Abstract
A newly discovered negative glucocorticoid response element (nGRE) mediates DNA-dependent transrepression by the glucocorticoid receptor (GR) across the genome and plays a major role in immunosuppressive therapy. The nGRE differs dramatically from activating response elements and the mechanism driving GR binding and transrepression is unknown. To unravel the mechanism of nGRE-mediated transrepression by the glucocorticoid receptor, we characterize the interaction between GR and a nGRE in the thymic stromal lymphopoetin (TSLP) promoter. We show using structural and mechanistic approaches that nGRE binding represents a new mode of sequence recognition by human GR and that nGREs prevent receptor dimerization through a unique GR-binding orientation and strong negative cooperativity, ensuring the presence of monomeric GR at repressive elements.
The glucocorticoid receptor (GR) is a ubiquitously expressed vertebrate nuclear receptor that controls the transcription of genes critical for metabolism, immunity, development, and responses to stress1-3. Glucocorticoids, widely prescribed for their powerful immunosuppressive and anti-inflammatory properties4, drive the both the transactivation and transrepression of GR target genes, with transactivation of target genes slightly more prevalent than repression5. Therapeutically beneficial GR-mediated immunosuppression is thought to occur primarily through indirect or “tethered” DNA-independent interactions of GR with other transcription factors such as NF-κB6 and Stat37 to repress pro-inflammatory genes8. In contrast, side effects of glucocorticoids are often attributed to direct gene activation9. Recently, a new role for direct, DNA-dependent transrepression by GR was discovered through the identification of widely-prevalent negative glucocorticoid response elements (nGREs)10. These elements differ in sequence from activating glucocorticoid response elements ((+)GREs) and selectively recruit the corepressors NCoR and SMRT to the promoters of nGRE-containing genes upon GR binding10. Functional nGREs have been identified within hundreds of promoters, including many key inflammatory and metabolic genes10. Furthermore, nGRE-containing genes such as insulin, the insulin receptor, and Bcl-2 are implicated in side effects associated with glucocorticoid therapy10. Identifying the repressive mechanism of GR at nGRE-containing genes may support the quest for dissociated GR ligands that separate beneficial effects of glucocorticoid agonists from their side effects.
Numerous mechanistic studies have shown that GR transactivation requires the presence of (+)GREs which allosterically mediate GR binding, recruitment of coactivators, and transcription11,12. These elements contain two inverted repeat AGAACA sequences separated by three nucleotides, with bold residues critical for GR binding13. The three nucleotide spacing between half-sites is strictly required to preserve dimerization potential of GR on the element14. In contrast, the nGRE consensus sequence, CTCC(n)0-2GGAGA, differs dramatically from activating sequences. The spacing required in the nGRE is variable, ranging from 0-2 nucleotides, suggesting that GR dimerization may not be necessary for nGRE-mediated transrepression.
Given the radically different sequence and organization of nGREs, it is unclear how GR binds to this element to repress the vast array of nGRE containing genes. To unravel the mechanism of nGRE-mediated transrepression by the glucocorticoid receptor, we characterize the interaction between GR and a nGRE in the thymic stromal lymphopoetin (TSLP) promoter. This nGRE, 850 base pairs upstream of the TSLP transcription start site, mediates the reduction of TSLP mRNA levels by 50 % in response to GR agonists10. TSLP regulates many critical immune processes15-17 and is implicated in disorders such as atopic dermatitis, asthma, irritable bowel syndrome, and arthritis18-22. Using this prototypical nGRE, we employ structural, biochemical, and cellular approaches to demonstrate that two GR monomers bind nGREs in an everted repeat orientation with strong negative cooperativity. When combined, the unique GR conformation and negative cooperativity, ensures the presence of monomeric GR at nGREs. This interaction mechanism represents a new mode of GR-DNA binding and a new paradigm for GR transrepression.
RESULTS
nGRE binding displays negative cooperativity
To initially characterize the affinity of GR for nGREs, we compared binding of recombinantly expressed glucocorticoid receptor DNA binding domain (DBD) to fluorescently-labeled activating and repressive GR response elements. The canonical (+)GRE, contains two nearly-identical inverted GR binding sites separated by 3bp which enables cooperative DNA binding and dimerization. We found that GR binds these elements with a Kd of 73 nM and a Hill slope of 1.4, indicating the expected positive cooperativity (Fig. 1a, Table 1). However, when testing GR binding to the TSLP nGRE, we observed a dramatically different binding curve, which qualitatively appeared as a two-site binding event (Fig. 1a).
Figure 1. The glucocorticoid receptor interacts with negative glucocorticoid response elements in a unique orientation preventing receptor dimerization.
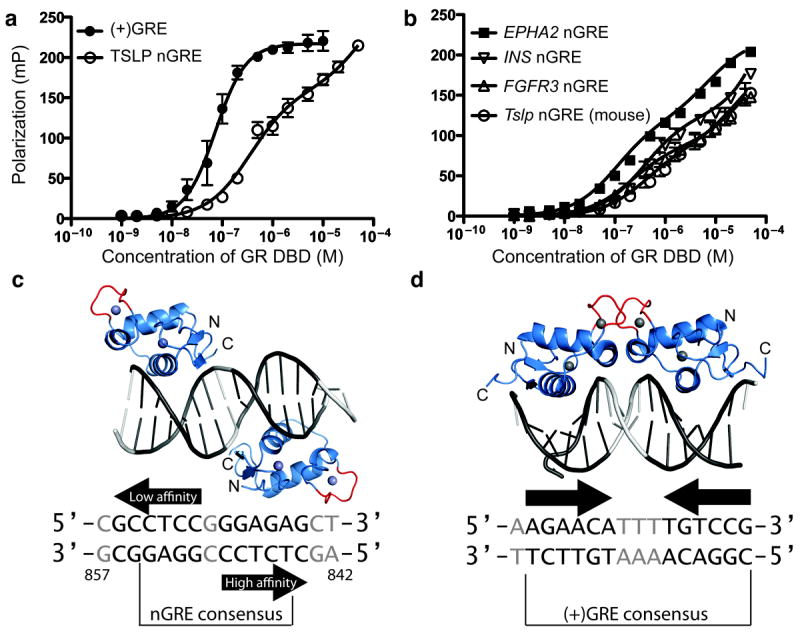
(a) GR binding to the TSLP nGRE is a two-site binding event with a lower Kd than cooperative GR binding to (+)GREs. (b) Additional human nGREs as well as the mouse Tslp nGRE show a similar binding profile with two GR binding sites and affinities comparable to the TSLP promoter. Binding data are represented as mean ± s.e.m from three replicates from at least two independent fluorescence polarization experiments. (c) Overall structure of GR DBD (blue) in complex with the TSLP nGRE (gray). Bases comprising the GR binding sites are in black, and Zn2+ is depicted as gray spheres. The GR dimerization interface (red) of each GR monomer is oriented away from the second monomer in an everted repeat conformation. (d) GR binds to a (+)GRE as a dimer in an inverted repeat conformation, enabling contact between dimerization loops (PDB 3FYL)12.
Table 1.
Interaction between GR DBD and nGREs, monitored by fluorescence polarization.
| F (DFn, DFd) for two-site binding | p-value for two-site binding | Kd, high ± s.e.m. (μM) | Kd, low ± s.e.m. (μM) | r2 | |
|---|---|---|---|---|---|
| (+)GRE | NC | NC | 0.07 ± .007 | NC | 0.92 |
| TSLP nGRE | 21.84 (1,92) | < 0.0001 | 0.36 ± 0.06 | 63 ± 86 | 0.97 |
| FGFR3 nGRE | 8.386 (1,92) | 0.0047 | 0.28 ± 0.08 | 42 ± 42 | 0.92 |
| INS nGRE | 43.26 (1,92) | < 0.0001 | 0.50 ± 0.07 | 444 ± 1976 | 0.98 |
| EPHA2 nGRE | 39.85 (1,43) | < 0.0001 | 0.14 ± 0.01 | 9.8 ± 2.1 | 0.99 |
| Tslp nGRE (mouse) | 3.457 (1,92) | 0.0662 | 0.52 ± 0.19 | 36 ± 37 | 0.92 |
| GR DBD R460D/D462R – TSLP nGRE | 10.16 (1,140) | 0.0018 | 0.14 ± 0.06 | 5.6 ± 1.5 | 0.95 |
| GR DBD K442A – TSLP nGRE | NC | NC | 5.14 ± 0.51 | NC | 0.97 |
| TSLP nGRE, high site mutation | 2.579 (1,92) | 0.1117 | 0.83 ± 0.2 | 14 ± 9.3 | 0.98 |
| TSLP nGRE, low site mutation | 16.83 (1,44) | 0.0002 | 0.26 ± 0.06 | 11 ± 1.2 | 0.99 |
| Tslp nGRE (mouse), IR0 | 8.968 (1,92) | 0.0035 | 0.79 ± 0.18 | 43 ± 57 | 0.97 |
| (+)GRE Half-site onlya | NC | NC | 0.38 ± 0.09 | NC | 0.92 |
| TSLP, low site only | NC | NC | 12.7 ± 0.8 | NC | 0.99 |
| GR DBD A458T – (+)GRE | 26.54 (2,44) | <0.0001 | 0.37 ± 0.06 | 10.9 ± 6.0 | 0.99 |
| GR DBD A458T – TSLP nGRE | NC | NC | 1.1 ± 0.08 | NC | 0.99 |
| GR DBD A458T – Tslp nGRE (mouse) | NC | NC | 1.5 ± 0.08 | NC | 0.99 |
| GR DBD A458T - INS nGRE | NC | NC | 1.1 ± 0.05 | NC | 0.99 |
| GR DBD R460D/D462R – (+)GRE | NC | NC | 0.08 ± 0.007 | NC | 0.97 |
| GR DBD A458T-low site only | NC | NC | 2.8 ± 0.3 | NC | 0.99 |
NC, no convergence of two-site binding model
Determined by competition vs. a consensus (+)GRE half-site (Ki is given instead of Kd)
To test the superiority of a two-site binding model for GR-nGRE interactions, we performed an extra-sum-of-squares F-test comparing a two-site binding event with a cooperative one-site binding event (Table 1). We found that the TSLP nGRE contains two distinct binding sites (p < .0001) with Kds of 363 nM and 63 μM. To establish this as a property of all nGREs, we confirmed this result on other nGREs from promoters of genes such as insulin and FGFR3 (Fig. 1b, Table 1). We found that relatively weaker binding of GR to nGREs compared to (+)GREs appears to be a general feature of nGREs and mirrors the affinity of GR for a canonical (+)GRE half-site (Table 1). While high-affinity site binding affinity was relatively constant among nGREs, affinity of the second site varied considerably, suggesting that flanking sequence of the low-affinity site may affect its ability to recruit GR.
Structure of the repressive GR DBD-nGRE complex
To discover the structural basis for this unusual mechanism of binding, we solved the crystal structure of the GR DNA binding domain (DBD) in complex with the TSLP nGRE to a resolution of 1.9 Å (Table 2). Surprisingly, the crystal structure showed two GR monomers bound to non-identical everted sites in a head-to-tail fashion, separated by 1 bp as predicted10 (Fig. 1c). In this orientation, the dimerization loop (or D-box) of each GR monomer is directed away from the other monomer and rotated by 180° around the DNA axis (Fig. 1c), abrogating the opportunity for DBD-mediated GR dimerization. In contrast, GR binds (+)GREs in a head-to-head orientation on the same side of DNA, allowing cooperative binding and dimerization (Fig. 1d). The everted repeat conformation found in the nGRE ensures monomeric binding by preventing DNA-mediated GR dimerization and may explain the element’s repressive character since monomeric GR is associated with gene repression23. To our knowledge, this unexpected everted repeat nuclear receptor-DNA binding geometry has been previously described only in the thyroid and retinoic acid receptors24,25.
Table 2.
Data collection and refinement statistics.
| GR DBD – TSLP nGRE | GR DBD R460D D462R – TSLP nGRE | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 39.3, 96.6, 104.0 | 38.7, 87.9, 103.2 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 1.9 (1.97 – 1.90)* | 2.55 (2.64 – 2.55)* |
| Rsym or Rmerge | 8.5 (54.5) | 9.8 (37.7) |
| I/σI | 26.7 (2.2) | 15.7 (2.8) |
| Completeness (%) | 99.7 (98.4) | 96.9 (83.3) |
| Redundancy | 6.3 (4.2) | 3.7 (2.8) |
| Refinement | ||
| Resolution (Å) | 1.90 | 2.55 |
| No. reflections | 31815 | 11685 |
| Rwork / Rfree | 20.7 / 23.5 | 19.5 / 24.8 |
| No. atoms | ||
| Protein | 1115 | 1110 |
| DNA | 650 | 650 |
| Water | 204 | 33 |
| B-factors | ||
| Protein | 39.7 | 32.6 |
| DNA | 56.5 | 45.5 |
| Water | 44.7 | 30.2 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.013 |
| Bond angles (°) | 1.15 | 1.78 |
Data collected from a single crystal; values in parentheses are for highest-resolution shell.
GR binds to nGREs as two monomers at nonequivalent sites
Based on the identification of low- and high-affinity sites in our GR–nGRE binding data, we hypothesized that each nGRE-bound GR monomer may make different contacts with DNA, resulting in differing affinities of each monomer for its binding site. Indeed, each of the two bound monomers uses different amino acid side chains to make base-specific contacts. One GR monomer makes three base-specific contacts, whereas a second monomer contacts only one base in a specific fashion (Supplementary Fig. 1). To assist in the determination of the high- and low-affinity sites, we used the PISA server26 to identify free energy gains from GR monomer–DNA interactions. The first DNA-monomer interaction, with three specific contacts, has a very favorable free energy change upon formation of the interface (ΔG = −9.5 kcal/mol). The second monomer shows a ΔG of only −5.9 kcal/mol, identifying the former site as the likely high-affinity site.
This suspected high-affinity GR DBD–nGRE DNA interaction involves three base specific contacts within the major groove (Fig. 2a): Val443 makes hydrophobic contacts with cytosine 846 and thymine 847 and Lys442 donates a hydrogen bond to N7 of guanine 849. Mutation of this guanine to adenine increases the Kd of GR for the high affinity site, confirming the identity of the high-affinity GR binding site (Table 1). In a previous study, the identical mutation ablates the repressive ability of the mouse TSLP nGRE10. Likewise, mutation of Lys442 significantly diminishes nGRE binding (Table 1). Unlike its DNA-reading function in (+)GRE structures, the Arg447 side chain is prevented from making base-specific contacts due to a steric clash with thymine 845 (Fig. 2b). The repositioned Arg447 instead make hydrophobic interactions with this base and ionic interactions with the cytosine 844 backbone phosphate. Mutation of thymine 845 to guanine, which would permit the “active” conformation of Arg447, abrogates transrepression10. The low-affinity GR DBD–DNA interaction involves only one sequence-specific contact: Arg447 contacts guanine 856, outside the nGRE consensus sequence (Supplementary Fig. 1a). Mutation of guanine 856 does not affect GR binding to the high-affinity site (Table 1), and Lys442 and Val443 do not sufficiently penetrate the major groove to facilitate sequence-specific DNA contacts (Supplementary Fig. 1b). As a result, the DNA major groove at the low-affinity site contains more waters than the high-affinity site. Recognition of the nGRE high-affinity site requires a more specific contacts and a greater hydrophobic interaction surface than either the low-affinity nGRE site or (+)GRE sequences, as confirmed by PISA26.
Figure 2. GR employs unique interactions to recognize the high-affinity site within nGREs.
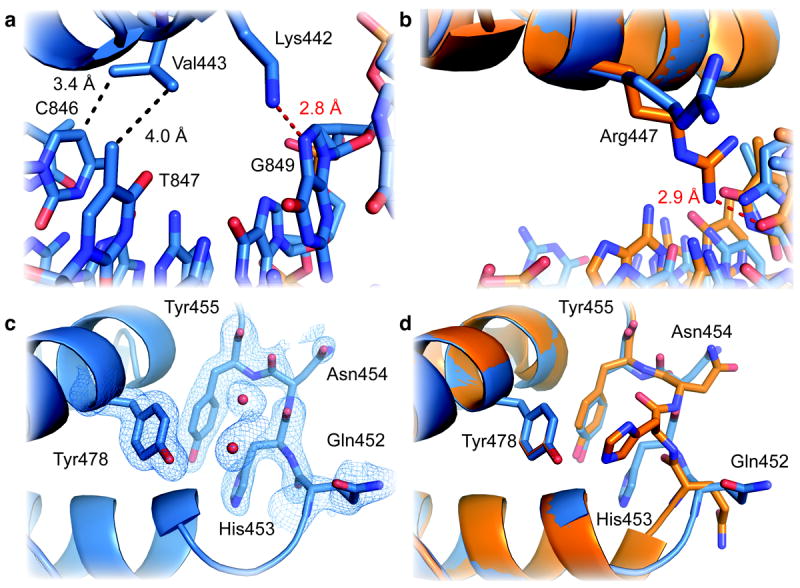
(a) Close up view of the high-affinity GR–TSLP nGRE interaction with side chains and nucleotide depicted as sticks (O, red; N, blue). Hydrogen bonds and van der Waals interactions are represented by red and black dashed lines, respectively. Three base-specific contacts are present between GR and the high-affinity nGRE binding site. Val443 makes two hydrophobic contacts, and Lys442 donates a hydrogen bond to guanine 849. (b) Arg447 makes unique non-specific interactions with DNA at the high-affinity nGRE binding site. In contrast, Arg447 makes base-specific contacts with a guanine base when bound to a (+)GRE (orange; PDB FYL)12. c) 2Fo-Fc electron density (blue mesh) contoured at 1σ showing the conformation of the lever arm residues in TSLP nGRE-bound GR alone and (d) superimposed on (+)GRE-bound GR (orange).
Taken together, these data demonstrate the mechanism by which GR recognizes the GGAG within the high-affinity nGRE binding site (Fig. 1c), and explains the strict conservation of one of these GGAG motifs present in the nGRE consensus10. The role of the low affinity GR site within the nGRE remains unclear. Despite an identical GGAG sequence present at the low-affinity site, GR binds this site very weakly (Table 1). The low affinity site is far more resistant to mutation than the high-affinity site, yet spacing between the low- and high-affinity sites affects both GR binding and transrepression (ref. 10, Table 1).
DNA-mediated allostery differs between activating and repressive GR response elements
Recent work comparing several GR – (+)GRE crystal structures demonstrated that DNA serves as an allosteric modulator of GR activity where the binding of the first GR monomer relays conformational information through DNA to promote the second binding event ultimately driving transactivation by favoring coactivator recruitment12,27. This positive cooperativity is so strong that detection of the intermediate state (monomeric GR on DNA) is often difficult28. In contrast, we found that GR binding to the TSLP nGRE exhibits unusually strong negative cooperativity, where binding of the first GR monomer impedes binding of the second (Supplemental Note, Supplementary Fig. 2). The GR nGRE complex also exhibits a different DNA shape than (+)GREs, with a narrow major groove compared to the average of 11 GR DBD structures solved12 in complex with 16 bp (+)GRE DNA constructs (Supplementary Fig. 3). B-factor analysis also reveals that nGRE and (+)GRE DNA undergo dramatically different structural perturbations upon GR binding (Supplementary Fig. 3). On (+)GREs, GR binding drives a constriction of the minor groove to facilitate direct protein protein contacts. In contrast, the GR nGRE interaction forces a narrower major groove and wider minor groove, which opposes the binding of a second GR monomer (Supplementary Fig. 3). Since monomeric GR is linked with transcriptional repression23, negative cooperativity may reinforce the repressive character of the nGRE. Alternatively, since recruitment of coactivators by steroid receptors may depend on cooperative binding on DNA response elements27, non-cooperative mechanisms of DNA binding may allow DNA-sensitive domains of GR to adopt alternate, repressive confirmations.
For example, the “lever arm,” which immediately follows the DNA reading helix, has been identified as being the critical structural motif sensitive to sequence-dependent conformational changes on (+)GREs12. When bound to a nGRE, these lever arm residues adopt a distinct conformation compared to (+)GRE-bound GR (Fig. 2c, 2d). Specifically, His453 adopts a “flipped” conformation in both monomers, interacting with Arg447 and Tyr455 rather than the “packed” conformation critical for activation from (+)GRE-containing promoter elements (Fig. 2d, Supplementary Fig. 4). The loss of sequence-specific contacts by Arg447 in the nGRE allows His453 to be stabilized in a “flipped” conformation by both a hydrogen bond and van der Waals contact from the repositioned Arg447 (Supplementary Fig. 4). Repositioning of Arg447 also eliminates half of a helical turn of the DNA reading helix, supporting the “flipped” conformation of the lever arm (Fig. 2d). The lever arm is the most dynamic portion of the GR DBD, with B-factors significantly higher than other regions of the protein, yet these residues display good electron density. This modulation of the lever arm by sequence-specific contacts illustrates the pivotal role of the lever arm in receptor activation and confirms the allosteric ability of DNA to drive receptor activation and repression.
Dimerization competes with nGRE binding and transrepression
Our structure and model of nGRE action predict that receptor dimerization opposes nGRE binding and therefore interferes with direct transrepression. Recent work has indicated that GR is unique among steroid receptors in that it exhibits no reversible self-association29 and is dependent on receptor-DNA interactions for dimerization30,31. To examine the effects of altered dimerization surfaces on nGRE binding and repression, we used two well-characterized GR mutants: A458T, often called the GRdim mutant, which is unable to support most glucocorticoid mediated gene activation, (+)GRE binding, or direct DNA repression in vivo10, and a double mutant containing the R460D and D462R mutations (R460D D462R), which has been shown to reduce GR dimerization and decrease activation of multiple (+)GREs32. Notably, this mutation was previously found to potentiate repression of Bcl-233, which was recently shown to harbor a consensus nGRE within its promoter10.
The A458T mutant bound to a (+)GRE in a clear 2-site binding event (Table 1), indicating a loss of cooperativity on this element. In this way, binding of the A458T mutant to (+)GREs strongly resembles binding of WT GR to nGREs. The R460D D462R mutant showed similar DNA binding as WT to (+)GRE sequences. Next, we tested each of these mutants for binding to nGREs. The A458T mutation differentially affected binding to each of the GR binding sites on the TSLP nGRE, improving low-affinity site binding but decreasing high-affinity site binding (Table 1). The net effect of this mutation is to decrease the affinity of GR for nGREs by 500 %, to nearly 3 μM. In contrast, the R460D D462R mutation improved binding at both sites on the TSLP nGRE. We then tested the ability of each variant to repress a reporter containing a constitutively active luciferase gene preceded by the nGRE-containing region of the TSLP promoter, as performed previously10. In line with our in vitro binding data, the A458T showed a modest ability to repress luciferase expression (Supplementary Fig. 5). Strikingly, the R460D D462R mutation was a more potent repressor of luciferase activity than WT GR (Fig. 3a). To observe the effects of this mutant on the GR dimerization interface, we solved the crystal structure of the GR R460D D462R mutant bound to the TSLP nGRE (Fig. 3b, Table 2). The structure of the R460D D462R mutant shows a less favorable dimerization interface (Fig. 3c–e), suggesting that the superior binding and repressive potential of the GR R460D D462R mutant is indeed due to decreased dimerization efficiency.
Figure 3. The R460D D462R mutant reduces receptor dimerization, enhancing GR-mediated transrepression at theTSLP nGRE.
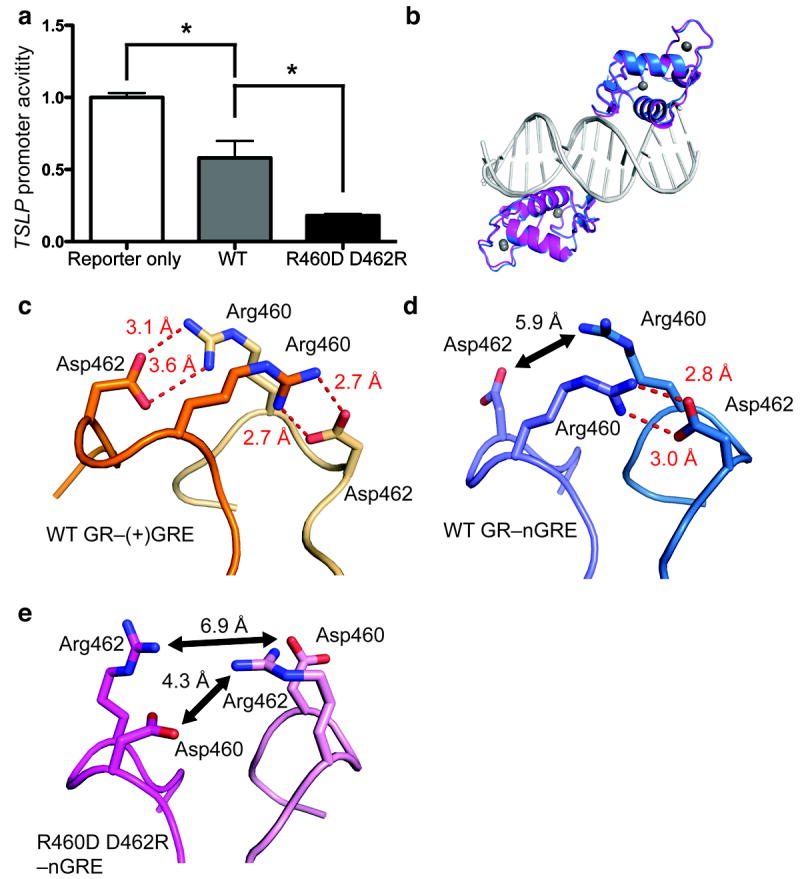
(a) Transfection of the R460D D462R mutant in HeLa cells potentiates downregulation of a constitutively active TSLP promoter compared to WT GR (5 ng each). Data are represented as the mean ± s.e.m of two independent experiments with five internal replicates each. (b) Superposition of GR (blue) and the GR R460D D462R mutant (magenta) bound to the TSLP nGRE (gray). (c) When bound to a (+)GRE element, Arg460 and Asp462 form two intermolecular salt bridges (red dashes) across the homodimer interface (PDB 3FYL). In the GR nGRE structure, crystal-packing interactions require the formation of a pseudo-continuous DNA helix and promote the formation of a pseudo-GR dimer across a two-fold symmetry axis (d). These interactions are necessary for crystal formation, but are not possible in solution-based binding assays. The R460D D462R mutation (e) ablates key dimerization contacts between GR monomers, disrupting symmetry-imposed dimerization contacts.
DISCUSSION
The glucocorticoid receptor controls the transcriptional activation and repression of thousands of genes. Multiple regulatory levels are required to achieve a coordinated response, including epigenetic and mRNA regulation, posttranslational modification, circadian rhythms, ligand availability, and target DNA sequence accessibility and binding13,34-37. Here, we demonstrate that DNA-binding orientation and sequence-specific contacts control repression of negative GR response element-containing genes. GR binds to these nGREs in a head-to-tail, rotated conformation that prevents DNA-mediated dimerization, in contrast to the DNA-mediated dimerization found on activating GR-binding sites. These unique nGRE sequences alter the conformation of GR residues critical for transcriptional activation, further illustrating the importance of DNA as an allosteric modulator of receptor activity.
A similar mechanism of allosteric modulation between repressive and activating response elements has been demonstrated with the transcription factor Pit-1. Like GR, Pit-1 is monomeric in solution and dimerizes in a DNA-dependent manner38. Pit-1 differentially represses and activates transcription of target genes based on spacing between DNA response elements, and this difference in DNA sequence allows recruitment of NCoR to repressive Pit-1 elements39. However, Pit-1 maintains similar protein-DNA contacts at both repressive and activating elements; repressive elements differ in that they contain two additional, conserved residues between half-sites39. Further, Pit-1 homodimerizes in both the transactivating and transrepressive complexes. In contrast, we demonstrate that nGREs have evolved to recognize GR using a new set of sequence specific criteria favoring monomeric binding over the cooperative binding observed in (+)GREs. This altered sequence generates a novel high-affinity GR binding site and affects the conformation of GR residues, such as His472, which are critical for receptor activation12. The comparison between Pit-1 and GR gives an excellent example of how different transcription factors adopt activating and repressive conformations via contrasting mechanisms. It is possible that other transcription factors have alternate DNA response elements that may differentially affect their function. Notably, the other 3-keto steroid receptors (the androgen, mineralocorticoid, and progesterone receptors) can recognize (+)GREs, but it is currently unknown whether these receptors can bind or mediate repression from nGREs.
In general, GR-dependent activation requires DNA-mediated receptor dimerization. We confirm that the GR dim mutation, A458T, ablates DNA-mediated cooperative binding to (+)GREs. Despite this, the GRdim mutation does not actively repress (+)GRE containing genes (e.g. it does not convert a (+)GRE into a repressive element), suggesting that the GRdim mutant is either incapable of stably binding (+)GREs as a monomer in vivo or that the presence of monomeric GR at (+)GRE elements is not sufficient for corepressor recruitment. This indicates that the nGRE sequence may be specific not only for monomeric binding of GR but also for arranging the receptor into a repressive conformation. The lever arm, previously implicated in receptor activation status12, adopts a distinct conformation in the nGRE-bound structures reported here, suggesting that it plays a critical role mediating not only GR transactivation but also transrepression.
Widespread clinical use of glucocorticoids has fueled the search for dissociated compounds, capable of minimizing side effects without compromising their anti-inflammatory function. One such GR ligand, Compound A (CpdA), has been shown to inhibit GR dimerization and consequently transactivation from (+)GRE containing genes40, yet still supports the transrepression of nGRE-containing genes such as POMC41,42. Thus, if the major effect of CpdA is to disrupt dimer formation, it is now clear why CpdA permits transrepression from nGREs while preventing transactivation, suggesting that the opposing effects of direct, DNA-dependent transrepression and transactivation are mediated by the dimerization status of the receptor.
Online Methods
Protein expression and purification
The DNA binding domain (DBD) of human glucocorticoid receptor (GR) alpha (residues 417-506, accession ADP91252) was cloned with a 6X-Histidine tag into the pMCSG7 vector. The DBD was expressed in BL-21(DE3)pLysS E. coli and induced with 0.3 mM IPTG for four hours at 30 °C. Cells were lysed in 1 M NaCl, 20 mM Tris-HCl (pH 7.4), 25 mM imidazole, and 5 % glycerol via sonication. Protein was purified via affinity chromatography (HisTrap) followed by TEV protease cleavage and dialysis to 100 mM NaCl, 20 mM Tris-HCl (pH 7.4), and 5 % glycerol. The DBD and affinity tag were separated by affinity chromatography (HisTrap), and further purified by gel filtration. For storage, protein was concentrated to 4 mg/ml, flash frozen in liquid N2, and stored at -80 °C. Mutations were made using the QuikChange Site-directed Mutagenesis kit (Stratagene).
Nucleic acid binding assays
Synthesized nucleic acid duplexes (Integrated DNA Technologies) were annealed in 10 mM NaCl, 20 mM Tris-HCl pH 8.0 by heating to 90 °C and slow cooling to room temperature. Fluorescence polarization was used to detect the formation of DBD-nucleic acid complexes. Indicated amounts of DBD were added to wells containing 10 nM of 6-FAM-labeled nucleic acids (Table 1). Reactions were performed in 100 mM NaCl, 20 mM Tris-HCl (pH 7.4), 5 % glycerol and measured with a Biotek Synergy plate-reader at an excitation/emission wavelength of 485/528 nm.
For each binding experiment, an F-test was used to compare a 2-site binding event to a one-site binding event with Hill slope, generating an F-statistic and p-value for a 2-site binding model. In table 1, these F-statistics (with numerator and denominator degrees of freedom) and p-values are shown in addition to Kd values for the low and high affinity DNA binding sites, and the coefficient of determination (r2) of the applicable fit. Complexes are with WT GR DBD unless otherwise noted, and Graphpad Prism 5 was used for binding data analysis and graph generation. Nucleotide sequences used in binding experiments are shown in Supplementary Table 1.
Reporter gene assays
A 400 bp region of the TSLP promoter surrounding the nGRE (chr5:110,406,332-110,406,745; GRCh37) was cloned between an SV40 enhancer and promoter upstream of firefly luciferase, similar to the construct described previously10. 50 ng of this construct, indicated amounts of receptor, and 1ng of constitutively active renilla luciferase were transfected with FuGene HD (Promega) in OptiMEM (Invitrogen) into HeLa cells cultured in AlphaMEM (Invitrogen) supplemented with 10 % charcoal stripped FBS (PAA). 24 hours after transfection, cells were treated with 1 μM dexamethasone, and after 18 hours, firefly and renilla luciferase were measured with the Dual-Glo assay system (Promega) on a Biotek Synergy plate-reader. Firefly luciferase activity was normalized to renilla luciferase for each well, and levels of all treatments were normalized to cells transfected only with the constitutively active nGRE construct and not treated with dexamethasone. An asterisk indicates p < 0.01 by ANOVA with Tukey’s multiple comparison test.
Structure determination
Crystals of the GR DBD-TSLP nGRE complex were grown by hanging-drop vapor diffusion in 15 % PEG 20000, 6 % glycerol, 7.5 % ethanol, and 0.1M HEPES (pH 7.5) with a protein concentration of 3.5 mg/ml and a 2:1 protein:DNA molar ratio. Crystals were cryoprotected in crystallant with 20 % PEG 20000 and 20 % glycerol and flash-frozen in liquid N2. Crystals of the GR DBD R460D D462R–TSLP nGRE complex were grown by hanging-drop vapor diffusion in 15 % PEG 2000 MME, 6 % glycerol, and 0.1M HEPES (pH 7.5), with a protein concentration of 3.5 mg/ml and a 2:1 protein:DNA molar ratio. These crystals were cryoprotected in crystallant with 25 % PEG 2000 MME and 20 % glycerol, and flash-frozen in liquid N2. Data were collected at 100 K and a wavelength of 1.00 Å at Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source (Argonne, IL) and processed using the HKL-2000 software. The structures were phased via molecular replacement using previously-solved structures12 of the GR-GRE complex in Phenix43. The structure was refined with phenix.refine43 and model building was performed in COOT44. 99 % of residues are Ramachandran favored or allowed regions for both the WT GR and R460D D462R structures, respectively, with 1 % outliers in both structures. Pymol was used to visualize the structure and generate figures45. 3DNA was used to analyze nucleic acid groove widths46. Amino acids are numbered according to the human GR sequence (ADP91252). Bases are numbered by position upstream of the TSLP (NM_033035) transcription start site, which is an additional 199 nt upstream of the translation start site (CCDS4101).
Supplementary Material
Acknowledgments
We thank N.M Choi, O. Laur, and the Emory Custom Cloning Core Facility for assistance with cloning the TSLP nGRE reporter construct. This work was supported with start-up funds from Emory University. W.H.H. was supported by an Emory-National Institute of Health Pharmacological Sciences graduate training grant (5T32GM008602).
Footnotes
Author Contributions
W.H.H. and E.A.O. designed the experiments, W.H.H. and C.E.Y. performed the experiments, and W.H.H. and E.A.O. wrote the manuscript.
Accession codes
Coordinates and structure factors of the human GR DBD and R460D D462R mutant bound to the human TSLP promoter have been deposited in the Protein Data Bank under accession numbers 4HN5 and 4HN6, respectively.
References
- 1.Chrousos GP. Stress and disorders of the stress system. Nat Rev Endocrinol. 2009;5:374–381. doi: 10.1038/nrendo.2009.106. [DOI] [PubMed] [Google Scholar]
- 2.Pikulkaew S, et al. The knockdown of maternal glucocorticoid receptor mRNA alters embryo development in zebrafish. Dev Dynam. 2011;240:874–889. doi: 10.1002/dvdy.22586. [DOI] [PubMed] [Google Scholar]
- 3.Rose AJ, Vegiopoulos A, Herzig S. Role of glucocorticoids and the glucocorticoid receptor in metabolism: Insights from genetic manipulations. J Steroid Biochem. 2010;122:10–20. doi: 10.1016/j.jsbmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Baschant U, Tuckermann J. The role of the glucocorticoid receptor in inflammation and immunity. J Steroid Biochem. 2010;120:69–75. doi: 10.1016/j.jsbmb.2010.03.058. [DOI] [PubMed] [Google Scholar]
- 5.Reddy TE, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–71. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auphan N, DiDonato J, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science. 1995;270:286–90. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 7.Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR Interaction Code: Predictive Value of Direct/Indirect DNA Recruitment for Transcription Outcome. Mol Cell. 2012;47:38–49. doi: 10.1016/j.molcel.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 8.Tuckermann JP, Reichardt HM, Arribas R. The DNA Binding-independent Function of the Glucocorticoid Receptor Mediates Repression of AP-1-dependent Genes in Skin. J Cell Biol. 1999;147:1365–1370. doi: 10.1083/jcb.147.7.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schäcke H, Döcke WD, Asadullah K. Mechanisms involved in the side effects of glucocorticoids. Phamacol Ther. 2002;96:23–43. doi: 10.1016/s0163-7258(02)00297-8. [DOI] [PubMed] [Google Scholar]
- 10.Surjit M, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–41. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Strähle U, Klock G, Schütz G. A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc Natl Acad Sci U S A. 1987;84:7871–7875. doi: 10.1073/pnas.84.22.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meijsing SH, et al. DNA Binding Site Sequence Directs Glucocorticoid Receptor Structure and Activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luisi BF, et al. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 15.Reardon C, et al. Thymic Stromal Lymphopoetin-Induced Expression of the Endogenous Inhibitory Enzyme SLPI Mediates Recovery from Colonic Inflammation. Immunity. 2011:223–235. doi: 10.1016/j.immuni.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochman Y, et al. Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7-induced signaling. Proc Natl Acad Sci U S A. 2010;107:19455–60. doi: 10.1073/pnas.1008271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siracusa MC, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011;477:229–33. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Shami A, Spolski R, Kelly J, Keane-Myers A, Leonard WJ. A role for TSLP in the development of inflammation in an asthma model. J Exp Med. 2005;202:829–39. doi: 10.1084/jem.20050199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyama K, et al. A possible role for TSLP in inflammatory arthritis. Biochem Biophys Res Commun. 2007;357:99–104. doi: 10.1016/j.bbrc.2007.03.081. [DOI] [PubMed] [Google Scholar]
- 20.Soumelis V, et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol. 2002;3:673–80. doi: 10.1038/ni805. [DOI] [PubMed] [Google Scholar]
- 21.Taylor BC, et al. TSLP regulates intestinal immunity and inflammation in mouse models of helminth infection and colitis. J Exp Med. 2009;206:655–67. doi: 10.1084/jem.20081499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo J, et al. Spontaneous atopic dermatitis in mice expressing an inducible thymic stromal lymphopoietin transgene specifically in the skin. J Exp Med. 2005;202:541–9. doi: 10.1084/jem.20041503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heck S, et al. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994;13:4087–95. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baniahmad a, Steiner C, Köhne A, Renkawitz R. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell. 1990;61:505–14. doi: 10.1016/0092-8674(90)90532-j. [DOI] [PubMed] [Google Scholar]
- 25.Tini M, Otulakowski G, Breitman ML, Tsui LC, Giguere V. An everted repeat mediates retinoic acid induction of the gamma F-crystallin gene: evidence of a direct role for retinoids in lens development. Genes Dev. 1993;7:295–307. doi: 10.1101/gad.7.2.295. [DOI] [PubMed] [Google Scholar]
- 26.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Heneghan AF, Connaghan-Jones KD, Miura MT, Bain DL. Coactivator assembly at the promoter: efficient recruitment of SRC2 is coupled to cooperative DNA binding by the progesterone receptor. Biochemistry. 2007;46:11023–32. doi: 10.1021/bi700850v. [DOI] [PubMed] [Google Scholar]
- 28.Tsai SY, et al. Molecular interactions of steroid hormone receptor with its enhancer element: evidence for receptor dimer formation. Cell. 1988;55:361–9. doi: 10.1016/0092-8674(88)90059-1. [DOI] [PubMed] [Google Scholar]
- 29.Robblee JP, Miura MT, Bain DL. Glucocorticoid Receptor-Promoter Interactions: Energetic Dissection Suggests a Framework for the Specificity of Steroid Receptor- Mediated Gene Regulation. Biochemistry. 2012;51:4463–4472. doi: 10.1021/bi3003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nwachukwu JC, Nettles KW. The nuclear receptor signalling scaffold: insights from full-length structures. EMBO J. 2012;31:251–253. doi: 10.1038/emboj.2011.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brélivet Y, Rochel N, Moras D. Structural analysis of nuclear receptors: From isolated domains to integral proteins. Mol Cell Endocrinol. 2012;348:466–473. doi: 10.1016/j.mce.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 32.Liu W, Wang J, Yu G, Pearce D. Steroid receptor transcriptional synergy is potentiated by disruption of the DNA-binding domain dimer interface. Mol Endocrinol. 1996;10:1399–406. doi: 10.1210/mend.10.11.8923466. [DOI] [PubMed] [Google Scholar]
- 33.Rogatsky I, Hittleman AB, Pearce D, Garabedian MJ. Distinct Glucocorticoid Receptor Transcriptional Regulatory Surfaces Mediate the Cytotoxic and Cytostatic Effects of Glucocorticoids. Mol Cell Biol. 1999;19:5036–5049. doi: 10.1128/mcb.19.7.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okret S, Poellinger L, Dong Y, Gustafsson JA. Down-regulation of glucocorticoid receptor mRNA by glucocorticoid hormones and recognition by the receptor of a specific binding sequence within a receptor cDNA clone. Proc Natl Acad Sci U S A. 1986;83:5899–5903. doi: 10.1073/pnas.83.16.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oster H, et al. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metabolism. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Irusen E, et al. p38 Mitogen-activated protein kinase-induced glucocorticoid receptor phosphorylation reduces its activity: Role in steroid-insensitive asthma. J Allergy Clin Immunol. 2002;109:649–657. doi: 10.1067/mai.2002.122465. [DOI] [PubMed] [Google Scholar]
- 38.Ingraham HA, et al. The POU-specific domain of Pit-1 is essential for sequence-specific, high affinity DNA binding and DNA-dependent Pit-1-Pit-1 interactions. Cell. 1990;61:1021–1033. doi: 10.1016/0092-8674(90)90067-o. [DOI] [PubMed] [Google Scholar]
- 39.Scully KM, et al. Allosteric Effects of Pit-1 DNA Sites on Long-Term Repression in Cell Type Specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 40.Dewint P, et al. A Plant-Derived Ligand Favoring Monomeric Glucocorticoid Receptor Conformation with Impaired Transactivation Potential Attenuates Collagen-Induced Arthritis. J Immunol. 2008;180:2608–2615. doi: 10.4049/jimmunol.180.4.2608. [DOI] [PubMed] [Google Scholar]
- 41.Drouin J, et al. Novel glucocorticoid receptor complex with DNA element of the hormone-repressed POMC gene. EMBO J. 1993;12:145–56. doi: 10.1002/j.1460-2075.1993.tb05640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson S, et al. Abrogation of glucocorticoid receptor dimerization correlates with dissociated glucocorticoid behavior of compound A. J Biol Chem. 2010;285:8061–75. doi: 10.1074/jbc.M109.087866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.The PyMOL Molecular Graphics System. Schrödinger, LLC; [Google Scholar]
- 46.Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


