Abstract
Cardiomyocyte phenotype changes significantly in 2D culture systems depending on the substrate composition and organization. Given the variety of substrates that are used both for basic cardiac cell culture studies and for regenerative medicine applications, there is a critical need to understand how the different matrices influence cardiac cell mechanics. In the current study, the mechanical properties of neonatal rat cardiomyocytes cultured in a subconfluent layer upon aligned and unaligned collagen and fibronectin matrices were assessed over a two week period using atomic force microscopy. The elastic modulus was estimated by fitting the Hertz model to force curve data and the percent relaxation was determined from stress relaxation curves. The Quasilinear Viscoelastic (QLV) and Standard Linear Solid (SLS) models were fit to the stress relaxation data. Cardiomyocyte cellular mechanical properties were found to be highly dependent on matrix composition and organization as well as time in culture. It was observed that the cells stiffened and relaxed less over the first 3 to 5 days in culture before reaching a plateau in their mechanical properties. After day 5, cells on aligned matrices were stiffer than cells on unaligned matrices and cells on fibronectin matrices were stiffer than cells on collagen matrices. No such significant trends in percent relaxation measurements were observed but the QLV model fit the data very well. These results were correlated with observed changes in cellular structure associated with culture on the different substrates and analyzed for cell-to-cell variability.
Introduction
Cardiomyocytes are the fundamental contractile muscle cells of the heart, accounting for over 90% of the myocardium volume (1). Native adult cardiomyocytes are rod-shaped, measuring approximately 100 μm by 25 μm, with contractile myofibrils running parallel to the long axis of the cell. Cardiomyocytes orient themselves end-to-end and join to form cardiac muscle fibers that are interconnected in branching, three-dimensional networks. At each cell-cell intersection, intercalated discs permit mechanical and electrical coupling so that when one portion of the cardiac muscle fiber network is stimulated, the impulse passes to other fibers and the entire structure contracts simultaneously.
There is a critical need to understand the cellular mechanisms of heart failure. As such, cardiomyocytes are often studied in a 2D culture environment (2). In traditional cell culture, an extracellular matrix (ECM) protein is spread over a substrate to promote cellular adhesion. The ECM proteins type I collagen, type III collagen, fibronectin, and laminin are commonly used in cardiomyocyte culture studies as these proteins are present in the native cellular microenvironment (1, 3). Cardiomyocyte behavior has been shown to change significantly depending on the substrate composition (4–8). For example, a combination of the naturally occurring ECM proteins and proteoglycans (termed cardiogel) promotes larger cardiomyocytes that are more strongly adhered and begin spontaneous contraction earlier than fibronectin or laminin alone (4, 8). The orientation of the ECM fibers has also been shown to greatly influence cardiomyocyte phenotype. Cells cultured on randomly oriented matrix proteins take on a spread and flattened morphology with disorganized myofibrils and diffuse intercellular junctions (9). Researchers have been able to promote a more in vivo-like phenotype by using highly organized culture conditions. In one method, cardiomyocyte shape is passively dictated by the shape of etched groves in which the cells are cultured (10–12). In the other method, cardiomyocytes actively take on an in vivo-like phenotype through interactions with underlying ECM proteins that are in an aligned orientation. Within this technique, many researchers simply use a cell scraper to align the ECM proteins (6, 13). While this method is quick and inexpensive, more complex methods such as printing matrix components in a line pattern have been shown to yield more precisely aligned cardiomyocytes with coordinated beating (9, 14).
While the effects of matrix composition (ECM protein) and orientation (unaligned vs. aligned) on cardiomyocyte phenotype have been well documented, changes in cytoskeletal architecture associated with phenotypical changes often lead to altered whole-cell mechanical properties. Because cardiomyocytes are very mechanically active cells, a great deal of research has focused on cardiomyocyte mechanics, but no researchers have yet investigated changes in cardiomyocyte mechanical properties with culture on these different types of matrices. Atomic force microscopy (AFM) is often used to study cell mechanics, with the majority of studies employing simple force-indentation curves to estimate the apparent cellular elastic modulus (15). More recently, stress relaxation experiments have been used to quantify viscoelastic properties of cells (16–19). Notably, several studies investigating cells in 2D culture environments observed a very high level of variability in whole-cell mechanical properties between cells from a single population (coefficient of variation 43% – 103% for AFM studies) (20–26). Previous AFM studies of cardiomyocyte whole-cell mechanical properties investigated topics such as changes with animal age (27) and comparison to other cell types (28). Other researchers have utilized AFM force mapping techniques to investigate subcellular cardiomyocyte mechanical properties (13, 29, 30). In many studies, cardiomyocyte contraction was chemically inhibited to simplify force measurements (27–29). Few studies have used actively beating cells.
The goal of this study was to characterize the mechanical properties of actively beating cardiomyocytes on aligned and unaligned collagen and fibronectin matrices in 2D culture using AFM techniques over a period of fifteen days. Cardiomyocytes cultured on aligned matrices take on a more uniform in-vivo-like phenotype including a more complex cytoskeleton. Therefore, we hypothesized that the cells on aligned substrates would exhibit higher elastic moduli and more homogeneous mechanical properties overall. The results of this study not only provide valuable insights about cardiomyocyte mechanics, they also highlight the need for researchers to be mindful in their substrate selection for cell culture studies. Additionally, this work represents the first stress relaxation experiments conducted on cardiomyocytes, where their true viscoelastic nature is assessed using standard biomechanical models. The fifteen day time course of this study also highlights the active response of the cardiomyocytes to their microenvironment over time. Finally, this study is the first to examine cell-to-cell variations in cardiomyocyte mechanical properties.
Materials and Methods
Substrates
Glass slides (12 mm diameter, Fisher Scientific, Pittsburgh, PA, USA) were oxygen plasma cleaned for 10 minutes, followed by UV sterilization in 70% ethanol for at least 1 hour. The slides were then dried and coated with thin layers of 1 mg/ml solutions of either type I collagen or fibronectin (BD Biosciences, Rockville, MD, USA). Each matrix solution was applied in both aligned and unaligned orientations to produce 4 samples: aligned collagen, unaligned collagen, aligned fibronectin, and unaligned fibronectin. A modified inkjet printer was utilized to align the substrate matrices within thin printed lines (14). For this process, 100 μl of the matrix solution was used as ink within HP 26 cartridges that were emptied and thoroughly cleaned prior to use. The matrix solutions were printed from the cartridges onto the substrates in a line pattern (line width ≈ 20 μm) in three successive prints using a HP DeskJet 500 printer. The entire printing apparatus was located within a tabletop biohazard cabinet to ensure aseptic technique. The unaligned samples were simply prepared by spreading 5 μl of the matrix solution over the slide surface. The matrix-coated slides were incubated at 37°C for approximately 24 hours before plating cells on them.
Cell Culture
To isolate neonatal rat cardiomyocytes, the hearts of 3 day old pups were removed, minced, and subjected to collagenase dissociation as described previously (6, 31, 32). Three-day-old Sprague-Dawley neonatal rats were euthanized by decapitation according to procedures approved by the Clemson University Institutional Animal Care and Use Committee (Protocol number AUP2010-032). The methods of euthanasia for neonatal animals are consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. The cardiomyocytes were collected as previously described. Simply, the heart was isolated and minced into 3 mm pieces with scissors and first digested with trypsin solution (0.14mg/ml with no EDTA) overnight, then shaken at 75 rpm in the collagenase solution (1 mg/ml Collagenase II, GIBCO; 0.24 unit/ml Neutral Protease, Worthington) for 1.5 hours. The fibroblasts were removed through incubating the cells in a 150-cm2 flask with culture medium (DMEM solution containing 20% Fetal bovine serum (FBS)) for 2 hours at 37 °C. The purified cardiomyocytes (~95% myocyte) solution were diluted to 100 K cells/ml, and then seeded into 35-mm glass-bottom culture dishes coated with laminin (20 μg/ml). Each matrix-coated slide received approximately 57,000 cardiomyocytes (50,000 cells/cm2) to create a near-confluent layer of cells. All samples were maintained in 24-well plates under standard conditions (37°C, 5% CO2). The cardiomyocytes were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Fisher), supplemented with 8% horse serum (Invitrogen Life Technologies, Chicago, IL, USA), 5% newborn bovine serum (Invitrogen), 1% penicillin/streptomycin (Fisher), sodium bicarbonate (Sigma-Aldrich, St. Louis, MO, USA), HEPES (pH 7.4) (Sigma-Aldrich), amphotericin B (Fisher), and proliferative cell inhibitor Cytosin β-D-arabinofuranoside (Sigma-Aldrich) to prevent fibroblast growth. The media was changed every 48 hours and cell growth was monitored daily via light microscopy. After 24 hours in culture, most cells had started contracting at a frequency of approximately 1 Hz. Figure 1 illustrates the structural differences between the cardiomyocytes on unaligned and aligned substrates after 5 days in culture.
Figure 1.
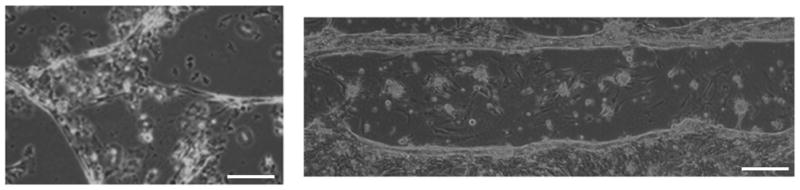
Representative brightfield microscopy images of day 5 cardiomyocytes on unaligned (left) and aligned (right) collagen matrices (scale bar = 20 μm).
AFM Indentation
Atomic force microscopy (AFM) cytoindentation experiments were performed 1 to 15 days after the cardiomyocytes were seeded on the slides. Specifically, a sample was tested each day on days 1, 2, 3, 5, 7, 9, and 15. For all AFM experiments, an Asylum Research MFP-3D AFM (Asylum Research, Santa Barbara, CA, USA) was operated in contact mode with a fluid cell. The cells remained on their slides throughout the study, with warm (37°C) media exchanged every 30 minutes. A 5 μm diameter borosilicate spherical-tipped AFM probe on a silicon-nitride cantilever with a spring constant of 0.12 N/m (Novascan, Ames, IA, USA) was used to mechanically probe individual cells. Before each experiment, the deflection sensitivity (nm/V) was determined by indenting onto a clean glass slide with media present. An optical microscope (Olympus IX81) with a 10x objective was used to position the tip of the cantilever over the center of a cell before data was collected. Each cardiomyocyte was indented five times to approximately 1 μm depth at 5 μm/sec (5 force curves). Each indentation was done immediately following cell contraction to ensure the force measurements represented relaxed cells, as cardiomyocytes have been shown to exhibit different elastic moduli in contraction and relaxation (13). Each cell was also subjected to two stress relaxation experiments. For each stress relaxation experiment, the cell was indented to a depth of 1 μm at the same speed of 5 μm/sec, followed by a 60 second hold period. For each sample, 20 cardiomyocytes from throughout the culture area were probed in this manner (5 force curves and 2 stress relaxation curves per cell).
Force Curve Analysis
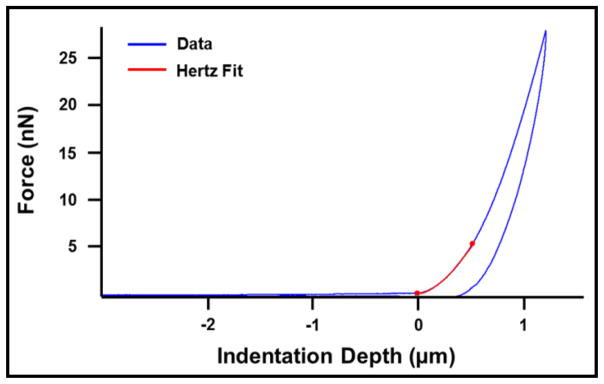
The force curves were exported from the AFM software and a custom MATLAB script (MathWorks, Natick, MA, USA) was used to normalize and shift the curves to a common zero point (“contact point”). The contact point was determined as the point where the slope of the curve changes by fitting the curve to a 2-region model (22). To obtain a measure of individual cell stiffness, the apparent elastic modulus of each cell was calculated by fitting the Hertz model to the data. This model was fit to the first 500 nm of indentation data. This indentation depth was chosen as approximately 10 percent of the average cell height as measured by contact mode AFM imaging (average cardiomyocyte height ≈ 5 μm), because the Hertz model only remains accurate within the 10 percent strain range (33). The Hertz model for a spherical indenter is defined by the following equation:
In this equation, F is the measured force (N), E is the apparent elastic modulus (Pa), υ is Poisson’s ratio (0.5) (34), R is the radius of the spherical indenter (2.5 μm), and δ is the indentation depth (m). Figure 2 shows a typical force curve with the overlying Hertz model fit to the first 500 nm of indentation.
Figure 2.
Sample cardiomyocyte force curve with Hertz model fit to the first 500 nm of indentation.
Stress Relaxation Curve Analysis
The stress relaxation curves were also exported from the AFM software and analyzed using custom MATLAB scripts. All curves were first shifted along the y-axis to move the baseline (minimum) force to zero. The curves were then normalized by setting the maximum force value to one, meaning all the relaxation data fell in the zero to one normalized force range. The normalized data can be described as a reduced relaxation function (G (t)) and the percentage of relaxation can be calculated as:
The percent relaxation during the 60 second hold was calculated in this manner for each stress relaxation curve. Each curve was also fit with two relaxation models: the Quasilinear Viscoelastic (QLV) model and the Standard Linear Solid (SLS) model. The QLV reduced relaxation function G (t) contains 3 parameters (c, τ1, and τ2) with a continuous relaxation spectrum (S (τ) = c/τ) between τ1 and τ2:
The constant c is unitless and represents a relative measure of viscous energy dissipation while τ1 and τ2 are time constants governing short and long term relaxation behavior, respectively (35). The SLS reduced relaxation function is:
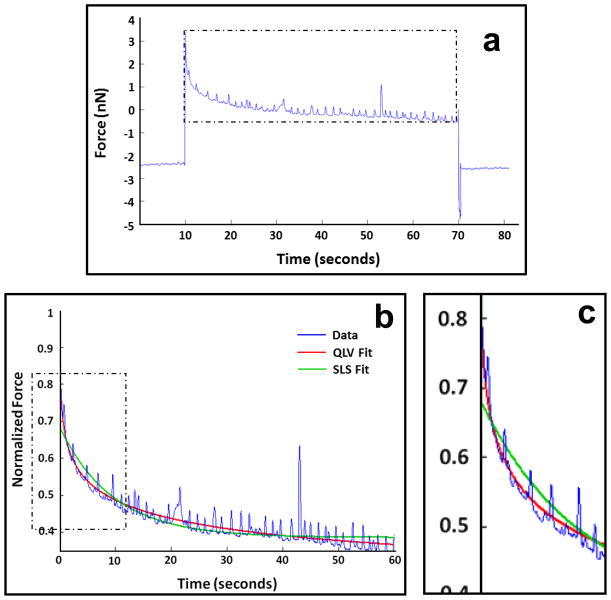
In this equation, ER is the reduced-relaxation modulus, τσ is the relaxation time for constant stress, and τε is the relaxation time for constant strain (35). Figure 3 shows typical cardiomyocyte stress relaxation data with QLV and SLS model fits.
Figure 3.
Sample cardiomyocyte stress relaxation curve. Spikes in data represent cellular contractions (beating). Raw data (a) and the relaxation portion of the data after normalization with overlying QLV and SLS model fits (b). Zoomed in section (c) shows how the SLS model fails to fit the fast initial relaxation in the data.
Statistics
Two-way analysis of variance (ANOVA) with replication was used to determine how the results (cellular elastic modulus and percent relaxation) were influenced by the factors sample type (aligned and unaligned collagen and fibronectin) and time (Days 1, 2, 3, 5, 7, 9, & 15). If the interaction between the two factors was found to be significant, each factor was then examined individually for significant differences between groups using a one-way ANOVA. Student’s t-tests were used to determine all statistically significant differences between samples on a given day and within samples across time points. For all tests, p-values less than 0.05 were considered statistically significant. Additionally, the coefficient of variation (COV = standard deviation / mean) was calculated within each sample as a measure of cell-to-cell variation and within each cell-loading session as a measure of data repeatability on a single point of indentation. The accuracy of the QLV and SLS models in predicting cellular stress relaxation behavior was accessed by calculating the coefficient of determination (R2) for each model fit.
Immunofluorescence
Day 5 cardiomyocytes on each sample were fixed and stained for nuclei, filamentous actin, and N-cadherin. Specifically, cells were fixed in 4 percent paraformaldehyde at room temperature for 10 minutes. Following 2 – 15 minute rises with PBS, the samples were incubated with a solution of 0.01M glycine and 0.1% Triton-X in PBS for 30 minutes, 5% bovine serum albumin (BSA) in PBS for 15 minutes, then 5% normal donkey serum / 1% BSA in PBS for 15 minutes. The samples were then treated with a mouse N-cadherin monoclonal primary antibody (1:100 in 1% BSA/PBS, Sigma-Aldrich) overnight at 4° C. The samples were rinsed with 1% BSA/PBS (2 × 15 minutes) and 5% normal donkey serum in 1% BSA/BPS (15 minutes) before application of a secondary donkey anti-mouse rhodamine (TRITC)-conjugated antibody (1:100 in 1% BSA/PBS, Jackson ImmunoResearch, West Grove, PA, USA) for 2 hours. Next, the samples were rinsed with 1% BSA/PBS (15 minutes) and twice with PBS. They were then incubated with Alexa Fluor 488 phalloidin (1:100 in PBS, Invitrogen) for 15 minutes. Finally, the samples were rinsed 3 times with PBS and mounted on microscope slides with SlowFade Gold antifade reagent with DAPI (Invitrogen). All samples were stored in the dark before microscopy. The samples were viewed using an Olympus IX81 spinning disk confocal microscope (Olympus, Tokyo, Japan) and digital images were collected and processed using MetaMorph Image Analysis software (Molecular Devices, Sunnyvale, CA, USA).
Results
Elastic Modulus
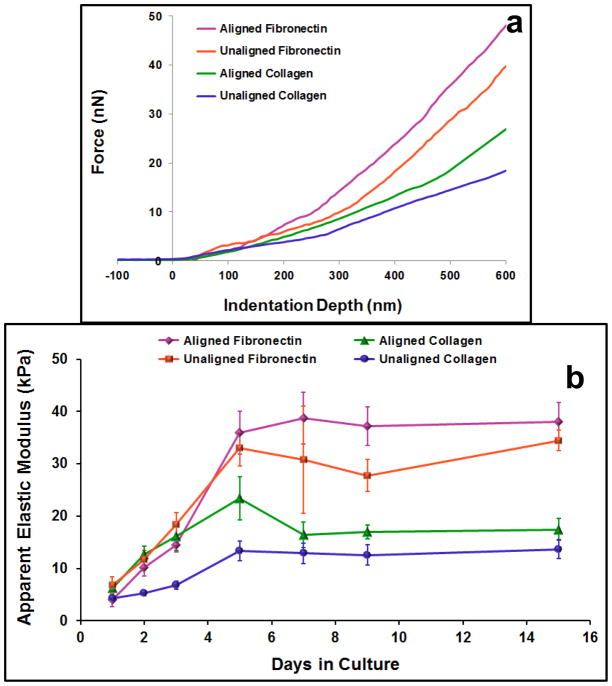
The mean apparent elastic moduli measures for cardiomyocytes on each substrate increased over the first 5 days in culture and then plateaued for the rest of the 15 day study (Figure 4). The results of the two-way ANOVA indicated that both sample type and time (p ≪ 0.05) were significant factors in determining cellular elastic moduli and the interaction between the factors was significant (p = 0.00008). The one-way ANOVA tests revealed that there were significant differences across time points within each sample type and across sample types within each time point (p ≤ 0.040). On day 1 in culture, the mean apparent elastic moduli for cells on each substrate ranged from 4.0 kPa to 6.8 kPa, with a significant difference between the cells on unaligned and aligned collagen substrates (p = 0.045). On days 2 and 3, cellular elastic modulus measures increased for each sample, with significant increases from day 1 to day 3 for each sample (p ≤ 0.005). On both of these days, cells on unaligned collagen were significantly less stiff than cells on the other substrates (p ≤ 0.015), which did not significantly differ from one another. Elastic moduli measures continued to increase from day 3 to day 5 (significant for all samples except aligned collagen, p ≤ 0.003). On day 5, the cells on unaligned collagen remained significantly less stiff than cells on all the other substrates (p ≤ 0.036). Additionally, the aligned collagen sample exhibited a significantly lower mean elastic modulus than the aligned fibronectin sample (p = 0.041). Within each sample, no more significant differences in elastic moduli measures with time were observed from day 5 through day 15. On day 7, aligned fibronectin samples remained significantly stiffer than unaligned and aligned collagen samples (p ≤ 0.0003). On both days 9 and 15, the unaligned and aligned collagen samples were significantly less stiff than the unaligned and aligned fibronectin samples (p ≤ 0.004). The final (day 15) mean elastic moduli measures were 13.7 kPa for unaligned collagen, 17.5 kPa for aligned collagen, 34.4 kPa for unaligned fibronectin, and 38.0 kPa for aligned fibronectin.
Figure 4.
Elastic moduli measures of cardiomyocytes on different substrates. Indentation portion of force curves for representative cells on each of the different substrates on day 5 in culture (a). The Hertz model was fit to the first 500 nm of indentation data to estimate the cellular apparent elastic modulus. Summary of all elastic moduli measures over 15 day culture period (b). Data presented as mean ± standard error. Elastic moduli measures for cells on all substrate types increased significantly over the first five days in culture. At the later time points, cells on fibronectin matrices were significantly stiffer than cells on collagen matrices.
The average cell-to-cell elastic modulus COVs were much greater than the corresponding repeated point measures for cardiomyocytes on each of the 4 sample types (Table 1). The levels of variation both from cell-to-cell and within repeated measures on a single cell were consistent across sample types. The average cell-to-cell COV for elastic moduli measures was relatively high at 57.2% for all samples. There was a drop in cell-to-cell variability from cells on unaligned fibronectin (68.2%) to cells on aligned fibronectin (55.5%), but this difference was not statistically significant. The average repeated point COV for elastic moduli measures was much smaller at 14.1%, indicating that the variation that was observed among elastic moduli measurements on different cells in a single sample was not due to the measurement technique, but was in fact due to differences in the cellular mechanical properties.
Table 1.
Cardiomyocyte cell-to-cell and repeated point coefficients of variation (COV) for elastic modulus and percent relaxation measures. Data from all days were averaged to get a measure for the average variation within each sample (substrate type).
| Samples | Average Cell-to-Cell COV (%) | Average Repeated Point COV (%) | |
|---|---|---|---|
| Elastic Modulus | Unaligned Collagen | 53.9 | 13.9 |
| Aligned Collagen | 51.3 | 14.9 | |
| Unaligned Fibronectin | 68.2 | 15.5 | |
| Aligned Fibronectin | 55.5 | 12.1 | |
| All (mean ± standard error) | 57.2 ± 6.2 | 14.1 ±1.5 | |
| Percent Relax | Unaligned Collagen | 37.3 | 7.3 |
| Aligned Collagen | 32.8 | 4.6 | |
| Unaligned Fibronectin | 39.3 | 7.4 | |
| Aligned Fibronectin | 40.2 | 6.3 | |
| All (mean ± standard error) | 37.4 ± 1.5 | 6.4 ± 0.6 |
Stress Relaxation
The mean percent relaxation measures for cardiomyocytes on each substrate decreased during the first 3 days of culture and then stayed relatively constant for the rest of the 15 day study (Figure 5). The results of the two-way ANOVA indicated that both sample type and time (p ≪ 0.05) were significant factors in determining cellular elastic moduli but there was no significant interaction between the factors (p = 0.895). On day 1 in culture, the mean percent relaxation measures for cells on each substrate ranged from 62.0% to 68.8%, with no significant differences between the different samples. Between days 1 and 2, the percent relaxation measures for all samples decreased (cells exhibited less relaxation during 60 second hold), but the drop was only significant for the cells on aligned fibronectin (p = 0.003). On day 2, the average percent relaxation measure for cells on unaligned collagen was significantly higher than that for aligned collagen (p = 0.017). From day 2 to day 3, percent relaxation measures continued to decrease, significantly for unaligned collagen (p = 0.040). The drop in percent relaxation measures between days 1 and 3 was significant for all samples (p ≤ 0.011). On day 3, there were no significant differences in percent relaxation measures among the different samples. Within each sample, no more significant differences in percent relaxation measures with time were observed from day 3 through day 15. On day 5, the unaligned collagen sample exhibited a significantly greater percent relaxation than the aligned collagen and aligned fibronectin samples (p ≤ 0.036). The only remaining statistically significant differences were between aligned and unaligned collagen on day 7 (p = 0.013) and aligned collagen and unaligned fibronectin on day 15 (p = 0.015). The final (day 15) mean percent relaxation measures during the 60 second hold were 52.6% for unaligned collagen, 46.0% for aligned collagen, 54.1% for unaligned fibronectin, and 50.6% for aligned fibronectin.
Figure 5.
Percent relaxation during 60 second hold for cardiomyocytes on different substrates. Normalized relaxation portion of curves for representative cells cultured on aligned collagen on days 1, 3, and 7 in culture (a). Summary of all percent relaxation measures over 15 day culture period (b). Data presented as mean ± standard error. Percent relaxation measures for cells on all substrate types decreased significantly over the first three days in culture. Although the unaligned substrates typically promoted higher percent relaxation measures than the aligned substrates at each time point, very few of these differences were statistically significant.
The levels of variation in percent relaxation measures both from cell-to-cell and within repeated measures on a single cell were consistent across sample types (no significant differences, Table 1). The average cell-to-cell COV for percent relaxation measures (37.4%) was lower than that for elastic moduli measures (57.2%) for all samples. The average repeated point COV for percent relaxation measures was very low at 6.4%, indicating that the variation that was observed among percent relaxation measurements on different cells in a single sample was not due to the measurement technique, but was in fact due to differences in the cellular mechanical properties.
In general, the QLV model fit the cardiomyocyte relaxation data very well (Figure 3). The mean coefficient of determination (R2) for QLV fits to all cardiomyocyte relaxation data was 0.89 ± 0.14 (standard deviation). Although the mean R2 for the SLS model fit was very similar (0.84 ± 0.13), it should be noted that the SLS model failed to fit the initial fast relaxation and then alternately overshot and undershot the remaining data (Figure 3). The R2 values would have been higher if the myocytes were in a relaxed state and not contracting during the hold period.
Immunofluorescence
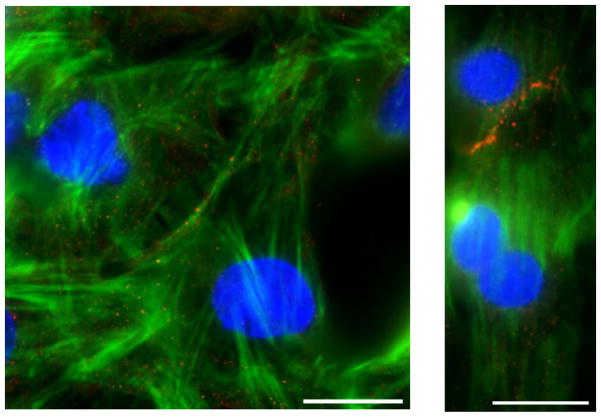
The cardiomyocytes cultured on the aligned collagen and fibronectin matrices typically lined up end-to-end and took on a phenotype similar to cells found in vivo (6). Cells on collagen and fibronectin matrices had similar morphological responses to substrate alignment. The immunofluorescence staining revealed that the actin fibers (components of myofibrils) were highly aligned within these cells and N-cadherin proteins (components of intercalated discs) were concentrated at the intersection between neighboring cells (Figure 6). In contrast to this, the cardiomyocytes on unaligned collagen and fibronectin matrices were typically spread in all directions with variable cell-cell interactions. Within these cells, the actin fibers were oriented in all directions and the N-cadherin was not more concentrated in any particular region.
Figure 6.
Representative immunofluorescence images of day 5 cardiomyocytes on an unaligned matrix (left) and an aligned matrix (right) (scale bar = 20 μm). Cells are stained for nuclei (blue), filamentous actin (green), and N-cadherin (red).
Discussion
The primary objective of this study was to characterize variations in cardiomyocyte mechanical properties resulting from diversified adhesion binding between the cells and their underlying matrices. To this end, neonatal cardiomyocytes were cultured on collagen and fibronectin matrices in aligned and unaligned orientations and their viscoelastic mechanical properties were evaluated between days one and fifteen in culture. Additionally, immunofluorescence straining was conducted to correlate the whole-cell mechanical property measurements with the varying internal cytoskeletal structure that cardiomyocytes are known to exhibit on these different types of substrates. The level of heterogeneity in mechanical property measurements between cardiomyocytes within each sample was also evaluated for further comparison of cellular behavior on the different matrices.
It is important to study cellular mechanical properties as cell growth, differentiation, migration, contractility, and apoptosis are all influenced by changes in cell shape and structural integrity (36–39). Any deviation in cell structural and mechanical properties can result in the breakdown of physiological function and may lead to disease (15, 40). In cardiomyocytes, mechanical properties are particularly indicative of force generation capabilities (27). In this study, elastic moduli measures increased over the first five days in culture before generally reaching a plateau for cardiomyocytes on all four sample types (aligned collagen, unaligned collagen, aligned fibronectin, unaligned fibronectin). This five day shift indicates that the cardiomyocytes were actively responding to their microenvironment, modifying their internal structure over this time period. From day five to day fifteen, the cardiomyocytes were the most stiff on aligned fibronectin (~37 kPa on day 15), followed by unaligned fibronectin (~32 kPa), aligned collagen (~18 kPa), and lastly unaligned collagen (~13 kPa) (Figure 4). Although most previous studies strictly investigated cardiomyocytes cultured on collagen matrices, our measures are on par with their AFM measurements of neonatal cardiomyocyte elastic moduli, including a previous study that found no significant changes in cardiomyocyte elastic moduli between days 5 and 20 in culture (13, 27, 41). This was the first study to show that aligned matrices promoted stiffer cardiomyocytes over unaligned matrices and that fibronectin matrices promoted stiffer cardiomyocytes over collagen matrices. These differences are most likely due to changes in the expression level and organization of cytoskeletal and contractile proteins within the cells on the different matrices resulting from diversified adhesion interactions (42). The differences in cardiomyocyte elastic moduli measures between cells cultured on collagen and fibronectin matrices were larger than the differences between aligned and unaligned matrices of the same matrix component, perhaps because there is a greater difference in integrin-ECM interactions with different matrix components. Specifically, cardiomyocytes bind to both type I collagen and fibronectin through integrin α3β1 but they additionally bind to fibronectin through integrin α5β1 (43). These additional cardiomyocyte-fibronectin integrin-based interactions would have resulted in stronger overall cell adhesion to the underlying substrate, which has previously been shown to correlate with increased cellular stiffness measurements (44).
Researchers have investigated viscoelastic mechanical properties of various types of adherent cells using AFM stress relaxation techniques, but this was the first time relaxation measurements were taken on cardiomyocytes (16–19, 45). The QLV model has been used extensively to model tissue behavior, but was only recently applied to cellular data (19, 46–48). The SLS model has previously been applied to both cells and tissues (19, 49, 50). In the present study, both models provided a good fit to the cardiomyocyte relaxation data with an average coefficient of determination (R2) of 0.89 for the QLV model and 0.84 for the SLS model. However, it should be noted that the SLS fit was less accurate in predicting the initial fast relaxation response of the cells than the QLV model. A comparison of R2 values for our data with those from a similar study involving vascular smooth muscle cells (QLV model R2 = 0.98, SLS model R2 = 0.91) highlights the reduction in fit in our data due to the cardiomyocyte contractions, but the same general trend of a better QLV model fit is consistent across cell types (19). Overall, the cardiomyocyte percent relaxation measures decreased over the first three days in culture for cells on all four sample types before reaching a plateau. On day 15, the cells on unaligned fibronectin exhibited the highest percentage of relaxation (54%), followed by unaligned collagen (53%), aligned fibronectin (51%), and lastly aligned collagen (46%) (Figure 5). In previous studies, disruption of the actin cytoskeleton within cells resulted in higher cellular percent relaxation measures as the fluid-like cytoplasm contributed more strongly to the relaxation behavior (19, 45). This suggests that the cardiomyocytes on all matrices developed a more robust actin network over the first 3 days in culture (percent relaxation decreased). Additionally, the cardiomyocytes on the aligned matrices (lower percent relaxation measures) likely had more robust actin networks than the cardiomyocytes on the unaligned matrices (higher percent relaxation measures), which is consistent with the immunofluorescence results. However, the cardiomyocytes exhibited more significant differences in their elastic moduli measures than in their percent relaxation measures when comparing behavior on the different matrices.
In order to correlate the elastic moduli and percent relaxation measurements with cellular morphology and internal actin content, cardiomyocytes on each of the substrates were imaged on day 5 using traditional brightfield and fluorescence microscopy techniques. Cardiomyocytes on both aligned substrates (collagen and fibronectin) generally positioned themselves end-to-end with an elongated morphology, much like native cells (Figure 1). Within the cells on the aligned matrices, the actin fibers were highly aligned and N-cadherin was localized between neighboring cells (Figure 6). Cardiomyocytes on both unaligned substrates exhibited a much more spread morphology and contained disorganized actin fibers and diffuse N-cadherin proteins. These morphological differences were anticipated, as several previous studies have outlined similar structural changes for cardiomyocytes cultured on aligned substrates (9–12, 14). In regards to the mechanical property measurements, these results give strength to the argument that highly oriented actin fibers within cells promote stiffer cells that relax less during a hold experiment (19, 51). In a previous study, force mapping revealed a significant correlation between elastic modulus and actin content within aligned cardiomyocytes (13). This suggests that the cardiomyocytes cultured upon aligned substrates not only took on an altered internal cytoskeletal arrangement, they likely exhibit increased overall actin content as well. In combination, these organizational factors explain the significantly altered whole-cell mechanical properties that were observed in this study.
Many previous studies investigating cellular mechanical properties in an in vitro culture environment have noted a high level of heterogeneity in whole-cell mechanical properties between cells from a single population (20–24). This heterogeneity has been observed in many cell populations (osteoblasts, chondrocytes, fibroblasts, endothelial cells, vascular smooth muscle cells) and with different measurement techniques (AFM, micropipette aspiration). It has been suggested that variations in cellular phenotype and local confluency are at least partially to blame for this high level of variability (21, 26). In order to more fully characterize differences in cardiomyocyte mechanical properties on the different substrates and to better understand this issue of mechanical heterogeneity in general, the level of variability in cardiomyocyte mechanical properties within each sample was assessed through calculation of the coefficient of variation. Since the aligned matrices provided a more uniform microenvironment for the cells, it was hypothesized that the cardiomyocytes cultured upon these matrices would take on a more homogeneous phenotype throughout the culture area, resulting in more homogeneous cellular mechanical properties. While the cells on the aligned matrices were very uniform in morphology and internal actin orientation from cell to cell, the level of variability in mechanical properties among the cells did not significantly decrease from the aligned samples to the unaligned samples, where the cells were highly variable in size and shape. For both elastic moduli and percent relaxation measures, the levels of heterogeneity (coefficients of variation) in cell-to-cell mechanical properties (average elastic moduli COV = 57%, average percent relaxation COV = 37%, Table 1) were within the typical range for AFM studies (COV = 30 – 128%) (22, 26). The levels of variability in repeated point measures (COV = 14% for elastic moduli, 6% for percent relaxation) were on par with previous studies as well (COV = 5%). Interestingly, for both cell-to-cell and repeated point measurements, the variability in percent relaxation measurements was consistently lower than the variability in elastic modulus measurements. The fact that variability measures did not decrease when the cells were more homogeneous in appearance (on aligned matrices) suggests that cell structure and mechanical properties may not be as closely correlated as anticipated and that other factors should be investigated as the cause of the high level of variability seen in cellular mechanical properties in 2D cell culture studies. For all cell types it is important to determine the source(s) of this heterogeneity to determine if the in vitro behavior we are observing is representative of cell behavior in vivo.
In summary, we more fully characterized diversified cardiomyocyte behavior when cultured upon aligned and unaligned matrices by measuring whole-cell viscoelastic mechanical properties for cardiomyocytes on four different matrix types over a fifteen day culture period. Over the first three to five days, the cardiomyocytes actively responded to their microenvironment by developing a more complex cytoskeleton (percent relaxation decreased) and stiffening (elastic modulus increased). Aligned matrices promoted cells with higher elastic moduli than cells on unaligned matrices. In a similar yet more significant fashion, fibronectin matrices promoted stiffer cells in comparison to collagen matrices. Percent relaxation differences among cells cultured upon the different matrices were less clear. Cardiomyocytes on aligned matrices were uniformly elongated with highly oriented actin fibers but their mechanical properties were just as variable from cell-to-cell within a single population as cardiomyocytes on unaligned matrices that were highly variable in appearance and internal actin orientation. These results should be considered for researchers planning cell culture studies where cell mechanics is an important factor.
Acknowledgments
The authors would like to thank Dr. Jason Hemmer for writing the custom MATLAB scripts that were used for data analysis. This research was supported by NIH K25 HL092228 and P20GM103499 grants.
Contributor Information
Sandra Deitch, Email: sdeitch@clemson.edu.
Bruce Z. Gao, Email: zgao@clemson.edu.
Delphine Dean, Email: finou@clemson.edu.
References
- 1.Walker CA, Spinale FG. The structure and function of the cardiac myocyte: A review of fundamental concepts. Journal of Thoracic and Cardiovascular Surgery. 1999;118(2):375–382. doi: 10.1016/S0022-5223(99)70233-3. [DOI] [PubMed] [Google Scholar]
- 2.Louch WE, Sheehan KA, Wolska BM. Methods in cardiomyocyte isolation, culture, and gene transfer. Journal of Molecular and Cellular Cardiology. 2011;51(3):288–298. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miner EC, Miller WL. A look between the cardiomyocytes: The extracellular matrix in heart failure. Mayo Clinic Proceedings. 2006;81(1):71–76. doi: 10.4065/81.1.71. [DOI] [PubMed] [Google Scholar]
- 4.VanWinkle WB, Snuggs MB, Buja LM. Cardiogel: A biosynthetic extracellular matrix for cardiomyocyte culture. In Vitro Cellular & Developmental Biology-Animal. 1996;32(8):478–485. doi: 10.1007/BF02723051. [DOI] [PubMed] [Google Scholar]
- 5.Borg TK, McFadden WA, Hastings JF, Goodwin RP, Goldsmith EC. The role of the extracellular matrix (ECM) in cardiac development. Faseb Journal. 2003;17(4):A359–A359. [Google Scholar]
- 6.Simpson DG, et al. Modulation of cardiac myocyte phenotype in-vitro by the composition and orientation of the extracellular-matrix. Journal of Cellular Physiology. 1994;161(1):89–105. doi: 10.1002/jcp.1041610112. [DOI] [PubMed] [Google Scholar]
- 7.Bullard TA, Borg TK, Price RL. The expression and role of protein kinase C in neonatal cardiac myocyte attachment, cell volume, and myofibril formation is dependent on the composition of the extracellular matrix. Microscopy and Microanalysis. 2005;11(3):224–234. doi: 10.1017/S1431927605050476. [DOI] [PubMed] [Google Scholar]
- 8.Bick RJ, Snuggs MB, Poindexter BJ, Buja LM, Van Winkle WB. Physical, contractile and calcium handling properties of neonatal cardiac myocytes cultured on different matrices. Cell Adhesion and Communication. 1998;6(4):301. doi: 10.3109/15419069809010789. [DOI] [PubMed] [Google Scholar]
- 9.McDevitt TC, et al. In vitro generation of differentiated cardiac myofibers on micropatterned laminin surfaces. Journal of Biomedical Materials Research. 2002;60(3):472–479. doi: 10.1002/jbm.1292. [DOI] [PubMed] [Google Scholar]
- 10.Lieberman M, Johnson EA, Roggevee Ae, Purdy JE. Synthetic strands of cardiac muscle - growth and physiological implication. Science. 1972;175(4024):909. doi: 10.1126/science.175.4024.909. [DOI] [PubMed] [Google Scholar]
- 11.Rohr S, Scholly DM, Kleber AG. Patterned growth of neonatal rat heart cells in culture - morphological and electrophysiological characterization. Circulation Research. 1991;68(1):114–130. doi: 10.1161/01.res.68.1.114. [DOI] [PubMed] [Google Scholar]
- 12.Thomas SP, et al. Synthetic strands of neonatal mouse cardiac myocytes - Structural and electrophysiological properties. Circulation Research. 2000;87(6):467–473. doi: 10.1161/01.res.87.6.467. [DOI] [PubMed] [Google Scholar]
- 13.Azeloglu EU, Costa KD. Cross-bridge cycling gives rise to spatiotemporal heterogeneity of dynamic subcellular mechanics in cardiac myocytes probed with atomic force microscopy. American Journal of Physiology-Heart and Circulatory Physiology. 2010;298(3):H853–H860. doi: 10.1152/ajpheart.00427.2009. [DOI] [PubMed] [Google Scholar]
- 14.Deitch S, Kunkle C, Cui X, Boland T, Dean D. Collagen matrix alignment using inkjet printer technology. Materials Research Society Symposium Proceedings; 2008. p. 1094. [Google Scholar]
- 15.Costa KD. Single-cell elastography: probing for disease with the atomic force microscope. Disease Markers. 2003;19(2–3):139–154. doi: 10.1155/2004/482680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darling EM, Zauscher S, Block JA, Guilak F. A thin-layer model for viscoelastic, stress-relaxation testing of cells using atomic force microscopy: Do cell properties reflect metastatic potential? Biophysical Journal. 2007;92(5):1784–1791. doi: 10.1529/biophysj.106.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darling EM, Zauscher S, Guilak F. Viscoelastic properties of zonal articular chondrocytes measured by atomic force microscopy. Osteoarthritis and Cartilage. 2006;14(6):571–579. doi: 10.1016/j.joca.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Okajima T, et al. Stress relaxation of HepG2 cells measured by atomic force microscopy. Nanotechnology. 2007;18(8) [Google Scholar]
- 19.Hemmer JD, et al. Role of cytoskeletal components in stress-relaxation behavior of adherent vascular smooth muscle cells. Journal of Biomechanical Engineering-Transactions of the Asme. 2009;131(4):9. doi: 10.1115/1.3049860. [DOI] [PubMed] [Google Scholar]
- 20.Charras GT, Horton MA. Single cell mechanotransduction and its modulation analyzed by atomic force microscope indentation. Biophysical Journal. 2002;82(6):2970–2981. doi: 10.1016/S0006-3495(02)75638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemmer JD, Dean D, Vertegel A, Langan E, LaBerge M. Effects of serum deprivation on the mechanical properties of adherent vascular smooth muscle cells. Proceedings of the Institution of Mechanical Engineers Part H-Journal of Engineering in Medicine. 2008;222(H5):761–772. doi: 10.1243/09544119JEIM371. [DOI] [PubMed] [Google Scholar]
- 22.Jaasma MJ, Jackson WM, Keaveny TM. Measurement and characterization of whole-cell mechanical behavior. Annals of Biomedical Engineering. 2006;34(5):748–758. doi: 10.1007/s10439-006-9081-0. [DOI] [PubMed] [Google Scholar]
- 23.Jones WR, et al. Alterations in the Young’s modulus and volumetric properties of chondrocytes isolated from normal and osteoarthritic human cartilage. Journal of Biomechanics. 1999;32(2):119–127. doi: 10.1016/s0021-9290(98)00166-3. [DOI] [PubMed] [Google Scholar]
- 24.Mahaffy RE, Shih CK, MacKintosh FC, Kas J. Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Physical Review Letters. 2000;85(4):880–883. doi: 10.1103/PhysRevLett.85.880. [DOI] [PubMed] [Google Scholar]
- 25.Sato M, Theret DP, Wheeler LT, Ohshima N, Nerem RM. Application of the micropipette technique to the measurement of cultured porcine aortic endothelial cell viscoelstic properties. Journal of Biomechanical Engineering-Transactions of the Asme. 1990;112(3):263–268. doi: 10.1115/1.2891183. [DOI] [PubMed] [Google Scholar]
- 26.Jaasma MJ, Jackson WM, Keaveny TM. The effects of morphology, confluency, and phenotype on whole-cell mechanical behavior. Annals of Biomedical Engineering. 2006;34(5):759–768. doi: 10.1007/s10439-005-9052-x. [DOI] [PubMed] [Google Scholar]
- 27.Lieber SC, et al. Aging increases stiffness of cardiac myocytes measured by atomic force microscopy nanoindentation. American Journal of Physiology-Heart and Circulatory Physiology. 2004;287(2):H645–H651. doi: 10.1152/ajpheart.00564.2003. [DOI] [PubMed] [Google Scholar]
- 28.Mathur AB, Collinsworth AM, Reichert WM, Kraus WE, Truskey GA. Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. Journal of Biomechanics. 2001;34(12):1545–1553. doi: 10.1016/s0021-9290(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann UG, Rotsch C, Parak WJ, Radmacher M. Investigating the cytoskeleton of chicken cardiocytes with the atomic force microscope. Journal of Structural Biology. 1997;119(2):84–91. doi: 10.1006/jsbi.1997.3868. [DOI] [PubMed] [Google Scholar]
- 30.Shroff SG, Saner DR, Lal R. Dynamic micromechanical properties of cultured rat atrial myocytes measured by atomic-force microscopy. American Journal of Physiology-Cell Physiology. 1995;269(1):C286–C292. doi: 10.1152/ajpcell.1995.269.1.C286. [DOI] [PubMed] [Google Scholar]
- 31.Borg TK, Rubin K, Lundgren E, Borg K, Obrink B. Recognition of extracellular-matrix components by neonatal and adult cardiac myocytes. Developmental Biology. 1984;104(1):86–96. doi: 10.1016/0012-1606(84)90038-1. [DOI] [PubMed] [Google Scholar]
- 32.Walsh KB, Sweet JK, Parks GE, Long KJ. Modulation of outward potassium currents in aligned cultures of neonatal rat ventricular myocytes during phorbol ester-induced hypertrophy. Journal of Molecular and Cellular Cardiology. 2001;33(6):1233–1247. doi: 10.1006/jmcc.2001.1386. [DOI] [PubMed] [Google Scholar]
- 33.Dimitriadis EK, Horkay F, Maresca J, Kachar B, Chadwick RS. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophysical Journal. 2002;82(5):2798–2810. doi: 10.1016/S0006-3495(02)75620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahaffy RE, Park S, Gerde E, Kas J, Shih CK. Quantitative analysis of the viscoelastic properties of thin regions of fibroblasts using atomic force microscopy. Biophysical Journal. 2004;86(3):1777–1793. doi: 10.1016/S0006-3495(04)74245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fung YC. Biomechanics: Mechanical Properties of Living Tissues. 2 Springer Science and Business Media; New York: 2004. [Google Scholar]
- 36.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(3):849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudreau N, Bissell MJ. Extracellular matrix signaling: integration of form and function in normal and malignant cells. Current Opinion in Cell Biology. 1998;10(5):640–646. doi: 10.1016/s0955-0674(98)80040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, Ingber DE. The structural and mechanical complexity of cell-growth control. Nature Cell Biology. 1999;1(5):E131–E138. doi: 10.1038/13043. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nature Cell Biology. 2002;4(4):E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 40.Ingber DE. Mechanobiology and diseases of mechanotransduction. Annals of Medicine. 2003;35(8):564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 41.Akiyama N, Ohnuki Y, Kunioka Y, Saeki Y, Yamada T. Transverse stiffness of myofibrils of skeletal and cardiac muscles studied by atomic force microscopy. Journal of Physiological Sciences. 2006;56(2):145–151. doi: 10.2170/physiolsci.RP003205. [DOI] [PubMed] [Google Scholar]
- 42.Worth NF, Rolfe BE, Song J, Campbell GR. Vascular smooth muscle cell phenotypic modulation in culture is associated with reorganisation of contractile and cytoskeletal proteins. Cell Motility and the Cytoskeleton. 2001;49(3):130–145. doi: 10.1002/cm.1027. [DOI] [PubMed] [Google Scholar]
- 43.Ross RS, Borg TK. Integrins and the myocardium. Circulation Research. 2001;88(11):1112–1119. doi: 10.1161/hh1101.091862. [DOI] [PubMed] [Google Scholar]
- 44.Simon A, et al. Characterization of dynamic cellular adhesion of osteoblasts using atomic force microscopy. Cytometry Part A. 2003;54A(1):36–47. doi: 10.1002/cyto.a.10052. [DOI] [PubMed] [Google Scholar]
- 45.Smith BA, Tolloczko B, Martin JG, Grutter P. Probing the viscoelastic behavior of cultured airway smooth muscle cells with atomic force microscopy: stiffening induced by contractile agonist. Biophysical Journal. 2005;88(4):2994–3007. doi: 10.1529/biophysj.104.046649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka TT, Fung YC. Elastic and inelastic properties of canine aorta and their variation along aortic tree. Journal of Biomechanics. 1974;7(4):357–370. doi: 10.1016/0021-9290(74)90031-1. [DOI] [PubMed] [Google Scholar]
- 47.Doehring TC, Carew EO, Vesely I. The effect of strain rate on the viscoelastic response of aortic valve tissue: A direct-fit approach. Annals of Biomedical Engineering. 2004;32(2):223–232. doi: 10.1023/b:abme.0000012742.01261.b0. [DOI] [PubMed] [Google Scholar]
- 48.Toms SR, Dakin GJ, Lemons JE, Eberhardt AW. Quasi-linear viscoelastic behavior of the human periodontal ligament. Journal of Biomechanics. 2002;35(10):1411–1415. doi: 10.1016/s0021-9290(02)00166-5. [DOI] [PubMed] [Google Scholar]
- 49.Engler AJ, Richert L, Wong JY, Picart C, Discher DE. Surface probe measurements of the elasticity of sectioned tissue, thin gels and polyelectrolyte multilayer films: Correlations between substrate stiffness and cell adhesion. Surface Science. 2004;570(1–2):142–154. [Google Scholar]
- 50.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nature Materials. 2003;2(11):715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 51.Nagayama K, Nagano Y, Sato M, Matsumoto T. Effiect of actin filament distribution on tensile properties of smooth muscle cells obtained from rat thoracic aortas. Journal of Biomechanics. 2006;39(2):293–301. doi: 10.1016/j.jbiomech.2004.11.019. [DOI] [PubMed] [Google Scholar]