Chemica Scripta. 1986;26B:91. [Google Scholar]
- 7.Ouzounis CA, Kunin V, Darzentas N, Goldovsky L. Res Microbiol. 2006;157:57. doi: 10.1016/j.resmic.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 8.Forster AC, Church GM. Mol Syst Biol. 2006:2. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Mol Cell. 2006;21:87. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Grosshans H, Lecointe F, Grosjean H, Hurt E, Simos G. J Biol Chem. 2001;276:46333. doi: 10.1074/jbc.M107141200. [DOI] [PubMed] [Google Scholar]
- 11.Gerlt JA, Babbitt PC, Jacobson MP, Almo SC. J Biol Chem. 2012;287:29. doi: 10.1074/jbc.R111.240945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christian T, Evilia C, Williams S, Hou YM. J Mol Biol. 2004;339:707. doi: 10.1016/j.jmb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Nucleic Acids Res. 2005;33:3955. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galperin MY, Koonin EV. J Biol Chem. 2012;287:21. doi: 10.1074/jbc.R111.241976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crécy-Lagard V. Nucleic Acids Res. 2009;37:2894. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crécy Lagard V. EMBO J. 2011;30:882. doi: 10.1038/emboj.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. EMBO J. 2011;30:873. doi: 10.1038/emboj.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davanloo P, Sprinzl M, Watanabe K, Albani M, Kersten H. Nucleic Acids Res. 1979;6:1571. doi: 10.1093/nar/6.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sengupta R, Vainauskas S, Yarian C, Sochacka E, Malkiewicz A, Guenther RH, Koshlap KM, Agris PF. Nucleic Acids Res. 2000;28:1374. doi: 10.1093/nar/28.6.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatterjee AK, Blaby I, Thiaville PC, Majumder M, Grosjean H, Yuan YA, Gupta R, de Crécy-Lagard V. RNA. 2012;18:421. doi: 10.1261/rna.030841.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wurm JP, Griese M, Bahr U, Held M, Heckel A, Karas A, Soppa J, Wohnert J. RNA. 2012;18:412. doi: 10.1261/rna.028498.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grosjean H, de Crécy-Lagard V, Marck C. FEBS Lett. 2010;584:252. doi: 10.1016/j.febslet.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 23.Silva FJ, Belda E, Talens SE. Nucleic Acids Res. 2006;34:6015. doi: 10.1093/nar/gkl739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Crécy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H. IUBMB Life. 2007;59:634. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
- 25.Fabret C, Dervyn E, Dalmais B, Guillot A, Marck C, Grosjean H, Noirot P. Mol Microbiol. 2011;80:1062. doi: 10.1111/j.1365-2958.2011.07630.x. [DOI] [PubMed] [Google Scholar]
- 26.van der Gulik P, Hoff W. J Mol Evol. 2011;73:59. doi: 10.1007/s00239-011-9470-3. [DOI] [PubMed] [Google Scholar]
- 27.Maniloff J. In: Molecular Biology and Pathogenicity of Mycoplasmas. Razin S, Hermann R, editors. Kluwer Academic/Plenum Publisher; New York: 2000. p. 31. [Google Scholar]
- 28.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, Fritchman JL, Weidman JF, Small KV, Sandusky M, Fuhrmann J, Nguyen D, Utterback TR, Saudek DM, Phillips CA, Merrick JM, Tomb JF, Dougherty BA, Bott KF, Hu PC, Lucier TS. Science. 1995;270:397. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 29.Sirand-Pugnet P, Citti C, Barré A, Blanchard A. Res Microbiol. 2007;158:754. doi: 10.1016/j.resmic.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Moya A, Pereto J, Gil R, Latorre A. Nat Rev Genet. 2008;9:218. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- 31.John PM. Curr Opin Microbiol. 2010;13:73. doi: 10.1016/j.mib.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCutcheon JP, Moran NA. Nat Rev Micro. 2012;10:13. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 33.Douglas AE. Cell Host Microbe. 2011;10:359. doi: 10.1016/j.chom.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M, Meyer F, Olsen G, Olson R, Osterman A, Overbeek R, McNeil L, Paarmann D, Paczian T, Parrello B, Pusch G, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marck C, Grosjean H. RNA. 2002;8:1189. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higgs PG, Ran W. Mol Biol Evol. 2008;25:2279. doi: 10.1093/molbev/msn173. [DOI] [PubMed] [Google Scholar]
- 37.Dong H, Nilsson L, Kurland CG. J Mol Biol. 1996;260:649. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 38.Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, Lee SH, Robertson HM, Kennedy RC, Elhaik E, Gerlach D, Kriventseva EV, Elsik CG, Graur D, Hill CA, Veenstra JA, Walenz B, Tubío JMC, Ribeiro JMC, Rozas J, Johnston JS, Reese JT, Popadic A, Tojo M, Raoult D, Reed DL, Tomoyasu Y, Kraus E, Mittapalli O, Margam VM, Li HM, Meyer JM, Johnson RM, Romero-Severson J, VanZee JP, Alvarez-Ponce D, Vieira FG, Aguadé M, Guirao-Rico S, Anzola JM, Yoon KS, Strycharz JP, Unger MF, Christley S, Lobo NF, Seufferheld MJ, Wang N, Dasch GA, Struchiner CJ, Madey G, Hannick LI, Bidwell S, Joardar V, Caler E, Shao R, Barker SC, Cameron S, Bruggner RV, Regier A, Johnson J, Viswanathan L, Utterback R, Sutton GG, Lawson D, Waterhouse RM, Venter JC, Strausberg RL, Berenbaum MR, Collins FH, Zdobnov EM, Pittendrigh BR. Proc Natl Acad Sci USA. 2010;107:12168. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bujnicki JM, Oudjama Y, Roovers M, Owczarek S, Caillet J, Droogmans L. RNA. 2004;10:1236. doi: 10.1261/rna.7470904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moukadiri I, Prado S, Piera J, Velázquez-Campoy A, Björk GR, Armengod ME. Nucleic Acids Res. 2009;37:7177. doi: 10.1093/nar/gkp762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Näsvall SJ, Chen P, Björk GR. RNA. 2007;13:2151. doi: 10.1261/rna.731007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasvall SJ, Chen P, Björk GR. RNA. 2004;10:1662. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuelsson T, Guindy YS, Lustig F, Boren T, Lagerkvist U. Proc Natl Acad Sci USA. 1987;84:3166. doi: 10.1073/pnas.84.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takai K, Okumura S, Hosono K, Yokoyama S, Takaku H. FEBS Lett. 1999;447:1. doi: 10.1016/s0014-5793(99)00255-0. [DOI] [PubMed] [Google Scholar]
- 45.Ledoux S, Olejniczak M, Uhlenbeck OC. Nat Struct Mol Biol. 2009;16:359. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takai K, Yokoyama S. Nucleic Acids Res. 2003;31:6383. doi: 10.1093/nar/gkg839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agris PF. Nucleic Acids Res. 2004;32:223. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atkins JF, Björk GR. Microbiol Mol Biol Rev. 2009;73:178. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grosjean H, Nicoghosian K, Haumont E, Söll D, Cedergren R. Nucleic Acids Res. 1985;13:5697. doi: 10.1093/nar/13.15.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 51.López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R. J Bacteriol. 2011;193:5587. doi: 10.1128/JB.05749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamames J, Gil R, Latorre A, Pereto J, Silva F, Moya A. BMC Evolutionary Biology. 2007;7:181. doi: 10.1186/1471-2148-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Douglas AE, Raven JA. Phil Trans R Soc Lond B Biol Sci. 2003;358:5. doi: 10.1098/rstb.2002.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blanchard A, Browning GF. Mycoplasmas: Molecular Biology, Pathogenicity and Strategies for control. Horizon Scientific Press; Norwich UK: 2005. [Google Scholar]
- 55.Drummond AD, Wilke CO. Nat Rev Genet. 2009;10:715. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, Boniecki MT, Jaffe JD, Imai BS, Yau PM, Luthey-Schulten ZA, Martinis SA. Proc Natl Acad Sci USA. 2011;108:9378. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyerovich M, Mamou G, Ben-Yehuda S. Proc Natl Acad Sci USA. 2010;107:11543. doi: 10.1073/pnas.0912989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novozhilov A, Koonin E. Biology Direct. 2009;4:44. doi: 10.1186/1745-6150-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lowe TM, Eddy SR. Nucl Acids Res. 1997;25:955. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benítez-Páez A, Villarroya M, Douthwaite S, Gabaldón T, Armengod ME. RNA. 2010;16:2131. doi: 10.1261/rna.2245910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Crécy-Lagard V. In: Practical Bioinformatics. Bujnicki J, editor. Springer-Verlag; Berlin Heidelberg: 2004. p. 169. [Google Scholar]
- 63.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. Nucleic Acids Res Symp Series. 2005;33:5691. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.