Abstract
It is unclear whether or not earlier age at menarche is associated with higher body mass index (BMI) because they share a common genetic underpinning. We investigated the impact of single nucleotide polymorphisms (SNPs) influencing menarche timing on peri-pubertal BMI. For 556 Fels Longitudinal Study children (277 boys/279 girls) born 1928–1992, a genetic risk score (GRS42) was computed as the sum of the number of risk alleles in 42 putative menarche SNPs. Serial BMI Z-scores within ±6.99 years from each individual’s age at peak height velocity (Age@PHV) were grouped into seven time points (−6yrs, −4yrs, −2yrs, Age@PHV, +2yrs, +4yrs, +6yrs). Heritability of BMI ranged from 0.53 to 0.85 across the time points. The effect of GRS42 on BMI Z-scores at each time point was modeled using variance components-based procedures. GRS42 had a significant (p < 0.05) effect at every time point; an increase of one risk allele was associated with an increase of 0.03–0.08 BMI Z-scores. A separate score (GRS29) was computed that excluded 13 of the menarche SNPs previously documented to also influence adiposity; significant main effects were observed at Age@PHV+4yrs and +6yrs. This finding supports a causal effect of advanced sexual development on post Age@PHV BMI. Significant positive GRS42 (or GRS29)-by-birth year interactions indicate that some genetic influences on BMI have amplified over the 20th century. This gene-by-environment interaction also suggests that children with a genetic predisposition to earlier sexual development might avoid elevated BMI through alteration of their nutritional environment.
Keywords: genetic variants, menarche, developmental timing, pleiotropy, body mass index
The average age of menarche in girls in the United States of America (USA) has decreased throughout the second half of the 20th century (Wattigney et al., 1999; Freedman et al., 2002; Anderson et al., 2003; Chumlea et al., 2003). Data from nationally representative surveys in the United States, for example, show a three-month advancement in the median age at menarche, from 12.75 to 12.5 years, in girls between the 1960’s and the early 1990’s (Anderson et al., 2003; Chumlea et al., 2003). An expert panel, convened to examine all the available published data on pubertal timing events in the USA (i.e., menarche and stages of breast, genitalia, and pubic hair development) concluded that there is enough evidence to support a trend toward earlier breast development onset and menarche in girls (Euling et al., 2008). This trend has occurred concurrently with an increase in dietary intake (Briefel and Johnson, 2004) and an upward shift in the adolescent and adulthood body mass index (BMI) distribution (Troiano et al., 1995; Komlos et al., 2009; Johnson et al., 2012), thereby indicating an association between BMI and pubertal timing events which may be confounded by diet, socioeconomic status, psychosocial stress, and other time-varying factors.
At the individual level, children with advanced sexual development display greater adulthood BMI values and levels of adiposity (Lakshman et al., 2008; He et al., 2010) and, in addition, greater BMI values as early as five years of age (Davison et al., 2003; Mamun et al., 2009; Salsberry et al., 2009). Advanced sexual development may, therefore, be a consequence of high childhood BMI (Kaplowitz et al., 2001; Ahmed et al., 2009), which tracks throughout the life course (Deshmukh-Taskar et al., 2006). Teenage girls who suffer from eating disorders and undergo major weight loss are at increased risk of amenorrhea (Lock and Fitzpatrick, 2009); weight gain generally results in return of menstrual cycling in these girls (Swenne, 2004, 2008). These observations suggest, but do not prove, that BMI and adiposity are causally related to the timing of sexual development.
Because in energetically constrained environments, body fat is mobilized to support the energetic costs of pregnancy and lactation (Prentice et al., 1996; Dewey, 1997), low energy stores in sexually mature women would likely have posed a selective disadvantage during human evolution. This suggests a scenario in which peri-pubertal BMI level would be mechanistically tied to the timing of sexual maturation (i.e., menarche). It is known that both age at menarche and BMI are significantly heritable (Schousboe et al., 2003; Towne et al., 2005; Wardle et al., 2008), and that part of this heritability is common for both traits (Kaprio et al., 1995), but the question here is whether the same genes are operating on both phenotypes, such that the genetic factors promoting earlier sexual development are also promoting greater BMI. When genes affect more than one phenotype, they demonstrate pleiotropy. Genome-wide association studies (GWAS) have identified a large number of loci containing common genetic variants (i.e., single nucleotide polymorphisms, SNPs) that are associated with age at menarche (Elks et al., 2010) and BMI (Speliotes et al., 2010). To date, SNPs at a total of 42 loci have been identified to influence age at menarche in European ancestry women, with some of the same SNPs having pleiotropic effects on adulthood BMI (Elks et al., 2010). Whether these SNPs also influence BMI earlier in life, near the time of reproductive maturity, is however unclear. Further, the combined effect of a large number of menarche genetic variants on BMI in childhood and adolescence has not yet been investigated.
By assessing the effect of genetic variants for early menarche on BMI before, during, and after puberty, it is possible to make inferences about the temporal order of advanced maturation and the development of excess adiposity. Further, because alleles are transmitted from parent to offspring randomly at gamete formation, genotype is not confounded by environmental factors nor is it prone to reverse causation (Davey-Smith and Ebrahim, 2003; Lawlor et al., 2008); genetic factors related to age at menarche may influence BMI, but cannot be influenced by BMI. Nonetheless, because gene expression occurs within a cellular environment, which can be affected by numerous hormonal, dietary, and other factors (Hamilton, 2011; Jin and Robertson, 2011), genotype-phenotype associations are frequently modified by the environment (i.e., gene-environment interaction). The aforementioned secular trends exhibited in both earlier age at menarche and increasing BMI suggest that the relative influence of menarche SNPs on peri-pubertal BMI may depend on environmental factors that have changed over time.
The present study used data from the Fels Longitudinal Study, the world’s longest continuously-running, family-based study of human physical growth, to address a number of these gaps in knowledge. The first aim was to test the hypothesis that a genetic risk score for earlier menarche would influence serial BMI before, during, and after puberty. Relationships with height were also tested to allow us to infer whether effects of the menarche SNPs on BMI were primarily due to effects on weight or on height. The second aim was to determine whether any significant effects of the menarche SNPs remained after discounting a sub-group of menarche SNPs already known to have pleiotropic effects on adiposity. This second aim tests the hypothesis that sexual developmental timing and peri-pubertal BMI have a shared genetic control beyond the variants already known to influence adiposity. The final third aim was to test the hypothesis that the effects of menarche SNPs on peri-pubertal BMI differed by birth year.
METHODS
Study sample
The sample comprised 556 European American participants (277 boys, 279 girls) in the Fels Longitudinal Study who were born between 1928 and 1992. The Fels Longitudinal Study has been described in detail elsewhere (Roche, 1992), but in summary began in 1929 as a study of normative child growth and development, and continues today as a study of the early life antecedents of chronic diseases of aging. Infants living in Yellow Springs and nearby towns in southwestern Ohio, USA have been enrolled from 1929 onward. Mothers and other family members were also simultaneously enrolled. Participants were not selected on the basis of any pre-existing disease. Fels Longitudinal Study data were in fact the basis for the NCHS 1977 growth reference chart from birth to three years of age (Hamill et al., 1977). The 556 participants in the present study were distributed among 121 nuclear and extended families and in total comprised of 2641 relative pairs, of which 667 pairs were of first degree relatives, 560 pairs were of second degree relatives, 559 were of third degree relatives, and 849 pairs were of less closely related relatives.
The sample was selected on the basis of having at least one BMI recording between two and 18 years of age and also having been SNP genotyped. Scatter plots of BMI against age for the analysis sample and for all Fels Longitudinal Study participants with BMI data between two and 20 years of age (n = 1404) showed that the BMI distributions by age were similar. There were also no significant differences in the first and last BMI measurements (i.e., closest to ages two and 20 years), after being converted to Z-scores against the CDC 2000 reference (Kuczmarski et al., 2000), between the 556 children included in the present analysis and the remaining 848 children with BMI data but who were not SNP genotyped (p-values >0.4).
All protocols and informed consent documents used in the Fels Longitudinal Study were approved by the Wright State University Institutional Review Board. Parents of minors provided written consent for their children and minor children aged seven to 17 years provided their additional assent.
Genotype data
DNA was extracted from whole blood collected via venipuncture using standard procedures and stored at −80 degrees Celsius. Individuals were SNP genotyped using the genome-wide Illumina 610-Quad Bead-chip array (Applied Biosystems Incorporated, USA) at the Texas Biomedical Research Institute. SimWalk2 mistyping analysis (Sobel and Lange, 1996; Sobel et al., 2001, 2002) was used to determine genotypes that had a probability greater than 0.95 of being incorrectly called, and these Mendelian errors were removed by blanking these genotypes. Merlin (Abecasis et al., 2002) was used to impute missing genotypes using familial data. HapMap2 SNP genotypes were imputed using Mach1 (Li et al., 2009, 2010). Of the 42 independent SNPs identified in a recent GWAS of age at menarche (Elks et al., 2010), 41 SNPs were available. For the missing SNP (rs6762477 near the gene RBM6), a proxy (rs7061) in high linkage disequilibrium (r2 = 0.967) was used.
Genetic risk scores
For each of the 42 SNPs used in the present analysis, the allele that was reported to be associated with earlier menarche was considered the risk allele and then the number of risk alleles for each SNP (0, 1, or 2) was counted. A genetic risk score (GRS42) was computed as the summation of risk alleles across the 42 SNPs, so that a one unit increase corresponded to an increase of one risk allele. In addition, a separate genetic risk score (GRS29) was computed that excluded 13 of the 42 SNPs that have previously been implicated in adiposity development, either at the whole body level or at the mechanistic level. These SNPs were: rs9939609 (FTO), rs633715 (SEC16B), rs2002675 (TRA2B and ETV5), and rs2947411 (TMEM18) which were associated with childhood or adulthood BMI in GWAS (Frayling et al., 2007; Thorleifsson et al., 2009; Willer et al., 2009); rs4929923 (TRIM66), rs3914188 (ECE2), rs10899489 (GAB2), rs466639 (RXRG), rs13187289 (PHF15), and rs7359257 (IQCH) which were associated with BMI in adults in the GIANT consortium in a secondary analysis of the Elks et al (2010) paper; and rs6589964 (BSX), rs10423674 (CRTC1), and rs4840086 (PRDM13 and MCHR2) which are located in or near genes implicated in the regulation of energy homeostasis and body weight in animal models (Pissios et al., 2006; Sakkou et al., 2007; Altarejos et al., 2008; Wu et al., 2009). Examination of the effect of GRS29 on BMI during childhood and adolescence provides a test of whether the effects of GRS42 were solely due to the effects of known adiposity related variants.
Phenotype data
Serial weight and height measurements were collected at six monthly intervals from two to 20 years of age using standard anthropometric methods (Lohman et al., 1998). BMI (weight (kg)/height2 (m)) was calculated if weight and height were available at the same study assessment. Our goal was to compare the association of the sexual development genetic risk scores with BMI and height before, during, and after puberty, and we did so by using information on the age at peak height velocity (Age@PHV) to “center” the serial anthropometric data relative to this developmental landmark. A triple logistic model implemented in the statistical program AUXAL (version 3) was applied to each individual’s serial stature data and it was from these growth curves that Age@PHV was estimated (Bock et al., 2003).
The BMI and height measurements closest to Age@PHV were chosen, then the measurements closest to two, four, and six years before and after Age@PHV were chosen. A two year window around each of these seven time points was allowed, so that there were seven, mutually exclusive time points being compared: −6.99 to −5.00 years before PHV (called Age@PHV−6 years), −4.99 to −3.00 years before PHV (Age@PHV−4 years), −2.99 to −1.00 year before PHV (Age@PHV−2 years), −0.99 before to +0.99 year after PHV (Age@PHV), +1.00 to +2.99 years after PHV (Age@PHV+2 years), +3.00 to +4.99 years after PHV (Age@PHV+4 years), and +5.00 to +6.99 years after PHV (Age@PHV+6 years). Not all of the children had anthropometric data at each of the seven time points, although there were never less than 363 children with BMI and height at any time point. In total, 3,537 serial measurements of BMI and height were used in the present analysis (mean = 6.4 measurements per child). To account for the variation in the age of the measurements within a time point, the BMI and height data were converted to Z-scores according to the CDC 2000 reference (Kuczmarski et al., 2000).
Quantitative genetic analysis
The effects of GRS42 and GRS29 on BMI and height phenotypes were modeled in SOLAR using variance components based procedures that account for the non-independence among family members (Almasy and Blangero, 1998; Boerwinkle et al., 1986). Additive genetic effects and residual environmental effects were modeled as random effects, while GRS42 (or GRS29), sex, year of birth, exact age from Age@PHV, and also GRS42 (or GRS29)-by-sex and GRS42 (or GRS29)-by-year of birth interactions were modeled as fixed effects (Almasy and Blangero, 1998; Boerwinkle et al., 1986). Significance of individual covariate effects were tested using likelihood ratio tests comparing models where covariate effects were estimated against models where covariate effects were set to zero. The heritability (h2) of BMI and height phenotypes (i.e., the phenotypic variance attributable to additive genetic effects; h2 = σ2G/σ2P, where σ2G is the additive genetic variance and σ2P is the total phenotypic variance) has been studied extensively and was not the focus of this investigation, but as heritabilities are estimated from the above models we present them and their standard errors in Tables 2–4. Additional models that excluded GRS42 (or GRS29) terms and its interactions with sex and birth year were performed, and the proportion of the variance in each trait explained by GRS42 (or GRS29) and its interactions was calculated as the difference in explained variance between these models and the full aforementioned models.
Table 2.
Associations between a genetic risk score (GRS42) for advanced sexual development and BMI Z-scores1 from six years before to six years after age at peak height velocity (Age@PHV): Estimates from quantitative genetic models2.
| −6 years | −4 years | −2 years | Age@PHV | +2 years | +4 years | +6 years | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| N | 501 | 533 | 546 | 548 | 539 | 507 | 363 |
|
| |||||||
| B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | |
| p-value | p-value | p-value | p-value | p-value | p-value | p-value | |
| Heritability (h2) | 0.614 (0.099) | 0.551 (0.101) | 0.648 (0.099) | 0.669 (0.099) | 0.838 (0.082) | 0.732 (0.097) | 0.536 (0.153) |
| <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |
| GRS42 (estimate for boys born in 1928)3 | 0.031 (0.016) | 0.035 (0.016) | 0.038 (0.017) | 0.035 (0.016) | 0.049 (0.015) | 0.039 (0.016) | 0.075 (0.026) |
| 0.051 | 0.028 | 0.027 | 0.035 | 0.001 | 0.013 | 0.004 | |
| Sex: | |||||||
| Boys (referent) | -- | -- | -- | -- | -- | -- | -- |
| Girls | −0.057 (0.074) | −0.019 (0.076) | −0.031 (0.079) | −0.052 (0.077) | 0.123 (0.068) | 0.114 (0.072) | 0.019 (0.108) |
| 0.445 | 0.802 | 0.698 | 0.501 | 0.070 | 0.114 | 0.863 | |
| Year of birth (referent: 1928) | −0.002 (0.002) | −0.001 (0.002) | 0.001 (0.003) | 0.002 (0.003) | 0.002 (0.002) | 0.005 (0.002) | 0.006 (0.003) |
| 0.391 | 0.629 | 0.795 | 0.518 | 0.430 | 0.046 | 0.070 | |
| GRS42*sex | −0.012 (0.019) | −0.012 (0.020) | −0.024 (0.021) | −0.009 (0.020) | −0.036 (0.018) | −0.030 (0.019) | −0.054 (0.029) |
| 0.527 | 0.541 | 0.248 | 0.664 | 0.042 | 0.120 | 0.061 | |
| GRS42*year of birth | 0.001 (0.001) | 0.001 (0.001) | 0.001 (0.001) | 0.000 (0.001) | 0.000 (0.001) | 0.001 (0.001) | 0.001 (0.001) |
| 0.048 | 0.030 | 0.073 | 0.458 | 0.421 | 0.140 | 0.224 | |
|
| |||||||
| Variance explained by covariates (%) | 3.7 | 3.0 | 1.4 | 2.0 | 2.6 | 3.0 | 4.1 |
|
| |||||||
| Variance explained by GRS42 and its interactions (%) | 2.3 | 2.8 | 1.3 | 1.5 | 2.2 | 1.7 | 2.7 |
Z-scores were calculated according to the CDC 2000 growth reference.
Models were fitted in SOLAR using a maximum-likelihood variance components-based procedure.
B(SE) for GRS42 are per risk allele increase.
Table 4.
Associations between a genetic risk score (GRS29) for advanced sexual development (excluding known adiposity variants) and BMI Z-scores1 from six years before to six years after age at peak height velocity (Age@PHV): Estimates from quantitative genetic models2.
| −6 years | −4 years | −2 years | Age@PHV | +2 years | +4 years | +6 years | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| N | 501 | 533 | 546 | 548 | 539 | 507 | 363 |
|
| |||||||
| B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | |
| p-value | p-value | p-value | p-value | p-value | p-value | p-value | |
| Heritability (h2) | 0.606 (0.100) | 0.539 (0.103) | 0.630 (0.101) | 0.672 (0.101) | 0.837 (0.084) | 0.727 (0.097) | 0.525 (0.154) |
| <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |
| GRS29 (estimate for boys born in 1928)3 | 0.013 (0.018) | 0.009 (0.019) | 0.013 (0.020) | 0.021 (0.019) | 0.033 (0.017) | 0.039 (0.018) | 0.078 (0.028) |
| 0.454 | 0.628 | 0.503 | 0.277 | 0.052 | 0.030 | 0.006 | |
| Sex: | |||||||
| Boys (referent) | -- | -- | -- | -- | -- | -- | -- |
| Girls | −0.049 (0.074) | −0.010 (0.076) | −0.023 (0.080) | −0.046 (0.078) | 0.130 (0.068) | 0.122 (0.072) | 0.026 (0.108) |
| 0.512 | 0.892 | 0.769 | 0.557 | 0.058 | 0.091 | 0.812 | |
| Year of birth (referent: 1928) | −0.002 (0.002) | −0.001 (0.002) | 0.001 (0.003) | 0.002 (0.003) | 0.002 (0.002) | 0.005 (0.002) | 0.006 (0.003) |
| 0.456 | 0.681 | 0.795 | 0.508 | 0.412 | 0.048 | 0.045 | |
| GRS29*sex | −0.001 (0.023) | 0.002 (0.024) | −0.017 (0.025) | −0.016 (0.024) | −0.031 (0.021) | −0.039 (0.022) | −0.061 (0.032) |
| 0.966 | 0.946 | 0.481 | 0.519 | 0.145 | 0.078 | 0.061 | |
| GRS29*year of birth | 0.002 (0.001) | 0.002 (0.001) | 0.001 (0.001) | 0.001 (0.001) | 0.001 (0.001) | 0.001 (0.001) | 0.002 (0.001) |
| 0.019 | 0.014 | 0.067 | 0.353 | 0.209 | 0.041 | 0.104 | |
|
| |||||||
| Variance explained by covariates (%) | 3.4 | 2.3 | 1.1 | 0.9 | 1.5 | 3.4 | 4.3 |
|
| |||||||
| Variance explained by GRS29 and its interactions (%) | 1.9 | 2.0 | 1.0 | 0.4 | 1.2 | 2.1 | 3.0 |
Z-scores were calculated according to the CDC 2000 growth reference.
Models were fitted in SOLAR using a maximum-likelihood variance components-based procedure.
B(SE) for GRS29 are per risk allele increase.
Because each individual menarche SNP may have a different effect size on a phenotype, we wanted to be certain that a composite risk score (i.e., GRS42) is a valid representation of the cumulative effect of each SNP. For this validation analysis, we used the seven BMI Z-score phenotypes. For each phenotype, a model in which all 42 SNPs were included as covariates but were constrained to have equal effect was compared to a model in which the effect of each SNP was estimated separately. For all seven BMI Z-score phenotypes, there were no significant differences between the two models being compared using the likelihood ratio test (df = 41, all p-values > 0.057). Further, the variances explained by covariates of the two models being compared were always similar. These results confirm that the genetic risk score used in this analysis is not a biased representation of the aggregate SNP effects.
RESULTS
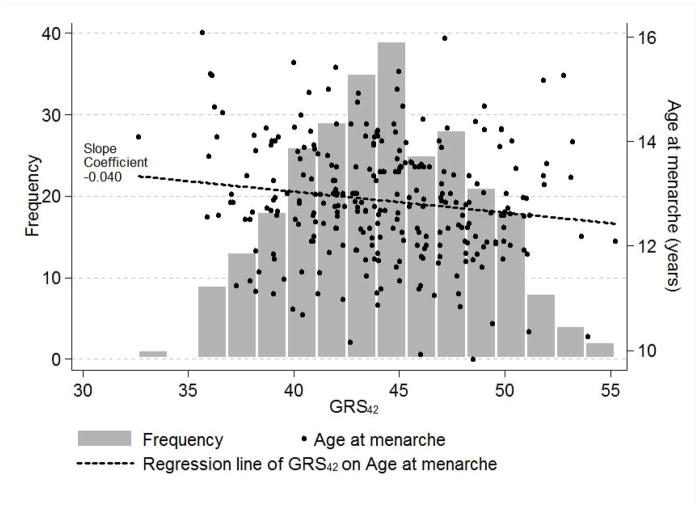
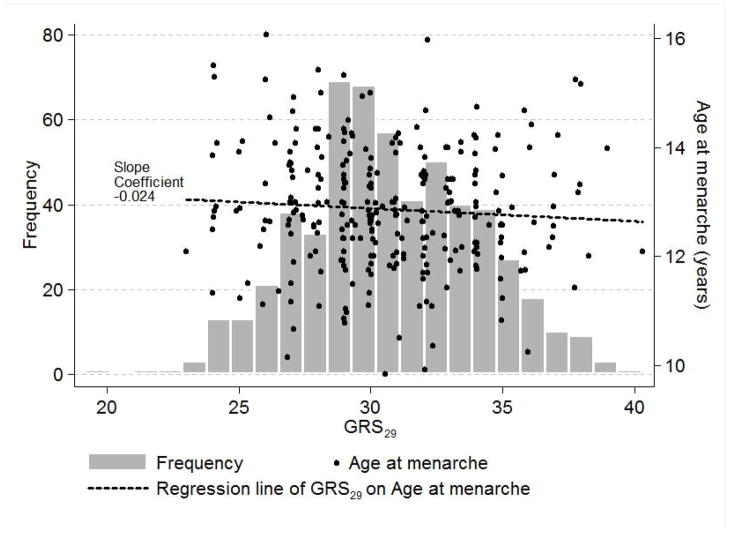
Mean Age@PHV was 12.5 years, with a mean difference between boys and girls of two years (mean in girls = 11.5 years, mean in boys = 13.5 years) (p < 0.001 using an independent samples t-test). Anthropometric data were collected between 3.5 to 20.0 years of chronological age (Table 1). Mean age at menarche for 276 girls (i.e., 99% of the analysis sample of 279 girls) was 12.9 years. In a crude analysis, age at menarche for these girls was regressed on GRS42 (mean = 44.4 SD = 3.9) and also GRS29 (mean = 30.8 SD = 3.3) to confirm phenotypic associations with age at menarche in Fels Longitudinal Study girls that could easily be plotted (Figs. 1a,b). A one risk allele increase in GRS42 was negatively associated with earlier age at menarche expressed as decimal years (beta = −0.040, p = 0.013), as was GRS29, to a lesser and non-significant extent (beta = −0.024, p = 0.241). The same regression models, but with Age@PHV as the outcome, showed that GRS42 and GRS29 had effects (albeit non-significant) on another measure of developmental timing in the expected direction (beta = −0.024, p = 0.139; beta = −0.022, p = 0.264, respectively). The effects of GRS42 and GRS29 on age at menarche in these girls were also analyzed in SOLAR in quantitative genetics models (unadjusted for covariates) that accounted for the familial structure of the sample. The effects of GRS42 and GRS29 on age at menarche (beta = −0.036, p = 0.025; beta = −0.025, p = 0.228, respectively) were similar to those obtained in the crude regression analyses. The h2 estimate of age at menarche was 0.44 (SE = 0.17) from the model including GRS42 and 0.48 (SE = 0.17) from the model including GRS29, and the variances explained by GRS42 and GRS29 were 3.1% and 1.3%, respectively.
Table 1.
Description of study data, by age from age at peak height velocity (Age@PHV).
| −6 years | −4 years | −2 years | Age@PHV | +2 years | +4 years | +6 years | |
|---|---|---|---|---|---|---|---|
| N | 501 | 533 | 546 | 548 | 539 | 507 | 363 |
|
| |||||||
| Mean (range) age (years) from PHV | −6.00 (−6.99 to −5.12) | −4.00 (−4.91 to −3.19) | −2.00 (−4.91 to −3.19) | −0.02 (−0.71 to 0.75) | 1.98 (1.17 to 2.95) | 3.96 (3.04 to 4.98) | 5.91 (5 to 6.99) |
| Mean (range) age (years) | 6.59 (3.48 to 10.01) | 8.54 (4.87 to 12.01) | 10.47 (6.97 to 14.00) | 12.46 (8.94 to 16.20) | 14.44 (10.59 to 18.18) | 16.34 (13.32 to 19.98) | 17.73 (14.97 to 20.00) |
| N (%) girls | 254 (50.7) | 270 (50.7) | 276 (50.5) | 275 (50.2) | 272 (50.5) | 265 (52.3) | 248 (68.3) |
| Mean (range) birth year | 1959 (1928 to 1992) | 1960 (1928 to 1992) | 1960 (1928 to 1992) | 1960 (1928 to 1992) | 1960 (1928 to 1992) | 1960 (1928 to 1992) | 1961 (1929 to 1991) |
| Mean (SD) BMI (kg/m2) | 15.63 (1.63) | 16.24 (2.10) | 17.22 (2.57) | 18.58 (2.95) | 20.29 (3.07) | 21.41 (3.08) | 21.36 (3.12) |
| Mean (SD) BMI Z-score1 | −0.07 (0.91) | −0.10 (0.95)* | −0.14 (1.01)* | −0.10 (0.99)* | 0.05 (0.92) | 0.04 (0.91) | 0.05 (0.90) |
| Mean (SD) weight (kg) | 22.53 (5.17) | 28.19 (6.68) | 34.91 (8.37) | 45.08 (10.74) | 57.37 (12.29) | 62.93 (12.51) | 64.97 (13.51) |
| Mean (SD) weight Z-score | 0.02 (0.89) | −0.00 (0.91) | −0.11 (0.97)* | 0.01 (0.95) | 0.33 (0.85)* | 0.26 (0.82)* | 0.17 (0.88)* |
| Mean (SD) height (cm) | 119.35 (10.17) | 130.96 (9.64) | 141.55 (9.55) | 154.90 (10.31) | 167.50 (10.53) | 170.94 (9.85) | 169.84 (9.79) |
| Mean (SD) height Z-score | 0.17 (0.89)* | 0.10 (0.88)* | 0.09 (0.89)* | 0.19 (0.95)* | 0.48 (0.88)* | 0.38 (0.91)* | 0.37 (0.90)* |
Z-scores were calculated according to the CDC 2000 growth reference.
Significantly different from the CDC 2000 median (i.e., zero) at p < 0.05.
Figure 1.
Figure 1a. The relationship of a genetic risk score (GRS42) for advanced sexual development to age at menarche in 276 Fels Longitudinal Study girls.
Figure 1b. The relationship of a genetic risk score (GRS29) for advanced sexual development to age at menarche in 276 Fels Longitudinal Study girls.
Quantitative genetic models indicated a significant positive association of GRS42 with BMI Z-score phenotypes at every time point (p-values < 0.05), except at Age@PHV−6 years where the association was marginal (p = 0.051) (Table 2). None of the sex or GRS42-by-sex interaction terms were significant except at Age@PHV+2 years. At Age@PHV−6 years and −4 years, the GRS42-by-year of birth interaction terms were significant and positive, indicating a stronger effect of GRS42 on pre-pubertal BMI as birth year increased. The amount of variance explained by the covariates ranged from 1.4 to 4.1%, with GRS42 and its interactions explaining between 1.3 to 2.8%. In the models for height Z-scores, a significant effect of GRS42 was observed only at Age@PHV; a one unit increase in GRS42 was associated with a 0.035 unit increase in height Z-score (Table 3).
Table 3.
Associations between a genetic risk score (GRS42) for advanced sexual development and height Z-scores1 from six years before to six years after age at peak height velocity (Age@PHV): Estimates from quantitative genetic models2.
| −6 years | −4 years | −2 years | Age@PHV | +2 years | +4 years | +6 years | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| N | 501 | 533 | 546 | 548 | 539 | 507 | 363 |
|
| |||||||
| B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | B(SE) | |
| p-value | p-value | p-value | p-value | p-value | p-value | p-value | |
| Heritability (h2) | 0.814 (0.069) | 0.777 (0.074) | 0.818 (0.082) | 0.798 (0.099) | 0.875 (0.072) | 0.893 (0.066) | 0.939 (0.053) |
| <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |
| GRS42 (estimate for boys born in 1928)3 | 0.004 (0.015) | 0.008 (0.014) | 0.023 (0.01) | 0.035 (0.015) | 0.011 (0.014) | −0.006 (0.014) | 0.014 (0.022) |
| 0.805 | 0.560 | 0.115 | 0.022 | 0.440 | 0.687 | 0.512 | |
| Sex: | |||||||
| Boys (referent) | -- | -- | -- | -- | -- | -- | -- |
| Girls | −0.073 (0.070) | −0.180 (0.067) | −0.055 (0.067) | −0.088 (0.073) | −0.135 (0.066) | −0.127 (0.070) | −0.009 (0.093) |
| 0.293 | 0.008 | 0.415 | 0.226 | 0.041 | 0.071 | 0.921 | |
| Year of birth (referent: 1928) | 0.003 (0.002) | 0.003 (0.002) | 0.003 (0.002) | 0.005 (0.002) | 0.003 (0.002) | 0.005 (0.002) | 0.003 (0.003) |
| 0.187 | 0.137 | 0.166 | 0.052 | 0.146 | 0.051 | 0.248 | |
| GRS42*sex | −0.006 (0.018) | −0.014 (0.017) | −0.027 (0.018) | −0.025 (0.019) | −0.020 (0.017) | −0.009 (0.019) | −0.022 (0.024) |
| 0.750 | 0.422 | 0.127 | 0.186 | 0.250 | 0.622 | 0.369 | |
| GRS42*year of birth | 0.000 (0.001) | 0.000 (0.001) | 0.000 (0.001) | 0.000 (0.001) | −0.001 (0.001) | −0.001 (0.001) | −0.001 (0.001) |
| 0.715 | 0.573 | 0.812 | 0.618 | 0.292 | 0.307 | 0.242 | |
|
| |||||||
| Variance explained by covariates (%) | 1.1 | 2.0 | 0.6 | 1.5 | 2.3 | 2.6 | 3.4 |
|
| |||||||
| Variance explained by GRS42 and its interactions (%) | 0.0 | 0.1 | 0.1 | 0.0 | 0.4 | 0.8 | 0.8 |
Z-scores were calculated according to the CDC 2000 growth reference.
Models were fitted in SOLAR using a maximum-likelihood variance components-based procedure.
B(SE) for GRS42 are per risk allele increase.
In sensitivity analyses (data not shown), models of the effects of GRS42 on BMI and height Z-score phenotypes stratified by sex were performed to ensure that our approach of pooling data from boys and girls together did not obscure possible sex differences. Beta estimates (of the effects of GRS42, year of birth, and GRS42-by-year of birth) were very similar for boys and girls when considered separately, with significant main effects of GRS42 on BMI Z-scores only at Age@PHV +2 and +6yrs in boys and Age@PHV in girls. In addition, a restricted risk score composed of 32 SNPs that retained significance in replication analysis in the Elks et al (2010) study was calculated and tested in sex combined models; estimated effects on BMI and height Z-score phenotypes were marginally smaller than in the presented GRS42 analysis and associated p-values were marginally larger, thereby indicating that part of the previously observed genetic signal in the GRS42 analysis had been lost by excluding 10 genome-wide significant but un-replicated menarche variants. The restricted risk score also explained less variance than GRS42, thereby providing additional support for our use of all 42 known menarche SNPs.
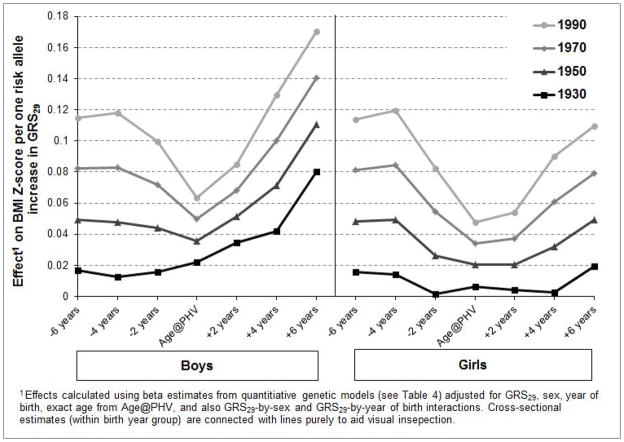
We then examined GRS29, which excluded 13 variants known to also influence adiposity, and observed significant main effects on BMI Z-scores at Age@PHV+4 years and +6 years (Table 4). There were significant GRS29-by-year of birth interaction effects at Age@PHV −6 years, −4 years, and +4 years, again indicating a stronger genetic effect as birth year increased. This amplified effect of GRS29 on childhood BMI Z-scores across the 20th century is shown in Fig 2. The secular trend was greatest at the most extreme time points (i.e., Age@PHV −6 years and +6 years) and was somewhat diminished at Age@PHV. In 1930, each additional risk allele was associated with an estimated increase in BMI Z-score at Age@PHV −6 years of approximately 0.02 units. This was compared to the corresponding statistic for 1990, where each additional risk allele was associated with an increase in BMI Z-score of approximately 0.12 units [i.e., (βyear of birth*62 + βGRS29*1 + βyear of birth-by-GRS29*62) − (βyear of birth*62 + βGRS29*2 + βyear of birth-by-GRS29*2)] after holding all other covariates equal. GRS29 had no significant effects on height Z-score phenotypes (data not shown).
Figure 2.
Amplified effects of a one risk allele increase in a genetic risk score (GRS29) for advanced sexual development on childhood BMI Z-scores across the 20th century.
DISCUSSION
In the present analysis, we tested the combined effect of a large number of well-replicated common genetic variants involved in female sexual development on peri-pubertal BMI and height in both boys and girls. Our primary finding was that these variants together demonstrated a significant main effect on BMI prior to, during, and after puberty, thereby indicating that sexual development and adiposity in childhood and adolescence share a common genetic basis. Even after discounting 13 SNPs previously known to influence adiposity, the remaining 29 SNPs together had significant main effects on post-pubertal BMI. Our data also show, for the first time, that the effect of menarche SNPs on pre-pubertal BMI was stronger in children born more recently compared to those born earlier in the century, thereby suggesting that the developmental genetic susceptibility to elevated BMI may have only been “uncovered” in the more obesogenic environments of the recent past. Our novel findings are that genetic factors regulating sexual developmental timing also influence peri-pubertal BMI, additional SNPs beyond those already identified as possible adiposity variants are involved, and that these genetic effects have changed over time.
By centering the serial anthropometric data on a common developmental landmark (i.e., Age@PHV) we ensured that significant effects of genetic risk scores on BMI and height at a given age were not artifacts due to variations in developmental timing. It is after all only logical that, for example, 16-year-old adolescents with earlier sexual development (and thus a greater genetic risk for earlier development) would have greater sizes because they would have experienced an increased growth velocity, associated with the pubertal growth spurt, for longer than adolescents with later sexual development. The approach we used made it possible to maximize the sample by presenting data for both sexes combined because boys and girls were assessed on a common developmental time scale.
The 42-SNP genetic risk score used in the present analysis together had a persistent significant main effect on BMI, whereas the 29-SNP genetic risk score only had significant main effects in the post-pubertal period. The main effects cannot, however, be interpreted on their own. GRS29-by-year of birth interactions were significant in the pre-pubertal period, so (as shown in Fig. 2) the interpretation here is that GRS29 did demonstrate a positive association with BMI across the age range being studied, but during the pre-pubertal period the effects were stronger in more recent birth years. The secular trend in the effect of GRS29 on BMI Z-scores was somewhat diminished at Age@PHV (Fig. 2), which may represent developmental timing genes not acting on BMI as much when height is increasing at its maximum velocity. In a study of the effects of one menarche SNP (rs314276 at the locus LIN28B) on BMI across the life course, Ong et al. (2011) found that significant associations were present only after age 15 years. The authors concluded that there were causal effects of sexual developmental timing on adolescent and adulthood BMI, despite the fact that the SNP they used has known pleiotropic effects on childhood BMI as well as menarche timing (Ong et al., 2009). The findings of the present study, which considered pleiotropy, support the notion that (at least in girls) earlier menarche leads to a causal increase in BMI soon after Age@PHV.
No significant effects of either genetic risk score on height were observed at any of the time points, with the exception of GRS42 at Age@PHV. Our interpretation is that children with a higher number of risk alleles for advanced sexual development had greater weights not different heights. This finding contrasts with that of Elks et al (2010) in which significant associations of individual menarche SNPs were found with adult height. The greater statistical power of the Elks et al (2010) study (N ~ 130,000) is a possible explanation for the difference in results. One published study has investigated the effects of a single menarche SNP (rs7759938 near the locus LIN28B) and also a single partially correlated SNP (rs314277) on linear growth and observed significant effects over the entire life course (Widen et al., 2010), but otherwise little is known about the effects of menarche SNPs on linear growth during childhood and adolescence.
In the 1970s it was proposed by Frisch and Revelle (1970) that there is a critical weight necessary for the onset of menses in girls, thereby explaining the earlier occurrence of menarche in girls with greater weights. Despite receiving much criticism (Johnston et al., 1975; Cameron, 1976; Scott and Johnston, 1982), the critical weight hypothesis paved the way for subsequent literature from the fields of human growth and development, epidemiology of eating disorders, and microbiology in mouse and human models, indicating that sexual developmental timing and BMI are mechanistically linked (Lock and Fitzpatrick, 2009; Swenne, 2004, 2008; Cheung et al., 1997; Chehab et al., 1997; Pierce and Leon, 2005). Genetic mutations in the structural gene for leptin (LEP) are, for example, known to be associated with obesity and failure of normal pubertal progression (Clement et al., 1998; Strobel et al., 1998). The present study adds to this literature by demonstrating that sexual developmental timing and BMI in childhood and adolescence have a shared genetic control beyond the genes already known to influence adiposity.
It is well known that high pre-pubertal BMI as well as obesity and high levels of measured adiposity are associated with advanced sexual development (Salsberry et al., 2009; Kaplowitz et al., 2001; Demerath et al., 2004; Buyken et al., 2009; Rosenfield et al., 2009), which is in turn is associated with high post-pubertal BMI and levels of adiposity, as well as increased risk for obesity and various co-morbidities (He et al., 2010; Pierce and Leon, 2005; Taeymans et al., 2008; Pierce et al., 2010, 2011). Few studies have, however, been able to assess the causal associations between the pace of sexual development and adiposity traits, whilst also adjusting for the myriad factors that confound these relationships, such as socioeconomic status (Laitinen et al., 2001), smoking (Flegal et al., 1995), and alcohol intake (Molarius, 2003). A strength of the present paper is that it examines the relationship of sexual developmental timing to BMI using an instrumental variable approach, in which the relationship between the instrumental variable (i.e., GRS42 and GRS29) and the outcome cannot be confounded by factors such as diet (Jacobson-Dickman and Lee, 2009) due to randomization of alleles at gamete formation; GRS42 and GRS29 had effects on BMI that were independent of variations in diet and any other potential confounding factors.
Evidence that the 29 menarche SNPs (not known to also influence adiposity) together had a greater influence on BMI as birth year increased was found. In 1930 a one risk allele increase in GRS29 approximated a 0.02 unit increase in BMI Z-score at Age@PHV −6 years, but in 1990 a one risk allele increase approximated a 0.12 unit increase in BMI Z-score at the same time point. Because the range of risk alleles for GRS29 in the present study was 19–40 and the standard deviation about the mean value was 3.3 risk alleles, a comparison of a modern day child with one risk allele at each of the 29 loci to a modern day child with no risk alleles, resulting in a BMI advantage of 3.48 Z-scores (i.e., 0.12*29), is improbable. A difference between children of just a few risk alleles causing a much smaller increase in BMI is far more probable. The interpretation of the gene-by-year of birth finding is that as the nutritional environment has changed (e.g., decreasing cost of energy-dense foods, reduction in the required physical activity levels for most occupations) genetic susceptibility to earlier maturation and obesity becomes more apparent. It is possible that over the examined time period, individuals with higher genetic burden for accelerated sexual development are for the first time “allowed” by liberalization of the environment to alter dietary intake and energy expenditure to support their genetic potential for rapid weight gain and earlier sexual development. A recent study in 1,474,065 Swedish 18–19 year old conscripts supports the idea that genetic influences on BMI may have increased over time; they found a secular increase in the heritability and additive genetic variance of BMI over the 20th century (Rokholm et al., 2011).
The sample size in the present study is an obvious limitation and may have limited our power and thus ability to detect statistically significant results. Nonetheless, the Fels Longitudinal Study offers a unique opportunity to assess the relationships between menarche SNPs and BMI and height because of the extensive longitudinal data collected on the participants. Multiple statistical tests were used which will have increased the risk of false-positive findings, but in a paper that presented only 21 separate models multiple testing is less of an issue than in large-scale genetic studies that often test thousands of associations. Other limitations include a sample composed exclusively of European-American children born in southwestern Ohio, which limits generalizability of the results to other ethnic groups, and available data ended with those born in 1992, thus the effects of SNPs on BMI in children born into the most recent obesogenic environment were not fully captured.
CONCLUSIONS
Common genetic variants influencing sexual developmental timing were associated with BMI before and after puberty in the present sample of European-American boys and girls. This finding of pleiotropy, due not only to known adiposity variants but other developmental variants as well, supports the previously documented phenotypic associations between adiposity and sexual maturation. Our results help to explain the concurrent secular trends from the 1950s onward in earlier sexual development and increasing adolescent BMI, by showing that the shared genetic underpinnings of developmental timing and peri-pubertal BMI have had a greater influence among children born later in the 20th century than those born earlier. This gene-by-environment interaction suggests that children with a genetic predisposition to earlier sexual development might avoid an elevated BMI through alteration of their nutritional environment, perhaps through changes in diet and physical activity. Research comparing the influence of behavioral change on children with high versus low genetic risk of advanced sexual development is a necessary next step.
Acknowledgments
Funding: This study was supported by grants from the National Institutes of Health: R01 HD012252 (Czerwinski) and R01 HD053685 (Demerath).
We acknowledge the life-long contributions of the Fels Longitudinal Study participants, and the study staff members, without whose commitment and enthusiasm the work of the study could never have been completed. In particular, we would like to thank Frances Tyleshevski for her help in the creation of the dataset and the past and present Lifespan Health Research Center data collection team for their contributions.
Footnotes
The authors have no conflicts of interest.
LITERATURE CITED
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin: rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Ahmed ML, Ong KK, Dunger DB. Childhood obesity and the timing of puberty. Trends Endocrinol Metab. 2009;20:237–42. doi: 10.1016/j.tem.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos JY, Goebel N, Conkright MD, Inoue H, Xie J, Arias CM, Sawchenko PE, Montminy M. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat Med. 2008;14:1112–17. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SE, Dallal GE, Must A. Relative weight and race influence average age at menarche: results from two nationally representative surveys of US girls studied 25 years apart. Pediatrics. 2003;111:844–50. doi: 10.1542/peds.111.4.844. [DOI] [PubMed] [Google Scholar]
- Bock RD, Du Toit SHC, Thissen D. AUXAL: auxological analysis of longitudinal measurements of human stature. Version 3. Chicago, IL: Scientific Software International; 2003. [Google Scholar]
- Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–94. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- Briefel RR, Johnson CL. Secular trends in dietary intake in the United States. Annu Rev Nutr. 2004;24:401–31. doi: 10.1146/annurev.nutr.23.011702.073349. [DOI] [PubMed] [Google Scholar]
- Buyken AE, Karaolis-Danckert N, Remer T. Association of prepubertal body composition in healthy girls and boys with the timing of early and late pubertal markers. Am J Clin Nutr. 2009;89:221–30. doi: 10.3945/ajcn.2008.26733. [DOI] [PubMed] [Google Scholar]
- Cameron N. Weight and skinfold variation at menarche and the critical body weight hypothesis. Ann Hum Biol. 1976;3:279–82. doi: 10.1080/03014467600001451. [DOI] [PubMed] [Google Scholar]
- Chehab FF, Mounzih K, Lu R, Lim ME. Early onset of reproductive function in normal female mice treated with leptin. Science. 1997;275:88–90. doi: 10.1126/science.275.5296.88. [DOI] [PubMed] [Google Scholar]
- Cheung CC, Thornton JE, Kuijper JL, Weigle DS, Clifton DK, Steiner RA. Leptin is a metabolic gate for the onset of puberty in the female rat. Endocrinol. 1997;138:855–8. doi: 10.1210/endo.138.2.5054. [DOI] [PubMed] [Google Scholar]
- Chumlea WC, Schubert CM, Roche AF, Kulin HE, Lee PA, Himes JH, Sun SS. Age at menarche and racial comparisons in US girls. Pediatrics. 2003;111:110–3. doi: 10.1542/peds.111.1.110. [DOI] [PubMed] [Google Scholar]
- Clement K, Vaisse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, Gourmelen M, Dina C, Chambaz J, Lacorte JM, Basdevant A, Bougneres P, Lebouc Y, Froguel P, Guy-Grand B. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- Davey-Smith G, Ebrahim S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- Davison KK, Susman EJ, Birch LL. Percent body fat at age 5 predicts earlier pubertal development among girls at age 9. Pediatrics. 2003;111:815–21. doi: 10.1542/peds.111.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerath EW, Li J, Sun SS, Chumlea WC, Remsberg KE, Czerwinski SA, Towne B, Siervogel RM. Fifty-year trends in serial body mass index during adolescence in girls: the Fels Longitudinal Study. Am J Clin Nutr. 2004;80:441–6. doi: 10.1093/ajcn/80.2.441. [DOI] [PubMed] [Google Scholar]
- Deshmukh-Taskar P, Nicklas TA, Morales M, Yang SJ, Zakeri I, Berenson GS. Tracking of overweight status from childhood to young adulthood: the Bogalusa Heart Study. Eur J Clin Nutr. 2006;60:48–57. doi: 10.1038/sj.ejcn.1602266. [DOI] [PubMed] [Google Scholar]
- Dewey KG. Energy and protein requirements during lactation. Ann Rev Nutr. 1997;17:19–36. doi: 10.1146/annurev.nutr.17.1.19. [DOI] [PubMed] [Google Scholar]
- Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, Lunetta KL, Visser JA, Byrne EM, Cousminer DL, Gudbjartsson DF, Esko T, Feenstra B, Hottenga JJ, Koller DL, Kutalik Z, Lin P, Mangino M, Marongiu M, McArdle PF, Smith AV, Stolk L, van Wingerden SH, Zhao JH, Albrecht E, Corre T, Ingelsson E, Hayward C, Magnusson PK, Smith EN, Ulivi S, Warrington NM, Zgaga L, Alavere H, Amin N, Aspelund T, Bandinelli S, Barroso I, Berenson GS, Bergmann S, Blackburn H, Boerwinkle E, Buring JE, Busonero F, Campbell H, Chanock SJ, Chen W, Cornelis MC, Couper D, Coviello AD, d’Adamo P, de Faire U, de Geus EJ, Deloukas P, Doring A, Smith GD, Easton DF, Eiriksdottir G, Emilsson V, Eriksson J, Ferrucci L, Folsom AR, Foroud T, Garcia M, Gasparini P, Geller F, Gieger C, Gudnason V, Hall P, Hankinson SE, Ferreli L, Heath AC, Hernandez DG, Hofman A, Hu FB, Illig T, Jarvelin MR, Johnson AD, Karasik D, Khaw KT, Kiel DP, Kilpelainen TO, Kolcic I, Kraft P, Launer LJ, Laven JS, Li S, Liu J, Levy D, Martin NG, McArdle WL, Melbye M, Mooser V, Murray JC, Murray SS, Nalls MA, Navarro P, Nelis M, Ness AR, Northstone K, Oostra BA, Peacock M, Palmer LJ, Palotie A, Pare G, Parker AN, Pedersen NL, Peltonen L, Pennell CE, Pharoah P, Polasek O, Plump AS, Pouta A, Porcu E, Rafnar T, Rice JP, Ring SM, Rivadeneira F, Rudan I, Sala C, Salomaa V, Sanna S, Schlessinger D, Schork NJ, Scuteri A, Segre AV, Shuldiner AR, Soranzo N, Sovio U, Srinivasan SR, Strachan DP, Tammesoo ML, Tikkanen E, Toniolo D, Tsui K, Tryggvadottir L, Tyrer J, Uda M, van Dam RM, van Meurs JB, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Weedon MN, Wichmann HE, Willemsen G, Wilson JF, Wright AF, Young L, Zhai G, Zhuang WV, Bierut LJ, Boomsma DI, Boyd HA, Crisponi L, Demerath EW, van Duijn CM, Econs MJ, Harris TB, Hunter DJ, Loos RJ, Metspalu A, Montgomery GW, Ridker PM, Spector TD, Streeten EA, Stefansson K, Thorsteinsdottir U, Uitterlinden AG, Widen E, Murabito JM, Ong KK, Murray A GIANT Consortium. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euling SY, Herman-Giddens ME, Lee PA, Selevan SG, Juul A, Sorensen TI, Dunkel L, Himes JH, Teilmann G, Swan SH. Examination of US puberty-timing data from 1940 to 1994 for secular trends: panel findings. Pediatrics. 2008;121:172–91. doi: 10.1542/peds.2007-1813D. [DOI] [PubMed] [Google Scholar]
- Flegal KM, Troiano RP, Pamuk ER, Kuczmarski RJ, Campbell SM. The influence of smoking cessation on the prevalence of overweight in the United States. New Eng J Med. 1995;333:1165–70. doi: 10.1056/NEJM199511023331801. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman DS, Khan LK, Serdula MK, Dietz WH, Srinivasan SR, Berenson GS. Relation of age at menarche to race, time period, and anthropometric dimensions: the Bogalusa Heart Study. Pediatrics. 2002;110:43. doi: 10.1542/peds.110.4.e43. [DOI] [PubMed] [Google Scholar]
- Frisch RE, Revelle R. Height and weight at menarche and a hypothesis of critical body weights and adolescent events. Science. 1970;169:397–9. doi: 10.1126/science.169.3943.397. [DOI] [PubMed] [Google Scholar]
- Hamill PV, Drizd TA, Johnson CL, Reed RB, Roche AF. NCHS growth curves for children birth-18 years. United States. Vital Health Stat. 1977;11(165):1–74. [PubMed] [Google Scholar]
- Hamilton JP. Epigenetics: principles and practice. Dig Dis. 2011;29:130–5. doi: 10.1159/000323874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Zhang C, Hunter DJ, Hankinson SE, Buck Louis GM, Hediger ML, Hu FB. Age at menarche and risk of type 2 diabetes: results from 2 large prospective cohort studies. Am J Epidemiol. 2010;171:334–4. doi: 10.1093/aje/kwp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson-Dickman E, Lee MM. The influence of endocrine disruptors on pubertal timing. Curr Opin Endocrinol Diabetes Obes. 2009;16:25–30. doi: 10.1097/med.0b013e328320d560. [DOI] [PubMed] [Google Scholar]
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607–17. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W, Soloway LE, Erickson D, Choh A, Lee M, Chumlea WC, Siervogel RM, Czerwinski SA, Towne B, Demerath EW. A changing pattern of childhood BMI growth during the 20th century: 70 years of data from the Fels Longitudinal Study. Am J Clin Nutr. 2012;95:1136–43. doi: 10.3945/ajcn.111.022269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston FE, Roche AF, Schell LM, Norman H, Wettenhall B. Critical weight at menarche. Critique of a hypothesis. Am J Dis Child. 1975;129:19–23. doi: 10.1001/archpedi.1975.02120380011003. [DOI] [PubMed] [Google Scholar]
- Kaplowitz PB, Slora EJ, Wasserman RC, Pedlow SE, Herman-Giddens ME. Earlier onset of puberty in girls: relation to increased body mass index and race. Pediatrics. 2001;108:347–53. doi: 10.1542/peds.108.2.347. [DOI] [PubMed] [Google Scholar]
- Kaprio J, Rimpela A, Winter T, Viken RJ, Rimpela M, Rose RJ. Common genetic influences on BMI and age at menarche. Hum Biol. 1995;67:739–53. [PubMed] [Google Scholar]
- Komlos J, Breitfelder A, Sunder M. The transition to post-industrial BMI values among US children. Am J Hum Biol. 2009;21:151–60. doi: 10.1002/ajhb.20806. [DOI] [PubMed] [Google Scholar]
- Kuczmarski RJ, Ogden CL, Grummer-Strawn LM, Flegal KM, Guo SS, Wei R, Mei Z, Curtin LR, Roche AF, Johnson CL. CDC growth charts: United States. Adv Data. 2000;314:1–27. [PubMed] [Google Scholar]
- Laitinen J, Power C, Jarvelin MR. Family social class, maternal body mass index, childhood body mass index, and age at menarche as predictors of adult obesity. Am J Clin Nutr. 2001;74:287–94. doi: 10.1093/ajcn/74.3.287. [DOI] [PubMed] [Google Scholar]
- Lakshman R, Forouhi N, Luben R, Bingham S, Khaw K, Wareham N, Ong KK. Association between age at menarche and risk of diabetes in adults: results from the EPIC-Norfolk cohort study. Diabetologia. 2008;51:781–6. doi: 10.1007/s00125-008-0948-5. [DOI] [PubMed] [Google Scholar]
- Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–63. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Ann Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock JD, Fitzpatrick KK. Anorexia nervosa. Clin Evid. 2009;2009:1011. [PMC free article] [PubMed] [Google Scholar]
- Lohman T, Roche A, Martorell R. Anthropometric standardization reference manual. Champaign, IL: Human Kinetics Publishers, Inc; 1998. [Google Scholar]
- Mamun AA, Hayatbakhsh MR, O’Callaghan M, Williams G, Najman J. Early overweight and pubertal maturation: pathways of association with young adults’ overweight: a longitudinal study. Int J Obes. 2009;33:14–20. doi: 10.1038/ijo.2008.220. [DOI] [PubMed] [Google Scholar]
- Molarius A. The contribution of lifestyle factors to socioeconomic differences in obesity in men and women: a population-based study in Sweden. Eur J Epidemiol. 2003;18:227–34. doi: 10.1023/A:1023376012627. [DOI] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, Gillson CJ, Glaser B, Golding J, Hardy R, Khaw KT, Kuh D, Luben R, Marcus M, McGeehin MA, Ness AR, Northstone K, Ring SM, Rubin C, Sims MA, Song K, Strachan DP, Vollenweider P, Waeber G, Waterworth DM, Wong A, Deloukas P, Barroso I, Mooser V, Loos RJ, Wareham NJ. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009;41:729–33. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Wills AK, Wong A, Wareham NJ, Loos RJ, Kuh D, Hardy R. Associations between the pubertal timing-related variant in LIN28B and BMI vary across the life course. J Clin Endocrinol Metab. 2011;96:125–9. doi: 10.1210/jc.2010-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce MB, Kuh D, Hardy R. The role of BMI across the life course in the relationship between age at menarche and diabetes, in a British Birth Cohort. Diabet Med. doi: 10.1111/j.1464-5491.2011.03489.x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce MB, Kuh D, Hardy R. Role of lifetime body mass index in the association between age at puberty and adult lipids: findings from men and women in a British birth cohort. Ann Epidemiol. 2010;20:676–82. doi: 10.1016/j.annepidem.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce MB, Leon DA. Age at menarche and adult BMI in the Aberdeen children of the 1950s cohort study. Am J Clin Nutr. 2005;82:733–9. doi: 10.1093/ajcn/82.4.733. [DOI] [PubMed] [Google Scholar]
- Pissios P, Bradley RL, Maratos-Flier E. Expanding the scales: The multiple roles of MCH in regulating energy balance and other biological functions. Endocr Rev. 2006;27:606–20. doi: 10.1210/er.2006-0021. [DOI] [PubMed] [Google Scholar]
- Prentice AM, Spaaij CJ, Goldberg GR, Poppitt SD, van Raaij JM, Totton M, Swann D, Black AE. Energy requirements of pregnant and lactating women. Eur J Clin Nutr. 1996;50:82–110. [PubMed] [Google Scholar]
- Roche AF. Growth, maturation and body composition: The Fels Longitudinal Study 1929–1991. Cambridge: Cambridge University Press; 1992. [Google Scholar]
- Rokholm B, Silventoinen K, Tynelius P, Gamborg M, Sorensen TI, Rasmussen F. Increasing genetic variance of body mass index during the Swedish obesity epidemic. PLoS One. 2011;6:27135. doi: 10.1371/journal.pone.0027135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfield RL, Lipton RB, Drum ML. Thelarche, pubarche, and menarche attainment in children with normal and elevated body mass index. Pediatrics. 2009;123:84–8. doi: 10.1542/peds.2008-0146. [DOI] [PubMed] [Google Scholar]
- Sakkou M, Wiedmer P, Anlag K, Hamm A, Seuntjens E, Ettwiller L, Tschop MH, Treier M. A role for brain-specific homeobox factor Bsx in the control of hyperphagia and locomotory behavior. Cell Metab. 2007;5:450–63. doi: 10.1016/j.cmet.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Salsberry PJ, Reagan PB, Pajer K. Growth differences by age of menarche in African American and white girls. Nurs Res. 2009;58:382–90. doi: 10.1097/NNR.0b013e3181b4b921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe K, Willemsen G, Kyvik KO, Mortensen J, Boomsma DI, Cornes BK, Davis CJ, Fagnani C, Hjelmborg J, Kaprio J, De Lange M, Luciano M, Martin NG, Pedersen N, Pietilainen KH, Rissanen A, Saarni S, Sorensen TI, Van Baal GC, Harris JR. Sex differences in heritability of BMI: a comparative study of results from twin studies in eight countries. Twin Res. 2003;6:409–21. doi: 10.1375/136905203770326411. [DOI] [PubMed] [Google Scholar]
- Scott EC, Johnston FE. Critical fat, menarche, and the maintenance of menstrual cycles: a critical review. J Adolesc Health Care. 1982;2:249–60. doi: 10.1016/s0197-0070(82)80059-4. [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–37. [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Sengul H, Weeks DE. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered. 2001;52:121–31. doi: 10.1159/000053366. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, MAGIC, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ Procardis Consortium. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42L:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel A, Issad T, Camoin L, Ozata M, Strosberg AD. A leptin missense mutation associated with hypogonadism and morbid obesity. Nat Genet. 1998;18:213–5. doi: 10.1038/ng0398-213. [DOI] [PubMed] [Google Scholar]
- Swenne I. Weight and growth requirements for menarche in teenage girls with eating disorders, weight loss and primary amenorrhea. Horm Res. 2008;69:146–51. doi: 10.1159/000112587. [DOI] [PubMed] [Google Scholar]
- Swenne I. Weight requirements for return of menstruations in teenage girls with eating disorders, weight loss and secondary amenorrhoea. Acta Paediatrica. 2004;93:1449–55. doi: 10.1080/08035250410033303. [DOI] [PubMed] [Google Scholar]
- Taeymans J, Hebbelinck M, Borms J, Clarys P, Abidi H, Duquet W. Tracking of adult adiposity in early, average and late maturing children: a thirty year longitudinal growth study. J Sport Med Phys Fitness. 2008;48:326–34. [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Towne B, Czerwinski SA, Demerath EW, Blangero J, Roche AF, Siervogel RM. Heritability of age at menarche in girls from the Fels Longitudinal Study. Am J Phys Anthropol. 2005;128:210–9. doi: 10.1002/ajpa.20106. [DOI] [PubMed] [Google Scholar]
- Troiano RP, Flegal KM, Kuczmarski RJ, Campbell SM, Johnson CL. Overweight prevalence and trends for children and adolescents. The National Health and Nutrition Examination Surveys, 1963 to 1991. Arch Pediatr Adolesc Med. 1995;149:1085–91. doi: 10.1001/archpedi.1995.02170230039005. [DOI] [PubMed] [Google Scholar]
- Wardle J, Carnell S, Haworth CM, Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr. 2008;87:398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- Wattigney WA, Srinivasan SR, Chen W, Greenlund KJ, Berenson GS. Secular trend of earlier onset of menarche with increasing obesity in black and white girls: the Bogalusa Heart Study. Ethn Dis. 1999;9:181–9. [PubMed] [Google Scholar]
- Widen E, Ripatti S, Cousminer DL, Surakka I, Lappalainen T, Jarvelin MR, Eriksson JG, Raitakari O, Salomaa V, Sovio U, Hartikainen AL, Pouta A, McCarthy MI, Osmond C, Kajantie E, Lehtimaki T, Viikari J, Kahonen M, Tyler-Smith C, Freimer N, Hirschhorn JN, Peltonen L, Palotie A. Distinct variants at LIN28B influence growth in height from birth to adulthood. Am J Hum Genet. 2010;86:773–82. doi: 10.1016/j.ajhg.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstrale M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN Wellcome Trust Case Control Consortium; Genetic Investigation of Anthropometric Traits Consortium. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Dumalska I, Morozova E, van den Pol A, Alreja M. Melanin-concentrating hormone directly inhibits GnRH neurons and blocks kisspeptin activation, linking energy balance to reproduction. Proc Natl Acad Sci USA. 2009;106:17217–22. doi: 10.1073/pnas.0908200106. [DOI] [PMC free article] [PubMed] [Google Scholar]