Abstract
Compounds that bind in the DNA minor groove have provided critical information on DNA molecular recognition, they have found extensive uses in biotechnology and they are providing clinically useful drugs against diseases as diverse as cancer and sleeping sickness. This review focuses on the development of clinically useful heterocyclic diamidine minor groove binders. These compounds have shown us that the classical model for minor groove binding in AT DNA sequences must be expanded in several ways: compounds with nonstandard shapes can bind strongly to the groove, water can be directly incorporated into the minor groove complex in an interfacial interaction, and the compounds can form cooperative stacked dimers to recognize GC and mixed AT/GC base pair sequences.
Keywords: DNA minor groove, heterocyclic amidines, DNA complex structures, surface plasmon resonance, stacked minor groove dimers
INTRODUCTION
Small molecules that bind specifically to the DNA minor groove (MG) have been both influential and experimentally useful in several areas. Many of these compounds, such as DB75, DAPI and Hoechst 33258 in Figure 1, have intense fluorescence when bound to DNA and their fluorescence quantum yield can vary significantly depending on which sequence of the minor groove they bind (Wilson et al., 1990; Karlsson et al., 2003; Stewart et al., 2005; Wheeler et al., 2012). This has made some minor groove binders useful cellular DNA stains as well as sensitive DNA probes for polyacrylamide gel electrophoresis (Karlsson et al., 2003; Stewart et al., 2005; Wheeler et al., 2012). Sequence-specific, minor-groove-binding compounds have also been developed with affinities rivaling transcription factors and the compounds have provided a wealth of information on the fundamental principles of DNA recognition (Lacy et al., 2002; Tidwell and Boykin, 2002; Dervan et al., 2005; Wilson et al., 2005; Nguyen et al., 2007; Raskatov et al., 2012a). Their effects on transcription factors or other cellular DNA targeted proteins have led to the development of MG binders as potential therapeutic agents and animal studies of some compounds are underway (Peixoto et al., 2008; Cai et al., 2009; Shinohara et al., 2010; Raskatov et al., 2012a, b).
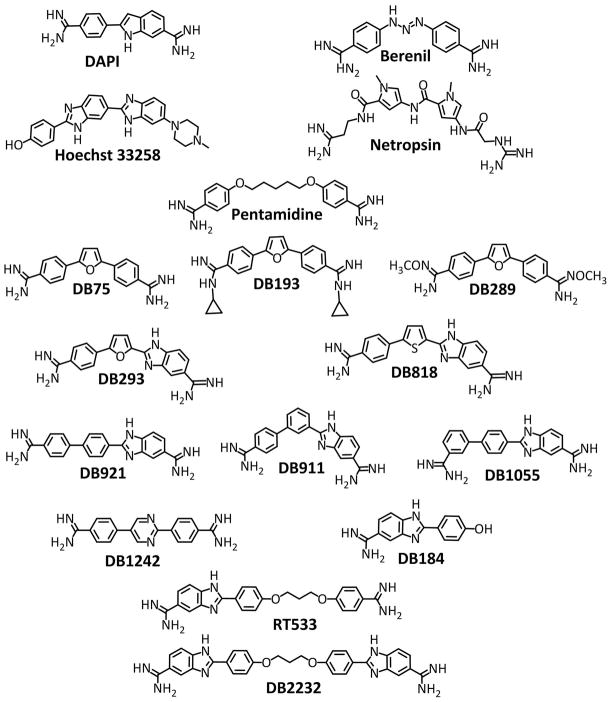
Figure 1.
Compound structures for the heterocyclic compounds discussed in this paper as well as some well-known reference compounds: DAPI, netropsin, Hoechst 33258, berenil and pentamidine. The heterocyclic diamidines are listed from top (DB75) to bottom as classical binders, nonclassical binders, and GC recognizing compounds.
Minor groove binding compounds are also used in humans and animals as effective antimicrobial drugs (Tidwell and Boykin, 2002; Wilson et al., 2005; Werbovetz, 2006; Soeiro et al., 2008; Wilson et al., 2008; Cai et al., 2009; Paine et al., 2010; Raskatov et al., 2012b). The groove-binding mode is preferred over intercalative binding for therapeutics due to increased sequence specificity and generally lower toxicity of groove binders. An additional advantage of MG binding agents for therapy is that they are not generally mutagenic. A therapeutically successful class of MG binders is based on arylamidines such as DB75, pentamidine and berenil (Fig. 1) that have biological activity against parasitic microorganisms (Tidwell and Boykin, 2002; Wilson et al., 2005; Paine et al., 2010). Pentamidine has been used clinically for over 60 years in humans against a number of microorganisms. It is particularly effective against human African trypanosomiasis (HAT) (Werbovetz, 2006; Paine et al., 2010). Although pentamidine has many successful uses, it is not orally available and has some limiting side effects. DB75, furamidine (Fig. 1), was identified as a highly active antitrypanosomal drug in animal studies. In a major breakthrough in HAT, treatment of an oral prodrug of DB75, DB289, pafuramidine (Fig. 1), was synthesized by Boykin and coworkers (Boykin et al., 1996; Bajic et al., 1996; Kumar et al., 1999) and found to be active in animals (Mdachi et al., 2009). The prodrug became the first HAT drug to enter a clinical trial pathway that had to comply with current requirements in drug testing. The clinical trials in Africa were highly successful with over 300 patients cured of HAT as the drug reached Phase III clinical trials (Paine et al., 2010). Unfortunately, in a separate and extended Phase I trial, renal toxicity emerged in a small percentage of the treated group following the last treatment with DB289. Although this has halted the HAT trials of the drug, many more active and specific compounds have been discovered including new compounds that show promising activity against stage II HAT (central nervous system). Because of these successful therapeutic applications of aryl diamidines, as well as, the wealth of information they have provided on DNA molecular recognition, binding induced conformational changes in DNA, and the energetics of binding and hydration, this review will focus on the aryl diamidine class of MG binders. An excellent earlier review in this series (Unit 8.4) focused on synthetic derivatives of minor groove binders with DNA as well as on the applications of the conjugates.
DNA GROOVE SHAPE AND CHEMISTRY
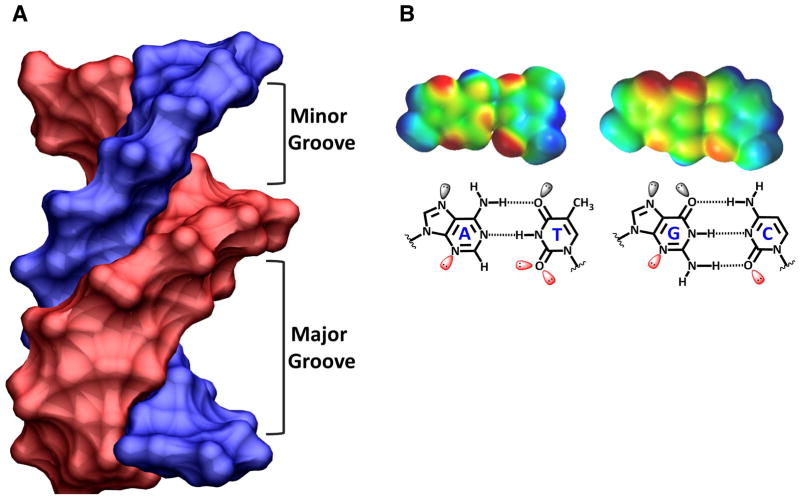
Molecules searching for a groove binding site on DNA find quite different, sequence-dependent, groove widths and functional groups in the minor and major grooves of DNA. A B-form DNA structure (Fig. 2A) and both an AT and a GC base pair (Fig. 2B) are shown to illustrate this point. In the minor groove, the purine N3 of A and G, as well as, the C2-C=O groups of C and T serve as H-bond acceptors. On the GC base pair, however, G has a C2-NH2 group, which is not present on A, which H-bonds with C and forms a steric block to binding of most agents deep into the groove. The groups in the major groove are positioned in a different array. The purine N7 is an H- bond acceptor while both GC and AT base pairs have C=O•••NH2 H-bonds, but the groups face in opposite directions on the base pairs (Fig. 2B). The C5-CH3 group on T has both steric and hydrophobic effects on binding of agents in the major groove, especially when several Ts are positioned closely in the sequence. The space-filling, electron-density maps at the top of Figure 2B clearly show the strong H-bond acceptors (in red - negative potential) and donors (blue - positive potential). The schematic illustrations in the bottom of Figure 2B define the atomic positions in the maps. These groups are arrayed along the bottom of the minor groove in a sequence dependent manner in a DNA double helix (Fig. 3).
Figure 2.
(A) A B-form DNA model with the chains colored blue and pink is tilted so that the width of both the minor and major grooves can be seen and compared. (B) Space-filling models of AT and GC base pairs (top) is shown with electrostatic potential coloring: from blue for positive to red for negative partial charges. The same base pairs are shown as line models (bottom) with the base pairs labeled and lone pair electrons shown on H-bond accepting groups.
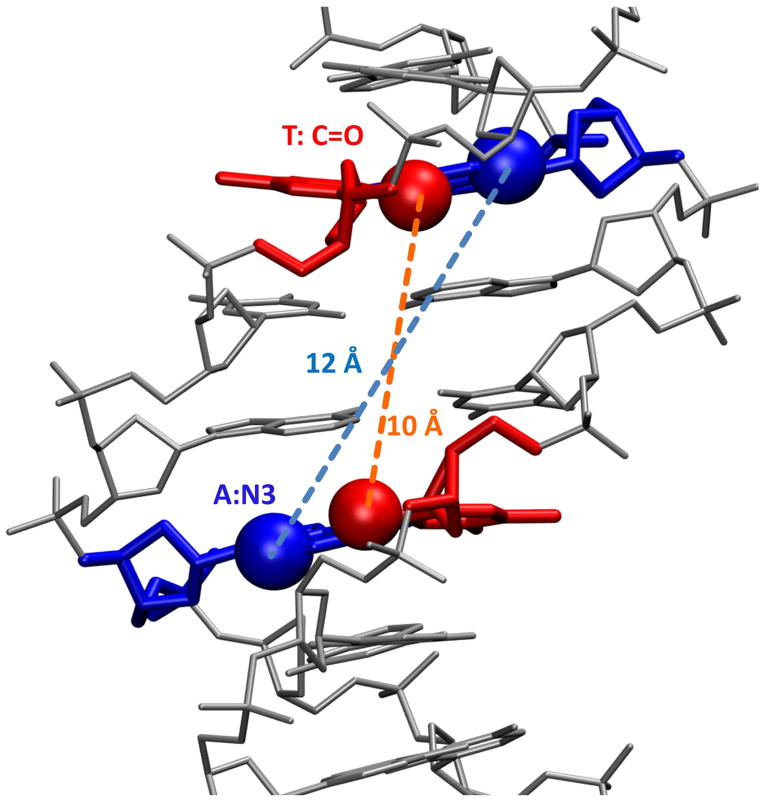
Figure 3.
The minor groove of a B-form DNA model with the central sequence 5′-AATT-3′ shown with the DNA as a gray stick model. The connecting distances between the 5′-As and 3′-Ts on opposite chains are shown.
In A-tract sequences (consecutive As on one strand and Ts on the other) or in shorter sequences such as the AATT in Figures 2A and 3, the minor groove generally assumes a narrow width that provides for tight binding of the heterocyclic-amidine system of the dications in Figure 1, while the groove in GC or mixed base pair sequences is wider (Neidle, 2001; Nguyen et al., 2009). In the narrow AT base pair sequences, single heterocyclic compounds can fit tightly and deeply into the groove such that groups on the inner face of the compounds can interact, typically through H-bonding and van der Waals interactions, with complementary groups on the edges of the AT base pairs (Wilson et al., 1990; Neidle, 2001; Tanious et al., 2007). The position of the H-bond acceptors change with the AT sequence, groove shape and curvature so that even sequences composed of only AT base pairs can be distinguished by bound agents. This is illustrated in the classical B-form structure in Figure 3, for example, where in the terminal base pairs of the central AATT sequence, the N3s of A are separated by over 12 Å while the keto groups of T are separated by less than 10 Å (Nguyen et al., 2009). Such differences dictate the sequences to which compounds with specific H-bond donating groups can bind with high affinity. It should be remembered, however, that there is a certain amount of flexibility in the DNA double helix as well as the unfused heterocycles that are characteristic of most minor groove binders. Induced fit interactions in the minor groove are, thus, quite common (Bostock-Smith, et al., 2001; Miao, et al., 2005; Tanious, et al., 2007; Chaires, 2008; Moretti et al., 2008).
In addition to the position of functional groups at the floor of the groove and the width, the minor groove has a helical curvature that can also vary slightly with sequence. This helical curvature is easily seen in the DNA duplex models at the top in Figures 2 and 3. Agents that bind to the minor groove must, thus, have a concave shape that complements that of the groove. Molecules that bind tightly and specifically to the groove (Fig. 1) will fit well within the walls of the groove (groove width), form optimum H-Bonds with base edges at the floor of the groove (functional groups) and match the local curvature of the groove (compound shape). Again, induced fit changes in both DNA and compounds with unfused heterocyclic systems give some flexibility to all of these recognition features.
The difference in groove width in A-tracts and GC or mixed sequences has been noted above. A number of groups have pointed out, however, that there are more minor, but quite significant, variations in local groove width with sequence (Bishop et al., 2011; Parker and Tullius, 2011). By using sequence dependent variations in hydroxyl radical cleavage of DNA, for example, Tullius and coworkers have established a large database of variations in the minor groove with local sequence (Bishop et al., 2011; Parker and Tullius, 2011). They also have software on the Web for prediction of the local groove width based on their cleavage database (ORChID software: [OH Radical Cleavage Intensity Database]; experimentally determined hydroxyl radical cleavage patterns of naked DNA molecules; includes an algorithm to predict the cleavage pattern of any DNA sequence (http://dna.bu.edu/orchid/). The web page is the direct link to the ORChID database and web server, where a user can predict cleavage patterns for any sequence, download the ORChID1 and ORChID2 code for large-scale predictions (Greenbaum et al., 2007). The Honig group has correlated the variations in groove width with electrostatic potential and the favorable binding affinity of different amino acid side chains, particularly arginine, into the minor groove (Rohs et al., 2009; Bishop et al., 2011). It should be emphasized, however, that the wider groove and different functional groups in GC containing sequences of DNA can also be effective binding sites for minor groove agents, particularly if they can bind as stacked dimers with H-bond accepting groups to interact with G-NH2 functional groups (this feature is discussed in more detail below).
CLASSICAL -AT- SPECIFIC MINOR GROOVE BINDERS
The dicationic polyamide, netropsin, in Figure 1 is the paradigm for classical AT specific MG binders. It was isolated in the early 1950s (Finlay et al., 1951) and found to have some activity against microorganisms and cancer cells. Zimmer and coworkers conducted an extensive series of studies on netropsin in the early 1970s and found its AT specificity, as well as, very different effects on DNA as compared to intercalators (reviewed in Zimmer, 1975). In 1974 Wartell et al. proposed a molecular model of netropsin bound into the minor groove in AT sequences (Wartell et al., 1974). Dickerson and coworkers determined the crystal structure of netropsin bound in the AATT minor groove of the oligomer duplex sequence, d(CGCGAATTCGCG)2 (Kopka et al., 1985) and all of these results firmly established the minor groove binding mode. The netropsin structure was the first detailed molecular level view of a DNA minor groove complex and the same DNA sequence has been used in many other studies of MG binders, including the heterocyclic amidines (Neidle, 2001). The netropsin results established much of the detailed molecular basis of the classical AT specific minor groove binding model: the need for a narrow groove, the steric hindrance that would be caused by the G-NH2 group, the molecular curvature and twist of the bound molecule to match the DNA groove and the advantage of H-bond donating and positively charged groups. This model supplies molecular detail but its overall structure is quite similar to that proposed earlier by Wartell et al. (1974).
Starting in the 1980s our laboratory, with that of Boykin and coworkers (Wilson et al., 1990; Wilson et al., 1993; Tanious, 1994), initiated an extensive series of studies on the minor groove complexes of aromatic diamidines. These studies clearly showed that there are a number of important features of the diamidines that lead to their strong MG binding to DNA as well as their biological activity (Tidwell and Boykin, 2002; Wilson et al., 2005; Nguyen et al., 2007; Wilson et al., 2008; Nguyen et al., 2009). The Neidle group has determined the DNA complex structures of a large number of diamidines (Neidle, 2001). Important molecular features in DNA MG binders, which have been derived from the initial studies of numerous DNA-targeted aromatic amidines, include a crescent shape that complements the helical minor groove, positively charged ends to enhance electrostatic interactions and solubility, H bonds donor/receptors for sequence recognition, and a somewhat flexible structure to allow thermodynamic optimization of the compound-DNA interactions. These features fit the classical model, but as will be shown below some of these requirements, especially the requirement for curvature matching and an AT sequence, can be relaxed in certain special cases. As noted above, the narrow width of the AT minor groove and lack of the third H-bond of GC base pairs is optimum for many MG binders such as those shown in Figure 1. In AT base pair sequences, aromatic amidines can fit into the groove such that groups on the inner face of the compounds can interact, through H-bonding and van der Waals interactions, with complementary groups on the edges of the AT base pairs. The torsional flexibility of the aromatic groups of the compounds lets them make optimum surface contacts with the DNA groups that form the walls of the minor groove receptor site. All of these features of the dicationic compounds increase their affinity and specificity for interaction with sequences of four or more AT base pairs (Neidle, 2001; Nguyen et al., 2007; Wilson et al., 2008).
Neidle and coworkers used the sequence d(CGCGAATTCGCG)2, as with the netropsin structure described above, with a large number of different heterocyclic amidines and their structural results have provided a wealth of detail on how these compounds selectively recognize the minor groove. They determined the structure of the furan-diphenyl-diamidine, DB75 (Fig. 1; Laughton et al., 1996), and a number of analogs (Neidle, 2001). A structure of a derivative with a cyclopropyl substituent on each amidine of DB75 from the Neidle group is shown as an example in Figure 4 (Trent et al., 1996). As with DB75 (Laughton et al., 1996; Nguyen et al., 2009), the amidines form H-bonds with the terminal T-C2 carbonyl groups on opposite chains of DNA (Fig. 4). The compound fits snugly between the walls of the minor groove while the cyclopropyl groups fit closer to the top of the groove and partially overlap with the GC base pairs at the ends of the AATT sequence. The compound excludes water from the groove, particularly from the AATT region, which, as described above, is strongly hydrated in the free DNA structure. It also binds to this sequence slightly better than DB75 and this is no doubt at least partially due to removal of the hydrophobic cyclopropyl groups from the water environment.
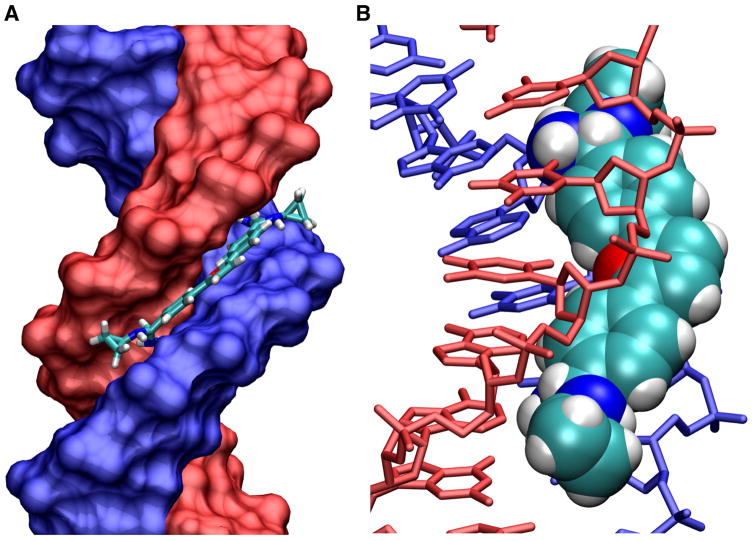
Figure 4.
(A) A space filling crystal model of a cyclopropyl substituted diamidine derivative (DB193 in Fig. 1) bound to an AATT sequences is shown with the DNA strands colored blue and red and the compound shown as a tube model. (B) The complex is shown tilted so that the compound (space filling model) can be seen bound in the DNA (tube model) minor groove.
The Neidle group has continued to provide critical structural information on minor groove binding agents including other alkyl-amidine derivatives of DB75 (Trent et al, 1996; Simpson et al, 2000). The alkyl derivatives from ethyl to cyclopropyl (Fig. 4) are essentially isomorphous. They assume the twist of the minor groove and have a curvature to closely match the groove. They fit between the walls of the narrow AATT groove and both unmodified amidine –NH groups form H-bonds with the edges of the AT base pairs at the floor of the groove, except for the cyclopropyl which forms a single H-bond. All of the alkyl groups fit into the minor groove near the top edge as in Figure 4.
A particularly informative study and structure was with the phenyl-thiophene-benzimidazole (DB818 in Fig. 1) analog of DB75 (Mallena et al., 2004). The phenyl-furan-benzimidazole compound, DB293, is similar to DB75, with conversion of one phenyl to benzimidazole and has a similar binding constant for AATT to DB75 (Wang et al., 2002). DB818 has only a one atom difference from DB293, thiophene to furan, and it was thus a surprise to find that it binds to AATT over 25 times more strongly than DB293. The increase in binding is entirely due to the binding enthalpy which is over 4 kcal/mol more favorable for DB818 while it has a less favorable entropy of binding (Mallena et al., 2004).
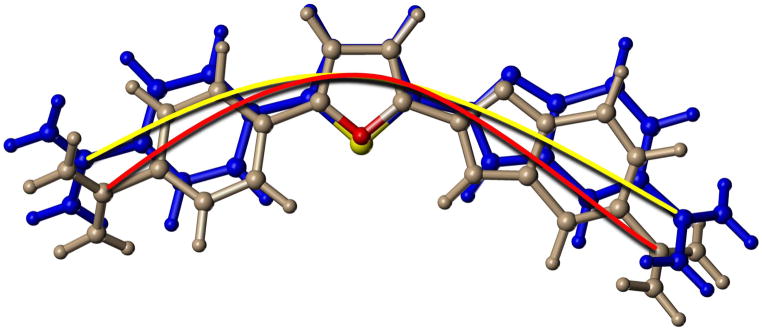
The crystal structure of DB818 and molecular modeling of DB293 for comparison provides an explanation for these observations. The larger size of sulfur and the resulting bond angle change from thiophene to phenyl, DB818, relative to furan-phenyl, DB293, results in an increased radius of curvature for DB818 (Fig. 5). This slightly flattens DB818 relative to DB293 (and other furans) which allows it to fit more optimally against the base pair edges at the floor of the groove. This allows the formation of a better benzimidazole H-bond in DB818 than in DB293 as well as more favorable van der Waals contacts with the floor and walls of the groove (Mallena et al., 2004). DB818, for a molecule of its size, can be viewed as an optimum classical AT specific-binding agent and serves as an excellent example for this class of minor groove compounds (Nguyen et al., 2009). The tight and favorable interactions of DB818 account for both its more favorable binding enthalpy and less favorable entropy. The smaller radius of curvature of DB293 (Fig. 5), due to the furan bond angle, pushes the benzimidazole slightly away from the floor of the minor groove and weakens the benzimidazole-base H-bond.
Figure 5.
An overlay tube model of optimized structures for DB293 (grey) and DB818 (blue) is shown with the furan O in red and the thiophene S in yellow. A red arc connects the amidine carbons of the furan, DB293, while a yellow arc connects the equivalent atoms in the thiophene, DB818. The slight difference in bond angles in the furan and thiophene, due to the size difference of O and S, creates a significant curvature difference between the compounds that gives DB818 a better binding constant for the DNA minor groove. The two arcs clearly show the differences in radius of curvatures for the compounds.
VARIATIONS ON THE CLASSICAL MINOR GROOVE MODEL
Minor Groove Binding and Intercalation of Small Compounds
As described above, the classical model of AT-specific, minor-groove complexes, starting with netropsin and continuing with the diamidines, successfully provided criteria for strong MG interactions. This model works quite well for the diamidine DB75 (Laughton et al., 1996), but we noticed that the compound could also assume a planar shape of the diphenylfuran core that was similar in size to intercalators. We asked the question of what would happen to the binding mode if some or all of the AT base pairs in the binding site were replaced by GC. The answer was quite clear, based on a broad array of biophysical methods, DB75 switches from a groove binder to an intercalator in GC containing sequences (Wilson et al., 1990; Tanious et al., 1994). The compound in 0.1 M NaCl buffer has a KD of ~100 nM in AT binding sites (minor groove) and this increases by over a factor of 100 in GC sequences (intercalation). The cost of opening an intercalation site and loss of favorable MG interactions clearly cause this significant reduction in affinity (Wilson et al., 1990, Tanious et al., 1994). Other similar small and relatively planar AT minor groove binders are also able to intercalate in GC sequences. Compounds that are non-planar or that are too large to fit well between base pairs do not form stable intercalation complexes.
Strong Binding of Compounds with Non-Standard Shapes to the MG
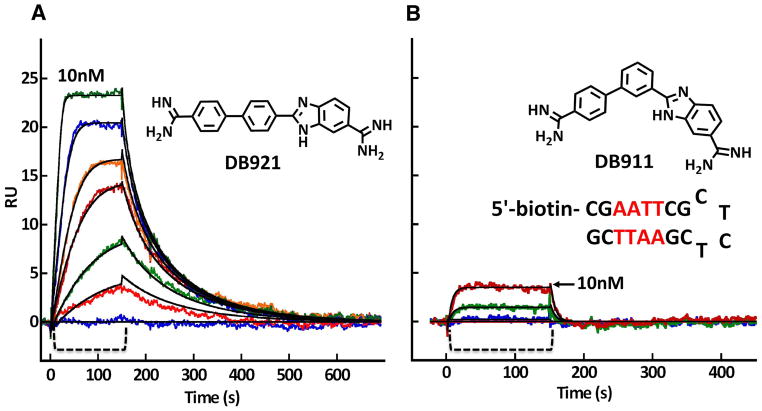
The finding of intercalation of some minor groove binding agents stimulated our interest in another question: how far can we go in the design of nonclassical minor groove binding agents that do not perfectly match the shape of the minor groove, one of the classical requirements. The results described above for DB818 emphasize the importance of shape matching. Understanding the limits of minor groove complex formation by compounds that do not fit the classical model is important for extending our knowledge of the broad aspects of DNA molecular recognition. New types of compounds that target DNA in different ways can also have many fundamental and applied uses. Two isomeric biphenyl-benzimidazole diamidine derivatives, DB911 and DB921 in Figure 1, have complementary and very interesting MG binding results. The central para-substituted phenyl of DB911 has a classical type curvature for MG binding, similar to that of DB75 and related minor groove binding agents. The central para-substituted phenyl of DB921, however, has a much more linear shape that clearly cannot match the curvature of the DNA minor groove. The biphenyl component of both compounds is expected to be significantly twisted. Based on the dogma established through classical groove binders, DB911 should bind in a similar manner to DB75 but DB921 should bind very weakly to the MG.
Biosensor-SPR experiments with these compounds, and an AATT binding site, provided very surprising results for the binding kinetics and affinities (Fig. 6). DB921 binds much more strongly than DB911 (KA of approximately 2 × 108 M−1 for DB921 and 2 × 107 M−1 for DB911) and has much slower dissociation kinetics (kd of 0.014 s−1 for DB921 and 0.24 s−1 for DB911). As expected, DB911 binds very similarly to DB75 but contrary to expectations, DB921 binds more, not less, strongly than these classical compounds. Isothermal titration calorimetry (ITC) of the two compounds showed that the stronger binding of DB921 was due to a more favorable binding enthalpy (Miao et al., 2005; Liu et al., 2011). The ΔH for binding of DB921 to the AATT site is −4.5 kcal/mol at 25 °C while the value for DB911 is only −2.5 kcal/mol. The –TΔS contributions to the binding free energy are more similar, −7.0 kcal/mol for DB921 and −7.5 kcal/mol for DB911. The overall binding, for both compounds, is entropy driven with the AATT site but the favorable binding enthalpy provides a much higher affinity for DB921.
Figure 6.
In a Biacore T100 biosensor surface plasmon resonance (SPR) instrument, the DNA hairpin shown in the figure was immobilized on a Biacore SA streptavidin coated sensor chip by biotin capture. After cleaning the instrument and chip with standard protocols (Nanjunda et al., 2011), buffer flow was started until a stable baseline was obtained. To monitor the compound-DNA association reaction, DB921 (A) and DB911 (B) was then injected over the DNA surface (the bracket region in both panels) at concentrations from 1- 100 nM and binding was monitored by the change in SPR signal (RU) observed in real time. After sample injection, buffer flow was again started and the dissociation reaction was observed. Fitting these curves with a global 1:1 kinetic fit model (black lines through the curves in both panels) provides the association and dissociation rate constants and the equilibrium constants for binding of both compounds (Nanjunda et al., 2011). The binding in the Figure is only shown to 10 nM to illustrate the much stronger binding of DB921. The flow rate in the experiment was 100 μL/min at 25 °C and 0.01 M MES buffer at pH 6.5 with 0.2 M NaCl.
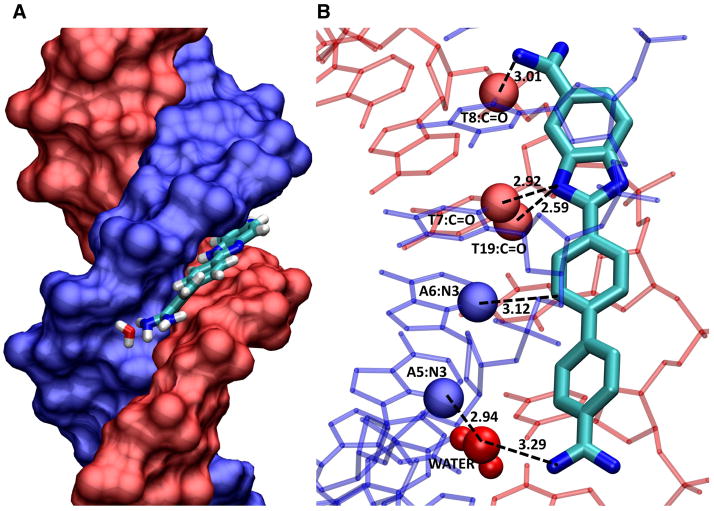
Insight into the reasons for these binding and thermodynamic differences was proved by a crystal structure from the Neidle group (Miao, et al., 2005; Nguyen et al., 2009) and by molecular dynamics calculations (Athri and Wilson, 2009). As expected, DB911 binds to the AATT site in agreement with the dogma from the classical model (see Fig. 4), but DB921 is clearly too linear to bind in this manner. In the crystal structure, the benzimidazole-amidine end of DB921 is anchored strongly to the floor of the minor groove by amidine and benzimidazole H-bonds to the base edges at the floor of the groove (Fig. 7). The central phenyl makes van der Waals contacts with the base edges and walls of the groove. The benzimidazole end of DB921 thus binds in a relatively classical manner. The terminal phenyl and especially the amidine attached to it, however, rise off of the floor of the groove due to the linear structure of DB921 as can be seen in Figure 7B. The molecule is “rescued” as a strong MG complex, however, by an interfacial water molecule that bridges the distance between the floor of the groove and the amidine. An amidine –NH H-bonds to the water O, while an– OH group on the same water forms an H-bond to an AN3 at the floor of the groove. This water is also H-bonded to a network of other waters that are in the same local area of the minor groove (Miao et al., 2005; Athri and Wilson, 2009; Liu et al., 2011). The ternary complex thus forms an optimized set of compound interactions with DNA both directly and through the water which forms a somewhat flexible linker that allows optimization of the interactions. The DB921-DNA and water interactions that stabilize the complex so strongly are illustrated in Figure 7. A combination of H-bonds, van der Waals and favorable electrostatic interactions with DB921, water and DNA, as well as, release of water from the compound and tightly bound water from the minor groove contribute to the affinity through the binding entropy.
Figure 7.
The figure is from a crystal structure by Neidle and coworkers (2B0K. pdb). (A) The DNA duplex is shown in space filling with one strand in blue and one in red. DB921 is shown as large tubes in the AATT minor groove sequence. The phenyl-amidine that rises off the floor of the groove points towards the reader in this view. The interfacial water that connects this end of the DB921 complex to the floor of the groove can be seen near the floor of the groove with important DNA contact atoms in space filling representation, T-C=O in red and AN3 in blue. H-bonds are shown as dashed lines. (B) The contacts that help with the strong affinity of DB921 are highlighted. Starting at the phenyl-amidine end (bottom of the diagram) there is the interfacial water-AN3 H-bond (this is the first A of AATT) and the water is closely H-bonded to other water molecules in the minor groove. The phenyl proton that is meta to the amidine points into the groove and makes a close contact with the interfacial water. The central phenyl has a close –CH ••AN3 contact that certainly provides a stabilizing interaction. The benzimidazole –NH points into the center of the groove and forms a bifurcated H-bond with the two middle T C2=O of AATT. The benzimidazole amidine –NH that points into the groove forms an H-bond with the last T C2=O of AATT. These contacts as well as the van der Waals contacts with the floor and walls of the groove and the amidine positive charges, result in a very high binding for DB921 binding to the AATT sequence. These features could not be seen without the structural model.
To evaluate how the bound water in the DB921 complex contributes to affinity, DB1055 (Fig. 1), which has the terminal amidine group of DB921 moved from the para to the meta position, was synthesized. As expected, crystallographic results show that this structural change removes the need for the interfacial water in the complex (Liu et al., 2011). DB1055 binds slightly more weakly than DB921 (loss of 0.5 kcal/mol in ΔG) with a significant decrease in the binding entropy relative to DB921, but with a more favorable ΔH of binding. The terminal phenylamidine of DB1055 forms a more rigid H-bonded system with the floor of the minor groove with somewhat better stacking with the groove walls than the water-linked complex of DB921. This leads to an improved binding enthalpy for DB1055 but a lower entropy with a slight loss in the Gibbs energy.
Changes such as that from DB921 to DB1055 have frequently been considered as a way to improve binding energetics through replacement of a linking water with a fixed compound group and a subsequent improvement of binding entropy. The results with DB1055 do show that it is able to replace the interfacial water of DB921, but with slightly less favorable energetics. This result illustrates that a dynamic and flexible linking water molecule can be energetically favorable in a complex. Clearly incorporation of water, at least near the end of DNA minor groove complexes, can be favorable and is a feature that deserves more attention.
EXTENSION TO GC BASE PAIR RECOGNITION
The Move of Minor Groove Binding From All AT to GC Recognition
The remarkable success with the human genome project not only opened a broad horizon of possibilities to further enhance our fundamental understanding of DNA and its associated biochemical processes but also to explore novel strategies to effectively use DNA as a practical therapeutic target. The decoding of the human genome at the nucleotide level also brought attention to the presence of a high GC-content (almost 40%) interspersed within the AT-rich genome. GC-rich sequences are dispersed within many biologically important stretches of DNA and are shown to play pivotal as well as accessory roles in various biological pathways (Zerial et al., 1986; Gardiner-Garden and Frommer, 1987; Gruss et al., 1991; Kim and Tinoco, 2000; Zhang et al., 2004 and Khuu et al, 2007). Another important guanine-mediated non-canonical DNA target that is gaining high therapeutic interest is the highly stable G-quadruplex structure (reviewed in Unit 17.2; Balasubramanian et al., 2011). In addition, locally repeated guanine sequences are found throughout the human genome and have been shown to fold into a plethora of G-quadruplex architectures (Burge et al., 2006; Patel, Phan and Kuryavyi, 2007; Balasubramanian et al., 2011; Petraccone et al., 2011). An overwhelming rise in the evidence of their potential role in several important biochemical pathways also provides an additional platform to develop G-quadruplex mediated therapeutic agents (Balasubramanian and Neidle, 2009). The complementary cytosine-rich strand of the duplex is shown to fold into i-motif structures under unique physiological conditions and has also gained prominence as a potential therapeutic platform (Brooks et al., 2010).
The insertion of GC-base pairs within AT-rich DNA significantly alters the physical and chemical characteristics of the sequence. GC-containing sequences have higher thermal stability because of their increased base-stacking potential and partly due to the additional H-bond within the GC base pair. As described above, GC-containing sequences also significantly alter the groove dimensions (both major and minor) of DNA. Contrary to the narrow and deep minor groove characteristics of AT-containing sequences, the grooves of GC-containing sequences are wider with decreased electronegative potential and have the third base pair in the minor groove. The fact that the G-NH2 group constitutes a critical negative recognition element for binding of many small molecules in the minor groove of DNA has now been unambiguously demonstrated using modified DNA bases in which the amino group has been either deleted from guanines and/or added to adenines (Waring and Bailly, 1994; Bailly and Waring, 1995). Given both the strategic position of the amino group in the minor groove and its potential to participate in hydrogen bonding, it was proposed that the introduction of an H-bond acceptor heteroatom in the pyrrole rings of netropsin might allow the drug to bind to GC-containing sequences (Goodsell and Dickerson, 1986; Lee et al., 1988, Lee et al., 1989; Mrksich et al., 1992; Dervan et al., 2005). The wider groove width of GC sequences can also favor the formation of stacked heterocyclic complexes within the groove and also to target both strands of DNA with improved selectivity. These concepts have been extensively exploited and developed into a series of polyamides that have increased selectivity for GC-containing sequences (Walker et al., 1997; Kielkopf et al., 1998; Reddy et al., 1999). Despite the elegant design strategy for GC-specific sequence recognition using polyamides, the biological activity of this class of minor groove binders has not yet led to clinically useful drugs although a number of promising developments have recently been described (Dervan et al., 2005; Franks et al., 2010; Edwards et al., 2011; Pandian et al. 2011; Raskatov et al., 2012b).
One of the major targets of heterocyclic dications is the mitochondrial kinetoplast-DNA. The AT-rich minicircles of kDNA are separated by short fragments of GC base pairs and are an optimum platform to develop small molecules to recognize mixed DNA sequences (Shapiro and Englund, 1995; Liu et al., 2005). Small molecules that have the appropriate features to recognize AT stretches as well as the GC motifs provide further understanding to develop selective ligands with increased therapeutic potential. In an effort to expand the repertoire of heterocyclic diamidines for duplex DNA recognition beyond the AT sequences, several new diamidines were rationally designed and developed using DB75 as the platform molecule. Systematic modifications were performed on the different ring systems of DB75 to give modules that were hypothesized to specifically recognize the potential H-bond donors and acceptors of GC motifs while still maintaining the AT-specific dicationic charge system. The remainder of this section will provide a brief overview of three representative classes of heterocyclic dications that were shown to recognize a wide range of DNA sequences with different GC composition. The synthetic accessibility and the excellent cell uptake of this class of minor groove binders provides alternate ways to better understand and establish newer DNA recognition principles and also to develop the next generation of ligands with better therapeutic potential.
Dimers of Dicationic Heterocyclic Amidines can Recognize GC Base Pairs
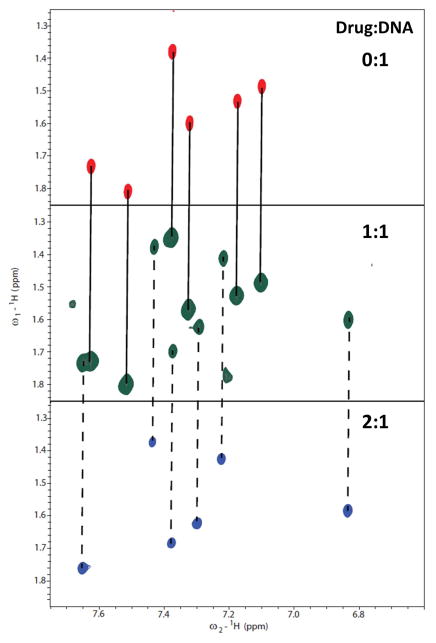
Interestingly, the initial breakthrough in recognizing a GC-containing sequence was provided by a simple substitution of one of the phenyl moieties of DB75 with a benzimidazole ring system, DB293 (Fig. 1). DNase I footprinting studies of DB293 showed a strong footprint with a mixed DNA sequence containing a single central GC step, 5′-ATGA-3′ (Wang et al., 2000; Wang et al., 2001). The larger benzimidazole ring of DB293 decreases the curvature of the ligand when compared to DB75 and also increases the stacking surface. The benzimidazole unprotonated nitrogen or the furan- oxygen atom can also potentially participate in H-bonding with the G-NH2 group. Thermal melting studies of DB293 with an oligomer containing the -ATGA- motif increased the thermal stability of the DNA by almost 30 °C suggesting a high stabilization potential by the ligand. Interestingly, the melting temperature values do not saturate until a compound:DNA ratio of 2:1 is reached and the intermediate ratio of 1:1 exhibited a biphasic transition in the melting curve with the two transition values coinciding with the Tm values of free DNA and the 2:1 complex. This behavior in the thermal melting studies suggests that the recognition of GC-containing DNA by DB293 is characterized by the formation of a stable dimer complex in the minor groove. Surface plasmon resonance studies with the -ATGA- oligomer further showed the formation of a strong, positively cooperative dimer by DB293 with the binding affinity of the second ligand ~25-fold stronger than the first ligand (K1 = 2.8 × 106 M−1; K2 = 7.3 × 107 M−1). Two dimensional proton NMR titrations of DB293 with an -ATGA- containing oligomer further confirmed the highly cooperative dimer formation (Fig. 8). The COSY spectra at the intermediate ratio of 1:1 contained peaks from the 2:1 complex and the free DNA establishing the positive cooperativity.
Figure 8.
Two-dimensional COSY spectra of the -ATGA- sequence and with DB293 complex showing the T-CH3 and T-H6 correlations at different ratios of DB293. The DNA sequence contains six thymine residues and all the six methyl-aromatic proton scalar couplings are observed in the absence of DB293 (0:1, top). At the intermediate ratio (1:1, middle), a doubling in the number of signals is observed suggesting the presence of two distinct species. One set of these peaks correspond to the free DNA (indicated with solid lines between 0:1 and 1:1). The other set of peaks correspond to the 2:1 complex based on the spectra at 2:1 ratio (bottom, indicated with broken lines between 1:1 and 2:1). The absence of any free DNA cross peaks at 2:1 highly suggests the complete saturation of free DNA at 2 molar equivalents of DB293. The presence of two distinct sets of signals at the intermediate ratio of 1:1 for the free DNA and the 2:1 species clearly shows the formation of a strong and highly cooperative stacked complex by DB293 with the –ATGA– sequence as also observed in SPR studies.
High resolution 2D-NMR studies of DB293 with the -ATGA- sequence further revealed several interesting DNA recognition features upon complex formation. The unsymmetrical phenyl-furan-benzimidazole units stacked in an antiparallel orientation in the minor groove of -ATGA- with several NOESY cross peaks with important minor groove protons. Strong cross peaks were also observed between the two stacked molecules of DB293 and both DNA strands suggesting the sequence-specific recognition of both strands. The discovery of the heterocyclic dication, DB293, forming a highly cooperative, antiparallel stacked dimer with a GC-containing mixed DNA was very significant because this was the first instance where a heterocyclic diamidine was shown to recognize a non-AT DNA sequence and other dications such as netropsin have not been discovered to form stacked dimers with any DNA sequences. The dimer with a local charge of +4 was unusual and a surprise (Wang et al., 2000; Wang et al., 2001). Several analogs of DB293 have been designed with better DNA binding affinities. The stacked-dimer recognition mode of these ligands is completely different from the traditional DNA recognition exhibited by lexitropsins and offers new ideas for developing new agents to recognize diverse motifs.
DB293 provided the initial breakthrough that was required for developing compounds targeting GC-containing sequences. The information obtained not only aids in developing better synthetic strategies for the ligands in a cost-effective manner but also provides useful information about targeting DNA sequences per se. The binding sequence for DB293 contained a GC base pair, however, it also contained multiple AT base pairs. Moreover, DB293 was also found to bind strongly to pure AT-rich sequences where it exhibited a classical monomer mode of binding (see above).
Heterocyclic Amidine Dimers that Recognize a Majority GC Base Pair Sequence
Compounds specifically developed to target GC-containing motifs should ideally exhibit very low to zero selectivity for AT-rich sequences. In addition, GC-specific ligands should be able to target a small number of continuous GC base pair steps -- a commonly observed theme in biologically important genomic regions. As mentioned earlier in this section, the minor groove characteristics of GC-containing sequences differ significantly from AT-rich DNA. To design compounds that are selective only to GC motifs therefore requires reevaluating the design strategies for ligands. Heterocyclic diamidines that have unusual shape and geometry that do not match the curvature of minor groove, surprisingly, have been shown to target AT-rich sequences with excellent binding affinities (see DB921 description above). Such compounds can be used as paradigms to test the hypothesis of linear-shaped ligands for multiple GC recognition. Binding of small molecules in the narrow minor groove of AT-rich sequences is generally entropically driven due to the release of the structured water from the spine of hydration upon complex formation. The wider grooves of GC-rich sequences and the bulky -NH2 group of guanines disrupt the formation of any such ordered water complexes. Therefore, designing small molecules to recognize such GC-rich sequences can be achieved by incorporating H-bonding partners at appropriate positions.
Using this shape-based design strategy, a series of linear heterocyclic diamidines that did not match the curvature of DNA minor groove were rationally designed, and nitrogen atoms were strategically placed at different positions that were proposed to be useful in recognizing important minor groove elements of GC-rich motifs (Munde et al., 2007). One compound that provided the necessary breakthrough in recognizing multiple GC base pairs using this design strategy was a linear, unfused, tricyclic diamidine, DB1242 (Fig. 1). The amidine-phenyl-pyrimidine-phenyl-amidine moieties of DB1242 were para-substituted to each other giving the overall molecule a very linear shape and with zero radius of curvature. DNase I footprinting studies showed the compound exhibited a strong footprint for a unique GC-rich sequence, 5′-GCTCG-3′ (Munde et al., 2007). Interestingly, no detectable footprint was seen for DB1242 with any of the AT-containing sequences even up to high ligand concentrations (5 μM). This was very promising since this was the first instance where a non-polyamide was shown to target a GC-rich motif and with very high selectivity over AT-containing sequences. This also suggested that DB1242 has a unique interaction mode with the -GCTCG-sequence considering the linear shape of the ligand.
Surface plasmon resonance studies of DB1242 with the cognate sequence interestingly showed a very strong and highly cooperative dimer formation (Munde et al., 2007). In this case, the binding affinity for the second DB1242 molecule was much stronger than the first (K1 = 2.0 × 104 M−1; K2 = 9.1 × 106 M−1). Moreover, the binding affinity of DB1242 for the AATT sequence was very weak (K = 3.1 × 105 M−1) as compared to the affinity of DB293 with the AATT sequence (K = 2.1 × 107 M−1). Based on the revised design principles, not only was DB1242 able to target sequences with multiple GC motifs but also was significantly selective over AT-rich sequences. Detailed molecular modeling studies of DB1242 with the -GCTCG- sequence revealed very important interactions between the two ligands and the DNA that favored the formation of a very uniquely stacked dimer in the minor groove in a sequence-dependent manner (Fig. 9, also Fig. 7 in Munde et al., 2007). The two molecules stack in an anti-parallel orientation with the terminal amidines of the two ligands extending to the edges of the binding site as well as being located deep in the minor groove. Several inter-ligand and DNA-ligand H-bonds are observed in the models that help in the overall complex stability. The amidine units of both ligands make strong H-bonds with the -C=O groups of cytosines throughout the binding site. Also, the inner facing nitrogens of both pyrimidine rings make H-bonds with the -NH2 group of different guanines in the minor groove. The overall stacked geometry of the two linear ligands match the curvature of the minor groove with the overlapped region of the ligands containing the phenyl rings optimally stacked on each other and the protruded amidine moieties making H-bonds with the outer facing nitrogen of pyrimidine rings on the other molecule of the stacked dimer (Fig. 9). The overall stabilization of the complex predominantly guided by H-bond interactions also explains the largely enthalpy-driven complex formation observed in calorimetry (Munde et al., 2007).
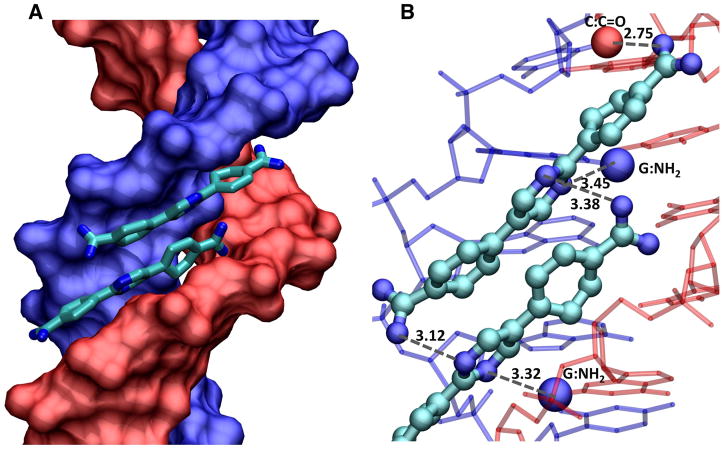
Figure 9.
A docked model for the stacked complex of DB1242 with the 5′-GCTCG-3′ sequence is shown. (A) The DNA helix is shown in a space filling model with one chain in blue and one in red. The two DB1242 molecules are shown as tube models with CPK color schemes. Hydrogens are not shown for clarity. (B) Several of the major interactions that help in the stabilization of the complex are highlighted. The amidine of the top molecule (bottom of the figure) makes strong contact with the outer nitrogen of the pyrimidine of the bottom molecule. The inner nitrogen of the same pyrimidine makes strong contacts with the G-NH2 in the minor groove with the 5′G of GCTCG (red colored DNA strand). Similar interactions are observed with the amidine of the bottom molecule to the pyrimidine nitrogens of the top molecule and to the G-NH2 of the first G of the 5′-CGAGC-3′ sequence of the complementary strand (blue colored DNA strand). The other amidine of the top molecule (top of the figure) makes a strong interaction with the carbonyl group in the minor groove of the first C in -CGAGC- (blue strand). All the DNA-ligand and the inter-ligand interactions help in the formation of a very strong and highly cooperative dimer in the wider minor groove of this DNA sequence.
Designed Compounds that Recognize Two AT Sites Separated by One or More GC Base Pairs
While DB293 and DB1242 provided the breakthrough in designing compounds that can target single as well as multiple GC units as non-covalent stacked motifs, covalently linking such molecules using simple linker systems would establish new DNA recognition principles. Such covalently linked monomer units can either fold into hairpin like structures, very similar to hairpin polyamides, and form a stacked complex within a single binding site, or can remain as linear elongated structures and recognize relatively longer DNA sequences with multiple binding sites. The idea behind such a design strategy is that even ligands with relatively weak affinity for a single site can synergistically bind to multiple target sites on a stretch of DNA and induce conformational changes that would be sufficient to elicit some level of desired biological response.
The AT-rich minicircles of kDNA are an excellent starting point to test for such compounds since the AT-rich sites are frequently separated by a small number of GC base pairs. The linking units, with enough conformational flexibility, can fold-back, recognize or “jump-across” the intervening GC base pairs. Considering the cost of synthesizing complex organic molecules can always be a limiting factor, simple, well-characterized systems that can target AT-rich sequences are excellent starting points. As shown above, the amidine-benzimidazole-phenyl (ABP) moieties are very effective in recognizing AT-rich sites and, therefore, were incorporated into the design strategy as the primary recognition units. A series of linked, symmetrical ABP units were rationally designed with varying linker lengths (-O-(CH2)n-O-, n = 3, 4, 5: Fig. 10) (Liu et al., 2012). Also, a series of DNA sequences mimicking the AT-rich minicircles of kDNA, with a varying number of intervening GC-base pairs, were designed for this purpose. Surface plasmon resonance and mass spectrometry results quite interestingly show that the number of GC base pairs can significantly influence the interaction mode (Liu et al., 2012). Detailed molecular modeling studies using experimental results provided an explanation for the different binding modes observed with different DNA sequences and the results are summarized below.
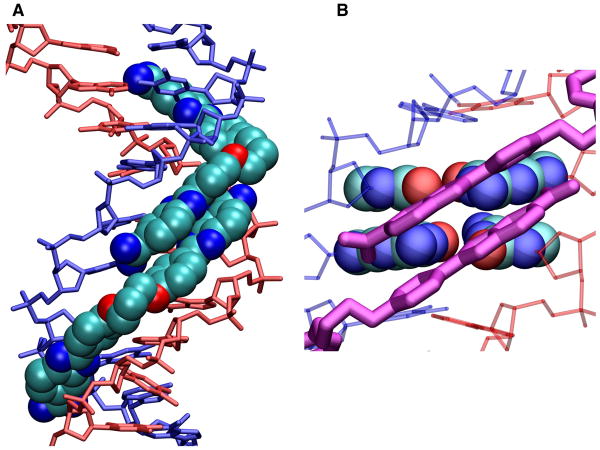
Figure 10.
A model for the stacked complex of DB2232 docked into the AATTGCAATT sequence is shown. (A) The DNA helix is shown as a tube model with one chain in blue and one in red. DB2232 is shown in space fill with carbons in light blue, nitrogens in dark blue and oxygens in red. Hydrogens are not shown for clarity. (B) The two central GC base pairs are shown in space filling representation with the same atom colors as in (A). The two stacked amidine-benzimidazole-phenyl modules are shown as purple tube models. The other DNA base pairs and backbone atoms are shown as thin red or blue tubes. The close interactions of the compounds with the GC base pairs and widened minor groove to accommodate the stacked modules are easily seen in this view. In the AATT sequences only single modules fit snugly into the narrow A-tract type groove structure. This model clearly provides the rationale for the 2:1 model of DB2232 at AATTGCAATT.
The control compound, DB184 (Fig. 1), with a mono-ABP moiety, as expected, binds as a weak monomer to a single-site DNA and as a non-cooperative dimer to the two-site sequences regardless of the number of GC base pairs (Liu et al., 2012). An asymmetric compound, RT533, with a single ABP module linked to a phenyl-amidine unit, had a 100-fold stronger affinity to the single AATT sequence when compared to the control compound, DB184. The phenyl-amidine moiety of RT533 can fold back and favorably stack on the benzimidazole-phenyl module resulting in increased affinity. With the sequences that have intervening GC base pairs, RT533 formed a 2:1 complex suggesting that the compound was binding to both AATT sites. With one and two intervening GC base pairs, RT533 shows negative cooperativity suggesting the two compounds bind too close at the two AATT sites to form a favorable, fold-back type complex. As the number of intervening GC base pairs was increased, the negative cooperativity decreased and with very long GC spacer lengths no cooperativity was observed.
The symmetric compounds with two ABP units exhibit a large variation in the binding mode and affinities with the different sequences (Liu et al., 2012). However, no significant differences in the affinities among the compounds are observed as the linker length is varied, and therefore, only the simplest molecule, DB2232 with an -O-(CH2)3-O- linker, is discussed here. The different modes of binding observed with DB2232 can be extrapolated to the other compounds with varying linker lengths. Interestingly, DB2232 binds very weakly to the single AATT sequence as a 1:1 complex when compared to the asymmetric RT533. This is because of the limited conformational flexibility imposed on DB2232 by the narrow minor groove of the AATT sequence. The two ABP units of DB2232 are too large to favorably stack on each other in the minor groove. With the two-site sequence separated by a single GC base pair, DB2232, however, forms a very strong 1:1 complex. This is very interesting since RT533 was able to form a 2:1 complex with the same sequence. Molecular modeling studies showed that DB2232 can bind to the AATTGAATT sequence with the two APB units stacking favorably in the two AATT sites while the linker spans over the single GC site (Liu et al., 2012).
As the number of GC base pairs is increased, quite surprising binding results are obtained. SPR and mass spectral studies show the formation of a very strong 2:1 complex with the AATTGCAATT sequence with the binding affinity of the second molecule much higher than the first ligand (K1 = 1.1 × 106 M−1; K2 = 9.4 × 108 M−1). Detailed molecular modeling studies provided a satisfactory explanation for the unusual dimer formation by DB2232 with this sequence (Fig. 10). The presence of the two GC base pairs between the AATT sites creates a wider groove that is energetically favorable to accommodate a stacked complex. The two molecules of DB2232 bind in an offset manner with the maximum overlap region of the two molecules located in the widest part of the minor groove, with the two GC base pairs (Fig. 10B) and the non-overlapped ABP moieties of DB2232 favorably located in the narrower AATT site. The offset orientation also puts the terminal amidines of the two molecules away from each other and decreases the electrostatic repulsion while forming optimum cation-π interactions with the inner phenyl rings of the stacked ligand. The complex is, thus, able to form a significant number of favorable interactions with base edges at the floor of the minor groove as well as with the groove walls. Other docked models of the 2:1 complex are less favorable than the overlapped model in Figure 10.
CONCLUSION AND PROSPECTS
From the discovery of netropsin in the early 1950s and the beginning studies with DNA by a number of pioneer investigators, the model of AT-specific minor groove binders was developed. Extensive synthetic efforts have provided a variety of heterocyclic diamidines that have provided clinically useful agents, extensive structural models for minor groove complexes and a greatly expanded understanding of DNA molecular recognition. The synthetic efforts also produced new types of derivatives that helped to show that the original model for AT specific minor groove binders was far too limited. The shape recognition requirement was shown to be too limited by very strong minor groove binding compounds that have a linear structure. It is highly likely, based on these results, that an entirely new class of minor groove binders can be developed that incorporate a bound water molecule into their DNA minor groove complex and form a very strong ternary complex. The requirement for an all AT base pair binding site has also been shown to be too limiting. Cooperative stacked dimers of a heterocyclic diamidine that actually have a total charge of +4 can bind into GC containing sequences better than they bind to pure AT sequences. Developments in this area are at the beginning stages and will certainly continue to produce new ways to recognize the minor groove. Developments of the compounds as transcription factor inhibitors and activators are also at an early stage and offer new agents and approaches in chemical biology. Finally, the heterocyclic diamidines have a record of clinical usefulness and they should continue to produce exciting new results in this critical area.
Acknowledgments
Our research in the area of minor groove binding agents for therapeutics and chemical biology has been generously supported by NIH for a number of years and is currently supported by NIAID grant AI064200. A number of collaborators and coworkers have made key contributions without which the research described in this overview would not have been possible. Although their names are listed in the references, we would like to call special attention to a few key scientists. Professor David Boykin and his coworkers have been equally responsible for all of the results and developments described in this paper. Without their compound design and synthesis efforts, these studies would not have been possible. The structures provided by Professor Stephen Neidle and his coworkers have frequently provided the missing link to help us understand puzzling results that continue to take our understanding of minor groove interactions to a new level. Professors Richard Tidwell and Moses Lee have also provided exciting new minor groove binders over many years that have helped to expand our understanding of minor groove interactions. The footprinting and biological studies of Drs. Christian Bailly and Marie-Hèlène David-Cordonnier have directed our attention to new minor groove recognition possibilities and provided the key information for GC base pair recognition. Not only are these collaborators outstanding scientists but they have also all become good friends and have certainly made our research much more enjoyable for many years.
LITERATURE CITED
- Athri P, Wilson WD. Molecular dynamics of water-mediated interactions of a linear benzimidazole-biphenyl diamidine with the DNA minor groove. J Am Chem Soc. 2009;131:7618–7625. doi: 10.1021/ja809249h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C, Tardy C, Wang L, Armitage B, Hopkins K, Kumar A, Schuster GB, Boykin DW, Wilson WD. Recognition of ATGA sequences by the unfused aromatic dication DB293 forming stacked dimers in the DNA minor groove. Biochemistry. 2001;40:9770–9779. doi: 10.1021/bi0108453. [DOI] [PubMed] [Google Scholar]
- Bailly C, Waring MJ. Transferring the purine 2-amino group from guanines to adenines in DNA changes the sequence-specific binding of antibiotics. Nucleic Acids Res. 1995;23:885–892. doi: 10.1093/nar/23.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic M, Kumar A, Boykin DW. Synthesis of 2,5-bis-(4-cyanophenyl)-furan. Heterocycl Commun. 1996;2:135–140. [Google Scholar]
- Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr Opin Chem Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop EP, Rohs R, Parker SCJ, West SM, Liu P, Mann RS, Honig B, Tullius TD. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS Chem Biol. 2011;6:1314–1320. doi: 10.1021/cb200155t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock-Smith CE, Harris SA, Laughton CA, Searle MS. Induced fit DNA recognition by a minor groove binding analogue of Hoechst 33258: fluctuations in DNA A tract structure investigated by NMR and molecular dynamics simulations. Nucleic Acids Res. 2001;29:693–702. doi: 10.1093/nar/29.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boykin DW, Kumar A, Bender BK, Hall JE, Tidwell RR. Anti-pneumocystis activity of bis-amidoximes and bis-O-alkylamidoximes prodrugs. Bioorg Med Chem, Lett. 1996;6:3017–3020. [Google Scholar]
- Brooks TA, Kendrick S, Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Gray PJ, Jr, Von Hoff DD. DNA minor groove binders: Back in the groove. Cancer Treat Rev. 2009;35:437–450. doi: 10.1016/j.ctrv.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Chaires JB. Allostery: DNA does it, too. ACS Chem Biol. 2008;3:207–209. doi: 10.1021/cb800070s. [DOI] [PubMed] [Google Scholar]
- Dervan PB, Doss RM, Marques MA. Programmable DNA binding oligomers for control of transcription. Curr Med Chem - Anticancer Agents. 2005;5:373–387. doi: 10.2174/1568011054222346. [DOI] [PubMed] [Google Scholar]
- Edwards TG, Koeller KJ, Slomczynska U, Fok K, Helmus M, Bashkin JK, Fisher C. HPV episome levels are potently decreased by pyrrole-imidazole polyamides. Antiviral Res. 2011:91177–186. doi: 10.1016/j.antiviral.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks A, Tronrud C, Kiakos K, Kluza J, Munde M, Brown T, Mackay H, Wilson WD, Hochhauser D, Hartley JA, Lee M. Targeting the ICB2 site of the topoisomerase IIalpha promoter with a formamido-pyrrole-imidazole-pyrrole H-pin polyamide. Bioorg Med Chem. 2010;18:5553–5561. doi: 10.1016/j.bmc.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Finlay AC, Hochstein FA, Sobin BA, Murphy FX. Netropsin, a new antibiotic produced by a Streptomyces. J Am Chem Soc. 1951;73:341–343. [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Goodsell D, Dickerson RE. Isohelical analysis of DNA groove-binding drugs. J Med Chem. 1986;29:727–733. doi: 10.1021/jm00155a023. [DOI] [PubMed] [Google Scholar]
- Greenbaum JA, Pang B, Tullius TD. Construction of a genome-scale structural map at single-nucleotide resolution. Genome Res. 2007;17:947–953. doi: 10.1101/gr.6073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A, Moretto V, Ehrlich SD, Duwat P, Dabert P. GC-rich DNA sequences block homologous recombination in vitro. J Biol Chem. 1991;266:6667–6669. [PubMed] [Google Scholar]
- Hamilton PL, Arya DP. Natural product DNA major groove binders. Nat Prod Rep. 2012;29:134–143. doi: 10.1039/c1np00054c. [DOI] [PubMed] [Google Scholar]
- Karlsson HJ, Eriksson M, Perzon E, Akerman B, Lincoln P, Westman G. Groove-binding unsymmetrical cyanine dyes for staining of DNA: Syntheses and characterization of the DNA-binding. Nucleic Acids Res. 2003;31:6227–6234. doi: 10.1093/nar/gkg821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuu P, Sandor M, DeYoung J, Ho PS. Phylogenomic analysis of the emergence of GC-rich transcription elements. Proc Natl Acad Sci USA. 2007;104:16528–16533. doi: 10.1073/pnas.0707203104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Baird EE, Dervan PB, Rees DC. Structural basis for G.C recognition in the DNA minor groove. Nat Struct Biol. 1998;5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]
- Kim CH, Tinoco I. A retroviral RNA kissing complex containing only two G C base pairs. Proc Natl Acad Sci USA. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka ML, Yoon C, Goodsell D, Pjura P, Dickerson RE. Binding of an antitumor drug to DNA. Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J Mol Biol. 1985;183:553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- Kumar A, Stephens CE, Boykin DW. Palladium catalyzed cross-coupling reactions for the synthesis of 2,5-disubstituted furans. Heterocycl Commun. 1999;5:301–304. [Google Scholar]
- Lacy ER, Madsen ER, Lee M, Wilson WD. Dicationic DNA minor groove binders as antimicrobial agents. In: Demeunynck M, Bailly C, Wilson WD, editors. DNA and RNA Binders, Vol. 1: From small molecules to drugs. WILEY-VCH; Weinheim: 2002. pp. 414–460. [Google Scholar]
- Laughton CA, Tanious F, Nunn CM, Boykin DW, Wilson WD, Neidle S. A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity. Biochemistry. 1996;35:5655–5661. doi: 10.1021/bi952162r. [DOI] [PubMed] [Google Scholar]
- Lee M, Hartley JA, Pon RT, Krowicki K, Lown JW. Sequence specific molecular recognition by a monocationic lexitropsin of the decadeoxyribonucleotide d-[CATGGCCATG]2: structural and dynamic aspects deduced from high field 1H-NMR studies. Nucleic Acids Res. 1988;16:665–684. doi: 10.1093/nar/16.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Krowicki K, Shea RG, Lown JW, Pon RT. Molecular recognition between oligopeptides and nucleic acids. Specificity of binding of a monocationic bis-furan lexitropsin to DNA deduced from footprinting and 1H NMR studies. J Mol Recognit. 1989;2:84–93. doi: 10.1002/jmr.300020206. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chai Y, Kumar A, Tidwell RR, Boykin DW, Wilson WD. Designed compounds for recognition of 10 base pairs of DNA with two at binding sites. J Am Chem Soc. 2012;134:5290–5299. doi: 10.1021/ja211628j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kumar A, DePauw S, Nhili R, David-Cordonnier MH, Lee MP, Ismail MA, Farahat AA, Say M, Chackal-Catoen S, Batista-Parra A, Neidle S, Boykin DW, Wilson WD. Water-me diated binding of agents that target the DNA minor groove. J Am Chem Soc. 2011;133:10171–10183. doi: 10.1021/ja202006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Mallena S, Lee MP, Bailly C, Neidle S, Kumar A, Boykin DW, Wilson WD. Thiophene-based diamidine forms a “super” at binding minor groove agent. J Am Chem Soc. 2004;126:13659–13669. doi: 10.1021/ja048175m. [DOI] [PubMed] [Google Scholar]
- Mdachi RE, Thuita JK, Kagira JM, Ngotho JM, Murilla GA, Ndung’u JM, Tidwell RR, Hall JE, Brun R. Efficacy of the novel diamidine compound 2,5-bis(4-amidinophenyl)- furan-fis-O-methlylamidoxime (pafuramidine, DB289) against Trypanosoma brucei rhodesiense infection in Vervet Monkeys after oral administration. Antimicrob Agents Chemother. 2009;53:953–957. doi: 10.1128/AAC.00831-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Lee MP, Parkinson GN, Batista-Parra A, Ismail MA, Neidle S, Boykin DW, Wilson WD. Out-of-shape DNA minor groove binders: Induced fit interactions of heterocyclic dications with the DNA minor groove. Biochemistry. 2005;44:14701–14708. doi: 10.1021/bi051791q. [DOI] [PubMed] [Google Scholar]
- Moretti R, Donato LJ, Brezinski ML, Stafford RL, Hoff H, Thorson JS, Dervan PB, Ansari AZ. Targeted chemical wedges reveal the role of allosteric DNA modulation in protein-DNA assembly. ACS Chem Biol. 2008;3:220–229. doi: 10.1021/cb700258r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrksich M, Wade WS, Dwyer TJ, Geierstanger BH, Wemmer DE, Dervan PB. Antiparallel side-by-side dimeric motif for sequence-specific recognition in the minor groove of DNA by the designed peptide 1-methylimidazole-2-carboxamide netropsin. Proc Natl Acad Sci USA. 1992;89:7586–7590. doi: 10.1073/pnas.89.16.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Padmanabhan PK, Sahani MH, Barrett MP, Madhubala R. Roles for mitochondria in pentamidine susceptibility and resistance in Leishmania donovani. Mol Biochem Parasitol. 2006;145:1–10. doi: 10.1016/j.molbiopara.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Munde M, Ismail MA, Arafa R, Peixoto P, Collar CJ, Liu Y, Hu L, David-Cordonnier MH, Lansiaux A, Bailly C, Boykin DW, Wilson WD. Design of DNA minor groove binding diamidines that recognize GC base pair sequences: a dimeric-hinge interaction motif. J Am Chem Soc. 2007;129:13732–13743. doi: 10.1021/ja074560a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjunda R, Munde M, Liu Y, Wilson WD. Real-time monitoring of nucleic acid interactions with biosensor-surface plasmon resonance. In: Wanunu M, Tor Y, editors. Methods for Studying Nucleic Acid/Drug Interactions. CRC Press; Boca Raton: 2012. pp. 91–122. [Google Scholar]
- Neidle S. DNA minor-groove recognition by small molecules. Nat Prod Rep. 2001;18:291–309. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- Nguyen B, Boykin DW, Wilson WD. DNA Minor Groove Interactions of Antiparasitic Diamidines: Re-Evaluation of the Crescent-Shape Concept in Groove-Binding. In: Lee M, Strekowski L, editors. Synthentic and Biophysical Studies of DNA Binding Compounds. Transworld Research Network; Kerala, India: 2007. pp. 39–66. [Google Scholar]
- Nguyen B, Neidle S, Wilson WD. A role for water molecules in DNA-ligand minor groove recognition. Acc Chem Res. 2009;42:11–21. doi: 10.1021/ar800016q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine MF, Wang MZ, Generaux CN, Boykin DW, Wilson WD, De Koning HP, Olson CA, Pohlig G, Burri C, Brun R, Murilla GA, Thuita JK, Barrett MP, Tidwell RR. Diamidines for human African trypanosomiasis. Curr Opin Investig Drugs. 2010;11:876–883. [PubMed] [Google Scholar]
- Pandian GN, Shinohara K, Ohtsuki A, Nakano Y, Masafumi M, Bando T, Nagase H, Yamada Y, Watanabe A, Terada N, Sato S, Morinaga H, Sugiyama H. Synthetic small molecules for epigenetic activation of pluripotency genes in mouse embryonic fibroblasts. ChemBio Chem. 2011;12:2822–2828. doi: 10.1002/cbic.201100597. [DOI] [PubMed] [Google Scholar]
- Parker SCJ, Tullius TD. DNA shape, genetic codes, and evolution. Curr Opin Struct Biol. 2011;21:342–347. doi: 10.1016/j.sbi.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto P, Liu Y, Depauw S, Hildebrand MP, Boykin DW, Bailly C, Wilson WD, David-Cordonnier MH. Direct inhibition of the DNA-binding activity of POU transcription factors Pit-1 and Brn-3 by selective binding of a phenyl-furan-benzimidazole dication. Nucleic Acids Res. 2008;36:3341–3353. doi: 10.1093/nar/gkn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petraccone L, Spink C, Trent JO, Garbett NC, Mekmaysy CS, Giancola C, Chaires JB. Structure and stability of higher-order human telomeric quadruplexes. J Am Chem Soc. 2011;133:20951–20961. doi: 10.1021/ja209192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskatov JA, Meier JL, Puckett JW, Yang F, Ramakrishnan P, Dervan PB. Modulation of NF-κB-dependent gene transcription using programmable DNA minor groove binders. Proc Natl Acad Sci USA. 2012a;109:1023–1028. doi: 10.1073/pnas.1118506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskatov JA, Hargrove AE, So AY, Dervan PB. Pharmacokinetics of Py-Im Polyamides Depend on Architecture: Cyclic versus Linear. J Am Chem Soc. 2012b;134:7995–7999. doi: 10.1021/ja302588v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BS, Sondhi SM, Lown JW. Synthetic DNA minor groove-binding drugs. Pharmacol Ther. 1999;84:1–111. doi: 10.1016/s0163-7258(99)00021-2. [DOI] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro TA, Englund PT. The structure and replication of kinetoplast DNA. Ann Rev Microbiol. 1995;49:117–143. doi: 10.1146/annurev.mi.49.100195.001001. [DOI] [PubMed] [Google Scholar]
- Shinohara K, Bando T, Sugiyama H. Anticancer activities of alkylating pyrrole-imidazole polyamides with specific sequence recognition. Anticancer Drugs. 2010;21:228–242. doi: 10.1097/CAD.0b013e328334d8f9. [DOI] [PubMed] [Google Scholar]
- Simpson IJ, Lee M, Kumar A, Boykin DW, Neidle S. DNA minor groove interactions and the biological activity of 2,5-bis. Bioorg Med Chem Lett. 2000;10:2593–2597. doi: 10.1016/s0960-894x(00)00511-4. [DOI] [PubMed] [Google Scholar]
- Stewart ML, Krishna S, Burchmore RJ, Brun R, de Koning HP, Boykin DW, Tidwell RR, Hall JE, Barrett MP. Detection of arsenical drug resistance in Trypanosoma brucei with a simple fluorescence test. Lancet. 2005;366:486–487. doi: 10.1016/S0140-6736(05)66793-1. [DOI] [PubMed] [Google Scholar]
- Soeiro MN, de Castro SL, de Souza EM, Batista DG, Silva CF, Boykin DW. Diamidine activity against trypanosomes: the state of the art. Curr Mol Pharmacol. 2008;1:151–161. doi: 10.2174/1874467210801020151. [DOI] [PubMed] [Google Scholar]
- Tanious FA, Laine W, Peixoto P, Bailly C, Goodwin KD, Lewis MA, Long EC, Georgiadis MM, Tidwell RR, Wilson WD. Unusually strong binding to the DNA minor groove by a highly twisted benzimidazole diphenylether: Induced fit and bound water. Biochemistry. 2007;46:6944–6956. doi: 10.1021/bi700288g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanious FA, Spychala J, Kumar A, Greene K, Boykin DW, Wilson WD. Different binding mode in AT and GC sequences for unfused-aromatic dications. J Biomol Struct Dyn. 1994;11:1063–1083. doi: 10.1080/07391102.1994.10508053. [DOI] [PubMed] [Google Scholar]
- Tanious F, Wilson WD, Wang L, Kumar A, Boykin DW, Marty C, Baldeyrou B, Bailly C. Cooperative dimerization of a heterocyclic diamidine determines sequence-specific DNA recognition. Biochemistry. 2003;42:13576–13586. doi: 10.1021/bi034852y. [DOI] [PubMed] [Google Scholar]
- Tidwell RR, Boykin DW. Dicationic DNA minor groove binders as antimicrobial agents. In: Demeunynck M, Bailly C, Wilson WD, editors. DNA and RNA Binders, Vol. 1: From small molecules to drugs. 2002. pp. 414–460. [Google Scholar]
- Trent JO, Clark GR, Kumar A, Wilson WD, Boykin DW, Hall JE, Tidwell RR, Blagburn BL, Neidle S. Targeting the minor groove of DNA: crystal structures of two complexes between furan derivatives of berenil and the DNA dodecamer d(CGCGAATTCGCG)2. J Med Chem. 1996;39:4554–4562. doi: 10.1021/jm9604484. [DOI] [PubMed] [Google Scholar]
- Walker WL, Kopka ML, Goodsell DS. Progress in the design of DNA sequence-specific lexitropsins. Biopolymers. 1997;44:323–334. doi: 10.1002/(SICI)1097-0282(1997)44:4<323::AID-BIP2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Wang L, Bailly C, Kumar A, Ding D, Bajic M, Boykin DW, Wilson WD. Specific molecular recognition of mixed nucleic acid sequences: an aromatic dication that binds in the DNA minor groove as a dimer. Proc Natl Acad Sci USA. 2000;97:12–16. doi: 10.1073/pnas.97.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Carrasco C, Kumar A, Stephens CE, Bailly C, Boykin DW, Wilson WD. Evaluation of the influence of compound structure on stacked-dimer formation in the DNA minor groove. Biochemistry. 2001;40:2511–2521. doi: 10.1021/bi002301r. [DOI] [PubMed] [Google Scholar]
- Wang L, Kumar A, Boykin DW, Bailly C, Wilson WD. Comparative thermodynamics for monomer and dimer sequence-dependent binding of a heterocyclic dication in the DNA minor groove. J Mol Biol. 2002;317:361–374. doi: 10.1006/jmbi.2002.5433. [DOI] [PubMed] [Google Scholar]
- Waring MJ, Bailly C. The purine 2-amino group as a critical recognition element for binding of small molecules to DNA. Gene. 1994;149:69–79. doi: 10.1016/0378-1119(94)90414-6. [DOI] [PubMed] [Google Scholar]
- Wartell RM, Larson JE, Wells RD. Netropsin. Specific probe for A-T regions of duplex deoxyribonucleic acid. J Biol Chem. 1974;249:6719–6731. [PubMed] [Google Scholar]
- Wheeler RJ, Gull K, Gluenz E. Detailed interrogation of trypanosome cell biology via differential organelle staining and automated image analysis. BMC Biol. 2012;10:1–17. doi: 10.1186/1741-7007-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbovetz K. Diamidines as antitrypanosomal, antileishmanial and antimalarial agents. Curr Opin Investig Drugs. 2006;7:147–157. [PubMed] [Google Scholar]
- Wilson WD. Reversible Interactions of Nucleic Acids with Small Molecules. In: Blackburn GM, Gait MJ, editors. Nucleic Acids in Chemistry and Biology. 2. Oxford University Press; New York: 1996. pp. 331–374. [Google Scholar]
- Wilson WD, Nguyen B, Tanious FA, Mathis A, Hall JE, Stephens CE, Boykin DW. Dications that target the DNA minor groove: Compound design and preparation, DNA interactions, cellular distribution and biological activity. Curr Med Chem - Anti-Cancer Agents. 2005;5:389–408. doi: 10.2174/1568011054222319. [DOI] [PubMed] [Google Scholar]
- Wilson WD, Ratmeyer L, Zhao M, Strekowski L, Boykin D. The search for structure-specific nucleic acid-interactive drugs: effects of compound structure on RNA versus DNA interaction strength. Biochemistry. 1993;32:4098–4104. doi: 10.1021/bi00066a035. [DOI] [PubMed] [Google Scholar]
- Wilson WD, Tanious FA, Buczak H, Venkatramanan MK, Das BP, Boykin DW. The Effects of Ligand Structure on Binding Mode and Specificity in the Interaction of Unfused Aromatic Cations with DNA. In: Pullman B, Jortner J, editors. Molecular Basis of Specificity in Nucleic Acid-Drug Interactions. Academic Press; New York: 1990. pp. 331–353. [Google Scholar]
- Wilson WD, Tanious F, Goodwin K, Georgiadis M, Laine W, Peixoto P, Bailly C, Tidwell RR. Induced fit and bound water provide strong interactions in the DNA minor groove complex of a highly twisted benzimidazole diamidine. Abstracts, 58th Southeast Regional Meeting of the American Chemical Society; Augusta, GA, United States. 2006. pp. 1–4. [Google Scholar]
- Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. Antiparasitic compounds that target DNA. Biochimie. 2008;90:999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Davis E, Ranjan N, Xue L, Hyde-Volpe D, Arya DP. Thermodynamics of nucleic acid “shape readout” by an aminosugar. Biochemistry. 2011;50:9088–9113. doi: 10.1021/bi201077h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, Salinas J, Filipski J, Bernardi G. Genomic localization of hepatitis B virus in a human hepatoma cell line. Nucleic Acids Res. 1986;14:8373–8386. doi: 10.1093/nar/14.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kasif S, Cantor CR, Broude NE. GC/AT-content spikes as genomic punctuation marks. Proc Natl Acad Sci USA. 2004;101:16855–16860. doi: 10.1073/pnas.0407821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C. Effects of the antibiotics netropsin and distamycin A on the structure and function of nucleic acids. Prog Nucleic Acid Res Mol Biol. 1975;15:285–318. doi: 10.1016/s0079-6603(08)60122-1. [DOI] [PubMed] [Google Scholar]